Abstract
A purification procedure for a new kind of extradiol dioxygenase, termed chlorocatechol 2,3-dioxygenase, that converts 3-chlorocatechol productively was developed. Structural and kinetic properties of the enzyme, which is part of the degradative pathway used for growth of Pseudomonas putida GJ31 with chlorobenzene, were investigated. The enzyme has a subunit molecular mass of 33.4 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Estimation of the native Mr value under nondenaturating conditions by gel filtration gave a molecular mass of 135 ± 10 kDa, indicating a homotetrameric enzyme structure (4 × 33.4 kDa). The pI of the enzyme was estimated to be 7.1 ± 0.1. The N-terminal amino acid sequence (43 residues) of the enzyme was determined and exhibits 70 to 42% identity with other extradiol dioxygenases. Fe(II) seems to be a cofactor of the enzyme, as it is for other catechol 2,3-dioxygenases. In contrast to other extradiol dioxygenases, the enzyme exhibited great sensitivity to temperatures above 40°C. The reactivity of this enzyme toward various substituted catechols, especially 3-chlorocatechol, was different from that observed for other catechol 2,3-dioxygenases. Stoichiometric displacement of chloride occurred from 3-chlorocatechol, leading to the production of 2-hydroxymuconate.
The microbial degradation of various chloroaromatics has been described to occur via chlorocatechols as central intermediates, which are further degraded through the modified ortho pathway (45, 49). Chlorocatechol 1,2-dioxygenase, chloromuconate cycloisomerase, dienelactone hydrolase, and maleylacetate reductase fulfill the convergence of the chlorocatechol and catechol degradative pathways.
Alternatively to the intradiol type of ring cleavage, the pathway is initiated by extradiol ring cleavage in some microorganisms. The catechol 2,3-dioxygenases of the meta pathway are able to convert catechol, both isomeric methylcatechols, and 4-chlorocatechol at respectable rates (10, 18, 27, 33, 38, 44, 47, 48). The further degradation of the ring cleavage product of 4-chlorocatechol seems to be a slow process, since all strains degrading a chloroaromatic compound via 4-chlorocatechol through the meta pathway grow slowly on these substrates (1, 2, 15, 16, 29).
However, when 3-chlorocatechol occurs in strains with a meta pathway, the catechol 2,3-dioxygenase is negatively influenced, either by 3-chlorocatechol itself, as a chelating compound resulting in a reversible inactivation (24), or by a reactive acylchloride, the product of the cleavage of 3-chlorocatechol, which causes irreversible inactivation of the enzyme (3). Auto-oxidation of accumulating 3-chlorocatechol leads to a general toxic effect on the cells; therefore, degradation of haloaromatics via meta cleavage of 3-chlorocatechol has been considered impossible.
Recently, we reported that Pseudomonas putida GJ31 degrades chlorobenzene with a generation time of 3 h via 3-chlorocatechol, using the meta pathway without any apparent toxic effects (26, 40). We now present data on the purification and characterization of the unusual meta-cleaving enzyme that converts 3-chlorocatechol productively. Comparison with various previously published catechol 2,3-dioxygenases was performed.
MATERIALS AND METHODS
Organism and culture conditions.
P. putida GJ31 was grown at 30°C in five separate 0.5-liter cultures with mineral medium (9). The growth substrate chlorobenzene was added via the vapor phase. After the cultures had been grown to an optical density at 546 nm of 1.6, the cells were harvested.
Preparation of cell extracts.
Cells were removed by centrifugation at 4,000 × g for 20 min at 4°C. The pellet was resuspended in Tris-HCl buffer (50 mM; pH 7.5) containing 1 mM ascorbate (buffer A). After another centrifugation at 4,000 × g for 20 min, the cells were suspended in 9 ml of the same buffer. Disruption took place at 4°C by one passage through a French pressure cell (140 MPa; Aminco, Silver Spring, Md.). Cell debris were removed by centrifugation at 100,000 × g for 60 min at 4°C.
Enzyme assays.
Catechol 2,3-dioxygenase was measured by a modification of the method of Nozaki (37). The reaction mixture contained 50 μmol of phosphate buffer (pH 7.4) and 0.1 μmol of catechol in a total volume of 1 ml. After addition of enzyme, the increase at 375 nm (corresponding to the formation of 2-hydroxymuconic semialdehyde at ɛ of 36,000 liters/mol · cm) was measured in a silica cuvette with a 1.0-cm light path. One unit of activity was defined as the amount of enzyme required to form 1 μmol of product per min under the conditions of the assay.
The relative velocity of turnover of substituted catechols by the catechol 2,3-dioxygenase of strain GJ31 was estimated with an incubation mixture containing, in a total volume of 1 ml, 0.01 μmol of substrate. The λmax and ɛ values of the products were estimated in this study.
The relative V values were determined as percentages with respect to the reaction with catechol (100%).
pH optimum.
The pH optimum was determined by using a substrate concentration (catechol) of 0.1 mM in 50 mM NaH2PO4-Na2HPO4 buffer (pH 5.0 to 8.8) and 50 mM glycine-NaOH buffer (pH 8.4 to 10.0). Since the molar extinction coefficient of the reaction product of catechol was markedly increased by increasing the pH, it was determined at each pH.
Formation of chloride from 3-chlorocatechol.
The liberation of chloride during turnover of 3-chlorocatechol was determined as follows. The assay mixture contained (in 1 ml) 50 μmol of phosphate buffer (pH 7.4), 0.05 to 0.2 μmol of 3-chlorocatechol, and 20 μl of purified enzyme (≈45 μg of protein). To remove any chloride from the enzyme stock solution, it was dialyzed against 100 mM Tris-H2SO4 (pH 7.5) containing 0.1 mM (NH4)2Fe(SO4)2. After addition of the enzyme, the assay mixture was incubated for 1 h. Chloride was quantitatively determined by the silver chloride method according to the method of Freier (11). To 800 μl of chloride-containing assay mixture, 100 μl of concentrated HNO3 and 100 μl of 0.1 N AgNO3 were added in sequence. After 10 min of incubation in the dark, the absorption at 546 nm was measured.
