ABSTRACT
Improved broad-spectrum influenza virus vaccines are desperately needed to provide protection against both drifted seasonal and emerging pandemic influenza A viruses (IAVs). Antibody-based protection from influenza A virus-induced morbidity and mortality is traditionally associated with neutralizing antibodies. As such, vaccine efforts have solely focused on the hemagglutinin (HA) as a vaccine target; however, the HA is mutation prone resulting in the need for annual vaccine reformulation. Broad-spectrum vaccines could be achieved through non-neutralizing antibodies that target conserved influenza virus antigens. Here, we describe six human monoclonal antibodies (mAbs) isolated from two H3N2-infected donors that showed robust binding against the conserved internal nucleoprotein (NP) or matrix protein 1 (M1) of IAV strains. Despite the capacity for potent antigen binding, substantial morbidity was observed in mice prophylactically treated with these mAbs and then challenged with A/Netherlands/602/2009 (H1N1) or A/Switzerland/9715293/2013 (H3N2) viruses. While our findings need to be confirmed with a larger number of mAbs and with polyclonal serum, these findings suggest that human NP and M1 antibodies that are elicited following IAV infection/vaccination do not protect from substantial weight loss in the mouse model and imply that protection afforded targeting these antigens following vaccination/infection is most likely the result of cellular-based immunity.
IMPORTANCE
Currently, many groups are focusing on isolating both neutralizing and non-neutralizing antibodies to the mutation-prone hemagglutinin as a tool to treat or prevent influenza virus infection. Less is known about the level of protection induced by non-neutralizing antibodies that target conserved internal influenza virus proteins. Such non-neutralizing antibodies could provide an alternative pathway to induce broad cross-reactive protection against multiple influenza virus serotypes and subtypes by partially overcoming influenza virus escape mediated by antigenic drift and shift. Accordingly, more information about the level of protection and potential mechanism(s) of action of non-neutralizing antibodies targeting internal influenza virus proteins could be useful for the design of broadly protective and universal influenza virus vaccines.
KEYWORDS: influenza, antibodies, nucleoprotein, matrix protein
INTRODUCTION
Influenza virus infections result in mild to severe respiratory illness in humans and are a major health burden worldwide, with considerable annual morbidity and mortality (1 – 4). Before the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, seasonal influenza virus infections resulted in around 3–5 million cases of severe disease and 290,000–650,000 global deaths annually (5). Moreover, influenza pandemics happen at unpredictable and irregular intervals (2). Vaccination is the cornerstone in preventing seasonal influenza virus infections, although vaccines are only effective when vaccine strains match circulating strains (6 – 8). Vaccines elicit a robust humoral response toward the immuno-dominant globular head domain of the major surface glycoprotein, the hemagglutinin (HA) (3, 9). Anti-HA antibodies can prevent influenza virus infection by blocking virus binding to its sialic acid receptor and subsequent viral entry and are, therefore, called neutralizing antibodies. Neutralizing antibody titers are a correlate of vaccine-elicited protection (10 – 12). These antibodies, including monoclonal antibodies (mAbs) with the same activity, can also protect mice from influenza virus infection and/or mortality via passive transfer in a prophylactic (13 – 22) and/or therapeutic setting (16 – 24). In spite of the effectiveness of neutralizing anti-HA head antibodies, their applicability as a vaccine target is restricted as these types of vaccines induce narrow, strain- or clade-specific immune responses (10, 11, 25). Specifically, the plastic globular head domain of HA undergoes constant antigenic drift, especially within epitopes recognized by neutralizing antibodies (antigenic sites), circumventing the established immunity induced by the previous year’s vaccine, and thereby forcing annual vaccine reformulation and re-administration (9, 26 – 28). Several neutralizing antibodies that target more conserved regions of the HA head domain have also been described, including mAbs that are directed to the receptor-binding site (RBS) (22, 29 – 36), to sites outside of the RBS (37), to the trimer interface (38 – 40), and to the vestigial esterase domain (16, 41). Nonetheless, the neutralization breadth of these HA head-directed antibodies is still mostly restricted to specific influenza virus subtypes, clearly highlighting the need for novel broader protective or universal vaccines that offer protection against different influenza virus strains.
Unlike neutralizing antibodies, non-neutralizing antibodies (noneAbs) cannot prevent viral infection in vitro (42). These antibodies are, therefore, not readily detectable in classical in vitro neutralization or hemagglutination inhibition assays. Hence, it was presumed that noneAbs could not contribute to protective immunity against influenza viruses. This has been challenged by increasing evidence that suggests that HA-directed noneAbs can provide some protection in vitro (10) and in vivo (8, 27, 43 – 47) against both seasonal and pandemic influenza viruses. The protective effects employed by noneAbs are thought to encompass mechanisms involving the fragment crystallizable (Fc) of antibodies, such as antibody-dependent cellular cytotoxicity (ADCC) (28, 42, 48 – 54), antibody-dependent complement activation (55 – 57), and antibody-dependent cellular phagocytosis (ADCP) (27, 58 – 60). Interestingly, Fc-dependent noneAbs target either conserved regions on the HA, primarily in the stalk region (10, 27, 42, 43, 52, 61 – 66), or the neuraminidase (NA) (42, 67 – 71). In addition, such noneAbs can also target internal influenza virus antigens, such as the matrix protein (M1), matrix protein 2 (M2, which has small ectodomain), and nucleoprotein (NP) (8, 42, 45, 71). These latter proteins are relatively well conserved and could, therefore, represent compelling candidates for the induction of broad protection (1, 9, 72). Indeed, noneAbs targeting internal influenza virus proteins induced by prior seasonal influenza virus infection have been reported to offer a level of protection against subsequent pandemic influenza virus infection in vivo (44). In agreement, broadly protective, heterosubtypic humoral immune responses directed toward the M2 protein were observed after vaccination in mice [(73) and reviewed in detail in reference (74)]. Similarly, in a mouse model, antibodies directed against NP provided protection, although weak, against influenza virus challenge (45, 46). These studies provide evidence that noneAbs might provide an alternative pathway to induce broad cross-reactive protection.
Given the broad cross-protection of some highly efficacious noneAbs that target conserved, sometimes internal, influenza virus proteins, efforts to create a universal vaccine should not ignore the contribution of these long-underappreciated antibodies in anti-influenza virus immune responses. Whereas protective noneAbs to HA and NA have been relatively well defined, less is known about the level of protection induced by noneAbs that target the internal proteins M1 or NP. Therefore, in this study, we analyzed the breadth and potential mechanism of protection of a panel of six human anti-M1 and anti-NP noneAbs that were cloned from plasmablasts isolated from two H3N2-infected individuals. First, a characterization of the binding profile of the mAbs was performed via a cell-based enzyme-linked immunosorbent assay (ELISA) followed by ADCC and ADCP in vitro bioreporter assays with influenza A virus (IAV)-infected cells. To translate the potential of in vitro noneAb-mediated Fc-effector mechanisms toward in vivo protection, prophylactic passive transfer challenge experiments in mice were subsequently performed. Information about the mechanisms of partial protection induced by these noneAbs could give important insights in the development of future broad-spectrum vaccines against both drifted seasonal and emerging pandemic influenza viruses.
RESULTS
Isolation of human mAbs from two H3N2-infected individuals
Plasmablasts (defined as CD19+IgD−CD38+CD20−CD71hi) were single sorted from peripheral blood mononuclear cells (PBMCs) collected from two individuals with confirmed H3N2 influenza virus infection on day 4 and day 5 after symptom onset during the 2017–2018 H3N2-predominant influenza virus season. Corresponding immunoglobulin heavy and light variable (IGHV and IGLV) chain genes were cloned into expression vectors and expressed as described previously (75). A total of 55 antibodies were isolated from these individuals. Most antibodies bound to HA; several also bound to NA. A total of six clonally distinct mAbs reactive to seasonal influenza virus vaccine but non-reactive to recombinant HA or NA were produced; three of the six antibodies were derived from subject 1718002 (1H01, 1F11, and 1B06), and three were from subject 1718003 (1F03, 1D11, and 1H06). Sequence analysis of the generated mAbs showed that they predominantly belonged to the IGHV3 family, which is compatible with the occurrence of this family in human repertoires; IGHV4 and 5 families included the remnant of the mAb population (Fig. S1A). The light chains were distributed among IGKV1-3 families, with the IGKV1 family representing 50%. The IGHV gene complementarity-determining region 3 (CDR3) lengths ranged from 13 to 20 amino acid residues (Fig. S1B). As expected, the CDR3 lengths of the IGLV gene were relatively similar and ranged from 11 to 13 residues (Fig. S1C). The variety of IGHV and IGLV families, in addition to IGHV CDR3 lengths, suggest distinct modes of interaction with the influenza virus proteins.
Human mAbs bind to the internal M1 and NP proteins of IAVs
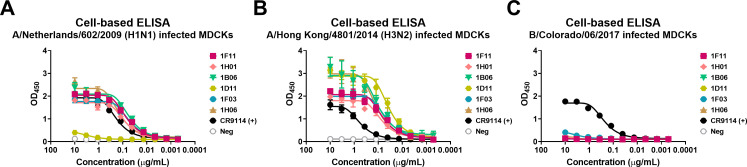
The mAbs were first screened for their breadth of binding to IAV- and influenza B virus (IBV)-infected cells via cell-based ELISAs. Three different viruses, A/Netherlands/602/2009 (H1N1), which represents a pandemic strain; A/Hong Kong/4801/2014 (H3N2), which is seemingly closely related to the strain that caused the infection; and B/Colorado/06/2017, which belongs to the B/Victoria/2/1987-like lineage, were chosen for this assay and for downstream in vitro characterization. mAbs CR9114, a cross-reactive influenza virus HA stem binding mAb (41), and 1D04, an anti-chikungunya virus (CHIKV) E1 glycoprotein mAb, were used as positive and negative controls, respectively. Five out of the six mAbs showed robust binding to the H1N1- and H3N2-infected cells, while none of the mAbs recognized the IBV-infected cells (Fig. 1).
Fig 1.
mAb reactivity to different influenza virus strains in a cell-based ELISA. Binding profiles of six human mAbs against A/Netherlands/602/2009 (H1N1) (A), A/Hong Kong/4801/2014 (H3N2) (B), and B/Colorado/06/2017 (C) viruses measured by ELISA. CR9114, a cross-reactive influenza virus HA stem binding mAb, was used as a positive control. 1D04, an anti-CHIKV E1 glycoprotein mAb, was used as a negative control. For all panels, OD450 values are shown. The non-linear regression for each group is indicated. Data are representative of one experiment with duplicate measurements.
