ABSTRACT
Nanobodies are single-domain antibodies derived from camelid animals. Here, we discovered three anti-SARS-CoV-2 nanobodies, namely, Nanosota-2, -3, and -4, from an alpaca immunized with SARS-CoV spike protein. We further characterized the antiviral activities of these Fc-tag-fused nanobodies. Notably, Nanosota-2 inhibits the prototypic SARS-CoV-2 strain in vitro (with an IC50 of 2 pM) and in mice (at a dosage of 4 mg/kg or administered 18 hours post-challenge). These potency metrics are the best among known SARS-CoV-2 entry inhibitors. Moreover, Nanosota-3 effectively inhibits the omicron variant, both in vitro and in mice, regardless of the administration route (intraperitoneal or intranasal). Furthermore, Nanosota-3 has been biochemically engineered to inhibit both early and currently circulating subvariants of omicron. Additionally, Nanosota-4 uniquely inhibits both SARS-CoV-1 and SARS-CoV-2. Cryo-EM data revealed that the three nanobodies bind to functionally critical and non-overlapping regions in the spike protein. Given their cost-effectiveness, ease of adaptation to new viral strains, and potential use as inhalers, the Nanosota series are powerful therapeutic tools against coronavirus pandemics.
IMPORTANCE
The COVID-19 pandemic exposed limitations of conventional antibodies as therapeutics, including high cost, limited potency, ineffectiveness against new viral variants, and primary reliance on injection-only delivery. Nanobodies are single-domain antibodies with therapeutic potentials. We discovered three anti-SARS-CoV-2 nanobodies, named Nanosota-2, -3, and -4, from an immunized alpaca. Nanosota-2 is super potent against prototypic SARS-CoV-2, Nanosota-3 is highly potent against the omicron variant, and Nanosota-4 is effective against both SARS-CoV-1 and SARS-CoV-2. In addition to their super potency and combined broad antiviral spectrum, these nanobodies are cost-effective, can be easily adapted to new viral variants through phage display, and can potentially be administered as inhalers. The Nanosota series are powerful therapeutic candidates to combat circulating SARS-CoV-2 and prepare for possible future coronavirus pandemics.
KEYWORDS: SARS-CoV-2, omicron, SARS-CoV-1, single-domain antibody from camelids, spike protein, receptor-binding domain, ACE2, virus neutralization, animal model, cryo-EM
INTRODUCTION
The COVID-19 pandemic was seriously complicated by the emergence of SARS-CoV-2 variants and the occurrence of breakthrough infections in vaccinated individuals (1, 2). While the pandemic has recently subsided, there is still a pressing need for potent, broad-spectrum, cost-effective, and easily administered therapeutics to combat the persistent circulation of SARS-CoV-2 and prepare for potential future coronavirus outbreaks. The virus-surface spike protein played a central role in the pandemic by mediating viral entry into host cells, triggering most of the host immune responses, and representing a prime target for antiviral interventions (3, 4). The receptor-binding domain (RBD) located at the tip of the spike protein is responsible for binding the host receptor ACE2 and is heavily targeted by the host’s immune defenses (5 – 7). The RBD can adopt either a standing-up conformation for receptor binding or a lying-down conformation to evade immune responses (8 – 10). The spike protein, particularly the RBD, undergoes extensive mutations in SARS-CoV-2 variants (1, 11). The immune evasiveness and rapid evolution of the RBD pose challenges in the development of potent and broad-spectrum entry inhibitors.
Conventional antibodies have traditionally been the primary therapeutic approach for targeting coronavirus entry (Table S1; Table S2). However, the limitations of these antibodies were exposed during the pandemic. First, conventional antibodies are expensive due to the high costs associated with production, storage, and transportation (12, 13). Second, their potency is typically limited, as they are effective when administered prior to or shortly after SARS-CoV-2 infection but not against actively replicating virus (Table S2) (14, 15). Third, they have limited antiviral spectrums, as viral mutations almost inevitably render them ineffective (9, 10). Finally, their administration is limited to injections, which poses practical challenges (16 – 18). Therefore, conventional antibodies are not feasible solutions for effectively combating a global pandemic like COVID-19.
Nanobodies are a type of single-domain antibodies derived from heavy chain-only antibodies found in camelid animals, such as llamas and alpacas (19, 20). Due to their single-domain structure, nanobodies have many advantages over conventional antibodies as antiviral therapeutics. Their small size enables them to access cryptic sites on viral targets, exhibit excellent tissue permeability, and possess ease of production and robust physical and chemical stabilities. They also have the ability to be rapidly adapted to viral variants using phage display techniques and can potentially be administered via inhalation (21 – 27). Moreover, nanobodies demonstrate low immunogenicity in humans (21, 22). A nanobody drug is clinically available to treat a blood-clotting disorder (28), validating the safety of nanobodies as human therapeutics. Several anti-SARS-CoV-2 nanobodies have been developed, demonstrating similar potency to conventional antibodies (Table S1). Among these was Nanosota-1, which we discovered through a naïve nanobody phage display library and subsequently optimized through in vitro affinity maturation using phage display (24). However, none of these nanobodies have exhibited extraordinary potency or broad-spectrum activities against SARS-CoV-2, both in vitro and in animal models (Table S1; Table S2).
In this study, we discovered three novel nanobodies, named Nanosota-2, -3, and -4, from an alpaca immunized with SARS-CoV-2 spike protein. We evaluated their anti-SARS-CoV-2 potency both in vitro and in animal models and also determined the structural basis for their anti-SARS-CoV-2 potency. Our study demonstrated that nanobodies can potentially achieve super potency and broad-spectrum activities against their viral targets and that the Nanosota series can serve as powerful therapeutics tools in battling the circulating SARS-CoV-2 and possibly future coronavirus pandemics.
RESULTS
Discovery of three novel nanobodies from an immunized alpaca
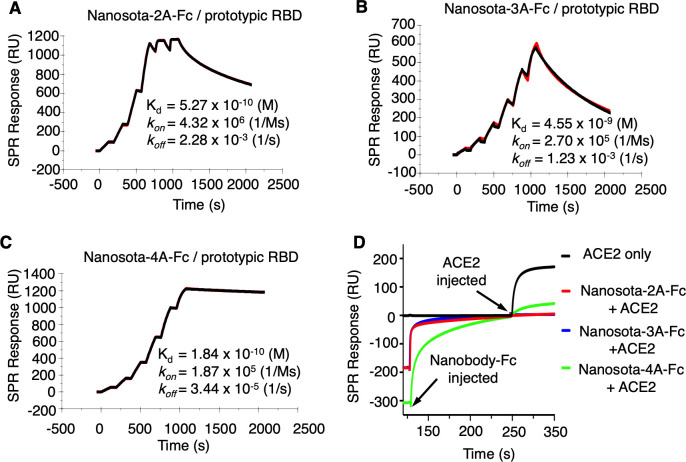
To discover potent anti-SARS-CoV-2 nanobodies, we immunized an alpaca with recombinant SARS-CoV-2 spike (prototypic strain), collected their peripheral blood mononuclear cells (PBMCs), and established an induced nanobody phage display library (Fig. 1). We screened this library using recombinant SARS-CoV-2 spike and identified three spike-binding nanobodies, named Nanosota-2A, Nanosota-3A, and Nanosota-4A. We then constructed human Fc-tagged dimeric nanobodies, named Nanosota-2A-Fc, Nanosota-3A-Fc, and Nanosota-4A-Fc, respectively. In our previous research, we demonstrated that the Fc-tagged nanobody, Nanosota-1C-Fc, could be produced in bacteria at high yields, exceeding 40 mg/L of cell culture without any optimization (24). In this study, we discovered that these three Fc-tagged nanobodies could be expressed in mammalian cells with comparable yields. We further assessed these mammalian cell-produced nanobodies in our current investigation. Using surface plasmon resonance (SPR), we determined the dissociation constants (K d) between each of the three Fc-tagged nanobodies and the prototypic SARS-CoV-2 RBD as follows: 5.27 × 10−10 M for Nanosota-2A-Fc, 4.55 × 10−9 M for Nanosota-3A-Fc, and 1.84 × 10−10 M for Nanosota-4A-Fc (Fig. 2). In comparison, recombinant human ACE2 binds to the same RBD with a K d of 4.4 × 10−8 M (6). Thus, these Fc-tagged nanobodies all have significantly higher affinity for the prototypic RBD than does ACE2.
