Abstract
Vibrio cholerae, the causative agent of the human diarrheal disease cholera, is a motile bacterium with a single polar flagellum. Motility has been implicated as a virulence determinant in some animal models of cholera, but the relationship between motility and virulence has not yet been clearly defined. We have begun to define the regulatory circuitry controlling motility. We have identified five V. cholerae flagellin genes, arranged in two chromosomal loci, flaAC and flaEDB; all five genes have their own promoters. The predicted gene products have a high degree of homology to each other. A strain containing a single mutation in flaA is nonmotile and lacks a flagellum, while strains containing multiple mutations in the other four flagellin genes, including a flaCEDB strain, remain motile. Measurement of fla promoter-lacZ fusions reveals that all five flagellin promoters are transcribed at high levels in both wild-type and flaA strains. Measurement of the flagellin promoter-lacZ fusions in Salmonella typhimurium indicates that the promoter for flaA is transcribed by the ς54 holoenzyme form of RNA polymerase while the flaE, flaD, and flaB promoters are transcribed by the ς28 holoenzyme. These results reveal that the V. cholerae flagellum is a complex structure with multiple flagellin subunits including FlaA, which is essential for flagellar synthesis and is differentially regulated from the other flagellins.
Cholera is a life-threatening diarrheal disease caused by Vibrio cholerae, a gram-negative curved rod that is highly motile by means of a single sheathed polar flagellum. The organism enters the host through the ingestion of contaminated food or water. Once in the intestine, V. cholerae swims toward and penetrates the mucus gel lining of the small intestine, eventually adhering to the apical surface of the intestinal epithelial cells (19). Adherent bacteria produce cholera toxin (CT), which activates adenylate cyclase in host epithelial cells, which in turn leads to the profuse watery diarrhea that is the hallmark of this disease (30, 37).
A number of virulence factors are coordinately regulated by the action of ToxR, a transcriptional regulatory protein implicated in the control of CT expression (40, 41). ToxR is known to activate expression of ToxT, a second transcriptional regulator (8), which activates the expression of both CT and the toxin-coregulated pilus (TCP), the primary intestinal colonization factor of V. cholerae (54). Laboratory conditions that stimulate ToxR-dependent expression of CT and TCP have been elucidated, but the true in vivo environmental conditions that influence virulence factor production are not known. Clearly, environmental cues present during the course of infection stimulate virulence factor production.
Motility has been identified as a virulence determinant of V. cholerae. Nonmotile mutants have been shown to be defective for adherence to isolated rabbit brush borders (13) and to cause less fluid accumulation in rabbit ligated ileal loops and less disease in the rabbit RITARD model (47); however, these observations are greatly influenced by changes in the particular biotype or mutant strain of V. cholerae evaluated and also by the animal model used. Interestingly, compared to isogenic motile strains, nonmotile mutants of live attenuated V. cholerae vaccines show reduced reactogenicity in humans while maintaining their ability to colonize the intestine (7, 24). Further, nonmotile mutants show no significant defect in their ability to colonize the infant mouse small intestine in competition assays (14, 47), a widely used model system that has accurately predicted the colonization properties of live attenuated cholera vaccines. Recently, genetic studies have suggested that virulence factor production and motility phenotypes are related. For example, some nonmotile mutants express higher levels of CT and TCP than wild-type strains do under noninducing laboratory conditions, while other “hyperswarming” mutants express little or no CT or TCP under inducing laboratory conditions (14) (the nature of hyperswarming, which is characterized by large swarm sizes in motility agar, remains to be determined). A toxR mutant has a similar hyperswarmer phenotype, perhaps indicative of a negative regulatory role for ToxR in motility. Bile has been shown to stimulate V. cholerae motility while simultaneously decreasing CT production in a ToxR-independent manner (16), indicating that other factors may contribute to the relationship of virulence and motility. Mutations affecting motility can also alter V. cholerae protease production and adherence to cultured cells (14). However, with the exception of motB mutants, these studies were performed with V. cholerae strains carrying unidentified motility mutations. Thus, the exact connection between motility and virulence gene expression has remained elusive.
Studies of the motility of two closely related Vibrio species, the human pathogen V. parahaemolyticus and the fish pathogen V. anguillarum, have revealed that these organisms have a polar flagellum composed of multiple flagellin subunits (33, 34). Mutations in several of the flagellin genes of V. anguillarum, although not causing significant motility defects, lead to significant defects in virulence, even after direct inoculation into the host (34, 42). This indicates that some of the flagellins may play additional roles in virulence not directly related to motility.
In the present study, we have identified and characterized multiple flagellin genes in V. cholerae. Our results reveal that the flagellum of V. cholerae, like those of V. parahaemolyticus and V. anguillarum, is composed of multiple flagellin subunits. We have identified a “core” flagellin essential for flagellar synthesis and have found that it is differentially regulated from the other flagellins; namely, its expression is controlled by RNA polymerase containing the alternate sigma factor ς54.
MATERIALS AND METHODS
Media.
Luria broth (LB) in both liquid medium and agar plates was routinely supplemented with 2 mM glutamine and supplemented with antibiotics when appropriate. Agar plates consisting of LB with 0.3% agar and 2 mM glutamine were used to measure motility. Evans blue-uranine indicator plates (31) supplemented with 2 mM glutamine were used to purify all constructed Salmonella typhimurium strains free of P22 phage. LB agar, made without NaCl and supplemented with 10% sucrose, was used to select for second recombinational events during construction of chromosomal deletions and insertions with vectors containing the sacB gene (see below).
Oligonucleotides and PCR.
Degenerate oligonucleotide primers based on conserved amino acid sequences of flagellin genes from multiple bacteria were used for PCR amplification of V. cholerae flagellin genes. The primers used were FLAX1 (GCGGATCCTCNATGGARCGNYTNTCNTC) and FLAX2 (GCGAATTCRTTNATRTANGTNGCNARCT), where N represents any nucleotide, R represents any purine, Y represents any pyrimidine, corresponding to the conserved amino acid sequences SMERLSS and ELATYIN, respectively; the underlined nucleotides represent restriction sites for BamHI and EcoRI. Degenerate oligonucleotide primers based on conserved amino acid sequences of putative GTP-binding proteins homologous to ORF1 from V. anguillarum (34) were used for PCR amplification of an internal fragment of a putative V. cholerae GTP-binding protein. The primers used were ORF1-1 (GCGAAGCTTTTRACNTTRAARCCNACNATG) and ORF1-2 (GCGAATTCTTTNCCNGCYTCYTTNGCNCC), corresponding to the conserved amino acid sequences LTLKPTM and GAKEAGK, respectively; the underlined nucleotides represent restriction sites for HindIII and EcoRI. PCR with degenerate primers was performed for 30 cycles of 45 s at 92°C, 1 min at 42°C, and 2 min 30 s at 72°C with TaqPlus DNA polymerase (Stratagene). Two fragments of approximately 600 bp and 2 kb in length were produced from V. cholerae Classical O1 strain O395 with the FLAX primer pair; the smaller fragment corresponded to the coding sequence for amino acids 28 to 227 of FlaA, and the larger fragment corresponded to the coding sequences of amino acids 28 to 377 of FlaD, the flaD-flaB intergenic region, and amino acids 1 to 226 of FlaB. One fragment of approximately 450 bp was produced with the ORF1 primer pair corresponding to the equivalent coding sequence of amino acids 197 to 341 in the Haemophilus influenzae homolog HI0393 (12).
We cloned the flaAC locus by amplifying overlapping PCR fragments. The oligonucleotides used to amplify the carboxyl-terminal coding region of flgL, the flgL-flaA intergenic region, and the amino-terminal coding region of flaA, were FLGLD1 (GCTCTAGAGGCTATCAGTTAGAGCGTAA) (based on the V. cholerae flgL sequence kindly provided by S. Mel; this primes immediately upstream of the flaAC locus sequence reported here) and FLAAU1 (GCGAATTCCATCGCACCTTCTGCGGTTTG) (corresponding to amino acids 76 to 82 of FlaA); the underlined nucleotides correspond to restriction sites for XbaI and EcoRI, respectively. The oligonucleotides used to amplify the carboxyl-terminal coding region of flaA, the flaA-flaC intergenic region, and the amino-terminal coding region for flaB were FLAAD1 (GCGAATTCCGCATGGGTGGCCAATCCTTT) (corresponding to amino acids 170 to 176 of FlaA) and FLAX2 (see above; this primed to the ELATYIN coding sequence in FlaC); the underlined sequence represents a restriction site for EcoRI. The oligonucleotides used to amplify the carboxyl-terminal coding region of flaC were FLACD1 (GCGAATTCGCTGACCGTGTTGCGATTCAAG) (corresponding to amino acids 107 to 113 of FlaC) and ORF1D1 (GCGAAGCTTTACAATGACTACATCCAATTC) (based on the deduced sequence of the V. cholerae putative GTP-binding protein ORF1 sequence [see above]); the underlined nucleotides correspond to restriction sites for EcoRI and HindIII, respectively. This oligonucleotide spuriously primed to a sequence downstream of IS1004.