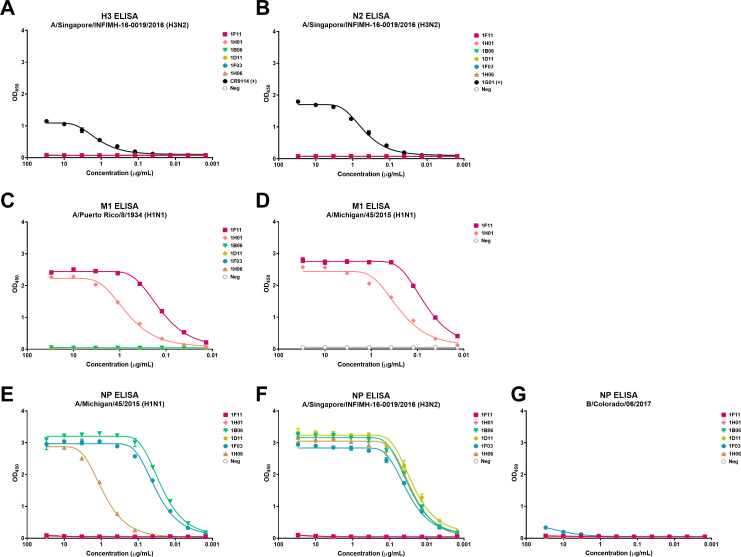
To determine which antigens these mAbs bind, we assessed binding to HA, NA, M1, and NP in an ELISA. Protein-based ELISAs revealed that none of these mAbs recognized recombinant H3 HA or N2 NA proteins from the A/Singapore/INFIMH-16-0019/2016 (H3N2) strain in contrast to the positive controls CR9114 (a pan anti-HA mAb) and 1G01 (a pan anti-NA mAb) (3) (Fig. 2A and B). Instead, two mAbs—1F11 and 1H01—displayed binding to recombinant M1 proteins derived from strains A/Puerto Rico/8/1934 (H1N1) and A/Michigan/45/2015 (H1N1) (Fig. 2C and D). The four remaining mAbs—1B06, 1D11, 1F03, and 1H06—bound the influenza virus NP of A/Singapore/INFIMH-16–0019/2016 (H3N2) (Fig. 2F). Three of those mAbs—1B06, 1F03, and 1H06—also bound to recombinant NP protein of A/Michigan/45/2015 (H1N1) (Fig. 2E). In agreement with the cell-based ELISAs, none of the NP-binding mAbs recognized the recombinant B/Colorado/06/2017 NP protein (Fig. 2G), indicating that these mAbs only recognize proteins of IAVs. Similarly, none of the HA, NA, M1, and NP recombinant proteins were recognized by the negative control mAb, 1D04 (Fig. 2). Taken together, these results indicate that the H3N2 infections in these two individuals induced robust plasmablast responses with cross-reactive mAbs specific for the internal M1 and NP proteins of IAVs.
Fig 2.
Binding profile of mAbs isolated from two H3N2-infected individuals to influenza virus proteins. Binding of the human mAbs against A/Singapore/INFIMH-16-0019/2016 H3 (A), A/Singapore/INFIMH-16-0019/2016 N2 (B), A/Puerto Rico/8/1934 M1 (C), A/Michigan/45/2015 M1 (D), A/Michigan/45/2015 NP (E), A/Singapore/INFIMH-16-0019/2016 NP (F), and B/Colorado/06/2017 NP (G). 1D04, an anti-CHIKV E1 glycoprotein mAb, functioned as a negative control. The OD450 values are displayed in each panel. The non-linear regression for each group is displayed. Data are representative of one experiment with duplicate measurements.
Anti-M1 and anti-NP antibodies are non-neutralizing in vitro
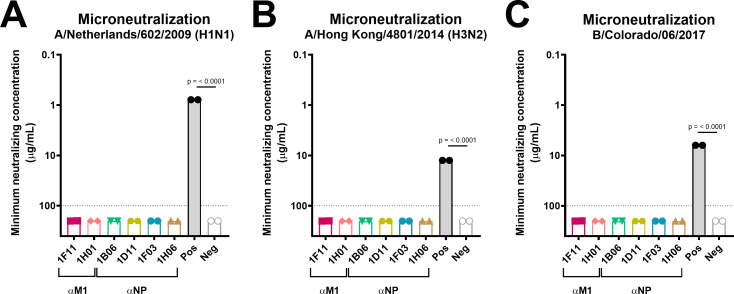
Next, we characterized the mAbs in an in vitro neutralization assay to evaluate their ability to neutralize diverse influenza viruses. Typically, mAbs directed against internal M1 and NP proteins are not able to prevent infection, since they cannot compete with the influenza virus for binding to the cell surface receptor and, consequently, do not show any activity in in vitro neutralization assays (8, 42, 76, 77). In order to assess possible antiviral effects downstream of entry, the mAbs remained in the overlay at all times during the incubation period. mAbs CR9114 (a pan anti-HA mAb) and 2E01 (an IBV NA mAb) (78) served as positive controls, whereas the mAb 1D04 functioned as the negative control. As expected, although all anti-M1 and anti-NP mAbs bound to IAV-infected cells (Fig. 1), none of them showed neutralizing activity and never reached a 50% inhibition endpoint, against any of the tested viruses (A/Netherlands/602/2009 (H1N1), A/Hong Kong/4801/2014 (H3N2), and B/Colorado/06/2017) (Fig. 3). Accordingly, the isolated broadly reactive anti-M1 and anti-NP mAbs were non-neutralizing.
Fig 3.
Neutralization activity assessment of IAVs and an IBV for anti-M1 and anti-NP mAbs. Neutralization of A/Netherlands/602/2009 (H1N1) (A), A/Hong Kong/4801/2014 (H3N2) (B), and B/Colorado/06/2017 (C) strains measured in microneutralization assays. The minimum neutralization concentration in micrograms per milliliter of the respective mAb is displayed and was established via a hemagglutination-based readout. The minimum neutralizing concentration represents the lowest antibody concentration at which no hemagglutination was detected. The data are presented as individual technical replicates of a single experiment, and the linear regression for each mAb group is represented. The dashed vertical gray line represents the assay detection limit (100 µg/mL). mAbs that did not reach this limit were assigned a value of 200 µg/mL. CR9114, which has known neutralizing activity against H1N1 and H3N2 viruses, or 2E01, which has known neutralizing activity against influenza B viruses, and a CHIKV E1 virus mAb, 1D04, were used as positive and negative controls for all experiments, respectively. P values indicate the statistical significance of the difference between the groups and the irrelevant antibody control assessed by one-way analysis of variance (ANOVA) followed by a multiple-comparison test. Only P values lower than 0.05 are presented.
Anti-NP, but not anti-M1, noneAbs exhibit ADCC and ADCP activity in vitro
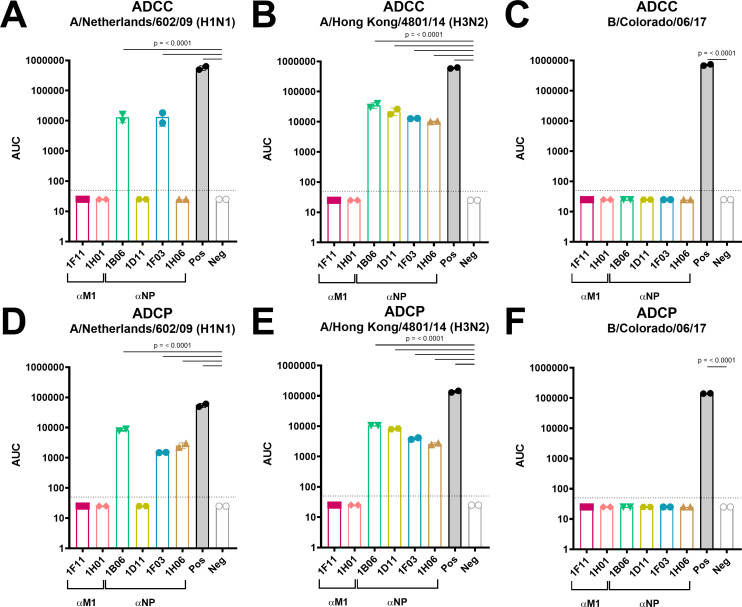
FcR-mediated effector functions are a pivotal mechanism for broadly protective noneAbs (45, 48, 49, 52, 79). Although internal proteins, patches of typically intracellular NP antigens are transiently expressed on the surface of influenza virus-infected cells, and the M1 has been reported to become readily accessible in cells that die following influenza virus infection (57, 77, 80). This makes NP and M1 proteins reachable targets for antibody-mediated Fc-receptor functions. Therefore, to examine potential mechanisms of the six isolated mAbs, in vitro ADCC bioreporter assays were performed and functioned as a surrogate for in vivo Fc-FcR based effector functions. Here, Madin Darby canine kidney (MDCK) cells were infected with A/Netherlands/602/2009 (H1N1), A/Hong Kong/4801/2014 (H3N2), or B/Colorado/06/2007 strains, and ADCC induction of mAbs in varying concentrations was evaluated. CR9114 and 1D04 functioned as a positive and negative control mAb, respectively. Indeed, all anti-NP mAbs induced a significant ADCC activity toward IAVs when compared to the negative control antibody (Fig. 4A and B). More precisely, two anti-NP mAbs—1B06 and 1F03—facilitated ADCC induction in response to the two influenza A virus strains, while the other two anti-NP mAbs—1D11 and 1H06—only mediated ADCC activity against A/Hong Kong/4801/2014 (H3N2). In agreement with the ELISA data, none of the anti-NP mAbs mediated ADCC activity against the B/Colorado/06/2017 strain (Fig. 4C). Notably, none of the anti-M1 mAbs displayed ADCC activity to any of the influenza viruses (Fig. 4A through C), which is consistent with the absence of studies reporting M1’s ADCC potency. The ADCC assays were repeated, and the data were shown in Fig. S3A through C.
Fig 4.
Functional in vitro characterization of ADCC and ADCP Fc-effector mechanisms mediated by anti-M1 and anti-NP mAbs. In vitro ADCC activity of anti-M1 and anti-NP mAbs against A/Netherlands/602/2009 (H1N1) (A), A/Hong Kong/4801/2014 (H3N2) (B), and B/Colorado/06/2017 (C). In vitro ADCP activity of anti-M1 and anti-NP mAbs against A/Netherlands/602/2009 (H1N1) (D), A/Hong Kong/4801/2014 (H3N2) (E), and B/Colorado/06/2017 (F). CR9114 functioned as a positive control, whereas the anti-CHIKV E1 mAb (1D04) served as a negative control. Data are presented as areas under the curve (AUCs) calculated with a lower detection limit of the average background plus three standard deviations (SDs). Every symbol represents the AUC of a single mAb and bars denote mean ± SD of one experiment conducted with two technical replicates. For statistical significance calculations, values were first log(y) transformed, and subsequently, a one-way ANOVA corrected for multiple comparison test was performed. Only P values lower than 0.05 are shown.
