Abstract
The cop operons of Helicobacter pylori and Helicobacter felis were cloned by gene library screening. Both operons contain open reading frames for a P-type ion pump (CopA) with homology to Cd2+ and Cu2+ ATPases and a putative ion binding protein (CopP), the latter representing a CopZ homolog of the copYZAB operon of Enterococcus hirae. The predicted CopA ATPases contained an N-terminal GMXCXXC ion binding motif and a membrane-associated CPC sequence. A synthetic N-terminal peptide of the H. pylori CopA ATPase bound to Cu2+ specifically, and gene disruption mutagenesis of CopA resulted in an enhanced growth sensitivity of H. pylori to Cu2+ but not to other divalent cations. As determined experimentally, H. pylori CopA contains four pairs of transmembrane segments (H1 to H8), with the ATP binding and phosphorylation domains lying between H6 and H7, as found for another putative transition metal pump of H. pylori (K. Melchers, T. Weitzenegger, A. Buhmann, W. Steinhilber, G. Sachs, and K. P. Schäfer, J. Biol. Chem. 271:446–457, 1996). The corresponding transmembrane segments of the H. felis CopA pump were identified by hydrophobicity analysis and via sequence similarity. To define functional domains, similarly oriented regions of the two enzymes were examined for sequence identity. Regions with high degrees of identity included the N-terminal Cu2+ binding domain, the regions of ATP binding and phosphorylation in the energy transduction domain, and a transport domain consisting of the last six transmembrane segments with conserved cysteines in H4, H6, and H7. The data suggest that H. pylori and H. felis employ conserved mechanisms of ATPase-dependent copper resistance.
Helicobacter pylori is a human gastric pathogen associated with chronic gastritis, peptic ulcers, and gastric cancer (8, 16, 27, 30, 64). Helicobacter felis, a related microorganism, was originally isolated from the stomach of a cat (42) but can also survive in the gastric environments of other mammalian species, such as mice. Mice infected with H. felis are often used as a model to study H. pylori colonization, pathogenicity, and eradication. Therefore, survival mechanisms of both H. pylori and H. felis are of interest. The specialized ecological niche of gastric Helicobacter spp., the mammalian stomach, is dominated by the gastric acid produced by the H+/K+ ATPase of the parietal cells but also presents a highly variable cationic environment for these microorganisms. In bacteria, cation cellular homeostasis as well as resistance to several transition metals is based on the action of both proton-cation antiporters and transition metal P-type ion pumps (36, 51). The ion pumps of H. pylori, as well as those of other gastric Helicobacter species, may participate in the survival mechanisms of the pathogenic bacterium.
Expression of these P-type ATPases is environmentally regulated. Expression of some of these pumps is controlled either by a two-component system, consisting of a sensor kinase and a response regulator with affinity for specific DNA sequences, or by small cytoplasmic cation binding proteins acting as repressors or activators of gene expression. An example of the former is the high-affinity K+ uptake ATPase encoded by the kdpABC operon of Escherichia coli. This operon was shown to be controlled by an adjacent operon containing two genes, kdpDE, defining the sensor kinase and the corresponding response regulator of a two-component regulatory system (10, 25, 44, 63). While the KdpD sensor kinase protein is membrane associated, the KdpE regulator protein is cytoplasmic. The KdpDE proteins regulate expression of the kdp operon, apparently as a function of cellular turgor pressure (26, 59). In contrast, expression of the two copper-transporting P-type ATPases in Enterococcus hirae, CopA and CopB (39, 55), is thought to be controlled by two small cytoplasmic proteins, CopY and CopZ (38). The copYZ genes, which encode small transition metal binding proteins, precede the structural P-type ATPase genes A and B as part of the enterococcal copYZAB operon. Their protein products were postulated to act as repressors (CopY) or activators (CopZ) regulating gene transcription of the cop operon depending on the availability of Cu2+ (38). The CopAB P-type ATPases of Enterococcus hirae belong to a family of transition metal ATPases containing N-terminal ion binding motifs and a membrane-associated CPX sequence, both suggested to play a role in ion binding and/or ion transport (5, 29, 51, 52, 57).
Of special interest is the membrane topology of P-type ATPases, which provides a structural basis for the ion transport pathway through the membrane. Hydropathy profiles, as well as other computer-aided methods for detection of amphipathic helices in membrane proteins, have been misleading, and therefore the determination of topology needs experimental evaluation (7, 50). The first bacterial P-type ATPase to have its transmembrane (TM) segments defined experimentally was the Mg2+ ATPase of Salmonella typhimurium (54). This membrane pump was shown to contain 10 membrane-spanning helices, with the large cytoplasmic ATP binding and phosphorylation loop being between H4 and H5. This ATPase therefore resembles the eukaryotic alkali metal P-type pumps which transport alkali cations as well as Mg2+ and Ca2+, the latter also being relatively small cations (21). The amino acid sequences of transition metal ATPases exhibit a different hydrophobicity profile, with two additional hydrophobic domains in the N-terminal region and four fewer in the C-terminal region. The core structure of six TM helices is likely to be similar (28). The first transition metal ATPase investigated experimentally for the number and orientation of TM segments was an H. pylori P-type pump (31) most closely related to the Cd2+ ATPase of Staphylococcus aureus, CadA, the latter previously being thought to contain only six TM segments (52, 53). However, the H. pylori ATPase was shown to contain a membrane domain of eight TM helices comprising the additional N-terminal pair of membrane spans followed by the core structure of six membrane spanning segments with only one pair of C-terminal TM segments (31).
Here we describe the isolation of the copAP operons of H. pylori 69A and H. felis ATCC 49179. They both carry two genes, one encoding another member of the bacterial transition metal ion ATPase family, CopA, and the other encoding a putative cation binding regulatory peptide of 66 amino acid residues, CopP. The small CopP peptide is homologous to CopZ, encoded by the E. hirae cop operon (38). In a previous study using a truncated variant of the ATPase cloned from H. pylori UA802, it was claimed that the pump is a Cu2+ export ATPase (13). In this study, the function of the H. pylori 69A-derived ATPase was analyzed by knockout mutagenesis of H. pylori copA, and we also investigated the N-terminal ion binding properties of the putative Cu2+ ATPase by ion affinity chromatography and electrospray ionization mass spectrometry (ESI-MS). Compared to the other P-type pump cloned from H. pylori 69A (31), the DNA-derived amino acid sequence of the CopA P-type pumps discussed here contains additional hydrophobic segments, and therefore a topological analysis was performed on the H. pylori CopA ATPase. The results obtained by experimental identification of membrane-inserted segments and the homology analysis of the CopA pumps cloned from the two related microorganisms, H. pylori and H. felis, allowed detection of domains of high amino acid sequence identity that have perhaps been conserved to transport copper across the cytoplasmic membrane in these gastric bacteria.
MATERIALS AND METHODS
Bacterial strains.
E. coli HB101 containing the H. pylori gene library was a gift of Rainer Haas (Tübingen, Germany). Replication of random pRH948-derived DNA fragments in vector pTZ18R for subsequent DNA sequencing was performed in E. coli KK186, a gift from I. Rasched (Konstanz, Germany). E. coli MM294, supplied by the American Type Culture Collection (ATCC 32625), was used for plasmid cloning experiments. E. coli XL1-Blue MRF′ and SOLR, both obtained from Stratagene, were used for amplification of H. felis genomic library in the Lambda ZAP II vector or in vivo excision of pBluescript phagemids with H. felis insertion DNA (pHF) from lambda vectors. H. pylori strains were from the American Type Culture Collection (ATCC 49503) or from Rainer Haas (clinical isolate 69A). H. felis was supplied by the American Type Culture Collection (ATCC 49179).
Culture conditions.
E. coli cells were grown in Luria-Bertani (LB) broth or agar plates supplemented with 50 μg of tetracycline per ml or 10 μg of kanamycin per ml as required. For experiments with the H. felis gene library, the E. coli strains used were grown as recommended by the supplier of the strains (Stratagene). H. pylori cells were grown in brain heart infusion (Difco) medium (BHI) supplemented with 6% horse serum and 0.25% yeast extract (Difco) in a CO2 incubator (10% CO2) at 37°C in 10-ml cell culture flasks. Growth was monitored by determination of the optical density at 578 nm of aliquots of the bacterial cultures at various time points. H. felis cells were grown in Columbia EH broth (Difco) supplemented with 6% horse serum in GasPak jars under microaerophilic conditions (Anaerocult C; Merck) at 37°C in a shaker incubator.
Selection of DNA oligonucleotide sequences for detection of P-type ATPases.
All known P-type ATPases from eukaryotic and bacterial cells contain a highly conserved DKTGT(I/L)T phosphorylation consensus sequence (11, 43). This sequence can be encoded by a pool of 2,304 different 20-base oligonucleotides (main pool) covering all possible sequences of the first six amino acids of the phosphorylation consensus sequence as well as the AC of the threonine codon. This pool of oligonucleotides was divided into five chemically synthesized oligonucleotide subpopulations (subpools I-405 to I-409). The DNA nucleotide sequences of the synthesized subpools consisted of GA(TC) AA(AG) AC(AGCT) GG(AGCT) AC(AGTC) with a 3′ extension of AT(TC) AC (for I-405), AT(CA) AC (for I-406), TT(AG) AC (for I-407), CT(TC) AC (for I-408), or CT(AG) AC (for I-409). The variable nucleotides are in parentheses. The DNA primers (MWG Biotech, Ebersberg, Germany) were labeled by using a digoxigenin (DIG) 3′-end labeling kit (Boehringer Mannheim). Each of the primers listed above was employed for genomic Southern blot analysis (58). Subsequently, primer I-408 was selected for Southern blot screening of the H. pylori 69A library in plasmid pRH160 and plaque screening of membrane filters obtained from the H. felis gene library in the Lambda ZAP II phage. I-408 was also used for initial DNA sequencing to verify that positive plasmids contained the DKTGT(I/L)(T) target sequence.
Preparation of DNA.
Genomic DNA of H. pylori or H. felis was prepared by standard protocols as previously described (1, 48). Plasmid DNA was isolated by anion-exchange chromatography (Qiagen).
Southern blot analysis of H. pylori genomic DNA with various DNA oligonucleotides.
The DNA of H. pylori was digested with the HindIII DNA endonuclease. The digested DNA was separated by agarose gel electrophoresis, denatured, and blotted onto nylon membranes. Each of the Southern membranes containing the restriction endonuclease-digested H. pylori DNA was hybridized with one of the DIG-labeled DNA oligonucleotide mixtures (I-405 to I-409) in accordance with a Boehringer Mannheim protocol. Hybridization was performed for >6 h in a solution containing 5× standard saline citrate (SSC) buffer (Maniatis) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× blocking agent, 0.02% (wt/vol) sodium dodecyl sulfate (SDS), and 0.1% (wt/vol) N-laurylsarcosine at 45°C. Hybridized blots were washed in 5× SSC buffer–0.01% (wt/vol) SDS at 45°C. Positive restriction fragments were detected by chemiluminescence (Boehringer Mannheim).