Protein determinations.
Protein concentrations were determined by the Bradford method (7), with crystalline bovine serum albumin as the standard. During enzyme purification, the eluted protein was detected with a Pharmacia UV-1 monitor at 254 nm.
Enzyme purification.
All enzyme purification steps were carried out at 4°C.
(i) Ammonium sulfate precipitation.
A cold saturated aqueous solution of (NH4)2SO4 (pH 7.0) containing 0.1 mM EDTA was added to crude extract (10 ml) with constant stirring to give 40% saturation. After 30 min of equilibration, the precipitate was collected by centrifugation at 8,000 × g for 20 min. The pellet was dissolved in buffer A to give a total volume of 2.5 ml, and the resulting protein solution was desalted by gel filtration through a PD10 column with buffer A.
(ii) Incubation with hydroxyapatite.
The protein solution from step i was added to a suspension of 5 ml of preequilibrated hydroxyapatite gel in 5 ml of buffer A. After 30 min of incubation, the suspension was separated by centrifugation at 8,000 × g for 2 min. The supernatant containing catechol 2,3-dioxygenase was collected, and the pellet was washed with another 5-ml portion of buffer A. After centrifugation, both supernatants were combined. Because of the significant loss of activity due to complexation of Fe(II) ions, the protein solution was supplemented with (NH4)2Fe(SO4)2 to give a concentration of 0.1 mM.
(iii) Ion-exchange chromatography on Mono-Q.
The pooled supernatants from step ii were applied at 0.5 ml/min to a Mono-Q column (5 by 10 mm) which had been preequilibrated with buffer A. After washing of the column with 30 ml of buffer A at 0.5 ml/min, catechol 2,3-dioxygenase was eluted with 20 ml of buffer A containing NaCl in a linear gradient from 0 to 1 M. Fractions of 0.5 ml were collected at a flow rate of 0.5 ml/min. Six fractions containing the highest level of activity were pooled.
(iv) Storage.
The combined fractions from step iii were stored at −20°C in 100 mM Tris-HCl buffer, pH 7.5, containing 50% glycerol, 150 mM NaCl, and 1 mM ascorbate.
Determination of Mr values.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to determine the subunit Mr values and the purity of the catechol 2,3-dioxygenase. It was performed by the method of Laemmli (25) on 1-mm-thick vertical slab gels (13.5 by 15.5 cm) containing 12.3% (wt/vol) acrylamide in the resolving gels. Electrophoresis was performed at 100 V for 1 h followed by 200 V for 6 h. The apparatus was cooled to 4°C. Protein was detected by silver staining (30). The calibration proteins were bovine albumin (Mr, 66,000), chicken ovalbumin (Mr, 45,000), glyceraldehyde-3-phosphate dehydrogenase (Mr, 36,000), carbonic anhydrase (Mr, 29,000), trypsinogen (Mr, 24,000), and α-lactalbumin (Mr, 14,200) (Sigma Chemical Co., St. Louis, Mo.).
The Mr value of the native protein was determined by means of gel filtration on a Superose 12 fast protein liquid chromatography column (1 by 30 cm) at a flow rate of 0.5 ml/min. The column buffer was 100 mM Tris-HCl, pH 7.5, containing 200 mM NaCl. Horse ferritin (Mr, 443,000), sweet potato β-amylase (Mr, 200,000), yeast alcohol dehydrogenase (Mr, 150,000), bovine albumin (Mr, 67,000), chicken ovalbumin (Mr, 43,000), and chymotrypsinogen A (Mr, 25,000) were used as the reference proteins (Boehringer, Mannheim, Germany, and Sigma). Standards and the purified enzyme were injected in 50-μl samples, and the proteins were detected by monitoring of the eluate at 254 nm.
Isoelectric focusing.
Isoelectric focusing was carried out on 3.8% (wt/vol) polyacrylamide gels containing 2% (vol/vol) ampholytes by the method of O’Farrell (39) with carrier ampholytes in a pH range of 6 to 8.
Absorption spectra.
Spectra were recorded on a Shimadzu Recording Spectrophotometer, model UV-240. Kinetic measurements at a single wavelength were carried out on a UVIKON 820 spectrophotometer (Fa. Kontron, Eching, Germany).
N-terminal amino acid sequencing and database search.
One hundred thirty micrograms of the purified enzyme was applied to an SDS-polyacrylamide gel (1.5 mm thick). The subunits were blotted onto an Amersham Hybond polyvinylidene difluoride membrane according to the Amersham protocol (staining was done with amido black). The N-terminal amino acid sequence was determined with an Applied Biosystems model 477A protein sequencer and an Applied Biosystems model 120A on-line high-performance liquid chromatograph (HPLC).
The N-terminal sequence was compared with sequences in the nonredundant SwissProt-PIR-SPUpdate-GenPept-GPUpdate database (as of 21 May 1997) by using the BLASTP program.
HPLC.
HPLC of substrates and metabolites was conducted as described previously by diode array detection, which allows a determination of the UV spectrum of the respective substrate or metabolite (19).
Chemicals.
2-Pyrone-6-carboxylic acid was prepared by cyclization of the condensation product obtained from diethyl oxalate and ethyl crotonate in the presence of potassium, according to the method of Wiley and Hart (51).
2-Hydroxymuconic acid was prepared in situ by alkaline hydrolysis of 2-pyrone-6-carboxylic acid. For this, an aqueous 10 mM solution of the pyrone was incubated with a fivefold molar excess of sodium hydroxide at 60°C for 5 min. The resulting solution containing the yellow trianion of 2-hydroxymuconic acid (46) was stored on ice prior to use.
Chlorocatechols and 4-fluorocatechol were taken from our laboratory stock. The other chemicals are available commercially.
RESULTS
Purification of catechol 2,3-dioxygenase.
Catechol 2,3-dioxygenase was purified to homogeneity from chlorobenzene-grown cells of P. putida GJ31. The results of a typical enzyme purification procedure are summarized in Table 1. During purification the specific activity of the enzyme increased to 6.48 U/mg of protein, indicating 21.6-fold purification with 72.5% recovery of activity. The specific activity of the GJ31 dioxygenase was very low compared to other catechol 2,3-dioxygenases.
