ABSTRACT
SARS-CoV-2 has a remarkable ability to respond to and evolve against the selection pressure by host immunity exemplified by the emergence of Omicron lineage. Here, we characterized the functional significance of mutations in Omicron (BA.1 and BA.5) spike. By systematic transfer of mutations in wild type (WT) spike, we assessed neutralization sensitivity, fusogenicity, and TMPRSS2-dependence for entry. The data revealed that the mutations in both S1 and S2 complement to make Omicron a highly resistant variant. Strikingly, the mutations in Omicron S2 modulated the neutralization sensitivity to N-terminal domain and receptor binding domain antibodies, but not to S2-specific neutralizing antibodies, suggesting that the mutations in S2 were primarily acquired to gain resistance to S1-antibodies. Although all six mutations in S2 appeared to act in concert, D796Y showed the greatest impact on neutralization sensitivity and rendered the WT virus >100-fold resistant to S309, COVA2-17, and 4A8. The S2 mutations greatly reduced the antigenicity to neutralizing antibodies due to reduced exposure of epitopes. In addition to the effect on potency, S2 mutations increased antigenic heterogeneity of Omicron spike protein and neutralization of pseudoviruses plateaued below 100%. In terms of the entry pathway, S1 or S2 mutations only partially altered the entry phenotype of WT, and both sets of mutations were required for a complete switch to the endosomal route and loss of syncytia formation. In particular, N856K and L981F in Omicron BA.1 compromised fusion capacity which helps explain why subsequent Omicron variants lost them in order to regain fusogenicity.
IMPORTANCE
The Omicron subvariants have substantially evaded host-neutralizing antibodies and adopted an endosomal route of entry. The virus has acquired several mutations in the receptor binding domain and N-terminal domain of S1 subunit, but remarkably, also incorporated mutations in S2 which are fixed in Omicron sub-lineage. Here, we found that the mutations in the S2 subunit affect the structural and biological properties such as neutralization escape, entry route, fusogenicity, and protease requirement. In vivo, these mutations may have significant roles in tropism and replication. A detailed understanding of the effects of S2 mutations on Spike function, immune evasion, and viral entry would inform the vaccine design, as well as therapeutic interventions aiming to block the essential proteases for virus entry. Thus, our study has identified the crucial role of S2 mutations in stabilizing the Omicron spike and modulating neutralization resistance to antibodies targeting the S1 subunit.
KEYWORDS: SARS-CoV-2, Omicron, spike protein, neutralizing antibodies, virus entry, spike conformation
INTRODUCTION
The most effective response to the COVID-19 pandemic has been the development of vaccines which helped fight the pandemic and reclaim normalcy. However, the emergence of new and highly transmissible variants has jeopardized the vaccine efficacy (1 – 7). Several SARS-CoV-2 variants categorized as variants of concern (VOC) and variants of interest have emerged over the past year that influenced the trajectory of the pandemic (2, 8 – 10). The recent variant Omicron (BA.1) and its newer sub-lineages (BA.2, BA.4, BA.5, BQ.1, and XBB.1) have acquired resistance to vaccine-induced immunity and have been infecting large population worldwide (11 – 15). It is highly plausible that there is still much more genetic space that SARS-CoV-2 can explore to navigate through host immunity without compromising transmissibility (16 – 19).
The spike protein is composed of S1 and S2 subunits in a trimer-of-heterodimers meta-stable state. The protein is conformationally flexible and capable of large structural transitions after receptor binding which leads to membrane fusion and virus entry (20, 21). S1 is a surface protein and harbors key functional domains which are well-exposed to the immune system (22 – 28). The antibodies are frequently directed against the receptor binding domain (RBD) and N-terminal domain (NTD) of S1 subunit which antagonize the binding of spike to cellular receptor. These virus neutralizing antibodies (NAbs) exert selection pressure on these domains giving rise to progeny with escape mutations (17, 29 – 31). The most resistant VOC prior to the emergence of BA.1 was Beta which acquired resistance to convalescent or vaccine sera through only 10 mutations; amongst which E484K is most important and can alone drive much of neutralization resistance (32 – 34). BA.1, however, acquired 28 mutations in S1 including 15 in RBD, and 6 mutations in S2. While the S2 subunit is conformationally stable in the pre-fusion state of spike, the S1 subunit is inherently dynamic (21, 35, 36). Three RBDs on each trimer can transition from closed “down” to open “up” conformation and determine the sensitivity to NAbs and enable binding to cellular receptor. Similarly, NTD, although heavily protected by glycans, has epitopes for NAbs and its conformational dynamics influence their binding to the “supersite” (26, 37, 38). The open and closed transitions of these domains create intrinsic heterogeneity for the virus to exploit against neutralization. The receptor binding motif (RBM) which is occluded in RBD’s “down” conformation is exposed in the “up” conformation and favors interaction with ACE-2 (21, 35, 36, 39). Binding to the receptor follows proteolytic cleavage of the site in S2 and triggers massive conformational transition and insertion of fusion peptide (FP) into the cellular membrane that leads to fusion of the virus and cell membranes (40, 41). Although S1 is flexible and dynamic the RBD-RBD, NTD-RBD interactions are tightly regulated by direct contacts or through the structural linkers (36, 42, 43). The NAb resistance mutations are selected based on the principle that they do not affect the interaction with receptor and fusion with the host cell but prevent binding of NAbs to Spike protein (44 – 46). BA.1 has acquired several mutations in NTD and RBD as well as subdomain-1 and -2 (SD-1 and SD-2). In addition to that, and unlike previously known VOCs, BA.1 acquired six mutations in S2. These mutations alter the conformational states of S1 subunit and also the S2 subunit which in turn impact antibody binding to their cognate epitopes (47).
In the present work, we have studied the effects of mutations in BA.1 spike on the virus’s ability to resist NAbs and on the entry pathway. The mutations in S2 subunit have persisted in all the Omicron variants suggesting that they have a strong functional role. As S2 is much less immunogenic compared to S1, the selection of mutations in S2 must have indirect overall effects on the conformation and function of the spike protein. Identification of the mutations and understanding the mechanism of NAb resistance is critical for vaccine design. Similarly, the pathway of virus entry has implications for pathogenicity. BA.1 is less pathogenic than earlier VOCs as it does not fuse primarily at the plasma membrane (PM) and forms much smaller syncytia, unlike Delta variant which is highly fusogenic and much more pathogenic (48, 49). While BA.1 is poorly replicating due to its altered entry pathway, the new sub-lineages of Omicron tend to reverse this phenotype by acquiring novel mutations (50, 51). Therefore, understanding the mechanism and relationship between neutralization resistance and virus entry of Omicron is important and can provide insights into SARS-CoV-2 pathogenicity and vaccine design.
RESULTS
Omicron BA.1 is resistant to vaccine sera, but BTI boosts neutralization titers
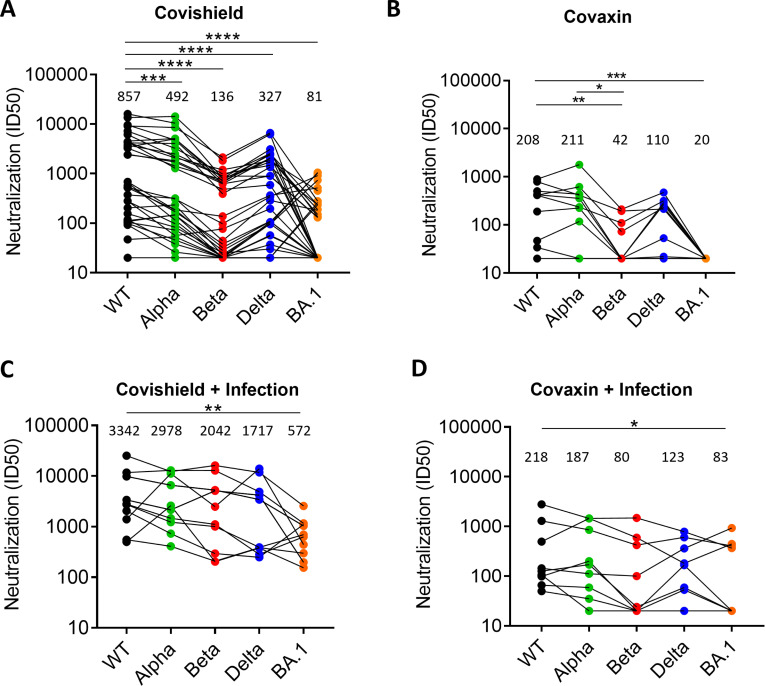
In India, two vaccines were approved in 2020 in the national program of vaccination—Covishield and Covaxin, and most of the population is vaccinated using these vaccines. Covishield is a chimpanzee adenovirus expressing the SARS-CoV-2 spike and was produced by the Serum Institute of India and Covaxin, a whole virus-inactivated vaccine that was developed by Bharat Biotech, India (52, 53). The sera from vaccinees were collected after two doses of the same vaccine between 3 and 5 weeks post-vaccination and used for the assessment of neutralization titers and geometric mean titer (GMT). The Covishield vaccine sera (n = 33) neutralized pseudovirus (PV) expressing wild type (WT) spike potently (GMT-857) compared to other VOCs (Fig. 1A). The neutralization titers significantly decreased for Alpha (P = 0.003), Beta (P < 0.0001), Delta (P < 0.0001), and Omicron BA.1 (P < 0.0001). BA.1 was the most resistant (GMT 81) followed by Beta (GMT 136), and only 13 and 17 out of 33 sera neutralized these variants, respectively, with serum neutralization titer of >50. Thus, as reported in several other studies E484K present in Beta probably drove resistance to serum NAbs, whereas E484K and several other mutations in the spike of BA.1 afforded the greatest resistance to neutralization. The Covaxin sera (n = 10) were weakly neutralizing compared to Covishield sera (Fig. 1A and B). The GMT of neutralization of Covaxin sera against the WT and Alpha variant was 208 and 211, respectively, whereas it was significantly reduced for Beta and Delta (Fig. 1B). None of the sera neutralized BA.1, but three sera neutralized the Beta variant at serum dilution of >100.