H. pylori 69A gene library.
The DNA library was constructed by Rainer Haas. It contains genomic DNA fragments of H. pylori 69A. The DNA fragments generated by partial Sau3A digestion were cloned into the BglII restriction site of the plasmid pRH160, also referred to as the pMin1 vector (18). The library was replicated in E. coli HB101. DNA inserts of pRH160, on average 3 × 103 to 4 × 103 bp in length, could be removed by digestion with EcoRI and XhoI. The plasmid harbors a tetracycline resistance gene (tetR).
H. felis gene library.
The H. felis library was prepared by Stratagene. Genomic DNA isolated from H. felis ATCC 49179 was sheared by pulling and pushing the DNA through a needle. DNA fragments obtained (5 to 10 kbp) were blunt ended with Klenow fragment of DNA polymerase. DNA was methylated by EcoRI methylase. Methylated DNA was cloned into the EcoRI cloning site of the Lambda ZAP II phage vector (Stratagene), providing the ability to excise the pBluescript SK(−) phagemid by using the M13 helper phage (Stratagene). A total of 3.7 × 106 primary plaques containing insertional H. felis DNA fragments, 5 to 10 kbp in length, were obtained. The library was amplified to a final titer of 1.3 × 1010 plaques/ml in E. coli XL1-Blue MRF′.
Screening of gene libraries for detection of P-type ATPases. (i) Southern blot screening and isolation of pRH vectors containing putative P-type ATPase genes of H. pylori.
Since the phosphorylation consensus target site selected for screening is present in all known P-type pumps, the H. pylori gene library was screened on isolated plasmid DNA mixtures to avoid contaminating signals from the E. coli genome. For preparation of distinct plasmid DNA mixtures, an aliquot of the library, 1.4 × 103 CFU, was diluted in LB broth medium supplemented with 50 μg of tetracycline per ml. The bacterial clones were used for inoculation of 20 cultures for subsequent mixed-plasmid preparations. Inoculated vials, each containing approximately 70 different plasmids of the library, were incubated overnight in a shaker incubator at 37°C. Mixed plasmid DNA was purified by ion-exchange chromatography (Qiagen). In addition, glycerol was added to an aliquot of each of the mixed bacterial suspensions, which were then stored at −70°C. The isolated plasmid DNA mixtures were subjected to digestion with restriction endonucleases EcoRI and XhoI. DNA fragments were separated on 1% (wt/vol) agarose gels and blotted onto nylon membranes (58). The membranes containing restriction enzyme-digested DNA of the mixed-plasmid preparations were hybridized with DIG-labeled DNA oligonucleotide I-408 according to the protocol of Boehringer Mannheim as described above for genomic Southern blot analysis. Positive plasmid mixtures were detected by chemiluminescence (Boehringer Mannheim). Aliquots taken from corresponding glycerol stocks were plated out on LB agar plates containing 50 μg of tetracycline per ml, and 60 colonies were selected for preparation of clonal plasmid DNA, using anion-exchange columns (Qiagen) for detection of the clones which had been hybridized in the mixed plasmid preparation. Plasmid DNAs were analyzed for sequences homologous to primer I-408 by another cycle of Southern blot analysis as described above. Three distinct positive DNA clones containing H. pylori 69A DNA insertions flanked by EcoRI and XhoI restriction sites of the pRH160 cloning vector were isolated, namely pRH514, pRH539, and pRH948. These plasmids were subjected to a cycle of DNA sequencing for detection of the DNA target sequence corresponding to the phosphorylation consensus sequence.
(ii) Plaque filter hybridization and in vivo excision of pHF vectors containing putative P-type ATPase genes of H. felis.
The H. felis gene library was screened by conventional plaque filter hybridization. Nylon membranes (Amersham) containing phage DNA from 2 × 103 plaques were prepared by standard protocols (48). DNA-containing membranes were hybridized with primer I-408 under the same conditions used for Southern blot screening of the H. pylori 69A gene library. From positive Lambda ZAP II phage clones, 10 pBluescript SK(−) plasmids (pHF1 to pHF10) containing the corresponding H. felis DNA fragments were isolated from E. coli SOLR by in vivo excision using M13 helper phages in accordance with the manufacturer’s protocol (Stratagene). As described for pRH vectors, detected pHF plasmids were subjected to a cycle of DNA sequencing for detection of the phosphorylation consensus sequences used as target sites for screening.
DNA sequencing.
DNA sequencing was performed at the Gesellschaft für Analyse-Technik und Consulting GmbH (GATC; Konstanz, Germany) using Sequenase 2.0 from Amersham. Biotin-labeled primers were obtained from MWG-Biotech (Eberberg, Germany). Plasmid DNA was subjected to a DNA sequencing protocol employing a GATC 1500 sequencing system for direct-blotting electrophoresis (47). Electroblotted DNA products were UV cross-linked to the nylon membranes at 254 nm for 2 min prior to colorimetric detection with streptavidin and alkaline phosphatase, as described in detail elsewhere (41). The sequencing data were manually digitized using a digitizer board connected to an Atari ST1040 personal computer and GFA-Basic software. Assembly of sequences obtained from the pRH948 shotgun library was done on an Apple Macintosh with DNA-Star/SeqMan software.
Determination of partial DNA sequences.
For determination of partial DNA sequences of I-408-positive clones, a biotin-labeled primer containing the I-408 primer sequence used for screening was synthesized. Sequencing reactions were carried out on DNA isolated from pRH or pHF vectors and were subjected to the direct blotting procedure. pRH539 as well as 6 of the pHF vectors (pHF3, -4, -5, -7, -9, and -10) did not give any results in the I-408-primed DNA sequencing reaction, whereas the two clones pRH514 and pRH948 and 4 of the 10 pHF vectors (pHF1, -2, -6, and -8) did. The DNA sequence information was used for synthesis of biotin-labeled primers of reverse orientation, allowing sequencing back through the phosphorylation consensus sequence. This reverse sequencing verified that pRH514, pRH948, and the four pHF vectors contained the phosphorylation consensus sequence. The partial sequences obtained by this procedure showed that pRH514 contained the H. pylori P-type ATPase isolated previously in our laboratory (31) whereas the target sequence of pRH948 was flanked by novel DNA sequences. DNA sequences found for the four pHF vectors, all containing the phosphorylation consensus sequences of P-type ATPases, were identical. pRH948 and pHF8 were selected for DNA sequencing of H. pylori or H. felis DNA insertions.
DNA sequencing of the insertions of pRH948 and pHF8.
A shotgun library was constructed from vector pRH948 or pHF8. We used a fast nebulization method to generate random, sequence-independent DNA fragments ranging from 0.5 to 1 kbp (41). Agarose gel-purified DNA fragments were blunt-end repaired with the Klenow fragment and T4 DNA polymerase (Boehringer Mannheim). The DNA was then ligated into the HindII site of plasmid pTZ18R and amplified in E. coli KK186. Single-stranded template DNA of several clones was prepared with M13KO7 helper phage. The sequencing procedure, using 5′-biotin-labeled universal and reverse sequencing primers for plasmid pTZ18R, was performed as described above, using Sequenase 2.0.
PCR.
DNA amplification of pRH948 plasmid DNA and chromosomal H. pylori DNA was performed. PCRs were carried out in a Perkin-Elmer Cetus thermal cycler. For each reaction, 1 μg of chromosomal DNA or 5 ng of plasmid DNA was mixed with 10 pg of primer DNA in a standard reaction volume of 50 μl. Twenty-five cycles of 1 min at 94°C, 1 min at 40°C, and 1.5 min at 72°C followed by an extension reaction at 72°C for 10 min were performed in the presence of 1 U of VentR polymerase in VentR buffer (both from New England BioLabs). PCR products were analyzed by agarose gel electrophoresis for determination of their molecular sizes. When necessary, PCR products were purified by using PCR purification kits (Qiagen).
Gene disruption mutagenesis of H. pylori copA.
For gene disruption mutagenesis of the copA gene, a BamHI-derived kanamycin resistance cassette from plasmid pUC4K was inserted into the copA gene of pRH948. A 498-bp DNA fragment of pRH948 corresponding to the N-terminal region spanning the amino acid sequence from Met-135 to His-300 and a DNA fragment of 732 bp encoding the C-terminal 244 amino acid residues (Gly-497 to His-741) of the Cop ion pump were amplified using primer pairs I-480–I-499 and I-481–I-500, respectively. PCR was carried out with the pairs of DNA primers given in brackets. The DNA sequences of the first pair of primers were ACCGAGTTGAATTCATGCATTGGGGG for I-480, including the ATG/Met-135 codon of cop and a preceding EcoRI restriction site (underlined), and GTCTACGGATCCGTGGCTATTGAATGT for I-499, containing a BamHI recognition site. The DNA sequences of the second pair of primers, used for amplification of the C-terminal Gly-497–to–His-741 region, were GTCTACGGATCCGGCATCAGCGCTAAAACAG, carrying a BamHI restriction site, for I-500 and CTGCAACTCAAGCTTATGATCCTTAATTTT for I-481, including a HindIII restriction site. The PCR products were cut with EcoRI-BamHI or BamHI-HindIII and cloned into EcoRI- and HindIII-digested pUC19 in E. coli MM294. The cloning product was opened by digestion with BamHI, and the kanamycin gene cassette, isolated by BamHI digestion of pUC4K, was inserted by ligation into the truncated cop gene of pUC19. Ligation of DNA fragments was performed by standard procedures (48).
Plasmid DNA was transformed into bacterial strain E. coli MM294 for replication of the cloning product, vector PY-123. Plasmid DNA of PY-123 containing the truncated copA gene with the internal kanamycin resistance cassette, localized in the direction of cop gene transcription as determined by EcoRI-HindIII digestion, was used for transformation of H. pylori ATCC 49503 cells.
Transformation of H. pylori by electroporation.
Cells of H. pylori ATCC 49503 grown in BHI broth medium were washed twice at 4°C in 10% (vol/vol) glycerol by centrifugation and resuspended in 10% (vol/vol) glycerol at about 109/ml. PY-123 DNA (2.5 μg) was added to 200 μl of H. pylori cell suspension. The mixture, after incubation on ice for 10 min, was transferred into a prechilled 0.2-cm cuvette (Bio-Rad) and subjected to single-pulse electroporation in a Bio-Rad Gene Pulser. Cells were transferred to BHI agar plates for a 24-h incubation under microaerophilic conditions at 37°C. The cells were subsequently transferred to BHI agar plates containing 10 μg of kanamycin sulfate per ml and incubated for 5 days as described above. Transformants obtained were grown in 10 ml of BHI-yeast extract-horse serum medium supplemented with 10 μg of kanamycin sulfate per ml.
Verification of copA knockout mutants of H. pylori by PCR.
Genomic DNAs of H. pylori mutant and parental strains were prepared according to standard methods (1, 48). These were subjected to PCR for analysis of the copA region. The DNA primers used for amplification were I-480 and I-481, as employed for construction of the hybrid copA-kanR plasmid PY-123, spanning a 1.8-kbp sequence of the cop gene. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
Effect of metal ions on growth of H. pylori.
