Abstract
Yersinia pestis produces a set of virulence proteins (Yops and LcrV) that are expressed at high levels and secreted by a type III secretion system (Ysc) upon bacterium-host cell contact, and four of the Yops are vectorially translocated into eukaryotic cells. YopD, YopB, and YopK are required for the translocation process. In vitro, induction and secretion occur at 37°C in the absence of calcium. LcrH (also called SycD), a protein required for the stability and secretion of YopD, had initially been identified as a negative regulator of Yop expression. In this study, we constructed a yopD mutation in both wild-type and secretion-defective (ysc) Y. pestis to determine if the lcrH phenotype could be attributed to the decreased stability of YopD. These mutants were constitutively induced for expression of Yops and LcrV, despite the presence of the secreted negative regulator LcrQ, demonstrating that YopD is involved in negative regulation, regardless of a functioning Ysc system. Normally, secretion of Yops and LcrV is blocked in the presence of calcium. The single yopD mutant was not completely effective in blocking secretion: LcrV was secreted equally well in the presence and absence of calcium, while there was partial secretion of Yops in the presence of calcium. YopD is probably not rate limiting for negative regulation, as increasing levels of YopD did not result in decreased Yop expression. Overexpression of LcrQ in the yopD mutant had no significant effect on Yop expression, whereas increased levels of LcrQ in the parent resulted in decreased levels of Yops. These results indicate that LcrQ requires YopD to function as a negative regulator.
Yersinia pestis, the causative agent of plague, has a plasmid-encoded low-calcium response (LCR) regulatory mechanism that governs the expression and secretion of a set of virulence proteins in response to environmental conditions (40). Contact with a eukaryotic cell triggers an increase in LCR gene expression and the vectorial translocation of four of the secreted Yops (Yersinia outer proteins) into the host cell where they disrupt cellular signaling and other biochemistry necessary for phagocytosis and the mobilization of an effective immune response (8, 40). The maximal Yop expression and secretion induced by bacterium-host cell contact is mimicked in vitro by the absence of Ca2+ at 37°C; in the presence of millimolar concentrations of Ca2+ Yop expression is down-regulated and secretion is completely blocked.
The Yops and LcrV are secreted from the bacterial cell without processing by a specific Yop secretion system (Ysc) that is encoded by the LCR virulence plasmid (8, 40). The Ysc system belongs to the recently identified class of type III, or contact-dependent, secretion systems of both animal and plant gram-negative pathogens. These systems are involved in direct delivery of bacterial proteins into the target cell (30, 52) and are extremely complex: at least 19 gene products are required for the Ysc (8, 40). Translocation into the eukaryotic cell involves YopB, YopK, and YopD (18, 19, 21, 22, 25, 41, 51, 58). YopB may form a pore in the target cell membrane, and YopK may modulate the size of this pore (19, 25). YopD’s function in translocation has not been determined.
There have been several gene products identified that play a role in LCR regulation. Thermal induction of LCR gene expression is mediated by the transcriptional activator LcrF (6, 24). LcrF exhibits sequence similarity to the AraC activator of Escherichia coli and has been shown to interact with several yop promoters (7, 29). The Ysc system has been shown to have a positive role on the LCR: expression of Yops in a ysc mutant is down-regulated, even under conditions that normally promote maximal expression (5, 10, 43–45). It is believed that this connection between contact-triggered secretion and transcriptional up-regulation is the secretion of a repressor (or antiactivator) component through the Ysc (37, 42). This is analogous to the export of an anti-sigma factor through the flagellar export apparatus (which has homology to inner membrane components of type III secretion systems) as a control of late-gene expression in flagellar biosynthesis (26, 27). Two secreted proteins, LcrE and LcrG, appear to regulate secretion of Yops and LcrV in response to environmental Ca2+: mutations inactivating either protein cause secretion at 37°C irrespective of Ca2+ levels, with a concomitant loss of negative control over Yop and LcrV expression (13, 55). LcrE (also called YopN) is thought to act at the bacterial surface, presumably as the Ca2+ sensor, although that activity has not been directly demonstrated (13). The majority of LcrG is located in the cytosol and is thought to block secretion from the cytoplasmic side (37). LcrV, in addition to functioning as an antihost protein, serves as a positive regulatory protein (4, 46, 56). LcrV is able to directly interact with LcrG and is thought to titrate LcrG away from the Ysc system, thus counteracting LcrG’s secretion block when elevated levels of LcrV are produced upon induction of the LCR (37).
The secreted protein LcrQ has previously been proposed to be the negative regulator whose secretion allows up-regulation of LCR expression (42). The level of LcrQ in the cell has been demonstrated to correlate to the level of Yop expression; i.e., Yop expression is maximal when LcrQ is depleted from the cell, while elevated levels of LcrQ result in down-regulation of Yop expression (42, 50). This is even true in a ysc lcrQ double mutant, which exhibits constitutive Yop expression, resulting in the conclusion that LcrQ secretion through the Ysc system is required for full induction of Yop expression (42). LcrQ is secreted from the cell quickly and efficiently both in vitro and after infection of HeLa cells (42). The regulatory function of LcrQ does not require translocation across the target cell membrane, as a yopB mutant had the same LCR induction as wild-type cells upon host cell contact (42). LcrQ has not been demonstrated to bind DNA, nor does its deduced amino acid sequence contain any DNA-binding motifs, and therefore, it most likely exerts its regulatory effect in an indirect manner (50).
The intracellular protein LcrH (also called SycD) was originally thought to be involved in the negative regulation of Yop and LcrV expression (4, 48). The phenotype of an lcrH mutant is constitutive induction of the LCR, as well as constitutive secretion of LcrV and partial secretion of YopM (4, 45, 48, 56). An lcrH mutation also caused a ysc strain to become constitutively induced for LCR expression (45). LcrH has subsequently been reported to be required for the stabilization and secretion of YopD (61). This raises the possibility that the lcrH phenotype is caused by the decreased stability of YopD. Additional evidence suggesting that YopD may be involved in regulation was the constitutive LCR induction caused by a Genblock insertion into the 3′ end of yopD (4). However, this effect was attributed to a destabilization of the lcrGVHyopBD transcript (4). In this study, we constructed an essentially complete deletion of the yopD open reading frame and characterized the resulting mutant for LcrV and Yop expression and secretion. Our data indicate that YopD is involved in the negative regulation of the LCR, regardless of a functioning Ysc system. In addition, we demonstrate that LcrQ’s negative regulatory function is dependent on the presence of YopD.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and growth conditions.
