Abstract
In the past decade, extracellular vesicles (EVs) have attracted substantial interest in biomedicine. With progress in the field, we have an increasing understanding of cellular responses to EVs. In this Technical Report, we describe the direct nanoinjection of EVs into the cytoplasm of single cells of different cell lines. By using robotic fluidic force microscopy (robotic FluidFM), nanoinjection of GFP positive EVs and EV‐like particles into single live HeLa, H9c2, MDA‐MB‐231 and LCLC‐103H cells proved to be feasible. This injection platform offered the advantage of high cell selectivity and efficiency. The nanoinjected EVs were initially localized in concentrated spot‐like regions within the cytoplasm. Later, they were transported towards the periphery of the cells. Based on our proof‐of‐principle data, robotic FluidFM is suitable for targeting single living cells by EVs and may lead to information about intracellular EV cargo delivery at a single‐cell level.
Keywords: cell‐to‐cell transport, intracellular delivery, mRNA injection, organelle‐specific targeting, robotic FluidFM, single‐cell, vesicle cargo, vesicle injection, vesicle release, vesicle trafficking
1. INTRODUCTION
Extracellular vesicles (EVs) are phospholipid bilayer enclosed structures released by all cells. EVs carry biomolecules such as nucleic acids (RNA and DNA), proteins, lipids, glycans and metabolites (Raposo & Stoorvogel, 2013). The release and uptake of these particles by various cells serve a number of functions, many of which may remain uncovered today. The EV field has significantly benefited from technological developments that enabled efficient separation, detection and characterization of EVs (Musto et al., 2021). Importantly, the delivery of proteins (Simpson et al., 2009) and RNAs (Valadi et al., 2007) by EVs was shown to have a role in cell‐to‐cell communication (Mathivanan et al., 2010) and cell signalling (Subra et al., 2010). EVs also have a role in pathological processes such as cancer cell proliferation, migration and metastasis formation (Kahlert & Kalluri, 2013). EV transfer protects the internal cargo molecules from extracellular enzymatic degradation and also helps to avoid dilution of the cargo. Transmembrane proteins in the lipid bilayer direct vesicles to specific cell types, rendering them promising candidates to deliver drugs or biomolecules (György et al., 2015; Oshchepkova et al., 2021). Thus, understanding how EVs are taken up and processed by cells has potential implications in EV‐based therapy and drug delivery systems.
EVs can be taken up by cells through different pathways depending on the cell type and the molecules decorating the outer surface of EVs (Mulcahy et al., 2014). Endocytosis (Doherty & McMahon, 2009), a process by which particles are internalized while being surrounded by the plasma membrane (Doherty & McMahon, 2009; Mulcahy et al., 2014), is responsible for EV uptake (Costa Verdera et al., 2017; Koumangoye et al., 2011; Mulcahy et al., 2014; Nabi & Le, 2003; Verdera et al., 2017). During endocytosis, EVs are contained in the endocytic membrane compartments. Upon fusion with the endomembranes, the cargo of EVs may be released into the cytosol. Alternatively, in some cases, vesicles can either bind to plasma membrane receptors inducing signalling or fuse with the plasma membrane and release their contents directly to the cytosol (Russell et al., 2019).
Mechanical injection of foreign substances into live cells is a commonly applied single‐cell manipulation technique. In most of the cases, cells are injected manually using glass micropipettes with only visual feedback about any potential damage to the cells (Wei & Xu, 2019). Correspondingly, this procedure is characterized by low throughput and low cell survival. To increase the efficiency of cell injection, force‐assisted techniques can be used, where the force between the micropipette and the cell can be monitored. A handful of such methods have been developed based on impedance measurements of the micropipette aperture or on the measurement of out‐of‐plane forces by the deformation of a resonator. Experiments were also carried out with atomic force microscopy‐based systems, where the tip of the cantilever was inserted into the cytoplasm and the material to be injected was let to desorb from the tip (Chen et al., 2007). Due to the passive nature of desorption, it is difficult to control the volume and concentration of the injected substances. Furthermore, the range of potential injectable substances is limited to those that can be immobilized appropriately to the tip.
Recently, the development of fluidic force microscopy revolutionized the field of single‐cell injection (Li et al., 2022; Meister et al., 2009). This technique is based on a microfluidic channel incorporated into an atomic force microscopy cantilever. The channel ends in an aperture on the side of the pyramidal tip, while its other terminus is connected to a pressure control device capable of varying the pressure between negative and positive values. This arrangement makes it possible to implement various cell manipulation workflows such as single‐cell adhesion measurements on eukaryotic (Potthoff et al., 2012; Sztilkovics et al., 2020) or prokaryotic cells (Potthoff et al., 2015), single‐cell sorting (Guillaume‐Gentil et al., 2014), nanolithography (Saftics et al., 2019), and most notably nanoinjection (Guillaume‐Gentil et al., 2013) and extraction (Guillaume‐Gentil et al., 2016, 2017) of single‐cells. The sharp end of the tip is responsible for penetrating the plasma membrane without clogging the aperture. When the tip comes into contact with the cell, its bending is detected through the displacement of a reflected laser beam from the back of the cantilever, according to the principle of atomic force microscopy. The bending is proportional with the force acting on the cantilever. Therefore, a real‐time force control is achieved during the injection process. Using a fluidic force microscopy device, O. Guillaume‐Gentil et al. successfully injected the nuclei of live HeLa cells with a fluorescent molecule (Guillaume‐Gentil et al., 2013). Using a dye that does not pass through the nuclear envelope, the authors showed that the nucleus can be selectively targeted for injection. The injected volumes were in the range of picolitres to tens of femtolitres, quantified on the basis of the fluorescent intensity of the injected dyes. The volume was primarily determined by the applied pressure and the time interval of injection but showed a large degree of variation due to fluctuations in cytoplasmic viscosity. Injection of GFP encoding plasmids resulted in successful transfection in 40% of cells. The authors also demonstrated the reverse process: a single‐cell extraction (Guillaume‐Gentil et al., 2016). In this case, after penetration of the plasma membrane by the fluidic force microscopy tip, negative pressure was induced at the aperture removing a fraction of the cytoplasm. The authors found that even after removing 4 pl of cytosol, representing a significant part of the entire volume of the cell, viability was as high as 82%. At the same time, cells proved to be more sensitive to extraction from the nucleus. The extracted nuclear material was deposited in microdroplets and further analysed by qPCR and mass spectroscopy (Guillaume‐Gentil et al., 2017).
The robotic system we applied in the current study has a large, motorized sample stage, allowing cell targeting over mm‐cm scale areas. The use of this innovative instrument has been successfully demonstrated in adhesion studies (Gerecsei et al., 2019; Sztilkovics et al., 2020) and patterning of cell adhesive surfaces (Saftics et al., 2019).
While the injection of live cells with solutions of dyes and DNA has been demonstrated already, this technology has not been exploited for the force‐controlled injection of EVs yet. Electroporation is a common method for the introduction of foreign substances into live cells based on nanometer‐sized pores generated in the cell membrane using electrical pulses (Weaver, 2000). It is also used to introduce cargo such as certain proteins or DNA into EVs (Lamichhane et al., 2015). However, the introduction of intact EVs into cells has not been demonstrated by this method, since the size of the created pores is too small for letting EVs pass through the membrane. Therefore, up till now, no method has been shown to mechanically deliver intact EVs into live cells in a way that does not rely on any natural vesicle uptake mechanism. Importantly, a common property of the natural mechanisms is that upon uptake, EVs are packaged into an endomembrane and do not come in contact directly with the cytosol.
In the current study, we used robotic fluidic force microscopy (robotic FluidFM) to inject suspensions of EVs and EV‐like particles directly into the cytosol of different cell lines. Next, the injected cells were fixed, and the plasma membrane was visualized by fluorescently labelled lactadherin. EV delivery to the cytoplasm was monitored by high‐resolution confocal microscopy. Analysis of the recorded images proved that vesicles were successfully delivered into the cytosol and that initially, they were localized in concentrated spot‐like areas. By injecting GFP encoding plasmids, we validated the injection parameters used in our experiments.
The EV delivery approach described here may pave the way for future studies investigating the intracellular processing and fate of EVs and other lipid nanovesicles inside single live cells.