To determine the MICs of various divalent cations for H. pylori wild-type strain ATCC 49503 or its copA-deficient derivative, cells were incubated in BHI broth medium in the absence or presence of various concentrations of Cu2+, Zn2+, Co2+, Ni2+, or Mg2+ as the Cl− salt. Growth of the bacterial cultures was monitored by measurement of optical density (578 nm) at various time points. Concentrations employed were 50 μM, 100 μM, 250 μM, 500 μM, and 1 mM for all cations listed above except for Mg2+. Additional concentrations used were 2.5, 7.5, 15, and 25 μM for CuCl2 and 2 mM, 4 mM, and 6 mM for NiCl2. Concentrations used to determine MICs of MgCl2 were 5, 25, 50, and 75 mM.
Synthesis of the N-terminal peptide of the H. pylori CopA ATPase (amino acid residues 1 to 52).
A 25-μmol quantity of the ATPase peptide from the N-terminal methionine to leucine-52 was synthesized by solid-phase peptide synthesis with an Abimed EPS221 automated peptide synthesizer (Abimed, Langenfeld, Germany), using the fluorenylmethoxycarbonyl (Fmoc) protection strategy (6, 12, 32). The Nα-protected leucine residue was carboxy-terminally linked by 4-hydroxymethylphenoxyacetic acid to a graft polymer of polyethylene glycol onto a polystyrene support (3) (NovaSyn TGA resin; Novabiochem, Bad Soden, Germany). Benzotriazole-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate (PyBOP) activator and Nα-protected amino acids were also purchased from Novabiochem (7a). Reagents and solvents were obtained from Aldrich (N-methylmorpholine), Merck (dimethylformamide), and Fluka (piperidine, trifluoroacetic acid, and triethylsilane).
Chain elongation was performed for 51 cycles in accordance with an operational cycle protocol that included Fmoc deprotection with 20% piperidine in dimethylformamide (45, 61) for 5 min at 20°C. Deprotection was followed by coupling. The time was increased from 40 min at the initial cycle to 50 min at the end of the synthesis. Coupling was performed in the presence of a 5-molar excess of PyBOP-activated (17) and Fmoc-protected amino acid relative to the peptide. Fmoc deprotection was monitored with a UVIDEC-100II UV spectrometer (Jasco, Gross-Umstadt, Germany). After final Fmoc deprotection, cleavage from the support and deprotection of side chains was carried out in a single step by treatment with trifluoroacetic acid for 2 h at 20°C, using a triethylsilane scavenger. The crude peptide was isolated by precipitation with t-butylmethyl ether (Fluka) and semipreparative high-performance liquid chromatography (Waters Bondapak C18 column) with a linear gradient of water and acetonitrile. The high-performance liquid chromatography-purified peptide was subjected to matrix-assisted laser desorption mass spectrometry (20, 45) and showed an average molecular mass of 5,851 Da, which was in good agreement with the molecular mass of 5,850.7 Da calculated from the amino acid sequence.
Metal ion binding by the N-terminal peptide of a synthetic ATPase.
Ni2+ affinity chromatography (Qiagen, Hilden, Germany) was used to study the ability of the first N-terminal 52 amino acids of the ATPase peptide (P95-030) to bind metal ions. To exchange the Ni2+ ions for other divalent cations, 1 ml of Ni2+-agarose suspension (equivalent to 500 μl of gel bed) was loaded onto a column (3-cm Econo; Bio-Rad), which was subsequently washed with 2 ml of H2O followed by 5 ml of 100 mM ethylenediaminetetraacetic acid (EDTA) to remove the Ni2+ ions and then equilibrated with 2 ml of H2O. The binding of the ions to be tested to the agarose matrix was carried out by incubating the agarose matrix with 2 ml of a 100 mM solution of either NiCl2, CuSO4, CoCl2, ZnCl2, CdCl2, or MgCl2. After transfer to a reaction tube, the beads were washed two times with 2 ml of H2O and three times with binding buffer, pH 7.8 (50 mM NaH2PO4, 0.3 M NaCl; pH adjusted with 1 N NaOH). A 100-μg sample of peptide P95-030 dissolved in 150 μl of binding buffer was added to 50 μl of the respective metal ion affinity matrix containing Ni2+, Cu2+, Co2+, Zn2+, Cd2+, or Mg2+ in a 1.5-ml reaction tube, and the mixture was incubated for 1 h at room temperature on a roller incubator. After this binding step, the supernatant was removed, and 30 μl was dried in a vacuum concentrator and resuspended in 40 μl of SDS-Tricine-polyacrylamide gel electrophoresis (PAGE) sample buffer. The matrix was washed once with 5 volumes of binding buffer and transferred to a 1.5-ml Mobitec column tube. Bound peptide was eluted with 3 ml of binding buffer adjusted to pH 4 with 1 N HCl and collected in two fractions of 1 and 2 ml.
PAGE of ATPase peptide.
The peptide of 52 amino acids (M1-L52) contained in the 1-ml fraction obtained by elution of the affinity agarose column was precipitated with 20% trichloroacetic acid and resuspended in 40 μl of SDS-Tricine gel sample buffer. The precipitates were separated by SDS-PAGE on a 15% polyacrylamide gel with Tricine buffer (49). The gels were stained with 0.025% Serva Blue G in 10% acetic acid for 1 h and destained in 10% acetic acid.
ESI-MS of the ATPase peptide.
ESI spectra of the N-terminal peptide of the synthetic ATPase were recorded on a Vestec 201 A single-quadrupole mass spectrometer (Vestec Corp., Houston, Tex.). The temperature of the ionization region was kept at approximately 40°C, while the voltage between the collimator and skimmer was maintained at 40 V during any measurement. Other instrumental conditions were as described previously (44). Peptide solutions (50 μM) were prepared in 5 mM ammonium acetate in water-methanol (9:1) at pH 4. A 10-molar excess of either NiCl2 or CuCl2 was added to peptide solutions, which were incubated for 30 min at 20°C and then immediately subjected to ESI-MS analyses. Sample delivery to the electrospray needle tip was performed with a Harvard-44 microinfusion pump (Harvard Apparatus, South Natick, Mass.) through a fused-silica capillary tube at a flow rate of 3 μl/min.
HK-M0 and HK-M1 vector construction and analysis.
Construction and function of the HK-M0 and HK-M1 cloning vectors have been described in detail elsewhere (2, 31). Coupled transcription-translation of HK-M0 and HK-M1 vectors containing putative TM segments of membrane proteins have been employed for analyses of membrane topology of the gastric H+/K+ ATPase (2), the sarcoplasmic Ca2+ ATPase (4), and a member of the bacterial transition ion P-type pumps cloned from H. pylori (31).
Hydropathy analysis was used for selection of putative membrane-spanning segments of CopA with the algorithms of Rao and Argos (46), Eisenberg et al. (9), and Klein et al. (22). Selected regions were amplified by PCR using DNA primers targeted to DNA sequences encoding the first 6 (sense) or last 6 (antisense) amino acids of possible TM segments. The primers were synthesized with 5′ extensions containing BglII (sense primers) or a HindIII (antisense primers) restriction sites for in-frame cloning into HK-M0 or HK-M1 vectors (Table 1). HK-M0 and HK-M1 plasmid constructs were used for synthesis of 35S-labeled fusion proteins by coupled transcription-translation with a reticulocyte lysate system (Promega). Reactions were carried out in the absence or presence of canine microsomes according to the manufacturer’s protocol (Promega). 35S-labeled fusion protein products were separated on SDS–10% polyacrylamide gels (24). The gels were then dried, and radioactivity was detected by using a phosphorimager and Ambis software (Image Acquisition and Analysis).
TABLE 1.
Sequences of the H. pylori CopA ATPase inserted into vector M0 or M1a
| Inserts | Sequence | Positions | Glycosylationb
|
Activityc | |
|---|---|---|---|---|---|
| M0 | M1 | ||||
| H1 | LALAVIFTLFVVYLSMGAMLS | L84 to S104 | + | − | SA, ST |
| H2 | HSNFLNACLQLIGTLIVMHLGR | H118 to R139 | ± | − | SA, ST |
| H3 | LIAIGTSAALISSLWQLYLVYT | L159 to T180 | + | − | SA, ST |
| H4 | WSYGHYYFESVCVILMFVMVGR | W183 to R205 | + | − | SA, ST |
| HXa | LVDSIVVGDILKVLPGSAIAV | L241 to V261 | − | + | |
| HXb | NQQIEVLVDSIVVGDILKVLPGSAIAVDG | N235 to G263 | − | + | |
| H5 | VSSVFVPSVIAIAILAFVVWLIIAA | V340 to A363 | + | − | SA, ST |
| H6a | FWWNFGIALEVFVSVLVISCPCALGLATPMSIL | F368 to L400 | ± | − | SA, ST |
| H6b | FWWNFGIALEVFVSVLVISCPCALGLA | F368 to A394 | ± | − | SA, ST |
| H6c | FWWNFGIALEVFVSVLVI | F368 to I385 | ± | − | SA, ST |
| H6d | IALEVFVSVLVISCPCALGLATPMSIL | I374 to L400 | ± | − | SA, ST |
| H6e | IALEVFVSVLVISCPCALGLA | I374 to A394 | − | − | ST |
| H6f | IALEVFVSVLVIS | I374 to I385 | ± | − | SA, ST |
| HY | LNDAPSLAMSDVAVVMK | L629 to K646 | − | + | |
| H7 | NLFWAFCYNSVFIPLACGVL | N684 to L703 | − | − | ST |
| H7a | KENLFWAFCYNSVFIPLACGVLKA | K682 to K705 | − | − | ST |
| H7b | KENLFWAFCYNSVFIPLACGVL | K682 to L705 | − | − | ST |
| H7c | KNIKENLFWAFCYNSVFIPLACGVL | K679 to L703 | − | − | ST |
| H7d | KNIKENLFWAFCYNSVFIPLACGVLKA | K679 to K705 | − | − | ST |
| H7e | NLFWAFCYNSVFIPLACGVLKA | N684 to K705 | − | − | ST |
| H8a | IMLSPAIAGLAMSLSSVSVVL | I708 to K728 | − | − | ST |
| H8b | IMLSPAIAGLAMSLSSVSVVLNSQRLRNFKIK | I708 to K739 | − | − | ST |
| HY-7 | LNDAPSLAMS/––––––––/VFIPLACGVL– | L629 to L703 | ± | ND | SAd |
| HY-8a | LNDAPSLAMS/––––––––/MSLSSVSVVL– | L629 to L728 | − | ND | SAd |
| HY-8b | LNDAPSLAMS/––––––––/SQRLRNFKIK– | L629 to K739 | − | ND | SAd |
In the fusion proteins, either HK-M0 or HK-M1 is followed by the listed copper ATPase insert sequences and the β-subunit segment. The last three columns present a summary of the results of all the different inserts expressed in the vectors HK-M0 (M0) and HK-M1 (M1).