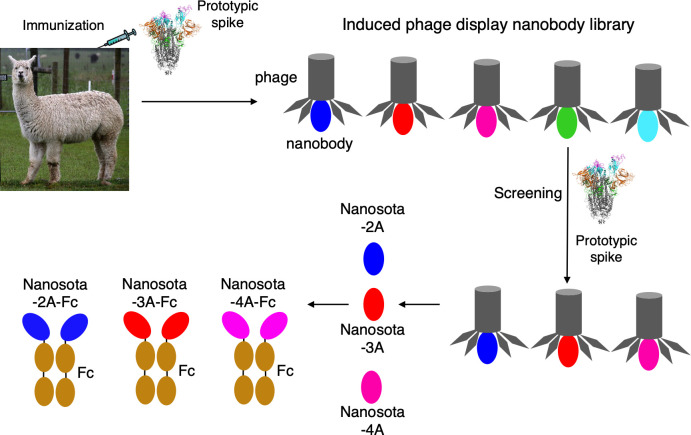
Fig 1.
Construction and screening of induced nanobody phage display library for anti-SARS-CoV-2 nanobodies. An alpaca was immunized with recombinant prototypic SARS-CoV-2 spike protein and the peripheral blood mononuclear cells were used to construct an induced nanobody phage display library. Panning against the library using the spike protein was performed to identify strong binders, which were then subjected to pseudovirus entry assay for selection of neutralizing nanobodies. Three neutralizing nanobodies (Nanosota-2A, Nanosota-3A, and Nanosota-4A) were discovered, and they all bind to the RBD. Fc-tagged nanobodies were constructed and prepared; their binding kinetics against prototypic SARS-CoV-2 RBD were determined using surface plasmon resonance (SPR). (Alpaca photo courtesy of Tony Hisgett; reprinted with permissionhttps://creativecommons.org/licenses/by/2.0/deed.en.)
Fig 2.
Binding kinetics between Fc-tagged nanobodies and prototypic SARS-CoV-2 RBD as measured by surface plasmon resonance (SPR). (A)–(C) Each of the Fc-tagged nanobodies (Nanosota-2A-Fc, Nanosota-3A-Fc, and Nanosota-4A-Fc) was immobilized to a protein A sensor chip. Prototypic SARS-CoV-2 RBD (His-tagged) was injected at different concentrations. The resulting data were analyzed using Biacore Evaluation Software. (D) Competition SPR analysis. His-tagged RBD was immobilized onto four CM5 sensor chips. Each of the Fc-tagged nanobodies was injected to one of the first three chips. In contrast, the fourth chip received only the running buffer. Following this, after the nanobodies had bound to the RBD, a mixture of recombinant human ACE2 and the corresponding nanobody was added to the first three chips. The fourth chip received only ACE2. Sensorgrams from all chips were then overlaid for comparison. No change in the resonance signal from nanobody-bound RBD indicated that ACE2 could not displace nanobody from binding to the RBD, as in the case of Nanosota-2A-Fc and Nanosota-3A-Fc. A slight increase in the resonance signal from nanobody-bound RBD suggested ACE2 largely could not displace nanobody from binding to the RBD, as in the case of Nanosota-4A-Fc.
The construction of the Fc-tagged nanobodies yielded several desirable properties. As we have recently demonstrated, single-domain nanobodies have a relatively short in vivo half-life of several hours, attributed to their molecular weight (~14 kDa) being below the kidney clearance threshold (~60 kDa). In contrast, Fc-tagged nanobodies exhibit an extended in vivo half-life of over 10 days due to their increased molecular weight (~78 kDa) (24). Despite this increase in size, Fc-tagged dimeric nanobodies only represent approximately half of the size of conventional antibodies and their single-domain antigen-binding site remains capable of accessing cryptic epitopes on targets (24).
Evaluation of anti-SARS-CoV-2 potency of the nanobodies in vitro
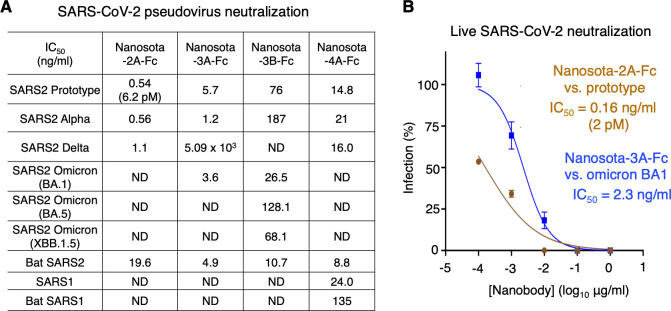
To evaluate the anti-SARS-CoV-2 potency of the nanobodies, we performed SARS-CoV-2 neutralization assays in vitro. Both SARS-CoV-2 pseudoviruses and live SARS-CoV-2 were used. For the pseudovirus assay, retroviruses pseudotyped with SARS-CoV-2 spike (i.e., SARS-CoV-2 pseudoviruses) were used to transduce human ACE2-expressing cells in the presence of the Fc-tagged nanobodies. To evaluate antiviral spectrums of the nanobodies, we included pseudoviruses corresponding to pre-omicron SARS-CoV-2 strains (prototype, alpha, and delta), omicron subvariants (early BA.1, later BA.5, and currently circulating XBB.1.5), and bat SARS2. Pseudoviruses corresponding to SARS-CoV-1 and bat SARS1 were also included, both of which are related to SARS-CoV-2 and use human ACE2 as their entry receptor (29 – 31). Our results showed that both Nanosota-2A-Fc and Nanosota-4A-Fc were potent against all SARS-CoV-2 strains, except for omicron; Nanosota-3A-Fc was potent against the prototype, alpha, omicron BA.1, and bat SARS2 but was weak against delta and ineffective against omicron BA.5 and XBB.1.5 (Fig. 3A). Interestingly, Nanosota-4A-Fc was also effective against SARS-CoV-1 and bat SARS1 (Fig. 3A). For the live SARS-CoV-2 assay, prototypic SARS-CoV-2 and omicron BA.1 infected Vero cells in the presence of Nanosota-2A-Fc and Nanosota-3A-Fc, respectively. Our results showed that Nanosota-2A-Fc and Nanosota-3A-Fc potently neutralized the prototypic strain and omicron BA.1, respectively (Fig. 3B). Of note, the IC50 values of Nanosota-2A-Fc against the prototypic SARS-CoV-2 pseudoviruses and live prototypic SARS-CoV-2 were 6.2 pM (0.5 ng/mL) and 2 pM (0.16 ng/mL), respectively, both of which are among the best of all known anti-SARS-CoV-2 antibodies and nanobodies (Table S1).
Fig 3.
Nanosota-2, -3, and -4 demonstrate super potency and broad-spectrum activities against SARS-CoV-2 in vitro. (A) Efficacy of Fc-tagged nanobodies in neutralizing SARS-CoV-2 and SARS-CoV-1 pseudoviruses. Retroviruses pseudotyped with coronavirus spike protein were used to transduce human ACE2-expressing cells in the presence of one of the Fc-tagged nanobodies at different concentrations. Entry efficiency was characterized as luciferase signal accompanying entry. The efficacy of each nanobody was expressed as the concentration capable of neutralizing pseudovirus entry by 50% (i.e., IC50). Bat SARS2 refers to strain BANAL236 strain; Bat SARS1 refers to strain Rs3367. ND means no obvious neutralization detected. Each experiment was repeated at least twice. (B) Efficacy of Fc-tagged nanobodies in neutralizing live infectious SARS-CoV-2 in vitro. Prototypic SARS-CoV-2 or omicron variant infected Vero cells in the presence of Nanosota-2A-Fc or Nanosota-3A-Fc, respectively, at different concentrations. Infection was characterized as the number of virus plaques formed in overlaid cells. Data are the mean ± SEM (n = 3). Nonlinear regression was performed using a log (inhibitor) versus normalized response curve. The efficacy of each nanobody was expressed as the concentration capable of reducing the relative percentage of infection by 50% (i.e., IC50). Each experiment was repeated at least twice.
