ABSTRACT
The persistence of replication-competent HIV-1 in people living with HIV (PLWH) is a barrier to a cure for HIV. Early-phase studies of clinical interventions to deplete the intact persistent HIV-1 reservoir are ongoing. However, the ability to distinguish intact proviruses is limited by sequence variation and the predominance of defective proviruses. In this study, we developed HIVepsilon-seq (HIVε-seq), a novel assay to analyze 33 samples consisting of unfractionated peripheral blood mononuclear cells and/or resting CD4+ T cells from antiretroviral therapy (ART)-treated durably suppressed PLWH, including samples from 17 female participants. HIVε-seq combines simplified target enrichment and long-read sequencing methodology with advanced bioinformatic analysis to increase the depth, characterization, and detection of intact persistent HIV-1 provirus. HIVε-seq detected persistent sequence-intact HIV-1 proviruses in samples from both male and female participants and represents a robust, scalable assay to detect intact proviral sequences that could be applied to studies of HIV cure interventions.
IMPORTANCE
The lack of a reliable method to accurately detect when replication-competent HIV has been cleared is a major challenge in developing a cure. This study introduces a new approach called the HIVepsilon-seq (HIVε-seq) assay, which uses long-read sequencing technology and bioinformatics to scrutinize the HIV genome at the nucleotide level, distinguishing between defective and intact HIV. This study included 30 participants on antiretroviral therapy, including 17 women, and was able to discriminate between defective and genetically intact viruses at the single DNA strand level. The HIVε-seq assay is an improvement over previous methods, as it requires minimal sample, less specialized lab equipment, and offers a shorter turnaround time. The HIVε-seq assay offers a promising new tool for researchers to measure the intact HIV reservoir, advancing efforts towards finding a cure for this devastating disease.
KEYWORDS: HIV, HIV persistence, third-generation sequencing
INTRODUCTION
As HIV-1 replicates, a diverse population of proviruses develops in patients including a mixture of defective and intact as well as integrated and non-integrated genomes (1). Some of these integrated proviruses establish a persistent reservoir. Cells harboring persistent replication-competent proviruses are difficult to detect due to their rarity and lack of discriminating biomarkers. Intact proviruses also selectively decay compared to defective proviruses (2). As over 95%–97% of proviruses are defective (3, 4), this further complicates the ability to accurately assess the success of strategies seeking to deplete the replication-competent reservoir (5 – 8).
The most definitive method to assess whether replication-competent provirus is present is an analytical treatment interruption (ATI). ATIs carry some risk to the participants and require intense monitoring to be performed safely (9, 10). Therefore, alternative analytical methods have been developed to detect replication-competent provirus. One of the most well-known and well-validated assays for determining whether a patient harbors replication-competent proviruses is the quantitative viral outgrowth assay (QVOA) (11, 12). While being an accurate method to detect viruses capable of recrudescence in the absence of ART therapy, this assay only provides a minimum estimate of the frequency of latent proviruses as it may under-represent replication-competent but non-inducible proviruses (13, 14). Additionally, this assay is resource and time-intensive and is not available in most research or clinical centers. The intact proviral DNA assay (IPDA) is a digital droplet PCR (ddPCR) assay that quantifies two amplicons that are highly enriched in intact HIV-1 proviruses. These two amplicons cover a total of 224 bp in the packaging signal (ψ) and the Rev response element (RRE) and utilize an unlabeled competitive probe designed to exclude hypermutation in the RRE region (3). The IPDA represents a significant improvement over assays that measure total viral DNA. The IPDA probes were carefully selected to cover areas that have a high likelihood of detecting common proviral defective patterns making it the best low cell input, moderate throughput assay to date for estimating intact reservoir size in ART-suppressed PWH initiating ART during chronic HIV infection. However, it still has a limited ability to discriminate with absolute certainty between intact and defective viruses as it interrogates a limited proportion of the viral sequence (approximately 2.5%), and proviral variation across the reservoirs of different individuals may result in variable specificity of the IPDA probes for intact viruses (15, 16). Additional methods that use short-read sequencing have also been used to characterize the reservoir with nearly full-length sequencing, but have limited depth, due to reliance on isolation of single genomes and multiple rounds of PCR, making their use in large-scale studies problematic (17 – 19).
The goal of this study was twofold: (i) develop a method to detect intact HIV-1 proviruses that allowed excellent sequence resolution with reduced cost to inform or reduce the need for ATIs, and (ii) apply this assay for in-depth characterization of proviruses derived from cells obtained from women participants, an under-represented population in cure and reservoir studies. Here, we introduce a simple workflow, termed HIVepsilon-seq (HIVε-Seq), to interrogate and characterize the HIV-1 proviral reservoir using long amplicons and bioinformatic analysis. We then applied this analysis to clinical samples obtained from a cohort of ART-treated, stably suppressed people living with HIV (PLWH).
Notably, our cohort includes samples from 17 women. Women comprise over half of PLWH worldwide and have distinct physiological and societal disparities that influence responses to interventions (20). Women are less likely to be on antiretroviral treatment and achieve viral suppression and have significantly higher morbidity and mortality (21, 22). Interaction with comorbidities, reproductive concerns, and differences in hormone levels are likely to affect not only response to antiretroviral treatment but also potentially curative therapies. However, women are dramatically underrepresented in clinical trials investigating curative strategies and studies characterizing persistent infection (20). Our study, therefore, sheds new light on a unique and important, yet under-studied, patient cohort in addition to providing a new method to interrogate the HIV-1 proviral reservoir, with significant utility for studies of potential curative interventions.
RESULTS
Participant cohort
Study participants (men and women) were recruited through the University of North Carolina (UNC) Global HIV Clinical Trials Unit, the UNC MACS/WIHS Combined Cohort Study (MWCCS, formerly WIHS) site, and the UNC CFAR HIV Clinical Cohort. This study was approved by the University of North Carolina Biomedical Institutional Review Board, and all participants provided informed consent. Participants were stably suppressed on ART (HIV-1 RNA < 50 copies/mL) for at least 12 months prior to enrollment (Table 1). Other inclusion criteria included a CD4 T cell count ≥ 300 cells/µL, negative hepatitis B surface antigen (HBsAg), and hepatitis C RNA prior to enrollment.
TABLE 1.