2. MATERIALS AND METHODS
2.1. Cell culture and related protocols
The human embryonic kidney HEK 293T‐PalmGFP cell line was a kind gift of Dr. Charles Lai, and the cells were cultured as described previously with minor modifications (Lai et al., 2015). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS) (BioSera, Nuaillé, France) and 100 U/mL penicillin 100 µg/mL streptomycin (Sigma‐Aldrich, St. Louis, MO, USA) in a humidified atmosphere at 37°C, 5% CO2.
Differentiation and expansion of bone marrow‐derived mast cells (BMMCs) were carried out as described earlier (Vukman et al., 2020). Briefly, bone marrow cells of the femoral and tibial bones of GFP transgenic mice (C57BL/6‐Tg(UBC‐GFP)30Scha/J, The Jackson Laboratory) were cultured in complete Iscove's modified Dulbecco's medium (IMDM, Gibco) in the presence of 10% heat‐inactivated FBS (Gibco), 100 U/mL penicillin/100 µg/mL streptomycin (Sigma‐Aldrich). As a source of murine IL‐3, cells were grown in 30% WEHI‐3 conditioned IMDM medium (TIB‐68; American Type Culture Collection, Manassas, VA, USA) for 4 weeks.
Non‐fluorescent human embryonic kidney HEK 293 cell line (ECACC 85120602) was cultured in DMEM (Gibco), supplemented with 10% FBS (Biowest SAS, Nuaillé, France and 100 U/mL penicillin 100 µg/mL streptomycin (Sigma‐Aldrich, St. Louis, MO, USA) in a humidified atmosphere at 37°C, 5% CO2.
HeLa cells used in the experiments (ECACC 93021013) were cultured in DMEM (Gibco), supplemented with 10% FBS (Biowest SAS, Nuaillé, France), 4 mM L‐glutamine, 100 U/mL penicillin and 100 µg/mL streptomycin in an incubator with a humidified atmosphere containing 5% CO2 at 37°C.
The LCLC‐103H human large cell lung carcinoma cell line was cultured in DMEM (Gibco), supplemented with 10% FBS (Biowest SAS, Nuaillé, France), 4 mM L‐glutamine, 1% MEM non‐essential amino acids, sodium‐pyruvate and 100 U/mL penicillin and 100 µg/mL streptomycin in an incubator with a humidified atmosphere containing 5% CO2 at 37°C.
The MDA‐MB‐231 human breast adenocarcinoma cell line was cultured in DMEM (Gibco), supplemented with 10% FBS (Biowest SAS, Nuaillé, France), 4 mM L‐glutamine, 1% MEM non‐essential amino acids, sodium‐pyruvate and 100 U/mL penicillin and 100 µg/mL streptomycin in an incubator with a humidified atmosphere containing 5% CO2 at 37°C.
The H9c2 (2‐1) BDIX rat cardiomyoblast cell line (ECACC 88092904) was cultured in DMEM (Gibco), supplemented with 10% FBS (Biowest SAS, Nuaillé, France), 4 mM L‐glutamine, 1% MEM non‐essential amino acids and 100 U/mL penicillin and 100 µg/mL streptomycin in an incubator with a humidified atmosphere containing 5% CO2 at 37°C as previously described (Koncz et al., 2023; Visnovitz et al., 2019).
All cell cultures were tested for mycoplasma infection by PCR, using the following PCR primes: GAAGAWATGCCWTATTTAGAAGATGG and CCRTTTTGACTYTTWCCACCMAGTGGTTGTTG (Koncz et al., 2023).
2.2. Preparation of EV containing samples
2.2.1. Separation of GFP positive small EVs (sEVs) released by bone marrow mast cells (BMMCs)
BMMCs of GFP‐transgenic mice were used as a source of fluorescent sEVs. The vesicles were separated as we described previously (Vukman et al., 2020). Briefly, BMMCs were incubated for 24 h in EV‐depleted complete culture medium in the presence of 0.5 µM calcium ionophore A23187 (Sigma‐Aldrich). Next, EVs were separated by a combination of differential centrifugation and multistep hydrostatic size filtration (Vukman et al., 2020). The green fluorescent internal cargo‐containing EVs were resuspended in PBS, re‐centrifuged, aliquoted, snap frozen in liquid nitrogen and stored at −80°C until use. For the nanoinjections, only the sEV fraction was used, the ratio of GFP positive sEVs was determined by high resolution flow cytometry. The major characteristics of the EVs have been described previously (Lamichhane et al., 2015).
2.2.2. Preparations of HEK 293T‐PalmGFP cell‐derived small extracellular vesicle like particles (sEVLPs)
The HEK 293T‐PalmGFP cells were incubated for 24 h in serum free culture medium. Next, the cells were removed by centrifugation (300 × g, 10 min) and filtration by gravity through a 5 µm pore size filter. Apoptotic bodies and other large EVs (lEVs) were eliminated using centrifugation (2000 × g, 30 min) and the supernatant was filtered by gravity through a 0.8 µm pore size filter. Membrane GFP‐positive medium‐sized EVs (mEVs) were enriched by centrifugation at 12,500 × g 40 min, re‐suspended in PBS and re‐centrifuged. The green fluorescent membrane enclosed mEVs were used to prepare GFP positive sEVLPs by intensive vortexing and sonication (20 min). The sEVLPs were filtered through a 0.2 µm pore size filter and were concentrated by centrifugation at 100,000 × g 70 min. The particles were aliquoted, snap frozen in liquid nitrogen and stored at −80°C until use. The HEK‐derived sEVLPs are characterized in detail by Charles Lai et al. (Lai et al., 2015).
2.2.3. Preparations of HEK 293 cell‐derived pMAXGFP plasmid containing sEVLPs
To demonstrate the release the plasmid cargo of nanoinjected vesicles, non‐fluorescent HEK 293 cell‐derived mEVs were separated as described above. The mEV pellet was resuspended in pMAXGFP plasmid solution (Lonza, 2 mg/mL in 5 mM Tris/HCL, pH 8.5). The mixture was frozen and thawed five times, then sonicated (20 min on ice) and vortexed intensively. Before nanoinjection, the sEVLPs were subjected to DNase digestion (Invitrogen) to degrade unpacked DNA. The DNase was inactivated by EDTA (2.3 mM). EV‐s larger than 500 nm diameter (which could potentially block the cantilever) were removed by centrifugation for 30 min at 10.000 × g, and the supernatant was used for injection.
2.3. Sample preparation and FluidFM injection
Before the nanoinjection experiments, cells were washed with pre‐warmed Dulbecco's phosphate‐buffered saline (DPBS, Sigma‐Aldrich) and detached by applying 0.05% (w/v) trypsin and 0.02% (w/v) EDTA solution for 2 min in an incubator. Afterwards, cells were centrifuged at 200 × g for 5 min and the pellet was re‐suspended in DMEM. Subsequently, 2 × 10 (Valadi et al., 2007) cells were deposited into a 35 mm Grid‐500 glass bottom μ‐dish (Ibidi, Gräfelfing, Germany) or 5 × 10 (Valadi et al., 2007) cells were deposited into a glass bottom WillCo‐dish (WillCo Wells, Amsterdam, The Netherlands). Before cell seeding, a glass coverslip with an engraved grid (Ibidi, Gräfelfing, Germany) was placed in the middle of the WillCo‐dish and fixed by applying a small droplet of silicone grease between the bottom of the dish and the coverslip. The coverslips were sterilized before use by sonication in 70% ethanol and then rinsed in sterile deionized water for 10 min. Samples were incubated at 37°C with 5% CO2 overnight. Directly before the FluidFM experiments, the samples were washed twice with pre‐warmed 10 mM HEPES‐HBSS buffer (4‐(2‐hydroxyethyl)−1‐piperazineethanesulfonic acid (HEPES) mixed with Hank's balanced salt solution (HBSS), pH 7.4) to remove floating and dead cells. Then the dish was filled with 2 mL of HEPES‐HBSS and placed into the sample holder of a robotic FluidFM device (FluidFM OMNIUM, Cytosurge, Glattbrugg, Switzerland). Of note, this robotic FluidFM device was called FluidFM BOT in earlier works. The vesicle suspension was vortexed before pipetting to minimize aggregation. Next, the nanosyringe probe (Cytosurge, CH) was filled with 1 µL of vesicle suspension prepared according to the above‐described protocol. Before use, the probe was coated with Sigmacote (Sigma‐Aldrich) according to the protocol of the manufacturer. This coating helps to avoid nonspecific binding of cells to the cantilever during injection. After placing the probe into the FluidFM device, the positioning of the laser was optimized so that the laser spot falls approximately at a 30 µm distance from the tip (Nagy et al., 2019). Then, the mirror projecting the laser onto the photodetectors was automatically tilted to its correct position. Subsequently, a positive pressure was introduced in the probe, filling the microchannel inside the cantilever. After placement of the probe into the sample containing the cells, approximately 50 cells were manually selected for injection in the Arya software (Cytosurge). Once the selection was done, the software automatically positioned the cantilever above the cells in a serial manner and injected all selected cells. Each injection consisted of the following steps: (i) The cantilever approached the cell from 10 µm height with force‐control until the signal induced by the bending reached a value of 600 mV corresponding to approximately 1 µN. This value is called the setpoint. The speed of the approach was 50 µm/s. (ii) Once the setpoint was reached, a 30–150 mBar positive pressure was induced in the cantilever for an injection time of 6–8 s. These parameters were varied by experiment. (iii) Once the injection time elapsed, the cantilever was retracted with a speed of 5 µm/s to a height of 20 µm.