Fig 1.
NAb response induced by vaccines to SARS-CoV-2 variants: The NAb response assessed in PV neutralization assay against five SARS-CoV-2 VOC induced by (A). Covishield sera (n = 33) (B). Covaxin sera (n = 10) (C). Covishield followed by breakthrough infection sera (n = 12) (D). Covaxin followed by breakthrough infection sera (n = 9). The GMT is shown for each PV above each column. The statistical significance of differences between the groups was calculated by using Wilcoxon matched-pairs signed rank test and two-tailed P value are indicated as **** (P < 0.0001) and *** (P < 0.003).
We also collected sera samples from vaccinated people who became infected with SARS-CoV-2 after >3 months post-vaccination. These individuals with breakthrough infection (BTI) were different from the only-vaccine group; therefore, the comparative analysis was cross-sectional. The sera were collected during the third wave in India in which BA.1 was the dominant variant circulating and Delta being the subdominant variant. The median neutralization titers in Covishield-BTI vaccinees (n = 12) were increased against all the variants tested (Fig. 1C). All the sera also neutralized BA.1 variant with moderate titers, although the titer was significantly lower than for WT virus (P = 0.008; Fig. 1C). The titers significantly increased in BTI group compared to vaccine group against Alpha, Beta, Delta, and BA.1 (Fig. S1). The neutralization titers in Covaxin-BTI vaccinees (n = 9) were also increased against all the variants tested, although, five sera still could not neutralize beta and BA.1 variants (Fig. 1D). Overall, these titers were much lower and showed high variance that precluded statistical analysis.
The neutralization resistance of BA.1 is contributed by both S1 and S2 subunit mutations
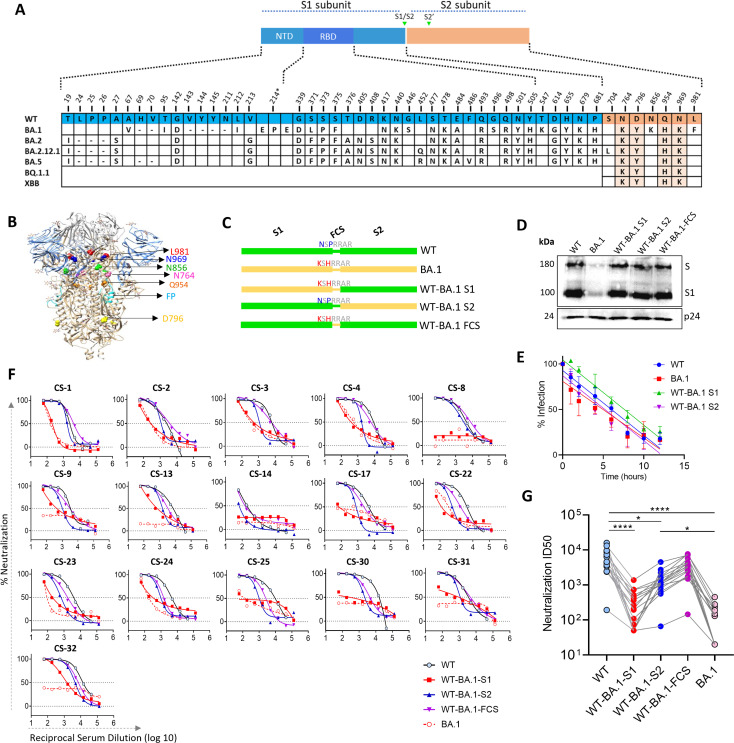
There are 34 mutations in Omicron-BA.1 spike, of which 28 are present in S1 and 6 are in S2 subunit (Fig. 2A and B). To investigate the contribution of these mutations to neutralization resistance of BA.1, we made the chimeric spikes by replacing S1 or S2 subunits of WT with that of BA.1. In the third construct, two BA.1 substitutions N679K and P681H near furin cleavage site (FCS) were also made in WT spike background (Fig. 2C). We prepared the PVs from these constructs and assessed their stability by analyzing Spike and HIV p24 protein expression in western blot. The data showed a similar expression of p24 across the PV panel. The expression of WT and chimeric Spike proteins was also similar; however, the BA.1 Spike expression was lower, which agreed with the findings reported earlier (Fig. 2D) (54). In the infectivity assay using 293T-hACE2/TMPRSS2 cells, the PVs showed similar time-dependent changes in infectivity after incubating at 37°C for various times (Fig. 2E). The data suggested that a similar amount of Spike was incorporated on the virions for the different constructs, and they did not differ in stability based on their biological function. Next, we assessed the neutralization sensitivity of WT and chimeric spike PVs bearing WT-BA.1S1, WT-BA.1S2, and WT-BA.1-FCS to potent Covishield sera. The PVs bearing WT or WT-BA.1-FCS spike were similarly sensitive to these sera, but the WT-BA.1S1 PV was resistant, and its degree of resistance was equivalent to BA.1 PV (Fig. 2F and G). Whereas WT-BA.1S2 PV was sensitive to all sera, its sensitivity was significantly reduced when compared with WT (P = 0.012; Fig. 2G). These results suggested that most of the neutralization resistance was driven by the mutations in S1 subunit and that is probably because they directly disrupt the sequence of epitopes or their conformations. The mutations in S2, albeit modestly, also contributed to resistance. The contribution of S2 mutations in neutralization resistance was surprising as S2 is poorly immunogenic, and no significant selection pressure on this subunit is expected.
Fig 2.
Neutralization sensitivity of WT and mutant spike-expressing PVs to Covishield vaccine sera: The BA.1 mutations in S1, S2 subunit, and two mutations near FCS (N679K and P681H) were separately transferred in WT spike. (A) The alignment of WT and Omicron Spike variants showing the mutations in Omicron with reference to WT. Four conserved mutations in Omicron sub-variants shown in orange (B). Six mutations in S2 subunit are highlighted on the Spike ectodomain structure, and their positions indicated on one protomer. (C) Schematic of the spike protein shown for each parental and mutant spike protein. All substitutions were done in WT background (D). Western blot analysis of PVs expressing various Spike proteins and capsid protein p24. Shown are the bands for S (whole Spike) and S1 subunit probed with mouse serum antibodies raised against WT and BA.1 RBD immunogen. p24 from PV capsid was a loading control and was probed with HIV-1 positive serum. The data are representative of three independent repeats. (E) Infection of PV in 293T-hACE2-TMPRSS2 cells after incubating at 37°C. The decrease in infection over time was plotted with linear regression (F). Neutralization of PVs displaying WT or mutant spike, by select high-titer Covishield vaccine sera with percent neutralization plotted as a function of serum dilution (G). Inhibitory dilution-50 (ID50) for the sera in F for parental and mutant viruses. The ID50 values were calculated from two independent assays. The statistical differences between the groups were calculated by using the Friedman test and comparisons done using Dunn’s multiple comparison test.
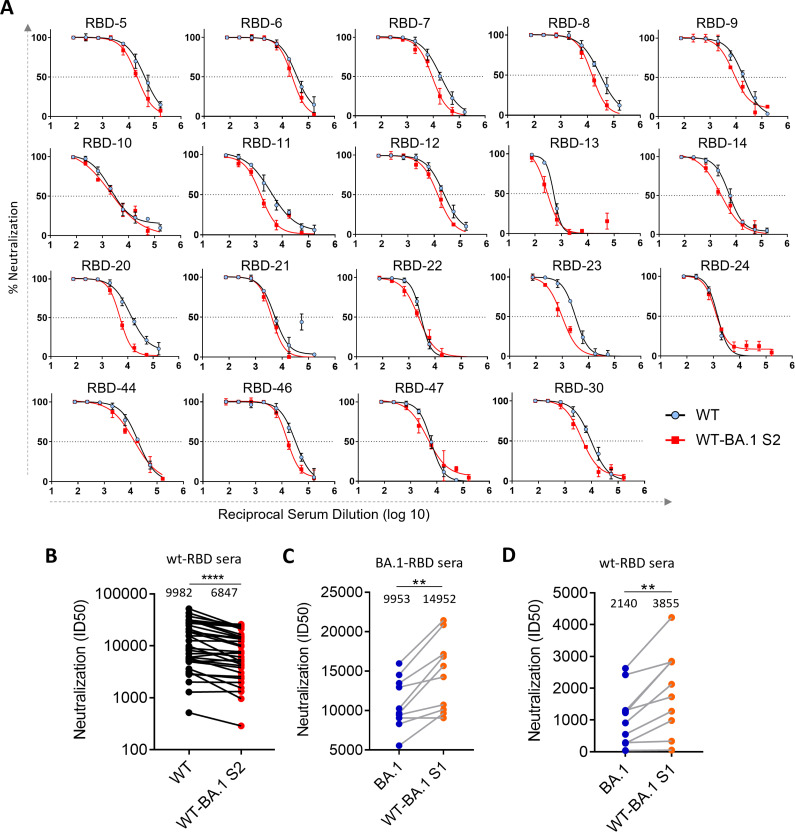
We considered two possibilities that might have driven the neutralization resistance via S2 mutations (i) there are low-titer antibodies in the sera targeting the S2 subunit and the mutations in S2 affected their binding to epitopes in that subunit. (ii) the mutations in S2 subunit indirectly altered the presentation of NAb epitopes in S1. To examine the second possibility, we compared the neutralization sensitivity of WT and WT-BA.1S2 PVs, both of which carry S1 of WT spike, to mouse sera raised by RBD immunogens (55, 56). These RBD immunogens were based on WT-spike sequence with or without stabilizing mutations (55, 56). The mice sera potently neutralized both WT and WT-BA.1S2 PVs; however, the neutralization potency against WT-BA.1S2 was moderately reduced for many of the sera (Fig. 3A and B). The GMT for the WT and WT-BA.1 S2 virus was 9,982 and 6,847, respectively (Fig. 3B). Taken together, the ID50 values were significantly reduced for WT-BA.1S2 compared to WT PV (P < 0.0001). To further probe the role of S2 mutations, we analyzed the neutralization of PV bearing BA.1 or WT-BA.1S1 spike (Fig. 2A). We used sera from mice that were immunized with BA.1- or WT-RBD antigen. The BA.1-specific sera neutralized both BA.1 and WT-BA.1S1 more potently than WT-RBD-specific sera. However, overall, the WT-BA.1S1 was significantly more sensitive to both panels of sera (P = 0.002; Fig. 3C and D). Taken together, these data suggested that the mutations in the S2 subunit modulated the neutralization sensitivity to RBD antibodies.