Summary of cohort
| Total participants | 30 |
|---|---|
| Sex at birth | |
| Female | 17 |
| Male | 13 |
| Median age (Q1, Q3) | |
| Female | 46 (38, 55) |
| Male | 49 (28, 52) |
| HIV infection at ART initiation | |
| Chronic | 26 |
| Acute | 4 |
| Median CD4 T cell count (Q1, Q3) | |
| Female | 893 (615, 1186) |
| Male | 707 (580, 737) |
| Median years of suppression (Q1, Q3) | |
| Female | 4.1 (2, 5.5) |
| Male | 6.4 (2.8, 7.9) |
| Race | |
| White | 13 |
| Black | 16 |
| Other | 1 |
| Ethnicity | |
| Hispanic | 1 |
| Non-Hispanic | 29 |
HIV-1 proviral sequencing with HIVε-seq
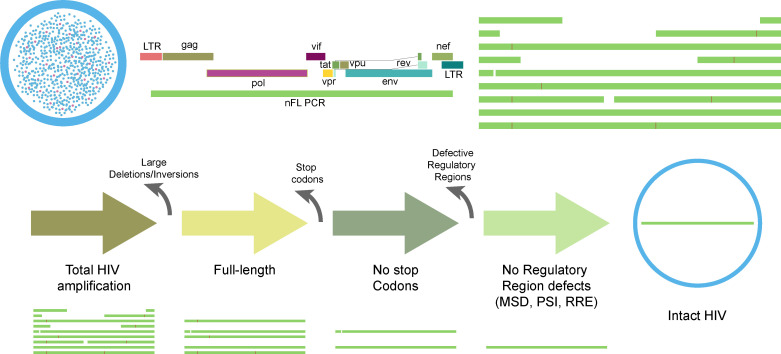
HIVε-seq is an efficient method for profiling the HIV-1 proviral reservoir (Fig. 1). In brief, DNA is isolated from pooled persistently infected cells, and nearly full-length mixed population HIV-1 is amplified. The amplified product is sequenced using Oxford Nanopore Technology (ONT) long-read sequencing. The resulting reads are delineated into HIV-1 proviral categories by stepwise removal of defective sequences. Reads are first aligned to the HIV-1 reference sequence, and those that contain both primer binding sites are characterized as HIV-1 amplification. Next, individual sequences mapping to less than 8,500 bp of the reference are removed, and the remaining sequences are characterized as full-length. Variant calling is performed on the full-length sequences, and sequences containing nucleotides associated with a putative stop codon are removed. A sample-specific, majority-base consensus sequence is then created, and all full-length sequences are aligned to it, and variants are called again. Sequences with stop codons in the context of the sample-specific reference are considered defective. The remaining sequences are categorized as not having stop codons. Lastly, select regulatory regions are interrogated for defects, and sequences that are predicted to have defective regulatory elements are removed. The remaining sequences are the inferred intact population. Each step in this analysis workflow was validated through the sequencing of reference samples and/or clinical isolates, described in detail below.
Fig 1.
HIVε-seq pipeline. Illustration of HIV ε-seq workflow to detect intact HIV proviral genomes. DNA is extracted from either PBMCs or CD4+ T cells, and nearly full-length PCR is performed on the extracted DNA. The reads are aligned to the HXB2 reference HIV sequence. Intact reads are inferred through the bioinformatic removal of reads containing large deletions/inversions, premature stop codons, or regulatory region defects.
Alignment consensus calling
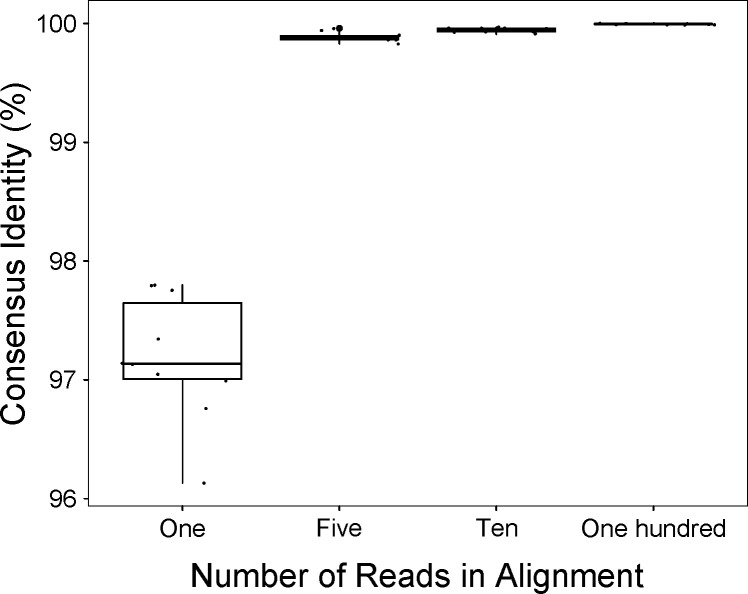
HIVε-seq uses an alignment, majority-base, column-wise, consensus approach to identify putative variants with sufficient support to provide confidence amid mixed population sequencing. To establish this approach, we first amplified proviral DNA from the latently infected cell line J89GFP (23), sequenced the product, and randomly selected 1, 5, 10, or 100 reads (10×) that aligned to the reference. We then created a consensus using the majority-base, column-wise approach and compared it to the consensus generated from the run containing over 100,000 aligned reads (Fig. 2). The median percent identity compared to the original consensus of a single read was 97.1% (BLASTN) (24). With the support of at least five reads, the median percent identity was 99.9%, compared to the sample reference sequence. This demonstrates that HIVε-seq can reproducibly resolve the complete HIV-1 proviral genome, with 99.9% consensus accuracy, based on a minimum read depth of five reads.
Fig 2.
Consensus congruence of amplified HIV provirus. Percent identity of a consensus sequence generated from randomly selected groups of reads containing 1, 5, 10, or 100 reads amplified from an integrated HIV-1 genome (J89GFP cell line).
Population-based, long amplification of rare persistent proviruses
We next interrogated clinical isolates of CD4 positive T cells or PBMCs from chronically infected stably suppressed participants. For each clinical sample, we interrogated approximately 266,000 cells for the presence of persistent proviruses using a single amplification reaction targeting conserved regions of the 5′ and 3′ long terminal repeats and 9,030 bp of a full-length HIV-1 provirus. The long-range PCR reactions were optimized starting with previously published methods and comparing polymerases, primer sets, and thermocycling parameters on DNA extracted from cell lines carrying integrated provirus and from clinical samples. Previous PCR methods were designed for stringent HIV-1 specificity to accommodate the single target requirements of Sanger or Illumina next generation sequencing including the use of limiting dilution, touch-down PCR, and high initial primer annealing temperatures. These conditions did not consistently amplify target provirus from cell lines or clinical samples due to a high drop-out rate (4, 14, 17, 25). A single PCR (i.e., non-nested) with a consistent primer binding temperature was found to increase sensitivity in samples with low viral load (details in the Materials and Methods section). Of note, we also found that visible bands on an agarose gel were not a reliable measure of subsequent detection of full-length provirus on the ONT platform.