Bacterial strains and plasmids are shown in Table 1. Synthetic oligonucleotides are shown in Table 2. Y. pestis strains were grown in heart infusion broth or on tryptose blood agar base medium (Difco Laboratories, Detroit, Mich.) for genetic manipulations. For physiological experiments, Y. pestis strains were grown in exponential phase at 26°C in the defined medium TMH (59) with or without Ca2+ for 7 to 9 generations and then were diluted to an absorbance at 620 nm (A620) of ca. 0.1 and grown to an A620 of ca. 0.2. Cultures were then either held at 26°C or shifted to 37°C for an additional 6 h before harvesting. Unless otherwise indicated, E. coli strains were grown in Luria-Bertani (LB) broth (53) or on LB agar plates. Bacteria containing antibiotic resistance markers were grown in the presence of the appropriate antibiotic(s) at a final concentration of 25 μg/ml (chloramphenicol), 50 μg/ml (kanamycin), or 100 μg/ml (ampicillin and streptomycin).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| SY327(λpir) | Δ(lac-pro) argE(Am) rif nalA recA56 λpir | 34 |
| DH5α | end-1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR [φ80 dlacΔ(lacZ)M15] | 20 |
| XL1-Blue | end-1 hsdR17(rK− mK+) supE44 thi-1 λ− recA1 gyrA96 (Nalr) relA1 (Δlac) [F+proAB+ lacIqZΔM15::Tn10 (Tcr)] | Stratagene |
| Y. pestis strainsb | ||
| KIM5-3001 | Smr pCD1 (Lcr+), pPCP1, pMT1 | 32 |
| KIM5-3001.2 | Smr pCD1 lcrD(Δ192–343), pPCP1, pMT1 | 44 |
| KIM5-3001.2.1 | Smr pCD1 lcrD(Δ192–343) yopD(Δ1–305), pPCP1, pMT1 | This study |
| KIM5-3001.16 | Smr pCD1 yopD(Δ1–305), pPCP1, pMT1 | This study |
| KIM8-3002 | Smr pCD1 (Lcr+), Pla−, pMT1 | 38 |
| KIM8-3002.2 | Smr pCD1 yopD(Δ1–305), Pla−, pMT1 | This study |
| Plasmids | ||
| pUK4134 | Suicide vector, Apr | 54 |
| pAW102 | 1.64-kb fragment of pCD1 containing ΔyopD (Δ1–305) cloned into pUK4134 | This study |
| pTrcH5 | lcrH cloned into pTrc99A (Pharmacia Biotech) | 12 |
| pET-24b | His tag fusion expression vector, Kmr | Novagen |
| pAW111 | yopD cloned into pET-24b | This study |
| pBAD33 | Expression vector (arabinose inducible), Cmr | 16 |
| pAW161 | yopD cloned into pBAD33 | This study |
| pAW162 | lcrQ cloned into pBAD33 | This study |
| pProEX-1 | His tag fusion expression vector, Apr | GibcoBRL |
| pAW171 | yopD cloned into pProEX-1 | This study |
| pGEX-3X | GST fusion expression vector, Apr | Pharmacia Biotech |
| pWGQ | lcrQ cloned into pGEX-3X | This study |
TABLE 2.
DNA oligonucleotides
| Primer | DNA sequence |
|---|---|
| LCRH1SDA | GCT AGA GCT CAG GAG GAA CAT ATG CAA CAA GAG ACG ACA GAC |
| LCRQ1-2 | CGT GGG ATC CAC ATG AAA ATC AAT ACT CTT CAA TCG |
| LCRQ1SDA | GCT AGA GCT CAG GAG GAA CAT ATG AAA ATC AAT ACT CTT CAA TCG |
| LCRQ2-1 | TTA TGA ATT CCC TCA GCC GTC AGC CGC CGT AT |
| LCRQ348B | GCT CTA GAT CAG CCG TCA GCC GCC GTA TC |
| YOPB296A | CGC GGA TCC GCT TGC TTC ACC TGA TAC ATT TG |
| YOPBΔ23B | GCC GGC CAA ATT ATG CAG CAT CAT GGG TTA TCA ACG CAC |
| YOPBΔ1169A | GTG CGT TGA TAA CCC ATG ATG CTG CAT AAT TTG GCC GGC |
| YOPD1SDA | CTA GCT AGC GGA GTG AAC ATA TGA CAA TAA ATA TCA AGA CAG AC |
| YOPD921B | GCT CTA GAT CAG ACA ACA CCA AAA GCG GC |
| YOPDHTAGB | CCG CTC GAG AAC ACC AAA AGC GGC TTT CAT G |
| YOPDOUTDB | CGC GGA TCC AGC GGC AAG TCA AAT ACC TG |
| YOPDΔ1A | GTT TAA GGA GGA ATA ACC GTC TGA CCA TTG ATG ACC |
| YOPDΔ915B | GGT CAT CAA TGG TCA GAC GGT TAT TCC TCC TTA AAC |
DNA methods.
Plasmid DNA was isolated by a standard alkaline lysis procedure or by the use of Qiagen spin or midi-prep columns (Qiagen, Inc., Studio City, Calif.). Cloning methods were essentially as described previously (53). DNA fragments were purified from agarose gels by use of a Qiaquick gel extraction kit (Qiagen, Inc.). Electroporation of DNA into E. coli and Y. pestis was done as previously described (39). Gene amplification was performed with Taq polymerase (either from GibcoBRL, Gaithersburg, Md., or Boehringer Mannheim, Indianapolis, Ind.) or PFU polymerase (Stratagene, La Jolla, Calif.) in a Perkin-Elmer GeneAmp model 2400 thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.). Unless stated otherwise, 30 cycles of amplification were used, with 1 cycle consisting of denaturing at 94°C for 15 s, annealing at 50°C for 15 s, and extension at 72°C for 15 s. Total bacterial DNA for gene amplification was prepared as previously described (31). Custom oligonucleotides were obtained either from Genosys Biotechnologies (The Woodlands, Tex.), Integrated DNA Technologies, Inc. (Coralville, Iowa), or the Macromolecular Structure Analysis Facility (University of Kentucky). DNA sequencing was performed at the Macromolecular Structure Analysis Facility.
Plasmid construction.
DNA used for cloning was obtained from pCD1 of Y. pestis by amplification with customized oligonucleotide primers. Some of the primers were designed using the Yersinia pseudotuberculosis DNA sequence, since portions of the region utilized have not yet been sequenced in Y. pestis, and LCR loci typically exhibit at least 95% sequence identity between the two species. Unless stated otherwise, the high-fidelity PFU polymerase was used for amplification of DNA, and the extension time of the cycles was increased to 1 min per kb amplified.
pAW102 (ΔyopD) was created by the blunt-end cloning of the DNA sequence flanking, but not including, yopD (ca. 950 bases upstream and ca. 700 bases downstream) into the EcoRV site of suicide vector pUK4134 (54). The only DNA present from the yopD gene was the last 6 nucleotides, coding for the C-terminal amino acid and the stop codon. The DNA fragment for cloning was obtained by recombinant PCR (23) in which internal complementary primers encoded sequence immediately upstream and downstream of the desired deletion of bases 1 through 915 of yopD. YOPDΔ915B and YOPDΔ1A were the two internal primers, and YOPB296A (which lay within upstream yopB) and YOPDOUTDB (which lay downstream of yopD) were the two outside primers; these primers were based on a Y. pseudotuberculosis sequence. The resulting fragment was 5′ end phosphorylated with T4 kinase (Promega, Madison, Wis.).
Plasmid pAW111 (encoding C-terminally histidine-tagged YopD) was constructed by amplifying yopD with YOPD1SDA and YOPDHTAGB, which contained the first 26 bases and bases 893 to 918 (with a G-to-C substitution at base 916 resulting in a conservative missense mutation of leucine to valine at the C-terminal position) of Y. pseudotuberculosis yopD, respectively, plus a 5′ end with an unencoded NdeI (YOPD1SDA) or XhoI (YOPDHTAGB) site. The PCR product was cleaved with NdeI and XhoI and cloned into the NdeI and XhoI sites of pET-24b (Novagen, Inc, Milwaukee, Wis.).
pAW171 (encoding N-terminally histidine-tagged YopD) was constructed by amplifying yopD with YOPD1SDA and YOPD921B (which contained the final 21 bases of Y. pseudotuberculosis yopD plus a 5′ end with an unencoded XbaI site). The PCR product was cleaved with NdeI and XbaI and cloned into the NdeI and XbaI sites of pProEX-1 (GibcoBRL).
Plasmid pAW161 (expressing yopD) was constructed by cloning lcrH and yopD into NheI- and XbaI-cleaved pBAD33 (16). The DNA fragment for cloning was obtained by recombinant PCR amplification (23) to delete bases 23 through 1169 of yopB, with YOPBΔ1169A and YOPBΔ23B as the two internal primers based on a Y. pseudotuberculosis sequence, and LCRH1SDA and YOPD921B based on Y. pestis and Y. pseudotuberculosis sequences, respectively, as the two outside primers, followed by digestion with NheI and XbaI (both sites engineered as nonencoded 5′ ends of the outside primer sequences).
