Abstract
Stress increases alcohol consumption in dependent animals and contributes to the development of alcohol use disorder. The nucleus of the solitary tract (NTS) is a critical brainstem region for integrating and relaying central and peripheral signals to regulate stress responses, but it is not known if it plays a role in alcohol dependence- or in stress-induced escalations in alcohol drinking in dependent mice. Here, we used RNA-sequencing and bioinformatics analyses to study molecular adaptations in the NTS of C57BL/6J male mice that underwent an ethanol drinking procedure that uses exposure to chronic intermittent ethanol (CIE) vapor, forced swim stress (FSS), or both conditions (CIE + FSS). Transcriptome profiling was performed at three different times after the last vapor cycle (0-hr, 72-hr, and 168-hr) to identify changes in gene expression associated with different stages of ethanol intoxication and withdrawal. In the CIE and CIE + FSS groups at 0-hr, there was upregulation of genes enriched for cellular response to type I interferon (IFN) and type I IFN- and cytokine-mediated signaling pathways, while the FSS group showed upregulation of neuronal genes. IFN signaling was the top gene network positively correlated with ethanol consumption levels in the CIE and CIE + FSS groups. Results from different analyses (differential gene expression, weighted gene coexpression network analysis, and rank-rank hypergeometric overlap) indicated that activation of type I IFN signaling would be expected to increase ethanol consumption. The CIE and CIE + FSS groups also shared an immune signature in the NTS as has been demonstrated in other brain regions after chronic ethanol exposure. A temporal-based clustering analysis revealed a unique expression pattern in the CIE + FSS group that suggests the interaction of these two stressors produces adaptations in synaptic and glial functions that may drive stress-induced drinking.
Keywords: Alcohol dependence, Chronic intermittent ethanol vapor, Voluntary ethanol consumption, Forced swim stress, Nucleus of the solitary tract, Transcriptome, WGCNA, Interferon and immune signaling
1. Introduction
Stress is a major contributor to the development or progression of alcohol and other substance use disorders (Becker, 2017; Lijffijt et al., 2014). Chronic alcohol exposure and withdrawal disrupt the function of the hypothalamic-pituitary-adrenocortical (HPA) axis, a principal neuroendocrine stress system (Lijffijt et al., 2014). Repeated alcohol-induced perturbations act as potent stressors, producing dysregulations in neuroendocrine responses and in brain stress circuits outside the hypothalamus that contribute to the development of alcohol use disorder (AUD) (Becker, 2017). In the brainstem, the nucleus of the solitary tract (NTS) is a critical regulator of stress responses (Herman, 2018). It acts as a hub for integrating inputs from the limbic forebrain (e.g., the prefrontal cortex and amygdala) with peripheral signals (e.g., glucocorticoids and visceral afferents) to influence behavioral, neuroendocrine (via the HPA axis), and autonomic stress responses (Myers et al., 2017). Unlike studies of the HPA or other brain stress systems, there are few studies of how ethanol affects the NTS beyond those showing that acute ethanol administration or binge-like drinking activate neurons in the NTS (Burnham and Thiele, 2017; Robinson et al., 2020; Ryabinin et al., 1997; Thiele et al., 2000). Although the NTS is highly interconnected with stress-regulating systems, the effects of chronic alcohol, repeated stress, or both stressors on this region are not known.
In the present study, we performed transcriptome profiling and bioinformatics analysis to investigate the molecular changes in the NTS from mice that underwent a model of alcohol drinking that incorporates exposure to repeated stress or to chronic intermittent ethanol (CIE) vapor or exposure to both conditions. This well-established model has shown that repeated, brief periods of forced swim stress (FSS) enhance ethanol consumption in dependent (CIE-treated) but not in nondependent mice (Anderson et al., 2016; Haun et al., 2022; Lopez et al., 2016). Our previous findings in prefrontal cortex identified a specific pattern of changes in neurotransmitter and ligand-gated ion channel related genes in mice treated with CIE + FSS (Farris et al., 2020). While the initial goal of the current study was to identify molecular changes in the NTS associated with stress-induced drinking in the CIE + FSS group, we found strong evidence of shared profiles in the CIE and CIE + FSS groups that could be important drivers of drinking escalations in dependent animals. We identified interferon (IFN) signaling as a critical pathway altered by CIE alone and by CIE + FSS. Our analyses predicted that activation of type I IFN signaling would increase ethanol consumption. We further identified a network of genes with a unique temporal expression pattern in the CIE + FSS group that indicate initial changes in synaptic and glial functions and long-term changes in cell survival and intracellular signaling in stress-induced drinking escalations.
2. Methods
2.1. Animals
We profiled the NTS from brains dissected previously from adult male C57BL/6J mice that were housed and treated (as described in the next section) at the Medical University of South Carolina (MUSC). Information on these animals can be found in our previous publication (Farris et al., 2020). We also used transcriptome data from the mPFC from these same mice (Farris et al., 2020). All procedures were conducted in accordance with the Institutional Animal Care and Use Committees at MUSC and the Guide for the Care and Use of Laboratory Animals adopted by the National Institutes of Health.
2.2. CIE-FSS drinking model
The CIE-FSS drinking model conducted previously at MUSC (Farris et al., 2020) is illustrated in Supplemental Fig. 1a and is also described in detail elsewhere (Anderson et al., 2016; Lopez et al., 2016). Briefly, adult male C57BL/6J mice were first trained to drink 15% (v/v) ethanol with access for 1-hr/day starting 3-hr after lights off. Once stable baseline drinking was established (~3 weeks), mice were separated into four treatment groups (equated for baseline levels of ethanol intake): AIR vapor (control), CIE vapor, FSS, and CIE + FSS. Ethanol drinking tests were performed in between CIE or AIR exposure cycles using the same drinking procedure used for baseline measurements. Mice were decapitated after their fifth and last CIE or AIR exposure at 0-hr, 72-hr, or 168-hr, and whole brains were immediately snap-frozen and shipped to UT Austin. The medial prefrontal cortex (mPFC) was dissected and transcriptionally profiled (Farris et al., 2020), and the NTS was isolated and transcriptionally profiled separately for the present study. Drinking data and blood ethanol concentrations (BECs) for these mice can be found in the mPFC study (Farris et al., 2020).
2.3. RNA-sequencing and bioinformatics analyses
Isolated total RNA from mouse NTS (Supplemental Fig. 1b) was submitted to the Genomic Sequencing and Analysis Facility at UT Austin. Sequencing libraries were constructed using a 3′ Tag-based approach (TagSeq), targeting the 3′ end of RNA fragments using ~16 ng/μL of each RNA sample. Samples were sequenced on the HiSeq 2500 (Illumina) platform with a read depth of approximately 7.6 million reads. Reads were mapped to the mouse reference genome (Gencode GRCm38. p6 release M25) using a STAR (version STAR_2.5.4 b) aligner. A total of 89 samples were included with an average of 2.6 million uniquely mapped reads (86.3%). Sequencing sample size (n) for each group: CIE + FSS: 0-hr (n = 7), 72-hr (n = 6), and 168-hr (n = 5); CIE: 0-hr (n = 8), 72-hr (n = 5), and 168-hr (n = 9); FSS: 0-hr (n = 8), 72-hr (n = 7), and 168-hr (n = 9); and CTL: 0-hr (n = 7), 72-hr (n = 8), and 168-hr (n = 10). On average, ~38,000 transcripts per sample were detected, representing ~22,000 protein-coding genes. Read quality was assessed using MultiQC (version 1.7). Raw counts were quantified using HTSeq (version 0.11.2). The raw data are available from the corresponding author upon request. NTS RNA-sequencing data were deposited in NCBI’s Gene Expression Omnibus and accessible at GSE202236. GSE202236 also includes the raw sequence data for the mPFC from Farris et al. (2020).