Overview of workflow for HIVepsilon-seq of the proviral population in bulk CD4 T cell DNA from ART-suppressed participants
Iterative sequence analysis of alignment-corrected, bulk-sequenced, HIV-1 reads that were generated using nanopore sequencing technology was used to distinguish between defective and intact proviral populations from the CD4 T cell and PBMC DNA. A novel methodology was developed to extract intact sequences from bulk amplified proviral product, which simplifies both benchwork and computation compared to methods that require a single genome prior to classification as intact. All coding regions and 98.7% of the genome are interrogated.
After sequencing, the resulting reads were systematically filtered in silico using the in-house developed algorithm to infer intact reads. Reads consistent with off-target amplification or common defective proviral phenotypes including large deletions, inversions, premature stop codons, and/or regulatory region defects were removed. The inferred intact reads are defined as those containing both primer binding sites, no large internal deletions or inversions, no premature stop codons, and no regulatory region defects.
To select for amplified HIV-1 proviral reads, all reads from the sequencing run were aligned to the HXB2 HIV-1 reference sequence (accession numbers: K03455 and M38432). Off-target reads that did not align to the reference sequence were removed from further analysis. Quality control was performed to select intact amplicons containing both primer binding sites. As the primer binding sites are located in the LTR, and binding sites for each primer are present at each end, this step excludes reads extending adjacent to the provirus, off-target amplification of host DNA, and abortive PCR products. Sequences containing both primer sites and that aligned to HXB2 were categorized as HIV-1 proviral amplification (step 1). Full-length sequences were then extracted from the pool of proviral amplification by selecting sequences that aligned to over 8,500 bp of the HXB2 reference sequence (step 2). Those that did not align to over 8,500 bp were defined as containing large internal deletions and were removed from further analysis.
Stop codon analysis
Full-length, HIV-1 proviral reads were interrogated for the presence of internal stop codons causing truncated proteins. To identify reads containing stop codons, the full-length reads from step 2 were aligned to the historic reference sequence HXB2. As an estimate of defective sequences, stop codons were called in relation to the HXB2 genome, and reads containing single nucleotide polymorphisms consistent with stop codons were temporarily removed from the alignment.
The remaining reads were then used to create a representative sample consensus sequence. We used aligned base positions with the support of over 10 reads to generate the representative sample consensus using a majority base method at each position of HXB2 genome. Locations with a depth of less than 10 reads were substituted with the base from HXB2. HXB2 was added to the sequence database in 2002 and is evolutionarily significantly different from modern sequences. Therefore, inferring stop codons in the context of HXB2 provides only an approximation of defective proviruses. Once we generated the representative sample consensus, all full-length reads from step 2 were aligned to the updated representative sample consensus and variants, and stop codons were called in relation to the sample consensus. In the quality control verification steps, we then found a significant number of compensatory mutations adjacent to the mutations predicted to cause stop codons. Therefore, a step was introduced to check and retain reads with compensatory mutations. Sequences that were retained in steps 1 and 2 and did not contain premature stop codons in any coding regions were characterized as amplified, full-length, no-stop codon sequences. APOBEC mutation density has historically been used as a predictor of defective proviral sequences when analyzing short fragments of HIV. As we are able to analyze the full genome for stop codons, we have not included an APOBEC mutational analysis.
Regulatory region analysis
The major splice donor (MSD), Ψ, and RRE are regulatory regions that are essential for HIV-1 replications (26 – 28). The regulatory region analysis was performed similar to the stop codon analysis in that reads were interrogated relative to alignment positions in HXB2. Reads with mutations in the MSD (located at 742–745 in HXB2) were considered defective. Deletions of >10 bp in the Ψ (679–810 HXB2) or 20 bp in the RRE (7775–8020 HXB2) were considered to be defective. Sequences from the first three steps and no defects in the assessed regulatory regions were characterized as intact.
Detection of inferred intact persistent HIV-1 in clinical samples using HIVε-seq
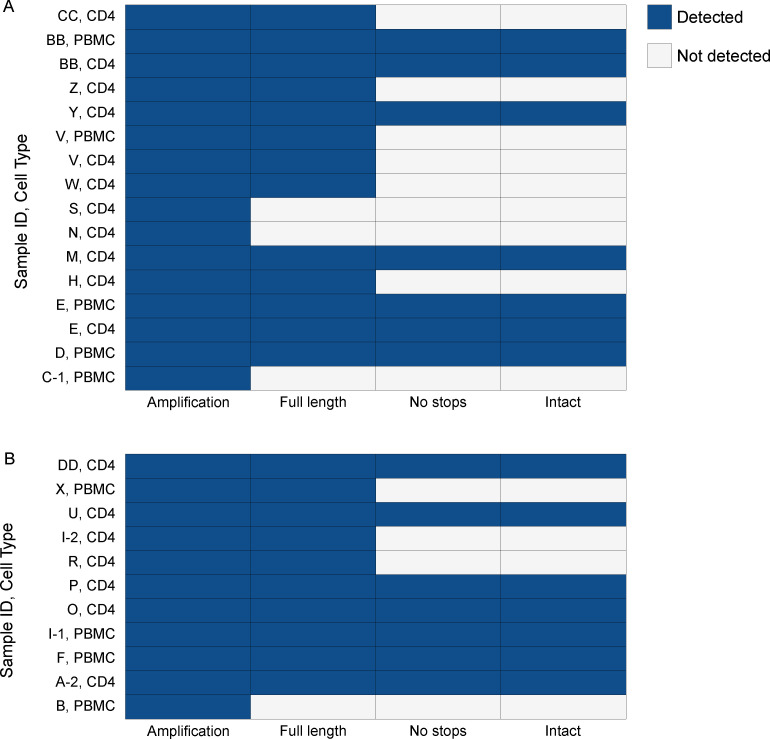
In total, 33 samples were analyzed representing 17 female and 13 male participants (Table 1). Three participants (A, C, and I) had repeat donations spanning 2, 13, and 4 months between the first and subsequent donation, respectively. Resting CD4+ T cells were available for 25 of the participants, and PBMCs were available for 12 of the participants. Sequences generated for each sample were classified using categorical indicators: full-length; full-length, no stops; or intact. Each sample was assessed twice, and the results are presented as an aggregate of the two runs for samples that had amplification (Fig. 3). HIVε-seq was performed on the equivalent of approximately 266,000 cells. We obtained amplification from 23 of the 33 samples analyzed. Amplification was detected for the matched PBMC and resting CD4+ T cells for three samples and had concordant highest levels of characterization. In total, 18 of the samples with amplification contained full-length virus. Full-length virus with no stop codons and intact provirus with no defects detected in the regulatory regions were found in cells from 12 participants. As all samples without stop codons were also found to contain inferred intact virus, further samples should be interrogated to determine if this trend is common and whether the regulatory region defects are sufficiently discriminatory to warrant inclusion in further analyses.
