ABSTRACT
The 2019 coronavirus disease (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) highlighted significant gaps in our mechanisms to prevent and control cross-species transmission of animal coronaviruses. Porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), and porcine delta coronavirus (PDCoV) are common porcine coronaviruses with similar clinical features. In the absence of effective drugs and methods of prevention and control, outbreaks of these viruses have led to significant economic losses in the global pig industry. Here, we report the effect of five fatty acids against porcine coronaviruses: sodium butyrate, lauric acid, palmitic acid, docosahexaenoic acid (DHA), and eicosapentaenoic acid (EPA). DHA and EPA reduced viral replication by attenuating the endoplasmic reticulum stress and inhibiting PEDV, TGEV, and PDCoV infections in vero cells, PK-15 cells, and LLC-PK1 cells in vitro, respectively. Additionally, DHA and EPA increased the host antioxidant levels and reduced inflammation. In conclusion, we report here for the first time the antiviral effects of DHA and EPA on porcine coronaviruses and provide a molecular basis for the development of new fatty acid-based therapies to control porcine coronavirus infection and transmission.
IMPORTANCE
Porcine epidemic diarrhea caused by porcine coronaviruses remains a major threat to the global swine industry. Fatty acids are extensively involved in the whole life of the virus. In this study, we found that docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) significantly reduced the viral load of porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), and porcine delta coronavirus (PDCoV) and acted on the replication of the viruses rather than attachment and entry. We further confirmed that DHA and EPA inhibited PEDV replication by alleviating the endoplasmic reticulum stress. Meanwhile, DHA and EPA alleviate PEDV-induced inflammation and reactive oxygen species (ROS) levels and enhance the cellular antioxidant capacity. These data indicate that DHA and EPA have antiviral effects on porcine coronaviruses and provide a molecular basis for the development of new fatty acid-based therapies to control porcine coronavirus infection and transmission.
KEYWORDS: porcine coronaviruses, docosahexaenoic acid, eicosapentaenoic acid, endoplasmic reticulum stress, anti-viral
INTRODUCTION
The harmful effects of coronaviruses on humans are well established. In particular, the spread of the coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has prompted great alarm (1, 2). Coronaviruses infect mammals, causing respiratory and gastrointestinal diseases as well as multiorgan dysfunction (3). Porcine enteric coronaviruses, including porcine epidemic diarrhea virus (PEDV), swine acute diarrhea syndrome coronavirus (SADS-CoV), transmissible gastroenteritis virus (TGEV), and porcine delta coronavirus (PDCoV), are endemic in swine herds and cause significant economic losses (4 – 7). Porcine enteric coronaviruses have similar epidemiological, clinical, and pathological features and are accompanied by diarrhea, vomiting, and dehydration, resulting in high morbidity and mortality in piglets (8).
The life cycle of viruses is inextricably linked to lipids. Viruses are intracellular parasites that exploit the metabolic machinery of their hosts to meet their own biosynthetic needs. Most positive-stranded RNA viruses regulate the host lipid metabolism, hijacking lipid droplets (LDs) to enhance fitness and replication/particle assembly (9). Fatty acids (FAs) are extensively involved in the viral life cycle; however, the roles of FAs in viral infections vary with different chain lengths and saturations. A classical swine fever virus (CSFV) infection induces increased FA biosynthesis and accumulation of free FAs. Curcumin and C75 were found to significantly inhibit fatty acid synthase (FASN) while reducing replication of dengue virus and CSFV (10). In contrast, oleic acid and palmitoleic acid significantly increased CSFV replication (11). Exogenous supplementation of infected cells with linoleic acid or arachidonic acid significantly inhibited the replication of human coronavirus HCoV-229E and the highly pathogenic Middle East respiratory syndrome (MERS) coronavirus (12). Medium-chain FAs and single triglycerides are used as feed additives to promote intestinal health and reduce pathogens in pigs. Indeed, the addition of medium-chain FAs to pig feed was shown to significantly reduce the detection of PEDV (13). However, different fatty acids have been less studied in porcine coronaviruses, and there is still a need to explore the role of fatty acids with different chain lengths and saturations on porcine coronaviruses.
Endoplasmic reticulum (ER) is an important membranous organelle in eukaryotic cells. Disturbance of ER homeostasis by multiple factors, such as microbial infection, calcium imbalance, and accumulation of unfolded/misfolded proteins, ultimately leads to ER stress (ERS), which triggers the adaptive unfolded protein response (UPR) mechanism to restore the normal ER function (14). ERS plays a critical role in the pathogenesis of various coronaviral diseases through its interactions with immune regulation, autophagy, and apoptosis pathways (15, 16). Both SARS-CoV-2 and enterovirus 71 infections are accompanied by the activation of ERS and the UPR (17, 18). In the case of a coronavirus infection, the formation of double membrane vesicle (DMV) replication complexes and infectious particles is dependent on ER-generated membranes (19). Regulation of the ER and inhibition of DMV production have been shown to have a broad effect on coronavirus replication in vitro. Fatty acids may regulate porcine coronavirus replication through ERs.
In the present study, the antiviral effects of five FAs [sodium butyrate (NaB), lauric acid (LA), palmitic acid (PA), docosahexaenoic acid (DHA), and eicosapentaenoic acid (EPA)] on PEDV, TGEV, and PDCoV were investigated by measuring the changes in viral mRNA, N protein, and viral yield. The results showed that the antiviral activity of DHA and EPA was exerted mainly by coincubation of the replication cycle of the virus. DHA and EPA significantly reduced the PEDV-induced ERS, increased the antioxidant capacity of host cells, and decreased the level of inflammation. Overall, DHA and EPA enhanced the host immunity and have potential as broad-spectrum anticoronavirus agents.