Fig 3.
Neutralization sensitivity of WT and WT-BA.1S2 to RBD sera: (A) The neutralization curves are shown for WT and WT-BA.1S2 PVs as a function of serum dilution. (B) The neutralization titers against WT and WT.BA.1S2 for mouse sera (n = 34) elicited by WT-RBD immunogen. The GMT titer is shown above each column. (C) The neutralization titers against BA.1 and WT-BA.1S1 PVs are shown for mouse sera (n = 10) elicited by BA.1-RBD immunogen. (D) Same as in C but with mouse sera elicited by WT-RBD (n = 10). The median titers are shown above each column. Two independent assays were done to confirm reproducibility.
The mutations in S2 affect the neutralization by monoclonal antibodies specific to NTD and RBD
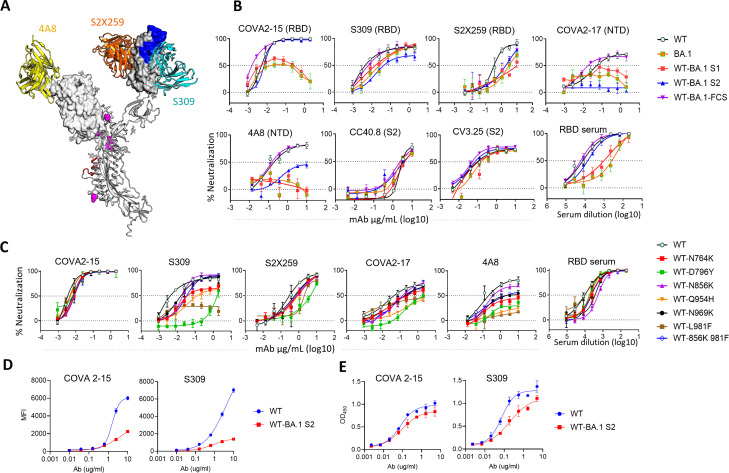
Next, we wanted to see the effect of mutations in S2 subunit on neutralization by antibodies that target NTD and distinct epitopes in RBD. We used a panel of PVs consisting of parental (WT and BA.1), chimeric (WT-BA.1S1 and WT-BA.1S2), FCS mutant (WT-BA.1-FCS), and WT virus with individual S2 mutations (Fig. 4). We used COVA2-17, 4A8 (NTD antibodies), and COVA2-15, S309, and S2 × 259 (RBD antibodies) to analyze the impact on these epitopes (Fig. 4A). In addition, we also used a pooled serum from mice, which were immunized with WT-RBD immunogens, and recognize multiple epitopes in RBD (56). The neutralization sensitivity of WT-BA.1S2 was unchanged for COVA2-15 compared to WT. However, the neutralization sensitivity to S309 and S2 × 259 was reduced for WT-BA.1S2 and some S2 mutants, particularly, WT-D796Y and WT-L981F (Fig. 4B and C). We also assessed the neutralization sensitivity of a double mutant WT-856K 981F, as these two mutations showed substantial effects on fusogenicity (see Fig. 7). This double mutant, however, showed similar neutralization sensitivity to WT. Thus, the resistance conferred by L981F alone was blunted by N856K suggesting epistatic effects. In the WT spike background, the D796Y alone enhanced neutralization resistance significantly to Covishield or human convalescent sera (Fig. S2A and B). Conversely, the reversion of this mutation in BA.1 spike significantly enhanced the neutralization by Spike- or BA.1-RBD sera confirming the role of this mutation in neutralization (Fig. S2C and D). For the polyclonal serum pool, WT-BA.1S2 and some of the point mutations also became slightly resistant (Fig. 4B and C). The NTD antibodies 4A8 and COVA2-17 neutralized WT virus potently, although the maximum neutralization achieved was in the range of 70%–80%. These antibodies did not neutralize BA.1 and WT-BA.1S1 due to sequence change in epitopes and directly affecting their binding. Interestingly, WT-BA.1S2 became completely resistant to COVA2-17 and partially resistant to 4A8 (Fig. 4B). The single S2 mutations also variably altered the neutralization pattern for NTD antibodies—several mutations reduced maximum neutralization to <50% (Fig. 4C). However, the neutralization by S2-specific antibodies (CC40.8 and CV3.25) was not changed (Fig. 4B). This data revealed that the mutations in S2 indirectly modulated the exposure of epitopes in NTD as well as RBD thereby affecting neutralization. As to how the S2 mutations might be influencing the neutralization, we further analyzed the antigenicity of WT and WT-BA.1S2 spikes by using flow-cytometry assay by expressing Spike on the cell surface or by ELISA using soluble spike protein. The S309 and COVA2-15 both bound better to WT than WT-BA.1S2 tracking with the neutralization results (Fig. 4D and E).
Fig 4.
Neutralization resistance to monoclonal NAbs conferred by Omicron mutations: (A) The model showing RBD and NTD domains on a Spike ectodomain from a published structure in complex with S309 Fab (PDB: 6WPS) (57). The Fab S2 × 259 [PDB:7RA8 (58)] and Fab 4A8 [PDB: 7C2L (24)] were superposed on this structure. The RBM is shown in blue which contains an epitope of COVA2-15 for which the high-resolution structure is unavailable. In S2 subunit, the FP is shown in red and mutations in magenta. The molecular graphics were created using Pymol software suite. For clarity, only one Spike protomer is shown. (B and C) The neutralization curves for various mAbs or a mouse serum pool elicited by WT-RBD antigen against a PV panel (D). The binding of RBD NAbs to WT and WT-BA.1S2 spikes expressed on 293T cells shown as mean fluorescence intensity (MFI) assayed using fluorescence activated cell sorting (FACS) (E). Binding of RBD NAbs to WT and WT-BA.1 S2 soluble ectodomain Spike protein with K986P, V987P (2P) stabilizing mutations, estimated by measuring OD at 450 nm in ELISA assay. Assays were repeated twice independently.