2.4. Differential gene expression and time-course cluster analyses
The R (version 3.6.1) package DESeq2 (version 1.22.2) identified differentially expressed genes (DEGs) between groups at each time using the default Wald Test or a likelihood ratio test (LRT) across different levels for clustering analysis using the DESeq function. For comparisons within time points, nominal p < 0.05 was selected to ascertain shared and nonshared changes in gene expression. For comparisons across time and groups, the clustering function degPatterns from the R package DEGReports (version 1.33) was used on regularized log transformation of the normalized counts of DEGs identified using LRT on the full DESeq model of ~Group + Time + Group:Time, rather than the reduced model ~ Group + Time. degPatterns uses hierarchical clustering based on pairwise correlations to identify groups of genes similarly expressed across time. This analysis produced clusters (n = 11) of genes with similar expression profiles across time and within groups. Statistically significant changes were limited to genes with FDR <0.05.
2.5. Weighted Gene Co-expression Network Analysis (WGCNA)
The R package WGCNA (Langfelder and Horvath, 2008) (version 1.69) is used to identify genes correlated with behavioral traits of interest. The general framework for WGCNA has been previously described (Zhang and Horvath, 2005). Briefly, we constructed a signed adjacency matrix by calculating Pearson correlations for all pairs of genes. To emphasize strong correlations on an exponential scale, we raised the adjacency to power B (which was recalculated for each time point comparison), so the resulting networks exhibited approximate scale-free topology (scale free topology fit = 0.85). To identify gene modules, all genes were hierarchically clustered based on connection strength determined using a topological overlap dissimilarity calculation. Resulting gene dendrograms were used for module detection using the dynamic tree cut method (minimum module size = 100). To determine module-trait relationships, Pearson correlations were calculated for module eigengene expression with CIE treatment status and BEC. Resulting p-values from module-trait correlations were adjusted for multiple comparisons using FDR <0.05.
2.6. Ingenuity Pathway Analysis (IPA)
Cluster gene lists were entered into Ingenuity Pathway Analysis (IPA) (Jiménez-Marín et al., 2009) (version 1-19-02) with log2 fold-change, p-value, and baseMean as inputs. We used a nominal p < 0.05 to identify DEGs. IPA identified enrichment for canonical pathways and molecular function based on the gene expression patterns submitted.
2.7. Gene ontology and cell-type enrichment analysis
DEGs were analyzed for enrichment of canonical gene ontologies and molecular pathways using the bioinformatics tool and R package Enrichr (Chen et al., 2013; Kuleshov et al., 2016) (version 3.0). Reported ontological categories and pathways were limited to the top six terms for each time within each experimental group (p < 0.05). DEGs (Group + Time) were compared to cell-type marker genes within the community-curated, single-cell RNA-sequencing dataset Panglao Database (Franzén et al., 2019) to determine the overrepresentation of the major CNS cell types using a Fisher’s exact test (p < 0.05).
2.8. Rank-rank hypergeometric overlap (RRHO) test
We used the RRHO test to compare patterns of gene regulation between the NTS and mPFC from the same animals (Farris et al., 2020). RRHO identifies overlap between expression profiles in a threshold-free manner to assess the degree and significance of overlap (Plaisier et al., 2010). Full differential expression lists were ranked by the −log10 (p-value) multiplied by the sign of the fold change from the differential gene expression analysis. The RRHO2 Bioconductor package (version 1.0) was used to evaluate the overlap of differential expression lists between the mPFC and NTS in the CIE + FSS group across time. Heatmaps visualize hypergeometric p-values for significant overlap between sets of genes, where smaller p-values correspond to warmer colors.
3. Results
3.1. Temporal changes in gene expression in the NTS after FSS, CIE, or CIE + FSS
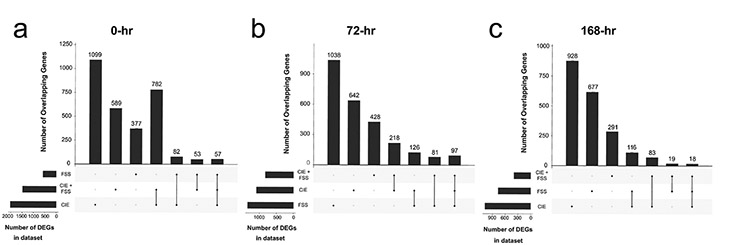
The number of DEGs was determined by comparing the control (AIR) condition with each treatment group (CIE, FSS, and CIE + FSS) at 0-hr, 72-hr, and 168-hr after the last CIE or AIR vapor exposure cycle (see Supplemental Fig. 2b). The number of DEGs with nominal p < 0.05 varied from 412 to 2020 genes (Fig. 1a-c, lower left corners). At 0-hr, the CIE and CIE + FSS groups had the largest numbers of DEGs and overlapping genes (782 common DEGs) (Fig. 1a). At 72-hr, the FSS and CIE groups contained the largest number of DEGs, while the CIE and CIE + FSS groups had more overlapping genes (Fig. 1b). At 168-hr, the CIE and FSS groups had the largest numbers of DEGs and overlapping genes (Fig. 1c). Many of the dysregulated genes were unique to each group and time point. DEGS unique to each treatment group and common between treatment groups compared with the air vapor control group are listed in Supplemental Table 1.
Fig. 1. Number of differentially expressed genes by group and time point.
The R package UpSetR (Version 1.4.0) was used to show the number of differentially expressed genes (DEGs) unique to each treatment group and common between treatment groups compared with the air vapor control group at p < 0.05. Number of DEGs by group at a) 0-hr, b) 72-hr, and c) 168-hr. Total number of DEGs per group is shown in the lower left corner of each plot. The filled circles indicate which group(s) contribute to the plotted number of overlapping DEGs above. FSS, forced swim stress; CIE, chronic intermittent ethanol vapor.
The three times were used to differentiate gene expression patterns related to different stages of intoxication and withdrawal (Jin et al., 2013). For example, 0-hr corresponds to immediate removal from the ethanol vapor inhalation chamber, when blood and brain alcohol levels are relatively high (e.g., 175–225 mg/dL). The 72-hr time corresponds to the early protracted phase of withdrawal, when tremor and convulsions have substantially subsided, but measures of negative affect (i.e., anxiety) may emerge (Heilig et al., 2010). This is also when mice resume access to their next drinking session and are likely in a highly anticipatory state. The 168-hr time is useful for evaluating persistent changes in gene expression after protracted abstinence.
There were striking similarities between the CIE and CIE + FSS treatments when evaluating cellular enrichment of DEGs. For example, upregulated DEGs in both groups at 0-hr were enriched for cellular response to type I IFN, type I IFN- and cytokine-mediated signaling pathways (Table 1), and for microglia at the cell-type level (Table 2). Both groups of ethanol-dependent mice also shared an immune signature, unlike the FSS group, which showed changes in expression of neuronal genes (Tables 1-2).
Table 1. Enrichment of DEGs from the NTS by treatment group and time, separated by upregulated (top) and downregulated (bottom) DEGs.