The oligonucleotide primer pairs used to amplify internal fragments of flaB, flaC, flaD, and flaE were FLAB1 (GCGAATTCCGCGCGATTCAAGAAGAAGTG) and FLAB2 (GCAAGCTTTAAGTCGTCACCTTGTTTGGC) (amplifying a fragment corresponding to amino acids 110 to 219 of FlaB with restriction sites for EcoRI and HindIII, respectively), FLAC1 (GCGGATCCATGGCGGTGAATGTAAACAC) and FLAC2 (GCGAATTCACGATTACGCTCATCATTCAA) (amplifying a fragment corresponding to amino acids 1 to 125 of FlaC with restriction sites for BamHI and EcoRI, respectively), FLAD1 (GCGGATTCTCAATGGAGCGTCTATCTTCA) and FLAD2 (GCGAATTCGTCAGCACCGATTTGGAACGA) (amplifying a fragment corresponding to amino acids 28 to 152 of FlaD with restriction sites for BamHI and EcoRI, respectively), and FLAE1 (GCGGATCCTGGTGAAGCCGCACAGCTTT) and FLAE2 (GCGAATTCTCAATCACCGAGACGGCGCG) (amplifying a fragment corresponding to amino acids 154 to 297 of FlaE with restriction sites for BamHI and EcoRI, respectively). The oligonucleotides used to amplify a second internal fragment of flaD were FLAD3 (GCGAATTCACCAATGCACAACAAACTTCA) and FLAD4 (GCAAGCTTGTCAGCACCGATTTGGAACGA) (amplifying a fragment corresponding to amino acids 22 to 152 of FlaD with restriction sites for EcoRI and HindIII, respectively). The oligonucleotides used to amplify the entire flaA gene were FLGL1 (see above) and FLAAU1 (GCGGATCCGTACCGTGAACTACTGCAATAAC) (amplifying a fragment corresponding to nucleotides 1 through 2010 of the reported flaAC locus sequence flanked with restriction sites for XbaI and BamHI). The oligonucleotides used to amplify the carboxyl terminus of FlaA as well as the flaA to flaC intergenic region for the construction of the ΔflaA1 allele were FLAA1 (GCGAATTCTCTGCAATCTCGTTATTGC) and FLAC3 (GCGGTACCTGCAAAAGAGGTGGTTTCAG), corresponding to nucleotides 1977 to 2953 of the flaAC locus sequence with restriction sites for EcoRI and KpnI, respectively. The oligonucleotides used to amplify the aminoglycoside 3′-phosphotransferase (Kanr) gene from pACYC177 (48) were KAN2 (GCCAATTGCAACTCAGCAAAAGTTCGAT) and KAN3 (GCCAATTGAACGGTCTGCGTTGTCGGGA), corresponding to nucleotides 1783 to 2872 of the published pACYC177 sequence; the underlined nucleotides represent restriction sites for MfeI.
The promoter region of each flagellin gene was PCR amplified with oligonucleotide primer pairs which contained XbaI and BglII sites to orient the promoters with respect to the lacZYA genes in the fusion vector. The oligonucleotide pairs used were FLGL1 (see above) and FLAAP1 (GCAGATCTCGATTCATTCATCGCACCT) for flaAp (corresponding to nucleotides 1 to 1115 of the flaAC locus sequence), FLACP1 (GCTCTAGACAGTTGCCAAACTCTGCAAT) and FLACP2 (GCAGATCTGTGTTTACATTCACCGCCAT) for flaCp (corresponding to nucleotides 1965 to 2573 of the flaAC locus sequence), FLAEP1 (GCTCTAGAAAGCTTGATTCCGTCGAGCT) and FLAEP2 (GCAGATCTCTCCAGAGACTGATTGAGCAT) for flaEp (corresponding to nucleotides 1 to 579 of the flaEDB locus sequence), FLADP1 (GCTCTAGATCTGTGCTCGCTCAAGCGAA) and FLADP2 (GCAGATCTACTATTGATTTTAAAGCCTG) for flaDp (corresponding to nucleotides 1567 to 2035 of the flaEDB locus sequence), and FLABP1 (GCTCTAGAATCAAGGACACCGATTTCGCG) and FLABP2 (GCAGATCTCGTGTTTACATTAATTGCCAT) for flaBp (corresponding to nucleotides 2921 to 3305 of the flaEDB locus sequence).
We have identified a V. cholerae gene encoding a ς54 activator, flrA; the sequence and characterization of this gene will be presented elsewhere (25). For the purposes of the present study, we wished to control overexpression of this protein by a translational fusion to the arabinose-inducible promoter, PBAD. PCR amplification of the flrA gene was performed with oligonucleotides FLRAMET (ATGCAGAGTTTAGCGAAACTA) (the underlined nucleotides are the initiating methionine codon) and FLRAU1 (GCGAAGCTTTGGGTTGGCTTCACGCACTA) (the underlined sequence represents a HindIII restriction site); these primers amplify a 1.7-kbp fragment which contains the entire flrA gene and extends partially into the downstream gene, flrB.
PCR with specific primers and V. cholerae O395 chromosomal DNA was performed for 30 cycles of 45 s at 92°C, 1 min at 50°C, and 1 min 30 s at 72°C with Vent DNA polymerase (New England Biolabs). For some reactions, the extension time was increased to 2 min 30 s at 72°C. For amplification with FLACD1 and ORF1D1 (see above), the annealing temperature was reduced to 42°C.
Plasmid construction.
PCR-amplified fragments obtained from the FLAX12 primers containing flagellin genes from the V. cholerae Classical strain O395 (see above) were digested with BamHI and EcoRI and ligated into pBR322 (56) that had been similarly digested, giving plasmids pKEK23 (internal fragment of flaA) and pKEK24 (′flaD-flaB′ fragment). These were subsequently used for sequencing (see below).
The Δ(flaD-B)1::Cmr mutation was constructed in several steps. The PCR-amplified internal fragment from flaD (FLAD12 [see above]) was digested with EcoRI and BamHI and ligated into pWSK30 (55) that had been similarly digested to form pKEK30. The PCR-amplified internal fragment from flaB (FLAB12 [see above]) was digested with HindIII and EcoRI and ligated into pKEK30 that had been similarly digested, to form pKEK31, which thus forms the Δ(flaD-B)1 deletion, which removes the coding sequences corresponding to amino acids 153 to 377 of FlaD and 1 to 109 of FlaB, as well as the flaD-flaB intergenic region. pKEK31 was then digested with EcoRI and ligated with a 1.05-kbp MfeI-digested PCR-derived chloramphenicol acetyltransferase (Cmr) gene fragment from pACYC184 (49), which has been described previously (CAT12) (26), to produce pKEK32, which carries Δ(flaD-B)1::Cmr. This mutation was PCR amplified with primers FLAD1 and FLAB2, and the resulting fragment was ligated into pCVD422 (9) that had been digested with SmaI, resulting in pKEK33. This mutation was integrated into the V. cholerae O395 chromosome as described below. Chromosomal DNA from the resultant strain KKV23 was digested to completion with HindIII, and ligated into the HindIII site of pWSK30 (55); selection for a Cmr transformant resulted in the isolation of pKEK52, which contains a ∼15-kbp chromosomal fragment that carries Δ(flaD-B)1::Cmr. This chromosomal fragment was digested with EcoRI and HindIII, and the resulting subclones were ligated into pBR322 (56) that had been digested with EcoRI and/or HindIII. One of the resulting plasmids, pKEK65, contains a 4-kbp HindIII-EcoRI fragment that encodes the complete flaE gene, the 5′ coding region of flaD, and a portion of Cmr (the Cmr gene contains a restriction site for EcoRI [49]). Another resulting plasmid was pKEK66, which contains a 2.5-kbp EcoRI fragment that includes the other portion of Cmr and the 3′ coding region of flaB as well as the coding sequence for flaG. These plasmids were used to sequence the flaEDB locus (see below).