Fig 3.
HIVe-seq characterization of proviral reservoir in PLWH on ART. Characterization of the persistent proviral reservoir in CD4+ T cells or PBMCs from women (A) and men (B) chronically infected with HIV-1. Amplification—reads with both PCR binding sites. Full-length—reads mapping to greater than 8,500 bp of HXB2 reference. No stops—full length reads with no stop codons detected in protein coding regions. Intact—no stop reads without known regulatory region defects.
Women living with HIV in this study were on average older, had a shorter duration of suppression, and had higher CD4 T cell counts than the men. In the HIVε-seq analysis, 5 of the 17 women (28%) analyzed had detectable intact virus, whereas 7 of the 13 men (54%) had detectable intact virus (Fig. 3; Tables 1 and 2).
TABLE 2.
Individual participant characteristics and anti-retroviral therapy regimens at the time of leukapheresis a
| Participant ID |
Sex at birth | Age at first leukapheresis | CD4 count | HIV status at treatment | ART at leukapheresis |
|---|---|---|---|---|---|
| A | Male | 66 | 610 | Chronic | DRV/ABC/3TC |
| B | Male | 25 | 706 | Acute | EVG/c/TDF/FTC |
| C | Female | 45 | 1,168 | Acute | EVG/c/TDF/FTC |
| D | Female | 45 | 1,788 | Chronic | RPV/TDF/FTC |
| E | Female | 55 | 704 | Chronic | RPV/TDF/FTC |
| F | Male | 41 | 737 | Chronic | RLT/TDF |
| G | Female | 34 | 948 | Chronic | EVG/c/TDF/FTC |
| H | Female | 61 | 934 | Chronic | ABC/DTG/3TC |
| I | Male | 50 | 732 | Chronic | 3TC/DDI/FPV/NVP/RTVb |
| J | Female | 38 | 1,238 | Chronic | RPV/TDF/FTC |
| K | Male | 37 | 922 | Chronic | RPV/TDF/FTC |
| L | Female | 48 | 580 | Chronic | NVP/TDF |
| M | Female | 44 | 1,203 | Chronic | EFV/FTC/TDF |
| N | Female | 54 | 484 | Chronic | EVG/c/FTC/TAF |
| O | Male | 30 | 382 | Chronic | DTG/ABC/3TC |
| P | Male | 52 | 1,277 | Chronic | EFV/FTC/TDF |
| Q | Male | 28 | 530 | Acute | RPV/TAF/FTC |
| R | Male | 49 | 707 | Chronic | EFV/FTC/TDF |
| S | Female | 38 | 844 | Chronic | ATZ/RTV/FTC/TDF |
| T | Male | 50 | 664 | Chronic | ABC/DTG/3TC |
| U | Male | 57 | 1,045 | Chronic | EVG/c/TDF/FTC |
| V | Female | 56 | 593 | Chronic | EVG/c/TDF/FTC |
| W | Female | 53 | 1,121 | Chronic | DTG/ABC/3TC |
| X | Male | 28 | 574 | Chronic | DRV/RTV/TDF/FTC |
| Y | Female | 50 | 636 | Chronic | ABC/3TC/DRV/RTV |
| Z | Female | 37 | 873 | Chronic | RTV/TDF/FTC/ATV |
| AA | Female | 31 | 1,224 | Chronic | FTC/TDF/DTG |
| BB | Female | 55 | 576 | Chronic | RPV/TDF/FTC |
| CC | Female | 55 | 893 | Chronic | FTC/RPV/TDF |
| DD | Male | 25 | 729 | Acute | ABC/DTG/3TC |
DRV—darunavir, ABC—abacavir, 3TC—lamivudine, EVG—elvitegravir, c—cobicistat, TDF—tenofovir, TDF DF—tenofovir disoproxil fumarate, FTC—emtricitabine, DTG—dolutegravir, RAL—raltegravir, NVP—nevirapine, EFV—efavirenz, RTV—ritonavir, and ATV—atazanavir.
Comparison of assays to measure intact provirus
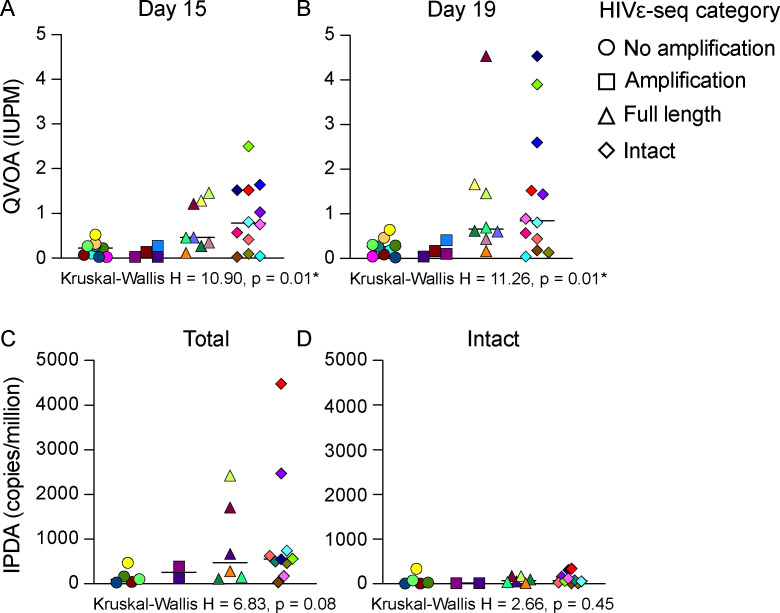
For the samples assessed using the HIVε-seq method, we also measured replication-competent HIV-1 using QVOA (Fig. 4; Table S1). A subset of these samples, for which we had sufficient cells, were also subjected to the IPDA (using five million resting CD4+ T cells per sample as starting material) to quantify intact proviral DNA (Fig. 4; Table S1). Intact virus was detected in all samples assessing the equivalent of up to 1 million cells for the IPDA and from 34 to 47 million resting CD4+ T cells for the QVOA. A comparison of results from HIVε-seq, QVOA, and IPDA found that HIV-1 DNA amplification was detected in all samples with an infected units per million (IUPM) by QVOA of 1 or higher for days 15 and 19 and was detected in samples with as little as 0.021 IUPM in QVOA. For 12 of the 24 samples with intact virus detected by QVOA and/or IPDA, intact virus was also observed for HIVε-seq. A Kruskal-Wallis test showed that there was a statistically significant difference in the IUPM at days 15 and 19 (H = 10.90 and 11.26, P = 0.01 and 0.01, respectively) between the four HIVε-seq categories (no amplification, amplification, full-length, and intact) (Fig. 4; Table S2). The Kruskal-Wallis test did not show a significant difference in the total or intact viral copies measured by IPDA (Fig. 4; Table S2). We then compared samples with intact virus detected by HIVε-seq vs those without using the Mann-Whitney U test to evaluate whether the level of IUPM measured by QVOA (day 15 or 19) or HIV copies per million cells measured by IPDA was higher in samples with intact virus (one-tailed P-value). The results indicated that samples with genetically intact virus by HIVε-seq had significantly higher IUPM by QVOA at days 15 and 19 (Z = −2.17, P = 0.02; Z = −1.85, P = 0.03, respectively) and total HIV DNA copies per million cells (Z = −1.80, P = 0.04) compared to those in which no genetically intact virus was found by HIVε-seq (Table S2). No significant difference was detected in the level of intact HIV copies per million by IPDA compared to the HIVε-seq results (Z = −0.87, P = 0.21; Table S2). HIVε-seq qualitatively characterizes the composition of the amplified proviral population, it is not quantitative. Intact virus was more likely to be detected by HIVε-seq in samples with higher IUPM (QVOA) and/or copies per million (IPDA) although it was also detected in samples near the limit of detection for these assays. These assays are expected to differ, as far fewer cells were interrogated in HIVε-seq (34–47 million resting CD4+ T cells for the QVOA vs 1 million cell equivalent for the IPDA vs 266,000 cells for HIVε-seq), giving differing limits of detection. HIVε-seq also assesses a greater proportion of the HIV genome than IPDA for deletions, stop codons, and regulatory region defects, which may result in differences in viruses characterized as intact. The HIVε-seq assay was more likely to detect intact virus in samples with higher HIV total DNA measure by IPDA. Overall, the QVOA assay was a better predictor of HIVε-seq readout than IPDA.
