Abstract
Candida albicans is an opportunistic fungal pathogen of humans. The cell wall of the organism defines the interface between the pathogen and host tissues and is likely to play an essential and pivotal role in the host-pathogen interaction. The components of the cell wall critical to this interaction are undefined. Immunoscreening of a lambda expression library with sera raised against mycelial cell walls of C. albicans was used to identify genes encoding cell surface proteins. One of the positive clones represented a candidal gene that was differentially expressed in response to changes in the pH of the culture medium. Maximal expression occurred at neutral pH, with no expression detected below pH 6.0. On the basis of the expression pattern, the corresponding gene was designated PRA1, for pH-regulated antigen. The protein predicted from the nucleotide sequence was 299 amino acids long with motifs characteristic of secreted glycoproteins. The predicted surface localization and N glycosylation of the protein were directly demonstrated by cell fractionation and immunoblot analysis. Deletion of the gene imparted a temperature-dependent defect in hypha formation, indicating a role in morphogenesis. The PRA1 protein was homologous to surface antigens of Aspergillus spp. which react with serum from aspergillosis patients, suggesting that the PRA1 protein may have a role in the host-parasite interaction during candidal infection.
Candida albicans is a dimorphic fungus of increasing medical importance. It commonly causes superficial infections of skin and mucosae but in immunocompromised patients can penetrate tissues and cause life-threatening systemic infections. The factors responsible for its pathogenesis are still not well understood, but several attributes related to the cell wall have been thought to contribute to C. albicans virulence (7, 12, 35).
The cell wall of C. albicans forms the interface between pathogen and host and thus is likely to play an essential and pivotal role in the host-parasite interaction. Besides its primary protective role in shielding the cell against external harm, the wall is involved in other functions, such as maintenance of cell shape, and consequently in the dimorphic process. Different studies have also shown the essential role that morphology-specific cell wall components, mainly mannoproteins, play in the adherence of the fungus to different host components (9, 21, 25, 45) and as inducers or modulators of the host immunogenic response (10, 43). Hydrolytic activities associated with the cell surface and external environment have also been implicated in tissue invasion and colonization (13, 23, 26, 30).
Both the molecular architecture and the functional components of the C. albicans cell wall vary between yeast and hyphal forms of the organism, as revealed by biochemical, immunological, and cytological studies. Molecular genetic approaches have identified several genes encoding hypha-specific cell surface proteins that may contribute to differences in cell wall structure or function (2, 22, 42). The aim of our work was to identify additional genes encoding cell wall proteins that might contribute to morphology-specific differences. The isolation of a number of morphology-specific cDNA clones by immunoscreening of a lambda expression library was reported previously (41). In this paper, we report the characterization of one of these clones that encodes a mannoprotein present on the cell surface of C. albicans. It shows extensive sequence conservation with antigens from other fungal species and is differentially expressed in response to changes in pH.
MATERIALS AND METHODS
Microorganisms and growth conditions.
The C. albicans strains used in this study are listed in Table 1. Cells were routinely grown in YPD (2% glucose, 1% yeast extract, 2% Bacto Peptone [Difco, Detroit, Mich.]) with shaking at the selected temperature. For specific experiments, cells were also cultured in medium 199 containing Earle’s salts and glutamine but lacking sodium bicarbonate (GIBCO-BRL), modified Lee’s medium (29) containing 0.5 g of proline per liter but lacking other amino acids, or SD minimal medium (2% glucose, 0.67% yeast nitrogen base without amino acids [Difco]). The medium 199 was buffered with 150 mM HEPES. Media were supplemented with uridine (25 μg/ml) as needed, and Urd− auxotrophs were selected on medium containing 5-fluoro-orotic acid as described previously (5).
TABLE 1.
Strains of C. albicans used
| Strain | Genotype | Reference or source |
|---|---|---|
| SC5314 | Wild type | 18 |
| CAI4 | Δura3::imm434/Δura3::imm434 | 15 |
| CAMB1 | Δpra1::hisG-URA3-hisG/PRA1 Δura3::imm434/Δura3::imm434 | This work |
| CAMB11 | Δpra1::hisG/PRA1 Δura3::imm434/Δura3::imm434 | This work |
| CAMB43 | Δpra1::hisG/Δpra1::hisG-URA3-hisG Δura3::imm434/Δura3::imm434 | This work |
| CAMB435 | Δpra1::hisG/Δpra1::hisG Δura3::imm434/Δura3::imm434 | This work |
| CAMB9 | Δpra1::hisG/PRA1-pUC18-URA3-Δpra1 Δura3::imm434/Δura3::imm434 | This work |
| SGY243 | ade2/ade2 Δura3::ADE2/Δura3::ADE2 | 24 |
| 26555 | Wild type | ATCCa |
| FC18 | Wild type | 48 |
| 699 | Wild type | D. Poulain |
ATCC, American Type Culture Collection.
For germ tube induction, cells were precultured in modified Lee’s medium as described previously (14) or in SD medium containing N-acetylglucosamine (2.5 mM). Alternatively, blastoconidia were inoculated in YPD and shaken at 180 rpm at 25°C until the culture had reached the late logarithmic-early stationary growth phase. Hyphae were formed after transfer of the cells to modified Lee’s medium (pH 6.8), SD medium containing N-acetylglucosamine (2.5 mM), or medium 199 (pH 6.8) and incubation at 37 or 42°C.