Evaluation of anti-SARS-CoV-2 potency of the nanobodies in mouse models
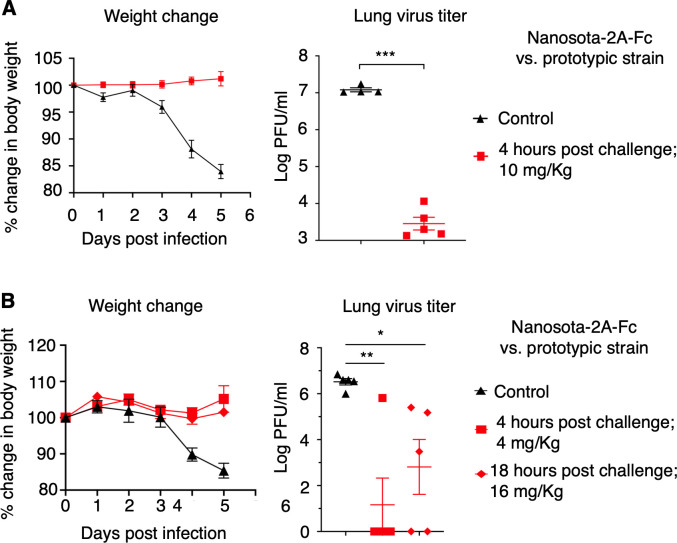
Subsequently, we evaluated the therapeutic efficacy of Nanosota-2A-Fc in a mouse model. Human-ACE2-transgenic mice were challenged with prototypic SARS-CoV-2 strain via intranasal inoculation. Four hours post-challenge, Nanosota-2A-Fc was administered at a dosage of 10 mg/kg weight via the intraperitoneal route (Fig. 4A). The mice were divided into two groups for analysis. One group was monitored for changes in body weight over a 5-day period, while the other group was examined for lung virus titers and lung pathology on day 2, when virus titers reached their peak. Mice in the control group were treated with PBS buffer. Our results showed that the control group of mice experienced significant weight loss, exhibited high titers of virus (~107 PFU/mL) in their lungs, and developed notable lung pathology (Fig. 4A; Fig. S1A). In contrast, the mice treated with Nanosota-2A-Fc did not experience any body weight loss, exhibited low titers of virus (<104 PFU/mL) in the lungs, and displayed less significant lung pathology (Fig. 4A; Fig. S1A). Thus, when administered four hours post-challenge at a dosage of 10 mg/kg weight, Nanosota-2A-Fc effectively treated mice infected with the prototypic SARS-CoV-2 strain.
Fig 4.
Nanosota-2 demonstrates super potency against prototypic SARS-CoV-2 in mouse model. The efficacy of Nanosota-2A-Fc was evaluated for treating the infection of prototypic SARS-CoV-2 in mice. (A) Nanosota-2A-Fc was administered at a dosage of 10 mg/kg weight and a time point of 4 h post-challenge. Human ACE2-transgenic mice were challenged via intranasal inoculation of prototypic SARS-CoV-2. In the treatment group (n = 5), mice received Nanosota-2A-Fc via intraperitoneal delivery. In the control group (n = 4), mice were administered PBS buffer. Body weight changes up to day 5 post-challenge were recorded and lung virus titers on day 2 post-challenge were measured. (B) Nanosota-2A-Fc was administered at a reduced dosage (4 mg/kg weight at 4 h post-challenge) or a delayed time point (18 h post-challenge at 16 mg/kg weight). n = 5 for both the treatment and control groups. Comparisons of lung virus titers between the control and treatment groups were performed using unpaired two-tailed Student’s t-test. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
To explore the limits of the therapeutic efficacy of Nanosota-2A-Fc, we repeated the above mouse challenge experiment with reduced nanobody dosage or delayed nanobody administration time (Fig. 4B). Under the former condition, mice were administered Nanosota-2A-Fc at a dosage of 4 mg/kg weight (at 4 hour post-challenge). Under the latter condition, mice were administered Nanosota-2A-Fc 18 hours post-challenge (at 16 mg/kg weight). Remarkably, even under these conditions, Nanosota-2A-Fc continued to effectively treat human-ACE2-transgenic mice infected with the prototypic SARS-CoV-2 strain (Fig. 4B; Fig. S1B). This demonstrates that Nanosota-2A-Fc can effectively combat prototypic SARS-CoV-2 infections even at lower dosages or with delayed administration times. Notably, the dosage and administration time of Nanosota-2A-Fc used in this study are among the lowest and latest, respectively, compared to other known anti-SARS-CoV-2 antibodies and nanobodies (Table S2).
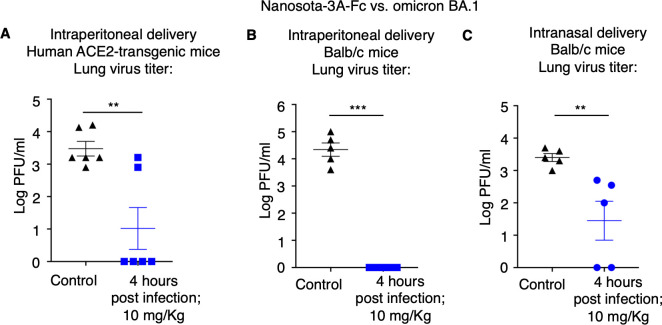
We also assessed the therapeutic efficacy of Nanosota-3A-Fc in two different mouse models challenged with SARS-CoV-2 omicron BA.1 subvariant. These two different models are human-ACE2-transgenic mice and wild-type mice, both of which are susceptible to omicron BA.1 (only the former is susceptible to the prototypic SARS-CoV-2 strain) (32). When administered at a dosage of 10 mg/kg weight, 4 hour post-challenge and via the intraperitoneal route, Nanosota-3A-Fc resulted in a significant reduction in lung virus titers in both mouse models (Fig. 5A; 5B). Therefore, Nanosota-3A-Fc effectively treated both mouse models infected with the omicron BA.1 subvariant when administered via the intraperitoneal route.
Fig 5.
Nanosota-3 demonstrates high potency against SARS-CoV-2 omicron in mouse models via different administration routes. The efficacy of Nanosota-3A-Fc was evaluated for treating the infection of SARS-CoV-2 omicron variant in mouse models and via different administration routes: (A) human ACE2-transgenic mice (n = 6) and intraperitoneal delivery; (B) Balb/c mice (n = 5) and intraperitoneal delivery; (C) Balb/c mice (n = 5) and intranasal delivery. Nanosota-3A-Fc was administered at 4 h post-challenge and a dosage of 10 mg/kg weight. In the control groups, mice were administered PBS buffer. Lung virus titers on day 3 post-challenge (for intraperitoneal delivery) or day 2 post-challenge (for intranasal delivery) were measured. Comparisons of lung virus titers between the control and treatment groups were performed using unpaired two-tailed Student’s t-test. Error bars represent SEM. **P < 0.01; ***P < 0.001.
To explore the potential of Nanosota-3A-Fc as an inhalation treatment, we conducted a similar experiment using mice, but this time administering the drug through the intranasal route. Balb/c mice were administered Nanosota-3A-Fc as nasal drops (at a dosage of 10 mg/kg weight and 4 hour post-challenge). Interestingly, even with this method, Nanosota-3A-Fc still resulted in a significant reduction in lung virus titers in the mouse model (Fig. 5C). Therefore, when administered through the intranasal route, Nanosota-3A-Fc effectively treated the mouse model infected with the omicron BA.1 subvariant.
Structural mechanisms for anti-SARS-CoV-2 potency of the nanobodies
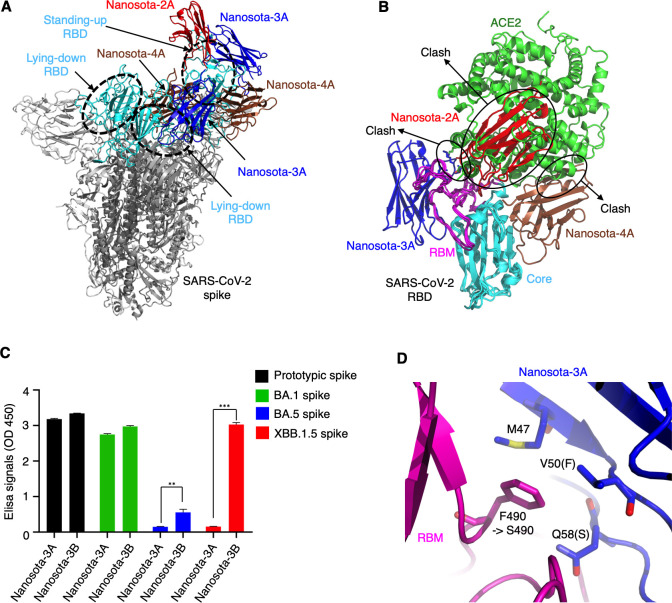
To investigate the structural mechanisms by which the three nanobodies inhibit SARS-CoV-2, we determined the cryo-EM structures of SARS-CoV-2 spike (prototypic strain) complexed with each of them (Nanosota-2A, Nanosota-3A and Nanosota-4A) (Fig. 6A; Fig. S2). The overall resolutions of the cryo-EM maps were 2.1 Å, 2.5 Å, and 3.4 Å, respectively (Table S3; Fig. S3; Fig. S4; Fig.S5). However, due to the mobility of the nanobodies when bound, the local resolutions of the maps in the nanobody regions were lower. As a result, we were able to construct an atomic model only for Nanosota-3A, based on a 3.2 Å local map (Fig. S6), whereas docking models were generated for Nanosota-2A and Nanosota-4A. Nanosota-2A exclusively binds to the standing-up RBD, whereas both Nanosota-3A and Nanosota-4A bind to both the standing-up and lying-down RBDs. By superimposing the three nanobodies on the same RBD, we observed that they bind to distinct regions on the RBD that do not overlap (Fig. 6B). Specifically, Nanosota-2A binds to the top portion of the RBD, and its binding site completely overlaps with the ACE2-binding region. In comparison, Nanosota-3A and Nanosota-4A bind to each side of the RBD, with their binding sites partially overlapping the ACE2-binding region. As a result, the binding of each nanobody to the RBD prevents ACE2 from attaching to it, providing an explanation for their ability to neutralize the entry of SARS-CoV-2.