TABLE 1.
Purification of the catechol 2,3-dioxygenase of P. putida GJ31
| Purification step | Vol (ml) | Total protein (mg) | Total activity (U) | Sp act (U mg−1) | Recovery of activity (%) | Purification factor |
|---|---|---|---|---|---|---|
| Crude extract | 10 | 500 | 150.0 | 0.30 | 100 | 1 |
| Ammonium sulfate precipitation (desalted) | 7.5 | 255 | 137.1 | 0.54 | 91.4 | 1.8 |
| Hydroxyapatite supernatanta | 18.5 | 21.5 | 81.4 | 3.79 | 54.3 | 12.6 |
| Mono-Q eluate | 3 | 16.8 | 108.8 | 6.48 | 72.5 | 21.6 |
Before addition of (NH4)2Fe(SO4)2.
In contrast to the enzyme from P. putida PaW1 (mt-2), the catechol 2,3-dioxygenase of strain GJ31 was sensitive to heat treatment. Heating to 40°C for 5 min completely destroyed the activity. In addition, the presence of acetone, which has been shown to be a stabilizing agent for some catechol 2,3-dioxygenases and phenylcatechol 2,3-dioxygenases, led to a dramatic decrease in activity.
Ammonium sulfate precipitation was shown to be a suitable and easy step to purify the protein without significant loss of activity. At a concentration of 40% saturation, the catechol 2,3-dioxygenase precipitated nearly quantitatively. After desalting of the redissolved pellet, batch incubation with hydroxyapatite was used. Almost all proteins other than catechol 2,3-dioxygenase were bound. Because of significant loss of activity due to complexation of Fe(II) ions by hydroxyapatite, the protein solution was supplemented with (NH4)2Fe(SO4)2 to give a concentration of 0.1 mM before the next purification step. The remaining contaminating proteins were removed by a Mono-Q chromatography step with an NaCl gradient from 0 to 1 M. Activity was eluted at approximately 150 mM NaCl.
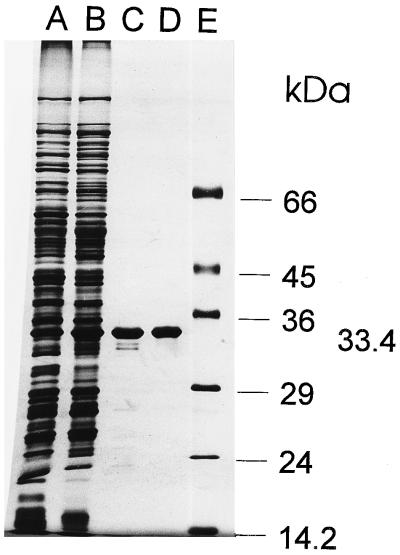
The purity of the catechol 2,3-dioxygenase activity was determined by SDS-PAGE (Fig. 1). After silver staining, the gel showed a single protein band. Omitting the 2-mercaptoethanol from the incubation buffer prior to electrophoresis had no effect on the apparent molecular mass of the subunit, suggesting that the quaternary structure is not stabilized by disulfide bonds.
FIG. 1.
Purification of the catechol 2,3-dioxygenase of P. putida GJ31 as monitored by SDS-PAGE. Lanes: A, crude extract (10 μg of protein); B, ammonium sulfate precipitate (10 μg of protein); C, hydroxyapatite supernatant (2 μg of protein); D, Mono-Q elution pool (2 μg of protein); E, molecular mass markers (ca. 1 μg each).
Structural properties.
The molecular mass of each of the subunits of the denatured protein, as determined by SDS-PAGE, was found to be 33.4 kDa. Estimation of the native Mr value of the catechol 2,3-dioxygenase by gel filtration gave a molecular mass of 135 ± 10 kDa on a Superose 12 column. On the basis of these results, catechol 2,3-dioxygenase from strain GJ31 should be a tetramer.
The isoelectric point of the purified enzyme was determined to be 7.1 ± 0.1.
Temperature and pH optima.
The temperature optimum of the reaction rate of catechol 2,3-dioxygenase was estimated to be 50°C. However, denaturation of the enzyme was significant at this temperature.
The dependence of the reaction rate on pH showed a bell-shaped curve with a surprisingly high pH optimum at 9.6. Similarly to the negative effect at 50°C, a rapid denaturation of the enzyme was found at the pH optimum.
N-terminal amino acid sequence.
A single amino acid was obtained in each cycle of automated Edman degradation, indicating that the protein was homogeneous and consisted of identical subunits.
The N-terminal amino acid sequence of catechol 2,3-dioxygenase was determined to be SIMRVGHVSI NVMDMAAAVK HYENVLGLKT TMQDNAGNVY LKK.
Inhibition and activation.
The effect of various compounds on the activity of catechol 2,3-dioxygenase was tested. Strong Fe2+ chelators, such as o-phenanthroline and α,α-dipyridyl, markedly inactivated catechol 2,3-dioxygenase at a 1 mM concentration (data not shown). The activation by ascorbate clearly showed that some part of the enzyme was in the oxidized form, i.e., inactive. The oxidizing agent H2O2 completely inactivated the enzyme. Some activity was restored if the sample was reduced with ascorbate. All of these results suggest that the active site is accessible to chelators and oxidizing and reducing agents.
To assign the metal of the catechol 2,3-dioxygenase, reconstitution studies were carried out. Enzyme (200 μg of protein) was dialyzed for 5 h at 4°C against 1 liter of Tris-HCl buffer (100 mM; pH 7.5). Aliquots of 10 μg of protein were incubated for 15 min at room temperature in the presence of 0.1 mM metal ion such as Mg2+, Co2+, Ni2+, Mn2+, Fe2+, and Fe3+. The resulting activities were measured by means of the standard assay and compared with the activity of a sample without an added heavy metal. Only Fe2+ led to a strong increase of activity. Mg2+ showed no effect, whereas the heavy metal ions Co2+, Ni2+, Mn2+, and Fe3+ resulted in a significant decrease in activity.