Fig 4.
HIVε-seq categories by viral levels measured with QVOA and IPDA. (A) Frequency of replication-competent persistent provirus in HIV-1 infected adults on ART with fully suppressed viremia on days 15 and 19 (B) measured by the QVOA assay or (C) frequency of total DNA and intact DNA (D) as measured by IPDA compared to HIVε-seq category. No amplification—no reads map to HXB2 reference. Amplification—reads that map to HXB2 and contain 5′ and 3′ PCR primer binding sites. Full-length—reads mapping to greater than 8,500 bp of HXB2 reference. No stops—full-length reads with no stop codons detected in protein-coding regions. Intact—no stop reads without known regulatory region defects. Colors denote individual participants. Asterisks indicate P-value less than 0.05.
Resistance mutation and subtype analysis
Majority base consensus sequences were generated for each position of HXB2 from reads that mapped to both primer sites, had a quality score above Q10, mapped to greater than 8,500 base pairs of HXB2, and did not contain mutations that indicated a stop codon relative to HXB2 for each sample. The sample-specific consensus sequence for which more than 10 reads were available to generate a consensus sequence was analyzed using the Stanford Database (29, 30).
All samples were found to be subtype B, which is consistent with the majority subtype found in the United States. Drug-resistance mutations were identified in three participants with some resistance to abacavir (ABC) and zidovudine (AZT) found in two of the participants (Table 3).
TABLE 3.
Drug-resistance mutations and genotype of HIV-1 in three participants a
| Sample ID | Mutations | Potential drug resistance | Subtype |
|---|---|---|---|
| A | NRTI—M41L, M184V, T215N | ABC, AZT, FTC, 3TC | B |
| Y | Prot—M461, I54V, V82A, K43T; NNRTI—K103N | ATV/r, LPV/r, EFV, NVP | B |
| DD | NRTI—M41L, T215S; INSTI—E157Q | ABC, AZT, TDF, EVG, RAL | B |
NRTI—nucleoside reverse transcriptase inhibitors, Prot— protease, and NNRTI—non-nucleoside reverse transcriptase inhibitors.
DISCUSSION
The novel HIVε-seq method demonstrates that a simplified wet-lab workflow combined with bioinformatic analysis can detect intact virus with lower starting input than QVOA in approximately half of the patient samples analyzed. In comparison with the IPDA method, HIVε-seq may be discordant due to deletions or truncated sequences in regions that are not included in the IPDA assay (the HIVε-seq assay surveys ~9,030 bp as opposed to 224 bp included in the IPDA assay). These assays may be complementary in some settings, as IPDA may over-report intact, replication-competent proviruses, while QVOA may underreport them. For example, in one sample with very high levels of intact virus detected by IPDA for which no amplification was detected by HIVε-seq, QVOA was quite low.
Overall, HIVε-seq results correlated more closely with the viral outgrowth assay than IPDA but were unable to detect amplification from samples with lower IUPM measured by outgrowth. It is possible that HIVε-seq could exclude some virus that is intact in culture with only a minor effect of a small truncation or functional defect at the level of viral protein. HIVε-seq can easily be scaled to include more cells per run or additional runs per sample. Increasing the input would likely increase the detection of intact virus in a greater proportion of samples. As the detection of intact provirus correlated with higher IUPM (QVOA) and proviral copies per million cells (IPDA), it may be useful to standardize the amount of HIV-1 DNA assayed rather than cell count. Further studies will clarify the sensitivity of the HIVε-seq assay.
Previously developed lab-based methods to assess the proviral reservoir are thought to under- or overestimate the presence of intact virus (2, 16). These molecular biology-based methods are a compromise between scalability, cost, sequence coverage, and sample requirements. Given the HIV-1 sequence diversity, the more primer/probe binding sites that are used, the higher the likelihood of drop out due to mismatch rather than genuine absence of viral DNA. Furthermore, each round of PCR adds to the likelihood of PCR-introduced error and drop out. HIVε-seq minimizes the use of primers and PCR cycles to reduce experimentally introduced errors. Individual nearly full-length genome sequencing has been successfully employed to characterize proviral composition in distinct cell subsets, but is lengthy, resource-intensive, and requires specialized equipment. HIVε-seq assesses 9,030 bp of HIV-1, similar to nearly full-length individual sequencing methods and can distinguish between defective and intact viruses with similar resolution to that of nearly full-length sequencing. The use of long-read sequencing supports the phasing of deleterious mutations and the removal of defective virus eliminating the need for serial dilutions.
This method also creates a participant-specific reference that could be used for drug resistance and genotyping. Of note, clinical diagnostic drug resistance results were not available for this cohort of stably aviremic PLWH to assess correlation.
As far as we are aware, this is the largest cohort of women to undergo characterization of their full-length proviral reservoir. By simplifying the method and removing the need for expensive, specialized equipment, this assay can be applied in more settings and to a broader range of patient populations including clinical trials and populations underrepresented in current studies such as women, children, and transgender individuals living with HIV.