The S2 mutations in Omicron BA.1 alter the exposure of epitopes in S1 subunit
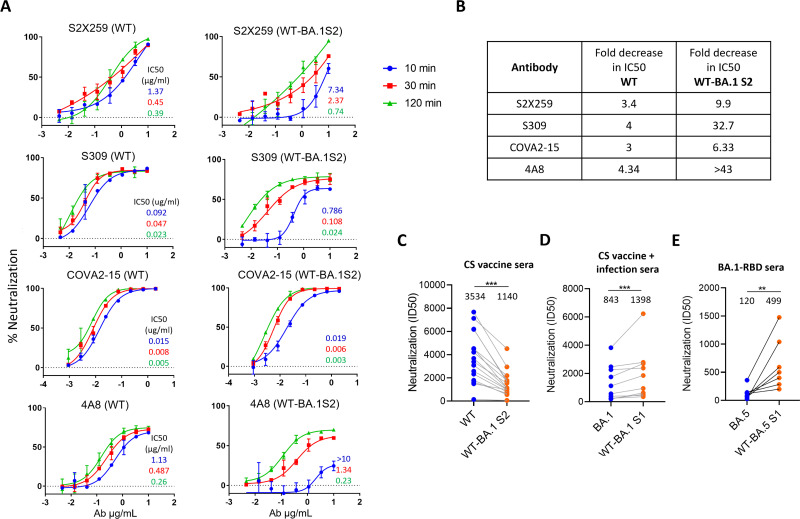
The BA.1 spike is relatively more stable and has a more compact configuration than WT or Delta spike (47, 59, 60). In addition to epitope disruptions by mutations in RBD and elsewhere in the S1 subunit, the slower transition from “down” to “up” conformation of RBD could be another reason why this variant is more resistant to RBD antibodies (47). The WT-BA.1S2 spike showed reduced sensitivity to monoclonal antibodies targeting RBD and NTD and also to the polyclonal sera raised by RBD immunogen. This result indicated that S2 mutations have an impact on conformational flexibility and on exposure of S1-epitopes. To understand the mechanism of reduced exposure, we assessed the kinetics of neutralization using a panel of antibodies including S2 × 259 (58). The epitope of S2 × 259 is well-characterized and exposed only in “up” conformation of RBD. The virus was incubated with the antibody for various time periods before adding to the target cells. S2 × 259 and S309 both neutralized WT virus efficiently within a 10-minute incubation period, and neutralization increased slightly at 30 and 120 minutes (Fig. 5A). In contrast, WT-BA.1S2 was neutralized poorly at 10 minutes, but neutralization substantially increased at longer incubation times (Fig. 5A). The reduction in S2 × 259 IC50 values from 10- to 120-minute incubation was by 3.4- and 9.9-fold for WT and WT-BA.1S2, respectively; for S309, fourfold and 32.7-fold, respectively. Similarly, 4A8 IC50 for WT and WT-BA.1S2 was reduced by 4.34- and >43-fold, respectively (Fig. 5B). However, for COVA2-15, which is a cluster-I antibody targeting RBM, the neutralization increased moderately, and only threefold and 6.33-fold reduction in IC50 were seen for WT and WT-BA.1S2, respectively (Fig. 5A and B). Although S309 is shown to engage both “up” and “down” conformations of RBD, our data indicated altered neutralization kinetics in the context of virus. Conversely, the same experiment was done with Omicron BA.1 and BA.5 by comparing with their mutants (reversion of S2 mutations to WT) WT-BA.1S1 and WT-BA.5S1, respectively. S309 neutralized WT-BA.1S1 and WT-BA.5S1 faster than their counterparts suggesting that the S2 mutations influenced the exposure of epitopes in RBD (Fig. S3). Next, the overall effect of reduced exposure on neutralization sensitivity was assessed using various sera samples. The Covishield vaccine sera neutralized WT-BA.1S2 less well than WT (Fig. 5C). Conversely, Omicron BA.1 or BA.5 with reverted S2 mutations (WT-BA.1S1 or WT-BA.5S1) was better neutralized than their parental PVs by the human sera from vaccine-and-infection individuals or BA.1-RBD-specific mouse sera (Fig. 5D and E).
Fig 5.
Kinetics of neutralization of WT and WT-BA.1S2 PVs by monoclonal NAbs: (A) Neutralization curves are shown for WT and WT-BA.1S2 PVs by NAbs incubated with virus for 10 or 30 or 120 minutes. The IC50 values (µg/mL) are shown in inset derived from respective neutralization curves and indicated by the same color as used for respective neutralization curves. The data shown represent the assays done at three independent times. (B) Values are the fold-decrease in IC50 from 10 to 120 minutes for WT and WT-BA.1S2 PVs against indicated antibodies. (C) Comparison of sensitivity of WT and WT-BA.1 S2 PV to Covishield vaccine sera. (D) Sensitivity of BA.1 and WT-BA.1S1 PV to Covishield vaccine followed by BTI sera (hybrid immunity). (E) Neutralization sensitivity of Omicron BA.5 and WT-BA.5S1 to mouse sera elicited by BA.1-RBD immunogen. The median serum titer is indicated for each PV variant.
The S2 mutations reduced the effective neutralization
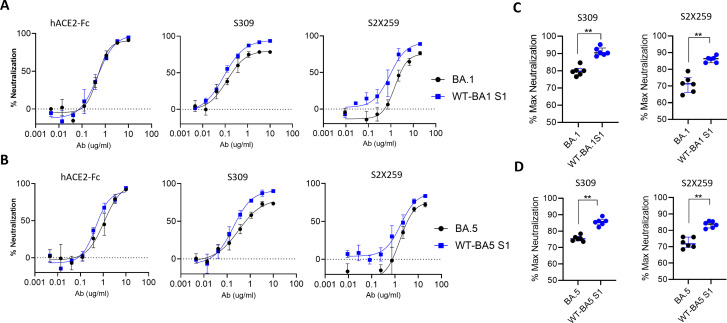
We observed that even single mutations in S2 region of WT spike reduced the maximum neutralization that S309 and S2 × 259 could achieve (Fig. 4B and C). Here, we assessed the effect of S2 mutations using BA.1 and BA.5 spikes, and their mutants in which S2 mutations reverted to WT. The neutralization assays were done to obtain a complete neutralization curve with a saturating neutralization plateau. S309 and S2 × 259 neutralized WT-BA.1S1 and WT-BA5S1 to a higher extent than their parental spikes BA.1 and BA.5, respectively (Fig. 6A and B). Whereas, the neutralization efficiency of hACE2-Fc was similar suggesting that exposure of RBM is not affected, but the conserved S309- and S2 × 259-epitope in RBD were less exposed for neutralization. The maximum neutralization at the highest concentration of S309 or S2 × 259 was significantly higher for WT-BA.1S1 and WT-BA5S1 compared to parental BA.1 and BA.5, respectively (Fig. 6C and D). This data suggested that S2 mutations regulate the exposure of RBD and also create heterogeneous antigenic forms of Spike protein that facilitate escape from neutralization not only by reducing the potency but also by reducing the efficacy of neutralization.
Fig 6.
Neutralization efficacy of Omicron subvariants and mutant PVs: Neutralization curves using HEK293T-ACE2/TMPRSS2 cells for (A). BA.1 and WT-BA.1S1 (B). BA5 and WT-BA5S1 against indicated neutralizing agents. (C and D) The percent maximum neutralization by S309 at 10 µg/mL or S2 × 259 at 20 µg/mL. The percent neutralization values for the indicated PVs were taken from six independent assays. The statistical difference between two PVs was calculated using Mann-Whitney test.
The route of viral entry and fusogenicity is determined by sequence changes in S1 and S2
A major adaptation in BA.1 compared to earlier VOCs is that it is less dependent on TMPRSS2 for the cleavage of S2’ site (second cleavage site in S2) and release of FP. Due to the non-requirement of TMPRSS2-mediated cleavage of BA.1 spike at the cell surface, it is not primed at the cell surface, and virus does not fuse at this location (49). The S2’ site is conserved across the variants, and we hypothesized that the mutations in S2 subunit of BA.1 might affect the cleavage by TMPRSS2 and thereby the fusion process at PM. We used the same panel of PVs as described in Fig. 2 to study the effect of S2 mutations on the entry-related features.
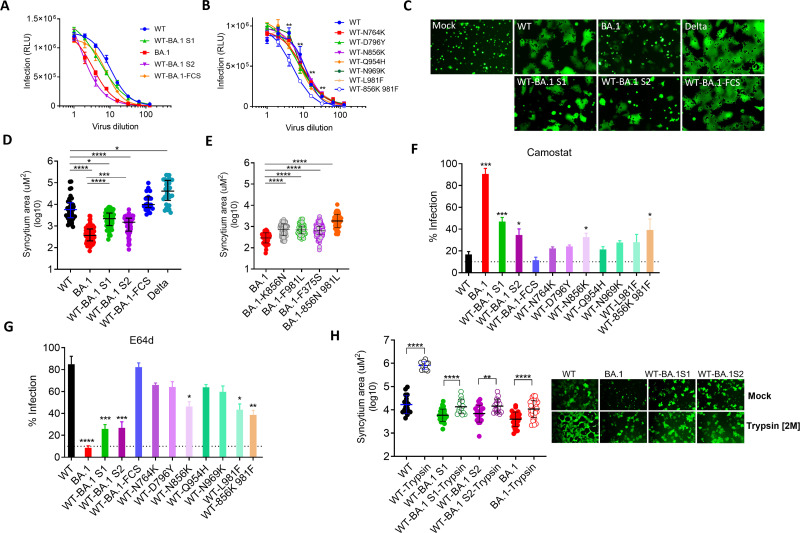
The infectivity of WT-BA.1S1 and WT-BA.1-FCS mutant (N679K, P681H) was slightly reduced compared to WT PV, but the infectivity of WT-BA.1S2 was markedly reduced and was comparable to BA.1 (Fig. 7A). However, individual S2 mutations in WT background did not affect the infectivity, but N856K and L981F together modestly reduced the infectivity suggesting that these two mutations have more impact than others (Fig. 7). The fusogenicity was highest for Delta followed by WT; and BA.1 showed least fusogenicity and displayed very few syncytia which were significantly smaller in size than WT (Fig. 7C). The chimeric spike PVs WT-BA.1S1 and WT-BA.1S2 showed reduced ability to fuse with receptor-expressing cells, and size of their syncytia also was significantly reduced (Fig. 7C and D). The fusogenicity of WT-BA.1-FCS mutant was slightly enhanced and was comparable to Delta. The fusogenicity of WT containing individual BA.1 S2 mutations showed similar fusogenicity as WT (Fig. S4A). Conversely, reversion of S2 mutations in BA.1 background was also evaluated. Here, the K856N and F981L individually or both combined clearly increased the fusogenicity of BA.1 spike; the double mutation appeared to exert an additive effect (Fig. 7E; Fig. S4B). The F375S change which was previously reported by Kimura et al. for increased fusogenicity of BA.1 was also confirmed for increased fusogenicity in this comparison (Fig. 7E; Fig. S4B). Furthermore, to assess the general effects of S2 mutations in Omicron sublineages, BA.5 variant was compared with its mutant counterpart in which all four S2 mutations were changed to WT. The fusogenicity of the mutant spike (WT-BA5S1) was significantly increased (Fig. S2E). Collectively, the data revealed that S2 mutations reduced the fusogenicity of Omicron lineage spikes.