Identification of 2-hydroxymuconate as the product of turnover of 3-chlorocatechol with the catechol 2,3-dioxygenase of P. putida GJ31.
To identify the reaction product of enzymatic turnover of 3-chlorocatechol, several incubations with purified catechol 2,3-dioxygenase from P. putida GJ31 were done.
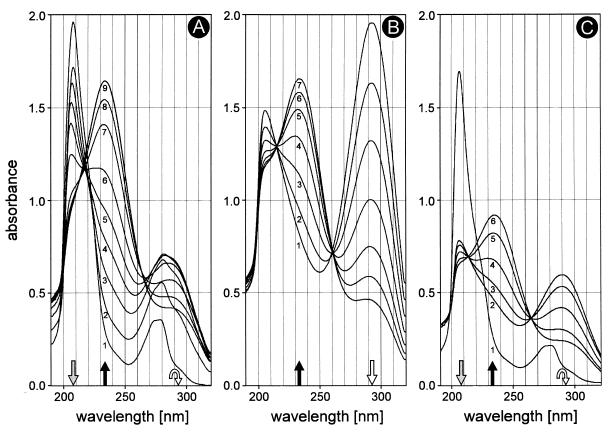
For the spectral characterization of the reaction product, 3-chlorocatechol was incubated with enzyme in phosphate buffer (pH 7.4). As shown in Fig. 2A, the absorbance of 3-chlorocatechol at 208 nm decreased during turnover. Simultaneously, a component which absorbs at 235 nm was formed. A second weaker absorption peak at 290 nm appeared and later decreased.
FIG. 2.
2-Hydroxymuconate and 4-oxalocrotonate as the products of enzymatic turnover of 3-chlorocatechol with the catechol 2,3-dioxygenase of P. putida GJ31. (A) Turnover of 3-chlorocatechol (0.2 μmol) with 8.4 μg of protein in a volume of 1 ml. Spectra were recorded at 0, 1, 3, 5, 7, 10, 15, 20, and 60 min. (B) Tautomerization of 2-hydroxymuconate (0.2 μmol per ml) to 4-oxalocrotonate in phosphate buffer (pH 7.4) in the absence of enzyme. Spectra were recorded at 2, 5, 10, 18, 28, 40, and 120 min. (C) Turnover of 3-chlorocatechol (0.1 μmol) in the presence of a large amount of enzyme (16.8 μg of protein) in a volume of 1 ml. Spectra were recorded at 0, 2, 5, 10, 20, and 30 min.
Similar spectral behavior was obtained by incubating chemically prepared 2-hydroxymuconic acid in phosphate buffer (pH 7.4) in the absence of enzyme (Fig. 2B). The absorption of the 2-hydroxymuconate (at 292 nm) decreased, and 4-oxalocrotonate (at 232 nm) was formed. The ratio of both tautomers present in equilibrium, i.e., the keto form 4-oxalocrotonate and the enol form 2-hydroxymuconate, was the same as in the dioxygenase assay mixture with 3-chlorocatechol.
2-Hydroxymuconate was determined to be the initial product from the conversion of 3-chlorocatechol when a large amount of enzyme was added to the incubation mixture. The formation of 2-hydroxymuconate from 3-chlorocatechol was completed before 4-oxalocrotonate accumulated in the assay mixture (Fig. 2C).
Data from HPLC analysis (retention times with different solvent systems and UV spectrum) of the first product formed from 3-chlorocatechol by the catechol 2,3-dioxygenase were found to be identical to those of an authentic sample of 2-hydroxymuconic acid produced by chemical synthesis.
Stoichiometry of chloride elimination during turnover of 3-chlorocatechol.
Chloride elimination during turnover of 3-chlorocatechol by catechol 2,3-dioxygenase from P. putida GJ31 was measured. Assay mixtures containing 3-chlorocatechol and the purified enzyme were incubated at room temperature until conversion was complete. Turnover of 3-chlorocatechol was accompanied by quantitative release of chloride (data not shown).
Substrate specificity.
The substrate range of catechol 2,3-dioxygenase was determined by incubating the enzyme with various potential substrates and determining the rate of appearance of products (Table 2). Various substituted catechols were oxidized by the catechol 2,3-dioxygenase of strain GJ31. The absorbance maxima of the products were observed to be in the range of 365 to 387 nm (the exception was 3-chlorocatechol), suggesting the occurrence of substituted 2-hydroxymuconic semialdehydes (typical meta-cleavage products), which can be explained by the cleavage of the respective substrate in a proximal extradiol manner between C-2 and C-3. Distal extradiol cleavage seemed to occur with 3,5-dichlorocatechol, since a yellow compound was formed, which does not occur by cleavage between C-2 and C-3.
TABLE 2.
Specificity of the catechol 2,3-dioxygenase of P. putida GJ31
| Substratea | Product λmax (nm) | ɛ (liters/mol · cm) | Relative V (%)b |
|---|---|---|---|
| Catechol | 375 | 36,000 | 100 |
| 3-Methylcatechol | 386 | 11,800 | 49 |
| 4-Methylcatechol | 381 | 25,500 | 97 |
| 3-Chlorocatechol | 290 | 12,500 | 130 |
| 4-Chlorocatechol | 380 | 42,000 | 70 |
| 3,5-Dichlorocatechol | 387 | 42,000 | 2 |
| 4,5-Dichlorocatechol | 380 | 30,000c | 7d |
| 4-Fluorocatechol | 385 | 37,500 | 123 |
The concentration of substrate used in the assay mixture for measurements of V was 10 μM. Doubling of the amount of enzyme in the assay mixture resulted in a doubling of velocity, indicating that the enzyme was saturated by the respective substrate; therefore, V is quasi-Vmax.
Activity determined with catechol (100%) to activity determined with substituted substrate.
The extinction coefficient was not available, so the value of 30,000 liters/mol · cm was used for a raft calculation of the conversion rate.
The rapid conversion for a short time period was accompanied by a drastic reduction in the turnover rate (overall turnover rate of only 10%).