Ontologies identified using Enrichr; CIE, chronic intermittent ethanol vapor; FSS, forced swim test.
| Time | Group | Name | Overlap | P-value |
|---|---|---|---|---|
| Upreg. | ||||
| 0-hr | CIE + FSS | cellular response to type I interferon (GO:0071357) | 19/65 | 4.95E-12 |
| type I interferon signaling pathway (GO:0060337) | 19/65 | 4.95E-12 | ||
| cytokine-mediated signaling pathway (GO:0019221) | 58/621 | 1.94E-09 | ||
| CIE | cytokine-mediated signaling pathway (GO:0019221) | 88/621 | 1.51E-16 | |
| cellular response to type I interferon (GO:0071357) | 21/65 | 1.58E-11 | ||
| type I interferon signaling pathway (GO:0060337) | 21/65 | 1.58E-11 | ||
| FSS | norepinephrine biosynthetic process (GO:0042421) | 3/6 | 5.15E-05 | |
| synaptic transmission, dopaminergic (GO:0001963) | 3/7 | 8.92E-05 | ||
| norepinephrine metabolic process (GO:0042415) | 3/7 | 8.92E-05 | ||
| 72-hr | CIE + FSS | synapse pruning (GO:0098883) | 5/9 | 9.07E-07 |
| cell junction disassembly (GO:0150146) | 4/6 | 4.73E-06 | ||
| cytokine-mediated signaling pathway (GO:0019221) | 33/621 | 1.90E-05 | ||
| CIE | cilium movement (GO:0003341) | 12/52 | 4.94E-07 | |
| axoneme assembly (GO:0035082) | 7/34 | 7.40E-05 | ||
| cellular response to type I interferon (GO:0071357) | 11/65 | 0.000184 | ||
| FSS | modulation of chemical synaptic transmission (GO:0050804) | 24/109 | 1.32E-11 | |
| chemical synaptic transmission (GO:0007268) | 40/306 | 1.19E-10 | ||
| anterograde trans-synaptic signaling (GO:0098916) | 34/244 | 5.43E-10 | ||
| 168-hr | CIE + FSS | positive regulation of smooth muscle cell migration (GO:0014911) | 3/17 | 0.000829 |
| regulation of cytokine-mediated signaling pathway (GO:0001959) | 5/74 | 0.001421 | ||
| collagen-activated tyrosine kinase receptor signaling pathway (GO:0038063) | 2/7 | 0.002505 | ||
| CIE | response to interferon-gamma (GO:0034341) | 12/80 | 2.59E-06 | |
| cell junction disassembly (GO:0150146) | 4/6 | 9.29E-06 | ||
| neutrophil activation involved in immune response (GO:0002283) | 32/485 | 1.02E-05 | ||
| FSS | cotranslational protein targeting to membrane (GO:0006613) | 7/94 | 0.001274 | |
| regulation of systemic arterial blood pressure by hormone (GO:0001990) | 3/16 | 0.002472 | ||
| membrane raft distribution (GO:0031580) | 2/5 | 0.002916 | ||
| Downreg. | ||||
| 0-hr | CIE + FSS | central nervous system development (GO:0007417) | 27/268 | 5.16E-07 |
| regulation of endothelial cell migration (GO:0010594) | 12/89 | 4.69E-05 | ||
| brain development (GO:0007420) | 16/150 | 5.34E-05 | ||
| CIE | positive regulation of neuron projection development (GO:0010976) | 17/88 | 4.92E-07 | |
| enzyme linked receptor protein signaling pathway (GO:0007167) | 21/140 | 1.91E-06 | ||
| commissural neuron axon guidance (GO:0071679) | 5/8 | 1.05E-05 | ||
| FSS | adenylate cyclase-modulating G protein-coupled receptor signaling pathway (GO:0007188) | 9/165 | 0.000698 | |
| embryonic skeletal system morphogenesis (GO:0048,704) | 4/31 | 0.001001 | ||
| negative regulation of insulin-like growth factor receptor signaling pathway (GO:0043,569) | 2/5 | 0.002036 | ||
| 72-hr | CIE + FSS | regulation of cholesterol esterification (GO:0010872) | 5/12 | 1.03E-06 |
| positive regulation of cholesterol esterification (GO:0010873) | 4/9 | 1.00E-05 | ||
| chylomicron assembly (GO:0034378) | 4/10 | 1.65E-05 | ||
| CIE | myelination (GO:0042552) | 7/48 | 0.0001 | |
| vascular transport (GO:0010232) | 9/84 | 0.000125 | ||
| transport across blood-brain barrier (GO:0150104) | 9/86 | 0.00015 | ||
| FSS | myelination (GO:0042552) | 8/48 | 2.79E-05 | |
| extracellular matrix organization (GO:0030198) | 21/300 | 3.66E-05 | ||
| supramolecular fiber organization (GO:0097435) | 23/351 | 4.44E-05 | ||
| 168-hr | CIE + FSS | chloride transport (GO:0006821) | 5/76 | 0.000475 |
| mammary gland epithelial cell differentiation (GO:0060644) | 2/8 | 0.001945 | ||
| inorganic anion transmembrane transport (GO:0098661) | 4/65 | 0.002279 | ||
| CIE | nervous system development (GO:0007399) | 30/447 | 1.42E-06 | |
| central nervous system development (GO:0007417) | 20/268 | 1.80E-05 | ||
| neural tube development (GO:0021915) | 5/16 | 3.56E-05 | ||
| FSS | regulation of Rho protein signal transduction (GO:0035023) | 8/73 | 0.000133 | |
| sphingolipid metabolic process (GO:0006665) | 10/116 | 0.000153 | ||
| membrane lipid biosynthetic process (GO:0046467) | 7/58 | 0.00019 | ||
Table 2. Cell-type enrichment of DEGs from the NTS by treatment group and time, separated by upregulated (top) and downregulated (bottom) DEGs.
Ontologies identified using Enrichr; CIE, chronic intermittent ethanol vapor; FSS, forced swim test.