Fig 7.
Entry of PVs bearing various spike proteins: (A and B) Serially diluted PVs were used for the infection of 293T-hACE2-TMPRSS2 cells. Infectivity [relative luminescence unit (RLU)] is shown against various dilutions of PV expressing parental and mutant spikes. The statistical significance for the difference in infectious titers between WT and WT-856K 981F at a given dilution was calculated using Mann-Whitney test. (C) HEK293T cells expressing Spike and GFP were added onto the monolayer of Vero-TMPRSS2. Shown are the images of syncytia after 2-hour incubation formed by Spike variants, or without Spike (Mock). (D and E) The size of the syncytia measured in µm2 (micrometer squared) plotted for indicated Spike variants. (F and G) Infectivity of PVs in the presence of TMPRSS2-inhibitor camostat or cathepsin-inhibitor E64D. The statistical significance was calculated with reference to WT. The statistical significance in D–G was calculated using Kruskal-Wallis test, and comparisons were done by Dunn’s multiple comparison post test. (H) Syncytia formation of Vero-TMPRSS2 by Spike-expressing HEK293T cells in the presence of trypsin. The images were taken after 2 hours of addition of spike-expressing 293T cells onto Vero-TMPRSS2. The statistical analysis was done by using Kruskal-Wallis test, and P values were calculated by Dunn’s multiple comparison post-test. All the experiments were repeated at least twice.
We next assessed the effect of mutations on the virus entry mechanism. We used protease inhibitors camostat and E64d as inhibitors of TMPRSS2 (route-i) and endosomal cathepsins (route-ii), respectively, to assess the dependence of these proteases on the entry of SARS-CoV-2 variants. The infectivity of WT virus was reduced to baseline by camostat but was not affected by E64d. Conversely, the infectivity of BA.1 was sensitive to E64d but not to camostat which suggested that WT mainly used TMPRSS2 and fused at PM, whereas BA.1 mainly used cellular cathepsins in the endosomal compartment. WT-BA.1S1 and WT-BA.1S2 both were partially sensitive to E64d and camostat. Compared to WT, the infectivity of WT-BA.1S1 and WT-BA.1S2 was significantly higher in the presence of camostat and significantly lower in the presence of E64d (Fig. 7F and G). The sensitivity of WT-BA.1-FCS to both the inhibitors was similar to WT. The infectivity of all six single S2 mutants in the presence of camostat was slightly higher than WT, but only for N856K and N856K/L981F, it was significantly higher than WT. Conversely, the infectivity of N856K, L981F, and N856K/L981F was significantly reduced by E64d treatment which suggested that these two mutations enhanced endosomal entry of BA.1 more than other mutations in S2 (Fig. 7F and G). The enhanced endocytosis of WT by these two mutations also corroborated the enhanced fusogenicity of reverse mutations in BA.1 spike (i.e., the opposite effect; Fig. 7E, S4B). Next, we assessed the fusogenicity of WT, BA.1, and WT-BA.1S2 in the presence of trypsin. Trypsin dramatically enhanced the fusogenicity of WT but modestly for BA.1 and WT-BA.1S2 (Fig. 7H). This data suggested that S2 mutations affected the S2’ cleavage by TMPRSS2 and trypsin can partially reverse that effect. Overall, the mutations in either of the subunit only partially switch the entry pathway from PM to endocytosis, but the mutations in both subunits complement for a complete switch to endosomal route of entry.
DISCUSSION
In the present study, we investigated the role of mutations in S1 and S2 subunits of omicron spike protein. Omicron is by far the most mutated variant of SARS-CoV-2 since the onset of the COVID-19 pandemic, and apart from acquiring neutralization resistance, it also employs a different entry pathway. As there are several mutations in omicron spike, it is unlikely that any single mutation or a smaller set of mutations would contribute to these phenotypes, instead it is a cumulative effect of several mutations (61). As expected, and because most neutralization epitopes lie in the S1 subunit, the mutations in S1 drove most of the resistance. However, mutations in S2 also afforded substantial resistance to neutralization. The S2 mutations are mostly buried in the spike structure, are not part of prominent epitopes in S2, and did not alter the neutralization by S2-specific NAbs (62, 63). Therefore, resistance imparted by S2 mutations must be due to their allosteric effects on S1 epitopes. The set of six S2 mutations in WT spike reduced its neutralization sensitivity to monoclonal or polyclonal antibodies targeting the RBD and also resulted in marked resistance to S309, 4A8, and COVA2-17. The individual S2 mutations in the WT background also variably reduced the sensitivity to RBD and NTD antibodies, particularly, D796Y. This mutation is located close to the FP and may alter the FP dynamics and change the energetics of RBD position for reduced accessibility. This appears to be an additional strategy employed by viruses to gain neutralization resistance to antibodies targeting RBDs (64, 65). Kemp et al. showed that D796H/D796Y frequently surfaced in viremia in an immunocompromised COVID-19 patient during plasma therapy (29). In another study by Schmidt et al., virus-acquired mutations, P792H and N801D, close to this region when passaged in the presence of neutralizing sera underscoring the importance of this domain in regulating spike conformation (30). Whether S2 mutations can also alter the glycosylation profile of viral Spike and influence the antibody binding to NTD needs further investigation.
Omicron spike protein represents a highly stabilized conformation due to improved contacts at the interface of S1 and S2 and at the interprotomeric S1-S1 and S2-S2 contacts which result in a compact configuration of Spike (59). The resistance to NAbs can be partially brought about by mutations in RBD and NTD, whereas mutations in S2 core are used as leverage to compact the overall configuration of the Spike and to reduce the exposure of neutralizing epitopes in RBD and NTD (47). Reduced exposure can provide more time for the virus to bind to ACE-2 before getting neutralized. The neutralization of WT-BA.1S2 by S309 and 4A8 was substantially increased when antibody and virus were incubated for longer period of time, whereas such increase in neutralization was much smaller for WT. The observation was further bolstered by reverting S2 mutations in BA.1 and BA.5 spike which led to increased neutralization by RBD-antibodies. Another important aspect of S2 mutations is to reduce the maximum neutralization capacity of antibodies (i.e., <100%). The S2 mutations in combination with other mutations in Omicron subvariants appear to enhance the Spike antigenic heterogeneity by retaining RBD in “down” conformations. This increases the fraction of un-neutralizable virus population reducing not only potency but also efficacy of neutralization. This provides an explanation for why Omicron and all its subvariants from BA.1 to XBB retained S2 mutations while changing the mutational landscape in S1.
Omicron not only adapted to resist host NAbs but also varies from earlier VOCs with respect to TMPRSS2 usage and the entry mechanism. In contrast to previous VOCs, Omicron takes the endosomal route for entry. The cleavage of S2’ is a critical event that allows the FP to insert into the host cell membrane to trigger membrane fusion (66). The mutations near this site affected its cleavage by TMPRSS2 and thereby fusion at the PM which was partially reversed by the addition of trypsin. Omicron entry depended on intracellular cathepsins but not TMPRSS2, whereas WT used TMPRSS2 and fused efficiently at PM (49, 67). This difference is substantially accounted for by the spike S2 mutations, particularly N856K and L981F, which substantially reduced the fusogenicity and attenuated the entry efficiency of Omicron. These two mutations are reverted to WT in Omicron subvariants (BA.2 to XBB), probably to regain entry efficiency. Indeed, the fusogenicity and replication rate is enhanced from BA.1 to BA.5 with optimization of the mutation profile (epistatic control) to adopt beneficial traits (68). The compensatory epistasis is more prevalent for maintaining ACE-2 affinity brought about by mutations within RBD (45). Recently, Kimura et al. showed the importance of S375F substitution for neutralization resistance to vaccine sera which also partially reduced Omicron’s fusogenicity (69). It is plausible that mutations in Omicron were selected for resistance to host NAbs by directly and indirectly modulating the epitopes in spike protein. As a result, multiple mutations stabilize spike trimer, retain affinity with ACE-2, but also alter the route of virus entry as an unintended consequence. These properties confer an advantage in terms of better transmissibility in the face of host defense and might expand the tissue tropism (70). In the continued course of evolution, the genetic drift shapes Omicron spike further to be more resistant and fusogenic as exemplified by sub-lineages BA.2, BA.4, and BA.5 (50). The N856K and L981F in WT reduced fusogenic capacity of spike and increased E64D sensitivity—the propensity for endosomal entry. The lack of these two mutations in BA.2 sub-variant of omicron may explain why it is more fusogenic than BA.1 (50). Thus, the present work delineated the role of mutations in spike-S2 subunit in the long-range interactions controlling the exposure of RBD and NTD and provide insights in conformational dynamics and allosteric modulation.
MATERIALS AND METHODS
Cells
Human embryonic kidney (HEK293T) cells were used for the transient expression of SARS-CoV-2 spike and HIV-1 proteins to prepare pseudotyped virus. HEK-293T cells expressing human angiotensin converting enzyme-2 (hACE2) or expressing hACE2 and TMPRSS2 were obtained from BEI Resources and used for the neutralization assays and infectivity assays. Vero-E6-TMPRSS2 (JCRB cell bank, JCRB #1818) and 293T-hACE2-TMPRSS2 cells were used in the cell-to-cell fusion assay. All cell lines were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% fetal bovine serum (FBS).