The entire workflow lasted 20–30 s, which resulted in a throughput of at least 50 cells per 30 min in an entirely automated way. Note, the cells are selected manually for injection, which adds another 10–15 min to the measurement time depending on the operator. In order to enable subsequent identification of the injected cells, only one cell in each zone of the grid on the coverslip was injected.
Of note, in the initial experiments used to determine the ideal injection settings, we used the HEK 293T‐PalmGFP cell‐derived sEVLP in order obtain fluorescent signal of all the injected particles. In later experiments, we used both sEVLPs and sEVs derived from BMMC cells, but in the latter case only the 30‐40% of particles were fluorescent (see Figure S3.)
2.4. Staining and confocal microscopy of fixed cells
2.4.1. Membrane staining by lactadherin
The phosphatidyl serine binding protein, lactadherin (Haematologic Technologies, USA) was fluorescently conjugated using a Cy5® Conjugation Kit (Abcam, UK) according to the manufacturer's instructions. The injected cells were fixed on the coverslips with 4% paraformaldehyde (PFA) in phosphate buffer saline (PBS) for 20 min at room temperature (RT) and then washed with PBS containing 50 mM glycine (three times for 5 min). Following a short blocking step (10% FBS in PBS for 20 min), fluorescently labelled lactadherin (3 ng/µL) was applied in PBS containing 10% FBS for 1 h at RT. After washing with PBS containing 10% FBS (5 min, RT) and PBS (two times for 5 min), the cells were postfixed with 4% PFA in PBS and were washed with PBS containing 50 mM glycine (three times for 5 min) (Koncz et al., 2023; Vukman et al., 2020).
2.4.2. Immunocytochemical detection
The lactadherin membrane‐labelled cells were permeabilized with PBS supplemented with 10% FBS and 0.5% TritonX100 (SERVA Electrophoresis) (binding‐buffer—BB) for 30 min at RT. Primary anti‐GFP antibodies were applied in 1:200 dilution in BB for 1 h at RT. For HEK 293T‐PalmGFP‐derived EVLPS, mouse anti‐GFP (Sigma‐Aldrich), while for murine BMMC sEVs, rabbit anti‐GFP (Sigma‐Aldrich) were used. After washing with BB (five times for 5 min at RT), goat anti‐mouse‐ATTO550 and goat anti‐rabbit‐ATTO550 secondary antibodies (Sigma‐Aldrich) were used, respectively, in PBS containing 1% FBS for 1 h at RT. After the washing steps (PBS two times for 5 min and then ultrapure water two times for 5 min), samples were mounted with Prolong Diamond with DAPI (Invitrogen) and were examined by a Leica SP8 Lightening confocal microscope.
The theoretical maximum resolution of the employed microscope in Lightning mode is 120 nm in lateral and 200 nm in axial direction.
2.5. Live cell confocal imaging
HeLa cells were monitored for 24 h after EV nanoinjection and during spontaneous EV uptake by live cell imaging. Cells were grown in 35 mm glass bottom Grid‐500 μ‐dishes (Ibidi). BMMC‐ derived GFP positive sEVs were injected into HeLa cells with the same method that we used during our experiments with fixed cells. Lysosomes in HeLa cells were detected by LysoTracker Deep Red (Invitrogen) according to the instructions of the manufacturer. Imaging of EV nanoinjected cells started approximately 3 h after injection. In the case of EV uptake of HeLa cells, 10‐well cell culture chamber‐slides (Greiner Bio‐One) were used. In these experiments, the same BMMC‐derived GFP positive sEVs were used as in the nanoinjection experiments. EVs were added to the cell culture medium after LysoTracker staining. Live cell imaging was performed by a Leica SP8 Lightening confocal microscope equipped with OkoLab environmental chamber. For co‐localization, LAS X software (Leica) was used, and Pearson's coefficients were determined to assess the correlation between lysosomal and GFP EV signals of individual cells.
2.6. Transmission electron microscopy (TEM)
Sample preparation for TEM was performed with minor changes to what has been described previously (Théry et al., 2006). EVs or EVLPs were attached to the surface of formvar coated grids (10 min, RT) and were fixed with 2% glutaraldehyde (10 min, RT). After the washing steps (three times for 5 min at RT), the particles were positively contrasted with 2% uranyl‐oxalate and negatively contrasted with 2% methyl cellulose containing 0.4% uranyl‐acetate. Samples were examined by a JEOL 1011 transmission electron microscope.
2.7. Immunogold electron microscopy
GFP positivity of sEV fraction of HEK293T‐PalmGFP cells was tested by Immunogold TEM technique (Figure S1). Samples were prepared as described previously (Koncz et al., 2023). EVs were attached to the surface of formvar coated nickel grids (10 min, RT) and were fixed with 4% formaldehyde (10 min, RT). After the washing steps (three times for 5 min at RT), 2% sucrose solution was used in PBS (1 h) for blocking. A primary antibody (monoclonal mouse anti‐GFP, Sigma‐Aldrich) was applied in blocking solution overnight at 4°C. After three washes (RT, 5 min) with blocking solution, secondary antibody (polyclonal anti‐mouse produced in goat with 5 nm gold particles, Sigma‐Aldrich) was applied in 2% sucrose for 1 h at RT. Unbound secondary antibodies were removed by three washes (for 5 min at RT). Samples were post‐fixed, positively contrasted and analysed as described for transmission electron microscopy.
2.8. Nanoparticle tracking analysis (NTA)
EVs‐ or EV‐like particles were diluted in PBS and were measured in 11 positions (two cycles per position) by ZetaView PMX120 (Particle Metrix) at 25°C using the following settings: shutter speed‐100, sensitivity‐80, frame rate‐30.
2.9. Flow cytometry
To detect sEV proteins, flow cytometry was performed as described previously by Vukman et al. (2020). Small EVs were bound to 4 µm aldehyde/sulphate latex beads (Life Technologies). Next, they were blocked with 100 mM glycine and 1% bovine serum albumin (Sigma‐Aldrich) in PBS. Samples were immunostained by anti‐CD9 Ab (PE, ThermoFisher), anti‐CD63 (APC, PE, eBioscience) anti‐CD81 (PE, ThermoFisher), or appropriate isotype controls (mouse IgG1, IgG2, IgG2b, BD Bioscience, Biolegend and SONY). The presence of phosphatidylserine on sEV surface was tested by lactadherin (Haematologic Technologies) labelled with Cy5 Fast Conjugation Kit, (Abcam) and annexinV‐FITC (SONY). FACSCalibur flow cytometer (BD Biosciences) and CellQuest software were used for analysis. Results are summarized in the Supplementary Information file (Figure S2).
High resolution flow cytometry was applied to determine the proportion of GFP‐positive BMMC‐derived sEVs. EVs were identified by their phospholipid membranes and were stained by BioxMLRed (Bioxol, Budapest, Hungary) lipophilic and auto‐quenchable dye as we described previously (Koncz et al., 2023). The vesicles were measured by Apogee flow cytometer. If a particle was positive for lipid staining, it was assessed as EV. The TritonX‐100® sensitive lipid/GFP double positive events were counted as GFP positive EVs. Limit of blank (LOB) and limit of detection (LOD) was assessed by measuring blank, GFP‐negative EVs (derived from wild type C57BL/6 mouse) and ApogeeMix (#1527) and calculated as described in Wood et al. (Wood et al., 2013) and Welsh et al. (Welsh et al., 2023). As the average particle size of BMMC‐derived EVs is between 100–300 nm (Vukman et al., 2020), configuration and threshold were set by the help of ApogeeMix (#1527) beads where the smallest detected particles were 80 nm polystyrene beads and the largest were 1300 nm silica beads. The data were analysed using FlowJo_V10 software (BD).