| Time | Group | Cell-type | Overlap | P-value |
|---|---|---|---|---|
| Upreg. | ||||
| 0-hr | CIE + FSS | Microglia | 24/158 | 1.91E-08 |
| Plasmacytoid Dendritic Cells | 20/150 | 2.39E-06 | ||
| Eosinophils | 18/127 | 3.14E-06 | ||
| CIE | Microglia | 65/158 | 2.23E-40 | |
| Macrophages | 71/204 | 2.84E-38 | ||
| Plasmacytoid Dendritic Cells | 55/150 | 3.09E-31 | ||
| FSS | Noradrenergic Neurons | 7/104 | 0.000627 | |
| Adrenergic Neurons | 6/106 | 0.003656 | ||
| Intercalated Cells | 6/118 | 0.006157 | ||
| 72-hr | CIE + FSS | Microglia | 27/158 | 9.84E-16 |
| Monocytes | 27/176 | 1.57E-14 | ||
| Plasmacytoid Dendritic Cells | 23/150 | 1.45E-12 | ||
| CIE | Ependymal Cells | 45/136 | 7.20E-34 | |
| Ciliated Cells | 29/104 | 6.73E-20 | ||
| Ionocytes | 17/107 | 3.71E-08 | ||
| FSS | Glutaminergic Neurons | 30/107 | 2.87E-17 | |
| Neuroendocrine Cells | 31/123 | 2.37E-16 | ||
| Immature Neurons | 31/136 | 4.91E-15 | ||
| 168-hr | CIE + FSS | Macrophages | 34/204 | 7.15E-17 |
| Microglia | 29/158 | 1.00E-15 | ||
| Oligodendrocytes | 30/178 | 3.58E-15 | ||
| CIE | Macrophages | 34/204 | 7.15E-17 | |
| Microglia | 29/158 | 1.00E-15 | ||
| Oligodendrocytes | 30/178 | 3.58E-15 | ||
| FSS | Proximal Tubule Cells | 10/164 | 0.000615 | |
| Ependymal Cells | 8/136 | 0.002654 | ||
| Epsilon Cells | 8/138 | 0.002905 | ||
| Downreg. | ||||
| 0-hr | CIE + FSS | Endothelial Cells | 34/246 | 3.59E-12 |
| Müller Cells | 24/147 | 1.71E-10 | ||
| Meningeal Cells | 21/115 | 2.80E-10 | ||
| CIE | Müller Cells | 30/147 | 5.38E-12 | |
| Radial Glia Cells | 26/114 | 9.91E-12 | ||
| Meningeal Cells | 24/115 | 4.40E-10 | ||
| FSS | Tanycytes | 7/121 | 0.00194 | |
| Gamma (PP) Cells | 7/128 | 0.002666 | ||
| Cholinergic Neurons | 6/102 | 0.003717 | ||
| 72-hr | CIE + FSS | Astrocytes | 10/154 | 0.000334 |
| Müller Cells | 9/147 | 0.001002 | ||
| Endothelial Cells | 12/246 | 0.001166 | ||
| CIE | Oligodendrocytes | 26/178 | 4.84E-14 | |
| Glycinergic Neurons | 16/100 | 9.55E-10 | ||
| Astrocytes | 19/154 | 2.42E-09 | ||
| FSS | Oligodendrocytes | 22/178 | 1.25E-09 | |
| Loop Of Henle Cells | 17/147 | 2.54E-07 | ||
| Endothelial Cells | 17/246 | 0.000231 | ||
| 168-hr | CIE + FSS | Interneurons | 14/305 | 0.004839 |
| Osteoblasts | 9/154 | 0.004936 | ||
| Endothelial Cells (Aorta) | 10/185 | 0.005403 | ||
| CIE | Bergmann Glia | 25/132 | 3.33E-15 | |
| Neuroendocrine Cells | 20/123 | 3.73E-11 | ||
| Purkinje Neurons | 22/155 | 6.19E-11 | ||
| FSS | Astrocytes | 9/154 | 6.91E-06 | |
| Oligodendrocytes | 6/178 | 0.004213 | ||
| Bergmann Glia | 5/132 | 0.005437 | ||
The DEGs that were unique to the CIE + FSS group may be important in stress-induced increases in ethanol consumption. There were 589, 428, and 285 unique DEGs at 0-hr, 72-hr, and 168-hr, respectively (Fig. 1a-c). Those at 0-hr were enriched for glutamate signaling and regulation of glial cell differentiation, while DEGs at 72-hr were enriched for steroid metabolic processes and cholesterol esterification (Table 3). At 168-hr, DEGs unique to CIE + FSS were enriched for death-inducing signaling complex and tyrosine kinase signaling (Table 3). These results indicate early adaptations in synaptic and glial cell functions and long-term changes in cell survival and intracellular signaling as potential drivers of stress-induced alcohol consumption in dependent mice.
Table 3. Biological pathways uniquely differentially regulated by CIE + FSS in the NTS.
Ontologies identified using Enrichr; CIE, chronic intermittent ethanol vapor; FSS, forced swim test.
| Time | Name | Overlap | P-value |
|---|---|---|---|
| 0-hr | glutamate receptor signaling pathway (GO:0007215) | 7/37 | 8.8491E-05 |
| regulation of glial cell differentiation (GO:0045685) | 3/7 | 0.00081403 | |
| vascular associated smooth muscle cell differentiation (GO:0035886) | 3/8 | 0.001274 | |
| 72-hr | positive regulation of steroid metabolic process (GO:0045940) | 5/13 | 2.23E-40 |
| positive regulation of cholesterol esterification (GO:0010873) | 4/10 | 2.84E-38 | |
| leukocyte tethering or rolling (GO:0050901) | 5/18 | 3.09E-31 | |
| 168-hr | collagen-activated tyrosine kinase receptor signaling pathway (GO:0038063) | 2/7 | 0.00405324 |
| death-inducing signaling complex assembly (GO:0071550) | 2/7 | 0.00405324 | |
| mammary gland epithelial cell differentiation (GO:0060644) | 2/8 | 0.00535363 |
The FSS treatment group did not show increased ethanol drinking, so its unique DEGs likely represent changes induced by stress alone (Fig. 1). These transcriptional changes in the NTS may inform other behavioral phenotypes altered by exposure to stress. For example, repeated FSS also alters performance in learning and memory tasks (Jin et al., 2013; Warner et al., 2013). Overall, the early and intermediate transcriptional changes after FSS indicate significant alterations in NTS synaptic function (Table 1). Given the abundant changes in immune-related genes observed in both the CIE and CIE + FSS groups, we specifically evaluated differential expression of immune genes in the FSS alone group. Several genes that were upregulated in the CIE and CIE + FSS groups were at some point downregulated in the FSS group (e.g., Ccl8, Il10ra, Il1r1, Ifi27, Il16, and Tnfrsf1a), suggesting that these drive drinking in dependent animals.
3.2. Immune regulatory gene networks are positively correlated with ethanol consumption at 0-hr and 72-hr
Genes tend to work in coordinated networks in response to different treatments. We used WGCNA module-trait relationships to identify gene networks related to ethanol intake across treatment groups. There were several modules at each time point that correlated negatively or positively with ethanol intake on the last test day (Test 4), blood ethanol concentration (BEC), and change in ethanol intake (Test 4 drinking compared with baseline).
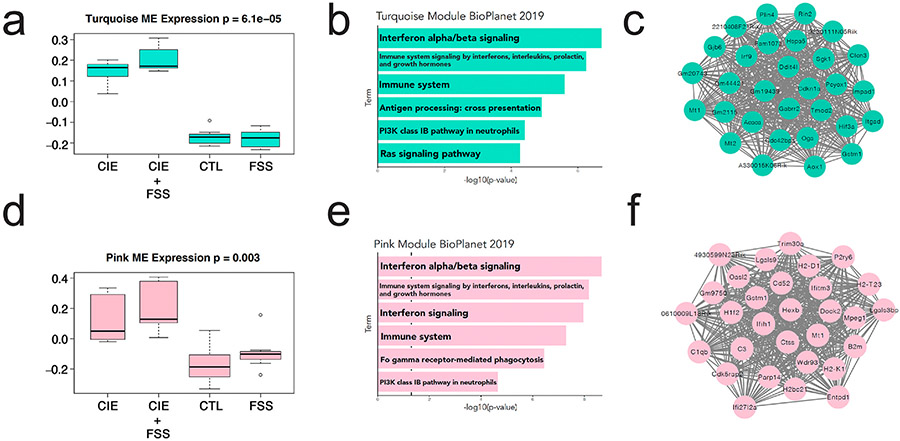
At 0-hr, the turquoise module was positively correlated with ethanol intake on the last test day (r2 = 0.73, p = 8e-06), while the brown, tan, and red modules were negatively correlated (Supplemental Fig. 2a). Turquoise eigengene expression was upregulated in both the CIE and CIE + FSS groups (Fig. 2a), consistent with these genes being positively correlated with ethanol consumption on the final drinking session. Mice in these treatment groups consumed the highest levels of ethanol compared with the FSS or air vapor control groups (Farris et al., 2020). The turquoise module was enriched for IFN α/β signaling, immune signaling by interferons, interleukins, prolactin and growth hormones, the immune system, and antigen processing (Fig. 2b). The top 30 hub genes (genes with the highest number of connections within the network) were significantly upregulated in the CIE + FSS group relative to control (Fig. 2c).