Fig 6.
Structural basis for anti-SARS-CoV-2 potency of discovered nanobodies and structure-guided in vitro affinity maturation of Nanosota-3. The cryo-EM structures of prototypic SARS-CoV-2 spike complexed with each of the three nanobodies were determined. (A) Superimposition of spike-bound Nanosota-2A, -3A, and -4A onto the structure of the same spike protein (with 1 RBD up). Spike trimer is colored in gray, with three RBDs (one up and two down) colored in cyan. Nanosota-2A, -3A, and -4A are colored in red, blue, and brown, respectively. (B) Superimposition of RBD-bound Nanosota-2A, -3A, -4A, and human ACE2 onto the structure of the same RBD. The three nanobodies are colored the same as in (A). The core structure of the RBD is colored in cyan, and the receptor-binding motif (RBM) is colored in magenta. ACE2 (PDB 6M0J) is colored in green. The clashes between each nanobody and ACE2 are labeled. (C) Binding between Nanosota-3 and the spike proteins of different SARS-CoV-2 variants using ELISA. Nanosota-3A is the original version of Nanosota-3, which binds to the spikes of the prototypic strain and omicron BA.1 subvariant, but not the spike of the currently circulating XBB.1.5. Structure-guided in vitro affinity maturation of Nanosota-3A using phage display generated Nanosota-3B, which binds the XBB.1.5 spike. Comparisons of the ELISA results between Nanosota-3A and Nanosota-3B were performed using unpaired two-tailed Student’s t-test. Error bars represent SEM. **P < 0.01; ***P < 0.001. (D) Detailed structure at the interface between prototypic RBD and Nanosota-3A. The RBD is colored in magenta and Nanosota-3A in blue. RBD residue 490 mutated from a phenylalanine in the prototypic strain (also the BA.1 subvariant) to a serine in XBB.1.5 subvariant. Three Nanosota-3A residues (Met47, Val50, and Asn58) surrounding RBD residue 490 were subjected to random mutagenesis in in vitro affinity maturation to generate Nanosota-3B. Compared to Nanosota-3A, Nanosota-3B contains two mutations, V50F and Q58S.
To validate the above structural information, we conducted competition SPR experiments to see if any of the three nanobodies would compete with ACE2 for RBD binding. Our results indicated that when each of the three Fc-tagged nanobodies bound to the RBD, it either completely blocked ACE2 from binding to the RBD (in the case of Nanosota-2A and Nanosota-3A) or mostly did so (for Nanosota-4A) (Fig. 2D).
Structure-guided engineering of Nanosota-3 to expand its antiviral spectrum
The above structural information provided a basis for the rational engineering of Nanosota-3A, with the goal of creating a highly effective entry inhibitor against the currently circulating omicron XBB.1.5 subvariant. By conducting ELISA between Nanosota-3A and the spikes from different omicron subvariants, we observed that Nanosota-3A binds to the prototypic spike and BA.1 spike but not XBB.1.5 spike (Fig. 5C); these results align with the pseudovirus neutralization data (Fig. 3A). Examination of the structural interface between Nanosota-3A and the RBD led to the identification of a mutation from BA.1 to XBB.1.5 that potentially disrupted the binding of Nanosota-3A to the XBB.1.5 spike (Fig. 6D). Specifically, while both the prototypic RBD and the BA.1 RBD contained a phenylalanine at position 490, the XBB.1.5 RBD featured a serine at the same position. To overcome the F490S mutation in the XBB.1.5 RBD, random mutations were introduced to three residues (Met47, Val50, and Gln58) surrounding this mutation in Nanosota-3A. Through phage display, a mutant nanobody named Nanosota-3B was successfully selected, which exhibited a high affinity binding to XBB.1.5 spike. Compared to Nanosota-3A, Nanosota-3B contained two mutations: V50F and Q58S. Nanosota-3B-Fc (Nanosota-3B with an added C-terminal Fc tag) effectively neutralized the entry of XBB.1.5 pseudoviruses (Fig. 3A). While the engineering of the nanobody did not specifically target the BA.5 spike, Nanosota-3B also displayed a moderate, yet significantly improved, affinity toward the BA.5 spike (Fig. 6C). Moreover, Nanosota-3B-Fc efficiently neutralized the entry of BA.5 pseudoviruses (Fig. 3A). Detailed structural mechanisms underlying the ability of Nanosota-3B to inhibit BA.5 and XBB.1.5 pseudoviruses will be examined in future structural studies.
DISCUSSION
The COVID-19 pandemic has revealed various limitations of conventional antibodies as therapeutics, such as their high cost, limited effectiveness, inability to combat new viral variants, and the requirement for injection-based administration. Given these factors, antibody drugs are not practical options during a global pandemic like COVID-19. Over the past 20 years, aside from SARS-CoV-2, two other highly pathogenic coronaviruses, namely, SARS-CoV-1 and MERS-CoV, have emerged from animal reservoirs (33, 34). SARS-CoV-1 is genetically related to SARS-CoV-2 and uses the same receptor, ACE2, as SARS-CoV-2 does (5, 29). This recent history of coronavirus epidemics raises concerns about the potential recurrence of new coronavirus pandemics, particularly those that also target ACE2 as a receptor. Consequently, there is an urgent need for highly potent, broad-spectrum, cost-effective, and easily administered therapeutics to fight the circulating SARS-CoV-2 and prepare for potential future coronavirus pandemics.
There are three notable features of nanobody therapeutics that set them apart from conventional antibodies. First, nanobodies (both monomer and Fc-tagged dimer) may be administered through the intranasal route (25 – 27) (Fig. 5C). This feature can greatly expand the applicability of nanobody therapeutics. Second, nanobodies (both monomer and Fc-tagged dimer) can be produced in large quantities from either bacteria or mammalian cells. They also exhibit stability even at high temperatures (24). These features make nanobodies cost-effective for production, transportation, and storage. Third, nanobodies can be easily adapted to target new viral variants through in vitro affinity maturation using phage display (24) (Fig. 6C and D). This feature greatly broadens the potential antiviral spectrums of nanobodies. Furthermore, nanobodies have low immunogenicity in humans, and their safety as human therapeutics has been affirmed by the FDA’s approval of a nanobody drug for the treatment of a blood clotting disorder (24). Despite these advantages, the main uncertainty surrounding nanobodies lies in their ability to achieve high potency and broad-spectrum activities, particularly against new viral variants. This study has identified three super potent and broad-spectrum nanobodies, Nanosota-2, -3, and -4, thereby establishing nanobodies as ideal therapeutics against global viral pandemics.
All three nanobodies, Nanosota-2, -3, and -4, exhibit remarkable potency against SARS-CoV-2. These nanobodies were discovered from an immunized alpaca. Unlike Nanosota-1, which we previously discovered from a naïve camelid nanobody phage display library and optimized using phage display, Nanosota-2, -3, and -4 demonstrated exceptional potency against their target without the need for in vitro affinity maturation. In particular, Nanosota-2 effectively inhibits the infection of prototypic SARS-CoV-2 in vitro at a low IC50 (2 pM or 0.16 ng/mL against live virus) and in mice at a low dosage (4 mg/kg weight) or with a late administration time (18 hour post-challenge). These potency metrics are the best of known anti-SARS-CoV-2 entry inhibitors. Similarly, Nanosota-3 also displays high potency in inhibiting the omicron variant in vitro and in mouse models, whereas Nanosota-4 is effective against both SARS-CoV-1 and SARS-CoV-2. Cryo-EM data revealed two factors contributing to the extraordinary anti-SARS-CoV-2 potency of these nanobodies. First, they bind to the RBD with high affinity, overpowering their competitor ACE2. Second, they target functionally critical regions on the RBD. The binding site of Nanosota-2 on the RBD completely overlaps with the ACE2-binding site, explaining its super potency. Although the binding sites of Nanosota-3 and Nanosota-4 on the RBD only partially overlap with the ACE2-binding site, they can bind to the RBD in both standing-up and lying-down conformations, explaining their high potency. Interestingly, in order to bind the lying-down RBD, Nanosota-4A fits into a cavity in the trimeric spike (Fig. S7A), which would not accommodate conventional antibodies (Fig. S7B). This example showcased that nanobodies, because of their small size, can bind to some epitopes not accessible to conventional antibodies. It is worth noting that achieving therapeutic efficacy with antibodies or nanobodies is generally more challenging than prophylactic efficacy. Therefore, the strong therapeutic performance of these Nanosota series suggests they might also have promising prophylactic potential, even though this aspect was not explored in our current study.