2.10. Transfection
The pMAXGFP control plasmid was purchased from Lonza (Lonza, Cologne, Germany). HeLa (4 × 10 (Subra et al., 2010)) cells were transfected with 0.5 µg pmaxGFP and 0.5 µL of Lipofectamine™ 2000 (Thermo Fisher Scientific) to achieve a 1:1 DNA : Lipofectamine ratio in 2 mL of culture medium, according to the manufacturer's protocol. In short, the premixed transfection solution was added into the Petri dish with cultured HeLa cells and incubated for 4 h in an incubator under standard conditions (37°C, 5% CO2). After incubation, the medium containing the transfection solution was discarded and fresh cell culture medium was added.
2.11. RNA injection
The solution used for the nanoinjection consisted of 20 ng/µL GFP mRNA (Miltenyi Biotec GmbH, 130‐101‐114) dissolved in 10 mM Tris pH7.5, 0.1 mM EDTA.
3. RESULTS AND DISCUSSION
In this work we aimed at nanoinjecting GFP containing sEVLPs, GFP containing EVs, mRNA, GFP‐coding plasmids, and GFP‐coding plasmid containing EVLPs into the cytosol of single live cells and investigating further the injected cells by fluorescent microscopy.
The sematic steps of GFP containing sEVLPs (green arrows) and GFP‐coding plasmid containing EVLPs (black arrows) are shown in Figure 1.
FIGURE 1.

Schematic illustration of preparation of the sEVLP (green arrows) and GFP‐coding plasmid containing EVLP (black arrows).
First, we separated EVs and sEVLPs and examined their size and shape by transmission electron microscopy. We studied both the sEVs of BMMC cells and the HEK 293T‐PalmGFP cell‐derived sEVLPs, and we found that both preparations had vesicles with a cup shape morphology (Figure 2a,b). We determined the size distribution of the vesicles with NTA. Both the sEVs of BMMC cells and the HEK 293T‐PalmGFP derived sEVLPs were in the range of 50–300 nm, which was in agreement with the TEM measurements (Figure 2c,d). We also confirmed the presence of EV membrane proteins and phosphatidylserine (Figure S1) on the vesicular surface.
FIGURE 2.

Transmission electron microscopy of BMMC derived sEVs (a) and sEVLPs of HEK 293T‐PalmGFP cells (b). Representative size distribution histograms of BMMC derived sEVs (c) and sEVLPs of HEK 293T‐PalmGFP cells (d) from NTA measurements. (Note: NTA measurements required to dilute the original vesicle solutions 100×).
Of note, in the case of the sEVs of HEK 293T‐PalmGFP cells, only 1% of the vesicles were GFP‐positive (See Figure S3). Therefore, we decided to us the ∼100% GFP‐positive plasma membrane‐derived sEVLP in the initial experiments.
After, we carried out force‐controlled injection experiments on HeLa cells using the FluidFM technology with Nanosyringe type FluidFM probes (see Figure 3 for more details). In order to track the possible post‐injection transfer of EVs between cells, we injected one to three cells in each square of the gridded glass slide, as shown in Figure 4a. We selected those individual cells for nanoinjection, which were easily identified in subsequent microscopic experiments. Since the individual squares (Figure 4b) have an area of 500 × 500 µm, it is reasonable to assume that if EVs were visible in multiple adjacent cells within a square, this could only occur because of EV transport across the plasma membrane.
FIGURE 3.
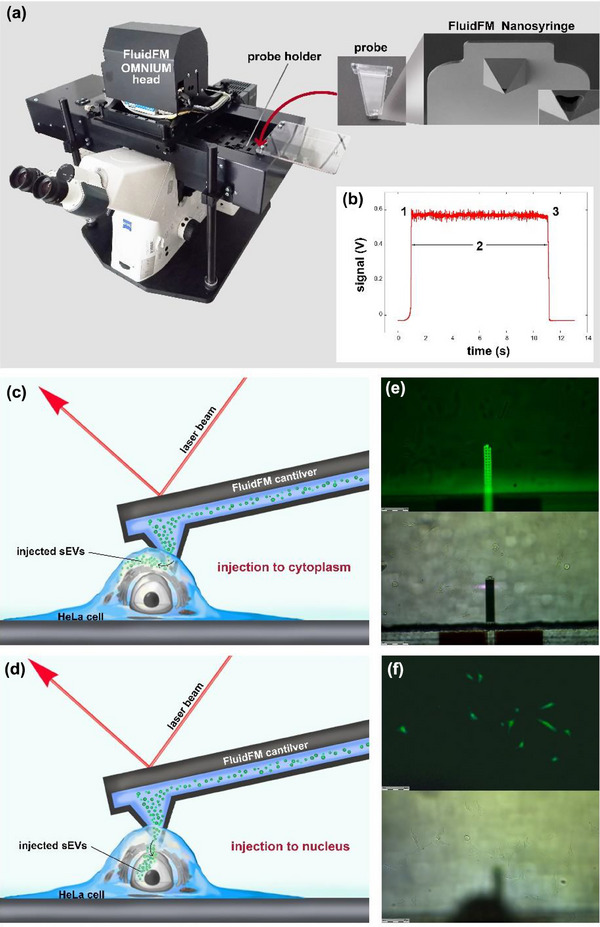
Summary of the FluidFM measurements. (a) Photo of the FluidFM appliance, probe and the FluidFM Nanosyringe. (b) Change of the voltage signal during injection of a live cell. At point ‘1’, contact is established, and the cantilever penetrates into the cell. During phase ‘2’, the setpoint is kept constant and a positive pressure is applied in the cantilever. At point ‘3’, the cantilever is retracted and leaves the cell. (c) Schematic illustration of the injection of sEVs to the cytoplasm by FluidFM. (d) Schematic illustration of the injection of sEVs to the nucleus by FluidFM. (e) Microscopic image of the FluidFM cantilever in fluorescent mode (upper part) and bright field mode (bottom part). (f) Microscopic image of the cells in fluorescent mode (upper part) and bright field mode (bottom part).
FIGURE 4.

Localisation and microscopic analysis of the injected cells. Microscopic images reveal inter‐ and intra‐cellular trafficking of the injected vesicles and confirm viability of the injected cells after transfection and injection. HeLa cells were grown on the surface of gridded glass coverslips (a) and typically a single cell was injected in each square (b). Based on the recorded coordinates, we were able to identify the injected cells during the later microscopic analysis (c). Localisation and intercellular transfer of GFP positive particles (HEK 293T‐PalmGFP cell‐derived sEV‐like particles) 1 h after the nanoinjection (d,e). HeLa cells were injected by applying 40 mbar (d) and 50 mbar (e) pressure. The presence of GFP was validated by immunocytochemistry using unlabelled anti‐GFP primary and ATTO550‐labelled secondary antibody. DAPI was used for detection of DNA and Cy5‐ conjugated lactadherin was used for membrane staining. In the donor cells (*) green, and in the acceptor cells yellow arrows point to GFP containing sEV‐like particles in large (approx. 0.5‐1 µm) membrane closed vesicles, and red the arrow shows similar DNA containing large vesicle. Nanoinjection of BMMC‐GFP derived sEVs into HeLa (f), LCLC‐103H (g), MDA‐MB‐231 (h) and H9c2 (i) cells. The presence of GFP was validated by immunocytochemistry using unlabelled anti‐GFP primary and ATTO550‐labelled secondary antibodies. DAPI was used for detection of DNA and Cy5‐conjugated lactadherin was used for membrane staining. HeLa cells 24 h after the injection of GFP mRNA (j). Viability of HeLa cells after transfection and nanoinjection of EVs was tested by assessing the GFP expression of the pMAXGFP plasmid (Lonza) (k,l). HeLa cells were transfected using Lipofectamine 2000 and the cells expressed GFP at different levels (k). Instead of traditional transfection, the same plasmid was nanoinjected into HeLa cells, the injected cell went through mitotic cell divisions, and the daughter cells showed bright GFP fluorescence (l). HeLa cell 24 h after the nanoinjection of HEK 293 cell‐derived pMAXGFP plasmid containing sEVLPs (m).