Plasmid pAW162 (expressing lcrQ) was constructed by amplifying lcrQ with primers LCRQ1SDA and LCRQ348B derived from the Y. pseudotuberculosis lcrQ sequence and containing unencoded NheI and XbaI restriction sites in the 5′ ends, respectively. The PCR product was cleaved with NheI and XbaI and cloned into NheI- and XbaI-cleaved pBAD33.
pWGQ (encoding glutathione S-transferase [GST]-LcrQ) was constructed by amplifying lcrQ with primers LCRQ1-2 and LCRQ2-2 based on the Y. pestis lcrQ sequence and containing 5′ unencoded EcoRI and BamHI restriction sites, respectively. The PCR product was cleaved with EcoRI and BamHI and cloned into EcoRI- and BamHI-cleaved pGEX-3X (Pharmacia Biotech, Piscataway, N.J.). Amplification of the DNA for pWGQ was performed with Taq polymerase, and the annealing, extension, and denaturation times for each cycle were 30 s (14).
Construction of mutants.
Plasmid pAW102 was used for the deletion of the yopD gene. The plasmid was electroporated into Smr Y. pestis, and the yopD deletion was introduced into pCD1 by allelic exchange as previously described (54). The presence of second crossovers was screened for by PCR, and the presence of mutations was confirmed by DNA sequence analysis. The yopD mutation was introduced into Y. pestis KIM5-3001, KIM8-3002, and KIM5-3001.2, resulting in Y. pestis KIM5-3001.16, KIM8-3002.2, and KIM5-3001.2.1, respectively.
Cell fractionation.
Cell pellets and culture supernatants of Y. pestis grown in TMH were separated by centrifugation at 4,000 × g for 16 min at 4°C. Cell pellets were washed once with ice-cold phosphate-buffered saline (PBS) (135 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4; pH 7.4). Whole-cell fractions were prepared by being resuspended in 1× protein sample buffer (SB) (2). Cellular extracts were made by disintegration of PBS-resuspended cell pellets in the ice-cold cell of a French press (20,000 lb/in2). Low-speed centrifugation (8,000 × g, 5 min, 4°C) removed unbroken cells and large debris. The total soluble fraction (cytoplasmic plus periplasmic) was separated from membranes by ultracentrifugation at 513,000 × g in a TLA120.2 rotor for 10 min at 4°C, using a Beckman Optima TLX ultracentrifuge (Beckman Instruments, Fullerton, Calif.). Culture supernatant proteins were precipitated either with 5% (wt/vol) trichloroacetic acid (TCA) for 4 h to overnight on ice or with 80% (wt/vol) ammonium sulfate overnight at 4°C. The TCA-precipitated samples were collected by centrifugation at 20,800 × g for 30 min at 4°C, neutralized, and resuspended in 1× SB. The ammonium sulfate-precipitated samples were resuspended in PBS, dialyzed against PBS at 4°C by using either a Slide-A-Lyzer 2,000-molecular-weight-cutoff cassette (Pierce, Rockford, Ill.) or Spectra/Por 4 dialysis tubing (Spectrum, Laguna Hills, Calif.), and added to an equal volume of 2× SB.
Antibody preparation.
Plasmid pAW111, which encodes a C-terminally polyhistidine-tagged YopD (HT-YopD) behind the T7 promoter of pET-24b (Novagen), and plasmid pTrcH5 (12), which contains the lcrH gene behind the trc promoter of pTrc99A (Pharmacia Biotech), were introduced into E. coli BL21(DE3). Cultures (200 ml) were grown at 37°C in tryptone-phosphate medium (35) to an A600 of ca. 0.6, at which point high-level expression of the proteins was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 1.0 mM followed by incubation at 37°C for an additional 7.5 h. The cells were harvested by centrifugation at 11,300 × g for 10 min at 4°C. Cell pellets were resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl) and lysed by passage through a French pressure cell (20,000 lb/in2). Unlysed cells, large debris, and inclusion bodies were removed by centrifugation at 8,000 × g for 5 min at 4°C. The soluble fraction was loaded on a 1-ml Talon resin column (Clontech, Palo Alto, Calif.), washed with 10 column volumes of lysis buffer, and then washed with 10 column volumes of lysis buffer with 6 M guanidinium-HCl to dissociate LcrH bound to HT-YopD (HT-YopD remains bound to the Talon resin in the presence of 6 M guanidinium-HCl). HT-YopD was eluted from the column in lysis buffer with 50 to 200 mM imidazole, with subsequent dialysis against lysis buffer to remove the imidazole. HT-YopD was purified to homogeneity by this procedure, as assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining (data not shown).
Plasmid pWGQ, which encodes an N-terminal GST fusion to LcrQ expressed from the promoter of pGEX-3X, was introduced into E. coli DH5α. Cultures (100 ml) were grown at 37°C in tryptone-phosphate medium to an A620 of ca. 0.4, at which point high-level expression was induced by the addition of IPTG to 1.0 mM followed by incubation at 37°C overnight. The cells were harvested by centrifugation at 11,300 × g for 20 min at 4°C. Cell pellets were resuspended in PBS and lysed by passage through a chilled French pressure cell (20,000 lb/in2). The soluble fraction, which contained the majority of GST-LcrQ, was obtained by ultracentrifugation at 417,000 × g for 15 min in a TLA100.4 rotor at 4°C. GST-LcrQ was purified on a glutathione column (Pharmacia Biotech) as recommended by the manufacturer.
Anti-HT-YopD and anti-GST-LcrQ antibodies were raised in female New Zealand White rabbits as previously described (36). Immunoglobulin G was affinity purified by chromatography on an Affi-Prep-protein A column (Bio-Rad, Hercules, Calif.) as recommended by the manufacturer. Anti-HT-YopD and anti-GST-LcrQ antibody preparations were used for immunoblot analysis at dilutions of 1:40,000 and 1:20,000, respectively.
Protein electrophoresis and immunodetection.
Proteins were separated by SDS-PAGE, with 12 or 15% (wt/vol) polyacrylamide gels as indicated, by the method of Laemmli (28). Samples containing whole-cell fractions were boiled for 5 min, while all other fractions were heated to ca. 95°C for 3 to 5 min prior to being loaded on the gels. Samples were loaded so as to contain amounts of the fractions derived from the same volume of original culture. Proteins separated by SDS-PAGE were transferred to an Immobilon-P membrane (Millipore Corp., Bedford, Mass.) with carbonate buffer (pH 9.9) (37). Specific proteins were visualized on the membranes as previously described (43) by using the polyclonal antibodies specific for the proteins and a secondary goat anti-rabbit antibody (Sigma Chemical, St. Louis, Mo.) conjugated to alkaline phosphatase.
RESULTS
Construction of a Y. pestis yopD null mutant.
To test the hypothesis that YopD destabilization causes the negative regulatory phenotype originally attributed to lcrH, a yopD deletion mutant was constructed (Fig. 1A). Recombinant PCR mutagenesis (23) was used to delete essentially the entire open reading frame to ensure that a partial gene product could not be expressed. yopD occurs at the end of the lcrGVHyopBD operon, so there were no downstream genes to disrupt. However, the potential ρ-independent termination signal (4) was left intact to ensure proper processing of the transcript. The only yopD sequence retained was the 6 3′-terminal nucleotides in case that DNA sequence is important for the termination signal. The yopD deletion was introduced into pCD1 of Smr Y. pestis KIM8-3002 by allelic exchange, resulting in the strain KIM8-3002.2.