Nanosota-2, -3, and -4 also demonstrate broad anti-SARS-CoV-2 spectrums. All three nanobodies potently inhibit pre-omicron SARS-CoV-2 variants and bat SARS2 (except for Nanosota-3 against the delta variant). Additionally, Nanosota-3 and Nanosota-4 effectively inhibit the SARS-CoV-2 omicron variant and SARS-CoV-1, respectively. Hence, the combined antiviral spectrums of the three nanobodies cover pre-omicron SARS-CoV-2 variants, SARS-CoV-2 omicron variant, bat SARS2, and SARS-CoV-1. Since these nanobodies bind to non-overlapping regions on the SARS-CoV-2 RBD, they can be used in combination as a cocktail to battle these viruses and their variants. It is worth noting that the original version of Nanosota-3 was only effective against the early omicron subvariant BA.1. However, through in vitro affinity maturation, we successfully engineered Nanosota-3 to also become an effective inhibitor of the currently circulating omicron subvariant XBB.1.5. Indeed, because of their single-chain structure, nanobodies can be conveniently adapted to new mutations in viral RBDs through in vitro affinity maturation using phage display. This adaptability of nanobodies significantly broadens their antiviral spectrums. The current study serves as proof of concept, demonstrating that these nanobodies can be engineered and optimized to potentially target all ACE2-recognizing coronaviruses related to SARS-CoV-2 and SARS-CoV-1.
In summary, our research offers a thorough and innovative exploration into how nanobodies can achieve super therapeutic potency and broad antiviral spectrums. Our work covers alpaca immunization, construction and screening of a nanobody phage display library, extensive pseudovirus entry assay to test discovered nanobodies against almost all of the major SARS-CoV-2 strains, live SARS-CoV-2 infection in vitro, extensive animal testing of discovered nanobodies against selected SARS-CoV-2 strains using different nanobody dosages, administration times and administration routes, cryo-EM structure determinations of SARS-CoV-2 spike protein complexed with each of the discovered nanobodies, and adaptation of a discovered nanobody to new SARS-CoV-2 variants. The current study is among the most comprehensive and systematic studies researching anti-SARS-CoV-2 nanobodies. Moreover, the three discovered nanobodies each have their novel features: Nanosota-2 inhibits prototypic SARS-CoV-2 with super potency; Nanosota-3 originally inhibited earlier omicron subvariants and then was adapted to inhibit late omicron subvariants through a novel combination of structure-based design and phage display; Nanosota-4 uniquely inhibits both SARS-CoV-1 and SARS-CoV-2 through binding to a conserved epitope on the spike protein inaccessible to conventional antibodies. Further development and characterization of these nanobodies will be conducted in future studies, such as using two or all of them in combination treatment of SARS-CoV-2 infections both in vitro and in animal models. Overall, the Nanosota series are powerful therapeutic tools for battles against circulating SARS-CoV-2 and potential future coronavirus pandemics.
MATERIALS AND METHODS
Cell lines, plasmids, and viruses
HEK293T cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 units/mL penicillin, and 100 µg/mL streptomycin (Life Technologies). ss320 E. coli (Lucigen) and TG1 E. coli (Lucigen) were grown in 2YT medium. Vero E6 cells (American Type Culture Collection) were grown in Eagle’s minimal essential medium (EMEM) supplemented with penicillin (100 units/ml), streptomycin (100 µg/mL), and 10% fetal bovine serum (FBS). Original SARS-CoV-2 spike gene (Wuhan strain; GenBank: QHD43416.1), bat SARS2 spike gene (strain BANAL-20-236; GenBank: MZ937003.2), SARS-CoV-1 spike gene (strain Tor2; GenBank: AFR58742.1) and bat SARS1 spike gene (strain Rs3367; GenBank: AGZ48818.1) were synthesized (GenScript). Mutations were introduced to the original SARS-CoV-2 spike gene to generate the prototypic SARS-CoV-2 spike gene (encoding the spike protein from Wuhan strain plus D614G mutation) and the spike gene of alpha variant (strain B.1.1.7; GISAID: EPI_ISL_6135157), delta variant (strain B.1.617.2; GenBank: UEM53021.1), omicron BA.1 subvariant (strain BA.1; GISAID: EPI_ISL_6590782.2), omicron BA.5 subvariant (strain BA.5; GISAID: EPI_ISL_12954165), and omicron XBB.1.5 subvariant (strain XBB.1.5; GISAID: EPI_ISL_17774216). Each of the spike gene was cloned into the pcDNA3.1(+) vector.
SARS-CoV-2 spike ectodomain (residues 14–1211) and SARS-CoV-2 RBD (residues 319–529) were subcloned into Lenti-CMV vector (Vigene Biosciences) with an N-terminal tissue plasminogen activator (tPA) signal peptide and a C-terminal His tag. For the spike ectodomain construct, D614G and six proline mutations were introduced to the S2 subunit region to stabilize the spike protein in its prefusion state (9, 35). Nanosota-2A-Fc, Nanosota-3A-Fc, and Nanosota-4A-Fc were constructed in the same way as the RBD except that a C-terminal human IgG1 Fc tag replaced the His tag. Infectious prototypic SARS-CoV-2 (US_WA-1 isolate) and omicron variant (B.1.1.529 strain) were obtained from CDC (Atlanta) and Dr. Mehul Suthar (Emory University), respectively. Experiments involving infectious viruses were conducted at the University of Iowa in approved biosafety level 3 laboratories.
Construction of induced nanobody phage display library
Induced nanobody phage display libraries were constructed as previously described (36). Briefly, an alpaca was immunized 6 times with 125 µg of purified SARS-CoV-2 spike ectodomain in Gerbu adjuvant. Following immunization, blood was drawn and peripheral blood mononuclear cells (PBMCs) were isolated by centrifugation from 50 mL of blood using Sepmate centrifugal devices according to the manufacturer’s protocol (Stemcell Technologies). A cDNA library was made by reverse transcription using oligo dT primers and Superscript IV reverse transcriptase (Thermo Scientific). A nested PCR strategy was used to amplify coding regions of VHH fragments. The resulting PCR fragments were cloned into a modified pADL22 vector (Antibody Design Labs), and the phage library was produced with a library size of 5 × 109 following the manufacturer’s protocols (Antibody Design Labs).
Screening of induced nanobody phage display library
To identify anti-SARS-CoV-2 nanobodies, the above nanobody phage display library was used in bio-panning as previously described (24). Briefly, 5 µg purified prototypic SARS-CoV-2 spike ectodomain was used for one round of panning to obtain the SARS-CoV-2 spike-targeting nanobodies. After washing, the retained phages were eluted using 500 µL 100 mM triethylamine, neutralized with 250 µL 1 M Tris-HCl pH 7.5, and then used to infect ss320 E. coli. Ninety-six single colonies were picked, and expressions of nanobodies were induced by 1 mM IPTG. The supernatants were subjected to ELISA for identification of strong binders. Forty-eight strong binders were identified, and their genes were subjected to DNA sequencing. Six unique nanobody sequences were identified. Of these, three bound to the RBD and neutralized SARS-CoV-2 pseudovirus entry. The other three, however, did not bind to the RBD or neutralize SARS-CoV-2 pseudovirus entry. The three RBD-binding nanobodies were named Nanosota-2A, Nanosota-3A, and Nanosota-4A, and they were further evaluated in the current study.
Protein expression and purification
Nanosota-2A, Nanosota-3A, and Nanosota-4A were purified as previously described (24). Briefly, the nanobodies (all with a C-terminal His tag) were each purified from the periplasm of ss320 E. coli after induction by 1 mM IPTG. The E. coli cells were collected and re-suspended in 15 mL TES buffer (0.2 M Tris pH 8, 0.5 mM EDTA, 0.5 M sucrose), shaken on ice for 1 h, diluted with 40 mL ¼ TES buffer, and then shaken on ice for another hour. The proteins in the supernatant were sequentially purified using a Ni-NTA column and a Superdex200 gel filtration column (Cytiva).
Prototypic SARS-CoV-2 spike ectodomain (with a His tag), SARS-CoV-2 RBD (with a His tag), and Fc-tagged nanobodies were prepared from 293F mammalian cells as previously described (37). Briefly, lentiviral particles were packaged using the plasmid encoding one of the above proteins and then infected 293F cells for selection of stable cell lines in the presence of Puromycin (Gibco). The proteins were harvested from the supernatants of cell culture medium, purified on Ni-NTA column for His-tagged proteins or on Protein A column for Fc-tagged proteins, and purified further on Superdex200 gel filtration column (Cytiva).