Since the diameter of the injected particles can reach up to 300–400 nm, and the aperture of the Nanosyringe is approximately 600 nm in size (Figure 3a), the possible clogging of the probes was constantly monitored. Before injection of the target cells, we verified the flow in the Nanosyringe by injecting a cell outside the area of interest using 1000 mbars for 10 s. Using such a high pressure, the injected cell visually expanded providing immediate feedback about the flow. After the cells of interest were injected, another high‐pressure injection was carried out to verify that the Nanosyringe remained functional and did not clog. This was necessary since injections with mild parameters (30–150 mbars) did not result in visible cellular expansion or in any other morphological changes of the cells. While usually there is a fluorescent tracer mixed with the injection liquid, here we aimed to prevent co‐injection of fluorescent tracers to avoid non‐EV related fluorescence during the successive imaging.
By employing the above nanoinjection settings, the estimated volume injected into a single cell is around 100–200 fL. In order to estimate the number of injected vesicles in such tiny volumes, we performed TEM experiments with vesicle suspensions deposited on a TEM grid in droplets with known volumes. Based on the applied volumes of the TEM samples, the surface of the grids and the injected volumes, we estimated that approximately 10–100 vesicles were injected into a single live HeLa cell. Microscopic images visualizing the injected cells confirmed this finding. Of note, given the employed idle pressure (1 mbar), the area of the aperture (0.2 µm2) and the time of probe transfer (<1 s) to the targeted cells, possible leakage of vesicles from the probes directly to the extracellular solution around the cells was negligible.
After nanoinjection, the cells were fixed using the previously described protocol and kept at 4°C until staining and imaging with confocal microscopy. By confocal microscopy, we could recognize both types of nanoinjected vesicles (sEVs of BMMC cells and HEK 293T‐PalmGFP cell‐derived sEVLPs) in the cells. We validated the presence of GFP with an anti‐GFP primary and an anti‐mouse secondary antibody (Figure 4f‐i). Interestingly, the injected particles were lumped together in a single spot of <5 µm within the cytoplasm in the case of HeLa, MDA‐MB‐231 and LCLC‐103H (Figure 4f‐h). Thus, the injected vesicles did not spread within the cytosol, rather they remained localized in a single EV cluster within the cell. According to this observation, when the pressure is switched on at the Nanosyringe tip inside the cell, probably due to the viscosity of the cytosol and the vesicle suspension, the EV particles do not spread within the cytoplasm. However, later the deposited cluster of EVs is actively processed by the cell. In H9c2 cells, we found some dissemination of the injected vesicles, and we hypothesize that the extensive filamentous network of the cardiomyoblast H9c2 cells enabled spreading of the injected vesicles (Figure 4i).
To get a glimpse into this process, additional injections were carried out. In these experiments, after the injection, the cells were incubated at 37°C at 5% CO2 for 1 h. This extra incubation step allowed the cells to process the introduced vesicles, and the intracellular localization of the sEVLPs clearly changed after the 1 h incubation. Instead of being present in a single lump of particles, they were distributed in several spots (Figure 4d,e), and were often located close to the periphery of the cytoplasm (Figure 4d,e). We hypothesize that cells recognized the injected particles and surrounded them by endomembranes to form membrane enclosed particles (ranging from 0.8 to 2 µm in diameter) for transport towards the plasma membrane for a possible release (Figure 5).
FIGURE 5.

Microscopic images showing the injected vesicles from LCLC‐103H surrounded by membrane (a), and MDA‐MB‐231 (b) and released from cells in membrane surrounded vesicles in the case of H9c2 (c). Red colour indicates the membrane staining by lactadherin while green represents GFP.
The proposed release of the nanoinjected EVs is supported by our finding that in addition to EVs found within the injected cells (Figure 4d,e, cells labeled with asterisks), we also observed green fluorescent EVs in multiple, adjacent cells within a single square of the coverslip grid (Figure 4d,e). This observation suggests that within the 1 h incubation period, packages of EVs can be transported between cells via a hitherto unknown biological process to be investigated in more detail in a later work.
In order to test the further technical capabilities of our injection protocols, we also injected HeLa cells with GFP mRNA. After the injection, we cultured the cells for one day, and successfully confirmed the translation of the mRNA to protein (Figure 4j). During these experiments, 51.7% of the injected cells were confirmed to be fluorescent the next day.
In order to further verify the parameters of the employed protocols and to confirm cell viability after injection, HeLa cells were either transfected or injected by a GFP expressing plasmid in parallel experiments. HeLa cells transfected by lipofectamine and GFP plasmids, showed variable expression of GFP (see Figure 4k). Figure 4l shows that upon injection of the GFP plasmid, only the injected cell and its daughter cells showed GFP fluorescence also proving that the nanoinjected cells were viable and were capable of undergoing mitosis.
To prove that the cargo of the injected vesicles can be released into the cytoplasm, we injected HeLa cells with sEVLPs carrying a GFP‐coding plasmid (see the particle preparation in Figure 1 black arrows). After nanoinjection, the cells were cultured in an incubator for one day, and the cells were fixed. Importantly, we found that the cytoplasm of the injected cells was fluorescent suggesting that the cargo was indeed released, and the GFP‐encoding plasmid was expressed in the cells (Figure 4m).
The fate of nanoinjected sEVs and their lysosomal degradation was followed by live cell imaging. Nanoinjected cells were stained by LysoTracker and were monitored by confocal microscopy for 24 h. Imaging of the nanoinjected cells started approximately 3 h after the injection. The fate of nanoinjected EVs was also compared with the one after cellular EV uptake. At the beginning of live cell imaging, nanoinjected EVs were distributed in the cytoplasm as several discrete fluorescent dots in line with what we observed in the case of the fixed cells. In contrast to the physiological EV uptake, where the number and the intensity of the GFP positive cytoplasmic dots increased with time, the level of the injected intracellular GFP containing EVs remained constant in the studied confocal plane (Figure 6a). During cellular EV uptake, together with the increasing GFP signal, the GFP‐lysosome co‐localization increased showing a saturation kinetics. This saturation was not present in the case of the nanoinjected EVs, and a co‐localisation of GFP signal with lysosomes was not detected. Video recordings of representative cells are available as supplementary files: EV uptake (Video S1) and nanoinjected HeLa cells (Video S2). Given that 3D reconstruction could not be performed after live imaging and we could only observe a single confocal plane, displacement of the nanoinjected EVs could not be followed as in the case of fixed and 3D reconstructed cells.
FIGURE 6.

Live cell imaging and EV co‐localization with lysosomes. HeLa cells were grown in 10‐well cell culture chamber‐slides (EV uptake) and 35 mm diameter Petri dishes with gridded coverslip bottom (injected cells). Representative cells at different time points (0‐6‐12‐18 h after starting live cell imaging) are shown. The estimated boundaries of the cells are indicated with dashed white lines, inserts show EVs and lysosomes with increased magnification. (a) Lactadherin‐based membrane labelling and 3D reconstruction was not possible during live cell imaging. Lactadherin binds to phosphatidyl‐serine (PS) molecules which are not available on the surface of living cells. Images obtained by transmitted light are not confocal images in our system, thus, they cannot be used for 3D reconstruction. Therefore, the plasma membrane cannot be precisely localized. Pearson's correlation was calculated to study co‐localization of lysosomes and GFP containing EVs. Scale bars represent 10 µm. (b) Depending on cellular movements at each time point, co‐localization analysis was performed on 16–44 individual cells during EV uptake and 12–14 cells after EV nanoinjection. Figure shows data points, mean and ±SEM (****: p < 0.0001).
An obvious emerging question relates to the applicability of the platform described in this manuscript. This single cell nanoinjection technique may enable organelle‐specific targeting of nanoinjected EVs along with their cargo (Figure 7). Cells can be engineered to release EVs with surface molecules (Danilushkina et al., 2023; Vukman et al., 2020; Wang et al., 2023) that are known to have a role in organelle targeting (Pfeffer & Aivazian, 2004; Saminathan et al., 2022; Wu et al., 2010) (e.g., various Rab proteins or proteins with known signal sequences). Alternatively, targeting molecules can be conjugated to the surface of EVs using click chemistry (Murphy et al., 2019; Ruan et al., 2023; Smyth et al., 2014) or the nanoinjected EVs can be decorated with targeting proteins by generating an artificial protein corona (Musicò et al., 2023; Tóth et al., 2021).