RESULTS
Antiviral effects of the five FAs on PEDV, TGEV, and PDCoV
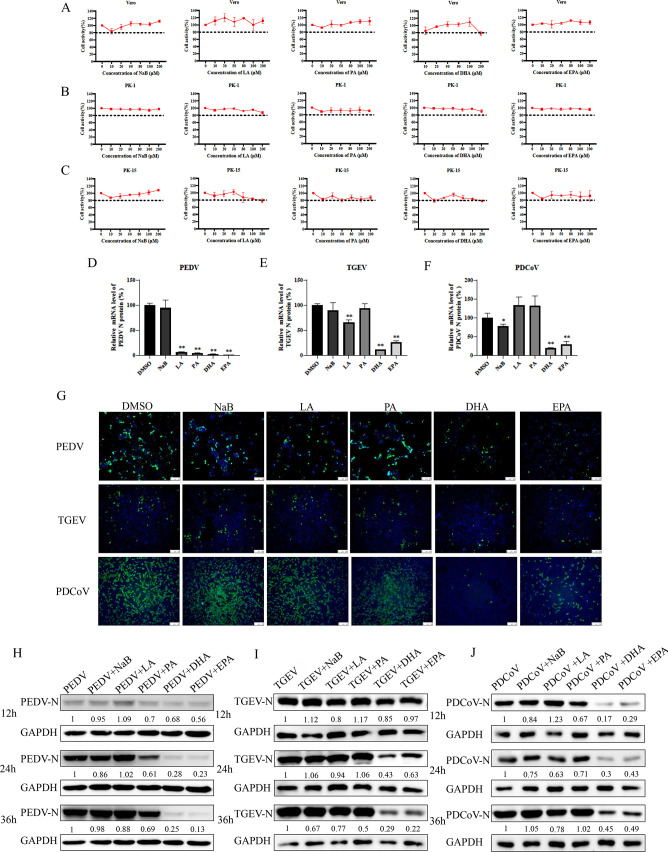
FAs at concentrations below 200 µM showed no significant cytotoxicity (Fig. 1A through C). To observe antiviral activity, 100 µM of each FA was added to vero cells, LLC-PK1 cells, and PK-15 cells at the time of virus infection (Fig. 1D through F) and qRT-PCR was used to examine the expression of the viral N gene. At the gene level, NaB showed weak inhibition of TGEV and PDCoV but had no significant effect on PEDV; LA significantly inhibited PEDV and PDCoV; PA was effective against PEDV; and DHA and EPA inhibited all three porcine coronaviruses. The addition of DHA or EPA significantly reduced the strong fluorescence signal under infection with PEDV, TGEV, or PDCoV, while NaB, LA, and PA had no significant effect, as observed by an immunofluorescence assay (IFA) (Fig. 1G). Western blotting results also showed significant antiviral activity of DHA and EPA against all three coronaviruses (Fig. 1H through J).
Fig 1.
Cytotoxicity and antiviral effects of five fatty acids. (A–C) Vero cells, PK-15 cells, and LLC-PK1 cells were treated with 0–200 μM of NaB, LA, PA, DHA, or EPA for 24 h. The relative cell viability was evaluated by the Cell Counting Kit-8 (CCK-8) according to the manufacturer’s instructions. (D–F) Vero cells, PK-15 cells, or LLC-PK1 cells were infected with TGEV, PEDV, or PDCoV [multiplicity of infection (MOI) = 0.01], respectively, for 1 h and treated with NaB, LA, PA, DHA, or EPA. The virus N gene load at 12 h was quantified by qRT-PCR. (G) At 24 h postinfection, TGEV, PEDV, or PDCoV (MOI = 0.1) replication in PK-15, vero, or LLC-PK1 cells was determined by IFA. Cells were fixed with 4% paraformaldehyde and stained with an anti-TGEV-N, anti-PEDV-N, or anti-PDCoV-N antibody and a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (green); nuclei were stained blue with 4′,6-diamidino-2-phenylindole (DAPI). (H–J) Vero cells, PK-15 cells, or LLC-PK1 cells were infected with TGEV, PEDV, or PDCoV (MOI = 0.01), respectively, for 1 h and treated with 100 µM of fatty acids. Cells were collected after 12 h, 24 h, and 36 h for western blotting analysis. Results are presented as mean ± SD, and the asterisks indicate significance compared to dimethylsulfoxide (DMSO)-treated cells (Student’s t test, *P < 0.05; **P < 0.01).
Fig 2.
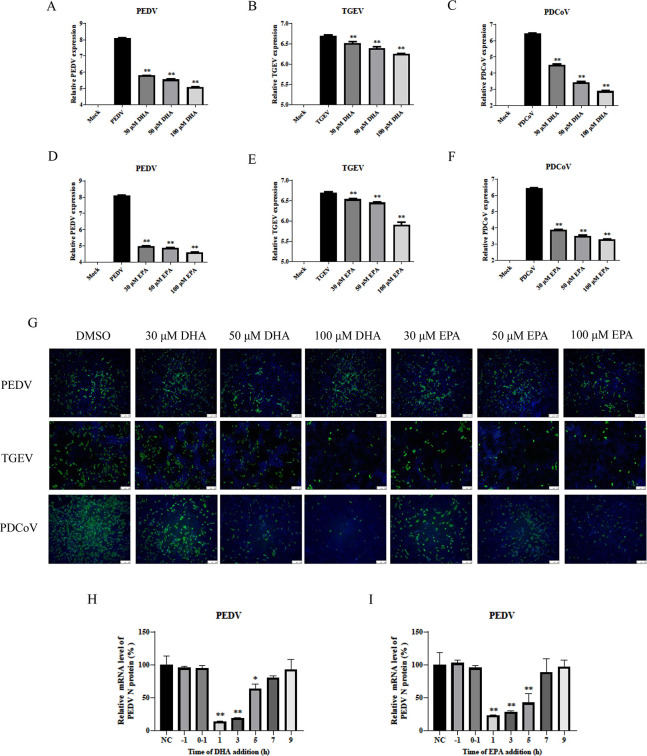
DHA and EPA inhibited PEDV in a dose-dependent manner. (A–F) PEDV, TGEV, and PDCoV were infected in vero cells, PK-15 cells, and LLC-PK1 cells, respectively; 30 µM, 50 µM, and 100 µM of DHA and EPA were added, and replication in vero cells, PK-15 cells, or LLC-PK1 cells was determined by qPCR at 12 h. (G) Replication of TGEV, PEDV, or PDCoV (MOI = 0.1) in vero cells, PK-15 cells, or LLC-PK1 cells was determined by IFA at 24 h. Cells were fixed with 4% paraformaldehyde and stained with anti-TGEV-N, anti-PEDV-N, or anti-PDCoV-N antibody and FITC-conjugated secondary antibody (green); nuclei were stained blue with DAPI. (H–I) In postinfection experiments, cells were first infected with virus for 1 h, and then DHA or EPA was added at different time points (1, 3, 5, 7, and 9 h). Cells were collected after 12 h for qRT-PCR. P values <0.05 were considered statistically significant. Bars represent standard deviations. Results were presented as mean ± SD, and the asterisks indicate significance relative to controls (Student’s t test, *P < 0.05; **P < 0.01).