Fig. 2. Weighted Gene Co-expression Network Analysis (WGCNA) modules correlated with ethanol intake at 0-hr and 72-hr.
a) Turquoise module eigengene (ME) expression by group (Summary output of the network analysis is available in Supplemental Tables 2-4). b) Enrichment of turquoise module using BioPlanet repository (Huang et al., 2019) Supplemental Table 5c) Top 30 hub genes for turquoise module. d) Pink module eigengene expression by group (Summary output of the network analysis is available in Supplemental Tables 2-4). e) Enrichment of pink module using BioPlanet repository (Huang et al., 2019) Supplemental Table 5f) Top 30 hub genes for pink module. CTL, air vapor control; FSS, forced swim stress; CIE, chronic intermittent ethanol vapor.
At 72-hr, the pink module was the only one positively correlated with ethanol consumption (r2 = 0.59, p = 0.002) (Supplemental Fig. 2b). Pink eigengene expression was upregulated in the CIE and CIE + FSS groups (Fig. 2d). Like the turquoise module at 0-hr, the pink module was enriched for IFN α/β signaling and immune signaling by interferons, interleukins, prolactin, and growth hormones, and the immune system (Fig. 2e). The top 30 hub genes from the pink module include several that are known to regulate alcohol drinking in mice, such as B2m and Ctss (Blednov et al., 2012) (Fig. 2f).
3.3. Clustering analysis identifies a unique subset of genes related to cell-to-cell signaling in the CIE + FSS group
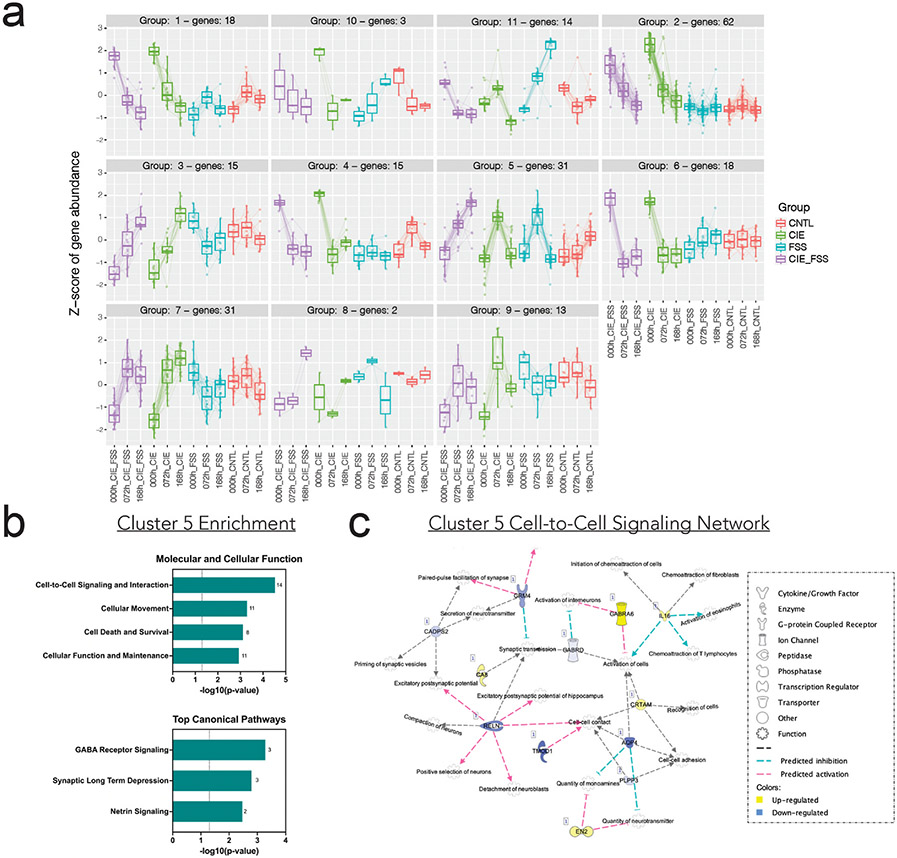
Using WGCNA, none of the modules were uniquely correlated with the CIE + FSS group. This is likely because gene expression changes that are unique to this group are below the detection limit (minimum module size = 100 genes). Thus, we used the degPatterns function from R package DEGreports to identify temporal gene expression signatures unique to the CIE + FSS group. We identified 224 significantly DEGs using the LRT with an adjusted p < 0.05. The function degPatterns then clustered these DEGs into 11 different clusters with similar expression profiles (Fig. 3a).
Fig. 3. Clustering analysis of significantly differentially expressed genes identified temporal gene expression signatures unique to each treatment group.
a) Cluster plots of differentially expressed genes (DEGs) across time within groups plotted against the Z-score of gene abundance. b) Enrichment analysis of Cluster 5: Molecular and Cellular Function and Top Canonical Pathways using IPA. c) Cell-to-Cell Signaling and Interaction Network visualized with IPA software.
The CIE and CIE + FSS DEGs typically showed very similar expression patterns across time, except for cluster 5. Here, gene expression in the CIE + FSS group increased sequentially across the three times, whereas the same genes showed more transient changes in the other treatment groups. Cluster 5 gene membership is listed in Supplemental Table 6. The DEGs in this cluster were enriched for cell-to-cell signaling and interaction, cellular movement, and other critical cellular processes (Fig. 3b). Top canonical pathways were GABA receptor signaling, synaptic long term depression, and netrin signaling (Fig. 3b). The cell-to-cell signaling network had the largest number of DEGs, many of which are involved in immune function (Il16), synaptic signaling (Gabra6, Gabrd, Ca8, Grm4), and cell-to-cell contact (Reln, Aqp4, Tmod1) (Fig. 3c). Cluster 5 enrichment highlights the unique cellular changes in the NTS associated with stress-induced alcohol consumption in dependent mice.
3.4. Overlapping expression profiles between the NTS and mPFC
In our previous study, we identified unique transcriptional changes in mouse mPFC associated with CIE + FSS (Farris et al., 2020). The mPFC sends projections directly to the NTS (Gasparini et al., 2020) and may act to depress NTS activity (Owens et al., 1999). Therefore, we examined correlated gene expression patterns between the NTS and mPFC in the same CIE + FSS group of mice using the RRHO test (Cahill et al., 2018; Plaisier et al., 2010). The RRHO algorithm assessed the overlap in DEGs between the mPFC and NTS in the CIE + FSS group to identify concordant and discordant patterns in gene expression between two datasets (i.e., genes that are coordinately up- or down-regulated, or oppositely up- or down-regulated between the two brain regions).
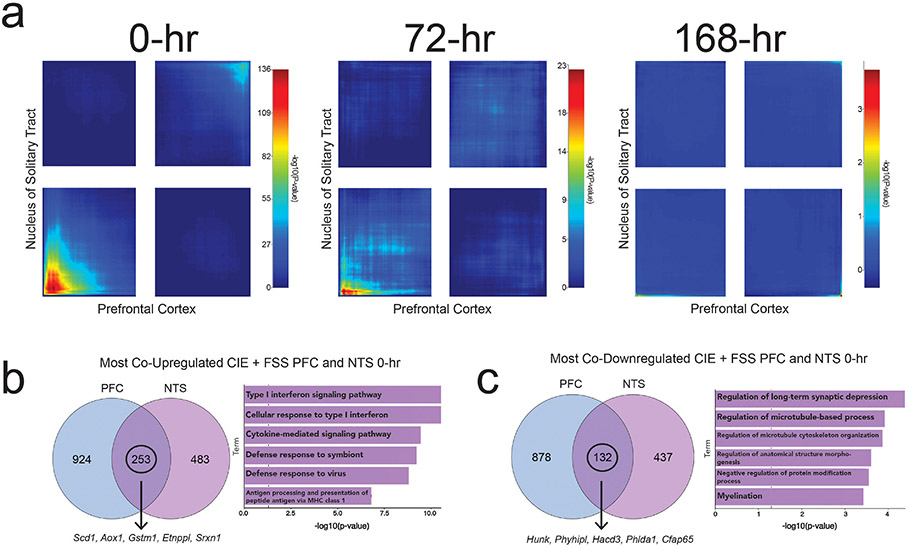
The largest overlap in genes upregulated in the mPFC and NTS was at 0-hr (max −log10 (p-value) = 136) (Fig. 4a). There were 253 significantly upregulated DEGs in both regions in the CIE + FSS group (Fig. 4b). Enrichment analysis again revealed a type I IFN signaling signature (p = 2.88 × 10™11) (Fig. 4b). There was weaker overlap in genes coordinately downregulated at 0-hr (max ™log10 (p-value) = 55) (Fig. 4a), suggesting that CIE + FSS leads to more common upregulated DEGs across these regions. There were 142 coordinately downregulated DEGs in the mPFC and NTS that were enriched for regulation of long-term synaptic depression and regulation of microtubule-based process (Fig. 4c), suggesting synaptic and structural changes in both regions.