Serum samples and ethical approval
Serum samples were collected from individuals who were given two doses of Covishield or Covaxin vaccine. Sera were also collected from covaxin or covishield-vaccinated individuals after the BTIs in the third wave in February 2022 in India. Written informed consent forms were collected from all participants before collecting blood samples for sera. The study was approved by Institutional Ethics Committee of CSIR-Institute of Microbial Technology (IEC August 2020#2) and CSIR-Institute of Genomics and Integrative Biology (Approval no. CSIR-IGIB/IHEC/2020–21/01).
Spike gene constructs
The SARS-CoV-2 spike constructs expressing the spike protein of Wuhan strain with D614G, Omicron BA.1, or BA.5 spike were based on the codon-optimized spike sequence of SARS-CoV-2 and were generated by GenScript Inc. Wuhan strain sequence with D614G was termed as WT in this paper. All the spike constructs had 19 amino acid deletion at the C-terminal end for better cell-surface expression (71). The chimeric spike constructs were made between WT and Omicron genes where S1 or S2 of WT spike were replaced by corresponding fragments of Omicron. S1 or S2 of Omicron fragments were PCR amplified by using specific primers. Similarly, vector backbone of WT spike excluding S1 or S2 was amplified by reverse primers with the overlap of 18 bases between insert-amplifying and vector-backbone- amplifying primers. The PCR fragments were digested by DpnI to remove template DNA and purified by gel-extraction. The vector and insert PCR fragments were then ligated by using In-Fusion cloning kit (CloneTech, Inc.). Two BA.1 mutations, N679K and P681H, adjacent to FCS were added in WT spike by using specific primers containing that sequence change and site-directed mutagenesis was done by overlapping PCR and ligation of insert and vector backbone was done by using In-Fusion cloning kit.
Antibodies
The codon-optimized DNA sequences of heavy and light chain of S309, 4A8, CC40.8, CV3.25, and S2 × 259 were purchased from GenScript and cloned in pcDNA3.4. The expression plasmids of COVA2-15 and COVA2-17 were gifted by Rogier Sanders and Marit van Gils and described in Brouwer et al. (72). The expi293 cells were co-transfected by Heavy and Light chain plasmids in 1:1 ratio by using polyethylenimine, and supernatants were harvested after 4 days post-transfection. The antibodies were purified from supernatants by using Protein A/G beads and finally stored in phosphate buffered saline (PBS) for use in experiments.
SARS-CoV-2 PV preparation and neutralization assay
PV neutralization assays were performed by using HIV-1-based PV as described previously with some modifications (56). Briefly, HEK293T cells were transiently transfected with plasmid DNA pHIV-1 NL4·3Δenv-Luc and Spike-Δ19-D614G by using a profection mammalian transfection kit (Promega Inc.). After 48 hours, the culture supernatant was harvested and filtered using 0.22 µm filter, and stored at −80°C until further use. 293T-hACE-2 (BEI resources, NIH, Catalog No. NR-52511) cells expressing the ACE2 receptors were cultured in DMEM (Gibco) supplemented with 5% FBS, penicillin−streptomycin (100 U/mL). Vaccine sera were heat inactivated at 56°C for 30 minutes and serially diluted in a growth medium starting from 1:20 for neutralization of PV infection. The PV was incubated with serially diluted sera in a total volume of 100 µL for 1 hour at 37°C. The cells were then trypsinized, and 1 × 104 cells/well were added to make up the final volume of 200 µL/well. The plates were further incubated for 48 hours in a humidified incubator at 37°C with 5% CO2. After incubation, neutralization was measured as indicator of luciferase activity in the cells [relative luminescence unit (RLU)] by using nano-Glo luciferase substrate (Promega Inc.). Luminescence was measured by using Cytation-5 multimode reader (BioTech Inc.). The luciferase activity, measured as RLU, in the absence of sera was used as 100% infection. The serum dilution that resulted in half-maximal neutralization of PV (ID50) relative to no-serum control was determined from neutralization curves. All the assays involving the use of SARS-CoV-2 PVs were conducted after the approval by Institutional Biosafety Committee (CSIR/IMTECH/IBSC/2020/J21).
Fusogenicity of spike protein
HEK293T cells were seeded in six-well plate each well containing 4 × 105 cells. The next day, cells reached ~40%–50% confluency and were co-transfected by using spike-expressing and GFP-expressing constructs. On the same day, in separate 96-well plate, Vero-TMPRSS2 cells were seeded each well receiving 2 × 104 cells. After 24-hour post-transfection, 293T cells were checked under a fluorescent microscope for qualitative assessment to confirm if the transfection of each spike construct was equivalent across various spike constructs. The cells were then trypsinized, and 5 × 103 cells/well were added on the monolayer of Vero-TMPRSS2 cells in 96-well plate. The plate was incubated at 37°C and periodically checked under a microscope for syncytia formation. The size of the syncytia was calculated using NIS Elements software (Nikon Instruments Inc.). The same experiment in the presence of trypsin was done using growth media without FBS. The DMEM containing trypsin was added in Vero cells. The spike-expressing cells were trypsinized, resuspended in plain DMEM, and added onto the monolayer of Vero cells. The images were recorded after 2 hours of incubation.
Infectivity assay
1 × 104 293T-hACE2-TMPRSS2 cells/well were seeded in 96-well plate. The next day, PVs stored at −80°C were thawed at room temperature. The serial dilution of virus was done in growth medium starting from neat up to 128-fold dilution. The growth medium from 293T-hACE2-TMPRSS2 cells were removed, and 100 µL virus was slowly added. The plate was incubated for 48 hour at 37°C in a CO2 incubator, and luciferase activity was quantified by adding nano-glo luciferase substrate (Promega). The luminescence was recorded by using Cytation 5 plate reader (BioTek inc). Based on the titration curve, the dilution at which 2 × 106 RLU was obtained was identified for each variant and was considered to contain equivalent infectious units. With this normalization, the infectivity of PV was measured in serial dilution of PV in 293T-hACE2-TMPRSS2 cells.
Infectivity in the presence of protease inhibitors
The sensitivity to protease inhibitors—camostat or E64d—was assessed in 293T-ACE2-TMPRSS2 cells. 2 × 104 cells/well were plated in 96-well plate. The next day, protease inhibitors were serially diluted starting from 40 µM in 100 µL growth media. The media on the cells was replaced with the media containing inhibitors was added on the cells and incubated for 2 hours. 100 µL virus stock was then added on top to make the final volume 200 µL, and the plate was further incubated for 48 hours. The infection was measured by assaying the activity of intracellular luciferase enzyme by using nano-go luciferase system (Promega). The infectivity of each spike variant in the absence of inhibitor was recorded as 100%, and the relative infectivity in the presence of inhibitor was calculated with reference to no-inhibitor control of Spike.
RBD-based immunogen and mice sera
All mice sera raised against RBD immunogens were used from our previously published study (55, 56). The immunogens were based on WT-RBD or BA.1-RBD or Spike ectodomain and were expressed in expi293 cells and purified by using Ni-NTA column followed by tag removal by digesting with protease. Animals were immunized with 10 µg of RBD-based subunit vaccine candidates mixed with adjuvants in 50 µL injection volume intramuscularly, with the prime dose at day 0 and a boost on day 21. After 2 weeks of complete immunization, blood was collected, and serum was separated and stored at −80°C until further use. All animal studies were approved by the Institutional Animal Ethics committee of Indian Institute of Science.
Flow cytometry of transfected cells expressing spike proteins
HEK293T cells (0.6 × 106 cells/well in six-well plate) were transfected with spike-expressing construct. After 24 hours, cells were trypsinized and washed with PBS and subsequently resuspended in FACS buffer (PBS containing 5% FBS). The cells were stained with spike-specific monoclonal antibodies (COVA 2–15, S309, and 4A8) fourfold serially diluted in FACS buffer starting from 10 µg/mL. The cells were incubated with antibodies for 1 hour at 4°C. The cells were washed twice with FACS buffer followed by incubation with 100 µL goat anti-Human IgG, Alexa Fluor 488, at 1:500 dilution of original stock for 45 minutes at 4°C. The cells were washed twice and resuspended in FACS buffer, and the fluorescence intensity was measured using BD FACSVerse. The data were analyzed in FlowJo software (Treestar).
ACKNOWLEDGMENTS
We thank Dr. Paul Bieniasz for providing SARS-CoV-2 Spike expressing plasmid and pHIV-1-NL4.3-nanoLuc construct. Drs. Rogier Sanders and Marit van Gils for providing H and L chain constructs of mAbs COVA2-15 and COVA2-17, Dr. Jayanta Bhattacharya for HIV positive serum. We thank BEI Resources for providing HEK293T-ACE2 and HEK293T-ACE2/TMPRSS2 cells.
This work was funded by a grant from Science and Engineering Research Board (IPA/2020/000168 to RPR) and Council of Scientific and Industrial Research (CSIR) to K.G.T. and R.P.R. R.V. acknowledges infrastructural support from the following programs of the Government of India: DST-FIST, UGC Center for Advanced Study, MHRD-FAST, the DBT-IISc Partnership Program, and of a JC Bose Fellowship from DST. D.C. and K.K. thank CSIR and IISc respectively for doctoral fellowship. R.P.R. is the recipient of DBT-Ramalingaswami fellowship. R.P. acknowledges the support of CSIR-IGIB grant (MLP-2005) and Fondation Botnar (CLP-0031).