Dose dependency of antiviral effects of DHA and EPA
To understand in detail the efficacy of DHA or EPA against coronaviruses, we treated vero cells, LLC-PK1 cells, and PK-15 cells with 30 µM, 50 µM, or 100 µM of DHA or EPA and observed their inhibitory effects on PEDV, TGEV, and PDCoV infection. The intensity of inhibition of viral replication rose with increasing concentrations of DHA and EPA, as observed by qRT-PCR (Fig. 2A through F) and IFA (Fig. 2G). These results suggested that treatment with either DHA or EPA led to a significant reduction in viral load in a dose-dependent manner. To characterize the effects of DHA and EPA on PEDV infection, we examined the effect of dosing time on viral mRNA levels. DHA and EPA both reduced the viral mRNA levels of PEDV in the coincubation experiment. Furthermore, the degree of viral inhibition decreased as the duration of treatment with DHA or EPA decreased, indicating that DHA and EPA had inhibitory effects on viral replication that became more pronounced as the maintenance time increased (Fig. 2H and I).
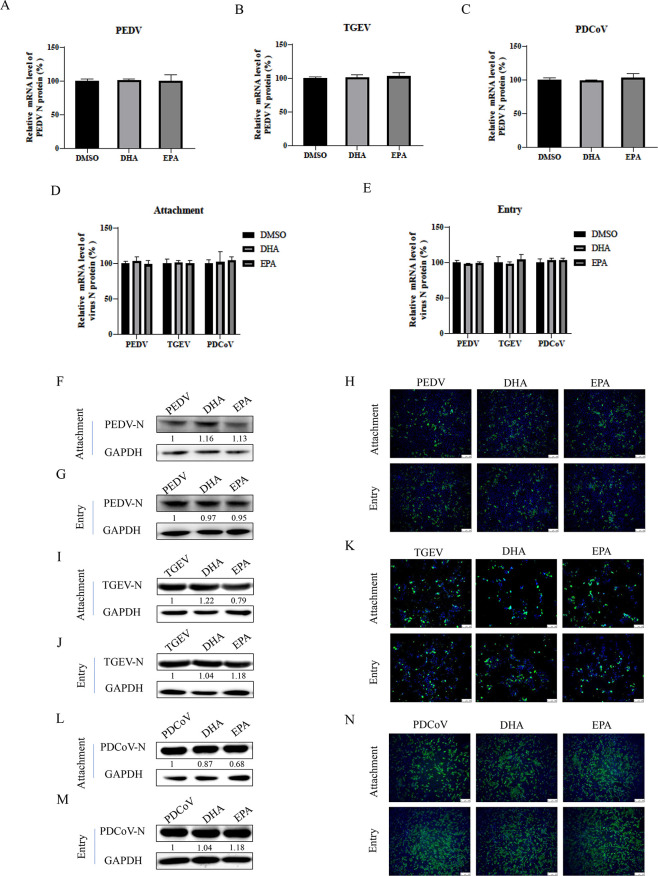
Attachment and entry of PEDV, TGEV, and PDCoV unaffected by DHA or EPA
To determine whether DHA or EPA had a direct viricidal effect on each coronavirus, we incubated PEDV, TGEV, or PDCoV [multiplicity of infection (MOI) = 0.05] with 100 µM of DHA or EPA for 1 h at room temperature prior to infection of vero cells, LLC-PK1 cells, and PK-15 cells. The mRNA levels of each viral N protein gene were measured by qRT-PCR at 12 h postinfection. We found that neither DHA nor EPA directly inhibited the infectivity of PEDV, TGEV, or PDCoV (Fig. 3A through C). To investigate the stage of action of DHA and EPA against PEDV, TGEV, and PDCoV, we performed virus attachment and entry experiments. After vero cells, LLC-PK1 cells, and PK-15 cells were infected with PEDV, TGEV, or PDCoV (MOI = 10), DHA or EPA was added and the cells were incubated at 4°C for 1 h to complete attachment, followed by incubation at 37°C for 1 h for invasion. The results of qRT-PCR showed that addition of DHA or EPA had no effect on the mRNA levels of the viral N protein gene from PEDV, TGEV, or PDCoV following completion of viral attachment and entry (Fig. 3D and E). Consistently, western blotting and IFA results also showed no significant effects of DHA or EPA on N protein levels of PEDV (Fig. 3F through H), TGEV (Fig. 3I through K), or PDCoV (Fig. 3L through N). These results demonstrated that DHA and EPA had no effect on the attachment and entry of PEDV, TGEV, and PDCoV and that their main effects are in the viral replication phase.
Fig 3.
DHA or EPA has no viricidal effect on TGEV, PEDV, and PDCoV. (A–C) PEDV, TGEV, or PDCoV (MOI = 0.1) with 100 µM of DHA or EPA for 1 h at room temperature before infection. Cells were collected after 12 h for qRT-PCR. (D and E) DHA or EPA (100 µM) was added to vero cells infected with virus (MOI = 10), and the cells were collected after incubation at 4°C for 1 h or at 37°C for 1 h. The expression levels of viral mRNAs were analyzed by qRT-PCR. (F–N) The N protein expression level was analyzed by western blot and indirect immunofluorescence assays. P values <0.05 were considered statistically significant. Bars represent standard deviations. Results were presented as mean ± SD, and the asterisks indicate significance relative to controls (Student’s t test, *P < 0.05; **P < 0.01).