Fig. 4. Overlapping differentially expressed genes in the CIE + FSS group between NTS and mPFC.
a) Rank-rank hypergeometric overlap (RRHO) plots of overlapping differentially expressed genes at 0-hr, 72-hr, and 168-hr between the nucleus of the solitary tract (NTS) and prefrontal cortex (mPFC) in the CIE + FSS group. b) Venn diagram of coordinately upregulated genes from the lower left quadrant of 0-hr RRHO plot (left). Arrow points to the top 5 overlapping genes in the rank-rank gene list (see Supplemental Table 7 for full list of genes). Enrichment of 253 coordinately upregulated genes in the NTS and mPFC (right). c) Venn diagram of coordinately downregulated genes from the upper right quadrant of 0-hr RRHO plot (left). Arrow points to the top 5 overlapping genes in the rank-rank gene list (see Supplemental Table 7 for full list of genes). Enrichment of 132 coordinately downregulated genes in NTS and mPFC (right). CIE, chronic intermittent ethanol vapor; FSS, forced swim stress.
Co-upregulated genes also showed a significant overlap at 72-hr (max ™log10 (p-value) = 23) (Fig. 4a), though to a lesser degree than at 0-hr. At 168-hr, there was no significant overlap for co-regulated or discordantly regulated genes (Fig. 4a). Thus, CIE + FSS treatment produced similar expression patterns across two distinct brain regions at early and intermediate time points but not after protracted withdrawal.
4. Discussion
We identified molecular networks in the NTS associated with FSS, CIE, or CIE + FSS using a systems genomics approach that integrates co-expression network analysis, temporal clustering analysis, and behavioral evidence. While a primary goal was to identify molecular changes that drive stress-induced drinking in dependent animals, the highly shared signatures in the CIE and CIE + FSS groups emerged as strong candidates of drinking escalation in dependent mice. Different analyses showed consistent upregulation of type I IFN and cytokine signaling in the NTS in both groups of alcohol-dependent animals. For example, DEGs in both the CIE alone and CIE + FSS groups at 0-hr were enriched for type I IFN signaling. These findings provide new evidence that chronic alcohol perturbs immune signaling in the NTS as has been demonstrated in other brain regions (Grantham et al., 2020) and that chronic alcohol and stress also alter immune signaling in this region. Activation of the neuroimmune system has also been linked with stress responses in other substance use disorders (Smiley and Wood, 2022). Interestingly, the largest overlap in upregulated genes in the mPFC and NTS from the CIE + FSS group at 0-hr were also enriched for type 1 IFN signaling.
There were very few persistent gene expression changes. Only three genes (Ccl5, Ly86, and Ttyh1) were differentially expressed across all three time points in the CIE and CIE + FSS groups. Ccl5 was the only upregulated gene across all times, suggesting long-term alterations in its transcriptional regulation. Ccl5 encodes C─C motif chemokine 5 and induces migration of phagocytes across the blood brain barrier to sites of inflammation (Ubogu et al., 2006) and recruits peripheral macrophages into the CNS in response to chronic alcohol consumption (Lowe et al., 2020). The persistent upregulation of Ccl5 in both treatment groups and its role in alcohol-induced neuroimmune activation make it an interesting target for future study.
Co-expression network analysis is useful for identifying gene networks correlated with ethanol intake that may play a role in excessive drinking (Osterndorff-Kahanek et al., 2015; Smith et al., 2016, 2020). Genes of interest in the 0-hr turquoise hub network that are correlated with increased ethanol consumption in the CIE and CIE + FSS groups include neuronal genes Gabrr2 and Clcn3, glucocorticoid-inducible genes Mt1, Mt2, and Sgk1, immune-related genes Irf9, Itgad, and Gstm1, and extracellular matrix (ECM)-related genes Tmod2, Gjb6, and Plin4. Several of these have been linked with alcohol consumption. For example, Gabrr2 encodes the GABA-A receptor subunit ρ2, which is linked to alcohol-related behaviors in mice and to alcohol dependence in humans (Xuei et al., 2010). Clcn3 is upregulated in the nucleus accumbens of a genetic rodent model of excessive ethanol consumption (Rodd et al., 2008). Metallothionein 1 and 2 genes (Mt1 and Mt2) encode glucocorticoid-inducible astrocytic proteins that have neuroprotective functions (Chung et al., 2008; Hidalgo et al., 1997; Køhler et al., 2003; Waller et al., 2018), and Mt2 expression in the brain is associated with preference for ethanol in mice (Loney et al., 2003). The ECM genes are also of interest considering the critical function of the ECM and its known sensitivity to ethanol (Erickson et al., 2018; Lasek, 2016; Smith, 2017).
Hub genes in the pink module at 72-hr also include immune genes (e.g., B2m and Ctss) that have been shown to regulate ethanol consumption in mice (Blednov et al., 2012). Several of the immune genes associated with escalations in drinking are localized in microglia, including C1qb, C3, and Ifitm3 (Baker et al., 2017; Coleman et al., 2020; Lacagnina et al., 2017; Warden et al., 2020). Mt1 and Gstm1 found in the turquoise module at 0-hr are also hub genes in the pink module at 72-hr, suggesting their expression in the NTS may be critical for escalations in alcohol consumption. Interestingly, Gstm1-promotes astrocyte-driven microglial activation during brain inflammation (Kano et al., 2019) and may be involved in microglial-mediated drinking escalations in alcohol-dependent animals (Warden et al., 2020).
Next, we performed temporal clustering analysis to identify a group of dysregulated genes in the NTS that were unique to the CIE + FSS condition (Fig. 3c). Notably, none of these genes from this group were dysregulated in the mPFC (Farris et al., 2020), which instead showed upregulation of genes involved in transmitter-gated ion channel activity. Several genes within this cluster have been implicated in AUD, including Gabra6 and Gabrd. Gabra6 is linked to alcohol dependence (Li et al., 2014; Loh et al., 1999; Radel et al., 2005) and to variations in stress response (Gonda et al., 2019; Uhart et al., 2004) in humans. The GABAergic system is widely known as a critical regulator of stress responses and is a common drug target for stress-related psychiatric disorders (Hou et al., 2020). Gabra6 and Gabrd are also both upregulated in the brains of high alcohol-drinking P rats (McClintick et al., 2016). Their altered expression in the NTS suggests they also have a role in stress-induced escalations in alcohol consumption.