The Δ(flaE-D)1::Kanr mutation was constructed in several steps. The PCR-derived internal fragment of flaE (FLAE12 [see above]) was digested with EcoRI and BamHI and ligated into pWSK30 (55) that had been similarly digested, to give pKEK19. The PCR-derived internal fragment of flaD (FLAD34 [see above]) was digested with HindIII and EcoRI and then ligated into pKEK19 that had been similarly digested, resulting in pKEK20, which thus contains Δ(flaE-D)1; this mutation removes coding sequences corresponding to amino acids 296 to 378 of FlaE and 1 to 21 of FlaD, as well as the flaED intergenic region. The Kanr PCR-derived fragment (KAN23 [see above]) was digested with MfeI and ligated into pKEK20 which had been digested with EcoRI, resulting in pKEK21 which carries Δ(flaE-D)1::Kanr. This mutation was PCR amplified with FLAE1 and FLAD4 and ligated into the SmaI site of pCVD422 (9), resulting in pKEK22. This mutation was integrated into the V. cholerae chromosome as described below.
To make the ΔflaA1::Cmr mutation, the PCR-derived FLAA1-FLAC3 fragment (see above) was digested with EcoRI and KpnI and ligated into pWSK30 (55) that had been similarly digested, to form pKEK90. Then the PCR-derived internal fragment of flaA (pKEK23 [see above]) was digested with BamHI and EcoRI and ligated into pKEK90 that had been similarly digested, resulting in pKEK91, which thus contains ΔflaA1, a deletion of amino acids 228 to 372 of FlaA. pKEK91 was then digested with EcoRI and ligated with the MfeI-digested Cmr fragment (CAT12 [26]) to give pKEK92, which contains ΔflaA1::Cmr. pKEK92 was digested with BssHII, which removes the entire ΔflaA1::Cmr fragment; this fragment was made blunt ended with the Klenow fragment of DNA polymerase and ligated into pCVD442 (9) that had been digested with SmaI to form pKEK93, which was used to cross the mutation back onto the V. cholerae chromosome (see below).
Promoter-lacZ fusions to the five flagellin promoters were constructed by first modifying the lacZ fusion vector pRS551 (53) by creating unique XbaI and BglII restriction sites between the unique EcoRI and BamHI restriction sites (25) to form pKEK75. The PCR-derived flagellin promoter fragments (see above) FLGL1FLAAP1 (flaAp), FLABP12 (flaBp), FLACP12 (flaCp), FLADP12 (flaDp), and FLAEP12 (flaEp) were digested with XbaI and BglII and ligated into pKEK75 that had been similarly digested, resulting in pKEK80, pKEK79, pKEK76, pKEK77, and pKEK81, respectively.
To construct the suicide vectors containing internal gene fragments, the PCR-derived internal fragments of flaA (FLAX12), flaB (FLAB12), flaC (FLAC12), flaD (FLAD12), and flaE (FLAE12) were digested with EcoRI and ligated into pGP704 (39) that had been digested with EcoRV and EcoRI, to form pKEK27, pKEK29, pKEK81, pKEK28, and pKEK88, respectively; these plasmids were used to create insertional mutations in these respective genes.
The PCR-derived fragment FLGL1-FLAAU1 (see above) containing the complete flaA gene was digested with XbaI and BamHI and ligated into pACYC177 (48) that had been digested with NheI and BamHI, to form pKEK89, which was used to complement a flaA strain (see below). The PCR fragment containing the complete flrA gene which was amplified with FLRAMET and FLRAU1 (see above) was digested with HindIII and ligated into pBAD24 (17) that had been digested with NcoI, made blunt ended with the Klenow fragment of DNA polymerase, and digested with HindIII. The resulting plasmid, pKEK94, is an in-frame translational fusion of flrA to an initiating methionine codon under the control of the PBAD promoter.
Bacterial strains.
Escherichia coli DH5α (18) was used for all cloning manipulations, unless the vector being used was a derivative of pGP704 (39) or pCVD442 (9), which contain the R6K origin of replication and therefore require the product of the pir gene for replication, in which case E. coli DH5αλpir or SM10λpir (39) was used. For construction of the flagellin promoter-lacZ chromosomal fusions in S. typhimurium, E. coli TE2680 and TE1335 (10) were used in intermediate steps (see below).
The V. cholerae and S. typhimurium strains used in this study are listed in Table 1. All V. cholerae strains used are isogenic with the O1 Classical strain O395, referred to as wild type. To construct strains containing mutations in single flagellin genes, plasmids pKEK27 (flaA), pKEK29 (flaB), pKEK81 (flaC), pKEK28 (flaD), and pKEK88 (flaE) were mated by conjugation from E. coli SM10λpir into V. cholerae O395, selecting for streptomycin and ampicillin resistance. Since these plasmids contain internal gene fragments cloned into a suicide vector which requires the pir gene product for replication, the resulting strains have chromosomal insertions caused by the integration of the plasmids through homologous recombination (39). The strains formed were KKV12 [flaA1::pGP704 (Ampr)], KKV22 [flaB1::pGP704(Ampr)], KKV171 [flaC1::pGP704(Ampr)], KKV7 [flaD1::pGP704(Ampr)], and KKV6 [flaE1::pGP704(Ampr)]. pKEK81 was conjugated in a similar manner into strains KKV8, KKV23, and KKV34 to form strains KKV172, KKV173, and KKV174, respectively. Correct insertion into the target flagellin gene was confirmed by Southern blot analysis.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| V. cholerae | ||
| O395 | Wild type (Classical Ogawa) | 37 |
| CG842 | O395, ΔlacZ | 14 |
| KKV6 | flaE1::pGP704 | This study |
| KKV7 | flaD1::pGP704 | This study |
| KKV8 | Δ(flaE-D)1::Kanr | This study |
| KKV12 | flaA1::pGP704 | This study |
| KKV22 | flaB1::pGP704 | This study |
| KKV23 | Δ(flaD-B)1::Cmr | This study |
| KKV34 | Δ(flaE-D)1::Kanr, Δ(flaD-B)1::Cmr | This study |
| KKV62 | ΔtoxR1, ΔlacZ | This study |
| KKV90 | ΔflaA1::Cmr; ΔlacZ | This study |
| KKV171 | flaC1::pGP704 | This study |
| KKV172 | flaC1::pGP704, Δ(flaE-D)1::Kanr | This study |
| KKV173 | flaC1::pGP704, Δ(flaD-B)1::Cmr | This study |
| KKV174 | flaC1::pGP704, Δ(flaE-D)1::Kanr, Δ(flaD-B)1::Cmr | This study |
| S. typhimurium | ||
| 14028 | Wild type | American Type Culture Collection |
| KK1 | ntrA209::Tn10 | 26 |
| KK80 | Δ(ntrB-C)1 | 26 |
| KK105 | fliA5059::Tn10dTc | P22.TH1479 × 14028 |
| KK140 | putPA1303[Kanr-′lacZYA] | 26 |
| KK156 | putPA1303[Kanr-flaDp-′lacZYA] | This study |
| KK157 | ntrA209::Tn10, putPA1303[Kanr-flaDp-′lacZYA] | P22.KK156 × KK1 |
| KK158 | fliA5059::Tn10dTc, putPA1303[Kanr-flaDp-′lacZYA] | P22.KK156 × KK105 |
| KK159 | putPA1303[Kanr-flaBp-′lacZYA] | This study |
| KK160 | ntrA209::Tn10, putPA1303[Kanr-flaBp-′lacZYA] | P22.KK159 × KK1 |
| KK161 | fliA5059::Tn10dTc, putPA1303[Kanr-flaDp-′lacZYA] | P22.KK159 × KK105 |
| KK164 | putPA1303[Kanr-flaAp-′lacZYA] | This study |
| KK165 | ntrA209::Tn10, putPA1303[Kanr-flaAp-′lacZYA] | P22.KK164 × KK1 |
| KK166 | fliA5059::Tn10dTc, putPA1303[Kanr-flaAp-′lacZYA] | P22.KK164 × KK105 |
| KK167 | putPA1303[Kanr-flaEp-′lacZYA] | This study |
| KK168 | ntrA209::Tn10, putPA1303[Kanr-flaEp-′lacZYA] | P22.KK167 × KK1 |
| KK169 | fliA5059::Tn10dTc, putPA1303[Kanr-flaEp-′lacZYA] | P22.KK167 × KK105 |
| KK173 | putPA1303[Kanr-flaCp-′lacZYA] | This study |
| KK174 | ntrA209::Tn10, putPA1303[Kanr-flaCp-′lacZYA] | P22.KK173 × KK1 |
| KK175 | fliA5059::Tn10dTc, putPA1303[Kanr-flaCp-′lacZYA] | P22.KK173 × KK105 |
| KK188 | Δ(ntrB-C)1, putPA1303[Kanr-glnAp-′lacZYA] | P22.SK3041 × KK80 |
| KK189 | Δ(ntrB-C)1; ntrA209::Tn10, putPA1303[Kanr-glnAp-′lacZYA] | P22.SK284 × KK188 |
| SK284 | ntrA209::Tn10, hisF645 | 27 |
| SK3041 | putPA1303[Kanr-glnAp-′lacZYA] | 23 |
| TH1479 | fliA5059::Tn10dTc | K. Hughes |
V. cholerae strains containing chromosomal deletions and insertions were made by the following steps: plasmids pKEK22 [Δ(flaE-D)1::Kanr], pKEK33 [Δ(flaD-B)1::Cmr], and pKEK93 (ΔflaA1::Cmr) were mated into strains O395, CG842, KKV23, or KKV34 from E. coli SM10λpir (39) by selecting for streptomycin and ampicillin resistance. Single colonies were grown for successive generations in LB with no antibiotic selection and then plated on LB plus 10% sucrose at 30°C. The integrated plasmid contains the sacB gene (9), whose expression is lethal on this medium and thus selects for a second recombinational event. Sucrose-resistant colonies were tested for antibiotic resistance; Cmr or Kanr strains that were also Amps were chosen. Confirmation of correct chromosomal integration for all resultant strains was obtained either by sequencing the flanking DNA (see below) or by Southern blotting and PCR. The same procedure was used to obtain KKV62 (ΔtoxR1); the donor plasmid pMD60 contains an in-frame deletion of toxR in pCVD442 (a kind gift of M. Dziejman, this mutation was derived from pVM16 [41] and removes coding sequences for amino acids 55 to 206 of ToxR [9]).