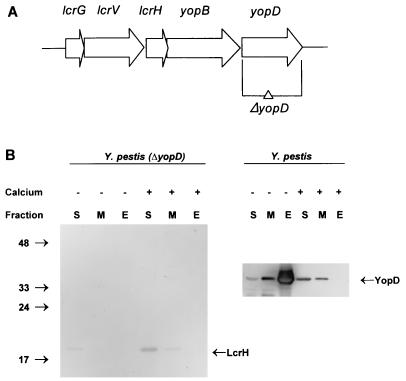
FIG. 1.
Construction of a complete ΔyopD mutation in Y. pestis. (A) Schematic representation of the lcrGVHyopBD operon of Y. pestis LCR plasmid pCD1. The positions and directions of the genes of this operon are shown as horizontal arrows. The DNA region deleted to create the yopD mutation is indicated (ΔyopD). (B) Immunoblot analysis of YopD and LcrH expressed in Y. pestis KIM8-3002.2 (ΔyopD) and Y. pestis KIM8-3002 (parent). Bacteria were grown at 37°C in the absence (−) or presence (+) of 2.5 mM Ca2+. Proteins from equal numbers of cells (0.025 A620 unit · ml) were separated by SDS-PAGE in a 15% (wt/vol) polyacrylamide gel. Antibodies were used to detect YopD and LcrH in the soluble (S) (cytoplasmic and periplasmic) fraction, the membrane (M) fraction, and the extracellular (E) fraction. The positions of the molecular mass markers (in kilodaltons) and the LCR-related proteins are indicated.
To test the phenotype of the yopD mutant, the mutant and parent Y. pestis strains were grown in the defined medium TMH at 26°C and then shifted to 37°C for 6 h in either the presence or absence of Ca2+. The yopD deletion mutant showed a growth phenotype comparable to that of the lcrH mutant: it underwent growth restriction when shifted from 26 to 37°C, irrespective of the presence of Ca2+ (growth restriction is a phenomenon observed in vitro in which the growth yield of Y. pestis is reduced during maximal induction of the LCR; it is not considered to play a role in vivo). As expected, there were no proteins recognized by the anti-YopD antibody in the deletion mutant (Fig. 1B). LcrH served as an internal control: its presence in the mutant demonstrated that the LCR proteins were still being expressed. In addition, the presence of equal levels of LcrH in the parent and yopD mutant strains (Fig. 2) showed that the yopD mutation was not exerting its phenotype by destabilizing the lcrGVHyopBD transcript. This also showed that LcrH does not require YopD to prevent it from being degraded, in contrast to YopD which does require LcrH for stability (61, 62). LcrH expression does not appear to be significantly up-regulated under inductive conditions. This is similar to the findings of a previous report, which noted that there was a 2.3-fold decrease in the level of LcrH expression in the presence of Ca2+ from that in the absence of Ca2+, while LcrV expression showed a 3.4-fold decrease (47). This difference was attributed to constitutive expression of LcrH from its own promoter in addition to expression from the Ca2+-regulated promoter. As found in the enteropathogenic Yersinia (21, 51, 58), the Y. pestis yopD mutant was also unable to efficiently translocate Yops into eukaryotic cells, despite the fact that Yops were effectively secreted (11), further indicating that the yopD gene was disrupted.
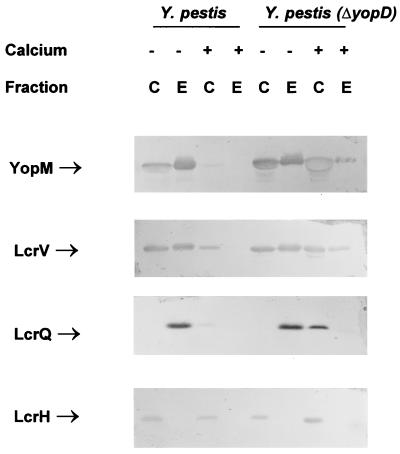
FIG. 2.
Immunoblot analysis of YopM, LcrV, LcrQ, and LcrH expressed in Y. pestis KIM8-3002 (parent) and Y. pestis KIM8-3002.2 (ΔyopD). Bacteria were grown at 37°C in the absence (−) or presence (+) of 2.5 mM Ca2+. Proteins from equal numbers of cells (0.025 A620 unit · ml) were separated by SDS-PAGE in a 12 or 15% (wt/vol) polyacrylamide gel. Antibodies were used to detect YopM, LcrV, LcrQ, and LcrH in both the whole-cell (C) (cytoplasmic, periplasmic, and membrane) fraction and the extracellular (E) fraction. The positions of the LCR-related proteins are indicated. The band corresponding to YopM in the whole-cell fraction of Y. pestis (ΔyopD) grown in the presence of Ca2+ has a hollow appearance, which occurs when a large quantity of protein is analyzed.
The yopD deletion was also introduced into a Y. pestis strain containing the pPCP1 plasmid to ensure that the absence of the Pla protease in the original yopD mutant was not affecting the observed results. The suicide vector containing the yopD deletion was introduced into pCD1 of Smr Y. pestis KIM5-3001 by allelic exchange, resulting in the strain KIM5-3001.16. The growth of this strain and its inability to express the YopD protein were characteristics similar to those of Y. pestis KIM8-3002.2 (data not shown).
Expression and secretion of LCR gene products.
To determine the role YopD plays in the LCR, Yop expression and secretion were examined in both Y. pestis KIM8-3002.2 and its parent by immunoblot analysis. The Y. pestis strains were grown in TMH with or without Ca2+ at 26°C and then shifted to 37°C for 6 h. After 6 h, all sample handling occurred at 4°C to minimize additional protein synthesis. The protein profiles of whole cells (containing the cytoplasmic, periplasmic, and membrane fractions) and culture supernatant (obtained by TCA precipitation) were determined with antibodies raised against YopM (36), LcrV (37), and LcrQ (Fig. 2). Similar results were seen if the culture supernatant fraction was obtained by ammonium sulfate precipitation (data not shown). The expression of YopM and LcrV was up-regulated in the mutant when grown in the presence of Ca2+, indicating that YopD is required for negative regulation. The results were similar when other Yops were analyzed (YopE and YopH) (data not shown). When the samples were examined for the presence of LcrQ, this putative negative regulator was still detected in the cellular fraction of the mutant grown in the presence of Ca2+ and in fact appeared to be at a higher concentration than in the parent Yersinia. This represents a situation where LcrQ does not cause down-regulation, suggesting that YopD is required for LcrQ to exhibit its negative regulatory activity. The elevated level of LcrQ likely reflects some effect of the LCR on LcrQ expression. There was no difference in Yop expression between wild-type and ΔyopD Y. pestis under normally inductive conditions (the absence of Ca2+).
The absence of YopD had a variable effect on secretion in the presence of Ca2+, when the Ysc system is normally blocked. In the yopD mutant, LcrV was not strongly blocked for secretion; i.e., there were similar levels of LcrV present in the whole-cell and culture supernatant fractions, regardless of the presence of Ca2+. However, the secretion of YopM (Fig. 2), as well as other Yops (data not shown), was not completely unblocked, but instead there was partial secretion in the presence of Ca2+. We have observed that YopM overexpression in the wild-type cell can result in partial (i.e., weak) secretion of YopM in the presence of Ca2+ (3). Therefore, YopM’s partial secretion in the yopD mutant could be due to the up-regulation of expression as opposed to YopD playing a direct role in YopM secretion. Importantly, it is evident that secretion of LcrV differs from that of the other Yops, at least as far as YopD is concerned.