A clear pattern emerged across all bioinformatic analyses pointing to type I IFN signaling as an important mediator of escalations in drinking in dependent animals. Based on these findings, we propose that activation of type I IFN signaling would be expected to drive alcohol consumption in B6 male mice. This prediction is also consistent with our work showing that poly(I:C) (a TLR3 agonist and type I IFN activator) increases drinking in B6 male mice (Warden et al., 2019).
To our knowledge, our findings are the first to demonstrate a role for immune-related genes in ethanol-dependent mice in the NTS, a critical stress-regulating brain region that has been understudied in alcohol research. The overlapping profiles in the CIE and CIE + FSS groups identified novel findings with translational potential in alcohol-dependent animals. In particular, type 1 IFN signaling in the CIE and CIE + FSS groups emerged as a key gene network by multi-level analyses. Finally, we identified a network of genes enriched for cell-to-cell signaling, cellular movement, and other critical cellular processes that were unique to the CIE + FSS condition and may contribute to stress-induced escalations in drinking in dependent mice.
Supplementary Material
Acknowledgement
We thank Jody Mayfield for critiquing and revising the manuscript.
Funding
This research was funded by the National Institutes of Health (NIH)/National Institute of Alcohol Abuse and Alcoholism (NIAAA): U01AA020926 (RDM), R01AA012404 (RDM),U24AA20929 (MFL), and U01AA014095 (HCB).
Footnotes
CRediT authorship contribution statement
Emily K. Grantham: Conceptualization, Formal analysis, Investigation, Writing – original draft. Gayatri R. Tiwari: Formal analysis. Olga Ponomareva: Investigation. R. Adron Harris: Writing – review & editing, Funding acquisition. Marcello F. Lopez: Investigation, Writing – review & editing. Howard C. Becker: Conceptualization, Writing – review & editing, Funding acquisition. R. Dayne Mayfield: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing, Visualization, Project administration, Supervision, Funding acquisition.
Declaration of competing interest
We declare this manuscript is original, has not been published before, and is not being considered for publication elsewhere. We have no conflicts of interest to declare.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neuropharm.2023.109768.
Data availability
Data will be made available on request.
References
- Anderson RI, Lopez MF, Becker HC, 2016. Forced swim stress increases ethanol consumption in C57BL/6J mice with a history of chronic intermittent ethanol exposure. Psychopharmacology 233, 2035–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JA, Li J, Zhou D, Yang M, Cook MN, Jones BC, Mulligan MK, Hamre KM, Lu L, 2017. Analyses of differentially expressed genes after exposure to acute stress, acute ethanol, or a combination of both in mice. Alcohol 58, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HC, 2017. Influence of stress associated with chronic alcohol exposure on drinking. Neuropharmacology 122, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Ponomarev I, Geil C, Bergeson S, Koob GF, Harris RA, 2012. Neuroimmune regulation of alcohol consumption: behavioral validation of genes obtained from genomic studies. Addiction Biol. 17, 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham NW, Thiele TE, 2017. Voluntary binge-like ethanol consumption site-specifically increases c-fos immunoexpression in male C57BL6/J mice. Neuroscience 367, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill KM, Huo Z, Tseng GC, Logan RW, Seney ML, 2018. Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci. Rep 8, 9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RS, Penkowa M, Dittmann J, King CE, Bartlett C, Asmussen JW, Hidalgo J, Carrasco J, Leung YKJ, Walker AK, Fung SJ, Dunlop SA, Fitzgerald M, Beazley LD, Chuah MI, Vickers JC, West AK, 2008. Redefining the role of metallothionein within the injured brain: extracellular metallothioneins play an important role in the astrocyte-neuron response to injury. J. Biol. Chem 283, 15349–15358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman LG Jr., Zou J, Crews FT, 2020. Microglial depletion and repopulation in brain slice culture normalizes sensitized proinflammatory signaling. J. Neuroinflammation 17, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson EK, Farris SP, Blednov YA, Mayfield RD, Harris RA, 2018. Astrocyte-specific transcriptome responses to chronic ethanol consumption. Pharmacogenomics J. 18, 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Tiwari GR, Ponomareva O, Lopez MF, Mayfield RD, Becker HC, 2020. Transcriptome analysis of alcohol drinking in non-dependent and dependent mice following repeated cycles of forced swim stress exposure. Brain Sci. 10 10.3390/brainsci10050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén O, Gan L-M, Björkegren JLM, 2019. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019. 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini S, Howland JM, Thatcher AJ, Geerling JC, 2020. Central afferents to the nucleus of the solitary tract in rats and mice. J. Comp. Neurol 528, 2708–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda X, Petschner P, Eszlari N, Sutori S, Gal Z, Koncz S, Anderson IM, Deakin B, Juhasz G, Bagdy G, 2019. Effects of different stressors are modulated by different neurobiological systems: the role of GABA-A versus CB1 receptor gene variants in anxiety and depression. Front. Cell. Neurosci 13, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham EK, Warden AS, McCarthy GS, DaCosta A, Mason S, Blednov Y, Mayfield RD, Harris RA, 2020. Role of toll-like receptor 7 (TLR7) in voluntary alcohol consumption. Brain Behav. Immun 89, 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun HL, Lebonville CL, Solomon MG, Griffin WC, Lopez MF, Becker HC, 2022. Dynorphin/kappa opioid receptor activity within the extended amygdala contributes to stress-enhanced alcohol drinking in mice. Biol. Psychiatr 91, 1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Egli M, Crabbe JC, Becker HC, 2010. Acute withdrawal, protracted abstinence and negative affect in alcoholism: are they linked? Addict. Biol 15, 169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JP, 2018. Regulation of hypothalamo-pituitary-adrenocortical responses to stressors by the nucleus of the solitary tract/dorsal vagal complex. Cell. Mol. Neurobiol 38, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo J, Belloso E, Hernandez J, Gasull T, Molinero A, 1997. Role of glucocorticoids on rat brain metallothionein-I and -iii response to stress. Stress 1, 231–240. [DOI] [PubMed] [Google Scholar]
- Hou X, Rong C, Wang F, Liu X, Sun Y, Zhang H-T, 2020. GABAergic system in stress: implications of GABAergic neuron subpopulations and the gut-vagus-brain pathway. Neural Plast. 2020, 8858415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Grishagin I, Wang Y, Zhao T, Greene J, Obenauer JC, Ngan D, Nguyen D-T, Guha R, Jadhav A, Southall N, Simeonov A, Austin CP, 2019. The NCATS BioPlanet - an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front. Pharmacol 10, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Marín Á, Collado-Romero M, Ramirez-Boo M, Arce C, Garrido JJ, 2009. Biological pathway analysis by ArrayUnlock and ingenuity pathway analysis. BMC Proc. 3 (Suppl. 4), S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li L, Shi M, Li Z, Zhou J, Chen L, 2013. The longitudinal study of rat hippocampus influenced by stress: early adverse experience enhances hippocampal vulnerability and working memory deficit in adult rats. Behav. Brain Res 246, 116–124. [DOI] [PubMed] [Google Scholar]