+, presence of glycosylation; −, absence of glycosylation; ±, variable glycosylation; ND, not determined.
SA, signal anchor activity; ST, stop transfer activity.
Stop transfer activity not determined.
Materials.
Helper phage M13KO7 as well as plasmids pUC4k, pUC19, and pTZ18R were from Pharmacia. M13 helper phage (ExAssist) was from Stratagene. Restriction enzymes were from Boehringer Mannheim. Chemicals used were all of analytical grade or higher.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the GenBank and EMBL databases under accession no. U59625 (H. pylori sequence) and AJ001932 (H. felis sequence).
RESULTS
Genomic Southern blot analyses.
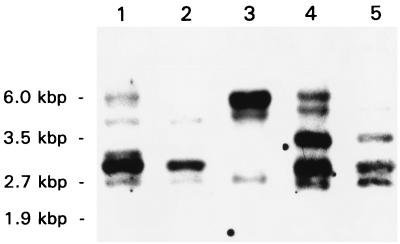
A DNA oligonucleotide mixture of 16 distinct 20-mer DNA molecules out of the 2,304 possible sequences encoding the DKTGT(I/L)T phosphorylation consensus sequence had been used previously to isolate an H. pylori P-type ATPase (31). In this study, five DNA oligonucleotide mixtures covering all possible DNA sequences of the phosphorylation target sequence, I-405 to I-409, were used for Southern blot analyses of H. pylori chromosomal DNA. Each of the synthesized DNA oligonucleotide mixtures, consisting of similar DNA sequences, was hybridized to membranes containing HindIII-restricted genomic DNA. In all cases, a pattern of up to five major hybridization bands was found, indicating a high degree of similarity but nonidentity (Fig. 1). This shows that even the phosphorylation site-specific DNA oligonucleotides of extended complexity are able to detect putative P-type ATPase DNA clones derived from H. pylori DNA. Since primer I-408 detected all of the major bands, this primer was selected for use in screening of the H. pylori 69A gene library. The same primer was used for screening of an H. felis gene library.
FIG. 1.
Fluorogram of a Southern blot of chromosomal H. pylori 69A DNA hybridized with various DNA oligonucleotides targeted to the phosphorylation signature sequences of P-type ATPases. The membranes containing HindIII-restricted DNA were hybridized with DIG-labeled primers I-405 (lane 1), I-406 (lane 2), I-407 (lane 3), I-408 (lane 4), and I-409 (lane 5), as described in Materials and Methods. Positive restriction fragments were detected by chemiluminescence. Molecular sizes are given in kilobase pairs.
Isolation of Helicobacter copAP operons.
Since E. coli DNA was also positive with each of the five DNA oligonucleotide probes (data not shown), the H. pylori 69A library was screened by subjecting mixed-plasmid preparations to Southern blot hybridization instead of the conventional colony filter hybridization technique. Using probe I-408, plasmid pRH948 as well as two additional DNA clones, pRH514 and pRH539, were isolated from the H. pylori genomic DNA library present in pRH160 in E. coli HB101. The lengths of the inserted H. pylori DNA sequences of the plasmids were determined by restriction analysis to be within the range of 3 to 5 kbp (data not shown).
DNA sequencing revealed that pRH948 contained a novel P-type ATPase gene, whereas the DNA sequence of pRH514 showed identity with the ion pump isolated previously (31), and pRH539 was not susceptible to the DNA sequencing procedure using primer I-408. Hence, vector pRH948 was selected for sequencing of the complete H. pylori DNA fragment by the direct blotting procedure as described in Materials and Methods.
Primer I-408 was also used to screen an H. felis gene library cloned in the λ ZAP II vector. Screening led to the isolation of 10 positive clones, pHF1 to pHF10. Partial sequencing showed that four of the clones isolated (pHF1, -2, -6, and -8) were identical, containing DNA sequences encoding a P-type ATPase. pHF8 was selected for DNA sequencing.
DNA sequence analysis of Helicobacter copAP operons and flanking DNA.
DNA sequencing showed that the H. pylori DNA sequence in pRH948 was 4,472 bp in length. About 2.3 kbp of the pRH948 3′ DNA sequence overlaps with a previously published sequence predicted to contain the copAP operon of H. pylori UA802 (13). Since this published sequence encodes a membrane pump lacking an N-terminal ion binding domain as well as the first pair of TM segments, it was assumed that this earlier-cloned version of the H. pylori CopA ion pump was N-terminally truncated (31). The DNA sequence inserted in pRH948 contained an additional 2.2 kbp of the nucleotide sequence region upstream of the DNA reported previously (13) before enclosing the N-terminal sequences not present in the first version of CopA. The missing sequences of the H. pylori UA802-encoded version of the CopA ion pump have been published recently (14). Sequence identity between the H. pylori 69A-derived DNA and the corresponding DNA region of H. pylori UA802 was found to be about 94%. The copAP locus, with a degree of DNA sequence identity similar to that of the former operons, is also present in H. pylori 26695 (60).
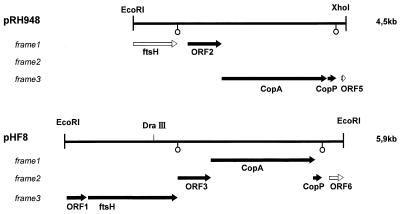
The sequence of the pRH948 DNA strand spanning the distance between the EcoRI cloning site and the XhoI recognition site of the vector backbone was found to predict five open reading frames (ORFs), whereas no ORF of any significant length was found on the reverse strand. The two ORFs located at the terminal regions of the inserted DNA fragment, ORF1 and ORF5, were interrupted by the cloning sites and therefore were incomplete. ORF1 encoding an N-terminally truncated version of the H. pylori FtsH protein, is also present upstream of the copAP operon in H. pylori UA802 (15). The next coding region (ORF2), which was separated from the preceding ftsH sequence by a putative transcriptional termination signal, predicted a protein of 237 amino acids. An N-terminal overlap of 173 amino acid residues of this amino acid sequence showed the closest identity to the phosphatidylserine synthase of Bacillus subtilis, a protein of 177 amino acids (40). ORF2 was followed by the largest ORF of pRH948, ORF3, which contains the P-type ATPase target sequence used for screening. The P-type gene of ORF3 (copA) is immediately followed by a small ORF4 encoding a peptide of 66 amino acids (CopP). The 3′-localized ORF of pRH948 (ORF5) is interrupted by vector sequences immediately downstream. The predicted N-terminal peptide of 27 amino acids, which is not identical to the gene product predicted by the corresponding region in H. pylori UA802 (13), is of unknown homology. In Fig. 2, a structural map of the pRH948 insertional DNA is given, showing the organization of the H. pylori copAP operon and of the ORFs flanking the operon. Putative transcription termination sequences are present in the DNA sequence downstream of the truncated ftsH region and also downstream of copP, separating ORF4 and ORF5. An AGGA Shine-Dalgarno consensus sequence is located 7 bases upstream of the copP gene, whereas copA, in the corresponding region, contained a DNA sequence with little similarity to the Shine-Dalgarno consensus motif (data not shown).
FIG. 2.
Organization of the copAP operons and flanking DNA cloned from H. pylori and H. felis. The figure shows the ORFs contained in plasmids pRH948 (H. pylori) and pHF8 (H. felis). The ORFs detected by translation of the DNA in the three possible frames (frames 1 to 3) are indicated by arrows. The ftsH gene and ORF5 of pRH948 are truncated (open arrows), as is ORF6 in pHF8. The locations of putative transcriptional termination sequences are indicated by open circles. Terminal EcoRI (pHF8) and XhoI-EcoRI (pRH948) restriction sites of plasmid insertional DNA as well as a unique DraIII site present in the DNA insertion of pRH948 are also displayed.
DNA sequencing of the H. felis 5,909 kbp insertion of pHF8 showed that this related organism also contained a cop operon consisting of genes encoding a P-type ATPase (CopA) and a small peptide with possible transition metal ion binding properties (CopP). Whereas the ORFs of copA and copP in H. pylori are separated by 3 nucleotides, the ATG start codon of copP in H. felis is located in another frame within the 3′ coding region of the copA gene. The DNA preceding the CopAP region in pHF8 contained three additional ORFs. The amino acid sequence of ORF1 displayed some similarity to ribosomal protein L11 methyltransferases of bacteria (data not shown). This ORF was followed by a gene encoding a protein of 638 amino acids with more than 90% sequence identity to the H. pylori FtsH protein (15). The ftsH locus is also contained in the corresponding region of H. pylori 69A, as described above. ORF3, downstream of the H. felis ftsH gene in pHF8 (214 amino acids), shows similarity to phosphatidylserine synthases in the first N-terminal 160 amino acids, as was found for ORF2 of the corresponding chromosomal H. pylori 69A DNA. ORF3 of pHF8 is followed by the copAP locus (ORF4 and ORF5 of pHF8). The 3′-localized ORF of the H. felis DNA fragment contained in pHF8 (ORF6) was truncated and is of unknown homology. The organization of the genes found on pHF8 DNA, including the copAP operon, is also depicted in Fig. 2.
Comparison of the DNA sequences contained in pRH948 and pHF8 shows that the sequence identity is about 55 to 70% within the region spanning the copAP operon and the ORF preceding the operon (ORF2) of pRH948 and ORF3 of pHF8). The degree of DNA sequence identity in the overlapping regions of the ftsH loci was found to be significantly higher (>90% [data not shown]). The results also show that the genes immediately preceding the copAP locus are the same in H. felis and H. pylori 69A as well as in H. pylori 26695 and UA802 (13–15, 60). The sequence data are in the EMBL and GenBank nucleotide sequence data libraries under accession no. U49625 (pRH948) and AJ001932 (pHF8).
Properties of CopP amino acid sequences.
The copP gene products of H. pylori 69A and H. felis both consist of 66 amino acids and have about 60% identity (Fig. 3). They both contain a CXXC-type transition metal binding motif, suggesting that this protein may act as a transition metal binding protein. This protein was also predicted from the copAP loci of H. pylori UA802 and H. pylori 26695 (13, 60). The CopP peptides of H. pylori and H. felis were found to exhibit homology with other ion binding proteins, such as the periplasmic mercury binding protein MerP of Serratia marcescens, MerP of Shigella flexneri, and especially the CopZ protein of Enterococcus hirae (33, 35, 38). Figure 3 displays a sequence alignment of enterococcal CopZ (38), CopP from H. felis (this study), and the CopP amino acid sequence variants cloned from H. pylori 69A (this study) and other H. pylori strains (13, 60). They all contain a CXXC motif in amino acid positions 12 to 15.
FIG. 3.