The concentration used for the determination of relative velocities was 10 μM, since a concentration of 100 μM used during the purification of the enzyme was found to result in lower rates due to substrate inhibition. With most substrates, the enzyme activity decreased with an increase in substrate concentration beyond a certain level. However, since a doubling of the amount of enzyme in the assay resulted in a doubling of the velocity, the enzyme was saturated by the respective substrate at a concentration of 10 μM.
The best substrate was found to be 3-chlorocatechol, indicating some adaptation of the enzyme to the metabolite of chlorobenzene. In contrast, the enzyme failed to cleave 3,4-dichloro- and 3,6-dichlorocatechol (data not shown). The turnover of 4,5-dichlorocatechol was characterized by a rapid initial reaction rate followed by a drastic decrease in the rate when 10 to 15% of the substrate was converted. In contrast, the rate with 3,5-dichlorocatechol was very slow but allowed complete turnover, as analyzed by HPLC.
DISCUSSION
Structure of catechol 2,3-dioxygenase.
We have described a purification procedure for the catechol 2,3-dioxygenase of strain GJ31. The isolation procedure allows purification of the enzyme to >95% homogeneity. After purification, the enzyme has a native molecular mass of 135 ± 10 kDa and consists of a single subunit type of 33.4 kDa, consistent with the enzyme existing as a tetramer of identical subunits. This is similar to the catechol 2,3-dioxygenase from P. putida PaW1 (mt-2), which has a molecular mass of 140 kDa and consists of four identical subunits (38). The N-terminal amino acid sequence of the catechol 2,3-dioxygenase of strain GJ31 shows around 70 to 42% identity to the published sequences of various catechol 2,3-dioxygenases (Fig. 3). The assignment of the metal as Fe2+ was based on chelation studies, the observation of inactivation by oxidants, and the requirement for Fe2+ in a reconstitution procedure. H2O2 inactivation at a concentration of 1 mM is typical for all iron-dependent extradiol dioxygenases, while manganese-dependent extradiol dioxygenases show only weak inactivation under these conditions (6, 43).
FIG. 3.
CLUSTAL V multiple-sequence alignment of the N-terminal amino acid sequence of the catechol 2,3-dioxygenase of P. putida GJ31 with those of other catechol 2,3-dioxygenases. The codes on the left are accession numbers. References are given at the end of each line.
The value of the pH optimum of the enzyme of strain GJ31 was found to be 9.6, which is quite different from the value of 6.5 obtained for the extradiol dioxygenase of P. putida PaW1 (mt-2) (34).
Substrate specificity.
It is shown here that the catechol 2,3-dioxygenase of strain GJ31 can convert a wide range of substrates. The substrate range of the enzyme is similar to that of XylE from strain PaW1, which can use methyl-substituted substrates and 4-chlorocatechol. However, XylE does not accept chlorine in the position immediately adjacent to the vicinal hydroxyl groups. The GJ31 dioxygenase is the only enzyme known to convert 3-chlorocatechol at a high rate, so it should be termed chlorocatechol 2,3-dioxygenase. The activities of the other listed catechol 2,3-dioxygenases indicate that some prefer 3-methylcatechol and others the analog substituted at the 4 position (Table 3). 3,5-Dichloro- and 4,5-dichlorocatechol were converted by the dioxygenase of strain GJ31 at a low rate, forming yellow products.
TABLE 3.
Comparison of the specificities of various catechol 2,3-dioxygenases with that of P. putida GJ31
| Substrate | Relative Vmax (%) by straina
|
||||||
|---|---|---|---|---|---|---|---|
| P. putida NCIB 10015 (48) | P. putida PaW1(mt-2) (3, 10, 27, 33, 38) | P. putida UCC (27) | Alcaligenes eutrophus (18) | Nocardia sp. (44) | A. vinelandii (47) | P. putida GJ31 | |
| Catechol | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 3-Methylcatechol | 131–178 | 57–64 | 180 | 139–150 | 88 | 49 | |
| 4-Methylcatechol | 310–350 | 77–86 | 40 | 30–43 | 516 | 97 | |
| 3-Chlorocatechol | 3–5 | 8b | 130 | ||||
| 4-Chlorocatechol | 51–80 | 19 | 16 | 70 | |||
| 3,5-Dichlorocatechol | 0.17–<1 | 1.5b | 2b | ||||
| 4,5-Dichlorocatechol | 7 | ||||||
| 4-Fluorocatechol | 123 | ||||||
References for strains are given in parentheses.
A yellow meta-cleavage product, indicating cleavage between C-1 and C-6, was obtained.
Products from 3-chlorocatechol.
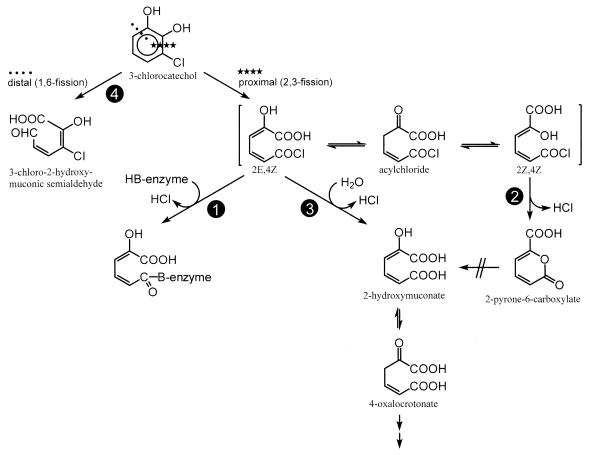
Surprisingly, P. putida GJ31 degraded chlorobenzene via 3-chlorocatechol and used a meta-cleavage pathway. 3-Halocatechols have been reported to be suicide substrates for the meta-fission enzyme catechol 2,3-dioxygenase of P. putida PaW1 (3) or to be inactivating chelators for the enzyme of P. putida F1 (24). In the first case, an acylhalide (5-chlorocarbonyl-2-hydroxy-penta-2,4-dienoic acid) has been proposed as the reactive intermediate which acylates the protein and destroys its enzymatic activity (Fig. 4, path 1).