Compared to other sequencing technologies, single-read accuracy from Oxford Nanopore Sequencing is around 95% with most inaccuracies being insertions or deletions. The error rate was minimized by using the PCR product, which does not include methylation (40% of nanopore sequencing errors), and by limiting our analysis to alignment positions and support from multiple reads. However, the reference-based analysis method is unable to accurately detect insertions and deletions. Therefore, HIVε-seq does not include the exclusion of frameshift mutations, which have been found to contribute to ~1% of defective viruses (4). Additionally, the limited amount of sample available for inclusion in the development of this proof-of-principle assay limited detection and direct comparison with other methods but could be addressed in future studies by including more cells in each reaction or performing additional reactions.
HIVε-seq is an important advancement in the development of assays to detect intact proviral sequences that have the potential to reinitiate new infection. This assay assesses for deletions/insertions, truncating stop codons, and regulatory region defects in nearly full-length proviral HIV-1 using alignment-based, long-read sequencing and represents an important tool that can be readily applied in clinical settings to assess the efficacy of HIV cure interventions and reduce the need for ATIs.
MATERIALS AND METHODS
Clinical samples
Peripheral blood mononuclear cells (PBMCs) were obtained by continuous-flow leukapheresis from durably suppressed on ART, HIV+ participants. Resting CD4+ T cells were isolated from PBMCs, and the purity was assessed as previously reported (31). A fraction of the PBMC and isolated resting CD4+ T cells were pelleted and immediately snap-frozen on dry ice in aliquots of 5 million cells per tube. Cells were stored at −80˚C until used for DNA isolation as described below.
Quantitative viral outgrowth assay
Replication-competent HIV-1 was recovered from purified resting CD4+ T cells as previously reported (7). Briefly, approximately 34–47 million resting CD4+ T cells were plated in replicate limiting dilutions of 2.5 million (18 cultures), 0.5 million (6 cultures), and 0.1 million (6 cultures) cells per well, activated with PHA (Remel, Lenexa, KS, USA) and a fivefold excess of allogeneic irradiated PBMCs from a seronegative donor, and 60 U/mL IL-2 for 24 hours. Cultures were washed and co-cultivated with CD8-depleted PBMCs collected from selected HIV-1 seronegative donors screened for adequate CCR5 expression. Culture supernatants were harvested on days 15 and 19 and assayed for virus production by p24 antigen capture ELISA (ABL, Rockville, MD, USA). Cultures were scored as positive if p24 was detected on day 15 and was increased in concentration on day 19. The number of resting CD4+ T cells in IUPM was estimated by a maximum likelihood method as previously described (32).
IPDA
DNA was extracted from total CD4 T cells using the Qiagen QiaAmp DNA extraction kit as described, including the RNAse A step (3). DNA was measured on the NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific). A total of 12–16 replicate wells of up to 800 ng DNA each were combined with 2× ddPCR Supermix for Probes (no dUTP, Bio-Rad) and primer/probe sets for regions in the packaging signal and envelope that favor amplification of intact proviral genomes, as described (3). Thermal cycling conditions with a 2°C ramp rate were: 10 minutes at 95°C followed by 45 cycles (30 seconds at 94°C and 60 seconds at 57°C per cycle) and 10 minutes at 98°C, followed by incubation at 12°C as described (3, 32). Droplets were read on the QX200 Droplet Reader (Bio-Rad Cat#1864003) using QuantaSoft Software Version 1.7.4.0917. Replicate wells were merged and analyzed in the QuantaSoft Analysis Pro Software. A correction for DNA shearing was applied based on a separate, duplicate measurement of two regions of the diploid host gene RPP30 using 7 ng of genomic DNA (3, 32).
DNA extraction for HIVε-seq
On the day of the participant’s leukapheresis, aliquots of 5 million PBMCs and 5 million purified resting CD4+ T cells were pelleted, snap-frozen, and stored at −80°C. DNA was then extracted from both PBMCs and resting CD4+ T cells using the Gentra PureGene (Qiagen) following the manufacturer’s instructions. DNA was eluted in 150 µL. The extracted DNA was then run on the Nanodrop 2000 spectrophotometer (Thermo Scientific) to determine the concentration and purity of the nucleic acid present. The extracted DNA was stored at −20°C until needed for further analysis.
PCR amplification
PCR was performed on DNA from 133,333 (4 µL) CD4+ T cells or PBMCs. The following primer set was used to amplify proviral sequences, Fwd (275F) ACAGGGACCTGAAAGCGAAAG and Rev (280R) CTAGTTACCAGAGTCACACAACAGACG (sequence homology of 100% for subtypes A, B, and C with a maximum difference of 1–2 bp from major subtype reference sequences) (14, 33). The polymerase chain reaction was carried out in a total volume of 25 µL with 1× buffer, 0.2 mM deoxynucleotide triphosphates (dNTPs), 2 µM each primer, and 0.625 U PrimeSTAR GXL DNA Polymerase. The following cycling conditions were used: 35 cycles of 98°C for 10 seconds, 60°C for 15 seconds, and 68°C for 10 minutes. Human DNA without HIV infection (Promega G0341) was used as a negative control, and DNA isolated from J89 cells [kind gift from D. N. Levy (23) ] was used as a positive control. Two aliquots were analyzed from each sample for a total of approximately 266,000 cells. PCR samples were purified using Ampure XP beads according to the manufacturer’s instructions.
Nanopore sequencing
Nanopore Native barcoding kit (EXP-NBD104/114) was used to barcode and prepare the purified PCR products for sequencing up to 12 samples per run as per the manufacturer’s instructions. The samples were loaded on a Minion flow cell (9.4.1) and run on a Gridion sequencing machine. Two assays were performed for each sample.
Bioinformatic analysis
Sequences were basecalled and demultiplexed using Guppy (v4.2.2, High accuracy model) and then mapped to HXB2 master sequence from the Los Alamos National Laboratory, which was trimmed to bases 631–9,719 to prevent erroneous alignment to the terminal repeats, using minimap2 (v2.17) (34). Sequences with a quality score above Q10 and that mapped to both the 5′ and 3′ primer binding sites were selected to exclude aborted PCR fragments. Sequences that mapped to greater than 8,500 bp of the reference sequence were selected to remove those with large deletions. Furthermore, reads with deletions larger than 250 bp were removed. Bcftools mpileup/call (v1.9) was used to call the variants in the bam file, and then, the variants were converted to consequences using the bcftools csq tool (35). As a conservative approach, sequences containing variants that resulted in a premature stop codon in any of the proteins were removed. The remaining sequences were used to generate a reference sequence specific for the sample if more than 100 sequences were remaining in the file. The pipeline was repeated substituting the sample-specific reference sequence in place of HXB2. After stop codons were called the second time, compensatory mutations that negate the stop codon were assessed, and those sequences were not removed. The MSD, PSI, and RRE regulatory regions were assessed for intactness by first converting the bam files to a tsv format (36) and then querying for sequences with any mutation in MSD (742–745 HXB2), greater than 10 bp deleted in the PSI/Ψ (679–810 HXB2), and greater than 20 bp in RRE (7,775–8,020 HXB2) (37). The sequences were then categorized into HIV-1 amplification (contains both primer sites, Q10), full-length (aligned to over 8,500 bp), defective, contains stop codons (contains stop codons relative to sample reference sequence), or full-length defective regulatory region (MSD, PSI, and RRE). The pipeline can be viewed at github.com (https://github.com/Psy-Fer/HIVE-seq).