Although less intensively investigated in the setting of influenza viruses, another antibody-mediated Fc-effector function is ADCP. Recent studies reported that antibody-mediated phagocytosis significantly contributed to the elimination of influenza virus-infected cells and, hence, protection against influenza virus infection (58, 60, 64, 81). In addition, this Fc-mediated mechanism has been shown to contribute to protection conferred by anti-influenza virus noneAbs (27, 42, 59). Therefore, we subsequently assessed the ability of anti-M1 and anti-NP noneAbs to induce ADCP activity in an in vitro ADCP bioreporter assay. The positive and negative control mAbs included CR9114 and 1D04, respectively. Interestingly, similar results were found to the ADCC bioreporter assays (Fig. 4A through C). Specifically, anti-NP mAbs induced a significant level of ADCP in response to influenza A, but not B, virus-infected cells when compared to the negative control antibody (Fig. 4D through F). In agreement with the binding profiles, mAb 1D11 only conferred ADCP activities against the A/Hong Kong/4801/2014 (H3N2) strain (Fig. 4E). In contrast, anti-NP mAbs 1B06, 1F03, and 1H06 showed heterosubtypic activity against both tested IAV strains (Fig. 4D and E). However, ADCP activity could not be observed for anti-M1 mAbs (Fig. 4D through F), which could indicate that these mAbs potentially function via alternative Fc-dependent mechanisms and/or Fc receptors.
Anti-M1 and anti-NP mAbs show low protectiveness in vivo
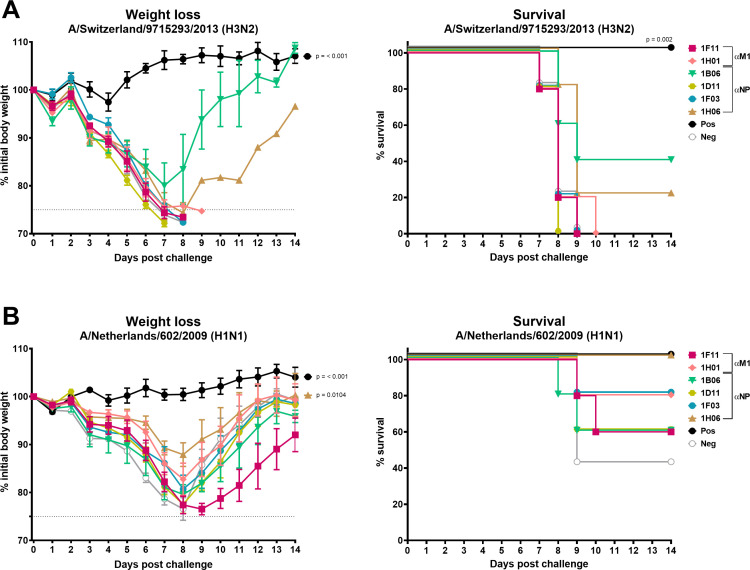
To evaluate whether in vitro ADCC and/or ADCP activity of the anti-NP mAbs is sufficient to provide protection against influenza A viral challenge in vivo, a prophylactic passive transfer challenge experiment in mice was performed. The anti-M1 mAbs were also included to investigate their protective capacity in vivo, as they displayed reactivity to diverse IAV-infected cells (Fig. 1). Mice were intraperitoneally injected with 10 mg/kg of mAb 3 h prior to intranasal A/Netherlands/602/2009 (H1N1) or mouse-adapted A/Switzerland/9715293/2013 (H3N2) challenge, and for a period of 2 weeks, weight loss and survival were monitored daily. The anti-NA mAb, 1G01, and anti-SARS-CoV-2 mAb, 1C11, served as positive and negative controls, respectively. All mice displayed high mAb titers, which is indicative of successful transfer, and hence, no mice were excluded from the experiments (Fig. S2A and B). Following challenge with the A/Switzerland/9715293/2013 (H3N2) virus, we found that all mice administered anti-M1 and the negative control, 1C11, mAbs crossed the 25% weight loss humane endpoint and were euthanized and scored dead (Fig. 5A). Of the anti-NP mAbs, all mice administered 1D11 and 1F03 did not survive. However, mice administered 1B06 or 1H06 showed 40% or 20% survival, respectively. Similar results were obtained in a biological replicate, in which 40% or 80% of the mice survived the A/Switzerland/9715293/2013 challenge when administered the anti-NP mAb 1B06 or 1H06, respectively (Fig. S4A and B). For the A/Netherlands/602/2009 (H1N1) challenge, we observed that mice administered the anti-M1 mAbs, 1F11 and 1H01; the anti-NP mAbs, 1B06, 1D11, or 1F03; or the negative control mAb, 1C11, lost a substantial amount of weight (Fig. 5B). It should be noted that the A/Netherlands/602/2009 (H1N1) challenge was performed at a lower lethal dose than anticipated, as 40% of the negative control mice survived the challenge. As the anti-NP mAbs 1B06 and 1H06 were partially protective in both challenge experiments, the A/Netherlands/602/2009 challenge was repeated with these mAbs (Fig. S4C and D). Upon repeating the challenge at 5 murine 50% lethal doses (mLD50), all mice administered the anti-NP mAbs, 1B06 and 1H06, and the negative control mAb lost weight rapidly and were euthanized by day 7 post-infection. Collectively, the data indicate that our noneAbs that target either internal M1 or NP proteins do not robustly protect mice from influenza virus-induced morbidity and mortality.
Fig 5.
Assessment of anti-M1 and anti-NP mAb protection in a prophylactic setting in vivo. Morbidity in A/Switzerland/9715293/2013 (H3N2) (A) and A/Netherlands/602/2009 (H1N1) (B) infected mice following intraperitoneal administration of anti-M1 (1F11 and 1H01) or anti-NP (1B06, 1D11, 1F03, and 1H06) mAbs. Five animals per experimental group were used. Weight loss and survival were monitored for 14 days post challenge. The percentage (%) of weight loss and survival is shown. Error bars indicate the standard error of the mean (SEM). 1G01, a pan NA mAb, functioned as a positive control, whereas the SARS-CoV-2 mAb, 1C11, served as a negative control. The dashed vertical gray line represents a 75% cutoff of the initial weight, which was defined as the humane endpoint. P values indicate the statistical significance of the difference between the groups and the irrelevant antibody control assessed by one-way ANOVA followed by a multiple-comparison test for weight loss or a log-rank (Mantel-Cox) test for survival. For weight loss comparisons, the maximum weight loss in percentage for each mouse was used. Mice that reached the human endpoint were assigned 75%. Only P values lower than 0.05 are displayed.
DISCUSSION
Classical antibody-based protection from influenza A virus-induced morbidity and mortality correlates with neutralizing antibodies that prevent virus binding to sialic acid receptors and subsequent viral entry (82). Hence, studies of antibody-induced immunity to influenza viruses historically focused on the highly variable HA as a target (83, 84). However, recently, this has been challenged by the discovery of the importance of another subset of antibodies, namely, noneAbs (42, 48, 62, 76). Whereas the protective role of noneAbs against HA and NA is relatively well defined (11, 27, 42, 61, 85), the relevance and mechanism(s) by which noneAbs against internal proteins, like M1 and NP, exactly operate in the setting of influenza virus infection remained largely elusive. Therefore, in this study, we characterized a panel of six novel human anti-M1 and anti-NP mAbs that showed robust binding to IAVs. Both the internal M1 and NP influenza virus proteins represent highly conserved targets for noneAbs (8, 42). While none of the mAbs had neutralizing activity in an in vitro neutralization assay, all anti-NP mAbs were able to mediate ADCC and/or ADCP activity in response to diverse IAV infections in reporter assays. In an attempt to translate the potential of in vitro noneAb-mediated Fc-effector mechanisms toward in vivo protection against IAVs, a prophylactic passive transfer challenge experiment in mice was performed. Notably, anti-IAV M1 mAbs did not robustly protect mice from influenza virus-associated morbidity and mortality. In contrast, the anti-IAV NP noneAbs, 1B06 and 1H06, partially protected against A/Switzerland/9715293/2013 (H3N2) challenge. Hence, our study suggests that noneAbs that bind internal IAV antigens, particularly the M1 protein, only provide limited protection in vivo. These findings, although based on a relatively small panel of mAbs, can provide important information for the design of broadly protective and universal influenza virus vaccines. An important caveat here is that human antibodies were used in the mouse model and crosstalk between human Fc and murine FcRs may not have been optimal. However, low-affinity non-neutralizing human anti-H7 mAbs showed protection in the mouse model despite this caveat (27). We are, therefore, confident that this system would detect a robust Fc-mediated protective effect.
Our work found that anti-M1 and anti-NP mAbs had robust binding to the two tested IAV strains, A/Netherlands/602/2009 (H1N1; pandemic) and A/Hong Kong/4801/2014 (H3N2), while no measurable reactivity was found toward the B/Colorado/06/2017 virus strain. Internal M1 and NP proteins exhibit over 90% amino acid sequence identity across a range of IAV subtypes and strains sampled from both humans and other host species (28, 72, 76, 86 – 90). In contrast, comparative analysis of the influenza A virus NP amino acid sequence with that of the influenza B virus NP revealed only a low 37% direct homology in the aligned regions (91). Similarly, a 25.4% direct homology was found between influenza A and B virus M1 amino acid sequences (92). Thus, the influenza virus internal proteins are only highly conserved within one influenza virus type, and it is, consequently, not surprising that the mAbs assessed in this study and others (77) do not provide cross-reactivity between diverse types.
Upon further assessment of the anti-M1 and anti-NP mAbs, we identified that the anti-NP mAbs induced significant levels of ADCC and ADCP activities when measured with IAV-infected cells in reporter assays compared to the negative control antibody, while the anti-M1 mAbs did not mediate any of these activities. These data might implicate that the anti-M1 mAbs conferred partial protection against the low dose A/Netherlands/602/2009 (H1N1) viral challenge via a different mechanism of action that has not been tested in this study, such as antibody-mediated complement-dependent cytotoxicity (CDC). Indeed, the Fc fragment of anti-M2e noneAbs has been demonstrated to bind to complement and thereby trigger the complement cascade leading to CDC (55, 56). Similarly, stalk (93) and NP (57) reactive mAbs promote complement-mediated virolysis upon binding to IAV-infected cells. In contrast, another anti-NP mAb was unable to mediate CDC following the recognition of IAV-infected cells (77). This discrepancy may be due to differences in the experimental conditions used or the inherent biological effect of the divergent anti-NP mAbs. Alternatively, the results might also be skewed by the M1 epitope accessibility on only dead, and not living, influenza virus-infected cells (2, 57, 77, 80). Accordingly, it might be conceivable that the in vitro ADCC and ADCP bioreporter assays were performed in the absence of accessible M1 proteins. Therefore, future experiments with a longer infection period and/or with M1 transfected cells might clarify which Fc-effector function(s) is mediated by mAbs targeting the internal M1 protein. In addition, it should be noted that the ADCC assays used in these studies are based on human FcRs, but mice—as mentioned above—express murine FcRs, which in our experiments perhaps would reduce the interaction between human mAbs and mouse FcRs that are required for protection in the mouse model. Besides, to verify the binding of mAbs 1F11 and 1H01 to influenza A virus internal M1 proteins, fluorescence-activated cell sorting analysis with living cells could be performed. Collectively, these findings firmly indicate that the antigen specificity was responsible for the different Fc-effector profiles obtained for anti-M1 and anti-NP mAbs, in which future studies are required to elucidate on which Fc-effector mechanism or mechanisms anti-M1 mAbs depend for providing in vivo partial protection against the low dose A/Netherlands/602/2009 (H1N1) viral challenge.