Protoplasts were obtained and regenerated as previously described (31).
Saccharomyces cerevisiae W303-1B was grown in YPD at 28°C, and Escherichia coli XL1Blue (Stratagene, La Jolla, Calif.) was used for most transformations.
Isolation of the PRA1 gene.
Positive cDNA clones were isolated by immunoscreening of a lambda expression library with rabbit polyclonal antiserum raised against cell walls of mycelial cells as previously described (41). The 1.0-kb cDNA insert in one of the positive clones, 8M, was amplified by PCR using commercial lambda primers and employed for hybridization-screening of a λ GEM-12 genomic library (4). Positive plaques were characterized by restriction endonuclease mapping and Southern blot hybridization. A 4.3-kb SacI genomic DNA fragment containing the full-length gene was isolated from one genomic clone, 11-1, and subcloned in both orientations in pBSK+ (Stratagene) to generate plasmids pMBW2 and pMBW3.
DNA sequence analysis.
Portions of the genomic insert in plasmids pMBW2 and pMBW3 were subcloned into either pUC18 or pBSK+ cloning vector. The nucleotide sequence was determined by the dideoxy-chain termination method using T7 polymerase and either universal sequencing primers or custom-synthesized oligonucleotide primers. Reaction products were analyzed on a model 370A sequencer (Applied Biosystems).
Sequence analyses were carried out with the software system PC/Gene, release 6.85 (IntelliGenetics, Inc., Geneva, Switzerland), and the PSORT program (33) from the National Institute for Basic Biology (Okazaki, Japan). Homology searches were conducted with the BLAST (1) and BLOCKS (19) algorithms. Multiple-sequence alignments were performed with MACAW (40) and CLUSTAL W (20).
Southern and Northern blot analysis.
Southern and Northern blot hybridizations were performed as previously described (38). C. albicans genomic DNA was prepared by the method of Scherer and Stevens (39). RNA was prepared by the method of Langford and Gallwitz (28). Transcript sizes were determined by comparison with rRNA species and the C. albicans actin gene mRNA identified in control hybridizations.
Pulsed-field gel electrophoresis.
Preparation of chromosomal DNA and pulsed-field gel electrophoresis were carried out as previously described (34). Hybridization of chromosomal DNA was done after blotting of the DNA onto a nylon membrane. The 8M cDNA was used as a hybridization probe.
Strain constructions.
To construct a PRA1 null mutant, plasmid pMBW3 was digested with NdeI-BstII to delete 0.6 kb from the PRA1 open reading frame and blunt-end ligated with a BglII-SalI fragment from plasmid pMB7 (15) containing the hisG-URA-hisG cassette. The resulting plasmid, pMBW8, was digested with SacI, releasing a PRA1 gene deletion-disruption fragment. Approximately 15 μg of this DNA was used to transform the Urd− strain CAI4 by the method of Gietz et al. (17). Transformed cells were selected as Urd+, and a representative clone was designated CAMB1. Spontaneous Urd− derivatives of CAMB1 were selected on medium containing 5-fluoro-orotic acid (5). One of the Urd− derivatives which had undergone intrachromosomal recombination, CAMB11, was used for targeted disruption of the second allele of PRA1. Preliminary analysis of these transformants was conducted by PCR amplification using primers from PRA1 (5′-CCGATTGATCTGTCGTGTAATGC-3′ and 5′-GGCCCCTGATCAGAGCCACT-3′) and URA3 (5′-CAATGGCACTACAGCAACTTTCAAC-3′). The PRA1 primers can amplify only wild-type alleles, since the first primer lies within the deleted region. Loss of this amplification product was indicative of the double deletion. Amplification with the URA3 primer and the second PRA1 primer served as a positive control. A representative null mutant, CAMB43, was chosen, and spontaneous Urd− segregants were selected by resistance to 5-fluoro-orotic acid. One of these Urd− null mutants was designated CAMB435. In each strain, the structure of the PRA1 locus was established by Southern blot analysis.
Strain CAMB9, a Urd+ Pra1+ derivative of CAMB435, was generated by transformation of CAMB435 with plasmid pMBW9. Plasmid pMBW9 was constructed by ligation of a 2.1-kb XbaI-EcoRV fragment containing the URA3 gene into the SpeI-SmaI sites of plasmid pMBW3. pMBW9 was digested with BamHI to target integration to the PRA1 locus and used to transform strain CAMB435 to Urd+. The site of integration and structure of the locus were verified by Southern blot analysis.
Overexpression of PRA1.
S. cerevisiae ADH-PRA1, a mutant with constitutive expression of PRA1, was obtained by transformation of S. cerevisiae W303-1B with plasmid pADH-PRA1. To construct plasmid pADH-PRA1, the PRA1 coding region was PCR amplified with primers designed to introduce a BglII site at position −1 with respect to the first nucleotide of the coding region and an XhoI site at the 3′ end. The primer sequences were 5′-CAGAGATCTATGAATTATTTATTGTTTTGT-3′ and 5′-CAGCTCGAGGTCTACAAGCGATTTTGC-3′. The PCR product was digested with BglII and XhoI and ligated with the 12.1-kb BglII-XhoI fragment of plasmid YPB1-ADHpL (2) to generate pADH-PRA1. This plasmid was used to transform S. cerevisiae W303-1B, and constitutive expression of PRA1 in Urd+ isolates was verified by Northern blot analysis.