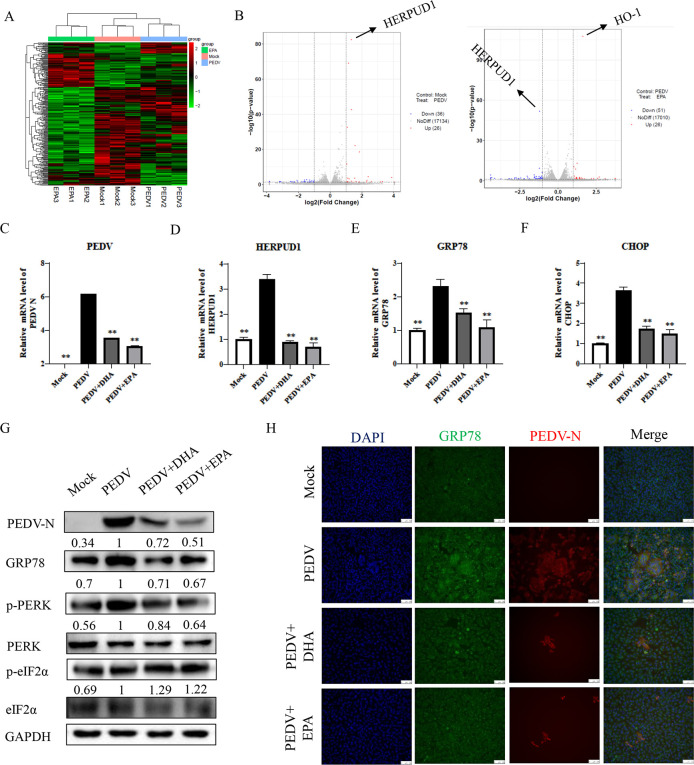
Alleviation of PEDV-induced ERS with DHA or EPA
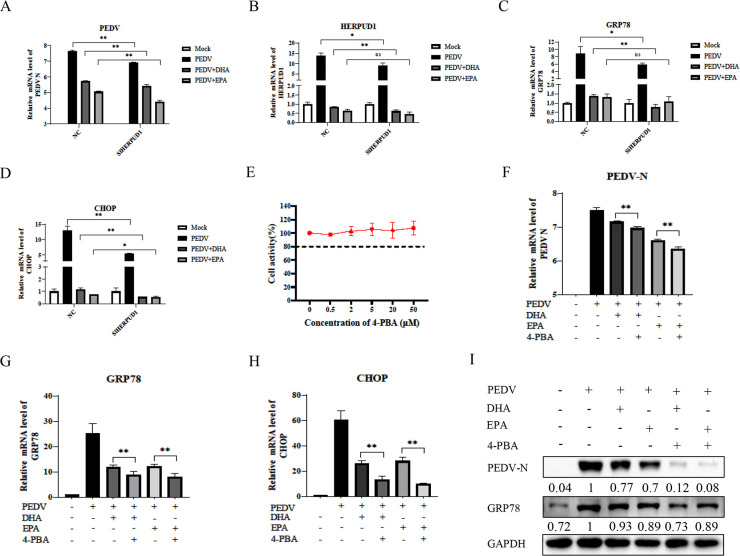
We used vero cells that were infected by PEDV followed by 100 µM EPA addition, and cell samples were harvested after 12 h for transcriptomic analysis. Using transcriptomic analysis of vero cells, we determined that PEDV infection led to upregulation of 26 genes and downregulation of 36 genes, whereas EPA treatment of vero cells led to upregulation of 26 genes and downregulation of 51 genes. The most significantly upregulated gene in the PEDV-infected group was HERPUD1, which showed significantly decreased expression under EPA treatment. HERPUD1 is a quality control protein and an indicator of occurrence of ERS, and its significant downregulation by EPA indicated that EPA has a regulatory effect on the ER. In addition, in the EPA-treated vero cells infected with PEDV, we also observed significant upregulation of HO-1. HO-1, as a key protein in the antioxidant pathway, plays an important role in the protection of organisms (Fig. 4A and B). Next, we performed a qRT-PCR analysis of the expression of the ERS-related genes HERPUD1, GRP78, and CHOP of vero cells at 24 h post-PEDV infection. While PEDV infection significantly upregulated all three ERS-related genes, cotreatment with DHA or EPA attenuated this upregulation (Fig. 4C through F). We found consistent results in western blotting and IFA assays of PEDV-N and GRP78 protein levels. Additionally, we examined the phosphorylation level of protein kinase R-like ER kinase (PERK) and found that PEDV promoted PERK phosphorylation, while treatment with DHA or EPA decreased it, suggesting that DHA and EPA may inhibit the replication of PEDV by attenuating the PERK pathway of ERS (Fig. 4G and H).
Fig 4.
The effect of DHA and EPA on PEDV-induced ERS. (A and B) Total cell extracts from uninfected cells, vero cells infected with PEDV (MOI = 0.1) for 12 h, or vero cells infected with PEDV (MOI = 0.1) and EPA for 12 h were subjected to a transcriptomic analysis. Volcano plots show pairwise ratio comparisons and the corresponding P values obtained from Student’s t tests, which were derived from the mean of two independent experiments for each condition and three technical replicates per sample. Blue and red colors indicate differentially expressed mRNA (ratio >0, P value of −log10 (P) ≥ 1.3). (C–H) PEDV-infected vero cells treated with DHA and EPA were collected at 24 h. The expression levels of PEDV-N and ER-related mRNAs were analyzed by qRT-PCR, western blot, and indirect immunofluorescence assays. Results were presented as mean ± SD, and the asterisks indicate significance relative to controls (Student’s t test, *P < 0.05; **P < 0.01).
DHA- and EPA-mediated inhibition of PEDV replication by alleviation of ERS
PEDV infection caused severe ERS and the fact that the expression of PEDV and ERS-related genes is also decreased after HERPUD1 silencing suggests that ERS is involved in PEDV replication and that PEDV replication decreases with ERS inhibition (Fig. 5A through D). Next, we sought to examine the effect of the ERS inhibitor 4-PBA on PEDV and ERS. We first verified that the host vero cells were viable under 4-PBA treatment, showing that 50 µM 4-PBA did not significantly damage the cells (Fig. 5E). We used 4-PBA to co-treat vero cells with DHA or EPA upon PEDV infection to observe changes in ERS and showed that 4-PBA significantly reduced the gene and protein expressions of PEDV-N, GRP78, and CHOP and showed synergistic effects with DHA and EPA (Fig. 5F through I).
Fig 5.
Effects of HERPUD1 and ERS agonists and inhibitors on PEDV replication and DHA/EPA effect. (A–D) Vero cells were infected with PEDV to control plasmid and siRNA for HERPUD1 treated with DHA or EPA, cells were collected at 24 h, and the expression levels of PEDV-N and ERS-related mRNAs were analyzed by qRT-PCR. (E) Vero cells were treated with 0–50 μM 4-PBA for 24 h. The relative cell viability was evaluated by the CCK-8 kit according to the manufacturer’s instructions. (F–I) PEDV-infected vero cells treated with DHA, EPA, and 4-PBA were collected at 24 h. Expression levels of PEDV-N and ERS-related mRNAs were analyzed by qRT-PCR and western blot. Results were presented as mean ± SD, and the asterisks indicate significance relative to controls (Student’s t test, *P < 0.05; **P < 0.01).