Statistical analysis
The Mann-Whitney test and Kruskal-Wallis tests were performed using the SPSS software package (IBM SPSS Statistics for Windows version 28, SPSS Inc., Chicago, IL, USA). The descriptive statistics and scatter plots were created using Prism software (Graphpad Prism version 9 for Mac, GraphPad Software, www.graphpad.com).
Drug resistance and subtype analysis
Consensus sequences from samples with intact sequences were submitted to the Stanford drug resistance database (29) (HIVDB version 9.1) and mutations associated with reduced susceptibility to common treatments are listed in Table 3.
ACKNOWLEDGMENTS
The authors thank the participants who made this study possible. We thank C. Baker for clinical data compilation support; J. Kuruc for clinical coordination; Y. Park and the staff of the UNC Blood Bank, UNC CTRC, and Blood Centers of the Pacific (BCP) for clinical support; and the CFAR Biostatistics Core for statistical support.
The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH). This work was supported by the National Institutes of Health (Grant numbers NIH R01AI134363 and 1R01AI170226 to N.M.A., NIH F30AI145588 to S.D.F., U01‐HL146194 to A.A.A., and NIH UM1AI164567 to D.M.M.). Clinical data for some of the participants in this manuscript were collected by the MACS/WIHS Combined Cohort Study (MWCCS). MWCCS (principal investigators): Data Analysis and Coordination Center (Gypsyamber D’Souza, Stephen Gange, and Elizabeth Topper), U01-HL146193 and UNC CRS (A.A.A. and Michelle Floris-Moore), and U01-HL146194. The MWCCS is funded primarily by the National Heart, Lung, and Blood Institute (NHLBI), with additional co-funding from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD), National Institute on Aging (NIA), National Institute of Dental & Craniofacial Research (NIDCR), National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Neurological Disorders and Stroke (NINDS), National Institute of Mental Health (NIMH), National Institute on Drug Abuse (NIDA), National Institute of Nursing Research (NINR), National Cancer Institute (NCI), National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institute on Deafness and Other Communication Disorders (NIDCD), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute on Minority Health and Health Disparities (NIMHD), and in coordination and alignment with the research priorities of the National Institutes of Health, Office of AIDS Research (OAR). MWCCS data collection is also supported by P30-AI-050410 (UNC CFAR). The authors gratefully acknowledge the contributions of the study participants and dedication of the staff at the MWCCS sites.
K.B., J.M.F., I.W.D., and N.M.A. designed the research; K.B., J.M.F., I.W.D., N.M.A., S.D.F., K.S.J., and J.K. performed the research; J.M.H., B.B., S.L.C., and M.A.S. contributed new reagents/analytic tools; K.B., J.M.F., I.W.D., N.M.A., and S.D.F. analyzed the data; C.R., A.A.A, and D.M.M. recruited study participants and provided clinical support; and K.B., J.M.F., I.W.D., and N.M.A. wrote the manuscript with input from all authors.
Contributor Information
Nancie M. Archin, Email: nancie_archin@med.unc.edu.
Frank Kirchhoff, Ulm University Medical Center, Ulm, Baden-Württemberg, Germany .
DATA AVAILABILITY
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00705-23.
Table S1 (Quantitative viral outgrowth assay, intact proviral DNA assay and HIVepsilon-seq results) and Table S2 (Mann-Whitney U test and Kruskal Wallis U test).
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Margolis DM, Archin NM. 2017. Proviral latency, persistent human immunodeficiency virus infection, and the development of latency reversing agents. J Infect Dis 215:S111–S118. doi: 10.1093/infdis/jiw618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deeks SG, Archin N, Cannon P, Collins S, Jones RB, de Jong M, Lambotte O, Lamplough R, Ndung’u T, Sugarman J, Tiemessen CT, Vandekerckhove L, Lewin SR, International AIDS Society (IAS) Global Scientific Strategy working group . 2021. Research priorities for an HIV cure: international AIDS society global scientific strategy 2021. Nat Med 27:2085–2098. doi: 10.1038/s41591-021-01590-5 [DOI] [PubMed] [Google Scholar]
- 3. Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. doi: 10.1038/s41586-019-0898-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hiener B, Horsburgh BA, Eden J-S, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2017. Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep 21:813–822. doi: 10.1016/j.celrep.2017.09.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Z, Simonetti FR, Siliciano RF, Laird GM. 2018. Measuring replication competent HIV-1: advances and challenges in defining the latent reservoir. Retrovirology 15:21. doi: 10.1186/s12977-018-0404-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Falcinelli SD, Ceriani C, Margolis DM, Archin NM. 2019. New frontiers in measuring and characterizing the HIV reservoir. Front Microbiol 10:2878. doi: 10.3389/fmicb.2019.02878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, Gay CL, Eron JJ, Margolis DM, Bosch RJ, Archin NM. 2015. Precise quantitation of the latent HIV-1 reservoir: implications for eradication strategies. J Infect Dis 212:1361–1365. doi: 10.1093/infdis/jiv218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdel-Mohsen M, Richman D, Siliciano RF, Nussenzweig MC, Howell BJ, Martinez-Picado J, Chomont N, Bar KJ, Yu XG, Lichterfeld M, Alcami J, Hazuda D, Bushman F, Siliciano JD, Betts MR, Spivak AM, Planelles V, Hahn BH, Smith DM, Ho Y-C, Buzon MJ, Gaebler C, Paiardini M, Li Q, Estes JD, Hope TJ, Kostman J, Mounzer K, Caskey M, Fox L, Frank I, Riley JL, Tebas P, Montaner LJ, BEAT-HIV Delaney Collaboratory to Cure HIV-1 infection . 2020. Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat Med 26:1339–1350. doi: 10.1038/s41591-020-1022-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henderson GE, Peay HL, Kroon E, Cadigan RJ, Meagher K, Jupimai T, Gilbertson A, Fisher J, Ormsby NQ, Chomchey N, Phanuphak N, Ananworanich J, Rennie S. 2018. Ethics of treatment interruption trials in HIV cure research: addressing the conundrum of risk/benefit assessment. J Med Ethics 44:270–276. doi: 10.1136/medethics-2017-104433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lau JSY, Smith MZ, Allan B, Martinez C, Power J, Lewin SR, McMahon JH. 2020. Perspectives on analytical treatment interruptions in people living with HIV and their health care providers in the landscape of HIV cure-focused studies. AIDS Res Hum Retroviruses 36:260–267. doi: 10.1089/AID.2019.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9:727–728. doi: 10.1038/nm880 [DOI] [PubMed] [Google Scholar]