Upon assessment of the anti-M1 and anti-NP mAbs in a prophylactic model, we found that all mice passively immunized with anti-M1 noneAbs succumbed to A/Switzerland/9715293/2013 (H3N2) infection. In contrast, these anti-M1, as well as the four anti-NP, reduced morbidity following challenge with A/Netherlands/602/2009 (H1N1). Although it should be highlighted that the A/Netherlands/602/2009 challenge was performed at a lower lethal dose than anticipated, as 40% of the negative control mice survived challenge. When the H1N1 study was repeated with an actual dose of 5× mLD50 for the anti-NP noneAbs, 1B06 and 1H06, as well as a negative control mAb, all mice showed severe weight loss and reached the 25% weight loss humane endpoint around day 7. This indicates that anti-NP noneAbs are unable to protect mice from A/Netherlands/602/2009 (H1N1) virus-induced mortality when a lethal dose of 5× mLD50 is used. For the A/Switzerland/9715293/2013 (H3N2) challenge, however, anti-NP mAbs 1B06 and 1H06 were able to partially protect mice against morbidity and mortality, and this may be because the mAbs were isolated from an influenza A H3N2-infected patient. This is reminiscent of the mouse mAbs 1H5 (a broadly reactive H7 HA head mAb) (85), 07-5E01 (a cross-reactive H7 HA stalk mAb) (27), 41-5D06 (a heterosubtypic H7 HA head binding mAb) (27), and 2D1 (a H7 HA stem reactive mAb) (11), which do not neutralize avian influenza A (H7N9) viruses but do significantly protect in vivo via, at least in part, Fc-dependent effector functions. Importantly, the observed protection in our H3N2 study was not as robust as seen for these and some other noneAbs that target HA (11, 27, 62 – 64, 85) or NA (3, 69) proteins. It, therefore, seems that noneAbs that target external, rather than internal, proteins on the IAV surface confer better protection in vivo. These differing protective profiles might be due to differential protein expression levels and/or protein accessibility on living and/or dead influenza virus-infected cells. Indeed, when comparing the affinity of our anti-M1 and anti-NP mAbs with other non-neutralizing HA stalk antibodies that better protect in vivo (27, 62), it becomes clear that our antibodies reveal lower minimum binding concentrations excluding the possibility that differences in antibody affinity were responsible for the divergent protective capacities conferred in vivo by our anti-M1 and anti-NP and the reported anti-HA noneAbs. Another possible explanation for the differing protective profiles is related to a limitation of our study. DBA2/j mice have a CD94 deficiency and, as a result, dysregulated NK cells, which may be why the noneAbs were not as protective following A/Switzerland/9715293/2013 (H3N2) challenge compared to the low dose A/Netherlands/602/2009 (H1N1) challenge (94). Another limitation with our animal studies is the challenge dose. We challenged with a virus dose that induces significant weight loss in challenged mice, and perhaps, the protective effects of these mAbs would be better assessed with a lower challenge dose. As mentioned above, one of our anti-NP mAbs—1H06—was able to completely protect in vivo when a low H1N1 challenge dose was used (an average maximum weight loss of 14% was observed). The protection afforded seemed to be contingent on in vitro ADCP activity as this mAb did not mediate ADCC activity against A/Netherlands/602/2009 (H1N1) virus-infected cells. As such, further studies that investigate different and especially lower virus doses for prophylactic passive transfer studies in mice would be critical to gain a better understanding of the potential cross-protective profile of the described anti-M1 and anti-NP mAbs.
Anti-M1 and anti-NP immune responses are primarily associated with the induction of broadly reactive cellular but not humoral immune responses. Along these lines, we and others (8, 45, 77, 95) have demonstrated that anti-M1 and anti-NP mAbs do not neutralize influenza viruses, which is seen as the main correlate of mAb-mediated protection. In contrast, broad-spectrum T-cells that primarily recognize conserved internal influenza virus antigens can provide protection by curbing influenza viral spread and limiting transmission (96 – 99). T-cell responses targeting these epitopes have been demonstrated to be highly protective in mice (100 – 102), and both CD4+ and CD8+ T-cells have been described as correlates of protection in humans (99, 103 – 106). In fact, two studies performed in mice (9) and ferrets (1) illustrated that a combined prime-boost vaccination regimen of sequential exposure to chimeric HAs [cHAs; contain a highly variable head, but a conserved stalk domain (107 – 110)] with a replication-deficient chimpanzee adenovirus-vectored vaccine expressing both NP and M1 proteins, called ChAdOx1 NP + M1 (111), protected against H3N2 infection through the induction of a robust anti-HA stalk humoral in combination with a long-lived cellular immune response. Of note, in the former study, the combination of both sets of vaccination strategies offered augmented protection against influenza virus challenge in comparison to vaccination with the monovalent (cHA or M1 + NP) viral vectors (9). These and corresponding (112, 113) data implicate a potential synergy between vaccine-induced humoral and cellular immune responses. Hence, efforts focusing on developing a universal influenza virus vaccine should consider the development of vaccines that are able to harness both types of adaptive immunity. More precisely, if such a vaccination platform would be able to elicit a humoral immune response that consists of a cocktail of broadly neutralizing and non-neutralizing antibodies that target non-overlapping but protective epitopes, influenza virus immune escape could be avoided as a consequence of complementary or alternative mechanisms induced by these two different types of antibodies [detailed in reference (6)]. Accordingly, it would be interesting and worthwhile to examine the interplay between neutralizing and non-neutralizing mAbs, in addition to distinct noneAbs that mediate different Fc-effector mechanisms, to gain a better understanding on which antibody patterns would be able to induce synergistical protection and would be suitable candidates for the design of future universal anti-influenza virus vaccines.
In conclusion, we demonstrate that on a monoclonal basis in the mouse model, broadly reactive non-neutralizing anti-IAV M1 antibodies do not significantly protect against an influenza virus challenge. Anti-IAV NP antibodies, on the other hand, were more protective than anti-IAV M1 antibodies and had increased levels of Fc-effector functions. These results need to be confirmed with a larger panel of mAbs, as well as with polyclonal serum, and ideally in a mouse model with humanized FcRs. Our results, nonetheless, suggest that efforts focusing on developing a universal influenza virus vaccine that incorporate M1 and NP antigens should perhaps focus on the cellular immune responses induced following vaccination with these antigens since protection from mAbs against NP and M1 is not robust.
MATERIALS AND METHODS
Patients
Human PBMCs were collected from two individuals enrolled in a prospective observational cohort study at the Barnes Jewish Hospital Emergency Department (EDFLU study) (114). The Institutional Review Board of Washington University in Saint Louis (Missouri, USA) approved the EDFLU study (approval #2017-10-220). Subject 1718002 was 35 years old, female, and had a history of asthma. She was infected with sequencing-confirmed H3N2 IAV and presented on the fourth day of symptomatic illness when the PBMC and serum samples were obtained. She had not received the 2017–2018 seasonal influenza vaccine prior to disease onset but reported that she received other seasonal influenza vaccines in the past 5 years. Subject 1718003 was 34 years old, female, and had a history of diabetes, asthma, and congestive heart failure. She was infected with sequencing-confirmed H3N2 IAV and presented on the fifth day of symptomatic illness when PBMC and serum samples were obtained. She received the 2017–2018 seasonal influenza vaccine prior to disease onset and reported receiving other seasonal influenza vaccines in the past 5 years. Both subjects required admission to hospital for their illness but were successfully treated and discharged within a week of arrival in the emergency department. A total of 55 antibodies were isolated from these two individuals (22 mAbs from subject 1718002 and 33 mAbs from subject 1718003). Most antibodies bound to HA; several also bound to NA. The remaining antibodies were screened for binding to NP and M1.
Cells
MDCK (CCL-34, American Type Culture Collection, Manassas, USA) cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Waltham, USA) accompanied with penicillin-streptomycin (Pen/Strep) antibiotics solution (100 U/mL of Pen and 100 µg/mL Strep; Gibco), 1 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Gibco) and fetal calf serum (10%; Corning, Corning, USA) resulting in complete DMEM at 37°C in a humidified incubator with 5% carbon dioxide (CO2). Expi293F cells (A14527, Thermo Fisher Scientific, Waltham, USA) were grown in Expi293 Expression Medium (Gibco) in 1-L Erlenmeyer shaker flasks (Corning) at 37°C and 125 revolutions per minute (rpm) in a humidified incubator with 8% CO2. Single-use aliquots of ADCC bioassay effector Fcgamma(γ)RIIIa (CD16a) and ADCP bioassay effector FcγRIIa-H (CD32a) cells (Promega, Madison, USA) were thawed directly prior to use.
Viruses
The IAV strains, A/Netherlands/602/2009 (H1N1), A/Hong Kong/4801/2014 (H3N2), and mouse-adapted A/Switzerland/9715293/2013 (H3N2), were propagated in 10-day-old specific pathogen-free (SPF) embryonated chicken eggs (Charles River Laboratories, Massachusetts, USA) for 2 days at 37°C. The IBV strain, B/Colorado/06/2017, was grown in 10-day-old SPF embryonated chicken eggs for 3 days at 33°C. Eggs were subsequently inactivated over night at 4°C. Following inactivation, allantoic fluid was harvested and centrifuged (Centrifuge 5810 R, 15 amp version, Eppendorf, Hamburg, Germany) at 4,000 × g for 10 min at 4°C to pellet cellular debris, and eventually, allantoic fluid was aliquoted and stored at −80°C prior to performing plaque assays to determine virus titers, as described previously (1, 115).