DHA and EPA mediated reductions in PEDV-induced inflammation and ROS levels and improved the antioxidant capacity
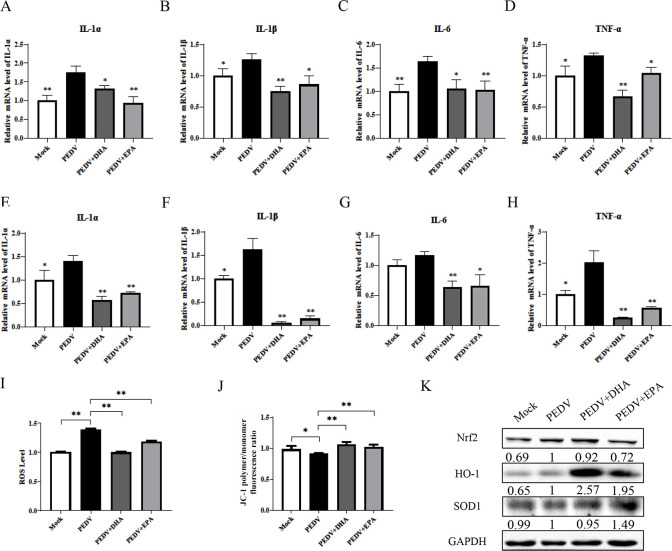
PEDV infection induced increases in inflammation-related factors interleukin IL−1α, IL-1β, IL-6, and tumor necrosis factor (TNF-α) in IPEC-J2 cells (Fig. 6A through D) and vero cells (Fig. 6E through H), which were significantly alleviated by cotreatment with DHA or EPA. In terms of antioxidant capacity, treatment with DHA or EPA significantly reduced the reactive oxygen species (ROS) levels (Fig. 6I), improved the mitochondrial function (Fig. 6J), and significantly increased the protein expression of HO-1 and SOD1 but had no significant effect on that of Nrf2 (Fig. 6K) in vero cells.
Fig 6.
Effects of DHA/EPA on the host inflammatory response and oxidation levels induced by PEDV infection. (A–H) The expression levels of inflammation-related mRNAs in vero and IPEC-J2 cells were analyzed by qRT-PCR. (I and J) Reactive oxygen species (ROS) and mitochondrial membrane potential were detected by flow cytometry after PEDV infection and DHA/EPA supplementation. (K) The expression level of peroxiredoxins was analyzed by qRT-PCR and western blot. Results were presented as mean ± SD, and the asterisks indicate significance relative to controls (Student’s t test, *P < 0.05; **P < 0.01).
DISCUSSION
MERS-CoV and the recently emergent SARS-CoV-2 all cause severe respiratory syndromes in humans and are highly infectious. Lipids are an integral part of viral life and are a target of intensive antiviral research. Viruses need to mobilize large amounts of lipids to enter the host, and changes in the microenvironment of lipid rafts facilitate viral entry (20). Upon entry into the host, the virus hijacks and reorganizes the host’s lipid metabolism, creating conditions conducive to its own replication. Viral infection significantly increases host lipid levels, and increased levels of triglycerides, LDs, and expression of genes related to lipid synthesis lead to significant changes in viral load (9, 21). Different lipids also play unique roles. LDs are oxidized to produce ATP and carry proteins and FAs for viral replication (22). A large amount of viral replication is accompanied by a rise in expression of sterol-regulatory element binding proteins, fatty acid synthase (FASN), but can be significantly inhibited using gene silencing or inhibitors of siRNA or C75 (18, 23, 24). Fatty acid-synthesizing and catabolic enzymes regulate fatty acid levels, which in turn affect viral replication in the host.
FAs are extensively involved in the viral life cycle, and LDs can carry acyl-CoA diacylglycerol acyltransferase and FASN to provide FAs for viral replication. NaB was shown to have both anti-inflammatory and antioxidant effects (25, 26). LA has antibacterial effects, and its suppression of inflammation levels to some extent suggests that it also improves immunity (27). In this study, we observed that NaB and LA had some effect on the mRNA levels of the PEDV N gene. It was previously reported that PA can act as a substrate to promote lipid synthesis and inflammatory responses, leading to increased viral replication (28). However, we observed the opposite effect of PA, which may be due to dosage issues or saturation of viral replication. As polyunsaturated FAs, DHA and EPA may improve physical health to some extent by increasing the host antioxidant levels and anti-inflammatory activity, but they have not been previously reported to have antiviral activity (29).
All three porcine coronaviruses, PEDV, TGEV, and PDCoV, showed prominent concentration-dependent inhibition by DHA and EPA. Previously, it was shown that polyunsaturated FAs act by blocking the entry of SARS-CoV-2 (12). Verification of the phase of action of DHA and EPA on viral replication in this study showed no significant role of DHA or EPA in the attachment or entry of any of the porcine coronaviruses. This result suggested that DHA and EPA might not exert their effects by regulating lipids on membranes, mobilizing lipid rafts, or affecting membrane receptors. Similarly, DHA and EPA showed no direct effect on PEDV infectivity, suggesting that DHA and EPA are unable to bind or directly interact with viral proteins. When we tested the addition of DHA and EPA at different times to explore their mechanism and phase of action, we found that the inhibitory effect became more pronounced as the maintenance time of the DHA and EPA coincubation period after virus entry was increased. This finding demonstrated that DHA and EPA exerted their inhibitory effects on PEDV in the replication phase.
ERS is an important component of the viral life cycle. Viral infection activates ERS, leading to massive accumulation of misfolded proteins. HERPUD1, as a stabilizing protein of the ER, can be utilized to reflect the ER status (30), and its significant downregulation by EPA indicated that EPA may have a regulatory effect on the ER. Meanwhile, a gene ontology (GO) analysis showed that CHOP-ATF3 was significantly activated under PEDV infection, indicating that ERS and its related pathways play an important role in PEDV infection. The host regulates ERS through the UPR, in which three pathways, PERK, ATF6, and IRE, are particularly critical. The major activation pathways vary among different viruses. For example, flaviviruses mainly activate the ATF6 pathway (10), while PEDV mainly activates the PERK pathway (31). We observed that DHA and EPA attenuated the marked increase in PERK phosphorylation and reduced the expression of GRP78, CHOP, and HERPUD1, suggesting that DHA and EPA inhibit viral replication by suppressing ERS (32). The ERS inhibitor 4-PBA significantly decreased the PEDV viral load after coactivation with DHA or EPA, indicating that the alleviation of ERS inhibited PEDV replication to some extent.