- 12. Siliciano JD, Siliciano RF. 2005. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 304:3–15. doi: 10.1385/1-59259-907-9:003 [DOI] [PubMed] [Google Scholar]
- 13. Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O’Doherty U, Palmer S, Deeks SG, Siliciano JD, Douek DC. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 9:e1003174. doi: 10.1371/journal.ppat.1003174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaebler C, Falcinelli SD, Stoffel E, Read J, Murtagh R, Oliveira TY, Ramos V, Lorenzi JCC, Kirchherr J, James KS, Allard B, Baker C, Kuruc JD, Caskey M, Archin NM, Siliciano RF, Margolis DM, Nussenzweig MC, Silvestri G. 2021. Sequence evaluation and comparative analysis of novel assays for intact proviral HIV-1 DNA. J Virol 95. doi: 10.1128/JVI.01986-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kinloch NN, Ren Y, Conce Alberto WD, Dong W, Khadka P, Huang SH, Mota TM, Wilson A, Shahid A, Kirkby D, Harris M, Kovacs C, Benko E, Ostrowski MA, Del Rio Estrada PM, Wimpelberg A, Cannon C, Hardy WD, MacLaren L, Goldstein H, Brumme CJ, Lee GQ, Lynch RM, Brumme ZL, Jones RB. 2021. HIV-1 diversity considerations in the application of the intact proviral DNA assay (IPDA). Nat Commun 12. doi: 10.1038/s41467-020-20442-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee GQ, Lichterfeld M. 2022. Near-full-length single-genome HIV-1 DNA sequencing. Methods Mol Biol 2407:357–364. doi: 10.1007/978-1-0716-1871-4_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joseph KW, Halvas EK, Brandt LD, Patro SC, Rausch JW, Chopra A, Mallal S, Kearney MF, Coffin JM, Mellors JW. 2022. Deep sequencing analysis of individual HIV-1 proviruses reveals frequent asymmetric long terminal repeats. J Virol 96:e0012222. doi: 10.1128/jvi.00122-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaebler C, Lorenzi JCC, Oliveira TY, Nogueira L, Ramos V, Lu CL, Pai JA, Mendoza P, Jankovic M, Caskey M, Nussenzweig MC. 2019. Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J Exp Med 216:2253–2264. doi: 10.1084/jem.20190896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adimora AA, Ramirez C, Poteat T, Archin NM, Averitt D, Auerbach JD, Agwu AL, Currier J, Gandhi M. 2021. HIV and women in the USA: what we know and where to go from here. Lancet 397:1107–1115. doi: 10.1016/S0140-6736(21)00396-2 [DOI] [PubMed] [Google Scholar]
- 21. Girum T, Wasie A, Lentiro K, Muktar E, Shumbej T, Difer M, Shegaze M, Worku A. 2018. Gender disparity in epidemiological trend of HIV/AIDS infection and treatment in Ethiopia. Arch Public Health 76:51. doi: 10.1186/s13690-018-0299-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beer L, Mattson CL, Bradley H, Skarbinski J, Medical Monitoring P. 2016. Understanding cross-sectional racial, ethnic, and gender disparities in antiretroviral use and viral suppression among HIV patients in the United States. Medicine (Baltimore) 95:e3171. doi: 10.1097/MD.0000000000003171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kutsch O, Benveniste EN, Shaw GM, Levy DN. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J Virol 76:8776–8786. doi: 10.1128/jvi.76.17.8776-8786.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–9. doi: 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. White JA, Kufera JT, Bachmann N, Dai W, Simonetti FR, Armstrong C, Lai J, Beg S, Siliciano JD, Siliciano RF, Douek DC. 2022. Measuring the latent reservoir for HIV-1: quantification bias in near full-length genome sequencing methods. PLoS Pathog 18:e1010845. doi: 10.1371/journal.ppat.1010845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emery A, Swanstrom R. 2021. HIV-1: to splice or not to splice, that is the question. Viruses 13:181. doi: 10.3390/v13020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mailler E, Bernacchi S, Marquet R, Paillart JC, Vivet-Boudou V, Smyth RP. 2016. The life-cycle of the HIV-1 Gag-RNA complex. Viruses 8:248. doi: 10.3390/v8090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mueller N, van Bel N, Berkhout B, Das AT. 2014. HIV-1 splicing at the major splice donor site is restricted by RNA structure. Virology 468–470:609–620. doi: 10.1016/j.virol.2014.09.018 [DOI] [PubMed] [Google Scholar]
- 29. Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 31:298–303. doi: 10.1093/nar/gkg100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu TF, Shafer RW. 2006. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 42:1608–1618. doi: 10.1086/503914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. 2009. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol 83:4749–4756. doi: 10.1128/JVI.02585-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Falcinelli SD, Kilpatrick KW, Read J, Murtagh R, Allard B, Ghofrani S, Kirchherr J, James KS, Stuelke E, Baker C, Kuruc JD, Eron JJ, Hudgens MG, Gay CL, Margolis DM, Archin NM. 2021. Longitudinal dynamics of intact HIV proviral DNA and outgrowth virus frequencies in a cohort of individuals receiving antiretroviral therapy. J Infect Dis 224:92–100. doi: 10.1093/infdis/jiaa718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leitner T, Korber B, Daniels M, Calef C, Foley B.. 2005. HIV-1 subtype and circulating recombinant form (CRF) reference sequences, 2005. Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 34. Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lindenbaum P. 2015. JVarkit: java-based utilities for bioinformatics. Figshare doi: 10.6084/m9.figshare.1425030.v1 [DOI] [Google Scholar]
- 37. Wright IA, Bale MJ, Shao W, Hu W-S, Coffin JM, Van Zyl GU, Kearney MF. 2021. HIVIntact: a python-based tool for HIV-1 genome intactness inference. Retrovirology 18:16. doi: 10.1186/s12977-021-00561-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (Quantitative viral outgrowth assay, intact proviral DNA assay and HIVepsilon-seq results) and Table S2 (Mann-Whitney U test and Kruskal Wallis U test).