Preparation of cell walls and isolation of membrane fractions from C. albicans.
Subcellular fractions were obtained as previously described (31). Briefly, blastoconidia and mycelial cells were collected by centrifugation at 3,000 × g for 10 min, washed twice with chilled distilled water, suspended in a small volume of 0.001 M phenylmethylsulfonyl fluoride in 0.01 M Tris-HCl buffer (pH 7.2), and broken by being shaken with glass beads. The procedure resulted in nearly complete cell breakage. The cell walls were sedimented (1,200 × g for 10 min) to remove membranous and cytoplasmic proteins, washed in chilled 0.001 M phenylmethylsulfonyl fluoride, and treated with boiling 2% (wt/vol) sodium dodecyl sulfate (SDS) to extract non-covalently bound proteins. The extracted cell walls were digested with Zymolyase, a β-1,3-glucanase, as previously described (31).
The crude supernatant remaining after removal of the cell walls was further fractionated by centrifugation at 40,000 × g for 40 min to obtain a pelleted mixed membrane preparation and a supernatant cytosol fraction.
Endoglycosidase H treatment.
Deglycosylation with endoglycosidase H was carried out as previously described (37).
PAGE and Western blotting techniques.
Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (27) in 10% (wt/vol) acrylamide gels loaded with 20 μg of protein. After transfer to nitrocellulose, proteins were immunodetected with either monospecific Pra1p antibodies (41), polyclonal antibodies against mycelial cell walls (41), or polyclonal antibodies raised against Aspergillus nidulans antigen AspNDI (kindly provided by F. Leal from University of Salamanca [8]). Detection was carried out with an ECL kit (Amersham) according to the manufacturer’s instructions.
Fluorescence microscopy.
The distribution of cell wall polysaccharides in C. albicans CAI4 and CAMB435 was analyzed by using commercially available fluorescence isothiocyanate-coupled lectins from Canavalia ensiformis (concanavalin A-fluorescein isothiocyanate [ConA-FITC]) to detect mannan and from Triticum vulgaris (wheat germ agglutinin [WGA]-FITC) to localize chitin. For ConA-FITC staining, cells were suspended in 20 mM Tris-HCl (pH 7.0)–0.15 M NaCl–1 mM each CaCl2, MnCl2, and MgCl2–0.5 mg of lectin conjugate per ml. For staining with WGA-FITC, cells were suspended in 20 mM Tris-HCl (pH 7.0) containing 1 mg of lectin per ml. In both cases, the cells were incubated in the dark for 30 min at room temperature, rinsed thoroughly with 20 mM Tris-HCl (pH 7.0), and observed with a Zeiss Photomicroscope III equipped for epifluorescence.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper has been submitted to GenBank under accession no. U84261.
RESULTS AND DISCUSSION
Isolation and sequence analysis of the PRA1 gene.
In a previous study, 18 cDNA clones which reacted with mycelium-specific antiserum raised against purified cell walls were isolated (41). Ten of those clones were related on the basis of cross-hybridization in Southern blots, and a representative of this group, 8M, was chosen for further analysis. The insert from the 8M cDNA was amplified by PCR and sequenced. It was found to contain an open reading frame of 897 bp starting 9 bp downstream from the adapters used in the construction of the library. On the basis of its expression pattern and relationship with other fungal proteins, the corresponding gene was designated PRA1 for pH-regulated antigen (see below).
To isolate a corresponding genomic clone, a C. albicans lambda library was screened and two independent clones, lambda 2-1 and lambda 11-1, were isolated. They contained a common 4.3-kb SacI fragment that hybridized with the cDNA clone. The genomic fragment was colinear with the genome as determined by restriction endonuclease mapping and appeared to represent a single locus. The only discrepancy between the cloned fragment and the genome was the location of a ClaI recognition site which proved to be a polymorphism in one allele (data not shown).
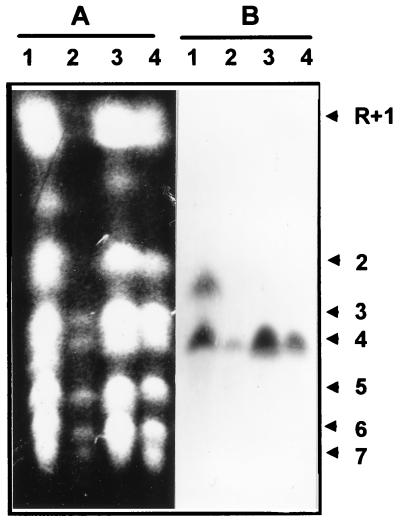
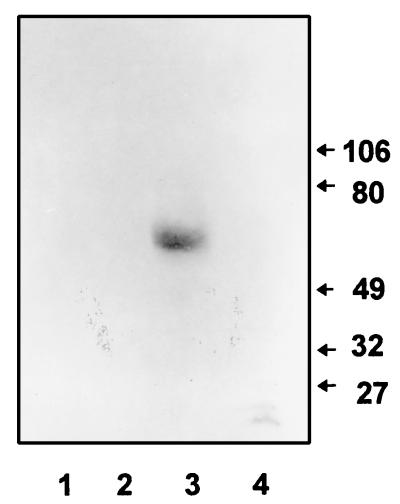
Karyotypic analysis also indicated that the cloned fragment represented a single locus on chromosome 4 (Fig. 1). A single chromosomal band hybridized with a PRA1 probe in three of the four strains tested. Only in strain ATCC 26555 did a second chromosomal band hybridize. This band lay between chromosomes 2 and 3 and was not apparent in the other strains. Interestingly, the signal intensity of this atypical band was approximately half that of chromosome 4, suggestive of partial aneuploidy in this strain.