Comparison of the amino acid sequences of CopZ from Enterococcus hirae and the CopP peptides from Helicobacter pylori (Hp) strains and Helicobacter felis (Hf). The CopP sequences of 66 amino acid residues encoded by plasmids pRH948 (H. pylori 69A) (this study) and pBHpC8 (H. pylori UA802) (13) or detected by H. pylori genome sequencing (H. pylori 26695) (60) have an overall identity of >95%. The CopP peptide predicted from the small ORF of the copAP DNA fragment of H. felis, also consisting of 66 amino acids, showed a lower degree of identity with the H. pylori peptides (about 60%), but a stretch of 10 identical amino acids is observed just after the CXXC motif. The degree of amino acid sequence identity of CopP peptides and E. hirae CopZ (69 amino acid residues) is still between 40 and 50%.
Properties of CopA amino acid sequences.
The P-type pumps predicted by the Helicobacter cop operons were 741 (CopA, H. pylori) or 732 (CopA, H. felis) amino acids in length, as shown in Fig. 4. The encoded proteins both contain a DKTGTLT phosphorylation signature sequence and a GDGVND ATP binding motif, as found in other P-type ion pumps (11, 43). There are also consensus sequences characteristic of transition metal P-type ATPases, such as an N-terminal GMXCXXC sequence motif and a CPC box in the membrane domain (52, 57). The amino acid sequence of the H. pylori 69A CopA protein exhibited 95% identity with the CopA protein products cloned from H. pylori 26695 (60) and H. pylori UA802, the latter containing a CPS motif instead of the CPC found in the H. pylori 69A and 26695 CopA sequences (13).
FIG. 4.
Comparison of the CopA amino acid sequences of H. pylori (HP) and H. felis (HF). The CopA amino acid sequences are 741 (HP) or 732 (HF) amino acid residues in length. The overall level of sequence identity between the two pumps is about 55%. TM segments of H. pylori CopA, determined experimentally in this study, are boxed and lightly shaded (TM1 to TM8). Also in boxes are the putative membrane-spanning segments of the H. felis pump. More darkly highlighted are conserved sequence boxes: the putative N-terminal Gly-Met-Thr-Cys-Thr/Ser-Ala-Cys metal ion binding motif, the membrane-associated Cys-Pro-Cys sequence, the Asp-Lys-Thr-Gly-Thr-Leu-Thr phosphorylation sequence, and a Gly-Asp-Gly-Leu/Val-Asn-Asp-Ala-Pro motif for ATP binding.
Of the proteins currently available in the database, the H. pylori 69A and H. felis CopA exhibited the highest degree of sequence homology of the proteins currently available in the database, to another ATPase of H. pylori cloned in our laboratory (31) and to the various bacterial and eukaryotic Cu2+ and Cd2+ ATPases. Prominent members of this family are CopA and CopB of Enterococcus hirae (39, 55); the Cd2+ ATPase of Staphylococcus aureus; CadA (37), a putative Synechococcus Cu2+ ATPase (19); and the human Menkes and Wilson gene products (5, 62). From the degree of amino acid sequence similarity and the occurrence of both GMXCXXC and CPC motifs, it was concluded that the cloned pump is a member of the transition metal ion pump family, recently also referred to as CPX-type ATPases (57).
Function of the CopA ATPase.
The function of the cloned CopA ATPase was determined by gene disruption mutagenesis of the ATPase gene contained in pRH948 and subsequent generation of H. pylori copA knockout mutants by homologous recombination, as described in Materials and Methods. This methodology is not yet available for H. felis. The wild-type strain subjected to copA mutagenesis was H. pylori ATCC 49503. In the mutant obtained, the copA ATPase gene lacked an entire segment of the pump, from amino acid 301 to 496, which was exchanged for a kanamycin resistance cassette, resulting in a kanamycin resistance phenotype in the recipients of the PY-123 construct. Neither the growth kinetics of the ATPase-deficient mutants nor their urease activities were different from that of the parental strain, H. pylori ATCC 49503, showing that this pump is not necessary for growth in vitro (data not shown). However, the enzyme might still be essential for survival in the stomach.
The mutant was susceptible to the same concentrations of Ni2+, Zn2+, Co2+, and Mg2+ as the wild-type strain but had a different susceptibility to Cu2+. The wild-type strain had a Cu2+ MIC of 50 μM, whereas the mutant had a Cu2+ MIC of 7.5 μM, suggesting that the ATPase can function as a Cu2+ export pump (Table 2). A corresponding change in copper sensitivity was obtained when the pRH948-encoded CPC-type ATPase was inactivated in the genome of H. pylori 69A by transposon shuttle mutagenesis (data not shown). Since a change in Cu2+ sensitivity was also found in H. pylori UA802 copA knockout mutants (13), it is evident that both of the natural CopA sequence variants, the CPC-type pump predicted by pRH948 and the CPS-containing sequence predicted from the copA gene cloned from H. pylori UA802, are able to export Cu2+.
TABLE 2.
MICs of divalent cations for H. pyloria
| Divalent cation | MIC of divalent cation for:
|
|
|---|---|---|
| H. pylori 49503 | H. pylori 49503/CopA− | |
| CuCl2 | 50 μM | 7.5 μM |
| ZnCl2 | 500 μM | 500 μM |
| NiCl2 | 4 mM | 4 mM |
| CoCl2 | 250 μM | 250 μM |
| MgCl2 | 50 mM | 50 mM |
To determine the MICs of various divalent cations for H. pylori, cells of the wild-type strain ATCC 49503 or its CopA-deficient derivative were incubated in BHI broth medium in the absence or presence of various concentrations of CuCl2, ZnCl2, CoCl2, or NiCl2. The growth MICs for the cations are shown.
Metal binding characteristics of the N-terminal region of the ATPase.
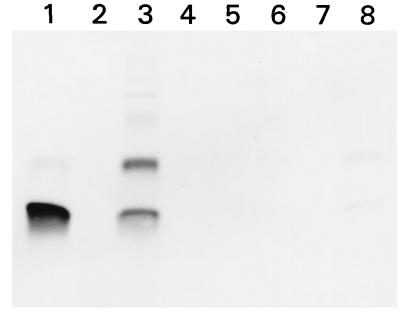
When the synthetic peptide representing the 52 N-terminal amino acids of H. pylori CopA containing the GMXCXXC ion binding motif was adsorbed to affinity agarose equilibrated with Ni2+, Co2+, Cd2+, Mg2+, Zn2+, or Cu2+, the peptide displayed binding mainly to Cu2+. A very weak binding reaction was observed with Zn2+. These data are shown in Fig. 5.
FIG. 5.
Ion binding affinity of a synthetic ATPase peptide (P95-030) as determined by metal ion affinity chromatography. A 100-μg portion of peptide P95-030 was bound to a divalent-cation column as described in Materials and Methods. Bound peptide was eluted and separated by SDS-PAGE, using 15% acrylamide and Tricine buffer under nonreducing conditions, and stained with Serva Blue G. Lane 1, 5 μg of peptide P95-030 (control); lanes 3 to 8, peptide eluted from the matrix after binding to divalent ions and contained in 1 ml of the 3-ml elution volume. The column matrix was equilibrated with CuCl2 (lane 3), NiCl2 (lane 4), CoCl2 (lane 5), CdCl2 (lane 6), MgCl2 (lane 7), or ZnCl2 (lane 8). The peptide was able to form dimers, presumably due to the formation of intermolecular Cys-Cys bonds (lanes 1, 3, and 8).
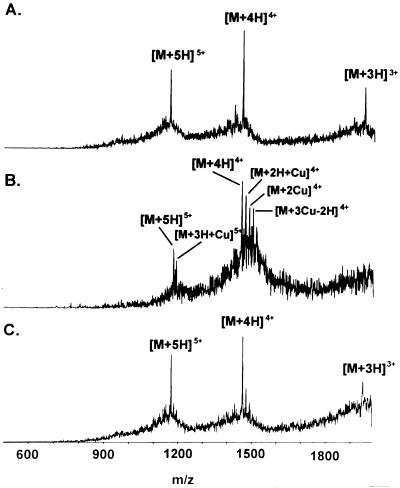
The spectra obtained by subjecting the N-terminal ATPase peptide to ESI-MS are depicted in Fig. 6. ESI-MS detected the three, four, and fivefold positively charged ions of the peptide. The peptide was preincubated with CuCl2 or NiCl2. When preincubated in the presence of CuCl2, adducts of the peptide carrying up to three copper atoms were observed, as demonstrated in Fig. 6B. Copper ion binding of the peptide is in agreement with the data obtained by ion affinity chromatography. The data show that the peptide was able to bind up to a threefold molar excess of copper ions. While the occurrence of copper adducts was significant, preincubation of the peptide with NiCl2 led to detection of faint peaks with m/z increments of the magnitude expected for peptides binding one or two nickel atoms (Fig. 6C). The weak binding of the peptide to Zn2+ observed in ion affinity chromatography was also detected by ESI-MS (data not shown).
FIG. 6.
Electrospray mass spectra of synthetic ATPase peptide (amino acids 1 to 52) in the absence and presence of Cu2+ and Ni2+. (A) ESI spectra of unmodified Cop ATPase peptide. Three, four, and fivefold positively charged ions of the peptide were detected. (B) After preincubation with CuCl2, 4.5-fold positively charged adducts of copper with the peptide were observed. (C) When the peptide was preincubated with NiCl2, complexes of the peptide with nickel seemed to be detectable also, as indicated by the very faint peaks immediately following the [M + 4H]4+ signal. Binding to Ni2+, therefore, was much less significant than binding to Cu2+ (C). M, peptide molecule; H, proton; m, mass; z, charge of molecule.
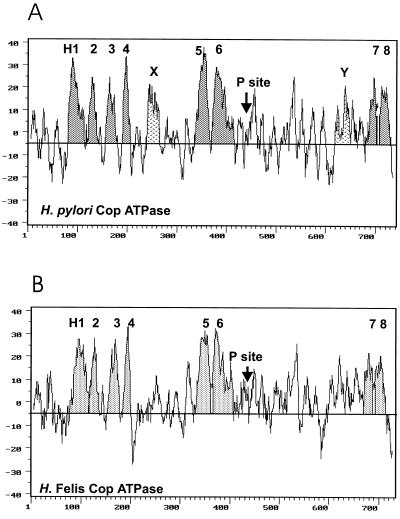
Analysis of membrane topology.
The Kyte-Doolittle hydropathy profiles (23) of the encoded CopA ion pump versions of H. pylori and H. felis predict several membrane spanning sequences (Fig. 7) and are very similar to those of the other transition metal P-type ATPases, particularly the various Cu2+ and Cd2+ ion pumps (5, 13, 14, 19, 36–39, 51, 52, 55–57) and the pRH439-encoded H. pylori P-type ATPase. The membrane domain of the latter ATPase was shown by in vitro translation to contain eight TM spanning sequences (31).
FIG. 7.
Kyte-Doolittle hydropathy profiles of H. pylori (A) and H. felis (B) CopA ATPases. (A) The putative TM segments determined by in vitro translation, H1 through H8, are highlighted. Also marked are hydrophobic regions HX and HY, which do not have membrane insertion activity. The localization of the conserved phosphorylation sequence is also shown (P site). (B) The membrane spanning segments of the H. felis pump are highlighted based on similarity to the CopA P-type ATPase sequence of H. pylori. As found for the H. pylori pump, the P site is between H6 and H7 in the CopA ATPase of H. felis.