FIG. 4.
Alternative reactions occurring when catechol 2,3-dioxygenases cleave 3-chlorocatechol. 1, Suicide inactivation when a nucleophilic group of the dioxygenases undergoes acylation; 2, Immediate and spontaneous cyclization occurring when the carbonyl group at C-6 of the ring fission product undergoes internal nucleophilic attack by the enolic hydroxyl at C-2 after configurational change; 3, Reaction of acylchloride with water to give 2-hydroxymuconate; 4, Distal extradiol cleavage between C-1 and C-6 to give a typical yellow meta-cleavage product of the 2-hydroxymuconic semialdehyde type.
In contrast, Kersten et al. (21) reported that a distal extradiol cleaving protocatechuate 4,5-dioxygenase in 4-hydroxybenzoate-grown cells of a nonfluorescent pseudomonad catalyzes 2-pyrone-4,6-dicarboxylic acid formation by nucleophilic displacement of methanol from 3-O-methylgallate. By the same mechanism, the enzyme catalyzes 2-pyrone-4,6-dicarboxylic acid formation by nucleophilic displacement of a halide ion from protocatechuates substituted with a halogen at the C-5 of the nucleus (22). This then allows growth with 5-chlorovanillate, which indicates that cyclization entailing nucleophilic displacement of halogen provides an effective alternative to the enzyme suicide inactivation that occurs when a nucleophilic group of the dioxygenase undergoes acylation. An important aspect of this mechanism is that the ring fission product remains bound to the enzyme during the complete configuration change that precedes nucleophilic displacement.
In analogy to the mechanism reported by Kersten et al. (22), 2-pyrone-6-carboxylate was expected to occur from 3-chlorocatechol by the action of the catechol 2,3-dioxygenase (Fig. 4, path 2). 2-Pyrone-6-carboxylate is formed when protocatechuate 3,4-dioxygenase and catechol 1,2-dioxygenase, which are intradiol dioxygenases, convert pyrogallol (46). Formation of the lactone has also been observed in the degradation of diphenylether (41, 42). Here, it resulted from the action of the extradiol cleaving 2,3-dihydroxybiphenyl-1,2-dioxygenase on 2,3-dihydroxydiphenylether. However, the 2-pyrone-6-carboxylate was not further degraded. 2-Pyrone-6-carboxylate has also been detected as a dead-end product in the degradation of the herbicide chloridazon by Phenylobacterium immobilis DSM 1986 (32).
In the crude extract of chlorobenzene-grown cells of strain GJ31, we did not detect any hydrolase activity towards 2-pyrone-6-carboxylate (26). The product identified after conversion of 3-chlorocatechol by catechol 2,3-dioxygenase was 2-hydroxymuconate. The production of 2-pyrone-6-carboxylate can occur only from the 2Z,4Z configuration. The change in the originally produced 2E,4Z configuration by the catechol 2,3-dioxygenase is therefore a prerequisite. The keto form as an intermediate is a plausible step for the change in the double bond. However, since a hydrolase activity for 2-pyrone-6-carboxylate was not detected in the crude extract of strain GJ31, and since the compound did not arise in the reaction of catechol 2,3-dioxygenase with 3-chlorocatechol, one can postulate that 2-pyrone-6-carboxylate will not be formed. This leaves the reaction of the acylchloride with water as the path for the degradation of 3-chlorocatechol in strain GJ31 (Fig. 4, path 3).
While the chlorocatechol 2,3-dioxygenase of strain GJ31 cleaved 3-chlorocatechol in a proximal manner, there are also some examples in the literature in which the cleavage occurred at a distal position between C-1 and C-6. Sala-Trepat and Evans (47) reported the cleavage of 3-chlorocatechol by catechol 2,3-dioxygenase from Azotobacter vinelandii 206 to give a chromophoric product with a λmax at 380 nm, indicating distal cleavage (Fig. 4, path 4). A typical yellow meta-cleavage product with a λmax at 388 nm was also reported to occur when the catechol 2,3-dioxygenase of A. vinelandii 206 and an Achromobacter sp. cleaved 3,5-dichlorocatechol (17). Just recently, Heiss et al. (13) observed that the 2,3-dihydroxybiphenyl dioxygenase from naphthalenesulfonate-degrading Sphingomonas sp. strain BN6 oxidized 3-chlorocatechol at a high rate, resulting in a product exhibiting the behavior of a typical muconic semialdehyde, i.e., 3-chloro-2-hydroxymuconic semialdehyde.
The chlorocatechol 2,3-dioxygenase of strain GJ31 is the first example of an extradiol dioxygenase that converts 3-chlorocatechol productively as part of growth via the meta pathway, while conversion by the enzymes of A. vinelandii and Sphingomonas sp. is a cooxidative reaction.
Dechlorination mechanisms in the degradation of chloroaromatic compounds.
The dechlorination mechanism used by strain GJ31 represents an alternative to the different dechlorination mechanisms which are normally used in the degradation of chlorobenzenes through the ortho pathway. These include the oxygenolytic removal of chlorine substituents at an early stage of the degradative pathway by the ring-activating dioxygenases prior to cleavage of the aromatic ring and, alternatively, degradation proceeding through chlorinated catechols as central metabolites. Chlorosubstituted aliphatic structures are then generated after ring cleavage, from which HCl is eliminated by chloromuconate cycloisomerases and maleylacetate reductases.
We are currently attempting to explain the ability of the chlorocatechol 2,3-dioxygenase of strain GJ31 to avoid the suicide inactivation found with the PaW1 enzyme. In addition, labelling experiments with the latter enzyme will help clarify the suicide inactivation mechanism.
ACKNOWLEDGMENTS
This work was financed partially by the European Union, under Environment Programme contract EV5V-CT92-0192, and by a grant from the Dutch IOP Environmental Biotechnology program.