HO-1 is a critical protein in the antioxidant pathway. It has been shown that HO-1 binds to IRF3 and promotes the production of IFN to resist IAV (33), demonstrating the important role of HO-1 in viral infection. In our study, DHA and EPA significantly increased the HO-1 expression, which was consistent with the transcriptomic results. Meanwhile, we observed that DHA and EPA alleviated inflammation, decreased the production of cytokines IL-1α, IL-1β, IL-6, and TNF-α, decreased the ROS levels (34), elevated the mitochondrial membrane potential, and improved the mitochondrial function.
In summary, we observed the effects of five FAs with different chain lengths and levels of saturation on the replication of PEDV, TGEV, and PDCoV in vero cells, PK-15 cells, or LLC-PK1 cells. DHA and EPA were found to inhibit all three swine coronaviruses by alleviating ERS, increasing the antioxidant capacity, and reducing the level of inflammation caused by viral infection. The findings of this study provide a new direction for research on vaccines and drugs to fight porcine coronavirus infection.
MATERIALS AND METHODS
Cells and viruses
Vero cells are transformed cells obtained from a normal adult African green monkey kidney in 1962. Vero cells are anchorage-dependent fibroblasts that do not express the antiviral protein interferon and support the proliferation of a variety of viruses. We cultured vero cells in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) at 37°C in a 5% CO2 incubator. IPEC-J2 is a porcine small intestinal epithelial cell and a major cell for PEDV infection. IPEC-J2 cells were cultured in DMEM supplemented with 10% (vol/vol) FBS. PEDV (isolate PEDV-AH2012) was propagated in vero cells and IPEC-J2 cells supplemented with DMEM containing 7.5 µg/mL trypsin (Sigma, St. Louis, MO, USA). The Lilly Laboratories porcine kidney proximal tubular epithelial (LLC-PK1) cell line was derived from a Hampshire pig kidney. Porcine kidney (PK)-15 cells were established from clones of PK-1 cells. LLC-PK1 cells and PK-15 cells were cultured in DMEM supplemented with 10% (vol/vol) FBS. PDCoV (strain NH isolate passage 10) and TGEV (strain H16) were cultivated in LLC-PK1 cells or PK-15 cells in DMEM containing 2% FBS. The cell line was chosen in accordance with the susceptibility of the virus.
Reagents and antibodies
Three of the selected FAs, namely, LA, DHA, and EPA, were diluted in dimethylsulfoxide (DMSO) to 100 mM, filter-sterilized, and stored at –80°C. NaB was diluted in water to 100 µM. PA and bovine serum albumin (BSA) chelate were diluted in phosphate-buffered saline (PBS) to 20 mM. Five fatty acids were used at a concentration of 100 µM, with DHA and EPA also used at 30 and 50 µM. DMSO and BSA were added as a control. The anti-PEDV-N, anti-PDCoV-N, and anti-TGEV-N antibodies were prepared in our laboratory. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from Proteintech (Wuhan, China). Horseradish peroxidase (HRP)-anti-rabbit IgG, HRP-anti-mouse IgG, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG, and FITC-conjugated goat anti-mouse IgG were purchased from KPL (Mountain View, CA, USA).
Cell viability assay
The cytotoxic effects of NaB, LA, PA, DHA, and EPA on vero cells, LLC-PK1 cells, and PK-15 cells were assessed using the Cell Counting Kit-8 (CCK-8, APExBIO). Cells were seeded onto a 96-well cell plate and cultured until they grew into a single layer. The culture medium was removed, washed with PBS twice, and replaced with a cell maintenance solution in the presence of serial dilutions of FAs. The CCK-8 solution (10 µL) was added to each well for 4 h at 37°C in the dark. Subsequently, the samples were analyzed by reading the optical density at 450 nm using a microplate reader (Bio-Rad, Hercules). All assays were performed in triplicate.
Antiviral activity of five FAs
To assess the antiviral effects of NaB, LA, PA, DHA, and EPA against PEDV, TGEV, or PDCoV infection, cells were washed with PBS twice and then infected at an MOI of 0.01 for 1 h. The cells were then washed with PBS twice to remove unbound virus particles and treated with different concentrations of FAs (diluted in a cell maintenance solution) for 12 h. The cells were collected for western blotting and quantitative real time (qRT)-PCR.
Virus replication assays under DHA and EPA
To assess the inhibitory effects of DHA and EPA on virus replication, cells were seeded onto a 24-well plate. In pretreatment experiments, cells were incubated with DHA or EPA for 1 h, after which the compound was removed and the cells were infected with PEDV, TGEV, and PDCoV. In the coincubation experiments, DHA, EPA, and virus were added simultaneously to cells, and 1 h later, the medium was removed, and cells were washed and cultured with DMEM. In the post-treatment experiments, cells were infected with virus for 1 h, washed to remove the viral inoculum, and DHA or EPA was added at 1, 3, 5, 7, and 9 h postinfection. In the pretreatment, coincubation, and postinfection experiments, cells were infected with PEDV at an MOI of 0.01 and DHA or EPA was added at a final concentration of 100 µM. The cells were collected 12 h postinfection with PEDV, TGEV, or PDCoV and were used for qRT-PCR. Cells were infected with PEDV and 50 µM 4-PBA and DHA or EPA were added, and cells were charged for 24 h to detect changes in ERS using qRT-PCR. As a negative control, cells were treated with the same volume of DMSO as that used for FAs.
Virus attachment assays under DHA and EPA
Cells were seeded onto a 12-well plate and cultured until they grew into a monolayer. Cells were then chilled at 4°C and infected with PEDV, TGEV, and PDCoV (MOI = 10) in the presence of DHA and EPA (100 µM) at 4°C for 1 h to allow the virus to attach to the cell surface. Unbound virus was removed with cold PBS. The viral genome load was then quantified by qRT-PCR amplification of the viral N gene and measurement of N protein levels by western blotting and IFA.
Virus entry assays under DHA and EPA
Cells were first grown in a 12-well plate to 90% confluency and then infected with PEDV, TGEV, and PDCoV (MOI = 10) with simultaneous addition of DHA or EPA to 100 µM at 37°C for 1 h to allow virus entry. They were then washed with PBS (pH = 2.0) three times to remove bound, non-internalized virion particles. The total protein was prepared from these cells for western blotting, IFA, and qRT-PCR analyses.