Yop protein expression and secretion were also checked for the Pla-containing Y. pestis KIM5-3001.16 and its parent by immunoblot analysis (data not shown). The patterns of Yop and LcrV expression in the presence and absence of Ca2+ were similar in the two different yopD mutants. However, some of the secreted proteins are sensitive to the Pla protease, causing difficulty in the interpretation of protein secretion. This was especially true for LcrQ, which is present in the culture supernatant fraction only as a degradation product in Y. pestis KIM5-3001 (data not shown).
Localization of YopD.
Soluble (periplasm and cytoplasm), total membrane, and supernatant fractions isolated from Y. pestis KIM8-3002 cultures were examined for the presence of YopD by immunoblot analysis (Fig. 1B). As expected, in the presence of Ca2+, when secretion is blocked and the LCR is repressed, no YopD was observed in the culture supernatant. Instead, there appeared to be comparable levels of YopD in both the soluble and membrane fractions. YopD has two predicted transmembrane spans (17), so it is not surprising to find it associated with the membrane. In contrast, in the absence of Ca2+, when secretion is not blocked and the LCR is maximally expressed, the majority of YopD was found in the culture supernatant. However, YopD could still be detected in the soluble fraction. This indicates that it is not necessary to completely deplete the cell of YopD to obtain increased LCR expression.
Expression of YopD in wild-type and yopD Y. pestis.
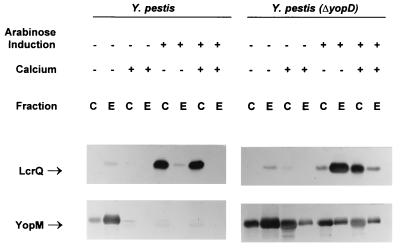
Complementation was performed to show that the phenotype of the yopD mutant was solely due to its mutation. lcrH and yopD were cloned in tandem behind the arabinose-inducible promoter of pBAD33 (16) by amplifying the region from pCD1 and using recombinant PCR mutagenesis (23) to remove the entire yopB gene. The rationale for including lcrH was that since the gene product had recently been demonstrated to be a chaperone for YopD (61), lcrH may be required for efficient YopD expression. Subsequent sequencing of the plasmid revealed a single-base deletion in the start codon for lcrH, resulting in the inability to express the gene. However, YopD was expressed in E. coli from the plasmid despite no LcrH expression (data not shown). The plasmid was introduced into wild-type and ΔyopD Y. pestis. Despite the fact that a very low level of expression is supposed to occur in the absence of induction of pBAD vectors (16), YopD was expressed without the addition of arabinose, although induction did result in an increase in YopD levels (Fig. 3).
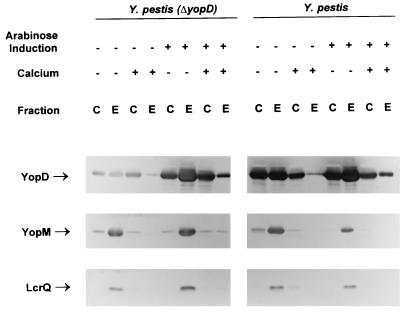
FIG. 3.
Immunoblot analysis of YopD, YopM, and LcrQ expressed in the complemented Y. pestis ΔyopD strain (KIM8-3002.2 carrying pAW161) and the parent Y. pestis with overexpression of YopD (KIM8-3002 carrying pAW161). Bacteria were grown at 37°C in both the absence (−) and presence (+) of 2.5 mM Ca2+. Expression of YopD from pAW161 was either uninduced (−) or induced (+) by the addition of arabinose to 0.05% (wt/vol). Proteins from equal numbers of cells (0.025 A620 unit · ml) were separated by SDS-PAGE in a 12 (YopD and YopM) or 15% (LcrQ) (wt/vol) polyacrylamide gel. Antibodies were used to detect YopD, YopM, and LcrQ in both the whole-cell (C) (cytoplasmic, periplasmic, and membrane) fraction and the extracellular (E) fraction. The positions of the LCR-related proteins are indicated.
In the yopD mutant, YopD expression from the plasmid without induction with arabinose resulted in lower-than-wild-type levels of YopD, but this amount of YopD was capable of complementing the mutant for Yop expression and secretion (Fig. 3). In addition, the growth phenotype of the complemented strain was similar to that of the wild type: it underwent growth restriction when shifted from 26 to 37°C only in the absence of Ca2+. When the level of YopD was increased, either by inducing the plasmid in the yopD mutant or expressing YopD from the plasmid in the wild-type strain without induction, the level of Yop expression did not decrease significantly (Fig. 3). Evidently, YopD is not the limiting factor for down-regulation. pAW171, encoding only a histidine-tagged YopD without the lcrH gene, provided similar complementation when expressed in the yopD mutant (data not shown). To confirm that additional LcrH could not overcome the yopD mutation, a plasmid containing the lcrH gene behind an IPTG-inducible promoter (pTrcH5) was used. This plasmid was unable to complement ΔyopD Y. pestis (data not shown).
Overexpressing YopD to significantly higher levels, such as when the plasmid was induced with arabinose in wild-type cells, did result in an apparent decrease in YopM expression. However, the growth rate of these cells significantly decreased over time (data not shown). This decrease in growth rate could be differentiated from the growth restriction normally seen under LCR-inductive conditions, since the level of YopM decreased rather than increased. It is possible that higher levels of YopD are toxic to the bacterial cell. This makes it difficult to interpret the significance of the lower level of YopM observed in these cells containing high levels of YopD.
Effect of the yopD mutation in a ysc strain.
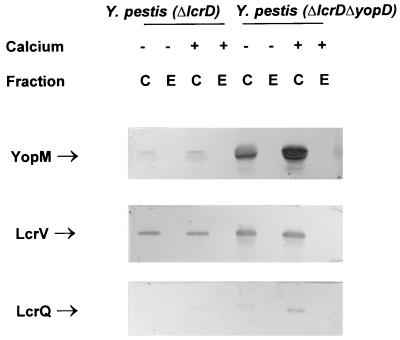
Yop expression in a ysc mutant is down-regulated, even under conditions that normally promote maximal expression, presumably through the inability to secrete a negative regulator (37, 42). A ysc lcrQ double mutant has previously been reported to show constitutive Yop expression (42). It was concluded that secretion of LcrQ is a prerequisite to achieve maximal induction of Yop expression. Since YopD also plays a role in negative regulation, a ysc yopD double mutant was constructed to determine the role of secretion on the action of YopD. The ΔyopD null mutation was moved into pCD1 of Y. pestis KIM5-3001.2 (ΔlcrD) by allelic exchange (LcrD is an essential component of the Ysc system [43, 44]). This strain contains pPCP1; however, the Pla protease does not present difficulties in interpreting results for intracellular proteins.
The ΔlcrD parent and ΔlcrD ΔyopD strains were analyzed for LCR expression and secretion (Fig. 4). As expected, the ΔlcrD parent strain had a low level of LCR expression and no detectable secretion. In contrast, the double mutant had high levels of cell-associated YopM and LcrV, with neither protein detected in the culture supernatant. These results indicate that the role YopD plays in negative regulation occurs irrespective of the Ysc system. Also, since no secretion was observed, not even the unblocked secretion of LcrV and partial secretion of YopM seen in the ΔyopD mutant, YopD’s observed role in secretion must depend on a functional Ysc. LcrQ was detected in the double mutant, as well as the ΔlcrD parent, irrespective of the presence of Ca2+ (Fig. 4). These results indicate that LcrQ secretion is not required for Yop expression to be up-regulated if YopD is absent, which supports the idea that LcrQ alone cannot down-regulate Yop expression.
FIG. 4.