FIGURE 7.

Examples of subcellular pathways potentially targeted by nanoinjected EVs (Gorvel et al., 1991; Goud et al., 2018; Jin et al., 2021; Koike & Jahn, 2017; Lőrincz et al., 2017; Pfeffer & Aivazian, 2004; Saminathan et al., 2022; Soldati et al., 1995; Zerial & McBride, 2001; Zhang et al., 2021). PM indicates plasma membrane.
An EV‐mediated direct organelle targeting cannot be achieved by the conventional endosomal route of EV uptake (Liu & Wang, 2023; O'Brien et al., 2022), therefore, the direct nanoinjection into the cytosol of cells may provide important novel experimental tool.
4. CONCLUSIONS
We successfully injected mRNAs, plasmids, sEVs of BMMC cells, HEK 293T‐PalmGFP cell‐derived sEVLPs and plasmid containing EVLPs to live HeLa cells using robotic fluidic force microscopy. sEVs of BMMC cells were also injected into MDA‐MB‐231, LCLC‐103H and H9c2 cells. Our observations using confocal microscopy showed that our injections resulted in the deposition of EVs or EVLP clusters in the cytoplasm. No immediate spread of the injected EVs or EV‐like particles in the cytoplasm was observed. Upon incubation of cells for 1 h after injection, we observed that the sEVs were transported in a membrane‐enclosed intracellular compartment towards the periphery of the cell. Live cell imaging provided evidence that at least part of the injected EVs remain in the cytoplasm even 24 h after the injection. In contrast to spontaneously uptaken EVs, the injected EVs did not co‐localize with and degraded by lysosomes. Furthermore, fluorescent EV packages also appeared in the adjacent non‐injected cells suggesting that intercellular trafficking of the EVs occurred within the incubation period. The presented data show that robotic fluidic force microscopy is suitable for the injection of EVs into single live cells, and this new experimental system may open up new research opportunities in the field of EV biology.
AUTHOR CONTRIBUTIONS
Frederic St‐Denis‐Bissonnette: Conceptualization; data curation; formal analysis; investigation; methodology; project administration; validation; visualization; writing—original draft; writing—review & editing. Sarah E. Cummings: Conceptualization; data curation; formal analysis; investigation; methodology; validation; visualization; writing—original draft; writing—review & editing. Shirley Qiu: Conceptualization; data curation; formal analysis; investigation; methodology; validation; visualization; writing—original draft; writing—review & editing. Andrew Stalker: Data curation; formal analysis; methodology; resources; software; validation; writing—review & editing. Gauri Muradia: Data curation; methodology. Jelica Mehic: Data curation; methodology. Karan Mediratta: Data curation; formal analysis; methodology; visualization; writing—review & editing. Shelby Kaczmarek: Methodology; writing—review & editing. Dylan Burger: Supervision; validation; writing—review & editing. Seung‐Hwan Lee: Validation; writing—review & editing. Lisheng Wang: Conceptualization; formal analysis; fundingAcquisition; investigation; methodology; project administration; resources; supervision; validation; visualization; writing—original draft; writing—review & editing. Jessie R. Lavoie: Conceptualization; data curation; formal analysis; fundingAcquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review & editing.
CONFLICT OF INTEREST STATEMENT
Edit I. Buzás is a member of the Advisory Board of Sphere Gene Therapeutics Inc. (Boston, USA).
Supporting information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
Szilvia Bősze kindly provided LCLC‐103H and MDA MB‐231 cells from the Research Group of Peptide Chemistry, Department of Organic Chemistry, Eötvös L. University, Budapest, Hungary. Help with the mRNA injection experiments of Cytosurge is greatly appreciated. This work was funded by the NVKP_16‐1‐2016‐0017, FK 138851, ÚNKP‐21‐5‐ELTE‐1122, TKP Health Program (TKP2022‐EGA‐04), ÚNKP‐23‐3‐I‐SE‐2, KDP‐2021 and KKP_19 (‘Élvonal’) grants of the Hungarian National Research, Development and Innovation Office (NKFIH). Lactadherin‐based membrane labelling technology development was funded by the Semmelweis Innovation Fund (STIA 2020 KFI). This work was also supported by the NKFIH OTKA120237, OTKA PD 131543, VEKOP‐2.3.2‐16‐2016‐00002, VEKOP‐2.3.3‐15‐2017‐00016, Hungarian Academy of Sciences Lendület, BO/00514/21, BO‐00514‐21‐8 and European Commission under Grant number H2020‐MSCA ITN‐2017‐722148 TRAIN EV. The project has also received funding from the EU's Horizon 2020 research and innovation program under grant agreement No. 739593. The study was also financed by the Higher Education Institutional Excellence Program of the Ministry of Human Capacities in Hungary within the framework of the ‘Therapeutic development’ thematic programs. This paper was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences (for B.P.). Figure 7 was generated in Biorender.
Kovács, K. D. , Visnovitz, T. , Gerecsei, T. , Peter, B. , Kurunczi, S. , Koncz, A. , Németh, K. , Lenzinger, D. , Vukman, K. V. , Balogh, A. , Rajmon, I. , Lőrincz, P. , Székács, I. , Buzás, E. I. , & Horvath, R. (2023). Nanoinjection of extracellular vesicles to single live cells by robotic fluidic force microscopy. Journal of Extracellular Vesicles, 12, e12388. 10.1002/jev2.12388
Contributor Information
Edit I. Buzás, Email: buzas.edit@med.semmelweis-univ.hu.
Robert Horvath, Email: horvath.robert@ek.hun-ren.hu.
REFERENCES
- Chen, X. , Kis, A. , Zettl, A. , & Bertozzi, C. R. (2007). A cell nanoinjector based on carbon nanotubes. Proceedings of the National Academy of Sciences of the United States of America, 104(20), 8218–8222. www.pnas.org/cgi/content/full/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Verdera, H. , Gitz‐Francois, J. J. , Schiffelers, R. M. , & Vader, P. (2017). Cellular uptake of extracellular vesicles is mediated by clathrin‐independent endocytosis and macropinocytosis. Journal of Controlled Release, 266, 100–108. 10.1016/j.jconrel.2017.09.019 [DOI] [PubMed] [Google Scholar]
- Danilushkina, A. A. , Emene, C. C. , Barlev, N. A. , & Gomzikova, M. O. (2023). Strategies for engineering of extracellular vesicles. International Journal of Molecular Sciences, 24(17), 13247. 10.3390/ijms241713247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, G. J. , & McMahon, H. T. (2009). Mechanisms of endocytosis. Annual Review of Biochemistry, 78(1), 857–902. 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- Gerecsei, T. , Erdődi, I. , Peter, B. , Hős, C. , Kurunczi, S. , Derényi, I. , Szabó, B. , & Horvath, R. (2019). Adhesion force measurements on functionalized microbeads: An in‐depth comparison of computer controlled micropipette and fluidic force microscopy. Journal of Colloid and Interface Science, 555, 245–253. 10.1016/j.jcis.2019.07.102 [DOI] [PubMed] [Google Scholar]
- Gorvel, J. P. , Chavrier, P. , Zerial, M. , & Gruenberg, J. (1991). rab5 controls early endosome fusion in vitro. Cell, 64(5), 915–925. 10.1016/0092-8674(91)90316-q [DOI] [PubMed] [Google Scholar]
- Goud, B. , Liu, S. , & Storrie, B. (2018). Rab proteins as major determinants of the Golgi complex structure. Small GTPases, 9(1–2), 66–75. 10.1080/21541248.2017.1384087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume‐Gentil, O. , Grindberg, R. V. , Kooger, R. , Dorwling‐Carter, L. , Martinez, V. , Ossola, D. , Pilhofer, M. , Zambelli, T. , & Vorholt, J. A. (2016). Tunable single‐cell extraction for molecular analyses. Cell, 166(2), 506–516. 10.1016/j.cell.2016.06.025 [DOI] [PubMed] [Google Scholar]