FIG. 1.
Separation of chromosomes of C. albicans ATCC 26555 (lanes 1), SGY243 (lanes 2), 996 (lanes 3), and FC18 (lanes 4). The gel was stained with ethidium bromide (A) and blotted onto nylon and hybridized with PRA1 (B). C. albicans chromosomal designations (on the right) are those proposed by Wickes et al. (48).
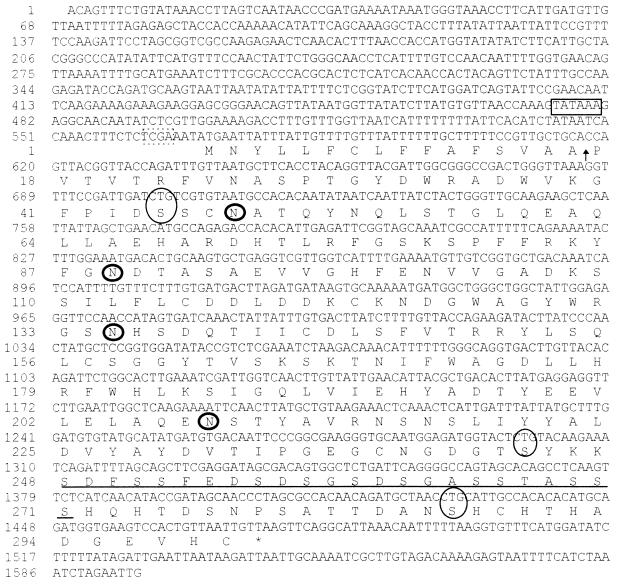
Approximately 1.6 kb of the SacI fragment from clone lambda 11-1 was sequenced in the region complementary to the cDNA. We identified a single, uninterrupted open reading frame identical in sequence to the cDNA (Fig. 2). The predicted protein was 299 amino acids in length and contained a putative signal sequence with a potential signal peptidase cleavage site between residues 15 and 16 (47). Cleavage at this site would result in a mature protein with a calculated molecular mass of 31.4 kDa. Also present were four potential N-glycosylation sites and a serine-rich region of potential O glycosylation, a motif often found in cell wall mannoproteins of S. cerevisiae and Yarrowia lipolytica (36, 46). No transmembrane domains, glycosylphosphatidylinositol attachment sites, or other motifs were evident.
FIG. 2.
Nucleotide sequence and deduced amino acid sequence of PRA1. A putative TATA sequence (boxed), the translation start site (boxed by a broken line), four potential N-glycosylation sites (circled with thick lines), noncanonical CTG codons (circled with thin lines), the predicted signal cleavage site (arrow), and the serine-rich regions (underlined) are indicated.
Regulation of PRA1 expression.
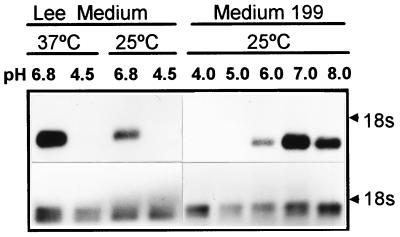
The 8M cDNA was isolated from an expression library on the basis of its reactivity with hypha-specific antiserum. This suggested that PRA1 was differentially expressed in relation to either cell morphology or the culture conditions used to induce hypha formation. Differential expression of PRA1 was evidenced by Northern blot analysis. A single, 1.1-kb transcript was detected in RNA extracted from cells grown in the medium of Lee et al. (29) adjusted to pH 6.8, but not pH 4.5, irrespective of temperature and cell morphology (Fig. 3). A similar pattern was observed with medium 199. PRA1 mRNA was not detected in cells cultured at pH 4.0 or 5.0 but was readily detected in cells cultured at pH 6.0. Maximal expression occurred around pH 7.0 (Fig. 3). Ambient pH was not the sole factor influencing expression. Temporal analysis demonstrated that PRA1 mRNA was not detectable for the first 3 h following inoculation into medium at the permissive pH, despite extensive cell growth, and expression was not detected in YPD medium buffered at pH 7.0 (data not shown). The delayed expression in some media and lack of expression in rich medium indicated that pH is not the sole determinant of expression.
FIG. 3.
Northern blot analysis of the effects of temperature and pH on PRA1 expression. Strain SC5314 was inoculated in the medium of Lee et al. (29) or medium 199 adjusted to the indicated pH and incubated for 3 h at 37 or 25°C. The results of hybridization with the 8M cDNA insert (top panel) and of hybridization with actin DNA (bottom panel) are shown. The electrophoretic position of 18s rRNA is indicated on the right.
Sequence comparisons.
As a first step in elucidating the function of PRA1, sequence comparisons were performed to potentially identify homologous proteins with known activity. A BLAST search of the GenBank and EMBL databases found two homologs previously identified as immunodominant antigens in aspergillosis. These were the AspFII protein of Aspergillus fumigatus (3) and the AspNDI protein of A. nidulans (8), which were 41.6 and 46.9% identical, respectively, to Pra1p (Fig. 4). Homology extended along their entire lengths and included conserved secretory signal sequences, four consensus N-glycosylation sites, and seven cysteine residues. A serine-rich region was present in AspFII and Pra1p but not in AspNDI. In this context, it should be noted that approximately 55% of the immunopositive cDNA clones we isolated were PRA1, suggesting that the C. albicans protein is also highly immunogenic. If it is similarly immunodominant in candidiasis patients, then it may have potential as a diagnostic antigen.