Immunoblot analysis of YopM, LcrV, and LcrQ expressed in the ysc mutant Y. pestis KIM5-3001.2 (ΔlcrD) and the double mutant Y. pestis KIM5-3001.2 (ΔlcrD ΔyopD). Bacteria were grown at 37°C in the absence (−) or presence (+) of 2.5 mM Ca2+. Proteins from equal numbers of cells (0.025 A620 unit · ml) were separated by SDS-PAGE in a 12 (YopM and LcrV) or 15% (LcrQ) (wt/vol) polyacrylamide gel. Antibodies were used to detect YopM, LcrV, and LcrQ in both the whole-cell (C) (cytoplasmic, periplasmic, and membrane) fraction and the extracellular (E) fraction. The positions of the LCR-related proteins are indicated.
LcrQ’s regulatory function is dependent on YopD.
The level of the negative regulator LcrQ within the cell has been proposed to correlate inversely to the level of Yop expression; i.e., Yop expression is maximal when LcrQ is depleted from the cell, whereas elevated levels of LcrQ result in down-regulation of Yop expression (42, 50). To determine if increased levels of LcrQ could overcome a deletion of essentially the entire yopD gene, lcrQ was cloned behind the arabinose-inducible promoter of pBAD33 and introduced into both Y. pestis and Y. pestis (ΔyopD). No increase in LcrQ levels was observed without induction with arabinose (Fig. 5 versus Fig. 2), indicating tight control of expression by the promoter. Yop expression and secretion were monitored by observing YopM levels in whole-cell and culture supernatant fractions of cells either induced or not induced with arabinose (Fig. 5). The level of Yop expression was constitutively down-regulated when LcrQ was overexpressed in wild-type Y. pestis, and no secretion was detected. This is in agreement with previously reported accounts for overexpression of LcrQ (50). In contrast, the increased levels of LcrQ were not sufficient to overcome the constitutively induced ΔyopD phenotype: the level of YopM expression was only slightly reduced upon strong overexpression of LcrQ (Fig. 5). This further supports the idea that LcrQ requires YopD to exert its negative regulatory function. However, it is possible that the small decrease in YopM expression associated with LcrQ overexpression could reflect an inefficient or secondary role of LcrQ alone on Yop expression.
FIG. 5.
Immunoblot analysis of LcrQ and YopM expressed in Y. pestis KIM8-3002 (parent) and Y. pestis KIM8-3002.2 (ΔyopD) with overexpression of LcrQ from plasmid pAW162 in both strains. Bacteria were grown at 37°C in the absence (−) or presence (+) of 2.5 mM Ca2+. Expression of LcrQ from pAW161 was either uninduced (−) or induced (+) by the addition of arabinose to 0.2% (wt/vol). Proteins from equal numbers of cells (0.025 A620 unit · ml) were separated by SDS-PAGE in a 12 (YopM) or 15% (LcrQ) (wt/vol) polyacrylamide gel. Antibodies were used to detect LcrQ and YopM in both the whole-cell (C) (cytoplasmic, periplasmic, and membrane) fraction and the extracellular (E) fraction. The positions of the LCR-related proteins are indicated.
Interestingly, it was observed that the large majority of LcrQ was cell associated when the protein was overexpressed in the parent Y. pestis (Fig. 5). In fact, the amount of LcrQ seen in the culture supernatant was comparable to the amount that was secreted without the additional LcrQ expression, giving the impression that all additional LcrQ was not being secreted. In contrast, when LcrQ was overexpressed in the ΔyopD mutant, more was secreted than was observed in the parent strain. This suggests that YopD may function to limit or control the secretion of LcrQ. However, the failure of overexpressed LcrQ to shift the phenotype of the ΔyopD mutant cannot be attributed to its increased secretion, as there was still more LcrQ in the cellular fraction of the yopD strain than is ever present in the wild-type strain.
DISCUSSION
In this study, we tested the hypothesis that YopD, previously shown to be necessary for Yop translocation (21, 41, 51, 58), also is a terminal negative regulator of the LCR. Our yopD null mutation resulted in constitutively induced expression of LcrV and the Yops, and provision of YopD in trans complemented this regulatory phenotype. The yopD mutant exhibited this phenotype regardless of a functioning Ysc system, demonstrating that its regulatory effect is not at the level of secretion.
Our finding that YopD is a negative regulator helps explain the phenotype originally attributed to LcrH (4, 48). A mutation in lcrH, whether polar or nonpolar, resulted in constitutive induction of the LCR (4, 48). It is now known that LcrH (also called SycD), binds to YopD and apparently stabilizes it against degradation (61). Accordingly, we can infer that the phenotype of lcrH mutants is likely due to their drastically decreased net expression of YopD. There is a previous finding that would appear to be inconsistent with this interpretation: a constitutively repressed phenotype occurred when LcrH was overexpressed in a Y. pseudotuberculosis strain with a Genblock insertion in yopD (4). However, the insertion was at the end of yopD, and perhaps enough of the YopD protein was expressed to retain its regulatory function, provided there also was extra LcrH to ensure its stability.
YopD’s negative regulatory function probably operates through the action of LcrQ (also called YscM), which has previously been identified as being required for LCR down-regulation (42, 50). We showed here that YopD must be present for negative regulation to occur: in our yopD mutant, Yop expression was up-regulated, despite the fact that LcrQ was still present in the cell. In fact, overexpressing LcrQ in the yopD mutant could not overcome the observed phenotype. In addition, the inability to secrete LcrQ in the yopD ysc double mutant did not result in down-regulation of Yop expression. Therefore, we conclude that LcrQ is not sufficient for negative LCR regulation. Previous reports describing an lcrQ Y. pseudotuberculosis strain (42) indicate that YopD alone also is not sufficient, so we hypothesize that both LcrQ and YopD are necessary for LCR negative regulation.
Our data support the hypothesis that LcrQ is the limiting factor in determining whether negative regulation will occur. Previous studies documented an inverse correlation of intracellular amounts of LcrQ and expression of Yops (42, 50), and LcrQ was demonstrated to be quickly and efficiently secreted from the bacterial cell upon induction by contact with eukaryotic cells (42). We also found that the level of LcrQ in the cell seemed to directly correlate to the amount of negative LCR regulation, with the new finding that YopD had to be present. However, although YopD was necessary for negative regulation, altering its level had no significant effect on Yop expression. This is consistent with the observation that there still is YopD present in the cell under conditions of maximal LCR induction. Therefore, we hypothesize that LcrQ is rate limiting for LCR negative regulation.
YopD likely influences LcrQ function in some manner. We showed that YopD’s role is not due to an indirect effect on Yop secretion, as strong secretion still occurred in the yopD mutant, and a yopD ysc double mutant also showed constitutive induction. One potential role for YopD in the translocation process is as an extracellular chaperone. In this model, YopD would be expected to bind the translocated Yops, including YopH. Because LcrQ and the amino-terminal end of YopH exhibit 42% sequence identity (50), it may be that YopD also directly binds to LcrQ. Future work will determine whether there is a direct interaction between YopD and LcrQ, as this has not yet been shown.