The S. typhimurium strains used are isogenic with ATCC 14028, also referred to as wild type. Mutant S. typhimurium strains were constructed with the high-transducing phage P22 HT int. (51), and their construction is outlined in Table 1, listing first the paternal donor upon which the P22 lysate was made and then the recipient. The integration of the flap-lacZ chromosomal fusion cassettes inserted into the putPA locus has been described previously (10, 23, 26). pKEK94 was transformed into the appropriate S. typhimurium strains by electroporation.
Sequencing.
Cycle dideoxynucleotide sequencing was carried out with an ABI sequencing kit and the ABI sequencer model 373AStretch. Both strands were sequenced for all sequences reported here. The complete nucleotide sequence of the flaAC locus was obtained with specific oligonucleotide primers, pKEK23, and the amplified flaAC PCR products (see above) (Fig. 1). The complete nucleotide sequence of the flaEDB locus was obtained with specific oligonucleotide primers on pKEK65, pKEK66, and pKEK24. The partial sequence of the ORF1 homolog was obtained by cloning into pTZ19U (35) and using M13 primers.
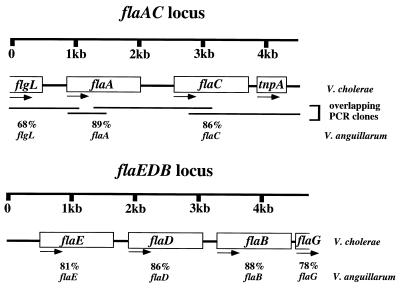
FIG. 1.
Schematic representation of the flagellin gene loci of V. cholerae. Genes are designated by open boxes, and arrows indicate the direction of transcription. PCR-derived fragments used to sequence the flaAC locus (see Materials and Methods) are indicated by lines. Below each gene, the percent amino acid identity to the corresponding gene of V. anguillarum is indicated. The tnpA gene lies within an insertion sequence element, IS1004, which has been described only in V. cholerae (4).
β-Galactosidase assays.
V. cholerae strains were grown in LB supplemented with 2 mM glutamine at 37°C. S. typhimurium strains were grown similarly, with the addition of 0.05% arabinose. The samples were assayed at an optical density at 600 nm of approximately 0.2 to 0.4, permeabilized with chloroform and sodium dodecyl sulfate, and assayed for β-galactosidase activity by the method of Miller (38).
Electron microscopy.
Strains were grown to the mid-log phase in LB 2 mM glutamine, centrifuged, and resuspended in 0.15 M NaCl. The samples were adhered to a carbon-coated grid and stained with 1% uranyl acetate before being subjected to microscopy.
Nucleotide sequence accession numbers.
The sequences described above were deposited into GenBank under accession no. AF007121, AF007122, and AF007294.
RESULTS
V. cholerae has five flagellin genes arranged into two loci, flaAC and flaEDB.
We used degenerate oligonucleotide primers designed to recognize flagellin gene sequences to PCR-amplify two V. cholerae chromosomal fragments encoding partial coding sequences for the flaA gene and the flaDB genes. We constructed a deletion-insertion of the flaDB locus [Δ(flaD-B)::Cmr] in vitro, recombined this mutation back into the V. cholerae chromosome (strain KKV23), and then isolated a large chromosomal fragment from this strain that conferred chloramphenicol resistance to obtain the entire flaEDB locus. Given the high homology of this locus to the equivalent genes from V. anguillarum (Fig. 1) (34), we reasoned that the flaA locus may be similarly homologous to that from V. anguillarum and were able to amplify sequences both upstream and downstream of the internal flaA sequence by overlapping PCR-derived fragments with primers designed to recognize a flgL homolog upstream of flaA and another flagellin gene downstream of flaA. Interestingly, we were able to amplify the sequence downstream of the flaC gene with an oligonucleotide primer specific to a V. cholerae putative GTP-binding protein (ORF1), which in V. anguillarum lies downstream of the flaC gene, but we amplified the corresponding fragment only with a reduced annealing temperature during PCR; the corresponding fragment revealed that the ORF1 primer annealed spuriously, and the gene immediately downstream of flaC is a transposase gene, tnpA, which lies within an insertion element IS1004 (4).
Complete sequencing of both strands of both loci revealed open reading frames (ORFs) for five flagellin genes (Fig. 1). As stated above, these loci have significant homology to the equivalent loci from V. anguillarum (34), and so we have named the flagellin genes according to their counterparts in V. anguillarum. The complete nucleotide sequence of the flaAC locus is shown in Fig. 2. It encodes two flagellin genes arranged in tandem. Upstream of flaA, a gene coding for a hook-associated protein (flgL) is located; the predicted protein product of the portion we sequenced shows 68% identity to flgL of V. anguillarum (which is similarly situated upstream of flaA in this organism) and 26% identity to that of E. coli. The predicted flaA and flaC gene products have homology to a large number of bacterial flagellin genes and are most homologous to the corresponding flaA and flaC gene products from V. anguillarum (89 and 86% identity, respectively). As stated above, downstream of flaC lies an insertion element, IS1004, which contains a transposase gene, tnpA. IS1004 has been detected and characterized only in V. cholerae (4); the gene encoding the putative GTP-binding protein is located in the corresponding location in V. anguillarum.
FIG. 2.
Nucleotide sequences of the flaA and flaC genes. Deduced amino acid sequences encoded by the ORFs are indicated. Also included are the partial coding sequence for the flgL gene upstream of flaA and the complete coding sequence of the tnpA gene downstream of flaC; tnpA lies within insertion sequence element IS1004 (4), whose boundaries are indicated by underlining of the first and last 10 bp. The putative ς54 promoter element in the flaA promoter and the putative ς28 promoter element in the flaC promoter are underlined. The boundaries of the deleted sequence of the ΔflaA1 mutation are shown by arrows.