FIG. 4.
Alignment of the amino acid sequences of Pra1p and related proteins. Identical residues present in at least five of the eight sequences (boxes), residues conserved specifically between Pra1p and the Aspergillus sp. antigens (lines above the sequences), and the zinc-binding motif conserved in metalloproteinases (thick bar) are shown. AspndI, A. nidulans antigen (6); AspfII, A. fumigatus antigen (3); ScAOB249, S. cerevisiae open reading frame; AspNPII, neutral protease of A. oryzae (46, 47); Pnclysin, penicillinolysin of Penicillium citricum (30); Afumep20, metalloproteinase from A. fumigatus (35); Aflmep20 metalloproteinase from Aspergillus flavus (35).
Unfortunately, the function of these Aspergillus proteins is unknown, as with a related S. cerevisae protein, ScAOB249, defined by an open reading frame on the left arm of chromosome XV (16). However, there was a clear, though weaker, relationship with the deuterolysin family of zinc metalloproteinases (Fig. 4). Sequence alignments showed the conserved positioning of critical cysteine residues involved in disulfide bond formation and tertiary structure of deuterolysins (44) and a conserved zinc-binding domain, suggesting a possible proteolytic role for Pra1p.
Another potentially interesting functional clue was provided by a recent GenBank submission of a partial cDNA sequence for a putative fibrinogen-binding protein of C. albicans (11). This sequence is virtually identical to that of PRA1 except for amino acid substitutions at positions 1, 18, 83, and 97 of the predicted protein and two silent base substitutions at the DNA level.
Construction of a PRA1 null mutant.
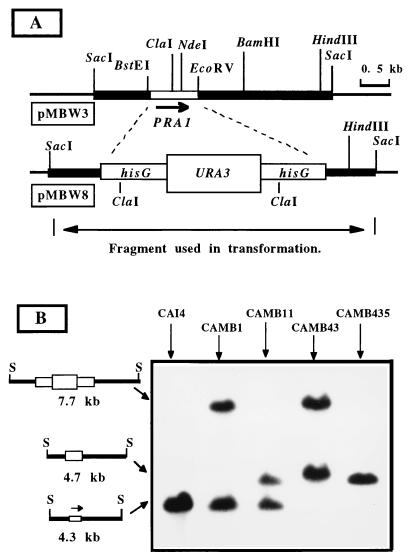
To further define the cellular function of PRA1, a null mutant was constructed to search for informative phenotypes. The experimental design of the deletion-disruption is depicted in Fig. 5A. All 10 of the first-round transformants we examined were heterozygous for the PRA1 deletion, as shown for a representative isolate, CAMB1 (Fig. 5B). CAMB11, a Urd− segregant of CAMB1 resulting from intrachromosomal excision of the URA3 marker, was transformed with the same deletion-disruption cassette to mutate the remaining wild-type allele. All 18 transformants examined by Southern blot analysis had undergone replacement of the previously disrupted allele. Consequently, a more extensive screening was performed with a PCR-based assay. Fifty-two additional transformants were screened, and two of them appeared be homozygous for the deletion. This was confirmed by Southern blot analysis, and one of these isolates, CAMB43, was used to select a Urd− segregant, strain CAMB435 (Fig. 5B).
FIG. 5.
Disruption of the PRA1 locus. (A) Restriction map of the 4.3-kb SacI genomic fragment containing the PRA1 gene (pMBW3) and the deletion-disruption construct (pMBW8). (B) Southern blot analysis of SacI-digested DNA from the parental strain CAI4, the Δpra1 heterozygote CAMB and its Urd− segregant, CAMB11, and the Δpra1/Δpra1 null mutant CAMB43 and its Urd− segregant, CAMB435. The blot was probed with the SacI fragment containing PRA1. The lengths and structures of the hybridizing fragments are shown on the left. S, SacI.
Phenotypic analysis of Δpra1 strains.
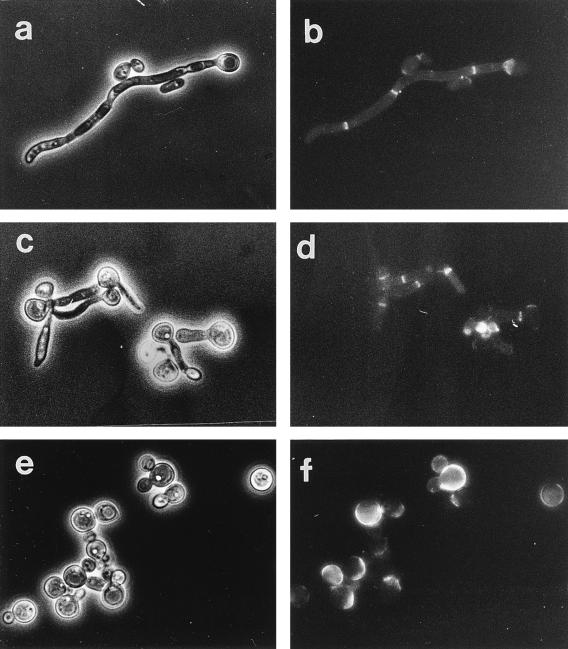
Successful recovery of the homozygous null mutant indicated that PRA1 was not essential. The specific growth rates of the parental strain, CAI4, and the null mutant, CAMB345, in SD medium at 37 and 42°C were similar, and the growth yields in stationary-phase cultures at 37 and 42°C were also similar (data not shown). Germ tube formation by the mutant was not affected in either rate or frequency when induced at 37°C in SD medium containing N-acetylglucosamine (data not shown). At 42°C, the parental strain formed germ tubes, but shorter ones than at 37°C (Fig. 6c and d). The null mutant, CAMB435, however, was unable to form germ tubes. Only a few cells formed pseudohyphae, and most cells were enlarged and spherical (Fig. 6e and f).