Antibodies
Antibodies were expressed by transient co-transfection of 17 µg of heavy and 33 µg of light chain plasmids in Expi293F (70 × 106 cells/50 mL) cells using the ExpiFectamine 293 Transfection kit (Thermo Fisher Scientific) as per the manufacturer’s recommendations and as has been previously described (74). Seven days post transfection, cell cultures were harvested by low-speed centrifugation at 4,000 × g for 30 min and filtered through a 0.22 µM sterile filtration unit. Next, mAbs were purified using protein A chromatography (GoldBio, Missouri, USA) and were eluted into a 50-mL Falcon tube (Denville Scientific, Inc., New Jersey, USA) each with 0.1 M glycine-hydrogen chloride (HCI; pH 2.7) and were directly neutralized with 1 M Tris-HCl (pH 9.0). mAbs were then concentrated and buffer exchanged with phosphate-buffered saline (PBS) using Amicon Ultra filter units (EMD Millipore) with 30-kDa cutoffs, and finally, mAb concentrations were determined at 280-nm absorbance with a Nanodrop spectrophotometer (Thermo Fisher Scientific).
Recombinant proteins
Recombinant H3 from A/Singapore/INFIMH-16-0019/2016 (H3N2), N2 from A/Singapore/INFIMH-16-0019/2016 (H3N2), and M1 from A/Michigan/45/2015 (H1N1) or A/Puerto Rico/8/1934 (H1N1) proteins were expressed and purified using the well-established baculovirus expression system as described previously (63, 116, 117). Recombinant NP proteins from A/Michigan/45/2015 (H1N1), A/Singapore/INFIMH-16-0019/2016 (H3N2), and B/Colorado/06/2017 were expressed via transient transfection of Expi293F cells according to the manufacturer’s instructions (Thermo Fisher Scientific).
Recombinant protein supernatants were harvested after 3 days, centrifuged at 4,000 × g for 20 min at 4°C, and passed through a 0.22-µm filter. Supernatants were subsequently mixed and incubated with nickel nitrilotriacetic acid (Qiagen, Hilden, Germany) agarose beads on a shaker (~125 rpm) for 2 h at room temperature (RT). Proteins were then purified via gravity flow columns (Qiagen), eluted as described previously (117), concentrated, and buffer exchanged using Amicon Ultra centrifugal units via three-time centrifugation at 4,000 × g for 30 min at 4°C and finally re-suspended in PBS. Purified proteins were quantified using the Quick Start Bradford 1× Dye Reagent (Bio-Rad; California, USA) and based on a standard curve. Recombinant proteins were analyzed via reducing sodium dodecyl sulphate-polyacrylamide gel electrophoresis to check for protein purity and integrity.
Cell-based ELISA
Ninety-six-well cell culture plates (Corning) were seeded with 100 µL/well of MDCK cells at a density of 2 × 105 cells/mL and incubated over night at 37°C with 5% CO2. The following day, cells were washed with PBS and infected with 100-µL virus inoculum at a multiplicity of infection (MOI) of 5 [corresponding to about 2 × 105 plaque-forming units (PFU) of virus; diluted in 1× minimal essential medium (MEM, Gibco)] for 16–20 h at 37°C with 5% CO2 for IAV or at 33°C with 5% CO2 for IBV strains. Cells were then fixed with 3.7% paraformaldehyde (Thermo Fisher Scientific; 200 µL/well) and stored at 4°C for 24 h. The subsequent day, plates were washed with PBS, and the cells were permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, Missouri, USA) diluted in PBS and incubated for 15 min at RT. Afterwards, plates were washed once and incubated for 1 h at RT with 200 µL/well blocking solution containing 3% milk in PBS. The mAbs were then diluted to a starting concentration of 10 µg/mL, serially diluted threefold across plates, and incubated for 2 h at RT. Plates were subsequently washed three times with PBS before adding the secondary anti-human IgG (Fab specific) horseradish peroxidase (HRP) (produced in goat, Sigma-Aldrich) antibody at a concentration of 1:3,000 in blocking solution for 1 h at RT. After incubation, plates were washed three times to remove residual secondary antibody and were subsequently developed by adding 3,3′-5,5′-tetramethylbenzidine substrate (Sigma-Aldrich; 100 µL/well). The reaction was stopped after 10 min with 4 M sulfuric acid (Thermo Fisher Scientific; 50 µL/well). The optical density (OD) of each plate was measured at 450 nm on a Synergy H1 microplate reader (BioTek, Vermont, USA), which represented the readout for this assay. Results were analyzed in Microsoft Excel and GraphPad Prism 9 (GraphPad Software, Inc., La Jolla, California, USA). To visualize the binding curves, a non-linear regression model was used based on one individual experiment with technical duplicates.
The ELISA to measure mAb titers in the pre-challenge sera from mice was performed similarly. Here, MDCK cells were infected with the A/Hong Kong/4801/2014 (H3N2) virus strain at a MOI of 5. The plates were treated and blocked as described above. Pre-diluted sera (1:10) were added to the first well to obtain a final concentration of 1:100 in blocking solution. The sera were then serially diluted threefold and incubated for 2 h at RT. Next, the secondary antibody, an anti-human IgG (Fab specific) HRP (produced in goat, Sigma-Aldrich) diluted at a concentration of 1:3,000 in blocking solution, was added. Subsequently, the ELISA was further processed as described above. Data are presented as the area under the curve (AUC) calculated with a cutoff value of the average of the OD values of the background plus three standard deviations (SDs).
Protein-based ELISA
Protein-based ELISAs were performed as described previously (3). In brief, 96-well nonsterile, flat-bottom, microtiter plates (Immunolon 4HBX; Thermo Fisher Scientific) were coated with a final concentration of 2 µg/mL of recombinant protein (50 µL/well) in PBS overnight at 4°C. The next day, plates were washed three times with PBS containing 0.1% Tween 20 (PBS-T; Fisher BioReagents, Pennsylvania, USA). Subsequently, plates were blocked for 1 h at RT with 220-µL blocking solution consisting of 3% goat serum (Life Technologies, Carlsbad, USA) and 0.5% milk in PBS-T. After blocking, mAbs were diluted to a starting concentration of 30 µg/mL in blocking solution and were serially diluted threefold in a final volume of 100 µL/well and incubated for 2 h at RT. The rest of the assay was conducted as described in the Cell-based ELISA section with the only alteration that the final washing step included four washes with PBS-T with intermitted shaking in between each washing step.
Microneutralization assay
MDCK cells were seeded in 96-well cell culture plates (Corning) at a density of 2 × 104 cells/well with a total volume of 100 µL in each well and were incubated over night at 37°C in a humidified incubator under 5% CO2. The following day, mAbs were diluted to a 100 µg/mL concentration and then twofold serially diluted across a 96-well plate in infection medium [1× MEM (Gibco), 2 mM L-glutamine (Gibco), 0.1% sodium bicarbonate (wt/vol, NaHCO3; Gibco), 0.01 M HEPES buffer (Gibco), Pen/Strep (100 U/mL penicillin, 100 µg/mL streptomycin; Gibco), 0.2% bovine serum albumin (MP Biomedicals, California, USA), and 1 µg/mL tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma-Aldrich)]. Next, 60 µL of 100× tissue culture infectious dose 50 of virus in infection medium and 60 µL of serially diluted mAb were incubated on a shaker for 1 h at RT. Prior to the end of the incubation time, MDCK cells were washed once with 200-µL PBS, and then, 100 µL of the incubated mAb-virus mixture was added for 1 h at 37°C with 5% CO2 for IAV or 33°C with 5% CO2 for IBV strains, respectively. Afterwards, the virus inoculum was aspirated, and MDCK cells were again washed with 200-µL PBS and incubated with 100 µL of mAb (at the same concentrations) for 48 h at 37°C under 5% CO2 for the IAV plates or for 72 h at 33°C under 5% CO2 for the IBV plates. To visualize the neutralizing potency of the mAbs, a classical hemagglutination assay was conducted. Results were analyzed in Microsoft Excel and GraphPad Prism 9. Data are displayed as the minimum neutralizing concentration, and this value represents the lowest antibody concentration at which no hemagglutination could be detected. The detection limit included 100 µg/mL, as this was the highest antibody concentration used for this assay, and mAbs that did not reach neutralizing activity at this limit were assigned a value of 200 µg/mL for graphing purposes. Each experiment was performed in duplicate. CR9114, a broadly cross-reactive influenza A and B virus HA stalk binding mAb with known neutralizing activity (41), and 2E01, an IBV NA mAb (78), were used as positive controls, and 1D04, an anti-CHIKV E1 virus mAb, was used as a negative control.
ADCC bioreporter assay
ADCC activity of the mAbs was tested by performing in vitro ADCC bioreporter assays as per the manufacturer’s protocol (Promega). In short, 2 × 105 MDCK cells/well (100 µL/well) were cultured over night at 37°C under 5% CO2 in white, flat-bottom, 96-well cell culture plates (Corning). The following day, the cells were washed with 200-µL PBS and infected with a virus inoculum at an MOI of 5 (equivalent to 200,000 PFU, diluted in 1× MEM: 100 µL/well). Plates were incubated for 16 h at 37°C with 5% CO2 for IAVs or at 33°C with 5% CO2 for IBVs. Next, mAbs were diluted to 30 µg/mL and twofold or threefold serially diluted in assay buffer consisting of Roswell Park Memorial Institute Medium 1640 (Gibco). An appropriate amount of human ADCC bioassay effector cells (Jurkat cells engineered to express FcγRIIIa, high affinity version H131, Promega) were then thawed in a 37°C water bath. Media were aspirated from the cell plates; plates were once washed with 100 µL/well PBS; and afterwards, 25 µL/well of assay buffer, 25 µL/well of serially diluted mAbs, and 25 µL/well of ADCC effector cells in a final concentration of 7.5 × 104 cells/25 µL were added. For the blank wells, 50 µL/well of assay buffer and 25 µL/well of bioeffector cells were added. Plates were incubated for 6 h at 37°C with 5% CO2 in a humidified incubator, followed by temperature equilibration for 15 min at RT and addition of 75 µL/well Bio-Glo Luciferase assay reagent (Promega, #G7940, G7941) for 10 min in the dark at RT. After incubation, the luciferase-induced luminescence was assessed using a Synergy H1 microplate reader (BioTek). Data were analyzed in Microsoft Excel and GraphPad Prism 9. Data are presented as the AUC calculated with a cutoff value of the average of the OD values of the background plus five SDs.
ADCP bioreporter assay
A commercial ADCP reporter assay kit (Promega) was used to investigate the ADCP activity of the mAbs. The protocol is similar to the ADCC bioreporter assay described above with the only difference being that human bioeffector FcγRIIa cells were used instead of the human FcγRIIIa cells.