The complete nucleotide sequence of the flaEDB locus is given in Fig. 3. This locus contains three flagellin genes arranged in tandem. Our sequence upstream of flaE extends to the HindIII site used to clone this fragment, which lies approximately 490 bp upstream of the start of translation, and this portion of the sequence did not reveal any open reading frames with significant homology to known genes. The predicted flaE, flaD, and flaB gene products have significant homology to numerous bacterial flagellins; they show the greatest homology to the flaE flaD and flaB gene products from V. anguillarum (81, 86, and 88% identity, respectively). Located downstream of flaB is an open reading frame encoding a homolog of the flaG gene product of V. anguillarum and V. parahaemolyticus (highest homology to the V. anguillarum FlaG, 78% identity in the portion we sequenced); these gene products have no known function.
FIG. 3.
Nucleotide sequences of the flaE, flaD, and flaB genes. Deduced amino acid sequences encoded by the ORFs are indicated. Also included is the partial coding sequence for the flaG gene downstream of flaB. The putative ς28 promoter elements in the flaE, flaD, and flaB promoters are underlined. The boundaries of the deleted sequences of the Δ(flaE-D)1 and the Δ(flaD-B)1 mutations are shown by arrows. The site of fusion iviVIII::pIVET5 (6), which detected an antisense transcript induced during infection, is shown by an arrow.
The V. cholerae flagellins are homologous to each other.
The predicted gene products of the five flagellin genes are similar in amino acid length (376 to 379 amino acids) and have calculated molecular masses of 40.4 kDa (FlaA), 39.5 kDa (FlaB), 39.9 kDa (FlaC), 39.9 kDa (FlaD), and 41.0 kDa (FlaE); therefore, they are predicted to be very similar in molecular mass. The gene products are highly homologous to each other (Fig. 4; Table 2), ranging from 61 to 82% identity. The predicted amino acid sequences have the highest homology at their amino and carboxyl termini.
FIG. 4.
Alignment of the deduced amino acid sequences of the five V. cholerae flagellin proteins. The sequences were aligned by Pileup (GCG Inc.). Amino acids which are identical in at least three of the flagellin genes are capitalized; amino acids which are identical in all five flagellin genes are denoted by an asterisk. The sequences used to design degenerate oligonucleotides for PCR are underlined (FLAX12; see Materials and Methods).
TABLE 2.
Percent identity between flagellin amino acid sequences
| Protein | % Identity to:
|
||||
|---|---|---|---|---|---|
| FlaA | FlaB | FlaC | FlaD | FlaE | |
| FlaA | 100 | 66 | 65 | 66 | 61 |
| FlaB | 100 | 70 | 71 | 61 | |
| FlaC | 100 | 82 | 64 | ||
| FlaD | 100 | 66 | |||
| FlaE | 100 | ||||
Because the flagellin genes code for a structural subunit of the flagellar filament, and because every bacterial flagellin studied thus far is located within the flagellum, we assumed that all five V. cholerae flagellin gene products are located within the flagellum. In fact, the highly homologous flagellin gene products FlaA, FlaB, FlaC, and FlaD from V. anguillarum were shown to be located within the flagellum (34). We were able to identify a single dominant species corresponding to an approximate molecular mass of 40 kDa in partially purified V. cholerae flagellar preparations separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). We assume that this band corresponds to the flagellin gene products, but a monoclonal antibody which cross-reacts with bacterial flagellins from the family Enterobacteriaciae (11) did not recognize the V. cholerae flagellins in a Western blot, and we were unable to obtain any separation between the five flagellins.
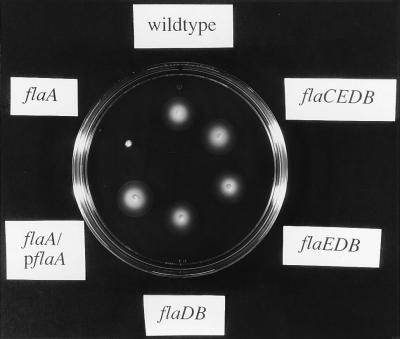
Only flaA is essential for motility.
We constructed V. cholerae strains containing chromosomal mutations in the various flagellin genes to assess their function (Table 1). A single mutation in flaA (KKV90 [Fig. 5] and KKV12 [data not shown]) results in a non-motile phenotype as assessed by swarm size in soft agar. The motility of the flaA strain KKV90 is recovered by complementation with a plasmid containing the entire flaA gene (pKEK89 [Fig. 5]). Single insertion mutations in flaB (KKV22), flaC (KKV171), flaD (KKV21), or flaE (KKV91) had no noticeable effect on the motility of V. cholerae in soft agar (data not shown).
FIG. 5.
Motility phenotypes of V. cholerae flagellin mutants. The bacteria were inoculated into motility agar (see Materials and Methods) at 37°C; motility is visualized by the swarm diameter. The strains shown (see Table 1) are O395 (wild type), KKV90 (flaA), KKV90 with pKEK89 (flaA/pflaA), KKV23 (flaDB), KKV34 (flaEDB), and KKV174 (flaCEDB).
We also constructed strains containing multiple mutations in the flaBCDE genes and assessed their motility phenotype in a similar manner. Strains containing mutations in flaED (KKV8), flaDB (KKV23), flaEDB (KKV34), and flaCEDB (KKV174) exhibited no obvious motility defect (Fig. 5 and data not shown).
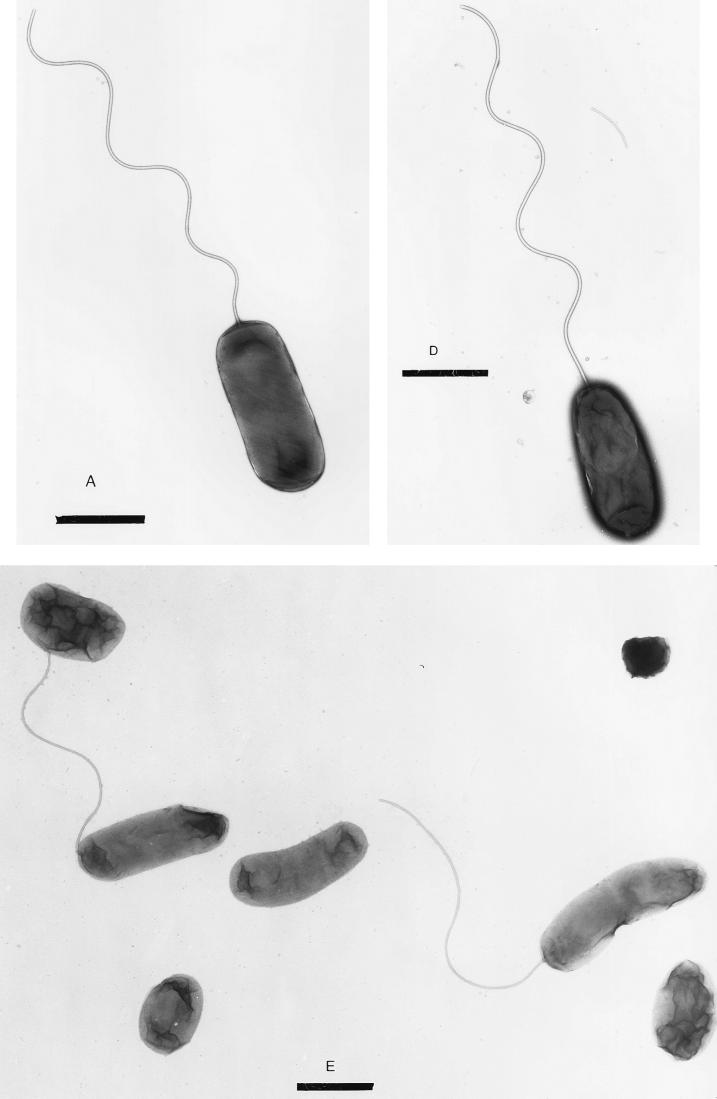
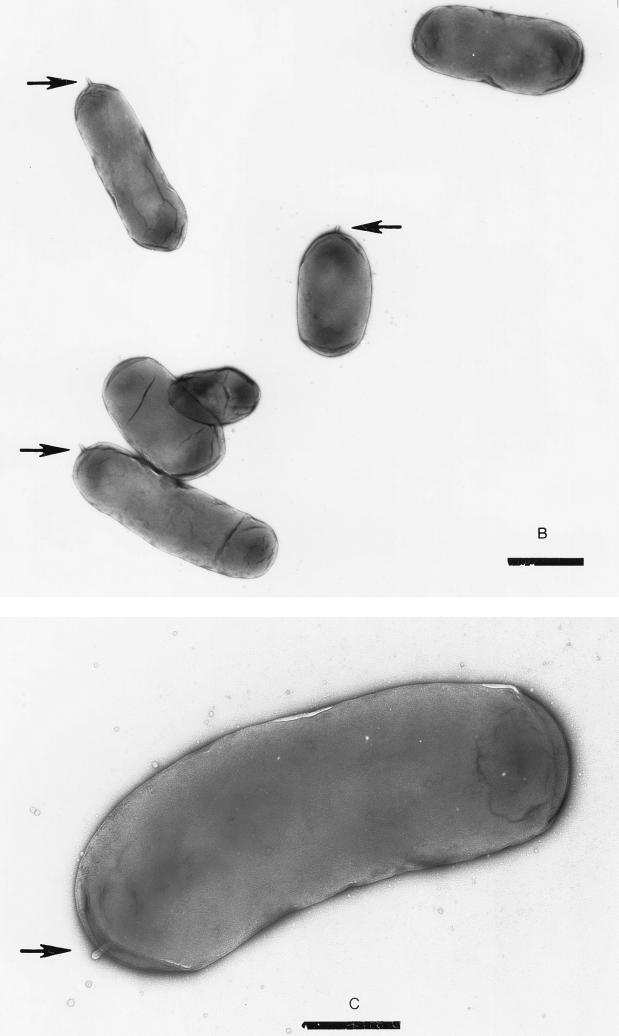
A flaA strain lacks a flagellum.