Interestingly, the existing data suggest that LcrQ and YopD specifically affect each other’s secretion to the culture medium. We found that when LcrQ was overexpressed, only a limited amount of it was released, even though the secretion machinery was not blocked by the absence of Ca2+. The amount of overexpressed LcrQ that was secreted increased in the yopD mutant, suggesting that LcrQ secretion is affected by YopD’s presence. Likewise, LcrQ is implicated in the control of YopD and LcrV secretion: an lcrQ mutant of Y. pseudotuberculosis secreted LcrV and all Yops normally in the absence of Ca2+, but in the presence of Ca2+, when secretion normally is blocked, LcrV and YopD were secreted (50). These phenomena might be manifestations of a hierarchy in the LCR induction and secretion mechanisms that could be reflected in the order in which the Yersinia proteins are secreted. A set of proteins would need to be secreted early to put in place the contact-induced translocation machinery. YopD and YopB would be among these proteins. Early LcrQ release would initiate contact-induced amplification of the antihost response while the existing pool of Yop toxins is being translocated into the host cell. Early LcrV release might be needed on two accounts: this would be one way of removing the inner secretion gate, LcrG, from its secretion-blocking location at the cytoplasmic face of the inner membrane (37); also, LcrV may play a role in translocation of Yops (38). This kind of hierarchy is supported by the phenotype of the partially secretion-defective Yersinia enterocolitica virG mutant, which, interestingly, was most severely defective in secretion of LcrV, YopB, and YopD (1). It would be of interest to examine LcrQ secretion in this mutant.
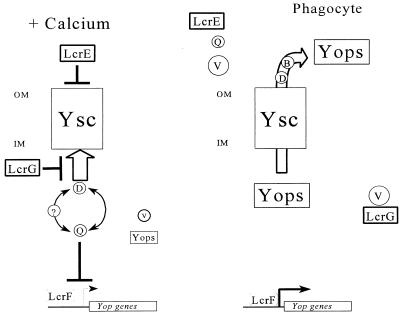
Our findings require a revision of the current model of LCR gene regulation to take into account YopD’s role (Fig. 6). In the presence of Ca2+, LcrE and LcrG block the secretory machinery from outside the cell and from the cytoplasmic face, respectively, as previously proposed (13, 37). This retains both LcrQ and YopD in the cell where they can function to down-regulate LcrV and Yop expression. The possibility of additional LCR components playing a role in gene regulation cannot be ruled out with the current data (denoted in Fig. 6 by ?). The secretion block also prevents the secretion of the low levels of LcrV and Yops that are in the cell in the presence of Ca2+. The continued, albeit low-level, expression of YopD along with other LCR gene products in the absence of an inductive signal is consistent with the model, since some YopD must be expressed to function as a negative regulator even though it is subject to the same regulation. (LcrQ appears to be more Ca2+ independent in its expression [42].) Upon contact with the eukaryotic cell or removal of Ca2+ from the medium, the secretion block is relieved. It has been proposed that this occurs by LcrE sensing the change in the environment (13), which then may allow the release of some secretion system-associated LcrG (37), accompanied by the secretion of low levels of Yops, as well as LcrQ and YopD. The secretion of LcrQ occurs very efficiently (42). The decrease in intracellular levels of the negative regulators allows an increase in the expression of the LCR gene products, including LcrV. The increased level of LcrV is then able to bind to LcrG and release it from the secretion system, thereby resulting in a complete unblocking of Yop secretion. This then results in full induction of the LCR until the time when the cellular concentration of LcrQ increases to the point where it can again function as a negative regulator. Since there are still low levels of YopD present in the cell under inductive conditions, its intracellular concentration might not need to be increased for negative regulation to be reestablished.
FIG. 6.
Model for regulation of the LCR in Y. pestis. In the presence of Ca2+ (+ Calcium), LcrE and LcrG block the Yop secretion system (Ysc) at the cell surface and in the cytoplasm, respectively. The secretion block prevents the secretion of LcrQ (Q) and YopD (D). These two proteins work in concert to cause a down-regulation of Yop expression. There may be other components (?) required that have not yet been identified. In the absence of Ca2+ and in vivo when the bacteria contact a eukaryotic cell, LcrE is released from the cell surface. This relieves the secretion block and allows secretion of both LcrQ and YopD, which results in induction of the yop genes, including lcrV. The increase in LcrV (V) levels titrates LcrG away from the Ysc, resulting in full LCR induction and secretion. YopD and YopB (B) are then involved in translocating the Yops into the eukaryotic cell. OM, outer membrane; IM, inner membrane.
There are at least two proteins required for negative control of LCR gene expression; however, the precise mechanism of this regulation is unknown. There is an LCR-specific transcriptional activator, LcrF (6, 24), whose activity could potentially be affected by the binding of a negative regulator. However, no direct interaction between LcrF and either LcrQ or YopD has yet been documented. In the flagellar system, where negative regulation of terminal loci in the regulatory cascade is due to the secreted anti-sigma factor FlgM (26, 27), the signal for induction of late-gene expression derives from monitoring the growing flagellar structure and not from an environmental cue (33). In contrast, the LCR regulon has previously been thought to respond to an extracellular signal, whether it be contact with a eukaryotic cell or the absence of Ca2+. However, the fact that YopD is known to be necessary for vectorial translocation of effector Yops into the eukaryotic cell raises the intriguing possibility that terminal regulation in the LCR is linked to the presence of a functional translocation mechanism at the interface between the bacterial and eukaryotic cells. Accordingly, regulation of Yop expression at the level of transcription could be tied to the bottom-line function of the LCR—i.e., the poisoning of the host cell by translocation of Yops.
One similar feature between the LCR and flagellar biogenesis systems is that the signal that elicits regulation within the bacterium most likely needs to be transmitted across both inner and outer membranes, although the method of transmitting the signal likely will differ, as the structures of the secretion machineries distal to the inner membrane in the two systems do not share homology. Flagellar late-gene expression is decreased as the filament grows, due to a decrease in the rate of export of flagellar products, which is thought to result in an increase in the intracellular concentration of FlgM (26, 27). It is not clear how LCR gene regulation is terminated after induction has occurred. The analogous situation would be that as Yops are translocated into the host cell, there is a point where the rate of secretion is slowed, thereby allowing the intracellular concentration of LcrQ to increase. Alternatively, the secretion channel may again become blocked when the bacterium dissociates from the eukaryotic cell. It could be that Yersinia uses two proteins for negative regulation instead of just one to make the system more sensitive to changes in LcrQ levels. In the flagellar system, low levels of FlgM are thought to exist in the cell during flagellar late-gene expression (i.e., under inductive conditions for synthesis of the flagellar filament) (26, 27), while LcrQ is not detected in the cell upon maximal induction of the LCR. This would then require that the intracellular pool of LcrQ accumulate before the system could be shut off. YopD could be responsible for allowing the cell to respond to even very small levels of LcrQ, as under inductive conditions, YopD is the most abundant Yop. The decrease or cessation of translocation may initiate the process of LCR down-regulation by focusing YopD’s function on its interaction with LcrQ.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grant AI21017.
We gratefully acknowledge Wendi Gardner, Kimberly McFarland, and Elżbieta Skrzypek for the construction of pWGQ and for the preparation of anti-GST-LcrQ antibodies; Matt Nilles for construction of Y. pestis KIM8-3002; and Ken Fields for verification of the translocation phenotype of the Y. pestis yopD mutant. We also acknowledge Mike Russ of the University of Kentucky Macromolecular Structure Analysis Facility for the synthesis of some of the oligonucleotides used in this study.
REFERENCES
- 1.Allaoui A, Scheen R, Lambert de Rouvroit C, Cornelis G R. VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to ExsB of Pseudomonas aeruginosa. J Bacteriol. 1995;177:4230–4237. doi: 10.1128/jb.177.15.4230-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. Analysis of proteins; p. 10.2.30. [Google Scholar]
- 3.Beiting, M., G. V. Plano, and S. C. Straley. Unpublished data.