Prophylactic passive transfer studies in mice
Animal experiments were performed following the Icahn School of Medicine Institutional Animal Care and Use Committee guidelines. Animals were given ad libitum access to food and water and were maintained in a 12-h light-dark cycle. Mouse passive transfer experiments to test the prophylactic efficacy of the mAbs were conducted as described earlier (3, 78). In brief, naïve six-week-old female BALB/c (for H1N1 studies) or DBA2/j (for H3N2 studies) mice (Jackson Laboratories) were intraperitoneally injected with 10 mg/kg of mAb (100 µL; 1F11, 1H01, 1B06, 1D11, 1F03, and 1H06; n = 5 mice/mAb tested). Positive control mice received 1G01, a pan NA mAb which has been documented to protect against lethal challenges with both influenza A and B viruses (3), and negative control mice received 1C11, an irrelevant human anti-SARS-CoV-2 mAb, or 8H9, an irrelevant mouse anti-H6 mAb, at an equivalent dose and volume. Three hours post mAb transfer, mice were bled to verify successful mAb transfer by ELISA as described above and were afterwards intranasally challenged with 1,600 PFU/50 µL of A/Netherlands/602/2009 (H1N1) (or 1,778.3 PFU of retitered virus/50 µL in repeated studies) or 1,600 PFU/50 µL mouse-adapted A/Switzerland/9715293/2013 (H3N2) virus strains under anesthesia (0.15 mg/kg/ketamine and 0.03 mg/kg/xylazine per mouse). Weight loss and survival were monitored daily for a period of 12–14 days, and data were analyzed in Microsoft Excel and GraphPad Prism 9. Mice that lost 25% or more of their day 0 body weight were euthanized, according to institutional guidelines.
Statistics and reproducibility
For line graphs, data are expressed as means. For bar graphs, data are expressed as individual values, and the average, presented as the mean. Error bars represent SD or SEM. Significance between antibody responses or weight loss data was analyzed by one-way analysis of variance (ANOVA) followed by a multiple-comparison test. Groups were compared to the irrelevant antibody control group. For the antibody response ADCC and ADCP data, the one-way ANOVA analysis was performed on log(y)-transformed AUC values. For the weight loss comparisons, the maximum weight loss in percentage for each mouse was used. Mice that reached the humane endpoint were assigned 75%. Survival curve comparisons were analyzed using a log-rank (Mantel-Cox) test. Data were considered statistically significant at P < 0.05. All statistical analyses were performed using GraphPad Prism 9.
ACKNOWLEDGMENTS
We thank George O’Dell for technical assistance.
This work was partially supported by the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C (F.K. and A.H.E.), NIAID Centers of Excellence for Influenza Research and Response (CEIRR) contract 75N93021C00014 (F.K. and A.H.E.), and NIAID CIVIC (75N93019C00051) (F.K. and A.H.E.) and NIAID R01 A146101. E.P.-M. was supported by a postdoctoral fellowship from Fundación Ramón Areces.
D.S., M.M., and F.K. conceived the experimental questions; W.F.R., D.S., E.P.-M., N.M.A.O., and M.M. conducted animal experiments and assays; A.E. provided antibodies; W.F.R. and D.S. analyzed data; W.F.R., F.K., and M.M. wrote the manuscript; all authors edited and reviewed the manuscript prior to submission.
Contributor Information
Florian Krammer, Email: florian.krammer@mssm.edu.
Mark T. Heise, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01646-22.
Fig. S1 to Fig. S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. McMahon M, Asthagiri Arunkumar G, Liu W-C, Stadlbauer D, Albrecht RA, Pavot V, Aramouni M, Lambe T, Gilbert SC, Krammer F. 2019. Vaccination with viral vectors expressing chimeric hemagglutinin, NP and M1 antigens protects ferrets against influenza virus challenge. Front Immunol 10:2005. doi: 10.3389/fimmu.2019.02005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krammer F. 2019. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol 19:383–397. doi: 10.1038/s41577-019-0143-6 [DOI] [PubMed] [Google Scholar]
- 3. Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, Strohmeier S, Yu W, Nachbagauer R, Mudd PA, Wilson IA, Ellebedy AH, Krammer F. 2019. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 366:499–504. doi: 10.1126/science.aay0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Vries RD, Nieuwkoop NJ, Pronk M, de Bruin E, Leroux-Roels G, Huijskens EGW, van Binnendijk RS, Krammer F, Koopmans MPG, Rimmelzwaan GF. 2017. Influenza virus-specific antibody dependent cellular cytoxicity induced by vaccination or natural infection. Vaccine 35:238–247. doi: 10.1016/j.vaccine.2016.11.082 [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization . 2021. Influenza (seasonal). Available from: https://www.who.int/en/news-room/fact-sheets/detail/influenza-(seasonal)
- 6. Gao R, Sheng Z, Sreenivasan CC, Wang D, Li F. 2020. Influenza A virus antibodies with antibody-dependent cellular cytotoxicity function. Viruses 12:276. doi: 10.3390/v12030276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Houser K, Subbarao K. 2015. Influenza vaccines: challenges and solutions. Cell Host Microbe 17:295–300. doi: 10.1016/j.chom.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carragher DM, Kaminski DA, Moquin A, Hartson L, Randall TD. 2008. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J Immunol 181:4168–4176. doi: 10.4049/jimmunol.181.6.4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Asthagiri Arunkumar G, McMahon M, Pavot V, Aramouni M, Ioannou A, Lambe T, Gilbert S, Krammer F. 2019. Vaccination with viral vectors expressing NP, M1 and chimeric hemagglutinin induces broad protection against influenza virus challenge in mice. Vaccine 37:5567–5577. doi: 10.1016/j.vaccine.2019.07.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okuya K, Yoshida R, Manzoor R, Saito S, Suzuki T, Sasaki M, Saito T, Kida Y, Mori-Kajihara A, Kondoh T, Sato M, Kajihara M, Miyamoto H, Ichii O, Higashi H, Takada A. 2020. Potential role of nonneutralizing IgA antibodies in cross-protective immunity against influenza A viruses of multiple hemagglutinin subtypes. J Virol 94:e00408-20. doi: 10.1128/JVI.00408-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang F, Xiao Y, Lu R, Chen B, Liu F, Wang L, Yao H, Wu N, Wu H. 2020. Generation of neutralizing and non-neutralizing monoclonal antibodies against H7N9 influenza virus. Emerg Microbes Infect 9:664–675. doi: 10.1080/22221751.2020.1742076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaiser J. 2006. A one-size-fits-all flu vaccine?. Science 312:380–382. doi: 10.1126/science.312.5772.380 [DOI] [PubMed] [Google Scholar]
- 13. Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng N-Y, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, Wilson PC. 2021. First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes. Sci Transl Med 13:eabg4535. doi: 10.1126/scitranslmed.abg4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christensen SR, Toulmin SA, Griesman T, Lamerato LE, Petrie JG, Martin ET, Monto AS, Hensley SE, Heise MT. 2019. Assessing the protective potential of H1N1 influenza virus hemagglutinin head and stalk antibodies in humans. J Virol 93:e02134–18. doi: 10.1128/JVI.02134-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meseda CA, Atukorale V, Soto J, Eichelberger MC, Gao J, Wang W, Weiss CD, Weir JP. 2018. Immunogenicity and protection against influenza H7N3 in mice by modified vaccinia virus ankara vectors expressing influenza virus hemagglutinin or neuraminidase. Sci Rep 8:5364. doi: 10.1038/s41598-018-23712-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bangaru S, Zhang H, Gilchuk IM, Voss TG, Irving RP, Gilchuk P, Matta P, Zhu X, Lang S, Nieusma T, Richt JA, Albrecht RA, Vanderven HA, Bombardi R, Kent SJ, Ward AB, Wilson IA, Crowe JE. 2018. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat Commun 9:2669. doi: 10.1038/s41467-018-04704-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benjamin E, Wang W, McAuliffe JM, Palmer-Hill FJ, Kallewaard NL, Chen Z, Suzich JA, Blair WS, Jin H, Zhu Q. 2014. A broadly neutralizing human monoclonal antibody directed against a novel conserved epitope on the influenza virus H3 Hemagglutinin globular head. J Virol 88:6743–6750. doi: 10.1128/JVI.03562-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JPM, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza a hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669 [DOI] [PubMed] [Google Scholar]
- 19. Wrammert J, Koutsonanos D, Li G-M, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208:181–193. doi: 10.1084/jem.20101352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, Palese P. 2010. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog 6:e1000796. doi: 10.1371/journal.ppat.1000796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen L, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273. doi: 10.1038/nsmb.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A, Palese P. 2009. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog 5:e1000350. doi: 10.1371/journal.ppat.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gerhard W, Mozdzanowska K, Furchner M, Washko G, Maiese K. 1997. Role of the B‐cell response in recovery of mice from primary influenza virus infection. Immunol Rev 159:95–103. doi: 10.1111/j.1600-065x.1997.tb01009.x [DOI] [PubMed] [Google Scholar]
- 24. Palladino G, Mozdzanowska K, Washko G, Gerhard W. 1995. Virus-neutralizing antibodies of immunoglobulin G (IgG) but not of IgM or IgA isotypes can cure influenza virus pneumonia in SCID mice. J Virol 69:2075–2081. doi: 10.1128/JVI.69.4.2075-2081.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krammer F, Palese P. 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14:167–182. doi: 10.1038/nrd4529 [DOI] [PubMed] [Google Scholar]
- 26. Nachbagauer R, Palese P. 2018. Development of next generation hemagglutinin-based broadly protective influenza virus vaccines. Curr Opin Immunol 53:51–57. doi: 10.1016/j.coi.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 27. Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng N-Y, Mullarkey CE, Ennis FA, Terajima M, Treanor JJ, Topham DJ, Subbarao K, Palese P, Krammer F, Wilson PC. 2016. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19:800–813. doi: 10.1016/j.chom.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jegaskanda S, Reading PC, Kent SJ. 2014. Influenza-specific antibody-dependent cellular cytotoxicity: toward a universal influenza vaccine. J Immunol 193:469–475. doi: 10.4049/jimmunol.1400432 [DOI] [PubMed] [Google Scholar]