Putative TM sequences of the pRH948-predicted H. pylori CopA pump were selected by various hydropathy-based algorithms and by their sequence homology to hydrophobic segments found in the H. felis CopA ATPase (Table 1 and Fig. 7). PCR-amplified copies of these segments of H. pylori CopA were assayed by coupled in vitro transcription-translation of the HK-M0–HK-M1 fusion proteins to determine their ability to membrane insert during translation. The N-terminal parts of the fusion proteins encoded by those vectors consist of either the first 101 (M0) or the first 139 (M1) amino acids of the α subunit of the gastric proton pump followed by the putative TM domain of the CopA ATPase. The C-terminal parts of the HK-M0–HK-M1 fusion proteins contain the 177 terminal amino acids of the β subunit of the gastric H+/K+ ATPase carrying five putative glycosylation sites. The presence of glycosylation due to signal anchor properties was evidenced by the increase in the Mr of the product when the HK-M0 vector with insert was translated in the presence of microsomal membranes. In turn, the presence of an insert encoding a stop transfer sequence was shown by inhibition of glycosylation of the HK-M1 fusion vector in the presence of microsomal membranes.
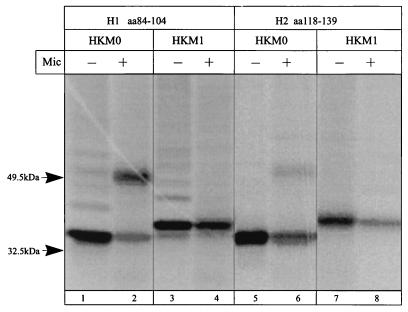
The first two hydrophobic regions, H1 (amino acid positions 84 to 104 [Fig. 8, lanes 1 to 4]) and H2 (amino acid positions 118 to 139 [Fig. 8, lanes 5 to 8]), were both signal anchor and stop transfer sequences. This should be the first pair of sequences in the TM domain of this ATPase (Table 1). The signal anchor activity of H2 is weaker; this could be explained by a reverse orientation in the vector compared to its natural folding.
FIG. 8.
Autoradiograph of an SDS-PAGE gel with products of in vitro transcription-translation, in the absence and presence of microsomes (Mic), of the HK-M0 and HK-M1 vectors containing the first hydrophobic domain (lanes 1 to 4) and the second hydrophobic domain (lanes 5 to 8) of the H. pylori CopA ATPase.
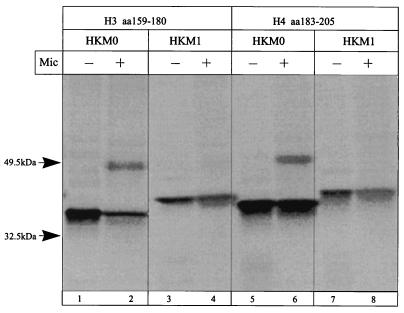
Sequences H3 (amino acid positions 159 to 180 [Fig. 9, lanes 1 to 4]) and H4 (amino acid positions 183 to 205 [Fig. 9, lanes 5 to 8]) are also both signal anchor and stop transfer sequences, indicating that H3 and H4 comprise the second pair of TM segments.
FIG. 9.
Autoradiograph of an SDS-PAGE gel with products of in vitro transcription-translation, in the absence and presence of microsomes (Mic), of the HK-M0 and HK-M1 vectors containing the third hydrophobic domain (lanes 1 to 4) and the fourth hydrophobic domain (lanes 5 to 8) of the H. pylori CopA ATPase.
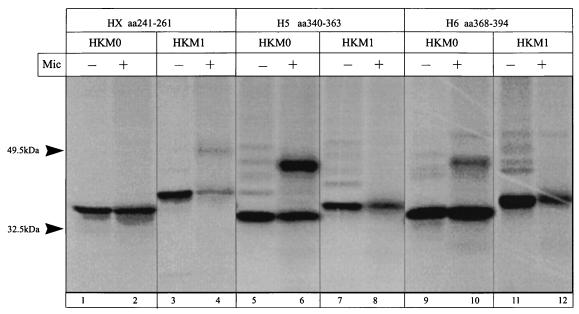
The next hydrophobic region following H4 in the H. pylori CopA ATPase is a sequence that has been named HX (amino acid positions 241 to 261 [Fig. 10, lanes 1 to 4]). Two segments of this region were translated in the vectors HK-M0 and HK-M1. Neither signal anchor nor stop transfer activity was found (Table 1). These results suggest that this region is not membrane inserted.
FIG. 10.
Autoradiograph of an SDS-PAGE gel with products of in vitro transcription-translation, in the absence and presence of microsomes (Mic), of the HK-M0 and HK-M1 vectors containing the HX hydrophobic domain (lanes 1 to 4), the fifth hydrophobic domain (lanes 5 to 8), and the sixth hydrophobic domain (lanes 9 to 12) of the H. pylori CopA ATPase.
The putative TM region H5 (amino acid positions 340 to 363 [Fig. 10, lanes 5 to 8]) promoted a strong glycosylated band in the HK-M0 vector, similar to the one obtained with the HK-M1 vector control, and also performed as a stop transfer sequence in HK-M1. Six overlapping segments coding for the putative TM domain H6 (between amino acids 368 and 400 [Table 1]) were inserted in both HK-M0 and HK-M1. Most of them were able to promote partial glycosylation of the H/K ATPase β-subunit-derived sequences in the HK-M0 vector, as did the segment comprising amino acid positions 368 to 394 (Fig. 10, lanes 9 and 10). All of them prevented the glycosylation of the β-subunit sequences in the HK-M1 vector, therefore acting as a stop transfer sequence, which is the membrane insertion activity expected for this hydrophobic segment in the topological model (Fig. 10, lanes 11 and 12). These results suggest that H5 and H6 form the third pair of antiparallel helices of the CopA membrane domain.
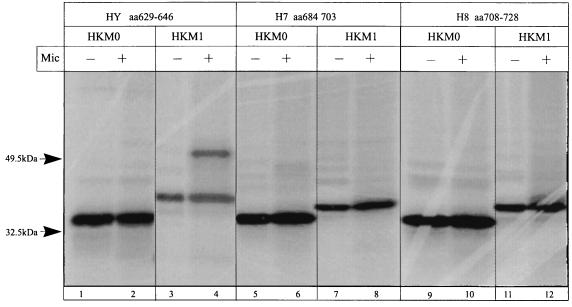
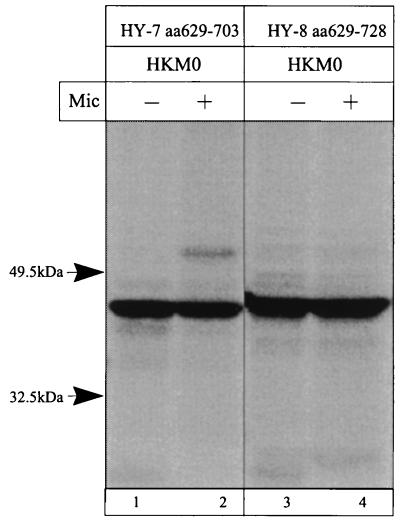
The C-terminal region contains three main hydrophobic regions, HY, H7, and H8, which were expressed in the HK-M0 and HK-M1 vectors (Table 1). The results of the translation showed that the region HY (amino acid positions 629 to 646) could not act as a signal anchor sequence. When this region was translated in the HK-M1 vector, partial inhibition of glycosylation was observed. Data are shown in Fig. 11, lanes 1 to 4. The hydrophobic segment H7 (amino acid positions 679 to 703), which was expected to act as a signal anchor, was only a stop transfer sequence (Table 1; Fig. 11, lanes 5 to 8). However, when the H7 segment is extended upstream, therefore including the HY region (amino acid positions 629 to 703), translation in the HK-M0 vector showed a glycosylated product (Fig. 12, lane 2), indicating that the sequence preceding the 684-to-703 segment is important for the membrane insertion of H7 as a signal anchor sequence. The putative TM domain H8 (amino acid positions 708 to 728) acts as a stop transfer sequence, as expected from the eight-segment model of transition ion pumps (31) (Table 1; Fig. 11, lanes 9 to 12). The addition of the H8 coding region to the HY-H7 fragment suppressed the glycosylated HK-M0–CopA ATPase (amino acid positions 629 to 728) fusion product when expressed in the HK-M0 vector (Fig. 12, lane 4). Hence, the HY region helps to direct the membrane insertion of H7, and the hydrophobic H8 sequence returns the C terminus to the cytoplasmic side. Thus, H7 and H8 constitute the fourth TM segment pair of this ATPase.
FIG. 11.
Autoradiograph of an SDS-PAGE gel with products of in vitro transcription-translation, in the absence and presence of microsomes (Mic), of the HK-M0 and HK-M1 vectors containing hydrophobic regions HY (lanes 1 to 4), H7 (lanes 5 to 8), and H8 (lanes 9 to 12) of the H. pylori CopA ATPase.
FIG. 12.
Autoradiograph of an SDS-PAGE gel with products of in vitro transcription-translation, in the absence and presence of microsomes (Mic), of the HK-M0 vector containing regions encompassing HY to H7 (lanes 1 and 2) and HY to H8 (lanes 3 and 4) of the H. pylori CopA ATPase.
Transmembrane segments of the CopA ATPase of H. felis and regions of high-level sequence homology.
The corresponding TM segments of the H. felis ATPase were determined by hydrophobicity analysis and by their sequence similarity to the H. pylori ATPase. The data suggested that the H1 to H8 TM helices of the H. felis pump are contained between positions L89 and L108 (H1), F119 and G136 (H2), L156 and T177 (H3), G184 and G200 (H4), V337 and L357 (H5), A465 and M483 (H6), N666 and A685 (H7), and I690 and L710 (H8) (Fig. 2). The locations as well as the orientations of TM segments found in the H. felis pump are predicted to be identical to those of its H. pylori counterparts.
The overall identity of the amino acid sequences of the H. felis and H. pylori CopA ATPases was about 55%; considerable differences in degrees of identity were found when the corresponding TM segments were compared. The identities found were about 50% for H1 and H2, 65% for H3, 85% for H4, 70% for H5, 90% for H6 and H7, and about 75% for H8 (Fig. 2). Thus, the three N-terminal helices, H1, H2, and H3, have reduced identity while segments H4 to H8 exhibit greater amino acid identity, suggesting a role for the C-terminal TM segments in ion transport. There are other regions of high-level identity (>80%), namely in the N-terminal segment of ion binding around the GMXXCXXC motif, the small cytoplasmic loop between H2 and H3, some sequences following H4, the sequences around the conserved phosphorylation and ATP binding motifs, and sequences preceding the last putative pair of TM helices.