REFERENCES
- 1.Arensdorf J J, Focht D D. Formation of chlorocatechol meta cleavage products by a pseudomonad during metabolism of monochlorobiphenyls. Appl Environ Microbiol. 1994;60:2884–2889. doi: 10.1128/aem.60.8.2884-2889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arensdorf J J, Focht D D. A meta cleavage pathway for 4-chlorobenzoate, an intermediate in the metabolism of 4-chlorobiphenyl by Pseudomonas cepacia P166. Appl Environ Microbiol. 1995;61:443–447. doi: 10.1128/aem.61.2.443-447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartels I, Knackmuss H-J, Reineke W. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl Environ Microbiol. 1984;47:500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartilson M, Shingler V. Nucleotide sequence and expression of the catechol 2,3-dioxygenase-encoding gene of phenol-catabolizing Pseudomonas CF600. Gene. 1989;85:233–238. doi: 10.1016/0378-1119(89)90487-3. [DOI] [PubMed] [Google Scholar]
- 5.Benjamin R C, Voss J A, Kunz D A. Nucleotide sequence of xylE from the TOL pDK1 plasmid and structural comparison with isofunctional catechol 2,3-dioxygenase genes from TOL pWWO and NAH7. J Bacteriol. 1991;173:2724–2728. doi: 10.1128/jb.173.8.2724-2728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boldt Y R, Sadowsky M J, Ellis L B M, Que L, Jr, Wackett L P. A manganese-dependent dioxygenase from Arthrobacter globiformis CM-2 belongs to the major extradiol dioxygenase family. J Bacteriol. 1995;177:1225–1232. doi: 10.1128/jb.177.5.1225-1232.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Carrington B, Lowe A, Shaw L E, Williams P A. The lower pathway operon for benzoate catabolism in biphenyl-utilizing Pseudomonas sp. strain IC and the nucleotide sequence of the bphE gene for catechol 2,3-dioxygenase. Microbiology. 1994;140:499–508. doi: 10.1099/00221287-140-3-499. [DOI] [PubMed] [Google Scholar]
- 9.Dorn E, Hellwig M, Reineke W, Knackmuss H-J. Isolation of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 10.Duggleby C J. Studies on some enzymes involved in the meta cleavage of catechol. Ph.D. thesis. Bangor, United Kingdom: University College North Wales; 1979. [Google Scholar]
- 11.Freier R K. Wasseranalyse (2. Aufl.) Berlin, Germany: Walter de Gruyter; 1974. pp. 45–46. [Google Scholar]
- 12.Ghosal D, You I S, Gunsalus I C. Nucleotide sequence and expression of gene nahH of plasmid NAH7 and homology with gene xylE of TOL pWWO. Gene. 1987;55:19–28. doi: 10.1016/0378-1119(87)90244-7. [DOI] [PubMed] [Google Scholar]
- 13.Heiss G, Stolz A, Kuhm A E, Müller C, Klein J, Altenbuchner J, Knackmuss H-J. Characterization of a 2,3-dihydroxybiphenyl dioxygenase from the naphthalenesulfonate-degrading bacterium strain BN6. J Bacteriol. 1995;177:5865–5871. doi: 10.1128/jb.177.20.5865-5871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann H, Müller C, Schmidt I, Mahnke J, Petruschka L, Hahnke K. Localization and organization of phenol degradation genes of Pseudomonas putida strain H. Mol Gen Genet. 1995;247:240–246. doi: 10.1007/BF00705655. [DOI] [PubMed] [Google Scholar]
- 15.Higson F K, Focht D D. Utilization of 3-chloro-2-methylbenzoic acid by Pseudomonas cepacia MB2 through the meta fission pathway. Appl Environ Microbiol. 1992;58:2501–2504. doi: 10.1128/aem.58.8.2501-2504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollender J, Dott W, Hopp J. Regulation of chloro- and methylphenol degradation in Comamonas testosteroni JH5. Appl Environ Microbiol. 1994;60:2330–2338. doi: 10.1128/aem.60.7.2330-2338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath R S. Co-metabolism of methyl- and chloro-substituted catechols by an Achromobacter sp. possessing a new meta-cleaving oxygenase. Biochem J. 1970;119:871–876. doi: 10.1042/bj1190871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes E J L, Bayly R C, Skurray R A. Evidence for isofunctional enzymes in the degradation of phenol, m- and p-toluate, and p-cresol via catechol meta-cleavage pathways in Alcaligenes eutrophus. J Bacteriol. 1984;158:79–83. doi: 10.1128/jb.158.1.79-83.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaschabek S R, Reineke W. Maleylacetate reductase of Pseudomonas sp. strain B13: dechlorination of chloromaleylacetates, metabolites in the degradation of chloroaromatic compounds. Arch Microbiol. 1992;158:412–417. doi: 10.1007/BF00276301. [DOI] [PubMed] [Google Scholar]
- 20.Keil H, Lebens M R, Williams P A. TOL plasmid pWW15 contains two nonhomologous, independently regulated catechol 2,3-oxygenase genes. J Bacteriol. 1985;163:248–255. doi: 10.1128/jb.163.1.248-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kersten P J, Dagley S, Whittaker J W, Arciero D, Lipscomb J D. 2-Pyrone-4,6-dicarboxylic acid, a catabolite of gallic acids in Pseudomonas species. J Bacteriol. 1982;152:1154–1162. doi: 10.1128/jb.152.3.1154-1162.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kersten P J, Chapman P J, Dagley S. Enzymatic release of halogens or methanol from some substituted protocatechuic acids. J Bacteriol. 1985;162:693–697. doi: 10.1128/jb.162.2.693-697.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitayama A, Achioku T, Yanagawa T, Kanou K, Kikuchi M, Ueda H, Suzuki E, Nishimura H, Nagamune T, Kawakami Y. Cloning and characterization of extradiol aromatic ring-cleavage dioxygenases of Pseudomonas aeruginosa JI104. J Ferment Bioeng. 1996;82:217–223. [Google Scholar]
- 24.Klecka G M, Gibson D T. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida by 3-chlorocatechol. Appl Environ Microbiol. 1981;41:1159–1165. doi: 10.1128/aem.41.5.1159-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Mars A E, Kasberg T, Kaschabek S R, van Agteren M H, Janssen D B, Reineke W. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J Bacteriol. 1997;179:4530–4537. doi: 10.1128/jb.179.14.4530-4537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClure N C, Venables W A. Adaptation of Pseudomonas putida mt-2 to growth on aromatic amines. J Gen Microbiol. 1986;132:2209–2218. doi: 10.1099/00221287-132-8-2209. [DOI] [PubMed] [Google Scholar]
- 28.McClure, N. C., C. P. Saint, and A. J. Weightman. 1991. Nucleotide sequence of 3-methylcatechol 2,3-dioxygenase gene tdnC from Pseudomonas putida UCC2. EMBL Data Library, accession no. X59790.