- 29. Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. 2014. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5:1–9. doi: 10.1038/ncomms4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hong M, Lee PS, Hoffman RMB, Zhu X, Krause JC, Laursen NS, Yoon S, Song L, Tussey L, Crowe JE, Ward AB, Wilson IA. 2013. Antibody recognition of the pandemic H1N1 influenza virus hemagglutinin receptor binding site. J Virol 87:12471–12480. doi: 10.1128/JVI.01388-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, Settembre EC, Dormitzer PR, Kepler TB, Zhang R, Moody MA, Haynes BF, Liao HX, Shaw DE, Harrison SC. 2013. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A 110:264–269. doi: 10.1073/pnas.1218256109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu R, Krause JC, McBride R, Paulson JC, Crowe JE, Wilson IA. 2013. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20:363–370. doi: 10.1038/nsmb.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532. doi: 10.1038/nature11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE. 2011. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol 85:10905–10908. doi: 10.1128/JVI.00700-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. 2011. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol 85:11048–11057. doi: 10.1128/JVI.05397-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao H-X, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108:14216–14221. doi: 10.1073/pnas.1111497108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, Harrison SC. 2018. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci U S A 115:168–173. doi: 10.1073/pnas.1715471115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y, Hu D, Wu Y, Ying T. 2020. Recent advances in “universal” influenza virus antibodies: the rise of a hidden trimeric interface in hemagglutinin globular head. Front Med 14:149–159. doi: 10.1007/s11684-020-0764-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, Gilchuk I, Turner HL, García-Sastre A, Li S, Ward AB, Wilson IA, Crowe JE. 2019. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177:1136–1152. doi: 10.1016/j.cell.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, McGee CE, Sempowski GD, Feng F, Urick P, Kepler TB, Takahashi Y, Harrison SC, Kelsoe G. 2019. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 177:1124–1135. doi: 10.1016/j.cell.2019.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sedova ES, Scherbinin DN, Lysenko AA, Alekseeva SV, Artemova EA, Shmarov MM. 2019. Non-neutralizing antibodies directed at conservative influenza antigens. Acta Naturae 11:22–32. doi: 10.32607/20758251-2019-11-4-22-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krammer F, Jul-Larsen A, Margine I, Hirsh A, Sjursen H, Zambon M, Cox RJ. 2014. An H7N1 influenza virus vaccine induces broadly reactive antibody responses against H7N9 in humans. Clin Vaccine Immunol 21:1153–1163. doi: 10.1128/CVI.00272-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang Y, Banner D, Kelvin AA, Huang SSH, Paige CJ, Corfe SA, Kane KP, Bleackley RC, Rowe T, Leon AJ, Kelvin DJ. 2012. Seasonal H1N1 influenza virus infection induces cross-protective pandemic H1N1 virus immunity through a CD8-independent, B cell-dependent mechanism. J Virol 86:2229–2238. doi: 10.1128/JVI.05540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. LaMere MW, Lam HT, Moquin A, Haynes L, Lund FE, Randall TD, Kaminski DA. 2011. Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. J Immunol 186:4331–4339. doi: 10.4049/jimmunol.1003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lamere MW, Moquin A, Lee FE-H, Misra RS, Blair PJ, Haynes L, Randall TD, Lund FE, Kaminski DA. 2011. Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance. J Virol 85:5027–5035. doi: 10.1128/JVI.00150-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rangel-Moreno J, Carragher DM, Misra RS, Kusser K, Hartson L, Moquin A, Lund FE, Randall TD. 2008. B cells promote resistance to heterosubtypic strains of influenza via multiple mechanisms. J Immunol 180:454–463. doi: 10.4049/jimmunol.180.1.454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sicca F, Neppelenbroek S, Huckriede A. 2018. Effector mechanisms of influenza-specific antibodies: neutralization and beyond. Expert Rev Vaccines 17:785–795. doi: 10.1080/14760584.2018.1516553 [DOI] [PubMed] [Google Scholar]
- 49. Jegaskanda S, Co MDT, Cruz J, Subbarao K, Ennis FA, Terajima M. 2017. Induction of H7N9-cross-reactive antibody-dependent cellular cytotoxicity antibodies by human seasonal influenza A viruses that are directed toward the nucleoprotein. J Infect Dis 215:818–823. doi: 10.1093/infdis/jiw629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Terajima M, Co MDT, Cruz J, Ennis FA. 2015. High antibody-dependent cellular cytotoxicity antibody titers to H5N1 and H7N9 avian influenza A viruses in healthy US adults and older children. J Infect Dis 212:1052–1060. doi: 10.1093/infdis/jiv181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simhadri VR, Dimitrova M, Mariano JL, Zenarruzabeitia O, Zhong W, Ozawa T, Muraguchi A, Kishi H, Eichelberger MC, Borrego F. 2015. A human anti-M2 antibody mediates antibody-dependent cell-mediated cytotoxicity (ADCC) and cytokine secretion by resting and cytokine-preactivated natural killer (NK) cells. PLoS One 10:e0124677. doi: 10.1371/journal.pone.0124677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jegaskanda S, Laurie KL, Amarasena TH, Winnall WR, Kramski M, De Rose R, Barr IG, Brooks AG, Reading PC, Kent SJ. 2013. Age-associated cross-reactive antibody-dependent cellular cytotoxicity toward 2009 pandemic influenza A virus subtype H1N1. J Infect Dis 208:1051–1061. doi: 10.1093/infdis/jit294 [DOI] [PubMed] [Google Scholar]
- 53. Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ. 2013. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol 87:5512–5522. doi: 10.1128/JVI.03030-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hashimoto G, Wright PF, Karzon DT. 1983. Antibody-dependent cell-mediated cytotoxicity against influenza virus-infected cells. J Infect Dis 148:785–794. doi: 10.1093/infdis/148.5.785 [DOI] [PubMed] [Google Scholar]
- 55. Kim Y-J, Kim K-H, Ko E-J, Kim M-C, Lee Y-N, Jung Y-J, Lee Y-T, Kwon Y-M, Song J-M, Kang S-M, Jung JU. 2018. Complement C3 plays a key role in inducing humoral and cellular immune responses to influenza virus strain-specific hemagglutinin-based or cross-protective M2 extracellular domain-based vaccination. J Virol 92:e00969-18. doi: 10.1128/JVI.00969-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang R, Song A, Levin J, Dennis D, Zhang NJ, Yoshida H, Koriazova L, Madura L, Shapiro L, Matsumoto A, Yoshida H, Mikayama T, Kubo RT, Sarawar S, Cheroutre H, Kato S. 2008. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Res 80:168–177. doi: 10.1016/j.antiviral.2008.06.002 [DOI] [PubMed] [Google Scholar]
- 57. Yewdell JW, Frank E, Gerhard W. 1981. Expression of influenza A virus internal antigens on the surface of infected P815 cells. J Immunol 126:1814–1819. doi: 10.4049/jimmunol.126.5.1814 [DOI] [PubMed] [Google Scholar]
- 58. Tay MZ, Wiehe K, Pollara J. 2019. Antibody-dependent cellular phagocytosis in antiviral immune responses. Front Immunol 10:332. doi: 10.3389/fimmu.2019.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. El Bakkouri K, Descamps F, De Filette M, Smet A, Festjens E, Birkett A, Van Rooijen N, Verbeek S, Fiers W, Saelens X. 2011. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: fc receptors and alveolar macrophages mediate protection. J Immunol 186:1022–1031. doi: 10.4049/jimmunol.0902147 [DOI] [PubMed] [Google Scholar]
- 60. Huber VC, Lynch JM, Bucher DJ, Le J, Metzger DW. 2001. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J Immunol 166:7381–7388. doi: 10.4049/jimmunol.166.12.7381 [DOI] [PubMed] [Google Scholar]
- 61. Hu Z, Zhao J, Zhao Y, Fan X, Hu J, Shi L, Wang X, Liu X, Hu S, Gu M, Cao Y, Liu X. 2019. Hemagglutinin-specific non-neutralizing antibody is essential for protection provided by Inactivated and viral-vectored H7N9 avian influenza vaccines in chickens. Front Vet Sci 6:482. doi: 10.3389/fvets.2019.00482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Asthagiri Arunkumar G, Ioannou A, Wohlbold TJ, Meade P, Aslam S, Amanat F, Ayllon J, García-Sastre A, Krammer F. 2019. Broadly cross-reactive, nonneutralizing antibodies against influenza B virus hemagglutinin demonstrate effector function-dependent protection against lethal viral challenge in mice. J Virol 93:93–18. doi: 10.1128/JVI.01696-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Amanat F, Meade P, Strohmeier S, Krammer F. 2019. Cross-reactive antibodies binding to H4 hemagglutinin protect against a lethal H4N6 influenza virus challenge in the mouse model. Emerg Microbes Infect 8:155–168. doi: 10.1080/22221751.2018.1564369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. DiLillo DJ, Palese P, Wilson PC, Ravetch JV. 2016. Broadly neutralizing anti-influenza antibodies require FC receptor engagement for in vivo protection. J Clin Invest 126:605–610. doi: 10.1172/JCI84428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Krammer F, Palese P. 2013. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3:521–530. doi: 10.1016/j.coviro.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gocník M, Fislová T, Mucha V, Sládková T, Russ G, Kostolanský F, Varečková E. 2008. Antibodies induced by the HA2 glycopolypeptide of influenza virus haemagglutinin improve recovery from influenza A virus infection. J Gen Virol 89:958–967. doi: 10.1099/vir.0.83524-0 [DOI] [PubMed] [Google Scholar]
- 67. Eichelberger MC, Monto AS. 2019. Neuraminidase, the forgotten surface antigen, emerges as an influenza vaccine target for broadened protection. J Infect Dis 219:S75–S80. doi: 10.1093/infdis/jiz017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, Monto AS, Garsin DA. 2018. Naction! how can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 9:e02332-17. doi: 10.1128/mBio.02332-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, Subramaniam S, Palese P, Krammer F. 2017. Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat Microbiol 2:1415–1424. doi: 10.1038/s41564-017-0011-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wohlbold T.J, Krammer F. 2014. In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses 6:2465–2494. doi: 10.3390/v6062465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mozdzanowska K, Maiese K, Furchner M, Gerhard W. 1999. Treatment of influenza virus-infected SCID mice with nonneutralizing antibodies specific for the transmembrane proteins matrix 2 and neuraminidase reduces the pulmonary virus titer but fails to clear the infection. Virology 254:138–146. doi: 10.1006/viro.1998.9534 [DOI] [PubMed] [Google Scholar]
- 72. Zheng M, Luo J, Chen Z. 2014. Development of universal influenza vaccines based on influenza virus M and NP genes. Infection 42:251–262. doi: 10.1007/s15010-013-0546-4 [DOI] [PubMed] [Google Scholar]