DISCUSSION
A DNA oligonucleotide mixture encoding the DKTGT(I/L)T phosphorylation consensus sequence had been used previously to isolate a putative transition metal P-type ATPase of H. pylori (31). Using a similar phosphorylation site screening strategy, we have isolated the copAP operons from gene libraries of H. pylori 69A and a related gastric microorganism, H. felis (ATCC 49179). Cloning of the copAP operons, therefore, provided evidence for the coexistence of at least two of the bacterial single-subunit P-type pumps in gastric Helicobacter species, the CopA ATPase and the pRH439-predicted pump (31). A third member of this class of membrane ATPases was observed in the genome of H. pylori 26695 (60).
The cop operons of both H. pylori and H. felis consist of only two genes, which encode a P-type pump, CopA, and a small peptide, CopP, with putative ion binding properties. Data obtained by insertion-deletion mutagenesis of the pRH948 P-type gene in H. pylori ATCC 49503 show that CopA may act as a copper export pump, as has been shown in H. pylori UA802 by using a sequence variant of the H. pylori copA gene (13) and in Enterococcus hirae for the CopB ATPase (38, 39, 56).
The predicted protein products of DNA sequences preceding the cloned copA gene are not homologous to proteins involved in ion transport or regulation of transport ATPase expression. In contrast, the copP gene immediately downstream of (in H. pylori) or overlapping with (in H. felis) copA DNA sequences predicts a protein homologous to the CopZ protein of the Enterococcus hirae Cu2+ ATPase operon (38). This suggests a role for the CopP peptide in regulation of H. pylori CopA ATPase expression rather than as a source of Cu2+ for the ATPase, as was suggested in a previous study (13). The presence of a CopZ-homologous protein in the Helicobacter copAP operon might also suggest that there are similarities in regulation of cop gene expression in the gastric microorganisms and Enterococcus hirae. On the other hand, in H. pylori and H. felis, copP is located downstream of a unique CopA ATPase-encoding gene, and the cloned copAP operons lack the equivalent of copY present in the enterococcal copYZAB operon, which is postulated to be a repressor of cop operon transcription (38). Given that the pRH948- and pHF8-predicted pumps analyzed here represent Cu2+ export ATPases, the Helicobacter operons also lack the physiological equivalent of the Enterococcus hirae copper import ATPase (38, 39, 55). However, there are additional genes contributing to transport of transition metal cations elsewhere in the genome of H. pylori, for example, the NixA Ni2+ transport protein (34, 60).
The ATPases predicted from the copAP operon, 741 (H. pylori CopA) and 732 (H. felis CopA) amino acids in length, exhibit a strong overall sequence similarity to the previously studied 75-kDa membrane ATPase of H. pylori (31) and to the Cd2+ and Cu2+ P-type pumps of bacteria as well as mammalian cells (5, 36–39, 52, 57, 62). As demonstrated for most of the members of this family of ion pumps, the cloned CopA ATPases have an N-terminal GMXCXXC ion binding motif (Cys box) and an intramembrane CPC sequence consistent with a role for this enzyme in transition metal transport (51, 52, 57). The GMXCXXC motif is a variant of a consensus sequence in the other putative transition metal ATPase isolated from H. pylori (31). The latter pump contains an N-terminal HXHXXXCXXC ion binding motif with affinity for Ni2+ ions, indicating, as does the presence of clusters of cysteine and histidine residues in the ATP binding and phosphorylation loop of the latter enzyme (31), that the two CopA P-type ATPases expressed in Helicobacter species and the membrane pump described previously have different ion specificities (31, 61).
Besides the data obtained by H. pylori copA gene knockout mutagenesis, evidence of a possible role for CopP in Cu2+-dependent gene regulation, and the homologies of the copA gene-predicted enzymes to the various Cd2+ and Cu2+ ATPases, there is additional preliminary evidence of a role for the cloned Helicobacter CopA pumps in binding as well as transport of copper ions. As shown in the data presented above, Cu2+, but not Co2+, Cd2+, or Mg2+, was significantly bound by the N-terminal peptide of the H. pylori 69A CopA ATPase predicted by vector pRH948. The selectivity of the peptide toward copper is most probably due to the presence of the GMXCXXC ion binding motif, as shown very recently for N-terminal domains of the human Wilson’s and Menkes Cu2+ transport ATPases (29). Weak binding of the H. pylori CopA peptide to Ni2+ and Zn2+ is probably not important for the physiology of H. pylori, since the CopA-deficient microorganism showed unchanged and high levels of resistance to both Ni2+ and Zn2+ (Table 2). The latter might be due to the existence of other proteins involved in metal ion resistance of H. pylori (31, 60). The N-terminal copper selectivity, as demonstrated for the H. pylori ATPase peptide, is in agreement with the increased sensitivity of H. pylori copA knockout mutants to Cu2+. The N-terminal region around the GMXCXXC box has a high degree of amino acid identity to various putative Cop ATPases, and a stretch of 22 amino acids in this region shows an identity of 91% when the CopA pumps of H. felis and H. pylori are compared, underlining a significant role of this domain in pump function. However, whether the N-terminal ion binding properties of P-type pumps contribute to the regulation of enzyme activity or to the transport of the ion itself is as yet unknown (28).
Structural features of these transition metal pumps must relate to their function as copper transport enzymes. Since the membrane domain contains the copper transport pathway, this domain was defined by in vitro translation scanning using membrane insertion detection vectors. In a previous study, this method was able, unequivocally, to detect eight membrane segments, H1 through H8, ordered pairwise along the polypeptide chain of the pRH439-predicted H. pylori ATPase, placing the ATP binding and phosphorylation loop between H6 and H7 (31). The latter ATPase, with the highest degree of homology to Cd2+ P-type pumps and binding affinity to Ni2+ (31, 61), is most probably a resistance pump transporting transition metals such as Cd2+ and Zn2+ (30a). A comparison of the hydrophobicity plots of the Cu2+ ATPases and the Ni2+-binding ATPase (31) showed generally similar profiles but two additional hydrophobic peaks in the H. pylori CopA ion pump, designated as HX and HY. In this study, the data obtained by in vitro translation showed that the H. pylori CopA ATPase contains three pairs of membrane-spanning helices (H1 to H6) in the N-terminal half of the enzyme and a fourth pair of antiparallel helices (H7 and H8) in the C-terminal region. To our knowledge, these results present the first experimental evidence that the membrane domain of Cu2+ P-type ATPases consists of eight TM segments. The additional hydrophobic peaks in the cloned H. pylori CopA ATPase, compared to the pRH439-encoded ATPase (31), did not act as signal anchors, and only the HY segment led to partial inhibition of glycosylation when translated in the HK-M1 vector. The H. felis CopA pump contains the corresponding H1 to H8 segments, as determined by hydropathy analysis and sequence similarity, but not the HX segment. The absence of this hydrophobic segment in the H. felis CopA ATPase is consistent with the finding that the HX segment of the corresponding H. pylori ATPase did not exhibit any membrane insertion properties. The HY segment was important for the membrane insertion properties of H7 in the H. pylori CopA ATPase and is retained in the H. felis CopA version. As in the other H. pylori P-type pump analyzed previously (31), H6 is followed by the large cytoplasmic energy transduction loop containing the phosphorylation site. On the basis of membrane topology, the cloned Helicobacter Cu2+ pumps, therefore, fall into a group of P-type ATPases containing eight TM segments.
The availability of CopA pumps from two distinct gastric Helicobacter species, H. pylori and H. felis, allowed a prediction of sequences related to the transport pathway for copper ions across the cytoplasmic membrane. Cysteine residues may play a role in ion transport, as found for the two cysteine residues located in the first TM region of the E. coli MerT mercuric ion transporter (35). In CopA, cysteines are thought to participate in Cu2+ binding sites and therefore may be involved in the transport of the ion through the membrane. A feature of these Helicobacter Cu2+ pumps is that they contain seven (for H. felis) or eight (for H. pylori) cysteine residues. Five of these cysteines are located at conserved positions in TM helices. These cysteines are in H4, in H6 as part of the conserved CPC motif, and in H7. When the Enterococcus hirae CopAB pumps (37) and the cloned Cu2+ pumps of H. pylori and H. felis are compared, only the cysteine residue in the CPX motif of H6 is conserved among the transition metal pumps, bringing into question the role of the other conserved cysteine residues present in the membrane domain of both the H. pylori and H. felis CopA pumps. However, the TM segments H7 and H8, as well as H4, H5, H6, and in part H3, are well conserved between the two ATPases. Along with the placement of the homologous cysteines, this conservation could be taken as preliminary evidence that TM segments H4 to H8 constitute the core structure of the ion transport domains of these ATPases. The first two TM sequences show less homology to each other and to the other P-type ATPases of H. pylori (31, 60). Therefore, H1 and H2 of the cloned CopA pumps, which represent the two additional N-terminal TM segments found in all transition metal ATPases, and perhaps H3 may be situated peripherally to this core structure, interacting mainly with phospholipid, which would account for their relative lack of conservation.
In conclusion, H. pylori and H. felis employ conserved mechanisms of copper resistance. These mechanisms include expression of a bicistronic copAP operon encoding a transition metal pump containing eight TM helices, CopA, and the peptide CopP, the latter most probably being a Cu2+ binding protein involved in copAP operon expression. The CopA ATPase, which provides the structural and functional basis for P-type Cu2+ export in Helicobacter species, uses conserved motifs such as the N-terminal domain with Cu2+ binding properties and conserved regions in the energy transduction domain. The CopA membrane domain shows the highest level of sequence identity in H4, H5, H6, and H7-H8 as well as in stretches of H3, suggesting that these six segments of the CopA ATPase may provide the TM pathway for copper ions across the H. pylori and H. felis inner membranes.
ACKNOWLEDGMENTS
We thank Anita Buhmann, Marion Eisenhauer, and Marina Rektorschek for excellent technical assistance.