FIG. 6.
Chitin distribution in CAI4 and CAMB435 after 6-h induction in SD medium with N-acetylglucosamine. The cells were labelled with WGA-FITC as described in Materials and Methods. Phase-contrast (a, c, and e) and fluorescence (b, d, and f) micrographs of CAMB435 at 37°C (a and b), CAI4 at 42°C (c and d), and CAMB435 at 42°C (e and f) are shown.
The cell wall of the mutant was not grossly altered in mannoprotein distribution, as demonstrated by staining with ConA-FITC (data not shown). However, chitin distribution was altered. At 37°C, the WGA-FITC staining patterns of CAMB435 and CAI4 were similar, with fluorescence concentrated in the septal region (Fig. 6a and b). At 42°C, the parental strain was unaltered, but in the mutant, fluorescence was randomly distributed throughout the cells (Fig. 6c to f).
To verify that the aberrant phenotypes of CAMB435 were due to the loss of PRA1 and not due to a random second-site mutation, a wild-type PRA1 allele was introduced into the null mutant. One such revertant, CAMB9, was incubated at 42°C and examined for morphological aberrations. The morphology of this strain was identical to that of CAI4, indicating that the phenotype of the null mutant was due specifically to the loss of PRA1 (data not shown).
Cell localization of PRA1.
Two pieces of indirect evidence suggested that PRA1 might encode a cell wall protein. Isolation of the gene was achieved by screening a mycelial library with an antiserum raised against cell wall material, and the putative protein contained motifs indicative of cell surface proteins. To provide direct evidence for the localization of Pra1p, cell fractionation studies were performed. Hyphal cells were separated into cytosolic, membrane, and cell wall fractions. The wall fraction was further separated into SDS-extractable and β-glucanase-extractable materials. Western blot analysis of those fractions (Fig. 7) demonstrated a single protein of approximately 60 kDa which was reactive with monospecific antibodies against Pra1p. This protein was detected only in the SDS extract of cell walls. It was not detectable in any of the other cell fractions or in the supernatant of regenerating protoplasts. Since it was separated from the cell wall by SDS extraction, Pra1p does not appear to be covalently linked to the wall.
FIG. 7.
Localization of Pra1p. A Western blot of mycelial fractions of strain ATCC 26555 was reacted with monospecific polyclonal antibodies against Pra1p. Lanes: 1, Zymolyase-solubilized cell wall fraction; 2, spent medium from protoplasts after 3 h of regeneration; 3, wall material solubilized with SDS; and 4, a mixed membrane preparation. Molecular masses (in kilodaltons) are indicated on the right.
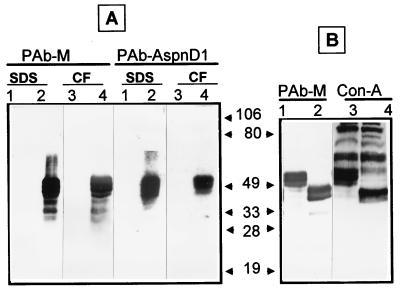
Native Pra1p was difficult to detect, reflecting either a low level of expression or low avidity of the antibody. To facilitate analysis of the protein, the gene was overexpressed in S. cerevisae by using plasmid pADH-PRA1. Yeast cells carrying this plasmid contained a readily detectable amount of Pra1p (Fig. 8). The protein was expressed as a series of bands of 50 to 60 kDa, the largest being similar in size to the native protein in C. albicans. As with C. albicans, the protein was present in the SDS extract of cell walls. However, unlike in C. albicans, the protein was also found in the culture supernatant. This suggests again the noncovalent association of the protein with the cell wall; however, it may also be an aberrant consequence of overexpression. The same set of proteins was also detected by use of antiserum against the AspNDI protein (8), substantiating the relationship between these proteins (Fig. 8).
FIG. 8.
Structure and localization of Pra1p. (A) Western blot of SDS extracts of cell walls (lanes 1 and 2) and culture supernatants (lanes 3 and 4) prepared from S. cerevisiae W303-1B (lanes 1 and 3) and W303-1B transformed with pADH-PRA1 (lanes 2 and 4). The blot was reacted with polyclonal antiserum against Pra1p (PAb-M) or AspNDI (PabAspnD1). CF, culture filtrates. (B) Effect of endoglycosidase H treatment. Culture filtrates from strain W303-1B transformed with pADH-PRA1 were separated by PAGE before (lanes 1 and 3) or after (lanes 2 and 4) endoglycosidase H treatment. The gel was blotted and either reacted with PAb-M or stained with ConA-peroxidase.