To prepare the complexes of the prototypic SARS-CoV-2 spike ectodomain and one of three nanobodies (Nanosota-2A, Nanosota-3A, and Nanosota-4A), the spike ectodomain and one of the nanobodies were incubated (with the nanobody in excess) at room temperature for 30 min before the mixture was subjected to Superose 6 increase 10/300 GL gel filtration column (Cytiva).
ELISA
To detect the binding between His-tagged SARS-CoV-2 spike ectodomain and HA-tagged nanobodies, ELISA was carried out as previously described (24). Briefly, ELISA plates were coated with recombinant SARS-CoV-2 spike ectodomain. They were then incubated either with the supernatant of ss320 E. coli expressing one of the nanobodies or with one of the purified recombinant nanobodies. Next, they were incubated with HRP-conjugated anti-HA antibody (1:1,000) (Sigma). Subsequently, ELISA substrate (Invitrogen) was added, and the reactions were stopped using 1 N H2SO4. The absorbance at 450 nM (A450) was measured using a Synergy LX Multi-Mode Reader (BioTek).
Surface plasmon resonance
To measure the binding affinity between each of the nanobodies and SARS-CoV-2 RBD, surface plasmon resonance assay was performed using a Biacore S200 system (Cytiva) as previously described (24). Briefly, each of the Fc-tagged nanobodies was immobilized on a protein A sensor chip (Cytiva). Serial dilutions of His-tagged SARS-CoV-2 RBD were injected at different concentrations from 2.5 to 80 nM. The resulting data were fitted to a 1:1 binding model using Biacore Evaluation Software (Cytiva). The analysis was conducted according to the Biacore handbook’s guidelines, specifically for interactions between a coated Fc-tagged antibody and a monomeric analyte that flows through. RU, resonance unit.
To assess the potential competition between human ACE2 and each of the three Fc-tagged nanobodies, competition SPR experiments were carried out. The prototypical SARS-CoV-2 RBD (with a His-tag) was immobilized onto four CM5 sensor chips (Cytiva) (800 RU for each chip). Subsequently, each Fc-tagged nanobody (at 6,250 nM), specifically Nanosota-2A-Fc, Nanosota-3A-Fc, or Nanosota-4A-Fc, was injected to the first three sensor chips. As a control, running buffer was injected to the fourth sensor chip. After the first three sensor chips were saturated with their respective nanobodies, a mixture of recombinant human ACE2 (His-tagged, at 6,250 nM) and the same individual nanobody (6,250 nM) were injected to each of the first three chips. On the control chip, only the ACE2 was injected. The resulting sensorgrams from all four chips were overlaid, setting the point at which ACE2 injection began as the baseline. Finally, the competitive binding between human ACE2 and each Fc-tagged nanobody was evaluated by comparing the SPR binding signals from the mixed nanobody/ACE2 injections to those from the ACE2-only injection.
Pseudovirus entry assay
The neutralizing potency of Nanosota-2A-Fc, Nanosota-3A-Fc, and Nanosota-4A-Fc against SARS-CoV-2 and SARS-CoV-1 pseudoviruses was evaluated using pseudovirus entry assay as previously described (37). Briefly, to prepare the pseudoviruses, HEK293T cells were co-transfected with a pcDNA3.1(+) plasmid encoding one of the coronavirus spike proteins, a helper plasmid psPAX2, and a reporter plasmid plenti-CMV-luc. Pseudoviruses were collected 72 h post transfection, incubated with nanobodies at different concentrations at 37°C for 1 h, and then used to transduce HEK293T cells stably expressing human ACE2. After another 60 h, cells were lysed. Aliquots of cell lysates were transferred to new plates, a luciferase substrate was added, and Relative Light Units (RLUs) were measured using an EnSpire plate reader (PerkinElmer). The efficacy of each nanobody was calculated and expressed as the concentration of the nanobody capable of inhibiting pseudovirus entry by 50% (IC50).
SARS-CoV-2 plaque reduction neutralization test
The neutralizing potency of Nanosota-2A-Fc against live SARS-CoV-2 infection and of Nanosota-3A-Fc against omicron BA.1 subvariant infection was evaluated using a plaque reduction neutralization test (PRNT) assay as described previously (24). Specifically, individual nanobody was serially diluted in DMEM and mixed with SARS-CoV-2 or omicron BA.1 subvariant (at a titer of 80 plaque-forming units or PFUs) at 37°C for an additional 45 min. The mixtures were then added to cells [Vero E6 for SARS-CoV-2 cells and Vero E6 overexpressing ACE2 and TMPRSS2 (A2T2) for omicron] at 37°C for an additional 45 min. After the culture medium was removed, the cells were overlaid with 0.6% agarose and cultured for 3 days. Plaques were visualized by 0.1% crystal violet staining. The efficacy of each nanobody was calculated and expressed as the concentration capable of reducing the number of virus plaques by 50% (i.e., IC50) compared to control serum-exposed virus.
SARS-CoV-2 challenge experiment in mouse models
The neutralizing potency of Nanosota-2A-Fc and Nanosota-3A-Fc against infectious SARS-CoV-2 in vivo was evaluated using SARS-CoV-2 challenge experiments in mouse models as previously described (24).
The efficacy of Nanosota-2A-Fc against prototypic SARS-CoV-2 was determined in K18-human-ACE2-transgenic mice (K18-Tg mice) (Jackson Laboratory). All mice were challenged via intranasal inoculation of prototypic SARS-CoV-2 (5 × 103 PFU/mouse) in a volume of 50 µL DMEM. In the treatment group (n = 10), mice received Nanosota-2A-Fc (10 mg/kg weight) via intraperitoneal delivery at 4 h post-challenge. In the control group (n = 8), mice were administered PBS buffer at 4 h post-challenge. The virus titers in the lungs of the mice were measured using a virus plaque assay as previously described (24). Briefly, half of the mice from each group were euthanized on day 2 post-challenge and lung tissue homogenate supernatants were collected. Twelve-well plates of Vero E6 cells were inoculated with serially diluted lung homogenates (in DMEM) and then incubated at 37°C in 5% CO2 for 1 h with gentle shaking every 15 min. Then, the inocula were removed and the plates were overlaid with 0.6% agarose containing 2% FBS. After 3 days, the overlays were removed and the plaques were visualized via staining with 0.1% crystal violet. Virus titers were quantified as PFU per mL tissue. The body weights of the other half of the mice from each group were monitored daily. Mouse lungs were collected on day 5 post-challenge and then were fixed in 10% formalin. The histopathology of the lung was examined by hematoxylin and eosin-stained tissue section.
To further determine the limits of its efficacy, Nanosota-2A-Fc was administered at lower dosage or at later treatment time point in the SARS-CoV-2 challenge experiment in K18-Tg mice. More specifically, mice were divided into three groups (n = 10 in each group): (i) low dosage treatment group—Nanosota-2A-Fc was administered 4 h post-challenge at 4 mg/kg weight; (ii) late treatment group—Nanosota-2A-Fc was administered 18 h post-challenge at 16 mg/kg weight; (iii) negative control group—PBS buffer was administered. The nanobody and PBS buffer were administered via intraperitoneal delivery. Lung virus titers, body weights, and lung histology data were collected in the same way as above.
The efficacy of Nanosota-3A-Fc against the omicron variant was determined in both K18-Tg mice (n = 6) and Balb/c mice (n = 5). More specifically, mice were challenged intranasally with omicron B.1.1.529 (105 PFU/mouse) and were then treated with either Nanosota-3A-Fc (10 mg/kg weight) or PBS buffer at 4 h post-challenge. The nanobody and PBS buffer were administered via intraperitoneal delivery. Lung virus titers on day 3 post-challenge were determined in the same way as above.
The efficacy of Nanosota-3A-Fc was further evaluated via intranasal delivery in the mouse model. Balb/c mice (n = 5) were challenged intranasally with 105 PFU omicron B.1.1.529 and were then treated with either Nanosota-3A-Fc (10 mg/kg weight in 50 µL PBS buffer) or 50 µL PBS buffer at 4 h post-challenge via intranasal delivery. The mice were sedated during the intranasal administration of the samples. Pipettes were used for the intranasal delivery of Nanosota-3A-Fc. Viral titers of infected lungs on second day post infection were determined in the same way as above.