- Kano S-I, Choi EY, Dohi E, Agarwal S, Chang DJ, Wilson AM, Lo BD, Rose IVL, Gonzalez S, Imai T, Sawa A, 2019. Glutathione -transferases promote proinflammatory astrocyte-microglia communication during brain inflammation. Sci. Signal 12 10.1126/scisignal.aar2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Køhler LB, Berezin V, Bock E, Penkowa M, 2003. The role of metallothionein II in neuronal differentiation and survival. Brain Res. 992, 128–136. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacagnina MJ, Rivera PD, Bilbo SD, 2017. Glial and neuroimmune mechanisms as critical modulators of drug use and Abuse. Neuropsychopharmacology 42, 156–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S, 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, 2016. Effects of ethanol on brain extracellular matrix: implications for alcohol use disorder. Alcohol Clin. Exp. Res 40, 2030–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Sulovari A, Cheng C, Zhao H, Kranzler HR, Gelernter J, 2014. Association of gamma-aminobutyric acid A receptor α2 gene (GABRA2) with alcohol use disorder. Neuropsychopharmacology 39, 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijffijt M, Hu K, Swann AC, 2014. Stress modulates illness-course of substance use disorders: a translational review. Front. Psychiatr 5, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E-W, Smith I, Murray R, McLaughlin M, McNulty S, Ball D, 1999. Association between variants at the GABAAβ2, GABAAα6 and GABAAγ2 gene cluster and alcohol dependence in a Scottish population. Mol. Psychiatr 10.1038/sj.mp.4000554. [DOI] [PubMed] [Google Scholar]
- Loney KD, Uddin KR, Singh SM, 2003. Strain-specific brain metallothionein II (MT-II) gene expression, its ethanol responsiveness, and association with ethanol preference in mice. Alcohol Clin. Exp. Res 27, 388–395. [DOI] [PubMed] [Google Scholar]
- Lopez MF, Anderson RI, Becker HC, 2016. Effect of different stressors on voluntary ethanol intake in ethanol-dependent and nondependent C57BL/6J mice. Alcohol 51, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe PP, Morel C, Ambade A, Iracheta-Vellve A, Kwiatkowski E, Satishchandran A, Furi I, Cho Y, Gyongyosi B, Catalano D, Lefebvre E, Fischer L, Seyedkazemi S, Schafer DP, Szabo G, 2020. Chronic alcohol-induced neuroinflammation involves CCR2/5-dependent peripheral macrophage infiltration and microglia alterations. J. Neuroinflammation 17, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, McBride WJ, Bell RL, Ding Z-M, Liu Y, Xuei X, Edenberg HJ, 2016. Gene expression changes in glutamate and GABA-A receptors, neuropeptides, ion channels, and cholesterol synthesis in the periaqueductal gray following binge-like alcohol drinking by adolescent alcohol-preferring (P) rats. Alcohol Clin. Exp. Res 40, 955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers B, Scheimann JR, Franco-Villanueva A, Herman JP, 2017. Ascending mechanisms of stress integration: implications for brainstem regulation of neuroendocrine and behavioral stress responses. Neurosci. Biobehav. Rev 74, 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterndorff-Kahanek EA, Becker HC, Lopez MF, Farris SP, Tiwari GR, Nunez YO, Harris RA, Mayfield RD, 2015. Chronic ethanol exposure produces time- and brain region-dependent changes in gene coexpression networks. PLoS One 10, e0121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens NC, Sartor DM, Verberne AJ, 1999. Medial prefrontal cortex depressor response: role of the solitary tract nucleus in the rat. Neuroscience 89, 1331–1346. [DOI] [PubMed] [Google Scholar]
- Plaisier SB, Taschereau R, Wong JA, Graeber TG, 2010. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkkunen M, Goldman D, 2005. Haplotype-based localization of an alcohol dependence gene to the 5q34 {gamma}-aminobutyric acid type A gene cluster. Arch. Gen. Psychiatr 62, 47–55. [DOI] [PubMed] [Google Scholar]
- Robinson SL, Dornellas APS, Burnham NW, Houck CA, Luhn KL, Bendrath SC, Companion MA, Brewton HW, Thomas RD, Navarro M, Thiele TE 2020. Distinct and overlapping patterns of acute ethanol-induced C-fos activation in two inbred replicate lines of mice selected for drinking to high blood ethanol concentrations. Brain Sci. 10 10.3390/brainsci10120988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ, 2008. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol. Biochem. Behav 89, 481–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabinin AE, Criado JR, Henriksen SJ, Bloom FE, Wilson MC, 1997. Differential sensitivity of c-Fos expression in hippocampus and other brain regions to moderate and low doses of alcohol. Mol. Psychiatr 2, 32–43. [DOI] [PubMed] [Google Scholar]
- Smiley CE, Wood SK, 2022. Stress- and drug-induced neuroimmune signaling as a therapeutic target for comorbid anxiety and substance use disorders. Pharmacol. Ther 239, 108212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ACW, 2017. A glitch in the matrix: aberrant extracellular matrix proteolysis contributes to alcohol seeking. Biol. Psychiatr 81, 900–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Lopez MF, Archer KJ, Wolen AR, Becker HC, Miles MF, 2016. Time-Course analysis of brain regional expression network responses to chronic intermittent ethanol and withdrawal: implications for mechanisms underlying excessive ethanol consumption. PLoS One 11, e0146257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Lopez MF, Wolen AR, Becker HC, Miles MF, 2020. Brain regional gene expression network analysis identifies unique interactions between chronic ethanol exposure and consumption. PLoS One 15, e0233319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele TE, Cubero I, van Dijk G, Mediavilla C, Bernstein IL, 2000. Ethanol-induced c-fos expression in catecholamine- and neuropeptide Y-producing neurons in rat brainstem. Alcohol Clin. Exp. Res 24, 802–809. [PubMed] [Google Scholar]
- Ubogu EE, Callahan MK, Tucky BH, Ransohoff RM, 2006. Determinants of CCL5-driven mononuclear cell migration across the blood-brain barrier. Implications for therapeutically modulating neuroinflammation. J. Neuroimmunol 179, 132–144. [DOI] [PubMed] [Google Scholar]
- Uhart M, McCaul ME, Oswald LM, Choi L, Wand GS, 2004. GABRA6 gene polymorphism and an attenuated stress response. Mol. Psychiatr 9, 998–1006. [DOI] [PubMed] [Google Scholar]
- Waller R, Murphy M, Garwood CJ, Jennings L, Heath PR, Chambers A, Matthews FE, Brayne C, Ince PG, Wharton SB, Simpson JE, on behalf of the Cognitive Function and Ageing Neuropathology Study Group, 2018. Metallothionein-I/II expression associates with the astrocyte DNA damage response and not Alzheimer-type pathology in the aging brain. Glia. 10.1002/glia.23465. [DOI] [PubMed] [Google Scholar]
- Warden AS, Azzam M, DaCosta A, Mason S, Blednov YA, Messing RO, Mayfield RD, Harris RA, 2019. Toll-like receptor 3 activation increases voluntary alcohol intake in C57BL/6J male mice. Brain Behav. Immun 77, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden AS, Wolfe SA, Khom S, Varodayan FP, Patel RR, Steinman MQ, Bajo M, Montgomery SE, Vlkolinsky R, Nadav T, Polis I, Roberts AJ, Mayfield RD, Harris RA, Roberto M, 2020. Microglia control escalation of drinking in alcohol-dependent mice: genomic and synaptic drivers. Biol. Psychiatr 88, 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner TA, Stafford NP, Rompala GR, Van Hoogenstyn AJ, Elgert E, Drugan RC, 2013. Intermittent swim stress causes Morris water maze performance deficits in a massed-learning trial procedure that are exacerbated by reboxetine. Pharmacol. Biochem. Behav 10.1016/j.pbb.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Xuei X, Flury-Wetherill L, Dick D, Goate A, Tischfield J, Nurnberger J Jr., Schuckit M, Kramer J, Kuperman S, Hesselbrock V, Porjesz B, Foroud T, Edenberg HJ, 2010. GABRR1 and GABRR2, encoding the GABA-A receptor subunits rho1 and rho2, are associated with alcohol dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet 153B, 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S, 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol 4, 17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.