R.P.R. conceived the study. R.P.R., S.K., R.D., D.C., K.K., R.S., and N.S. performed research. R.D. and D.C. performed molecular cloning. R.P.R., S.K., R.D., and N.S. analyzed the data. R.V. designed RBD-based immunogens for the immunogenicity in mice and supervised the immunogenicity studies. R.D. and R.S. expressed and purified Spike and RBD proteins and antibodies. R.S. performed the mouse immunizations and serum collection. S.S., A.T., S.J., and R.P. provided sera from COVID-19 human convalescent sera from the first wave of pandemic and vaccinated individuals. R.P.R. wrote original draft. K.K., D.C., R.S., and R.V. edited the paper.
Contributor Information
Rajesh P. Ringe, Email: rajeshringe@imtech.res.in.
Kanta Subbarao, The Peter Doherty Institute for Infection and Immunity, Melbourne, Victoria, Australia .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00922-23.
Supplemental figures.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Poland GA, Issa M, Sundsted K. 2022. Year 3 of COVID-19: harsh truths, brutal realities, and glimmers of hope. Mayo Clin Proc 97:2324–2332. doi: 10.1016/j.mayocp.2022.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrews N, Stowe J, Kirsebom F, Toffa S, Rickeard T, Gallagher E, Gower C, Kall M, Groves N, O’Connell AM, et al. 2022. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N Engl J Med 386:1532–1546. doi: 10.1056/NEJMoa2119451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lopez Bernal J, Gower C, Andrews N, Public Health England Delta Variant Vaccine Effectiveness Study Group . 2021. Effectiveness of COVID-19 vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med 385:e92. doi: 10.1056/NEJMc2113090 [DOI] [PubMed] [Google Scholar]
- 4. Keehner J, Horton LE, Binkin NJ, Laurent LC, Pride D, Longhurst CA, Abeles SR, Torriani FJ, SEARCH Alliance . 2021. Resurgence of SARS-CoV-2 infection in a highly vaccinated health system workforce. N Engl J Med 385:1330–1332. doi: 10.1056/NEJMc2112981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moore JP, Offit PA. 2021. SARS-CoV-2 vaccines and the growing threat of viral variants. JAMA 325:821–822. doi: 10.1001/jama.2021.1114 [DOI] [PubMed] [Google Scholar]
- 6. Bobrovitz N, Ware H, Ma X, Li Z, Hosseini R, Cao C, Selemon A, Whelan M, Premji Z, Issa H, Cheng B, Abu Raddad LJ, Buckeridge DL, Van Kerkhove MD, Piechotta V, Higdon MM, Wilder-Smith A, Bergeri I, Feikin DR, Arora RK, Patel MK, Subissi L. 2023. Protective effectiveness of previous SARS-CoV-2 infection and hybrid immunity against the Omicron variant and severe disease: a systematic review and meta-regression. Lancet Infect Dis 23:556–567. doi: 10.1016/S1473-3099(22)00801-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bok K, Sitar S, Graham BS, Mascola JR. 2021. Accelerated COVID-19 vaccine development: milestones, lessons, and prospects. Immunity 54:1636–1651. doi: 10.1016/j.immuni.2021.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. 2021. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184:2372–2383. doi: 10.1016/j.cell.2021.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lopez Bernal J, Gower C, Andrews N, Gallagher E, Simmons R, Thelwall S, Stowe J, Tessier E, Groves V, Dabrera G, Myers R, Campbell CNJ, Amirthalingam G, Edmunds M, Zambon M, Brown KE, Hopkins S, Chand M, Ramsay M. 2021. Effectiveness of COVID-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 385:e92. doi: 10.1056/NEJMc2113090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lopez Bernal J, Andrews N, Gower C, Robertson C, Stowe J, Tessier E, Simmons R, Cottrell S, Roberts R, O’Doherty M, Brown K, Cameron C, Stockton D, McMenamin J, Ramsay M. 2021. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on covid-19 related symptoms, hospital admissions, and mortality in older adults in England: test negative case-control study. BMJ 373:1088. doi: 10.1136/bmj.n1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan Y, Li X, Zhang L, Wan S, Zhang L, Zhou F. 2022. SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduct Target Ther 7:141. doi: 10.1038/s41392-022-00997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer A-S, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck H-M, Behrens GMN, Pöhlmann S. 2022. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 185:447–456. doi: 10.1016/j.cell.2021.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmann M, Zhang L, Pöhlmann S. 2022. Omicron: master of immune evasion maintains robust ACE2 binding. Signal Transduct Target Ther 7:118. doi: 10.1038/s41392-022-00965-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qu P, Evans JP, Faraone JN, Zheng Y-M, Carlin C, Anghelina M, Stevens P, Fernandez S, Jones D, Lozanski G, Panchal A, Saif LJ, Oltz EM, Xu K, Gumina RJ, Liu S-L. 2023. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 31:9–17. doi: 10.1016/j.chom.2022.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kurhade C, Zou J, Xia H, Liu M, Chang HC, Ren P, Xie X, Shi P-Y. 2023. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat Med 29:344–347. doi: 10.1038/s41591-022-02162-x [DOI] [PubMed] [Google Scholar]
- 16. Starr TN, Zepeda SK, Walls AC, Greaney AJ, Alkhovsky S, Veesler D, Bloom JD. 2022. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature 603:913–918. doi: 10.1038/s41586-022-04464-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. 2021. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29:463–476. doi: 10.1016/j.chom.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eguia RT, Crawford KHD, Stevens-Ayers T, Kelnhofer-Millevolte L, Greninger AL, Englund JA, Boeckh MJ, Bloom JD. 2021. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog 17:e1009453. doi: 10.1371/journal.ppat.1009453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Witte L, Baharani VA, Schmidt F, Wang Z, Cho A, Raspe R, Guzman-Cardozo C, Muecksch F, Canis M, Park DJ, Gaebler C, Caskey M, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2023. Epistasis lowers the genetic barrier to SARS-CoV-2 neutralizing antibody escape. Nat Commun 14:302. doi: 10.1038/s41467-023-35927-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofmann H, Pöhlmann S. 2004. Cellular entry of the SARS coronavirus. Trends Microbiol 12:466–472. doi: 10.1016/j.tim.2004.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He W-T, Limbo O, Smith C, Song G, Woehl J, et al. 2020. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369:956–963. doi: 10.1126/science.abc7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, et al. 2020. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584:443–449. doi: 10.1038/s41586-020-2548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, Chen Z, Guo Y, Zhang J, Li Y, Song X, Chen Y, Xia L, Fu L, Hou L, Xu J, Yu C, Li J, Zhou Q, Chen W. 2020. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science 369:650–655. doi: 10.1126/science.abc6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan J-W, Sahi V, Figueroa A, Guo XV, Cerutti G, Bimela J, Gorman J, Zhou T, Chen Z, Yuen K-Y, Kwong PD, Sodroski JG, Yin MT, Sheng Z, Huang Y, Shapiro L, Ho DD. 2020. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584:450–456. doi: 10.1038/s41586-020-2571-7 [DOI] [PubMed] [Google Scholar]
- 26. McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, et al. 2021. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184:2332–2347. doi: 10.1016/j.cell.2021.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li D, Sempowski GD, Saunders KO, Acharya P, Haynes BF. 2022. SARS-CoV-2 neutralizing antibodies for COVID-19 prevention and treatment. Annu Rev Med 73:1–16. doi: 10.1146/annurev-med-042420-113838 [DOI] [PubMed] [Google Scholar]
- 28. Wang Z, Muecksch F, Schaefer-Babajew D, Finkin S, Viant C, Gaebler C, Hoffmann H-H, Barnes CO, Cipolla M, Ramos V, et al. 2021. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature 595:426–431. doi: 10.1038/s41586-021-03696-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, et al. 2021. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592:277–282. doi: 10.1038/s41586-021-03291-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt F, Weisblum Y, Rutkowska M, Poston D, DaSilva J, Zhang F, Bednarski E, Cho A, Schaefer-Babajew DJ, Gaebler C, Caskey M, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2021. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature 600:512–516. doi: 10.1038/s41586-021-04005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann H-H, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2020. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9:e61312. doi: 10.7554/eLife.61312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, et al. 2021. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184:2348–2361. doi: 10.1016/j.cell.2021.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD. 2021. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593:130–135. doi: 10.1038/s41586-021-03398-2 [DOI] [PubMed] [Google Scholar]