We used electron microscopy to directly observe cells of the various flagellin mutant strains of V. cholerae. Unlike the wild-type strain O395, which has a single polar flagellum, the flaA strain KKV90 lacks a flagellum (Fig. 6A to C). Some flaA cells could be seen with a small appendage at one pole (Fig. 6B and C); this may correspond to the flagellar hook. Complementation of the flaA strain with the entire flaA gene on a plasmid (pKEK89) results in flagellated bacteria resembling the wild-type strain (Fig. 6D).
FIG. 6.
Transmission electron microscopy of V. cholerae flagellin mutants (see Table 1). V. cholerae strains in logarithmic growth were resuspended in 0.15 M NaCl, spread onto carbon-coated grids and stained with 1% uranyl acetate. Bars, 1 μm (A, B, D, and E) and 500 nm (C). (A) O395, wild type. (B and C) KKV90, flaA (arrows indicate small appendages, possibly flagellar hooks. (D) KKV166, flaA/pflaA (KKV90 with pKEK89). (E) KKV174, flaCEDB.
Single mutations in flaB, flaC, flaD, or flaE (KKV22, KKV17, KKV21, and KKV91, respectively) did not noticeably affect the flagella in strains containing these mutations (data not shown). Strains containing flaDB or flaEDB mutations (KKV23 and KKV34) exhibited a mixed population of flagellated and nonflagellated bacteria, with noticeable numbers of bacteria having apparently shortened or sheared flagella (data not shown). The flaCEDB strain KKV174 had some bacteria with apparently full-length flagella, but the appearance of large numbers of nonflagellated bacteria and bacteria with shortened flagella indicates that the flagella of this strain may be particularly fragile and are sheared during growth or preparation of samples for microscopy (Fig. 6E).
All five flagellin genes are transcribed during logarithmic growth in V. cholerae.
To determine if all five flagellin genes are expressed in V. cholerae, we constructed promoter-lacZ fusions of the flagellin promoters and measured the transcription of the plasmid-borne fusions in several background strains (Table 3). All five flagellin promoters are transcribed at relatively high levels in the wild-type strain CG842 (O395 ΔlacZ). These high levels of transcription are maintained in the flaA strain KKV90, as well as in the toxR strain KKV62.
TABLE 3.
All five flagellin promoters are transcribed simultaneously in V. choleraea
| Genotypeb | β-Galactosidase activity in strain with mutation:
|
||||
|---|---|---|---|---|---|
| flaAp-′lacZ | flaCp-′lacZ | flaEp-′lacZ | flaDp-′lacZ | flaBp-′lacZ | |
| Wild type | 9,641 | 11,047 | 3,417 | 14,639 | 16,298 |
| ΔflaA::Cmr | 11,063 | 13,789 | 6,833 | 11,341 | 15,833 |
| ΔtoxR | 9,436 | 15,441 | 5,101 | 13,846 | 13,407 |
Assays were performed as described in Materials and Methods. Strains were grown in LB supplemented with 2 mM glutamine; cultures were assayed in triplicate at an optical density at 600 nm of ∼0.2 to 0.4. Results are the average of three samples expressed in Miller units (38). Each strain harboring the vector pRS551 alone grown in this medium has 10 to 15 Miller units of activity, which can be considered background activity.
The actual strains used (Table 1) were CG842 (wild type), KKV90 (ΔflaA::Cmr), and KKV62 (ΔtoxR), harboring plasmids pKEK80 (flaAp-′lacZ), pKEK76 (flaCp-′lacZ), pKEK81 (flaEp-′lacZ), pKEK77 (flaDp-′lacZ), and pKEK79 (flaBp-′lacZ).
flaA has a ς54-dependent promoter, and the flaE, flaD, and flaB promoters are ς28 dependent.
To determine the regulatory characteristics of the various V. cholerae flagellin promoters, we measured transcription from the same fla promoter-lacZ fusions integrated into the chromosome of S. typhimurium, thus taking advantage of the extensive repertoire of genetic mutations in transcription components available in this organism (Table 4). The flaE, flaD, and flaB promoters were transcribed at relatively high levels in a wild-type S. typhimurium strain, and these high levels of transcription were dependent upon an intact fliA gene, which encodes ς28, but were independent of an intact ntrA gene, which encodes ς54. There is some residual transcription from the flaD promoter even in the absence of ς28, indicative of a second promoter which is independent of ς28. The flaC promoter was transcribed at low but significant levels in the wild-type strain, and this level of transcription remained essentially unaffected in fliA and ntrA strains. Promoter elements resembling the consensus ς28 binding site could be found in the flaB, flaC, flaD, and flaE promoters (Fig. 7).
TABLE 4.
The flaA promoter is transcribed by ς54-holoenzyme and the flaE, flaD, and flaB promoters are transcribed by ς28-holoenzyme in S. typhimuriuma
| Genotypeb | β-Galactosidase activity in strain with mutation:
|
|||||
|---|---|---|---|---|---|---|
| flaAp-′lacZ | flaCp-′lacZ | flaEp-′lacZ | flaDp-′lacZ | flaBp-′lacZ | glnAp-′lacZc | |
| Wild type | 23 | 651 | 2,125 | 6,649 | 9,049 | 508 |
| Wild type + FlrAd | 1,808 | 493 | 1,239 | 5,165 | 5,133 | 2,047 |
| ntrA | 27 | 241 | 1,285 | 2,260 | 2,374 | 503 |
| ntrA + FlrAd | 33 | 564 | 1,372 | 5,931 | 6,014 | 288 |
| fliA | 28 | 206 | 21 | 173 | 40 | |
| fliA + FlrAd | 2,311 | 164 | 26 | 174 | 42 | |
Assays were performed as described in Materials and Methods. Strains were grown in LB supplemented with 2 mM glutamine and 0.05% arabinose; cultures were assayed in triplicate at an optical density at 600 nm of ∼0.2 to 0.4. Results are the average of three samples expressed in Miller units (38). Strain KK140 (putPA::′lacZ) grown in this medium has 8 Miller units of activity, which can be considered background activity.
The actual strains used (Table 1) were KK164, KK173, KK167, KK156, and KK159 (wild type with flaAp-, flaCp-, flaEp-, flaDp-, and flaBp-′lacZ fusions, respectively), KK165, KK174, KK168, KK157, and KK160 (ntrA209::Tn10 with flaAp-, flaCp-, flaEp-, flaDp-, and flaBp-′lacZ fusions, respectively), and KK166, KK175, KK169, KK158, and KK161 (fliA5059::Tn10dTc with flaAp-, flaCp-, flaEp-, flaDp-, and flaBp-′lacZ fusions, respectively).
The genetic background in these reporter strains is Δ(ntrB-C); NtrC is the activator of ς54-dependent transcription of the glnA promoter, so we wished to measure activation by the heterologous activator FlrA in the absence of NtrC. Residual transcription in a ntrA Δ(ntrB-C) strain originates from a second ς70-dependent glnA promoter (32). The actual strains used (Table 1) were KK188 and KK189 [Δ(ntrB-C) and ntrA Δ(ntrB-C) with glnAp-′lacZ, respectively].