- Guillaume‐Gentil, O. , Potthoff, E. , Ossola, D. , Dörig, P. , Zambelli, T. , & Vorholt, J. A. (2013). Force‐controlled fluidic injection into single cell nuclei. Small (Weinheim an der Bergstrasse, Germany), 9(11), 1904–1907. 10.1002/smll.201202276 [DOI] [PubMed] [Google Scholar]
- Guillaume‐Gentil, O. , Rey, T. , Kiefer, P. , Ibáñez, A. J. , Steinhoff, R. , Brönnimann, R. , Dorwling‐Carter, L. , Zambelli, T. , Zenobi, R. , & Vorholt, J. A. (2017). Single‐cell mass spectrometry of metabolites extracted from live cells by fluidic force microscopy. Analytical Chemistry, 89(9), 5017–5023. 10.1021/acs.analchem.7b00367 [DOI] [PubMed] [Google Scholar]
- Guillaume‐Gentil, O. , Zambelli, T. , & Vorholt, J. A. (2014). Isolation of single mammalian cells from adherent cultures by fluidic force microscopy. Lab on a Chip, 14(2), 402–414. 10.1039/c3lc51174j [DOI] [PubMed] [Google Scholar]
- György, B. , Hung, M. E. , Breakefield, X. O. , & Leonard, J. N. (2015). Therapeutic applications of extracellular vesicles: Clinical promise and open questions. Annual Review of Pharmacology and Toxicology, 55, 439–464. 10.1146/annurev-pharmtox-010814-124630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , Tang, Y. , Yang, L. , Peng, X. , Li, B. , Fan, Q. , Wei, S. , Yang, S. , Li, X. , Wu, B. , Huang, M. , Tang, S. , Liu, J. , & Li, H. (2021). Rab GTPases: Central coordinators of membrane trafficking in cancer. Frontiers in Cell and Developmental Biology, 9, 648384. 10.3389/fcell.2021.648384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert, C. , & Kalluri, R. (2013). Exosomes in tumor microenvironment influence cancer progression and metastasis. Journal of Molecular Medicine, 91(4), 431–437. 10.1007/s00109-013-1020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike, S. , & Jahn, R. (2017). Probing and manipulating intracellular membrane traffic by microinjection of artificial vesicles. Proceedings of the National Academy of Sciences of the United States of America, 114(46), E9883–E9892. 10.1073/pnas.1713524114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, A. , Turiák, L. , Németh, K. , Lenzinger, D. , Bárkai, T. , Lőrincz, P. , Zelenyánszki, H. , Vukman, K. V. , Buzás, E. I. , & Visnovitz, T. (2023). Endoplasmin is a hypoxia‐inducible endoplasmic reticulum‐derived cargo of extracellular vesicles released by cardiac cell lines. Membranes, 13(4), 431. 10.3390/membranes13040431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumangoye, R. B. , Sakwe, A. M. , Goodwin, J. S. , Patel, T. , & Ochieng, J. (2011). Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS ONE, 6(9), e24234. 10.1371/journal.pone.0024234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C. P. , Kim, E. Y. , Badr, C. E. , Weissleder, R. , Mempel, T. R. , Tannous, B. A. , & Breakefield, X. O. (2015). Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature Communications, 6, 7029. 10.1038/ncomms8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane, T. N. , Raiker, R. S. , & Jay, S. M. (2015). Exogenous DNA loading into extracellular vesicles via electroporation is size‐dependent and enables limited gene delivery. Molecular Pharmaceutics, 12(10), 3650–3657. 10.1021/acs.molpharmaceut.5b00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Liu, L. , & Zambelli, T. (2022). FluidFM for single‐cell biophysics. Nano Research, 15(2), 773–786. 10.1007/s12274-021-3573-y [DOI] [Google Scholar]
- Liu, Y. J. , & Wang, C. (2023). A review of the regulatory mechanisms of extracellular vesicles‐mediated intercellular communication. Cell Communication and Signaling, 21(1), 77. 10.1186/s12964-023-01103-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz, P. , Tóth, S. , Benkő, P. , Lakatos, Z. , Boda, A. , Glatz, G. , Zobel, M. , Bisi, S. , Hegedűs, K. , Takáts, S. , Scita, G. , & Juhász, G. (2017). Rab2 promotes autophagic and endocytic lysosomal degradation. Journal of Cell Biology, 216(7), 1937–1947. 10.1083/jcb.201611027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan, S. , Ji, H. , & Simpson, R. J. (2010). Exosomes: Extracellular organelles important in intercellular communication. Journal of Proteomics, 73(10), 1907–1920. 10.1016/j.jprot.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Meister, A. , Gabi, M. , Behr, P. , Studer, P. , Vörös, J. , Niedermann, P. , Bitterli, J. , Polesel‐Maris, J. , Liley, M. , Heinzelmann, H. , & Zambelli, T. (2009). FluidFM: Combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano Letters, 9(6), 2501–2507. 10.1021/nl901384x [DOI] [PubMed] [Google Scholar]
- Mulcahy, L. A. , Pink, R. C. , & Carter, D. R. F. (2014). Routes and mechanisms of extracellular vesicle uptake. Journal of Extracellular Vesicles, 3, 1–14. 10.3402/jev.v3.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, D. E. , de Jong, O. G. , Brouwer, M. , Wood, M. J. , Lavieu, G. , Schiffelers, R. M. , & Vader, P. (2019). Extracellular vesicle‐based therapeutics: Natural versus engineered targeting and trafficking. Experimental and Molecular Medicine, 51(3), 1–12. 10.1038/s12276-019-0223-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musicò, A. , Zenatelli, R. , Romano, M. , Zendrini, A. , Alacqua, S. , Tassoni, S. , Paolini, L. , Urbinati, C. , Rusnati, M. , Bergese, P. , Pomarico, G. , & Radeghieri, A. (2023). Surface functionalization of extracellular vesicle nanoparticles with antibodies: A first study on the protein corona “variable”. Nanoscale Advances, 5(18), 4703–4717. 10.1039/D3NA00280B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musto, M. , Parisse, P. , Pachetti, M. , Memo, C. , Di Mauro, G. , Ballesteros, B. , Lozano, N. , Kostarelos, K. , Casalis, L. , & Ballerini, L. (2021). Shedding plasma membrane vesicles induced by graphene oxide nanoflakes in brain cultured astrocytes. Carbon, 176, 458–469. 10.1016/j.carbon.2021.01.142 [DOI] [Google Scholar]
- Nabi, I. R. , & Le, P. U. (2003). Caveolae/raft‐dependent endocytosis. Journal of Cell Biology, 161(4), 673–677. 10.1083/jcb.200302028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, Á. G. , Kámán, J. , Horváth, R. , & Bonyár, A. (2019). Publisher correction: Spring constant and sensitivity calibration of FluidFM micropipette cantilevers for force spectroscopy measurements. Scientific Reports, 9(1), 10287. 10.1038/s41598-019-54634-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, K. , Ughetto, S. , Mahjoum, S. , Nair, A. V. , & Breakefield, X. O. (2022). Uptake, functionality, and re‐release of extracellular vesicle‐encapsulated cargo. Cell Reports, 39(2), 110651. 10.1016/j.celrep.2022.110651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshchepkova, A. , Markov, O. , Evtushenko, E. , Chernonosov, A. , Kiseleva, E. , Morozova, K. , Matveeva, V. , Artemyeva, L. , Vlassov, V. , & Zenkova, M. (2021). Tropism of extracellular vesicles and cell‐derived nanovesicles to normal and cancer cells: New perspectives in tumor‐targeted nucleic acid delivery. Pharmaceutics, 13(11), 1911. 10.3390/pharmaceutics13111911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S. , & Aivazian, D. (2004). Targeting Rab GTPases to distinct membrane compartments. Nature reviews. Molecular Cell Biology, 5(11), 886–896. 10.1038/nrm1500 [DOI] [PubMed] [Google Scholar]
- Potthoff, E. , Guillaume‐Gentil, O. , Ossola, D. , Polesel‐Maris, J. , LeibundGut‐Landmann, S. , Zambelli, T. , & Vorholt, J. A. (2012). Rapid and serial quantification of adhesion forces of yeast and mammalian cells. PLoS ONE, 7(12), e52712. 10.1371/journal.pone.0052712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff, E. , Ossola, D. , Zambelli, T. , & Vorholt, J. A. (2015). Bacterial adhesion force quantification by fluidic force microscopy. Nanoscale, 7(9), 4070–4079. 10.1039/c4nr06495j [DOI] [PubMed] [Google Scholar]