This work was supported in part by the Bundesministerium für Forschung und Technologie (BMFT grant 514 4003 0310777) and in part by U.S. Veterans Administration research funds (SMI) and National Institutes of Health grants DK40615, DK41301, and DK17294.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Bamberg K, Sachs G. Topological analysis of the H,K-ATPase using in vitro translation. J Biol Chem. 1994;269:16909–16919. [PubMed] [Google Scholar]
- 3.Bayer E, Rapp W. New polymer supports for solid-liquid-phase peptide synthesis. Chem Pept Proteins. 1986;3:3–8. [Google Scholar]
- 4.Bayle D, Weeks D, Sachs G. The membrane topology of the rat sarcoplasmic and endoplasmic reticulum calcium ATPases by in vitro translation scanning. J Biol Chem. 1995;270:25678–25684. doi: 10.1074/jbc.270.43.25678. [DOI] [PubMed] [Google Scholar]
- 5.Bull P C, Thomas G R, Rommens J M, Forbes J R, Cox D W. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet. 1993;5:327–336. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- 6.Chang C D, Meierhofer J. Solid-phase peptide synthesis using mild base cleavage of Na-fluorenylmethyloxycarbonylamino acids, examplified by a synthesis of dihydrosomatostatin. Int J Pept Protein Res. 1978;11:246–249. doi: 10.1111/j.1399-3011.1978.tb02845.x. [DOI] [PubMed] [Google Scholar]
- 7.Clarke D M, Loo T W, MacLennan D H. The epitope for monoclonal antibody A20 (amino acids 870–890) is located on the luminal surface of the Ca-ATPase of sarcoplasmic reticulum. J Biol Chem. 1990;265:17405–17408. [PubMed] [Google Scholar]
- 7a.Coste J, Le-Nguyen D, Castro B. PyBOP: a new peptide coupling reagent devoid of toxic by-product. Tetrahedron Lett. 1990;31:205–208. [Google Scholar]
- 8.Cover T L, Blazer M J. Helicobacter pylori: a bacterial cause of gastritis, peptic ulcer disease, and gastric cancer. ASM News. 1995;61:21–26. [Google Scholar]
- 9.Eisenberg D, Schwarz E, Komaromy M, Wall R J. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 10.Epstein W. The Kdp system: a bacterial K+ transport ATPase. Curr Top Membr Transp. 1985;25:153–175. [Google Scholar]
- 11.Epstein W. Bacterial transport ATPases. In: Krulwich A, editor. Bacterial energetics. Vol. 12. San Diego, Calif: Academic Press Inc.; 1990. pp. 87–110. [Google Scholar]
- 12.Fields G B, Noble R L. Solid phase peptide synthesis utilizing 9-fluorenyl methoxy carbonyl amino acids. Int J Pept Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 13.Ge Z, Hiratsuka K, Taylor D E. Nucleotide sequence and mutational analysis indicate that two Helicobacter pylori genes encode a P-type ATPase and a cation-binding protein associated with copper transport. Mol Microbiol. 1995;15:97–106. doi: 10.1111/j.1365-2958.1995.tb02224.x. [DOI] [PubMed] [Google Scholar]
- 14.Ge Z, Taylor D E. Helicobacter pylori genes hpcopA and hpcopP constitute a cop operon involved in copper export. FEMS Microbiol Lett. 1996;145:181–188. doi: 10.1111/j.1574-6968.1996.tb08575.x. [DOI] [PubMed] [Google Scholar]
- 15.Ge Z, Taylor D E. Sequencing, expression, and genetic characterization of the Helicobacter pylori ftsH gene encoding a protein homologous to members of a novel ATPase family. J Bacteriol. 1996;178:6151–6157. doi: 10.1128/jb.178.21.6151-6157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham D Y, Go M F. Helicobacter pylori: current status. Gastroenterology. 1993;105:279–282. doi: 10.1016/0016-5085(93)90038-e. [DOI] [PubMed] [Google Scholar]
- 17.Hudson D. Methodological implications of simultaneous solid phase peptide synthesis. 1. Comparison of different coupling procedures. J Org Chem. 1988;53:617–624. [Google Scholar]
- 18.Kahrs A F, Odenbreit S, Schmitt W, Heuermann D, Meyer F T, Haas R. An improved TnMax mini-transposon system suitable for sequencing, shuttle mutagenesis and gene fusions. Gene. 1995;167:53–57. doi: 10.1016/0378-1119(95)00671-0. [DOI] [PubMed] [Google Scholar]
- 19.Kanamaru K, Kashiwagi S, Mizuno T. A copper-transporting P type ATPase found in the thylakoid membrane of cyanobacterium Synechococcus species PCC7942. Mol Microbiol. 1994;13:369–377. doi: 10.1111/j.1365-2958.1994.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 20.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 21.Karlish S J D, Goldshleger R, Jorgensen P L. Location of asn831 of the α chain of Na/K-ATPase at the cytoplasmic surface. Implication for topological models. J Biol Chem. 1993;268:3471–3478. [PubMed] [Google Scholar]
- 22.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Laimins L A, Rhoads D B, Altendorf K, Epstein W. Identification of the structural proteins of an ATP-driven potassium transport system in Escherichia coli. Proc Natl Acad Sci USA. 1978;75:3216–3219. doi: 10.1073/pnas.75.7.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laimins L A, Rhoads D B, Epstein W. Osmotic control of kdp operon expression in E. coli. Proc Natl Acad Sci USA. 1981;78:464–468. doi: 10.1073/pnas.78.1.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee A, Fox J, Hazell S. Pathogenicity of Helicobacter pylori: a perspective. Infect Immun. 1993;61:1601–1610. doi: 10.1128/iai.61.5.1601-1610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutsenko S, Kaplan J H. Organization of P-type ATPases: significance of structural diversity. Biochemistry. 1995;34:15607–15613. doi: 10.1021/bi00048a001. [DOI] [PubMed] [Google Scholar]
- 29.Lutsenko S, Petrukhin K, Cooper M J, Gillian C T, Kaplan J H. N-terminal domains of human copper-transporting adenosine triphosphatase (the Wilson’s and Menkes disease proteins) bind copper selectively in vivo and in vitro with stoichiometry of one copper per metal-binding repeat. J Biol Chem. 1997;272:18939–18944. doi: 10.1074/jbc.272.30.18939. [DOI] [PubMed] [Google Scholar]
- 30.Malfertheiner, P., and G. Bode. 1993. Helicobacter pylori and the pathogenesis of duodenal ulcer disease. Eur. J. Gastroenterol. 5(Suppl. 1):S1–S83.
- 30a.Melchers, K., et al. Unpublished data.
- 31.Melchers K, Weitzenegger T, Buhmann A, Steinhilber W, Sachs G, Schäfer K P. Cloning and membrane topology of a P type ATPase from Helicobacter pylori. J Biol Chem. 1996;271:446–457. doi: 10.1074/jbc.271.1.446. [DOI] [PubMed] [Google Scholar]
- 32.Merrifield R B. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 33.Misra T K, Brown N L, Fritzinger D C, Pridmore R D, Barnes W M, Haberstroh L, Silver S. Mercuric ion resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci USA. 1984;81:5974–5978. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mobley H L T, Garner R M, Bauerfeind P. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol Microbiol. 1995;16:97–109. doi: 10.1111/j.1365-2958.1995.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 35.Morby A P, Hobman J L, Brown N L. The role of cysteine residues in the transport of mercuric ions by the Tn501 MerT and MerP mercury-resistance proteins. Mol Microbiol. 1995;17:25–35. doi: 10.1111/j.1365-2958.1995.mmi_17010025.x. [DOI] [PubMed] [Google Scholar]
- 36.Nies D H, Silver S. Ion efflux systems involved in bacterial metal resistance. J Ind Microbiol. 1995;14:186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- 37.Nucifora G, Chu L, Misra T K, Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci USA. 1989;86:3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odermatt A, Solioz M. Two trans-acting metalloregulatory proteins controlling expression of the copper ATPases of Enterococcus hirae. J Biol Chem. 1995;270:4349–4354. doi: 10.1074/jbc.270.9.4349. [DOI] [PubMed] [Google Scholar]
- 39.Odermatt A, Suter H, Krapf R, Solioz M. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem. 1993;268:12775–12779. [PubMed] [Google Scholar]
- 40.Okada M, Matsuzaki H, Shibuya I, Matsumoto K. Cloning, sequencing, and expression in Escherichia coli of the Bacillus subtilis gene for phosphatidylserine synthase. J Bacteriol. 1994;176:7456–7461. doi: 10.1128/jb.176.24.7456-7461.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okupodu C M, Robertson D, Boss W F, Togasaki R K, Surzycki S J. Rapid isolation of nuclei from carrot suspension culture cells using a BioNebulizer. BioTechniques. 1994;16:154–159. [PubMed] [Google Scholar]
- 42.Paster B J, Lee A, Fox J G, Dewhirst F E, Tordoff L A, Fraser G J, O’Rourke J L, Taylor N S, Ferrero R. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int J Syst Bacteriol. 1991;41:31–38. doi: 10.1099/00207713-41-1-31. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen P L, Carafoli E. Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem Sci. 1987;12:146–150. [Google Scholar]
- 44.Polarek J W, Williams G, Epstein W. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J Bacteriol. 1992;174:2145–2151. doi: 10.1128/jb.174.7.2145-2151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Przybylski M, Glocker M O. Electrospray mass spectrometry of biomacromolecular complexes with noncovalent interactions—new analytical perspectives for supramolecular chemistry and molecular recognition process. Angew Chem Int Ed Engl. 1996;35:806–826. [Google Scholar]
- 46.Rao M J K, Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986;869:197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- 47.Richterich P, Heller C, Wurst H, Pohl F M. DNA sequencing with direct blotting electrophoresis and colorimetric detection. BioTechniques. 1989;7:52–59. [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Schägger H, Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 50.Shull G E, Schwartz A, Lingrel J B. Molecular cloning of the rat stomach (H+ + K+)-ATPase. Nature. 1985;316:691–695. [Google Scholar]
- 51.Silver S, Nucifora G, Chu L, Misra T K. Bacterial resistance ATPases: primary pumps for exporting toxic cations and anions. Trends Biochem Sci. 1989;14:76–80. doi: 10.1016/0968-0004(89)90048-0. [DOI] [PubMed] [Google Scholar]
- 52.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 53.Silver S, Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith D L, Tao T, Maguire M E. Membrane topology of a P-type ATPase. The MgtB magnesium transport protein of Salmonella typhimurium. J Biol Chem. 1993;268:22469–22479. [PubMed] [Google Scholar]
- 55.Solioz M, Mathews S, Fürst P. Cloning of the K+ ATPase of Streptococcus faecalis. J Biol Chem. 1987;262:7358–7362. [PubMed] [Google Scholar]
- 56.Solioz M, Odermatt A. Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae. J Biol Chem. 1995;270:9217–9221. doi: 10.1074/jbc.270.16.9217. [DOI] [PubMed] [Google Scholar]
- 57.Solioz M, Vulpe C. CPX-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem Sci. 1996;247:237–241. [PubMed] [Google Scholar]
- 58.Southern E. Detection of specific DNA sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 59.Sugiura A, Hirokawa K, Nakashima K, Mizuno T. Signal-sensing mechanisms of the putative osmosensor KdpD in Escherichia coli. Mol Microbiol. 1994;14:929–938. doi: 10.1111/j.1365-2958.1994.tb01328.x. [DOI] [PubMed] [Google Scholar]
- 60.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 61.Volz, J., F. U. Bosch, M. Wunderlin, M. Schuhmacher, K. Melchers, K. Bensch, W. Steinhilber, K. P. Schäfer, G. Toth, B. Penke, and M. Przybylski. Characterization of metal binding polypeptide domains by electrospray ionization mass spectrometry and metal chelate affinity chromatography. J. Chromatogr., in press. [DOI] [PubMed]
- 62.Vulpe C, Levinson B, Whitney S, Packman S, Gitschier J. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper transporting ATPase. Nat Genet. 1993;3:7–13. doi: 10.1038/ng0193-7. [DOI] [PubMed] [Google Scholar]
- 63.Walderhaug M O, Polarek J W, Voelkner P, Daniel J M, Hesse J E, Altendorf K, Epstein W. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J Bacteriol. 1992;174:2152–2159. doi: 10.1128/jb.174.7.2152-2159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warren J R, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]