These strains harbor plasmid pKEK94 (see Materials and Methods), which carries the gene encoding the ς54-activator FlrA from V. cholerae (25) under the control of the arabinose-inducible promoter PBAD. ς54 activators in high concentrations can activate ς54-dependent transcription from solution (45); in the present study, this protein serves to identify any ς54-dependent promoter.
FIG. 7.
Alignment of putative ς28 and ς54 promoter elements in V. cholerae fla promoters with ς28 and ς54 consensus promoter sequences. The consensus ς28 sequence for E. coli and S. typhimurium promoters is from reference 29, and the consensus ς54 sequence is from references 3 and 43.
The flaA promoter was not transcribed in the wild-type S. typhimurium strain or in the fliA and ntrA strains. ς54-dependent promoters require a transcriptional activator protein in addition to RNA polymerase containing ς54 (28), and these activator proteins are generally bound to enhancer elements located within the promoter region to increase their local concentration with respect to RNA polymerase. However, DNA binding is not essential to transcriptional activation, and these activator proteins can activate ς54-dependent transcription from solution when present in high enough concentrations (20, 21, 44, 45). We have identified a ς54-dependent transcriptional activator from V. cholerae, FlrA, which will be characterized elsewhere (25). When overexpressed from the arabinose-inducible promoter PBAD, this activator can activate transcription of the best-characterized ς54-dependent promoter, glnAp, from S. typhimurium (Table 4), and this increased level of activation is dependent upon an intact ntrA (ς54) gene. Overexpressed FlrA also activated the flaA promoter (an approximately 80-fold increase in transcription), and this high level of expression was dependent on an intact ntrA gene but independent of an intact fliA gene, consistent with flaA having a ς54-dependent promoter. FlrA had no significant effect on the transcription of any of the other fla promoters. A promoter element resembling the consensus ς54 binding site could be found in the flaA promoter (Fig. 7).
DISCUSSION
In the present study, we identified and characterized five flagellin genes in the human pathogen V. cholerae. Many flagellated bacterial species contain just one or two flagellin genes, which code for the structural subunit of the flagellar filament, so the presence of five separate genes in V. cholerae is puzzling, especially since the five predicted gene products have significant homology to each other (61 to 82% identity). In this respect, V. cholerae is similar to other Vibrio spp., notably the human pathogen V. parahaemolyticus (four polar flagellin genes [33]) and the fish pathogen V. anguillarum (five polar flagellin genes [34]), which has an identical arrangement of flagellin genes with the highest homology to those from V. cholerae.
Phenotypes of V. cholerae flagellin mutants revealed that the FlaA protein is essential for motility and that flaA strains are nonflagellated. Expression of the other four flagellins in a flaA strain remains high, indicating that although highly homologous, they cannot substitute for some essential function of the FlaA protein in assembling a flagellum. Substitution of function is also not easily obtained by mutation, because no revertant motile mutants arise in a flaA strain (data not shown). In contrast, a flaBCDE strain was motile and cells with flagella could be visualized, although the flagella of this strain appeared to be particularly fragile and easily broken. These results are consistent with the FlaA protein being required to form a flagellar core or scaffold into which the other flagellins are inserted to provide structural integrity. Interestingly, flaA mutants of V. anguillarum are flagellated but exhibit decreased motility (42); apparently these proteins, although highly homologous, do not have identical functions in the two bacteria.
We have shown that in S. typhimurium, the V. cholerae flaA gene is transcribed by RNA polymerase complexed with the alternate sigma factor ς54 (ς54 holoenzyme) while the flaE, flaD, and flaB genes are transcribed by RNA polymerase containing the flagellar sigma factor ς28. Neither ς54 nor ς28 regulates the expression of flaC, and so it remains unclear how this flagellin is regulated. We believe that the same differential regulation occurs in V. cholerae. We have created a mutation in the gene encoding ς54, rpoN (25). The V. cholerae rpoN mutant is nonflagellated, and the S. typhimurium ntrA (ς54) gene can complement this mutant for motility; likewise, the V. cholerae rpoN gene complements a S. typhimurium ntrA mutant for glutamine prototrophy. Thus, ς54 has maintained functional homology between these two organisms and must recognize the same promoter elements. We predict that ς28 has likewise maintained functional homology and activates the flaE, flaD, and flaB promoters in V. cholerae, although a ς28 homolog has yet to be identified. Interestingly, transcription of all five flagellin genes in V. cholerae is dependent on the presence of ς54, implicating a heirarchy where ς54-holoenzyme influences ς28-dependent transcription (25).
The ability to differentially regulate the flagellins within the flagellum may enable V. cholerae to produce flagella which are particularly suited for motility within a given environment (high-viscosity mucus, low or high osmolarity or pH, etc.) by changing helical pitch, thickness, or other flagellar parameters. We considered the possibility that the multiple flagellins were present to provide antigenic variation to the flagellum. The bacterial flagellum is a large target with repeating subunits, so that it generally induces a strong immune response, and it is well known that many pathogens use antigenic variation as a means of evading host immune defenses. For example, flagellar antigenic switching in S. typhimurium is a well-defined means of antigenic variation in which only one flagellin gene is exclusively expressed at a given time while the other remains silent (52). This particular antigenic variation is accomplished by genetic rearrangement. Selective expression of individual members of a “pool” of nonessential flagellins would be a suitable means of antigenic variation. However, in V. cholerae, such a mechanism is apparently not occurring, because the five flagellin promoters are expressed simultaneously. It must be noted, however, that our data addresses flagellin expression in vitro only; it remains to be determined if flagellin expression within the host is significantly different. Also, the presence of the insertion element IS1004, which contains a transposase gene, downstream of the flaC gene provides a mechanism whereby chromosomal rearrangements could occur by illegitimate recombination within this locus; the presence of the left arm of IS1004 within the O surface antigen locus rfb (4) suggests that such illegitimate recombination may be common in V. cholerae.
Another means by which pathogens evade immune response to flagellar antigens is by shutting off flagellar synthesis during colonization of the host. For example, in Bordetella bronchiseptica, the regulatory protein BvgA, which activates virulence gene expression, simultaneously represses flagellar gene synthesis, so that B. bronchiseptica is nonflagellated during infection (2). This may be an important means of immune system evasion, because B. bronchiseptica mutant strains that are flagellated during infection are more rapidly cleared (1). Alternatively, flagella are not needed during colonization, so that B. bronchiseptica may simply shut off synthesis for energetic reasons. Interestingly, the squid symbiont V. fischeri, which is closely related to V. cholerae, becomes aflagellate during colonization of the squid light organ (50). The likely reason for repressing flagellar synthesis in this case is to avoid unnecessary motility gene expression during a sessile phase of existence. In V. cholerae, genetic evidence suggests that motility phenotypes and the expression of some virulence genes are inversely related (14); i.e., some nonmotile mutants express higher levels of CT and TCP than does a wild-type strain under noninducing in vitro conditions, while toxR mutants, which express no CT or TCP, display a hyperswarmer phenotype. However, there is no direct evidence for repression of motility gene expression during infection. In this study, we were unable to detect any increase in fla gene transcription in a toxR mutant which might account for its hyperswarmer phenotype, and were also unable to detect any increase in CT and TCP expression by any of the fla mutants in vitro (data not shown).
Motility is important for full virulence of V. cholerae in the rabbit models of cholera (47), but various spontaneous nonmotile mutants show no defect for colonization in the infant mouse competition assay (14, 47). Consistent with these previous observations, the flaA mutant exhibited no defect for colonization of infant mice (data not shown). The attenuation of nonmotile mutants in rabbit animal models suggests that motility may be important for the organism to penetrate the mucus layer of the intestine and thus adhere to the apical surface of enterocytes. Perhaps the infant mouse has a sufficiently different (immature) mucus layer such that motility is not required to arrive at a permissive colonization site in this model.
Once the organisms colonize the intestinal surface, however, it might be advantageous to shut off flagellar synthesis to avoid immune system recognition and clearance, similar to B. bronchiseptica. Camilli and Mekalanos (6) identified a V. cholerae flagellin antisense transcript that was induced within the host during colonization. The reporter fusion they describe was inserted in a reverse orientation at a position corresponding to nucleotide 1677 of the flaEDB locus (Fig. 3) such that it would be measuring transcription originating in the flaE-flaD intergenic region or downstream within or past flaD. Notably, the antisense transcript would presumably regulate the expression of one of the ς28-dependent flagellins, flaE and/or flaD; such a mechanism may be required to more rapidly shut off flagellin synthesis.