Western blotting
Cells were lysed with RIPA buffer (Beyotime). Lysates were mixed with a 4× protein buffer (Solarbio) and boiled for 10 min. Proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane (NC, GE Amersham Biosciences). The membranes were blocked with 2% (wt/vol) nonfat milk for 2 h at room temperature, washed with PBST (PBS containing 0.1% Tween 20), and then incubated for 1 h with a primary antibody (anti-TGEV-N, anti-PEDV-N, anti-PDCoV-N, or GAPDH). After repeated washes with PBST, the membranes were further incubated for 1 h with a secondary antibody (HRP-anti-rabbit IgG or HRP-anti-mouse IgG). The target protein bands were then analyzed using a LI-COR Odyssey infrared image scanner.
IFA
Cells were fixed with 4% paraformaldehyde at room temperature for 30 min, washed with PBS three times, blocked with 5% nonfat dry milk in PBS for 1 h, and incubated with anti-TGEV-N, anti-PEDV-N, or anti-PDCoV-N antibody for 1 h at 37°C. The cells were then washed with PBS three times, incubated with an FITC-conjugated secondary antibody for 1 h at 37°C, and then incubated with 4′,6-diamidino-2-phenylindole (DAPI, Solarbio) for 10 min after washing with PBS three times. The stained cells were examined using a differential fluorescence microscope (Nikon).
QRT-PCR
Total RNA was extracted from the cells using a TRIzol reagent (Vazyme) in accordance with the manufacturer’s instructions. A HiScript II 1st strand cDNA synthesis kit (Vazyme) was used to synthesize cDNA. The gene expression was assessed in triplicate by SYBR (Vazyme) using a Light Cycler 96 real-time PCR system (Roche Diagnostics). Primer sequences are shown in Table 1. β-Actin or GAPDH was used as an internal control. Data were analyzed using the cycle threshold (2−∆∆Ct) method.
TABLE 1.
Sequences of the primers used for qRT-PCR
| Name | Sequence (5′−3′) |
|---|---|
| TGEV-N | Forward: GGAATAGGTAACAGGGATCAGC |
| Reverse: TCAGGAAGCTCTTTACGTTGG | |
| PEDV-N | Forward: ACAAATCCAGAGCCACTTCG |
| Reverse: TCGACAAATTCCGCATCTCC | |
| PDCoV-N | Forward: ACTGAGAAGACGGGTATGGC |
| Reverse: TTAGTTGGTTTGGTGGGTGG | |
| β-Actin-monkey | Forward: TGAAGTGTGACGTGGACATC |
| Reverse: CTTGATTTTCATCGTGCTGGG | |
| β-Actin-pig | Forward: CTCCATCATGAAGTGCGACGT |
| Reverse: GTGATCTCCTTCTGCATCCTGTC |
Transcriptomic analysis
Vero cells were infected with PEDV (0.1 MOI), or mock-infected, EPA (100 µM) was added, and the mixture was incubated at 37°C for 12 h in a DMEM medium. Total cellular RNAs from the virus-infected and mock-infected groups (n = 3) were collected. The gene expression following PEDV infection was analyzed using the RNA sequencing (RNA-Seq) technology. The differentially expressed genes (DEGs) between PEDV-infected and mock-infected samples were submitted to the DAVID server for pathway enrichment and cluster analysis. A gene ontology (GO) enrichment analysis of DEGs was implemented using the GOseq R package. KOBAS software was used to examine the statistical enrichment of DEGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Flow cytometry
Vero cells were incubated with CellROX Green (C10444, Invitrogen) at a ratio of 1:1,000 for 30 min to detect the levels of ROS. Data were acquired by flow cytometry on the BD FACSVerse (BD Biosciences, Brea, CA, USA) and analyzed using the BD FACSuite.
Statistical analysis
All data are presented as means ± standard deviation (SD) from triplicate experiments. The significance of differences between groups was analyzed using the Student’s t test with the GraphPad Prism 6 software. P values <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program of China (Grant No. 2021YFD1801104), the National Natural Science Foundation of China (32072808, 32272996, and 32202787), and the Jiangsu Agricultural Science and Technology Innovation Fund (CX(23)1029).
Contributor Information
Xiaojing Yang, Email: yangxj@njau.edu.cn.
Bin Li, Email: libinana@126.com.
Tom Gallagher, Loyola University Chicago, Maywood, Illinois, USA .
REFERENCES
- 1. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 580:265–269. doi: 10.1038/s41586-020-2202-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang CL, et al. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 588:E6. doi: 10.1038/s41586-020-2951-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24:490–502. doi: 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y. 2015. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis 62:575–580. doi: 10.1111/tbed.12399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xia L, Yang Y, Wang J, Jing Y, Yang Q. 2018. Impact of TGEV infection on the pig small intestine. Virol J 15:102. doi: 10.1186/s12985-018-1012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang D, Fang L, Xiao S. 2016. Porcine epidemic diarrhea in China. Virus Res 226:7–13. doi: 10.1016/j.virusres.2016.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan Z, Lu J, Wang N, He WT, Zhang L, Zhao W, Su S. 2020. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence 11:707–718. doi: 10.1080/21505594.2020.1771980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Q, Vlasova AN, Kenney SP, Saif LJ. 2019. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol 34:39–49. doi: 10.1016/j.coviro.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dias SSG, Soares VC, Ferreira AC, Sacramento CQ, Fintelman-Rodrigues N, Temerozo JR, Teixeira L, Nunes da Silva MA, Barreto E, Mattos M, de Freitas CS, Azevedo-Quintanilha IG, Manso PPA, Miranda MD, Siqueira MM, Hottz ED, Pão CRR, Bou-Habib DC, Barreto-Vieira DF, Bozza FA, Souza TML, Bozza PT, Fouchier RAM. 2020. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog 16:e1009127. doi: 10.1371/journal.ppat.1009127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao Y, Hu JH, Liang XD, Chen J, Liu CC, Liu YY, Cheng Y, Go YY, Zhou B. 2021. Curcumin inhibits classical swine fever virus replication by interfering with lipid metabolism. Vet Microbiol 259:109152. doi: 10.1016/j.vetmic.2021.109152 [DOI] [PubMed] [Google Scholar]