Due to the significant resources and effort required for animal testing, it is not feasible to test every nanobody against all SARS-CoV-2 variants. For this reason, we established certain priorities for our animal testing. First, we evaluated Nanosota-2A-Fc against the prototypic SARS-CoV-2 due to its pronounced effectiveness against this strain in pseudovirus entry assay and live SARS-CoV-2 infection assay in vitro. However, since the prototypic SARS-CoV-2 cannot infect Balb/c mice efficiently, we limited our testing of Nanosota-2A-Fc to K18-human-ACE2-transgenic mice. Second, we assessed Nanosota-3A-Fc against the omicron BA.1 variant because of its effectiveness against the omicron variant in vitro. Our interest in further developing Nanosota-3A-Fc was driven by the current clinical significance of the omicron variant. Consequently, we experimented with Nanosota-3A-Fc in mice using both intraperitoneal and intranasal delivery methods. Since the omicron BA.1 variant demonstrated a reasonable ability to infect both K18-human-ACE2-transgenic and Balb/c mice, we used both mouse models for the Nanosota-3A-Fc tests. Lastly, as our current study is centered on SARS-CoV-2, our plans to test Nanosota-4A-Fc against SARS-CoV-1 in mice are set for future research.
Cryo-EM grid preparation and data acquisition
The complexes of SARS-CoV-2 spike ectodomain and each of the nanobodies (4 µL at ~1.9 µM for the spike/Nanosota-2A complex, ~1.3 µM for the spike/Nanosota-3A complex, and ~1.1 µM for the spike/Nanosota-4A complex, respectively) were supplemented with 8 mM CHAPSO immediately before grid preparation. Each of the complexes was then applied to freshly glow-discharged Quantifoil R1.2/1.3 300-mesh copper grids (EM Sciences) and blotted for 4 s at 22°C under 100% chamber humidity and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI). Cryo-EM data were collected using EPU version 3.0 (ThermoFisher Scientific) on a Titan Krios electron microscope (ThermoFisher Scientific) equipped with a Falcon IV direct electron detector and with a Selectris-X energy filter (ThermoFisher Scientific) or using Latitude-S (Gatan) equipped with a K3 direct electron detector and with a Biocontinuum energy filter (Gatan). For the Falcon IV detector, the movies were collected at a nominal magnification of 165,000× (corresponding to 0.73 Å per pixel), slit width 10 eV, a dose rate of 5.9 e– per Å2 per second, and a total dose of 40 e–/Å2. For the K3 detector, the movies were collected at a nominal magnification of 75,300× (corresponding to 0.664 Å per pixel), slit width 20 eV, a dose rate of 25 e– per Å2 per second, and a total dose of 50 e–/Å2. The statistics of cryo-EM data collection are summarized in Table S3.
Image processing
Cryo-EM data were processed using cryoSPARC v3.3.2 (38), and the procedure is outlined in Fig. S3 through S5. Briefly, dose-fractionated movies were subjected to Patch motion correction with MotionCor2 (39) and Patch CTF estimation with CTFFIND-4.1.13 (40). Particles were then picked using both Blob picker and Template picker in cryoSPARC v3.3.2 and subjected to the Remove Duplicate Particles Tool. Junk particles were removed through three rounds of 2D classifications. Particles from the good 2D classes were used for Ab-initio Reconstruction of three or four maps. The initial models were set as the starting references for heterogeneous refinement (3D classification). A second round of Ab-initio Reconstruction and heterogeneous refinement was conducted for the spike/Nanosota-3A complex. The selected 3D classes were then subjected to further homogeneous, non-uniform and CTF refinements, generating the final maps. Particles in the good 3D class were then imported into RELION-4.0 (41) using the csparc2star.py module (UCSF pyem v0.5. Zenodo) and subjected to signal subtraction to keep only the receptor-binding subunit of the spike and Nanosota-3A in RELION-4.0, followed by masked 3D classification for the spike/Nanosota-3A complex. Particles with the subtracted signal (spike/Nanosota-3A and spike/Nanosota-4A) and the ones in the selected class from the masked 3D classification (spike/Nanosota-2A) were then subjected to local refinements to improve densities in cryoSPARC v3.3.2. Resolutions of the maps were determined by gold-standard Fourier shell correlation (FSC) at 0.143 between the two half-maps. Local resolution variations were estimated from the two half-maps in cryoSPARC v3.3.2.
Cryo-EM model building and refinement
Initial model building of the spike/nanobody complexes was performed in Coot-0.8.9 (42) using PDB 7TGX as the starting model. The initial model of each nanobody was predicted using SWISS-MODEL (https://swissmodel.expasy.org/) and then fitted into the density map. Several rounds of refinement in Phenix-1.16 (43) and manual building in Coot-0.8.9 were performed until the final reliable models were obtained. Standing-up RBDs and spike-bound nanobodies are generally flexible, and hence, they were fitted into the density as rigid bodies. Specially, in the local map of the receptor-binding subunit from the spike/Nanosota-3A complex, an atomic model was built at the interface between one lying-down RBD and Nanosota-3A. Model and map statistics are summarized in Table S3. Figures were generated using UCSF Chimera X v0.93 (44) and PyMol v2.5.2 (45).
In vitro affinity maturation of Nanosota-3A to generate Nanosota-3B
To engineer Nanosota-3 for expansion of its antiviral spectrum, in vitro affinity maturation of Nanosota-3A was performed. Briefly, random mutations were introduced to three residues (Met47, Val50, and Gln58) of Nanosota-3A surrounding Phe490 in the prototypic RBD. Specifically, random mutations were introduced to the PCR primers for generation of mutant Nanosota-3A genes. The mutant Nanosota-3A genes were then inserted into the PADL22c vector and electroporated into the TG1 cells for construction of a mutational library. Subsequently, mutant phages were selected for enhanced binding to XBB.1.5 spike protein as described above. After three rounds of selection, Nanosota-3B with two mutations (V50F and Q58S) was discovered, which showed the highest binding affinity for XBB.1.5 spike.
ACKNOWLEDGMENTS
The developments of Nanosota-2, Nanosota-3, and Nanosota-4 were supported by funding from the University of Minnesota (to F.L.), NIH grants R01AI089728 (to F.L.), R01AI157975 (to F.L., L.D., and S.P.), and U19AI171954 (to F.L., L.D., L.B., and S.P.). The pathology data on SARS-CoV-2-challenged mice were analyzed at Central Microscopy Research Facility at the University of Iowa. The University of Minnesota has filed a patent on Nanosota-2, -3, and -4 with F.L, G.Y., and B.L. as inventors. B.S. and B.E.W. are co-founders of Turkey Creek Biotechnology.
AFTER EPUB
[This article was published on 19 October 2023 without the attribution in the Fig. 1 legend. The attribution was added in the current version, posted on 30 November 2023.]
Contributor Information
Bin Liu, Email: liu00794@umn.edu.
Stanley Perlman, Email: stanley-perlman@uiowa.edu.
Fang Li, Email: lifang@umn.edu.
Kanta Subbarao, The Peter Doherty Institute for Infection and Immunity, Melbourne, Victoria, Australia .
ETHICS APPROVAL
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the mouse work was handled according to approved institutional animal care and use committee (IACUC) protocols of the University of Iowa (protocol number: 9051795). All of the alpaca work was performed by Turkey Creek Biotechnology (Waverly, TN) under their approved IACUC protocol.
DATA AVAILABILITY
The atomic models generated in this study have been deposited into the Protein Data Bank (PDB) with accession numbers 8G72 (SARS-CoV-2 spike complexed with Nanosota-2A, with 1 RBD up), 8G74 (SARS-CoV-2 spike complexed with Nanosota-3A, with 1 RBD up), 8G73 (SARS-CoV-2 spike complexed with Nanosota-3A, with 2 RBDs up), and 8G75 (SARS-CoV-2 spike complexed with Nanosota-4A, with 2RBDs up). The corresponding cryo-EM density maps generated in this study have been deposited into the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-29793 (SARS-CoV-2 spike complexed with Nanosota-2A, with 1 RBD up), EMD-29794 (local refinement of SARS-CoV-2 spike complexed with Nanosota-2A, with 1 RBD up), EMD-29796 (SARS-CoV-2 spike complexed with Nanosota-3A, with 1 RBD up), EMD-29795 (SARS-CoV-2 spike complexed with Nanosota-3A, with 2 RBDs up), and EMD-29797 (SARS-CoV-2 spike complexed with Nanosota-4A, with 2RBDs up).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01448-23.