We have no additional evidence for flagellar gene repression during V. cholerae colonization, but the existence of differential regulation of the flagellin genes provides a potential mechanism for quickly shutting off flagellar synthesis. RNA polymerase containing ς54 (ς54 holoenzyme) can initiate transcription only in conjunction with an activating protein, which generally responds to environmental signals and activates transcription only under inducing conditions (28). In contrast, ς28 holoenzyme is active in the absence of any activating proteins but requires the export of the anti-sigma factor FlgM, which occurs through a correctly assembled hook–basal-body complex (22). Shutting off ς54-dependent transcription only requires recognition of a change in environmental conditions, while shutting off ς28-dependent transcription requires a buildup of anti-sigma factor within the cell, which presumably is a slower response mechanism. Because the FlaA protein is essential for the assembly of a flagellum, shutting off flaA synthesis through the absence of ς54-dependent transcription would result in a nonmotile phenotype. We do not yet know the full extent of involvement of ς54 in flagellar gene synthesis in V. cholerae; ς54 may be required at multiple steps during flagellar synthesis, similar to the flagellar cascade of Caulobacter crescentus (5, 15). ς54 has also been shown to be required for flagellar synthesis in V. anguillarum (46), but it has not yet been determined whether it is directly involved in the transcription of flagellin genes, as it is in V. cholerae.
ACKNOWLEDGMENTS
We thank Andy Camilli, Michelle Dziejman, Kelly Hughes, Sydney Kustu, and Anne North for kindly providing strains and plasmids; Dan Steiger for providing DNA sequence support; Maria Ericsson for performing electron microscopy; Brian Akerley for performing analysis of flagellar preparations; and Cathy Lee for making constructive comments on the manuscript.
This study was supported by National Research Service Award AI09118-03 to K.E.K. and National Institutes of Health grant AI18045-13 to J.J.M.
REFERENCES
- 1.Akerley B J, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 2.Akerley B J, Miller J F. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J Bacteriol. 1993;175:3468–3479. doi: 10.1128/jb.175.11.3468-3479.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashraf S I, Kelly M t, Wang Y-K, Hoover T R. Genetic Analysis of the Rhizobium meliloti nifH promoter, using the P22 challenge phage system. J Bacteriol. 1997;179:2356–2362. doi: 10.1128/jb.179.7.2356-2362.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bik E M, Gouw R D, Mooi F R. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J Clin Microbiol. 1996;34:1453–1461. doi: 10.1128/jcm.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brun Y V, Marczynski G, Shapiro L. The expression of asymmetry during Caulobacter cell differentiation. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 6.Camilli A, Mekalanos J J. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coster T S, Killeen K P, Waldor M K, Beattie D, Spriggs D, Kenner J R, Trofa A, Sadoff J, Mekalanos J J, Taylor D N. Safety, immunogenicity and efficacy of a live attenuated Vibrio cholerae O139 vaccine prototype, Bengal-15. Lancet. 1995;345:949–952. doi: 10.1016/s0140-6736(95)90698-3. [DOI] [PubMed] [Google Scholar]
- 8.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott T. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J Bacteriol. 1992;174:245–253. doi: 10.1128/jb.174.1.245-253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng P. Identification of a common enterobacterial epitope with a monoclonal antibody. J Gen Microbiol. 1990;136:337–342. doi: 10.1099/00221287-136-2-337. [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitxHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 13.Freter R, O’Brien P C M. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: chemotactic responses of Vibrio cholerae and description of motile nonchemotactic mutants. Infect Immun. 1981;34:215–221. doi: 10.1128/iai.34.1.215-221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gober J W, Marques M V. Regulation of cellular differentiation in Caulobacter crescentus. Microbiol Rev. 1995;59:31–47. doi: 10.1128/mr.59.1.31-47.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta S, Chowdhury R. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect Immun. 1997;65:1131–1134. doi: 10.1128/iai.65.3.1131-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:577–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Holmgren J, Svennerholm A M. Mechanisms of disease and immunity in cholera: a review. J Infect Dis. 1989;136:S105–S112. doi: 10.1093/infdis/136.supplement.s105. [DOI] [PubMed] [Google Scholar]
- 20.Huala E, Ausubel F M. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J Bacteriol. 1989;171:3354–3365. doi: 10.1128/jb.171.6.3354-3365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huala E, Stigter J, Ausubel F M. The central domain of Rhizobium leguminosarum DctD functions independently to activate transcription. J Bacteriol. 1992;174:1428–1431. doi: 10.1128/jb.174.4.1428-1431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda T P, Shauger A E, Kustu S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J Mol Biol. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 24.Kenner J R, Coster T S, Taylor D N, Trofa A F, Barrera-Oro M, Hyman T, Adams J M, Beattie D T, Killeen K P, Spriggs D R, Mekalanos J J, Sadoff J C. Peru-15, an improved live attenuated vaccine candidate for Vibrio cholerae O1. J Infect Dis. 1995;172:1126–1129. doi: 10.1093/infdis/172.4.1126. [DOI] [PubMed] [Google Scholar]
- 25.Klose, K. E., and J. J. Mekalanos. Distinct roles of an alternate sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Submitted for publication. [DOI] [PubMed]
- 26.Klose K E, Mekalanos J J. Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect Immun. 1997;65:587–596. doi: 10.1128/iai.65.2.587-596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krajewska-Grynkiewicz K, Kustu S. Regulation of transcription of glnA, the structural gene encoding glutamine synthetase, in glnA::Mud1 (ApR, lac) fusion strains of Salmonella typhimurium. Mol Gen Genet. 1983;192:187–197. doi: 10.1007/BF00327665. [DOI] [PubMed] [Google Scholar]
- 28.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lospalluto J J, Finkelstein R A. Chemical and physical properties of cholera exo-enterotoxin (choleragen) and its spontaneously formed toxoid (choleragenoid) Biochim Biophys Acta. 1972;257:158–166. doi: 10.1016/0005-2795(72)90265-6. [DOI] [PubMed] [Google Scholar]
- 31.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 32.McCarter L, Krajewska-Grynkiewicz K, Trinh D, Wei G, Kustu S. Characterization of mutations that lie in the promoter-regulatory region for glnA, the structural gene encoding glutamine synthetase. Mol Gen Genet. 1984;197:150–160. doi: 10.1007/BF00327936. [DOI] [PubMed] [Google Scholar]
- 33.McCarter L L. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J Bacteriol. 1995;177:1595–1609. doi: 10.1128/jb.177.6.1595-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGee K, Hoerstedt P, Milton D L. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J Bacteriol. 1996;178:5188–5198. doi: 10.1128/jb.178.17.5188-5198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mead D A, Szczesna-Skorupa E, Kemper B. Single-stranded DNA ‘blue’ T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986;1:67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- 36.Mekalanos J J, Collier R J, Romig W R. Enzymic activity of cholera toxin. II. Relationships to proteolytic processing, disulfide bond reduction, and subunit composition. J Biol Chem. 1979;254:5855–5861. [PubMed] [Google Scholar]
- 37.Mekalanos J J, Swartz D J, Pearson G D, Harford N, Groyne F, de Wilde M. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 38.Miller J H. A short course in bacterial genetics. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 39.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 42.Milton D L, O’Toole R, Hoerstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morett E, Buck M. In vivo studies on the interaction of RNA polymerase-ς54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters: the role of NIFA in the formation of an open promoter complex. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 44.North A K, Klose K E, Stedman K M, Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryote-like Modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.North A K, Kustu S. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 46.O’Toole R, Milton D L, Wolf-Watz H. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol Microbiol. 1996;19:625–637. doi: 10.1046/j.1365-2958.1996.412927.x. [DOI] [PubMed] [Google Scholar]
- 47.Richardson K. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect Immun. 1991;59:2727–2736. doi: 10.1128/iai.59.8.2727-2736.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rose R E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988;16:356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose R E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988;16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruby E G, Asato L M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 51.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 52.Silverman M, Zieg J, Hilmen M, Simon M. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc Natl Acad Sci USA. 1979;76:391–395. doi: 10.1073/pnas.76.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 54.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang R F, Kushner S. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 56.Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988;70:399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]