- 11. Ma S, Mao Q, Chen W, Zhao M, Wu K, Song D, Li X, Zhu E, Fan S, Yi L, Ding H, Zhao M, Chen J. 2019. Serum lipidomics analysis of classical swine fever virus infection in piglets and emerging role of free fatty acids in virus replication in vitro. Front Cell Infect Microbiol 9:410. doi: 10.3389/fcimb.2019.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goc A, Niedzwiecki A, Rath M. 2021. Polyunsaturated omega-3 fatty acids inhibit ACE2-controlled SARS-CoV-2 binding and cellular entry. Sci Rep 11:5207. doi: 10.1038/s41598-021-84850-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jackman JA, Boyd RD, Elrod CC. 2020. Medium-chain fatty acids and monoglycerides as feed additives for pig production: towards gut health improvement and feed pathogen mitigation. J Anim Sci Biotechnol 11:44. doi: 10.1186/s40104-020-00446-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cybulsky AV. 2017. Endoplasmic reticulum stress, the unfolded protein response and autophagy in kidney diseases. Nat Rev Nephrol 13:681–696. doi: 10.1038/nrneph.2017.129 [DOI] [PubMed] [Google Scholar]
- 15. Bettigole SE, Glimcher LH. 2015. Endoplasmic reticulum stress in immunity. Annu Rev Immunol 33:107–138. doi: 10.1146/annurev-immunol-032414-112116 [DOI] [PubMed] [Google Scholar]
- 16. Fung TS, Liao Y, Liu DX. 2016. Regulation of stress responses and translational control by coronavirus. Viruses 8:184. doi: 10.3390/v8070184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ge M, Luo Z, Qiao Z, Zhou Y, Cheng X, Geng Q, Cai Y, Wan P, Xiong Y, Liu F, Wu K, Liu Y, Wu J. 2017. HERP binds TBK1 to activate innate immunity and repress virus replication in response to endoplasmic reticulum stress. J Immunol 199:3280–3292. doi: 10.4049/jimmunol.1700376 [DOI] [PubMed] [Google Scholar]
- 18. Tanner JE, Alfieri C. 2021. The fatty acid lipid metabolism nexus in COVID-19. Viruses 13:90. doi: 10.3390/v13010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gassen NC, Niemeyer D, Muth D, Corman VM, Martinelli S, Gassen A, Hafner K, Papies J, Mösbauer K, Zellner A, Zannas AS, Herrmann A, Holsboer F, Brack-Werner R, Boshart M, Müller-Myhsok B, Drosten C, Müller MA, Rein T. 2019. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection. Nat Commun 10:5770. doi: 10.1038/s41467-019-13659-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dou X, Li Y, Han J, Zarlenga DS, Zhu W, Ren X, Dong N, Li X, Li G. 2018. Cholesterol of lipid rafts is a key determinant for entry and post-entry control of porcine rotavirus infection. BMC Vet Res 14:45. doi: 10.1186/s12917-018-1366-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu B, Gao TT, Fu XY, Xu ZH, Ren H, Zhao P, Qi ZT, Qin ZL. 2021. PTEN lipid phosphatase activity enhances dengue virus production through Akt/FoxO1/Maf1 signaling. Virol Sin 36:412–423. doi: 10.1007/s12250-020-00291-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heaton NS, Randall G. 2011. Multifaceted roles for lipids in viral infection. Trends Microbiol 19:368–375. doi: 10.1016/j.tim.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee W, Ahn JH, Park HH, Kim HN, Kim H, Yoo Y, Shin H, Hong KS, Jang JG, Park CG, Choi EY, Bae J-S, Seo Y-K. 2020. COVID-19-activated SREBP2 disturbs cholesterol biosynthesis and leads to cytokine storm. Signal Transduct Target Ther 5:186. doi: 10.1038/s41392-020-00292-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chu J, Xing C, Du Y, Duan T, Liu S, Zhang P, Cheng C, Henley J, Liu X, Qian C, Yin B, Wang HY, Wang R-F. 2021. Pharmacological inhibition of fatty acid synthesis blocks SARS-CoV-2 replication. Nat Metab 3:1466–1475. doi: 10.1038/s42255-021-00479-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li G, Lin J, Zhang C, Gao H, Lu H, Gao X, Zhu R, Li Z, Li M, Liu Z. 2021. Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes 13:1968257. doi: 10.1080/19490976.2021.1968257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed] [Google Scholar]
- 27. Yu H, Byun Y, Chang PS. 2022. Lipase-catalyzed two-step esterification for solvent-free production of mixed lauric acid esters with antibacterial and antioxidative activities. Food Chem 366:130650. doi: 10.1016/j.foodchem.2021.130650 [DOI] [PubMed] [Google Scholar]
- 28. Zhang X, Zhuang J, Huang L, Zhang X. 2022. Palmitic amide triggers virus life cycle via enhancing host energy metabolism. Front Microbiol 13:924533. doi: 10.3389/fmicb.2022.924533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Das UN. 2020. Can bioactive lipids inactivate coronavirus (COVID-19)? Arch Med Res 51:282–286. doi: 10.1016/j.arcmed.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho DV, Chan JY. 2015. Induction of Herpud1 expression by ER stress is regulated by Nrf1. FEBS Lett 589:615–620. doi: 10.1016/j.febslet.2015.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Y, Zhang Y, Dong W, Gan S, Du J, Zhou X, Fang W, Wang X, Song H. 2023. Porcine epidemic diarrhea virus activates PERK-ROS axis to benefit its replication in vero E6 cells. Vet Res 54:9. doi: 10.1186/s13567-023-01139-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lepretti M, Martucciello S, Burgos Aceves MA, Putti R, Lionetti L. 2018. Omega-3 fatty acids and insulin resistance: focus on the regulation of mitochondria and endoplasmic reticulum stress. Nutrients 10:350. doi: 10.3390/nu10030350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma LL, Zhang P, Wang HQ, Li YF, Hu J, Jiang JD, Li YH. 2019. Heme oxygenase-1 agonist CoPP suppresses influenza virus replication through IRF3-mediated generation of IFN-alpha/beta. Virology 528:80–88. doi: 10.1016/j.virol.2018.11.016 [DOI] [PubMed] [Google Scholar]
- 34. Yuan C, Huang X, Zhai R, Ma Y, Xu A, Zhang P, Yang Q. 2021. In vitro antiviral activities of salinomycin on porcine epidemic diarrhea virus. Viruses 13:580. doi: 10.3390/v13040580 [DOI] [PMC free article] [PubMed] [Google Scholar]