Both native Pra1p and the heterologously expressed protein were much larger than the predicted 31 kDa. One explanation of this discrepancy is that the protein is highly glycosylated, consonant with the presence of several N-glycosylation consensus sites and a serine-rich region. The presence of N-linked carbohydrate chains was indicated by the effect of endoglycosidase H. Treatment with this enzyme decreased the size of Pra1p by about 15 kDa, indicating that at least 25% of the molecule consisted of N-linked oligosaccharides (Fig. 8). The endoglycosidase H-treated protein was still reactive with ConA, indicating the presence of either O-linked mannooligosaccharides or additional N-linked carbohydrate that was not susceptible to endoglycosidase H treatment. Nonetheless, the structure and localization of Pra1p were consistent with the sequence-based predictions and indicated that Pra1p is a cell surface glycoprotein.
In conclusion, we have identified a gene encoding a secretory protein localized to the wall of C. albicans that plays at least a limited role in morphogenesis. The role of Pra1p in this process is unclear but may be related to its potential proteolytic activity. The pH dependence of PRA1 expression is of interest, since pH has a major influence on C. albicans morphology in vitro (6) and two other genes required for morphogenesis, PHR1 and PHR2, are also pH regulated (32, 38). The morphological defect associated with loss of PRA1 reinforces the significance of pH in candidal morphogenesis and provides additional evidence that the pH effect is mediated through differential gene expression. The temperature dependence of the null mutant phenotype is also of interest in that temperature-sensitive defects in candidal hypha formation have not been previously described, and this suggests a new and useful screen for identifying morphological functions in this organism.
ACKNOWLEDGMENTS
This work was supported by grants from the Burroughs Wellcome Fund for Molecular Pathogenic Mycology; Public Health Service grant AI 371941 from the National Institute of Allergy and Infectious Diseases; grant 95/1602 from Fondo de Investigaciones Sanitarias de la Seguridad Social del Ministerio de Sanidad y Consumo, Madrid, Spain; and BIOMED grant BMH4-CT96-0310 from New Targets for Antifungal Therapy-Molecular Biology of Dimorphism in the Human Pathogen Candida albicans (Brussels, Belgium). M.S. was the recipient of predoctoral grants from the Direcció General de Universitat e Investigació de la Generalitat Valenciana, València, Spain.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bailey D A, Feldmann P J F, Bovey M, Gow N A R, Brown A J P. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee B, Kurup V P, Phadnis S, Greenberger P A, Fink J N. Molecular cloning and expression of a recombinant Aspergillus fumigatus protein AspfII with significant immunoglobulin E reactivity in allergic bronchopulmonary aspergillosis. J Lab Clin Med. 1996;127:253–262. doi: 10.1016/s0022-2143(96)90093-1. [DOI] [PubMed] [Google Scholar]
- 4.Birse C E, Irwin M Y, Fonzi W A, Sypherd P S. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 6.Buffo J, Herman M A, Soll D R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85:21–30. doi: 10.1007/BF00436698. [DOI] [PubMed] [Google Scholar]
- 7.Calderone R A, Braun P C. Adherence and receptor relationships of Candida albicans. Microbiol Rev. 1991;55:1–20. doi: 10.1128/mr.55.1.1-20.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calera J A, Ovejero M C, Lopez-Medrano R, Segurado M, Puente P, Leal F. Characterization of the Aspergillus nidulans Aspnd1 gene demonstrates that the ASPND1 antigen, which it encodes, and several Aspergillus fumigatus immunodominant antigens belong to the same family. Infect Immun. 1997;65:1335–1344. doi: 10.1128/iai.65.4.1335-1344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casanova M, Lopez Ribot J L, Martinez J P, Sentandreu R. Characterization of cell wall proteins from yeast and mycelial cells of Candida albicans by labelling with biotin: comparison with other techniques. Infect Immun. 1992;60:4898–4906. doi: 10.1128/iai.60.11.4898-4906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassone A. Cell wall of Candida albicans: its functions and its impact on the host. Curr Top Med Mycol. 1989;3:248–314. doi: 10.1007/978-1-4612-3624-5_10. [DOI] [PubMed] [Google Scholar]
- 11.Cervera, A., J. L. Lopez-Ribot, D. Gozalbo, J. P. Martinez, and M. Casanova. 1997. Unpublished results.