- 4.Bergman T, Håkansson S, Forsberg Å, Norlander L, Macellaro A, Bäckman A, Bölin I, Wolf-Watz H. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role of LcrH and LcrV. J Bacteriol. 1991;173:1607–1616. doi: 10.1128/jb.173.5.1607-1616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergman T, Erickson K, Golyov E, Persson C, Wolf-Watz H. The lcrB (yscN/U) gene cluster of Yersinia pseudotuberculosis is involved in Yop secretion and shows high homology to the spa gene clusters of Shigella flexneri and Salmonella typhimurium. J Bacteriol. 1994;176:2619–2626. doi: 10.1128/jb.176.9.2619-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis G R, Sluiters C, Delor I, Geib D, Keniga K, Lambert de Rouvroit C, Sory M-P, Vanootegehem J C, Michiels T. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol. 1991;5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis G R, Sluiters C, Lambert de Rouvroit C, Michiels T. Homology between VirF, the transcriptional activator of the Yersinia virulence region, and the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 9.Ferber D M, Brubaker R R. Plasmids in Yersinia pestis. Infect Immun. 1981;31:839–841. doi: 10.1128/iai.31.2.839-841.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields K A, Plano G V, Straley S C. A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J Bacteriol. 1994;176:569–579. doi: 10.1128/jb.176.3.569-579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields, K. A., and S. C. Straley. Unpublished data.
- 12.Fields K A, Williams A W, Straley S C. Failure to detect binding of LcrH to the V antigen of Yersinia pestis. Infect Immun. 1997;65:3954–3957. doi: 10.1128/iai.65.9.3954-3957.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg Å, Viitanen A-M, Skurnik M, Wolf-Watz H. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol. 1991;5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, W., E. Skrzypek, and S. C. Straley. Unpublished data.
- 15.Goguen J D, Yother J, Straley S C. Genetic analysis of the low calcium response in Yersinia pestis Mud1 (Ap lac) insertion mutants. J Bacteriol. 1984;160:842–848. doi: 10.1128/jb.160.3.842-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman L-M, Berlin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Håkansson S, Bergman T, Vanooteghem J-C, Cornelis G, Wolf-Watz H. YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect Immun. 1993;61:71–81. doi: 10.1128/iai.61.1.71-80.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Håkansson S, Galyov E E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 19.Håkansson S, Persson C, Schesser K, Galyov E E, Rosqvist R, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for translocation of Yop effector proteins across the target cell plasma membrane and displays contact-dependent membrane-disrupting activity. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D. Studies on transformation of E. coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Hartland E L, Green S P, Philips W A, Robins-Browne R M. Essential role of YopD in inhibition of the respiratory burst of macrophages by Yersinia enterocolitica. Infect Immun. 1994;62:4445–4453. doi: 10.1128/iai.62.10.4445-4453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartland E L, Bordun A-M, Robins-Browne R M. Contributions of YopB to virulence of Yersinia enterocolitica. Infect Immun. 1996;64:2308–2314. doi: 10.1128/iai.64.6.2308-2314.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 24.Hoe N P, Goguen J D. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol. 1993;175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmström A, Pettersson J, Rosqvist R, Håkansson S, Tafazoli F, Fällman M, Magnusson K-E, Wolf-Watz H, Forsberg Å. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol Microbiol. 1997;24:73–91. doi: 10.1046/j.1365-2958.1997.3211681.x. [DOI] [PubMed] [Google Scholar]
- 26.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 27.Kutsukake K. Excretion of the anti-sigma factor through a flagellar structure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lambert de Rouvroit C, Sluiters C, Cornelis G R. Role of the transcriptional activator VirF in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol Microbiol. 1992;6:395–409. [PubMed] [Google Scholar]
- 30.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 31.Levesque C, Piche L, Chantal L, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindler L E, Klempner M S, Straley S C. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect Immun. 1990;58:2569–2577. doi: 10.1128/iai.58.8.2569-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 34.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore J T, Uppal A, Maley F, Maley G F. Overcoming inclusion body formation in a high-level expression system. Protein Expr Purif. 1993;4:160–163. doi: 10.1006/prep.1993.1022. [DOI] [PubMed] [Google Scholar]
- 36.Nemeth J, Straley S C. Effect of Yersinia pestis YopM on experimental plague. Infect Immun. 1997;65:924–930. doi: 10.1128/iai.65.3.924-930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilles M L, Williams A W, Skrzypek E, Straley S C. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J Bacteriol. 1997;179:1307–1316. doi: 10.1128/jb.179.4.1307-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilles, M. L., and S. C. Straley. Unpublished data.
- 39.Perry R D, Pendrak M, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry R D, Fetherston J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Persson C, Nordfelth R, Holmström N, Håkansson S, Rosqvist R, Wolf-Watz H. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 42.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustavsson M, Magnusson K-E, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 43.Plano G V, Barve S S, Straley S C. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J Bacteriol. 1991;173:7293–7303. doi: 10.1128/jb.173.22.7293-7303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plano G V, Straley S C. Multiple effects of lcrD mutations in Yersinia pestis. J Bacteriol. 1993;175:3536–3545. doi: 10.1128/jb.175.11.3536-3545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plano G V, Straley S C. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis. J Bacteriol. 1995;177:3843–3854. doi: 10.1128/jb.177.13.3843-3854.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price S B, Cowan C, Perry R D, Straley S C. The Yersinia pestis V antigen is a regulatory protein necessary for Ca2+-dependent growth and maximal expression of low-Ca2+ response virulence genes. J Bacteriol. 1991;173:1491–1498. doi: 10.1128/jb.173.8.2649-2657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price S B, Leung K Y, Barve S S, Straley S C. Molecular analysis of lcrGVH, the V antigen operon of Yersinia pestis. J Bacteriol. 1989;171:5646–5653. doi: 10.1128/jb.171.10.5646-5653.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price S B, Straley S C. lcrH, a gene necessary for virulence of Yersinia pestis and for the normal response of Y. pestis to ATP and calcium. Infect Immun. 1989;57:1491–1498. doi: 10.1128/iai.57.5.1491-1498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Protsenko O A, Anisimov P I, Mozharov O T, Konnov N P, Popov Y A, Kokookshkin A M. Detection and characterization of Yersinia pestis plasmids determining pesticin I, fraction I antigen and mouse toxin synthesis. Genetika. 1983;19:1081–1090. [PubMed] [Google Scholar]
- 50.Rimpiläinen M, Forsberg Å, Wolf-Watz H. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis, shows extensive homology to YopH. J Bacteriol. 1992;174:3355–3363. doi: 10.1128/jb.174.10.3355-3363.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosqvist R, Magnusson K-E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salmond G P C, Reeves P J. Membrane traffic wardens and protein secretion in Gram-negative bacteria. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Skrzypek E, Haddix P L, Plano G V, Straley S C. New suicide vector for gene replacement in yersiniae and other gram-negative bacteria. Plasmid. 1993;29:160–163. doi: 10.1006/plas.1993.1019. [DOI] [PubMed] [Google Scholar]
- 55.Skrzypek E, Straley S C. LcrG, a secreted protein involved in negative regulation of the low-calcium response in Yersinia pestis. J Bacteriol. 1993;175:3520–3528. doi: 10.1128/jb.175.11.3520-3528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skrzypek E, Straley S C. Differential effects of deletions in lcrV on secretion of V antigen, regulation of the low-Ca2+ response, and virulence of Yersinia pestis. J Bacteriol. 1995;177:2530–2542. doi: 10.1128/jb.177.9.2530-2542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sodeinde O A, Goguen J D. Genetic analysis of the 9.5-kilobase virulence plasmid of Yersinia pestis. Infect Immun. 1988;56:2743–2748. doi: 10.1128/iai.56.10.2743-2748.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sory M-P, Cornelis G R. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 59.Straley S C, Bowmer W S. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect Immun. 1986;51:445–454. doi: 10.1128/iai.51.2.445-454.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Une T, Brubaker R R. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wattiau P, Bernier B, Deslée P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams, A. W., and S. C. Straley. Unpublished data.