- Raposo, G. , & Stoorvogel, W. (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. The Journal of Cell Biology, 200(4), 373–383. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, H. , Li, Y. , Wang, C. , Jiang, Y. , Han, Y. , Li, Y. , Zheng, D. , Ye, J. , Chen, G. , Yang, G. Y. , Deng, L. , Guo, M. , Zhang, X. , Tang, Y. , & Cui, W. (2023). Click chemistry extracellular vesicle/peptide/chemokine nanocarriers for treating central nervous system injuries. Acta Pharmaceutica Sinica. B, 13(5), 2202–2218. 10.1016/j.apsb.2022.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A. E. , Sneider, A. , Witwer, K. W. , Bergese, P. , Bhattacharyya, S. N. , Cocks, A. , Cocucci, E. , Erdbrügger, U. , Falcon‐Perez, J. M. , Freeman, D. W. , Gallagher, T. M. , Hu, S. , Huang, Y. , Jay, S. M. , Kano, S. I. , Lavieu, G. , Leszczynska, A. , Llorente, A. M. , Lu, Q. , … Vader, P. (2019). Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: An ISEV position paper arising from the ISEV membranes and EVs workshop. Journal of Extracellular Vesicles, 8(1), 1684862. 10.1080/20013078.2019.1684862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftics, A. , Türk, B. , Sulyok, A. , Nagy, N. , Gerecsei, T. , Szekacs, I. , Kurunczi, S. , & Horvath, R. (2019). Biomimetic dextran‐based hydrogel layers for cell micropatterning over large areas using the FluidFM BOT technology. Langmuir: The ACS Journal of Surfaces and Colloids, 35(6), 2412–2421. 10.1021/acs.langmuir.8b03249 [DOI] [PubMed] [Google Scholar]
- Saminathan, A. , Zajac, M. , Anees, P. , & Krishnan, Y. (2022). Organelle‐level precision with next‐generation targeting technologies. Nature Reviews Materials, 7(5), 355–371. 10.1038/s41578-021-00396-8 [DOI] [Google Scholar]
- Simpson, R. J. , Lim, J. W. E. , Moritz, R. L. , & Mathivanan, S. (2009). Exosomes: Proteomic insights and diagnostic potential. Expert Review of Proteomics, 6(3), 267–283. 10.1586/epr.09.17 [DOI] [PubMed] [Google Scholar]
- Smyth, T. , Petrova, K. , Payton, N. M. , Persaud, I. , Redzic, J. S. , Graner, M. W. , Smith‐Jones, P. , & Anchordoquy, T. J. (2014). Surface functionalization of exosomes using click chemistry. Bioconjugate Chemistry, 25(10), 1777–1784. 10.1021/bc500291r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati, T. , Rancaño, C. , Geissler, H. , & Pfeffer, S. R. (1995). Rab7 and Rab9 are recruited onto late endosomes by biochemically distinguishable processes. Journal of Biological Chemistry, 270(43), 25541–25548. 10.1074/jbc.270.43.25541 [DOI] [PubMed] [Google Scholar]
- Subra, C. , Grand, D. , Laulagnier, K. , Stella, A. , Lambeau, G. , Paillasse, M. , De Medina, P. , Monsarrat, B. , Perret, B. , Silvente‐Poirot, S. , Poirot, M. , & Record, M. (2010). Exosomes account for vesicle‐mediated transcellular transport of activatable phospholipases and prostaglandins. Journal of Lipid Research, 51(8), 2105–2120. 10.1194/jlr.M003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztilkovics, M. , Gerecsei, T. , Peter, B. , Saftics, A. , Kurunczi, S. , Szekacs, I. , Szabo, B. , & Horvath, R. (2020). Single‐cell adhesion force kinetics of cell populations from combined label‐free optical biosensor and robotic fluidic force microscopy. Scientific Reports, 10(1), 61. 10.1038/s41598-019-56898-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Amigorena, S. , Raposo, G. , & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology, Chapter 3: Unit 3 22. 10.1002/0471143030.cb0322s30 [DOI] [PubMed]
- Tóth, E. Á. , Turiák, L. , Visnovitz, T. , Cserép, C. , Mázló, A. , Sódar, B. W. , Försönits, A. I. , Petővári, G. , Sebestyén, A. , Komlósi, Z. , Drahos, L. , Kittel, Á. , Nagy, G. , Bácsi, A. , Dénes, Á. , Gho, Y. S. , Szabó‐Taylor, K. É. , & Buzás, E. I. (2021). Formation of a protein corona on the surface of extracellular vesicles in blood plasma. Journal of Extracellular Vesicles, 10(11), e12140. 10.1002/jev2.12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekström, K. , Bossios, A. , Sjöstrand, M. , Lee, J. J. , & Lötvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9(6), 654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Visnovitz, T. , Osteikoetxea, X. , Sódar, B. W. , Mihály, J. , Lőrincz, P. , Vukman, K. V. , Tóth, E. Á. , Koncz, A. , Székács, I. , Horváth, R. , Varga, Z. , & Buzás, E. I. (2019). An improved 96 well plate format lipid quantification assay for standardisation of experiments with extracellular vesicles. Journal of Extracellular Vesicles, 8(1), 1565263. 10.1080/20013078.2019.1565263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukman, K. V. , Ferencz, A. , Fehér, D. , Juhos, K. , Lőrincz, P. , Visnovitz, T. , Koncz, A. , Pálóczi, K. , Seregélyes, G. , Försönits, A. , Khamari, D. , Galinsoga, A. , Drahos, L. , & Buzás, E. I. (2020). An implanted device enables in vivo monitoring of extracellular vesicle‐mediated spread of pro‐inflammatory mast cell response in mice. Journal of Extracellular Vesicles, 10(1), e12023. 10.1002/jev2.12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Wang, D. , Ye, Z. , & Xu, J. (2023). Engineering extracellular vesicles as delivery systems in therapeutic applications. Advanced Science (Weinheim, Baden‐Wurttemberg, Germany), 10(17), e2300552. 10.1002/advs.202300552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, J. C. (2000). Electroporation of cells and tissues. IEEE Transactions on Plasma Science, 28(1), 24–33. 10.1109/27.842820 [DOI] [Google Scholar]
- Wei, Y. , & Xu, Q. (2019). A survey of force‐assisted robotic cell microinjection technologies. IEEE Transactions on Automation Science and Engineering, 16(2), 931–945. 10.1109/TASE.2018.2878867 [DOI] [Google Scholar]
- Welsh, J. A. , Arkesteijn, G. J. A. , Bremer, M. , Cimorelli, M. , Dignat‐George, F. , Giebel, B. , Görgens, A. , Hendrix, A. , Kuiper, M. , Lacroix, R. , Lannigan, J. , van Leeuwen, T. G. , Lozano‐Andrés, E. , Rao, S. , Robert, S. , de Rond, L. , Tang, V. A. , Tertel, T. , Yan, X. , … van der Pol, E. (2023). A compendium of single extracellular vesicle flow cytometry. Journal of Extracellular Vesicles, 12(2), e12299. 10.1002/jev2.12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. (2021). On your MARCKS, get set, deliver: Engineering extracellular vesicles. Molecular Therapy: The Journal of the American Society of Gene Therapy, 29(5), 1664–1665. 10.1016/j.ymthe.2021.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, B. , Jevremovic, D. , Béné, M. C. , Yan, M. , Jacobs, P. , Litwin, V. , & ICSH/ICCS Working Group . (2013). Validation of cell‐based fluorescence assays: Practice guidelines from the ICSH and ICCS ‐ part V ‐ assay performance criteria. Cytometry. Part B, Clinical cytometry, 84(5), 315–323. 10.1002/cyto.b.21108 [DOI] [PubMed] [Google Scholar]
- Wu, Y. W. , Oesterlin, L. K. , Tan, K. T. , Waldmann, H. , Alexandrov, K. , & Goody, R. S. (2010). Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nature Chemical Biology, 6(7), 534–540. 10.1038/nchembio.386 [DOI] [PubMed] [Google Scholar]
- Zerial, M. , & McBride, H. (2001). Rab proteins as membrane organizers. Nature Reviews. Molecular Cell Biology, 2(2), 107–117. 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Tan, J. , Miao, Y. , & Zhang, Q. (2021). The effect of extracellular vesicles on the regulation of mitochondria under hypoxia. Cell Death and Disease, 12(4), 358. 10.1038/s41419-021-03640-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information