Supplemental figures and tables.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, COVID-19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. doi: 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lipsitch M, Krammer F, Regev-Yochay G, Lustig Y, Balicer RD. 2022. SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat Rev Immunol 22:57–65. doi: 10.1038/s41577-021-00662-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li F. 2016. Structure, function, and evolution of Coronavirus spike proteins. Annu Rev Virol 3:237–261. doi: 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Du LY, He YX, Zhou YS, Liu SW, Zheng BJ, Jiang SB. 2009. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat Rev Microbiol 7:226–236. doi: 10.1038/nrmicro2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel Coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS Coronavirus. J Virol 94:e00127-20. doi: 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. doi: 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. doi: 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- 8. Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. 2020. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 117:11727–11734. doi: 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ye G, Liu B, Li F. 2022. Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain. Nat Commun 13:1214. doi: 10.1038/s41467-022-28882-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, Wang J, Wang Y, Niu X, Yang S, Liang H, Sun H, Li T, Yu Y, Cui Q, Liu S, Yang X, Du S, Zhang Z, Hao X, Shao F, Jin R, Wang X, Xiao J, Wang Y, Xie XS. 2022. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602:657–663. doi: 10.1038/s41586-021-04385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gobeil S-C, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P. 2021. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 373:eabi6226. doi: 10.1126/science.abi6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jovanoski N, Kuznik A, Becker U, Hussein M, Briggs A. 2022. Cost-effectiveness of casirivimab/imdevimab in patients with COVID-19 in the ambulatory setting. J Manag Care Spec Pharm 28:555–565. doi: 10.18553/jmcp.2022.21469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hernandez I, Bott SW, Patel AS, Wolf CG, Hospodar AR, Sampathkumar S, Shrank WH. 2018. Pricing of monoclonal antibody therapies: higher if used for cancer? Am J Manag Care 24:109–112. [PubMed] [Google Scholar]
- 14. Rubin R. 2021. Monoclonal antibodies for COVID-19 preexposure prophylaxis can’t come fast enough for some people. JAMA 326:1895. doi: 10.1001/jama.2021.19534 [DOI] [PubMed] [Google Scholar]
- 15. Tixagevimab and cilgavimab (evusheld) for pre-exposure prophylaxis of COVID-19. 2022. JAMA 327:384–385. doi: 10.1001/jama.2021.24931 [DOI] [PubMed] [Google Scholar]
- 16. Chames P, Van Regenmortel M, Weiss E, Baty D. 2009. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol 157:220–233. doi: 10.1111/j.1476-5381.2009.00190.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paul F, Cartron G. 2019. Infusion-related reactions to rituximab: frequency, mechanisms and predictors. Expert Rev Clin Immunol 15:383–389. doi: 10.1080/1744666X.2019.1562905 [DOI] [PubMed] [Google Scholar]
- 18. Brobst B, Borger J. 2022. Benefits and risks of administering monoclonal antibody therapy for Coronavirus (COVID-19), Statpearls. StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC, Treasure Island (FL). [PubMed] [Google Scholar]
- 19. Könning D, Zielonka S, Grzeschik J, Empting M, Valldorf B, Krah S, Schröter C, Sellmann C, Hock B, Kolmar H. 2017. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr Opin Struct Biol 45:10–16. doi: 10.1016/j.sbi.2016.10.019 [DOI] [PubMed] [Google Scholar]
- 20. De Meyer T, Muyldermans S, Depicker A. 2014. Nanobody-based products as research and diagnostic tools. Trends Biotechnol 32:263–270. doi: 10.1016/j.tibtech.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 21. Muyldermans S. 2013. Nanobodies: natural single-domain antibodies. Annu Rev Biochem 82:775–797. doi: 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- 22. Steeland S, Vandenbroucke RE, Libert C. 2016. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today 21:1076–1113. doi: 10.1016/j.drudis.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 23. Romao E, Morales-Yanez F, Hu Y, Crauwels M, De Pauw P, Hassanzadeh GG, Devoogdt N, Ackaert C, Vincke C, Muyldermans S. 2016. Identification of useful nanobodies by phage display of immune single domain libraries derived from camelid heavy chain antibodies. Curr Pharm Des 22:6500–6518. doi: 10.2174/1381612822666160923114417 [DOI] [PubMed] [Google Scholar]
- 24. Ye G, Gallant J, Zheng J, Massey C, Shi K, Tai W, Odle A, Vickers M, Shang J, Wan Y, Du L, Aihara H, Perlman S, LeBeau A, Li F. 2021. The development of nanosota-1 as anti-SARS-CoV-2 nanobody drug candidates. Elife 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu X, Cheng L, Fu M, Huang B, Zhu L, Xu S, Shi H, Zhang D, Yuan H, Nawaz W, Yang P, Hu Q, Liu Y, Wu Z. 2021. A potent bispecific nanobody protects hACE2 mice against SARS-CoV-2 infection via intranasal administration. Cell Rep 37:109869. doi: 10.1016/j.celrep.2021.109869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nambulli S, Xiang Y, Tilston-Lunel NL, Rennick LJ, Sang Z, Klimstra WB, Reed DS, Crossland NA, Shi Y, Duprex WP. 2021. Inhalable nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci Adv 7:eabh0319. doi: 10.1126/sciadv.abh0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesbølle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Deshpande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, Swaney DL, García-Sastre A, Ott M, Vignuzzi M, Walter P, Manglik A, QCRG Structural Biology Consortium . 2020. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive spike. Science 370:1473–1479. doi: 10.1126/science.abe3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scully M, Minkue Mi Edou J, Callewaert F. 2019. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura. N Engl J Med 380:e32. doi: 10.1056/NEJMc1902336 [DOI] [PubMed] [Google Scholar]
- 29. Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic Coronaviruses. Nat Rev Microbiol 17:181–192. doi: 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li F, Li WH, Farzan M, Harrison SC. 2005. Structure of SARS Coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868. doi: 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- 31. Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. 2013. Isolation and characterization of a bat SARS-like Coronavirus that uses the ACE2 receptor. Nature 503:535–538. doi: 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang W, Shi K, Geng Q, Ye G, Aihara H, Li F. 2022. Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant. Proc. Natl. Acad. Sci. U.S.A 119. doi: 10.1073/pnas.2206509119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling A-E, Humphrey CD, Shieh W-J, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang J-Y, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, the SARS Working Group . 2003. A novel Coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi: 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- 34. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. 2012. Isolation of a novel Coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 35. Hsieh CL, Goldsmith JA, Schaub JM, DiVenere AM, Kuo HC, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou CW, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, McLellan JS. 2020. Structure-based design of prefusion-stabilized SARS-Cov-2 spikes. Science 369:1501–1505. doi: 10.1126/science.abd0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pardon E, Laeremans T, Triest S, Rasmussen SGF, Wohlkönig A, Ruf A, Muyldermans S, Hol WGJ, Kobilka BK, Steyaert J. 2014. A general protocol for the generation of nanobodies for structural biology. Nat Protoc 9:674–693. doi: 10.1038/nprot.2014.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Geng Q, Tai W, Baxter VK, Shi J, Wan Y, Zhang X, Montgomery SA, Taft-Benz SA, Anderson EJ, Knight AC, Dinnon KH, Leist SR, Baric RS, Shang J, Hong S-W, Drelich A, Tseng C-TK, Jenkins M, Heise M, Du L, Li F, Subbarao K. 2021. Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog 17:e1009897. doi: 10.1371/journal.ppat.1009897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296. doi: 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 39. Rubinstein JL, Brubaker MA. 2015. Alignment of cryo-EM movies of individual particles by optimization of image translations. J Struct Biol 192:188–195. doi: 10.1016/j.jsb.2015.08.007 [DOI] [PubMed] [Google Scholar]
- 40. Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. doi: 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zivanov J, Nakane T, Scheres SHW. 2020. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in Relion-3.1. IUCrJ 7:253–267. doi: 10.1107/S2052252520000081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 43. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27:14–25. doi: 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. DeLano WL. 2002. Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40:82–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures and tables.
Data Availability Statement
The atomic models generated in this study have been deposited into the Protein Data Bank (PDB) with accession numbers 8G72 (SARS-CoV-2 spike complexed with Nanosota-2A, with 1 RBD up), 8G74 (SARS-CoV-2 spike complexed with Nanosota-3A, with 1 RBD up), 8G73 (SARS-CoV-2 spike complexed with Nanosota-3A, with 2 RBDs up), and 8G75 (SARS-CoV-2 spike complexed with Nanosota-4A, with 2RBDs up). The corresponding cryo-EM density maps generated in this study have been deposited into the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-29793 (SARS-CoV-2 spike complexed with Nanosota-2A, with 1 RBD up), EMD-29794 (local refinement of SARS-CoV-2 spike complexed with Nanosota-2A, with 1 RBD up), EMD-29796 (SARS-CoV-2 spike complexed with Nanosota-3A, with 1 RBD up), EMD-29795 (SARS-CoV-2 spike complexed with Nanosota-3A, with 2 RBDs up), and EMD-29797 (SARS-CoV-2 spike complexed with Nanosota-4A, with 2RBDs up).