- 73. Song J-M, Van Rooijen N, Bozja J, Compans RW, Kang S-M. 2011. Vaccination inducing broad and improved cross protection against multiple subtypes of influenza A virus. Proc Natl Acad Sci U S A 108:757–761. doi: 10.1073/pnas.1012199108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schotsaert M, De Filette M, Fiers W, Saelens X. 2009. Universal M2 ectodomain-based influenza A vaccines: preclinical and clinical developments. Expert Rev Vaccines 8:499–508. doi: 10.1586/erv.09.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453:667–671. doi: 10.1038/nature06890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Padilla-Quirarte HO, Lopez-Guerrero DV, Gutierrez-Xicotencatl L, Esquivel-Guadarrama F. 2019. Protective antibodies against influenza proteins. Front Immunol 10:1677. doi: 10.3389/fimmu.2019.01677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bodewes R, Geelhoed-Mieras MM, Wrammert J, Ahmed R, Wilson PC, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2013. In vitro assessment of the immunological significance of a human monoclonal antibody directed to the influenza a virus nucleoprotein. Clin Vaccine Immunol 20:1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Madsen A, Dai Y-N, McMahon M, Schmitz AJ, Turner JS, Tan J, Lei T, Alsoussi WB, Strohmeier S, Amor M, Mohammed BM, Mudd PA, Simon V, Cox RJ, Fremont DH, Krammer F, Ellebedy AH. 2020. Human antibodies targeting influenza B virus neuraminidase active site are broadly protective. Immunity 53:852–863. doi: 10.1016/j.immuni.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Boudreau CM, Alter G. 2019. Extra-neutralizing FcR-mediated antibody functions for a universal influenza vaccine. Front Immunol 10:440. doi: 10.3389/fimmu.2019.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Virelizier JL, Allison AC, Oxford JS, Schild GC. 1977. Early presence of ribonucleoprotein antigen on surface of influenza virus-infected cells. Nature 266:52–54. doi: 10.1038/266052a0 [DOI] [PubMed] [Google Scholar]
- 81. He W, Chen C-J, Mullarkey CE, Hamilton JR, Wong CK, Leon PE, Uccellini MB, Chromikova V, Henry C, Hoffman KW, Lim JK, Wilson PC, Miller MS, Krammer F, Palese P, Tan GS. 2017. Alveolar macrophages are critical for broadly-reactive antibody-mediated protection against influenza A virus in mice. Nat Commun 8:1–14. doi: 10.1038/s41467-017-00928-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hobson D, Curry RL, Beare AS, Ward-Gardner A. 1972. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond) 70:767–777. doi: 10.1017/s0022172400022610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ellebedy AH, Ahmed R. 2012. Re-engaging cross-reactive memory B cells: the influenza puzzle. Front Immunol 3:53. doi: 10.3389/fimmu.2012.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wilson PC, Andrews SF. 2012. Tools to therapeutically harness the human antibody response. Nat Rev Immunol 12:709–719. doi: 10.1038/nri3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tan GS, Leon PE, Albrecht RA, Margine I, Hirsh A, Bahl J, Krammer F. 2016. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog. 12:e1005578. doi: 10.1371/journal.ppat.1005578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xiaorong P, Haibo W, Xiuming P, Nanping W. 2015. Molecular phylogeny and evolutionary dynamics of matrix gene of avian influenza viruses in China. Infect Genet Evol 34:344–351. doi: 10.1016/j.meegid.2015.05.033 [DOI] [PubMed] [Google Scholar]
- 87. Furuse Y, Suzuki A, Kamigaki T, Oshitani H. 2009. Evolution of the M gene of the influenza A virus in different host species: large-scale sequence analysis. Virol J 6:1–13. doi: 10.1186/1743-422X-6-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Manzoor R, Sakoda Y, Mweene A, Tsuda Y, Kishida N, Bai G-R, Kameyama K-I, Isoda N, Soda K, Naito M, Kida H. 2008. Phylogenic analysis of the M genes of influenza viruses isolated from free-flying water birds from their northern territory to Hokkaido, Japan. Virus Genes 37:144–152. doi: 10.1007/s11262-008-0248-7 [DOI] [PubMed] [Google Scholar]
- 89. Ito T, Gorman OT, Kawaoka Y, Bean WJ, Webster RG. 1991. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol 65:5491–5498. doi: 10.1128/JVI.65.10.5491-5498.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yewdell JW, Bennink JR, Smith GL, Moss B. 1985. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc Natl Acad Sci U S A 82:1785–1789. doi: 10.1073/pnas.82.6.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Londo DR, Davis AR, Nayak DP. 1983. Complete nucleotide sequence of the nucleoprotein gene of influenza B virus. J Virol 47:642–648. doi: 10.1128/JVI.47.3.642-648.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Briedis DJ, Lamb RA, Choppin PW. 1982. Sequence of RNA segment 7 of the influenza B virus genome: partial amino acid homology between the membrane proteins (M1) of influenza A and B viruses and conservation of a second open reading frame. Virology 116:581–588. doi: 10.1016/0042-6822(82)90150-7 [DOI] [PubMed] [Google Scholar]
- 93. Terajima M, Cruz J, Co MDT, Lee J-H, Kaur K, Wrammert J, Wilson PC, Ennis FA. 2011. Complement-dependent lysis of influenza a virus-infected cells by broadly cross-reactive human monoclonal antibodies. J Virol 85:13463–13467. doi: 10.1128/JVI.05193-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Frank K, Paust S. 2020. Dynamic natural killer cell and T cell responses to influenza infection. Front Cell Infect Microbiol 10:425. doi: 10.3389/fcimb.2020.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dugan HL, Guthmiller JJ, Arevalo P, Huang M, Chen Y-Q, Neu KE, Henry C, Zheng N-Y, Lan LY-L, Tepora ME, Stovicek O, Bitar D, Palm A-KE, Stamper CT, Changrob S, Utset HA, Coughlan L, Krammer F, Cobey S, Wilson PC. 2020. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci Transl Med 12:573. doi: 10.1126/scitranslmed.abd3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sun W, Luo T, Liu W, Li J. 2020. Progress in the development of universal influenza vaccines. Viruses 12:1033. doi: 10.3390/v12091033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jang YH, Seong BL. 2019. The quest for a truly universal influenza vaccine. Front Cell Infect Microbiol 9:344. doi: 10.3389/fcimb.2019.00344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sebastian S, Lambe T. 2018. Clinical advances in viral-vectored influenza vaccines vaccines 6:29. Vaccines (Basel) 6:29. doi: 10.3390/vaccines6020029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. McMichael AJ, Gotch FM, Noble GR, Beare PA. 1983. Cytotoxic T-cell immunity to influenza. N Engl J Med 309:13–17. doi: 10.1056/NEJM198307073090103 [DOI] [PubMed] [Google Scholar]
- 100. Kreijtz J, Bodewes R, van Amerongen G, Kuiken T, Fouchier RAM, Osterhaus A, Rimmelzwaan GF. 2007. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25:612–620. doi: 10.1016/j.vaccine.2006.08.036 [DOI] [PubMed] [Google Scholar]
- 101. Ulmer JB, Fu TM, Deck RR, Friedman A, Guan L, DeWitt C, Liu X, Wang S, Liu MA, Donnelly JJ, Caulfield MJ. 1998. Protective CD4 and CD8 T cells against influenza virus induced by vaccination with nucleoprotein DNA. J Virol 72:5648–5653. doi: 10.1128/JVI.72.7.5648-5653.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schulman JL, Kilbourne ED. 1965. Induction of partial specific heterotypic immunity in mice by a single infection with influenza a virus. J Bacteriol 89:170–174. doi: 10.1128/jb.89.1.170-174.1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Clemens E, van de Sandt C, Wong S, Wakim L, Valkenburg S. 2018. Harnessing the power of T cells: the promising hope for a universal influenza vaccine. Vaccines 6:18. doi: 10.3390/vaccines6020018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. McElhaney JE, Kuchel GA, Zhou X, Swain SL, Haynes L. 2016. T-cell immunity to influenza in older adults: a pathophysiological framework for development of more effective vaccines. Front Immunol 7:41. doi: 10.3389/fimmu.2016.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hayward AC, Wang L, Goonetilleke N, Fragaszy EB, Bermingham A, Copas A, Dukes O, Millett ERC, Nazareth I, Nguyen-Van-Tam JS, Watson JM, Zambon M, Johnson AM, McMichael AJ. 2015. Natural T cell–mediated protection against seasonal and pandemic influenza. Results of the flu watch cohort study. Am J Respir Crit Care Med 191:1422–1431. doi: 10.1164/rccm.201411-1988OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wilkinson TM, Li CKF, Chui CSC, Huang AKY, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu X-N. 2012. Preexisting influenza-specific CD4 T cells correlate with disease protection against influenza challenge in humans. Nat Med 18:274–280. doi: 10.1038/nm.2612 [DOI] [PubMed] [Google Scholar]
- 107. Bernstein DI, Guptill J, Naficy A, Nachbagauer R, Berlanda-Scorza F, Feser J, Wilson PC, Solórzano A, Van der Wielen M, Walter EB, et al. 2020. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect Dis 20:80–91. doi: 10.1016/S1473-3099(19)30393-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Broecker F, Liu STH, Suntronwong N, Sun W, Bailey MJ, Nachbagauer R, Krammer F, Palese P. 2019. A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 4:31. doi: 10.1038/s41541-019-0126-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Krammer F, Palese P. 2019. Universal influenza virus vaccines that target the conserved Hemagglutinin stalk and conserved sites in the head domain. J Infect Dis 219:S62–S67. doi: 10.1093/infdis/jiy711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Liu W-C, Nachbagauer R, Stadlbauer D, Solórzano A, Berlanda-Scorza F, García-Sastre A, Palese P, Krammer F, Albrecht RA. 2019. Sequential immunization with live-attenuated chimeric hemagglutinin-based vaccines confers heterosubtypic immunity against influenza A viruses in a preclinical ferret model. Front Immunol 10:756. doi: 10.3389/fimmu.2019.00756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Antrobus RD, Coughlan L, Berthoud TK, Dicks MD, Hill AV, Lambe T, Gilbert SC. 2014. Clinical assessment of a novel recombinant simian adenovirus ChAdOx1 as a vectored vaccine expressing conserved influenza A antigens. Mol Ther 22:668–674. doi: 10.1038/mt.2013.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Laidlaw BJ, Decman V, Ali M-AA, Abt MC, Wolf AI, Monticelli LA, Mozdzanowska K, Angelosanto JM, Artis D, Erikson J, Wherry EJ. 2013. Cooperativity between CD8 T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog 9:e1003207. doi: 10.1371/journal.ppat.1003207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. McKinstry KK, Strutt TM, Kuang Y, Brown DM, Sell S, Dutton RW, Swain SL. 2012. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J Clin Invest 122:2847–2856. doi: 10.1172/JCI63689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Turner JS, Lei T, Schmitz AJ, Day A, Choreño-Parra JA, Jiménez-Alvarez L, Cruz-Lagunas A, House SL, Zúñiga J, Ellebedy AH, Mudd PA. 2020. Impaired cellular immune responses during the first week of severe acute influenza infection. J Infect Dis 222:1235–1244. doi: 10.1093/infdis/jiaa226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kirkpatrick Roubidoux E, McMahon M, Carreño JM, Capuano C, Jiang K, Simon V, van Bakel H, Wilson P, Krammer F. 2021. Identification and characterization of novel antibody epitopes on the N2 neuraminidase. mSphere 6:e00958-20. doi: 10.1128/mSphere.00958-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Margine I, Palese P, Krammer F. 2013. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J Vis Exp:e51112. doi: 10.3791/51112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P. 2012. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS One 7:e43603. doi: 10.1371/journal.pone.0043603 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to Fig. S4.