- 29.McCullar M V, Brenner V, Adams R H, Focht D D. Construction of a novel polychlorinated biphenyl-degrading bacterium: utilization of 3,4′-dichlorobiphenyl by Pseudomonas acidovorans M3GY. Appl Environ Microbiol. 1994;60:3833–3839. doi: 10.1128/aem.60.10.3833-3839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merril C R, Goldman D, Sedman S A, Ebert M H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981;211:1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- 31.Moon J, Chang H, Min K R, Kim Y. Cloning and sequencing of the catechol 2,3-dioxygenase gene of Alcaligenes sp. KF711. Biochem Biophys Res Commun. 1995;208:943–949. doi: 10.1006/bbrc.1995.1425. [DOI] [PubMed] [Google Scholar]
- 32.Müller R, Schmitt S, Lingens F. A novel non-heme iron-containing dioxygenase. Chloridazon-catechol dioxygenase from Phenylobacterium immobilis DSM 1986. Eur J Biochem. 1982;125:579–584. doi: 10.1111/j.1432-1033.1982.tb06722.x. [DOI] [PubMed] [Google Scholar]
- 33.Murray K, Duggleby C J, Sala-Trepat J M, Williams P A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972;28:301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakai C, Hori K, Kagamiyama H, Nakazawa T, Nozaki M. Purification, subunit structure, and partial amino acid sequence of metapyrocatechase. J Biol Chem. 1983;258:2916–2922. [PubMed] [Google Scholar]
- 35.Nakai C, Kagamiyama H, Nozaki M, Nakazawa T, Inouye S, Ebina Y, Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOL plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983;258:2923–2928. [PubMed] [Google Scholar]
- 36.Ng L C, Shingler V, Sze C C, Poh C L. Cloning and sequences of the first eight genes of the chromosomally encoded (methyl) phenol degradation pathway from Pseudomonas putida P35X. Gene. 1994;151:29–36. doi: 10.1016/0378-1119(94)90629-7. [DOI] [PubMed] [Google Scholar]
- 37.Nozaki M. Metapyrocatechase (Pseudomonas) Methods Enzymol. 1970;17A:522–525. [Google Scholar]
- 38.Nozaki M, Kotani S, Ono K, Senoh S. Metapyrocatechase. III. Substrate specificity and mode of ring fission. Biochim Biophys Acta. 1970;220:213–223. doi: 10.1016/0005-2744(70)90007-0. [DOI] [PubMed] [Google Scholar]
- 39.O’Farrell P H. High resolution two-dimensional electrophoresis. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 40.Oldenhuis R, Kuijk L, Lammers A, Janssen D B, Witholt B. Degradation of chlorinated and non-chlorinated aromatic solvents in soil suspensions by pure bacterial cultures. Appl Microbiol Biotechnol. 1989;30:211–217. [Google Scholar]
- 41.Pfeifer F, Schacht S, Klein J, Trüper H G. Degradation of diphenylether by Pseudomonas cepacia. Arch Microbiol. 1989;152:515–519. doi: 10.1007/BF00290914. [DOI] [PubMed] [Google Scholar]
- 42.Pfeifer F, Trüper H G, Klein J, Schacht S. Degradation of diphenylether by Pseudomonas cepacia Et4: enzymatic release of phenol from 2,3-dihydroxydiphenylether. Arch Microbiol. 1993;159:323–329. doi: 10.1007/BF00290914. [DOI] [PubMed] [Google Scholar]
- 43.Que L, Widom J, Crawford R L. 3,4-Dihydroxyphenylacetate 2,3-dioxygenase: a manganese(II) dioxygenase from Bacillus brevis. J Biol Chem. 1981;256:10941–10944. [PubMed] [Google Scholar]
- 44.Rast H G, Engelhardt G, Wallnöfer P. 2,3-Cleavage of substituted catechols in Nocardia sp. DSM 43251 (Rhodococcus rubrus). Zentralbl. Bakteriol. Mikrobiol. Hyg. 1 Abt. Orig C. 1980;1:224–236. [Google Scholar]
- 45.Reineke W, Knackmuss H-J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- 46.Saeki Y, Nozaki M, Senoh S. Cleavage of pyrogallol by non-heme iron-containing dioxygenases. J Biol Chem. 1980;255:8465–8471. [PubMed] [Google Scholar]
- 47.Sala-Trepat J M, Evans W C. The meta cleavage of catechol by Azotobacter species: 4-oxalocrotonate pathway. Eur J Biochem. 1971;20:400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 48.Sala-Trepat J M, Murray K, Williams P A. The metabolic divergence in the meta cleavage of catechols by Pseudomonas putida NCIB 10015. Physiological significance and evolutionary implications. Eur J Biochem. 1972;28:347–356. doi: 10.1111/j.1432-1033.1972.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 49.Schlömann M. Evolution of chlorocatechol catabolic pathways: conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation. 1994;5:301–321. doi: 10.1007/BF00696467. [DOI] [PubMed] [Google Scholar]
- 50.Shindo T, Ueda H, Suzuki E, Nishimura H. A catechol 2,3-dioxygenase gene as a reporter. Biosci Biotechnol Biochem. 1995;59:314–315. doi: 10.1271/bbb.59.314. [DOI] [PubMed] [Google Scholar]
- 51.Wiley R H, Hart A J. 2-Pyrones. IX. 2-Pyrone-6-carboxylic acid and its derivatives. J Am Chem Soc. 1954;76:1942–1944. [Google Scholar]