- 12.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1992;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 13.De Bernardis F, Agatensi L, Ross Y K, Emerson G W, Lorenzini R, Sullivan P A, Cassone A. Evidence for a role for secreted aspartate proteinase of Candida albicans in vulvovaginal candidiasis. J Infect Dis. 1990;161:1276–1283. doi: 10.1093/infdis/161.6.1276. [DOI] [PubMed] [Google Scholar]
- 14.Elorza M V, Marcilla A, Sentandreu R. Wall mannoproteins of the yeast and mycelial cells of Candida albicans: nature of the glycosidic bonds and polydispersity of their mannan moieties. J Gen Microbiol. 1988;134:2393–2403. doi: 10.1099/00221287-134-8-2393. [DOI] [PubMed] [Google Scholar]
- 15.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamo F J, Lafuente M J, Casamayor A, Aldea M, Casas C, Ario J, Herrero E, Gancedo C. Analysis of the DNA sequence of a 15.500 bp fragment of the left arm of chromosome XV from S. cerevisiae reveals a putative sugar transporter, a carboxypeptidase, precursor, and two new open reading frames. Yeast. 1996;12:709–714. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C709::AID-YEA957%3E3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillum A M, Tsay E Y H, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff S, Henikoff J G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogging D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;173:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 21.Hostetter M K, Lorenz J S, Preus L, Kendrick K E. The iC3b receptor on Candida albicans: subcellular localization and modulation of receptor expression by glucose. J Infect Dis. 1989;161:761–768. doi: 10.1093/infdis/161.4.761. [DOI] [PubMed] [Google Scholar]
- 22.Hoyer L L, Scherer S, Shatzman A R, Livi G P. Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol Microbiol. 1995;15:39–54. doi: 10.1111/j.1365-2958.1995.tb02219.x. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim A S, Mirbod F, Filler S G, Banno Y, Cole G T, Kitajima Y, Edwards J E, Jr, Nozawa Y, Ghannoum M A. Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect Immun. 1995;63:1993–1998. doi: 10.1128/iai.63.5.1993-1998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly R, Miller S M, Kurtz M B, Kirsch D R. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol Cell Biol. 1987;7:199–207. doi: 10.1128/mcb.7.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klotz S A, Smith R L. A fibronectin receptor on Candida albicans mediates adherence of the fungus to extracellular matrix. J Infect Dis. 1991;163:604–610. doi: 10.1093/infdis/163.3.604. [DOI] [PubMed] [Google Scholar]
- 26.Kwon Chung K J, Lehman D, Good C, Magee P T. Genetic evidence for role of extracellular proteinase in virulence of Candida albicans. Infect Immun. 1985;49:571–575. doi: 10.1128/iai.49.3.571-575.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Langford C J, Gallwitz D. Evidence for an intron-containing sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983;33:519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- 29.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 30.Macdonald F. Secretion of inducible proteinase by pathogenic Candida species. Sabouraudia. 1984;22:79–82. [PubMed] [Google Scholar]
- 31.Marcilla A, Mormeneo S, Elorza M V, Manclus J J, Sentandreu R. Wall formation by Candida albicans yeast cells: synthesis, secretion and incorporation of two types of mannoproteins. J Gen Microbiol. 1993;139:2985–2993. doi: 10.1099/00221287-139-12-2985. [DOI] [PubMed] [Google Scholar]
- 32.Muhlschlegel F A, Fonzi W A. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of expression. Mol Cell Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieto A, Sanz P, Sentandreu R, del Castillo Agudo L. Cloning and characterization of the SEC18 gene from Candida albicans. Yeast. 1993;9:875–887. doi: 10.1002/yea.320090808. [DOI] [PubMed] [Google Scholar]
- 35.Odds F C. Candida and candidosis. A review and bibliography. 2nd ed. London, United Kingdom: Bailliere Tindal; 1988. [Google Scholar]
- 36.Ramon A M, Gil R, Burgal M, Sentandreu R, Valentin E. A novel cell wall protein specific to the mycelial form of Yarrowia lipolytica. Yeast. 1996;12:1535–1548. doi: 10.1002/(SICI)1097-0061(199612)12:15%3C1535::AID-YEA59%3E3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 37.Sanz P, Herrero E, Sentandreu R. Role of glycosylation in the incorporation of intrinsic mannoproteins into cell walls of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1989;48:265–269. doi: 10.1016/0378-1097(89)90311-x. [DOI] [PubMed] [Google Scholar]
- 38.Saporito-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scherer S, Stevens D S. A Candida albicans dispersed, repeated gene family and its epidemiological applications. Proc Natl Acad Sci USA. 1988;85:1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 41.Sentandreu M, Elorza M V, Valentin E, Sentandreu R, Gozalbo D. Cloning of cDNAs coding for Candida albicans cell surface proteins. J Med Vet Mycol. 1995;33:105–111. [PubMed] [Google Scholar]
- 42.Staab J F, Ferrer C A, Sundstrom P. Developmental expression of a tandemly repeated, proline- and glutamine-rich amino acid motif on hyphal surfaces of Candida albicans. J Biol Chem. 1996;271:6298–6305. doi: 10.1074/jbc.271.11.6298. [DOI] [PubMed] [Google Scholar]
- 43.Sundstrom P M, Tam M R, Nichols E J, Kenny G E. Antigenic differences in the surface mannoproteins of Candida albicans as revealed by monoclonal antibodies. Infect Immun. 1988;56:601–606. doi: 10.1128/iai.56.3.601-606.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatsumi H, Ikegaya K, Murakami S, Kawabe H, Nakano E, Motai H. Elucidation of the thermal stability of the neutral proteinase II from Aspergillus oryzae. Biochim Biophys Acta. 1994;1208:179–185. doi: 10.1016/0167-4838(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 45.Tronchin G, Bouchara J P, Robert R. Dynamic changes of the cell wall surface of Candida albicans associated with germination and adherence. Eur J Cell Biol. 1989;50:285–290. [PubMed] [Google Scholar]
- 46.Van der Vaart J M, Caro L H P, Chapman J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickes B, Staudinger J, Magee B B, Kwon-Chung K-J, Magee P T, Scherer S. Physical and genetic mapping of Candida albicans: several genes previously assigned to chromosome 1 map to chromosome R, the rDNA-containing linkage group. Infect Immun. 1991;59:2480–2484. doi: 10.1128/iai.59.7.2480-2484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]