- 34. Dejnirattisai W, Zhou D, Ginn HM, Duyvesteyn HME, Supasa P, Case JB, Zhao Y, Walter TS, Mentzer AJ, Liu C, et al. 2021. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 184:2183–2200. doi: 10.1016/j.cell.2021.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsieh C-L, Goldsmith JA, Schaub JM, DiVenere AM, Kuo H-C, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou C-W, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, McLellan JS. 2020. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369:1501–1505. doi: 10.1126/science.abd0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 183:1735. doi: 10.1016/j.cell.2020.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suryadevara N, Shrihari S, Gilchuk P, VanBlargan LA, Binshtein E, Zost SJ, Nargi RS, Sutton RE, Winkler ES, Chen EC, Fouch ME, Davidson E, Doranz BJ, Chen RE, Shi P-Y, Carnahan RH, Thackray LB, Diamond MS, Crowe Jr JE. 2021. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 184:2316–2331. doi: 10.1016/j.cell.2021.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang G-Y, Ho DD, Sheng Z, Kwong PD, Shapiro L. 2021. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 29:819–833. doi: 10.1016/j.chom.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. 2020. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 588:327–330. doi: 10.1038/s41586-020-2772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pak AJ, Yu A, Ke Z, Briggs JAG, Voth GA. 2022. Cooperative multivalent receptor binding promotes exposure of the SARS-CoV-2 fusion machinery core. Nat Commun 13:1002. doi: 10.1038/s41467-022-28654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu S, Zheng X, Zhou B, Li J, Chen M, Deng R, Wong G, Lavillette D, Meng G. 2022. SARS-CoV-2 spike engagement of ACE2 primes S2' site cleavage and fusion initiation. Proc Natl Acad Sci U S A 119:e2111199119. doi: 10.1073/pnas.2111199119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Henderson R, Edwards RJ, Mansouri K, Janowska K, Stalls V, Gobeil SMC, Kopp M, Li D, Parks R, Hsu AL, Borgnia MJ, Haynes BF, Acharya P. 2020. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat Struct Mol Biol 27:925–933. doi: 10.1038/s41594-020-0479-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wieczór M, Tang PK, Orozco M, Cossio P. 2023. Omicron mutations increase interdomain interactions and reduce epitope exposure in the SARS-CoV-2 spike. iScience 26:105981. doi: 10.1016/j.isci.2023.105981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gobeil S-C, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P. 2021. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 373:eabi6226. doi: 10.1126/science.abi6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Starr TN, Greaney AJ, Stewart CM, Walls AC, Hannon WW, Veesler D, Bloom JD. 2022. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains. PLoS Pathog 18:e1010951. doi: 10.1371/journal.ppat.1010951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moulana A, Dupic T, Phillips AM, Chang J, Nieves S, Roffler AA, Greaney AJ, Starr TN, Bloom JD, Desai MM. 2022. Compensatory epistasis maintains ACE2 affinity in SARS-CoV-2 Omicron BA.1. Nat Commun 13:7011. doi: 10.1038/s41467-022-34506-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gobeil S-C, Henderson R, Stalls V, Janowska K, Huang X, May A, Speakman M, Beaudoin E, Manne K, Li D, Parks R, Barr M, Deyton M, Martin M, Mansouri K, Edwards RJ, Eaton A, Montefiori DC, Sempowski GD, Saunders KO, Wiehe K, Williams W, Korber B, Haynes BF, Acharya P. 2022. Structural diversity of the SARS-CoV-2 Omicron spike. Mol Cell 82:2050–2068. doi: 10.1016/j.molcel.2022.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, et al. 2022. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603:700–705. doi: 10.1038/s41586-022-04462-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meng B, Abdullahi A, Ferreira I, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, et al. 2022. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603:706–714. doi: 10.1038/s41586-022-04474-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, Uriu K, Tsuda M, Zahradnik J, Shirakawa K, et al. 2022. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 185:2103–2115. doi: 10.1016/j.cell.2022.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hoffmann M, Wong L-Y, Arora P, Zhang L, Rocha C, Odle A, Nehlmeier I, Kempf A, Richter A, Halwe NJ, Schön J, Ulrich L, Hoffmann D, Beer M, Drosten C, Perlman S, Pöhlmann S. 2023. Omicron subvariant BA.5 efficiently infects lung cells. Nat Commun 14:3500. doi: 10.1038/s41467-023-39147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kulkarni PS, Padmapriyadarsini C, Vekemans J, Bavdekar A, Gupta M, Kulkarni P, Garg BS, Gogtay NJ, Tambe M, Lalwani S, et al. 2021. A phase 2/3, participant-blind, observer-blind, randomised, controlled study to assess the safety and immunogenicity of SII-ChAdOx1 nCoV-19 (COVID-19 vaccine) in adults in India. EClinicalMedicine 42:101218. doi: 10.1016/j.eclinm.2021.101218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ella R, Reddy S, Blackwelder W, Potdar V, Yadav P, Sarangi V, Aileni VK, Kanungo S, Rai S, Reddy P, et al. 2021. Efficacy, safety, and lot-to-lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): interim results of a randomised, double-blind, controlled, phase 3 trial. Lancet 398:2173–2184. doi: 10.1016/S0140-6736(21)02000-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Q, Anang S, Iketani S, Guo Y, Liu L, Katsamba PS, Shapiro L, Ho DD, Sodroski JG. 2022. Functional properties of the spike glycoprotein of the emerging SARS-CoV-2 variant B.1.1.529. Cell Rep 39:110924. doi: 10.1016/j.celrep.2022.110924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ahmed S, Khan MS, Gayathri S, Singh R, Kumar S, Patel UR, Malladi SK, Rajmani RS, van Vuren PJ, Riddell S, et al. 2021. A stabilized, monomeric, receptor binding domain elicits high-titer neutralizing antibodies against all SARS-CoV-2 variants of concern. Front Immunol 12:765211. doi: 10.3389/fimmu.2021.765211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Malladi SK, Patel UR, Rajmani RS, Singh R, Pandey S, Kumar S, Khaleeq S, van Vuren PJ, Riddell S, Goldie S, et al. 2021. Immunogenicity and protective efficacy of a highly thermotolerant, trimeric SARS-CoV-2 receptor binding domain derivative. ACS Infect Dis 7:2546–2564. doi: 10.1021/acsinfecdis.1c00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583:290–295. doi: 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 58. Tortorici MA, Czudnochowski N, Starr TN, Marzi R, Walls AC, Zatta F, Bowen JE, Jaconi S, Di Iulio J, Wang Z, et al. 2021. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597:103–108. doi: 10.1038/s41586-021-03817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cui Z, Liu P, Wang N, Wang L, Fan K, Zhu Q, Wang K, Chen R, Feng R, Jia Z, Yang M, Xu G, Zhu B, Fu W, Chu T, Feng L, Wang Y, Pei X, Yang P, Xie XS, Cao L, Cao Y, Wang X. 2022. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 185:860–871. doi: 10.1016/j.cell.2022.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Tuttle KS, Marquez AC, Sekirov I, Subramaniam S. 2022. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 375:760–764. doi: 10.1126/science.abn7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cele S, Karim F, Lustig G, San JE, Hermanus T, Tegally H, Snyman J, Moyo-Gwete T, Wilkinson E, Bernstein M, et al. 2022. SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe 30:154–162. doi: 10.1016/j.chom.2022.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hurlburt NK, Homad LJ, Sinha I, Jennewein MF, MacCamy AJ, Wan Y-H, Boonyaratanakornkit J, Sholukh AM, Jackson AM, Zhou P, Burton DR, Andrabi R, Ozorowski G, Ward AB, Stamatatos L, Pancera M, McGuire AT. 2022. Structural definition of a pan-sarbecovirus neutralizing epitope on the spike S2 subunit. Commun Biol 5:342. doi: 10.1038/s42003-022-03262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou P, Yuan M, Song G, Beutler N, Shaabani N, Huang D, He W-T, Zhu X, Callaghan S, Yong P, Anzanello F, Peng L, Ricketts J, Parren M, Garcia E, Rawlings SA, Smith DM, Nemazee D, Teijaro JR, Rogers TF, Wilson IA, Burton DR, Andrabi R. 2022. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Sci Transl Med 14:eabi9215. doi: 10.1126/scitranslmed.abi9215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Anastassopoulou CG, Ketas TJ, Klasse PJ, Moore JP. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc Natl Acad Sci U S A 106:5318–5323. doi: 10.1073/pnas.0811713106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yewdell JW, Taylor A, Yellen A, Caton A, Gerhard W, Bächi T. 1993. Mutations in or near the fusion peptide of the influenza virus hemagglutinin affect an antigenic site in the globular region. J Virol 67:933–942. doi: 10.1128/JVI.67.2.933-942.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lavie M, Dubuisson J, Belouzard S. 2022. SARS-CoV-2 spike furin cleavage site and S2' basic residues modulate the entry process in a host cell-dependent manner. J Virol 96:e0047422. doi: 10.1128/jvi.00474-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, Cantoni D, Scott S, Logan N, Ashraf S, et al. 2022. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol 7:1709. doi: 10.1038/s41564-022-01241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kimura I, Yamasoba D, Tamura T, Nao N, Suzuki T, Oda Y, Mitoma S, Ito J, Nasser H, Zahradnik J, et al. 2022. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell 185:3992–4007. doi: 10.1016/j.cell.2022.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kimura I, Yamasoba D, Nasser H, Zahradnik J, Kosugi Y, Wu J, Nagata K, Uriu K, Tanaka YL, Ito J, et al. 2022. The SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant. iScience 25:105720. doi: 10.1016/j.isci.2022.105720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gupta R. 2022. SARS-CoV-2 Omicron spike mediated immune escape and tropism shift. Res Sq:rs.3.rs-1191837. doi: 10.21203/rs.3.rs-1191837/v1 [DOI] [Google Scholar]
- 71. Schmidt F, Weisblum Y, Muecksch F, Hoffmann HH, Michailidis E, Lorenzi JCC, Mendoza P, Rutkowska M, Bednarski E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Caskey M, Robbiani DF, Nussenzweig MC, Rice CM, Hatziioannou T, Bieniasz PD. 2020. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 217:e20201181. doi: 10.1084/jem.20201181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, et al. 2020. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369:643–650. doi: 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures.