ABSTRACT
Background
Immunoglobulin A nephropathy (IgAN) frequently leads to kidney failure. The urinary proteomics-based classifier IgAN237 may predict disease progression at the time of kidney biopsy. We studied whether IgAN237 also predicts progression later in the course of IgAN.
Methods
Urine from patients with biopsy-proven IgAN was analyzed using capillary electrophoresis–mass spectrometry at baseline (IgAN237-1, n = 103) and at follow-up (IgAN237-2, n = 89). Patients were categorized as “non-progressors” (IgAN237 ≤0.38) and “progressors” (IgAN237 >0.38). Estimated glomerular filtration rate (eGFR) and urinary albumin–creatinine ratio slopes were calculated.
Results
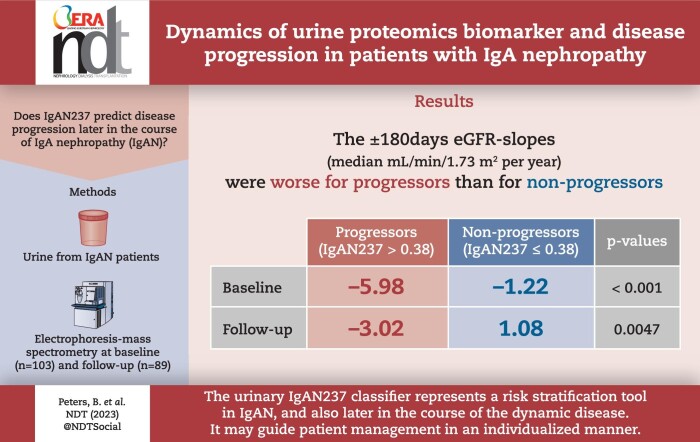
Median age at biopsy was 44 years, interval between biopsy and IgAN237-1 was 65 months and interval between IgAN237-1 and IgAN237-2 was 258 days (interquartile range 71–531). IgAN237-1 and IgAN237-2 values did not differ significantly and were correlated (rho = 0.44, P < .001). Twenty-eight percent and 26% of patients were progressors based on IgAN237-1 and IgAN237-2, respectively. IgAN237 inversely correlated with chronic eGFR slopes (rho = –0.278, P = .02 for score-1; rho = –0.409, P = .002 for score-2) and with ±180 days eGFR slopes (rho = –0.31, P = .009 and rho = –0.439, P = .001, respectively). The ±180 days eGFR slopes were worse for progressors than for non-progressors (median –5.98 versus –1.22 mL/min/1.73 m2 per year for IgAN237-1, P < .001; –3.02 vs 1.08 mL/min/1.73 m2 per year for IgAN237-2, P = .0047). In multiple regression analysis baseline progressor/non-progressor according to IgAN237 was an independent predictor of eGFR180days-slope (P = .001).
Conclusion
The urinary IgAN237 classifier represents a risk stratification tool in IgAN also later in the course of the dynamic disease. It may guide patient management in an individualized manner.
Keywords: biomarker, CKD, glomerulonephritis, IgA nephropathy, progression, urine proteomics
Graphical Abstract
Graphical Abstract.
KEY LEARNING POINTS.
What was known:
Immunoglobulin A nephropathy (IgAN) is the most common glomerulonephritis and only some patients respond to specific treatments while many develop end-stage kidney disease.
Good guidance predicting a progressive disease is currently lacking.
To serve this urgent need, this study aims at developing a biomarker-based algorithm that predicts disease progression in IgAN that enables personalized treatment.
This study adds:
Based on development of urine proteomic analysis a classifier was developed (IgAN237) to predict IgAN disease progression.
This clinical study aimed to confirm IgAN237 as a significant marker of progression to enable personalized follow-up and treatment of IgAN, both at the time of biopsy but also later in the disease course.
Potential impact:
The IgAN237 classifier predicts progressive loss of kidney function in IgAN patients significantly better than clinical parameters alone.
Even late after kidney biopsy IgAN237 guides the clinicians in deciding whether the IgAN is of the progressing or non-progressing type which enables individualized follow-up and treatment but also selection for trials.
INTRODUCTION
Immunoglobulin A nephropathy (IgAN) is the most common primary glomerulonephritis worldwide. However, prognosis is very variable and 20%–40% of patients develop kidney failure within 10–20 years after diagnosis [1, 2]. In this regard, risk stratification tools are needed to guide therapy and response to therapy. IgAN is diagnosed by kidney biopsy. Histologic grading using the MEST-C score can be used to assess prognosis [3, 4]. However, neither histology nor the International IgAN Prediction Tool calculators [5, 6] predict disease progression [5]. Furthermore, kidney biopsy is invasive and not well suited for repeated assessment of risk. Recommendations by Kidney Disease: Improving Global Outcomes (KDIGO) emphasize general supportive measures and renin–angiotensin system blockade as the initial treatment [5]. Patients who remain at high risk of progressive disease despite maximal supportive care may be considered for a 6-month course of glucocorticoid therapy [5]. An exception for more intense immunosuppressive therapy is rapidly progressive IgAN [5]. In recent years, promising results on renal outcomes have been shown with non-specific kidney protection with dapagliflozin [7] or empagliflozin [8] or immunosuppression with oral methylprednisolone [9] or budesonide [10].
Urinary proteomics using capillary electrophoresis coupled to mass spectrometry (CE-MS) has emerged as a risk stratification tool for various clinical conditions, including chronic kidney disease (CKD) from diverse causes [11–13]. Urinary proteome panels may provide diagnostic and risk stratification information for IgAN [14–17]. In urine samples obtained at the time of kidney biopsy, the international multicenter project “Personalized Treatment in IgA Nephropathy” (PersTIgAN) identified a urine peptidomics classifier (IgAN237) specific for IgAN. In addition, it could be used for risk stratification of IgAN that predicted the risk of progression defined as the change in estimated glomerular filtration rate (eGFR) [16]. This classifier improved prediction of kidney function loss in IgAN patients over clinical parameters alone.
The aim of the present prospective study was to evaluate the dynamics of IgAN237 over time and determine whether it predicts disease progression in later stages of IgAN, at different timepoints from kidney biopsy, to potentially guide personalized treatment.
MATERIALS AND METHODS
Study population and study design
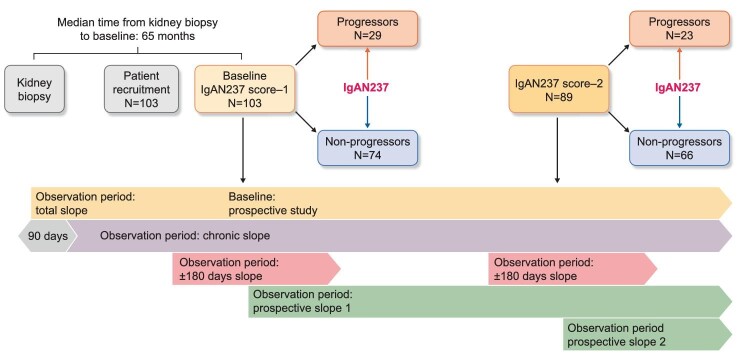
Urine samples and clinical data were obtained from 103 patients from seven centers in Europe (Leipzig, Germany, and in Sweden Skövde, Umeå, Östersund, Sundsvall, Gothenburg and Linköping) (Fig. 1). Inclusion criteria were adult patients (≥18 years) at baseline with biopsy-proven IgAN in native kidneys (Supplementary methods). The study was approved by the ethics committee of the Saxonian Chamber of Physicians (No: EK-BR-36/20-1) in Germany and by the Swedish Ethical Review Authority (Dnr 2019-02970).
Figure 1:
Study design and patient flow.
This clinical study was performed independent of time of biopsy. In addition, numerous biopsies were performed before the latest Oxford classifications (including the MEST-C score) [4] were published and therefore histological data are not fully adapted to those criteria. The histological data may have some inter-operator variability while the IgAN237 classifier is a quantitative test for the rate of progression [16].
After signed consent, participants provided urine sample for IgAN237 classifier assessment at baseline and after 6 months of follow-up. Based on a prior study, patients with IgAN237 values >0.38 were defined as progressors and those with values ≤0.38 as non-progressors [16]. Retrospective and prospective information for at least 12 months after baseline was collected, including serum creatinine for eGFR calculation using the CKD Epidemiology Collaboration (CKD-EPI, mL/min/1.73 m2; assuming Caucasian population) formula [18], and urine albumin–creatinine ratio (UACR g/mol).
eGFR and UACR slopes
The change in renal function was measured as eGFR slopes and the change in kidney injury as UACR slopes. Four different eGFR and UACR slopes were calculated using linear regression and the principle of the least square deviation formula for a timeline (Fig. 1 and Supplementary data, Fig. S1):
The “total slope” (eGFRTotal-slope, UACRTotal-slope) was calculated from all available eGFR and UACR values (retrospectively and prospectively) including those within the first 90 days after biopsy until the last available datapoint [19].
The “chronic slope” (eGFRChron-slope, UACRChron-slope) was calculated from all values from Day 90 after biopsy until the latest available value of eGFR and UACR, respectively (retrospectively and prospectively) [19]. This was done to exclude potential eGFR changes close to the biopsy due to acute treatment decisions (i.e. start of renin–angiotensin–aldosterone system blockade).
The “±180 days slope” (eGFR180days-slope, UACR180days-slope) was estimated from all eGFR and UACR values available from 180 days before to 180 days after the IgAN237 score-1. The same was done for the period around IgAN237 score-2 samples. In cases with <180 days follow-up for either slope, data were missing.
The “prospective slope” (eGFRProsp-slope, UACRProsp-slope) was estimated from all prospective eGFR and UACR values available after the assessment of IgAN237 score-1 and after the assessment of IgAN237 score-2, respectively.
Estimation of creatinine-based eGFR slopes using serum creatinine becomes less accurate when values exceed 90 or are below 15 mL/min/1.73 m2 [18, 20]. Therefore, eGFR slopes were only estimated for patients with an eGFR between 15 and 90 mL/min/1.73 m2 at baseline. All results related to the eGFR slopes were excluded for the patient if eGFR at baseline was either >90 or <15 mL/min/1.73 m2. Treatment for IgAN was decided locally and there was no common treatment protocol.
Urine proteome profiling
For IgAN237 assessment, a baseline urine sample (8–10 mL) was obtained in 103 patients and a follow-up sample in 89 of these patients more than 6 months after the baseline sample. Urine was collected in boric acid containing urine tubes (Urine Monovette®, Sarstedt, Germany) and transferred within 3 days to be stored at –80°C at the Biobank Northern Sweden. From there, samples were shipped on dry ice to Mosaiques Diagnostics, Hannover/Germany for urine proteomic analysis. German samples were sent directly to Mosaiques Diagnostics.
Sample preparation and CE-MS analysis
Urine sample preparation and CE-MS was performed as previously described [16]. In brief, urine samples were thawed and 700 μL was diluted with 700 μL of alkaline buffer (2 M urea, 10 mM NH4OH, 0.02% SDS). Subsequently, high molecular weight proteins were eliminated by ultrafiltration using a Centristat 20 kDa cut-off centrifugal filter device (Sartorius, Göttingen, Germany). PD 10 gel filtration columns (GE Healthcare Bio Sciences, Uppsala, Sweden) were used to remove urea, electrolytes and salts as well as to enrich polypeptides in the obtained filtrate. The samples were lyophilized and stored at 4°C until testing. Shortly before CE-MS analysis, the samples were re-suspended in 10 μL HPLC-grade H2O. The CE-MS analysis was performed as previously described [16] using P/ACE MDQ capillary electrophoresis system (Beckman Coulter, Fullerton, CA, USA) coupled with a Micro-TOF II MS (Bruker Daltronic, Bremen, Germany). The raw data were processed using the MosaFinder software [21] for deconvolution of mass spectra ion peaks into single masses. Data were calibrated using internal standards as reference data. Reference signals of 29 abundant peptides were used for calibration of signal intensity using linear regression as previously described [22]. The obtained peak list characterizes each detected polypeptide by its calibrated molecular mass, calibrated CE migration time and normalized signal intensity. All detected peptides were deposited, matched and annotated in a Microsoft SQL database allowing further statistical analysis.
Sample classification
Samples were classified according to the previously developed IgAN237 classifier [16]. IgAN237 is a support vector machine-based classification model that allows the classification of samples in the high-dimensional parameter space. The application of the IgAN237 classifier to CE-MS data of unknown urine samples provides a classification score based on the amplitudes of 237 peptide biomarkers. A classification threshold (cut-off) of 0.38 was defined using receiver operating characteristic curve analysis based on the total cross-validated training dataset for classifier developed described by Rudnicki et al. [16]. The defined cut-off corresponded to the best sensitivity/specificity pair based on the Youden index. IgAN237 is adimensional. Patients with IgAN237 values >0.38 were defined as progressors, while patients with IgAN-classifier value ≤0.38 were defined as non-progressors.
Statistical analyses
Descriptive statistics are presented as frequencies with percentages for categorical variables and mean with standard deviation or median with range for continuous variables. Paired tests with each patient as her/his own control were used to examine changes in eGFR, UACR and U-proteomics. Mann–Whitney U test, t-test and Fisher's exact test were used for group comparisons and Wilcoxon non-parametric test for paired comparisons. Spearman bivariate correlation coefficient was calculated to avoid significant effects of outlier data (rho-value). Multiple linear regression was performed with IgAN237 as the dependent factor and the significant variables from the Spearman analysis as independent variables; age and sex were also included in the model. A stepwise model was used to evaluate the importance of variables that might be associated with IgAN237 as the dependent factor (≤0.38 or >0.38). Logistic regression was performed in this case since the U-proteomics was not normally distributed, and we wanted to verify the results from linear regression. Because U-proteomics measurements 1 and 2 were dependent (repeated measurements on the same group of patients), data analyses were performed separately for the two samples when analyzing the association between IgAN237 and eGFR and UACR slopes. A two-tailed P-value <.05 was set as the significance level.
RESULTS
Clinical and histopathological characteristics
Baseline U-proteomics data were obtained 65 months (0-606) after a kidney biopsy that diagnosed IgAN. Based on clinical history, IgA vasculitis (IgAV) was diagnosed in 17/103 (16.5%) and IgAN in 86/103 (83.5%), respectively. Based on criteria by Saha et al. [2], 74 of 103 (72%) participants had a concomitant condition at the time of biopsy or later that would be considered secondary IgAN (Supplementary data, Table S1). In addition, 60 patients suffered from other diseases such as lactose intolerance, pulmonary obstructive diseases, upper respiratory or intestinal allergic reactions. Table 1 shows demographic, analytical and key kidney biopsy data. Mean eGFR at baseline and at follow-up was 56.4 ± 27.4 and 54.5 ± 28.6 mL/min/1.73 m2, respectively.
Table 1:
Demographic data and kidney biopsy findings.
| N (%) with data | Mean | SD | Median (range) | |
|---|---|---|---|---|
| Male | 65 (63) | |||
| Age at diagnosis (1st biopsy) (years) | 103 | 44.6 | 16.3 | 44 (11 to 92) |
| Age at baseline (years) | 103 | 53.9 | 15.4 | 54 (20 to 92) |
| eGFR at baseline (mL/min/1.73 m2) | 94 | 56.4 | 27.4 | 52.5 (8.9 to 117) |
| eGFR at follow-up (mL/min/1.73 m2) | 77 | 54.5 | 28.6 | 51.6 (8 to 125) |
| Number of antihypertensive drugs at baseline (N) | 103 | 2.0 | 1.38 | 2 (0 to 6) |
| Biopsy: glomerular sclerosis (% of glomeruli) | 85 | 19 | 16 | 14 (0 to 70) |
| Biopsy: interstitial fibrosis (% of kidney surface) | 93 | 23 | 18 | 25 (0 to 70) |
| Biopsy: crescents (% of biopsies with crescents)a | 25/91 (27.5%) | |||
| Baseline (at IgAN237-1) | ||||
| Time between biopsy and urine sample 1 (months) | 103 | 119.1 | 130.6 | 65 (0 to 606) |
| IgAN237-1 (value) | 103 | –0.0116 | 0.802 | 0.024 (–2.163 to 2.038) |
| Hematuria [N (%)] | 85 [38 (45%)] | |||
| UACR (g/mol) | 94 | 48.2 | 83.6 | 19.0 (0.5 to 655) |
| UACRTotal-slope (g/mol/year) | 96 | –28.1 | 107 | –2.79 (–745 to 18.8) |
| UACRChron-slope (g/mol/year) | 95 | –5.36 | 27.0 | –0.40 (–226 to 30.7) |
| UACRProsp-slope (g/mol/year) | 87 | –5.98 | 55.4 | 0.00 (–226 to 324) |
| UACR180days-slope (g/mol/year) | 87 | –11.1 | 122 | 0.00 (–1085 to 163) |
| Follow-up (at IgAN237-2) | ||||
| Time between urine sample 1 and 2 (days) | 89 | 271 | 78 | 258 (71 to 531) |
| IgAN237-2 (value) | 89 | –0.069 | 0.741 | 0.054 (–2.119 to 1.721) |
| Hematuria [N (%)] | 46 [18 (39%)] | |||
| UACR (g/mol) | 64 | 38.0 | 47.9 | 21.6 (0.30 to 250) |
| UACRTotal-slope (g/mol/year) | 85 | –25.2 | 100 | –2.74 (–745 to 18.8) |
| UACRChron-slope (g/mol/year) | 84 | –3.67 | 14.8 | –0.38 (–67.9 to 30.7) |
| UACRProsp-slope (g/mol/year) | 62 | 15.7 | 283 | 0.00 (–1160 to 1840) |
| UACR180days-slope (g/mol/year) | 62 | –9.98 | 55.3 | –0.16 (–238 to 147) |
N = number; SD = standard deviation.
Baseline is the timepoint when the first urine sample for assessment of IgAN237 was collected.
In one biopsy, one endocapillary finding was mentioned.
aIn those with crescent findings, few affected glomeruli were noted.
Supplementary data, Table S2 shows the distribution of participants in eGFR categories. Data from participants with eGFR 15–90 mL/min/1.73 m2 were used to calculate eGFR slopes. Key therapeutic agents are summarized in Supplementary data, Section S3. Antihypertensive drugs were prescribed for 90% of the patients. Most patients 89/102 (87%) were prescribed either angiotensin-converting enzyme (ACE) inhibitors or angiotensin-receptor blockers (ARBs) and 10/89 received both ACE inhibitors and ARBs. Sodium-glucose cotransporter 2 inhibitors were not prescribed. Prednisolone was prescribed to 29 patients during the disease. During the study period (from baseline sampling) prednisolone was prescribed to seven, immunosuppression only to three, and a combination of prednisolone and immunosuppression to three participants.
Urinary proteomics
Baseline IgAN237-1 (mean value –0.0116 ± 0.802) and follow-up IgAN237-2 (mean –0.069 ± 0.741) were separated by a median 258 days (range 71–531) (Table 1). Their values did not differ significantly (mean change –0.054 ± 0.857, P = .94, paired test) and were correlated (rho = 0.44, P < .001). Men had higher IgAN237 (P = .037), lower baseline eGFR and more declined Total and Chronic eGFR slopes (P ≤ .020) than women both for IgAN237-1 and IgAN237-2 analyses.
Table 2:
eGFR slopes for participants with eGFR 15–90 mL/min/1.73 m2.
| N | Mean | SD | Median (range) | |
|---|---|---|---|---|
| Baseline (at IgAN237-1) | ||||
| eGFR (mL/min/1.73 m2) | 74 | 48.4 | 19.4 | 47.9 (16.6 to 86.5) |
| eGFRTotal-slope (mL/min/1.73 m2/year) | 70 | –0.2 | 3.5 | –0.5 (–8.0 to 13.8) |
| eGFRChron-slope (mL/min/1.73 m2/year) | 70 | –0.9 | 3.4 | –0.8 (–8.9 to 16.7) |
| eGFRProsp-slope (mL/min/1.73 m2/year) | 70 | –1.4 | 7.1 | –1.1 (–19.8 to 24.1) |
| eGFR180days-slope (mL/min/1.73 m2/year) | 70 | –3.2 | 6.8 | –2.8 (–21.8 to 20.5) |
| Follow-up (at IgAN237-2) | ||||
| eGFR (mL/min/1.73 m2) | 57 | 47.9 | 20.4 | 45.6 (16.7 to 89.2) |
| eGFRTotal-slope (mL/min/1.73 m2/year) | 55 | 0.3 | 3.7 | –0.5 (–7.0 to 13.8) |
| eGFRChron-slope (mL/min/1.73 m2/year) | 55 | –0.8 | 3.5 | –0.8 (–8.1 to 16.7) |
| eGFRProsp-slope (mL/min/1.73 m2/year) | 53 | 2.6 | 14.4 | 0.3 (–42.5 to 62.2) |
| eGFR180days-slope (mL/min/1.73 m2/year) | 53 | –0.1 | 8.5 | –0.5 (–22.2 to 18.2) |
N = number of patients with available analyses; SD = standard deviation.
Both IgAN237-1 and IgAN237-2 were correlated with the percentage of glomerular sclerosis (rho = 0.250, P = .021, and rho = 0.389, P < .001, respectively) and UACR (rho = 0.311, P = .002, and rho = 0.341, P = .006, respectively). IgAN237-2 was also correlated with baseline UACR (rho = 0.341, P = .006) and UACRTotal-slope (rho = 0.226, P = .037).
Dynamics of IgAN237-based identification of progressors and non-progressors over time
IgAN237 value was >0.38 in 29/103 (28%) of baseline samples and in 23/89 (26%) of follow-up samples, identifying them as progressors. Categorization was concordant for both time-points in 73/89 (82%) patients (17 progressor/progressor and 56 non-progressor/non-progressor). Ten of 89 patients (11%) changed from progressor to non-progressor and 6/89 (7%) from non-progressor to progressor.
IgAN237 decreased more from baseline to the follow-up sample in those categorized as progressors at baseline (n = 27) than in non-progressors (n = 62): –0.469 ± 0.864 vs 0.127 ± 0.794 (P = .002). The shift from progressor to non-progressor was associated with a negative change in IgAN237 of –1.173 ± 0.971 (P = .005), while shift from non-progressor to progressor was associated with a positive change in IgAN237 of +0.797 ± 0.404 (P = .028).
eGFR slopes
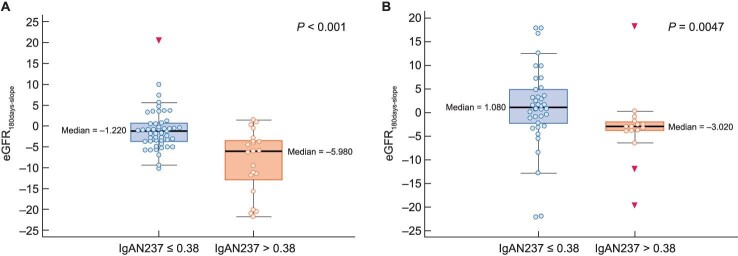
eGFR slopes were calculated in participants with baseline eGFR values 15–90 mL/min/1.73 m2, i.e. in 70 participants with baseline urine sample and in 53–55 with follow-up urine sample and available data (Table 2). In this population, participants categorized at baseline as progressors (IgAN237-1 >0.38) were younger (mean 53 versus 60 years, P = .036) (Table 3A) and showed more declined (worsened) eGFRTotal-slope (mean –1.92 vs 0.57, P = .010), eGFRChron-slope (mean –2.73 vs –0.15, P = .003) and eGFR180days-slope (mean –8.47 vs –0.91, P < .001) than those categorized as non-progressors (Table 3A, Fig. 2A). Progressors also had higher baseline UACR (P < .001) and a more increased (worsened) UACR180days-slope (P = .019) (Table 3A). In a binary logistic stepwise regression analysis, the IgAN237-1 (progressor versus non-progressor) was related to the eGFR180days-slope (P < .001).
Table 3:
Characteristics of patients categorized as progressors or non-progressors according to IgAN237s and eGFR values at baseline between 15 and 90 mL/min/1.73 m2.
| Non-progressors | Progressors | T-test | Mann–Whitney test | |||||
|---|---|---|---|---|---|---|---|---|
| Variables | N | Mean | SD | N | Mean | SD | P-value | P-value |
| (A) Progressors and non-progressors at baseline | ||||||||
| IgAN237-1 | 52 | –0.418 | 0.586 | 22 | 0.863 | 0.455 | <.001 | <.001 |
| Male (%) | 52 | 56 | 22 | 77 | .067* | |||
| Age at diagnosis (1st biopsy) (years) | 52 | 48.1 | 16.0 | 22 | 46.1 | 14.7 | .601 | .696 |
| Age at baseline (IgAN237-1) (years) | 52 | 59.7 | 14.0 | 22 | 52.7 | 13.4 | .052 | .036 |
| Baseline eGFR (mL/min/1.73 m2) | 52 | 49.9 | 17.9 | 22 | 44.7 | 22.7 | .292 | .208 |
| Baseline UACR (g/mol) | 48 | 43.0 | 97.8 | 20 | 86.6 | 75.3 | .08 | <.001 |
| Hematuria [N (%)] | 45 | 47% | 18 | 44% | .549* | |||
| Number of antihypertensive drugs at baseline (N) | 52 | 2.1 | 1.4 | 22 | 2.3 | 1.3 | .621 | .534 |
| Biopsy: glomerular sclerosis (% of glomeruli) | 38 | 19.3 | 14.4 | 21 | 27.3 | 17.4 | .065 | .069 |
| Biopsy: interstitial fibrosis (% of kidney surface) | 43 | 25.9 | 18.6 | 22 | 30 | 13.2 | .357 | .439 |
| Biopsy: crescents (% of biopsies with crescents) | 41 | 29% | 22 | 18% | .258* | |||
| eGFRTotal-slope (mL/min/1.73 m2/year) | 49 | 0.57 | 3.54 | 21 | –1.92 | 2.82 | .006 | .01 |
| eGFRChron-slope (mL/min/1.73 m2/year) | 49 | –0.15 | 3.38 | 21 | –2.73 | 2.80 | .003 | .003 |
| eGFRProsp-slope (mL/min/1.73 m2/year) | 49 | –0.73 | 6.60 | 21 | –3.12 | 7.98 | .197 | .04 |
| eGFR180days-slope (mL/min/1.73 m2/year) | 49 | –0.91 | 5.07 | 21 | –8.47 | 7.59 | <.001 | <.001 |
| UACRTotal-slope (g/mol/year) | 48 | –48.9 | 148 | 21 | –0.03 | 7.66 | .136 | .064 |
| UACRChron-slope (g/mol/year) | 48 | –8.56 | 35.3 | 21 | 2.1 | 11.7 | .179 | .091 |
| UACRProsp-slope (g/mol/year) | 46 | –14.7 | 47.5 | 18 | –3.0 | 49.2 | .383 | .66 |
| UACR180days-slope (g/mol/year) | 46 | –31.5 | 160 | 18 | 25.1 | 65.0 | .153 | .019 |
| (B) Progressors and non-progressors at follow-up | ||||||||
| IgAN237-1 (value) | 41 | –0.406 | 0.566 | 16 | 0.819 | 0.319 | <.001 | <.001 |
| Sex (male, %) | 41 | 56% | 16 | 69% | .285* | |||
| Age at diagnosis (1st biopsy) (years) | 41 | 51.3 | 15.4 | 16 | 40.9 | 13.3 | .022 | .015 |
| Age at follow-up (IgAN237-1) (years) | 41 | 62.9 | 10.8 | 16 | 51.5 | 15.1 | .002 | .011 |
| Follow-up eGFR (mL/min/1.73 m2) | 41 | 49.3 | 20.6 | 16 | 44.2 | 20.0 | .402 | .36 |
| Follow-up UACR (g/mol) | 35 | 25.2 | 29.8 | 13 | 71.0 | 59.2 | .018 | .003 |
| Hematuria [N (%)] | 37 | 41% | 13 | 46% | .486* | |||
| Number of antihypertensive drugs at baseline (N) | 41 | 2.05 | 1.26 | 16 | 1.75 | 1.18 | .418 | .337 |
| Biopsy: glomerular sclerosis (% of glomeruli) | 33 | 18.9 | 15.3 | 13 | 28.2 | 17.3 | .08 | .067 |
| Biopsy: interstitial fibrosis (% of kidney surface) | 36 | 24.5 | 18.1 | 13 | 28.2 | 18.3 | .531 | .497 |
| Biopsy: crescents (% of biopsies with crescents) | 34 | 29% | 13 | 23% | .482* | |||
| eGFRTotal-slope (mL/min/1.73 m2/year) | 40 | 0.65 | 4.01 | 15 | –0.70 | 2.77 | .238 | .303 |
| eGFRChron-slope (mL/min/1.73 m2/year) | 40 | –0.18 | 3.74 | 15 | –2.29 | 2.18 | .045 | .014 |
| eGFRProsp-slope (mL/min/1.73 m2/year) | 38 | 1.60 | 15.6 | 15 | 5.11 | 11.3 | .431 | .813 |
| eGFR180days-slope (mL/min/1.73 m2/year) | 38 | 1.15 | 8.61 | 15 | –3.20 | 7.74 | .095 | .005 |
| UACRTotal-slope (g/mol/year) | 40 | –42.4 | 141 | 16 | –8.42 | 37.5 | .349 | .095 |
| UACRChron-slope (g/mol/year) | 40 | –5.72 | 16.4 | 16 | 3.14 | 12.9 | .058 | .157 |
| UACRProsp-slope (g/mol/year) | 33 | 9.74 | 61.4 | 13 | 11.4 | 57.2 | .935 | .76 |
| UACR180days-slope (g/mol/year) | 33 | –12.8 | 48.9 | 13 | 21.1 | 51.1 | .043 | .095 |
N = number; SD = standard deviation; * = Fisher's exact test.
Figure 2:
Boxplot of comparisons of progressors (IgAN237 >0.38) vs non-progressors (IgAN237 <0.38) and eGFR180days-slope in IgAN237-1 (A) and IgAN237-2 (B).
Participants categorized at follow-up as progressors based on the IgAN237-2 were also younger (mean 51.5 versus 63 years, P = .011) and had more declining eGFRChron-slope (mean –2.3 vs –0.18, P = .014) and eGFR180days-slope (mean –3.2 vs 1.1, P = .005) (Table 3B, Fig. 2B).
As expected, baseline UACR was related to eGFR (rho = –0.326, P = .007), eGFRChron-slope (rho = –0.338, P = .005), eGFRProsp-slope (rho = –0.375, P = .002) and eGFR180days-slope (rho = –0.418, P < .001) (Supplementary data, Table S4A and S4B). Furthermore, the UACRTotal-slope was related to all eGFR slopes (rho > –0.301, P ≤ .012).
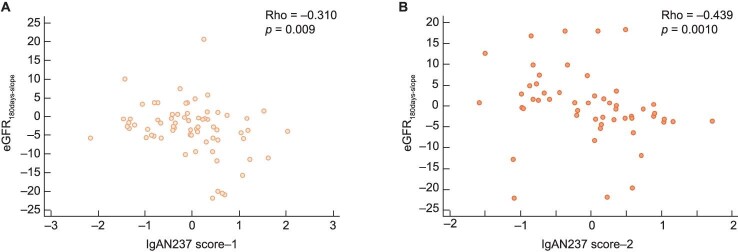
Both baseline IgAN237-1 and -2 were inversely related to their respective eGFRChron-slope (rho = –0.278, P = .020, and rho = –0.409, P = .002, respectively) and eGFR180days-slope (rho = –0.310, P = .009, and rho = –0.439, P = .001, respectively) (Fig. 3A and B and Supplementary data, Table S4A and B).
Figure 3:
Correlation between IgAN237-1 (A) and IgAN237-2 (B) with eGFR180days-slope.
For participants with baseline IgAN237 data, the eGFR180days-slope was the most declining slope and further analyses assessed predictors of this slope (see below).
Predictors of eGFR slopes
To evaluate whether there were specific variables that could predict progression, multiple regression analysis was performed for baseline data, given that the number of participants was larger at this stage, with the most declining slope, eGFR180days-slope as dependent variable. The variables sex, age at baseline, eGFR at baseline, UACR at baseline, biopsy glomerulosclerosis and interstitial fibrosis were tested in the first model. Baseline age was an independent predictor (P = .045) of eGFR180days-slope (Supplementary data, Table S5). When adding the variable progressor/non-progressor according to IgAN237 to the multiple regression analysis, the baseline progressor/non-progressor status according to IgAN237-1 was the only independent predictor of eGFR180days-slope (P = .001) (Supplementary data, Table S6).
Those with primary versus secondary IgAN were younger at time of diagnosis (P = .023) but did not differ in IgAN237-1 and IgAN237-2, eGFR value, eGFR slopes, antihypertensive medication, glomerular sclerosis or tubular atrophy (interstitial fibrosis). Those with IgAV had a lower UACR (13 ± 24 vs 62 ± 98, P = .006) and a lower extent of glomerular sclerosis (10 ± 12 vs 24 ± 16, P = .009) compared with those with IgAN, but there were no differences in IgAN237-1 and IgAN237-2, eGFR value, eGFR slopes, antihypertensive medication or tubular atrophy.
DISCUSSION
In a previous study, we showed that a high value of IgAN237 provided prognostic information for IgAN progression over a 3-year period when analyzed at the time of kidney biopsy [16]. To our knowledge, the present study is the first to present prospective clinical data that could verify that high IgAN237 also provides prognostic information when analyzed later in the course of the disease, i.e. a median of >5 years after initial treatment had been provided based on kidney biopsy results and risk of progression at the time of kidney biopsy. This implies that IgAN237 may eventually be tested as a risk stratification tool for randomized controlled trials that enroll patients who were diagnosed with IgAN in the past but have not had a recent kidney biopsy and are no longer treatment naïve.
There was no obvious difference in classifier levels of IgAN versus IgAV or between primary and secondary IgAN. Thus, the prognostic value of IgAN237 seems to cover the different subtypes of IgAN. Two timepoints were studied for IgAN237 and prediction of outcomes was tested for the first IgAN237 value, as a higher number of participants was available and the follow-up after the second timepoint was short. There was a correlation between IgAN237-1 and IgAN237-2 and no significant change over time was noted in the whole group, supporting the stability of the biomarker for most participants. The magnitude of the change from IgAN237-1 to IgAN237-2 was inversely related to the baseline data, which indicates more active pathophysiological processes and improvement in the most severe cases, potentially as a result of treatment or natural history flare and remission cycles.
Notably, almost 30% of patients had an IgAN237 of >0.38, indicating a progressive disease, at both sampling timepoints, suggesting that they might benefit from change of therapeutic approach or enrollment in clinical trials for high-risk patients. However, the study was not designed to address the response of IgAN237 to therapy. Some patients converted from progressors into non-progressors, and vice versa, which indicates a dynamic disease. Based on these results, we support regular follow-up at least for laboratory values of these patients at least every sixth month. The use of the IgAN237 classifier as an additional risk stratification tool may help when prognostic uncertainties exist; factors such as the presence of severe permanent kidney damage already at time of biopsy, male sex and age [23–25] are not modifiable by therapy.
Among limitations, the sample size, although large for a prospective IgAN study, may have been more informative if larger. In this regard, recent clinical trials have enrolled a far larger number of participants than prior IgAN trials [7, 8] and offer the opportunity to test the response of IgAN237 to therapy if biobanked samples can be accessed. Moreover, GFR was estimated and not measured. The estimation of GFR is here based on serum creatinine which is influenced not only by age, sex and race but also by diet and variables such as muscle mass, wasting and fluid intake. However, the manuscript also has strengths, as data were prospectively assessed it is a relative large multinational and multicenter IgAN population that was heterogenous from several points of view, ranging from time to biopsy to local treatment practices to the presence of coexistent conditions or vasculitis features. This heterogeneity, the lack of overlap between individual participants in the current and prior IgAN237 study, and just partial overlap of participating centers and countries increase the external validity of the data. However, data on race were not available and it is likely that non-Caucasians were underrepresented. In this regard, validation in Asian or US cohorts would be desirable. Although there is no significant difference between IgAV and IgAN in IgAN237 classifier and eGFR slopes we acknowledge a limitation of the small subsets in the present study and recommend larger studies of the separate groups.
In conclusion, the present study demonstrates that the IgAN237 classifier may provide predictive information for progressive kidney disease in IgAN patients, not only at the time of biopsy [16] but also several years post-biopsy, after treatment had already been prescribed based on initial biopsy results and risk evaluation that may have changed the course of the disease. This makes the IgAN237 classifier clinically useful for risk stratification in different clinical contexts, such as enrolment of high-risk patients in clinical trials to eventual decisions on initiation and/or modification of treatment and follow-up for IgAN patients in a personalized manner. Prospective studies should explore IgAN237 performance for each clinical context.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the patients who participated, the staff that performed data collection enabling the present study and the members of the PersTIgAN Working Group. Thanks also to the Clinical Research Center in Umeå, Sweden and the Biobank Northern Sweden for the data catch and centralization, collection, distributing and shipping the urine samples to Mosaiques Diagnostics GmbH in Germany. Thanks to Dr Michael Ott, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden and Prof. Dr Gregor Guron, Department of Molecular and Clinical Medicine, Institute of Medicine, the Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden for recruiting of patients and scientific discussion.
APPENDIX
PersTIgAN working group:
Division of Nephrology and KfH Renal Unit, Hospital St Georg, Leipzig, Germany: Joachim Beige, Ralph Wendt, Ulrike Schmidt
Mosaiques Diagnostics GmbH, Hannover, Germany: Justyna Siwy, Petra Zürbig, Harald Mischak, Annika Durban, Julia Raad, Igor Golovko
University of Toronto and Toronto General Research Institute, Toronto, Canada: Heather Reich, Ping Lam, Stuart Yang
Instituto de Investigación Sanitaria Fundación Jiménez Díaz (IIS-FJD UAM), Madrid, Spain: Ana Belen Sanz, Beatriz Fernandez-Fernandez, Jorge Enrique Rojas-Rivera, Maria Vanessa Perez-Gomez, Alberto Ortiz, Maria Dolores Sanchez-Niño, Jinny Sanchez-Rodriguez
Medical University Innsbruck, Innsbruck, Austria: Michael Rudnicki, Julia Kerschbaum, Johannes Leierer, Gert Mayer
Umea University, Dept Public Health and Clinical Medicine, Umea, Sweden: Bernd Stegmayr
Skaraborg Hospital, Department of Nephrology, Skövde, Sweden: Björn Peters
Notes
Members of the PersTIgAN Working Group are listed in the Appendix.
Contributor Information
Björn Peters, Department of Molecular and Clinical Medicine, Institute of Medicine, the Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden; Department of Nephrology, Skaraborg Hospital, Skövde, Sweden.
Joachim Beige, Kuratorium for Dialysis and Transplantation, Neu Isenburg/Leipzig, Germany; Division of Nephrology, Rheumatology and Endocrinology, Martin-Luther University Halle-Wittenberg, Halle/Saale, ., Germany.
Justyna Siwy, Mosaiques diagnostics GmbH, Hannover, Germany.
Michael Rudnicki, Department of Internal Medicine IV – Nephrology and Hypertension, Medical University Innsbruck, Innsbruck, Austria.
Ralph Wendt, Division of Nephrology and KfH Renal Unit, Hospital St Georg, Leipzig, Germany.
Alberto Ortiz, Instituto de Investigación Sanitaria Fundación Jiménez Díaz (IIS-FJD UAM), Madrid, Spain.
Ana Belen Sanz, Instituto de Investigación Sanitaria Fundación Jiménez Díaz (IIS-FJD UAM), Madrid, Spain.
Harald Mischak, Mosaiques diagnostics GmbH, Hannover, Germany.
Heather N Reich, Department of Medicine, University of Toronto and Division of Nephrology, University Health Network, Toronto, Ontario, Canada; Gabor Zellerman Chair in Nephrology Research, University of Toronto, Toronto, Ontario, Canada.
Salmir Nasic, Department of Molecular and Clinical Medicine, Institute of Medicine, the Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden; Research and Development Centre at Skaraborg Hospital, Skövde, Sweden.
Dana Mahmood, Department of Public Health and Clinical Medicine, Unit Östersund, Umeå University, Umea, Sweden.
Anders Persson, Department of Public Health and Clinical Medicine, Unit Sundsvall, Umeå University, Umea, Sweden.
Anders Fernström, Department of Nephrology and Department of Health, Medicine and Caring Sciences, Linköping University, Linköping, Sweden.
Maria Weiner, Department of Nephrology and Department of Health, Medicine and Caring Sciences, Linköping University, Linköping, Sweden.
Bernd Stegmayr, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
the PersTIgAN Working Group:
Joachim Beige, Ralph Wendt, Ulrike Schmidt, Justyna Siwy, Petra Zürbig, Harald Mischak, Annika Durban, Julia Raad, Igor Golovko, Heather Reich, Ping Lam, Stuart Yang, Ana Belen Sanz, Beatriz Fernandez-Fernandez, Jorge Enrique Rojas-Rivera, Maria Vanessa Perez-Gomez, Alberto Ortiz, Maria Dolores Sanchez-Niño, Jinny Sanchez-Rodriguez, Michael Rudnicki, Julia Kerschbaum, Johannes Leierer, Gert Mayer, Bernd Stegmayr, and Björn Peters
FUNDING
This work was supported by the ERA-NET PerMed programme (Ref. No. ERAPERMED2018-217 PersTIgAN) co-funded by the European Commission and the national funding agencies (see below). Germany (J.S., R.W., H.M. and J.B.): this project was supported by the Federal Ministry of Education and Research (BMBF) under grant number 01KU1922A for H.M. and J.S. and 01KU1922B 9899073 for R.W. and J.B. Spain (A.O., A.B.S. and M.V.P.-G.): these authors are supported by FIS/FEDER funds under grant number AC18/00 071 and AC18/00064. Salary support Ramon y Cajal program to A.B.S. Austria (M.R., J.K. and J.L.): this project was funded by the Austrian Science Fund under grant number I 4239-B to M.R. Sweden (B.P. and B.S.): this project was funded by the Swedish Research Council (2018-05615) and the Research Fund (FoU) at Skaraborg Hospital, Skövde, Sweden (VGSKAS-930079, 939 545, 968 165, 981 343). Canada (H.N.R.): this project was supported by the Canadian Institutes of Health Research through the ERA-Net PerMed initiative to H.N.R. H.N.R.’s work is funded by the Gabor Zellerman Chair in Nephrology Research at the University of Toronto.
CONFLICT OF INTEREST STATEMENT
J.S. is employed by Mosaiques Diagnostics GmbH. R.W. reports personal advisory and/or lecture fees from AstraZeneca, CSL Vifor and Boehringer Ingelheim outside the submitted work. A.O. has received grants from Sanofi and consultancy or speaker fees or travel support from Advicciene, Astellas, AstraZeneca, Amicus, Amgen, Boehringer Ingelheim, Fresenius Medical Care, GSK, Bayer, Sanofi-Genzyme, Menarini, Mundipharma, Kyowa Kirin, Lilly, Alexion, Freeline, Idorsia, Chiesi, Otsuka, Novo-Nordisk, Sysmex and Vifor Fresenius Medical Care Renal Pharma, and is Director of the Catedra Mundipharma-UAM of diabetic kidney disease and the Catedra Astrazeneca-UAM of chronic kidney disease and electrolytes outside the submitted work. H.M. is the co-founder and co-owner of Mosaiques Diagnostics GmbH. H.N.R. reports other financial activities from Calliditas and from Omeros outside the submitted work. All other authors have nothing to disclose. The results presented in this article have not been published previously in whole or part, except in abstract format.
DATA AVAILABILITY STATEMENT
The data underlying this article will be shared on reasonable request to the corresponding author.
REFERENCES
- 1. Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med 2013;368:2402–14. 10.1056/NEJMra1206793 [DOI] [PubMed] [Google Scholar]
- 2. Saha MK, Julian BA, Novak J et al. Secondary IgA nephropathy. Kidney Int 2018;94:674–81. 10.1016/j.kint.2018.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cattran DC, Coppo R, Cook HT et al. The Oxford classification of IgA nephropathy: rationale, clinicopathological correlations, and classification. Kidney Int 2009;76:534–45. 10.1038/ki.2009.243 [DOI] [PubMed] [Google Scholar]
- 4. Trimarchi H, Barratt J, Cattran DC et al. Oxford Classification of IgA nephropathy 2016: an update from the IgA Nephropathy Classification Working Group. Kidney Int 2017;91:1014–21. 10.1016/j.kint.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 5. Rovin BH, Adler SG, Barratt J et al. Executive summary of the KDIGO 2021 Guideline for the Management of Glomerular Diseases. Kidney Int 2021;100:753–79. 10.1016/j.kint.2021.05.015 [DOI] [PubMed] [Google Scholar]
- 6. Barbour SJ, Coppo R, Zhang H et al. Application of the International IgA Nephropathy Prediction Tool one or two years post-biopsy. Kidney Int 2022;102:160–72. 10.1016/j.kint.2022.02.042 [DOI] [PubMed] [Google Scholar]
- 7. Wheeler DC, Toto RD, Stefansson BV et al. A pre-specified analysis of the DAPA-CKD trial demonstrates the effects of dapagliflozin on major adverse kidney events in patients with IgA nephropathy. Kidney Int 2021;100:215–24. 10.1016/j.kint.2021.03.033 [DOI] [PubMed] [Google Scholar]
- 8. Group E-KC, Herrington WG, Staplin N et al. Empagliflozin in patients with chronic kidney disease. N Engl J Med 2022;388:117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lv J, Wong MG, Hladunewich MA et al. Effect of oral methylprednisolone on decline in kidney function or kidney failure in patients with IgA nephropathy: the TESTING randomized clinical trial. JAMA 2022;327:1888–98. 10.1001/jama.2022.5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barratt J, Lafayette R, Kristensen J et al. Results from part A of the multi-center, double-blind, randomized, placebo-controlled NefIgArd trial, which evaluated targeted-release formulation of budesonide for the treatment of primary immunoglobulin A nephropathy. Kidney Int 2023;103:391–402. 10.1016/j.kint.2022.09.017 [DOI] [PubMed] [Google Scholar]
- 11. Tofte N, Lindhardt M, Adamova K et al. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): a prospective observational study and embedded randomised placebo-controlled trial. Lancet Diabetes Endocrinol 2020;8:301–12. 10.1016/S2213-8587(20)30026-7 [DOI] [PubMed] [Google Scholar]
- 12. Pontillo C, Mischak H. Urinary peptide-based classifier CKD273: towards clinical application in chronic kidney disease. Clin Kidney J 2017;10:192–201. 10.1093/ckj/sfx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodriguez-Ortiz ME, Pontillo C, Rodriguez M et al. Novel urinary biomarkers for improved prediction of progressive eGFR loss in early chronic kidney disease stages and in high risk individuals without chronic kidney disease. Sci Rep 2018;8:15940. 10.1038/s41598-018-34386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haubitz M, Wittke S, Weissinger EM et al. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int 2005;67:2313–20. 10.1111/j.1523-1755.2005.00335.x [DOI] [PubMed] [Google Scholar]
- 15. Julian BA, Wittke S, Novak J et al. Electrophoretic methods for analysis of urinary polypeptides in IgA-associated renal diseases. Electrophoresis 2007;28:4469–83. 10.1002/elps.200700237 [DOI] [PubMed] [Google Scholar]
- 16. Rudnicki M, Siwy J, Wendt R et al. Urine proteomics for prediction of disease progression in patients with IgA nephropathy. Nephrol Dial Transplant 2021;37:42–52. 10.1093/ndt/gfaa307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siwy J, Zurbig P, Argiles A et al. Noninvasive diagnosis of chronic kidney diseases using urinary proteome analysis. Nephrol Dial Transplant 2017;32:2079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levey AS, Stevens LA, Schmid CH et al. A new equation to estimate glomerular filtration rate. Ann Intern Med 2009;150:604–12. 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inker LA, Heerspink HJL, Tighiouart H et al. GFR slope as a surrogate end point for kidney disease progression in clinical trials: a meta-analysis of treatment effects of randomized controlled trials. J Am Soc Nephrol 2019;30:1735–45. 10.1681/ASN.2019010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Earley A, Miskulin D, Lamb EJ et al. Estimating equations for glomerular filtration rate in the era of creatinine standardization: a systematic review. Ann Intern Med 2012;156:785–95. 10.7326/0003-4819-156-11-201203200-00391 [DOI] [PubMed] [Google Scholar]
- 21. Latosinska A, Siwy J, Mischak H et al. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: the past, the present, and the future. Electrophoresis 2019;40:2294–308. [DOI] [PubMed] [Google Scholar]
- 22. Jantos-Siwy J, Schiffer E, Brand K et al. Quantitative urinary proteome analysis for biomarker evaluation in chronic kidney disease. J Proteome Res 2009;8:268–81. 10.1021/pr800401m [DOI] [PubMed] [Google Scholar]
- 23. Goto M, Wakai K, Kawamura T et al. A scoring system to predict renal outcome in IgA nephropathy: a nationwide 10-year prospective cohort study. Nephrol Dial Transplant 2009;24:3068–74. 10.1093/ndt/gfp273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Audemard-Verger A, Pillebout E, Baldolli A et al. Impact of aging on phenotype and prognosis in IgA vasculitis. Rheumatology (Oxford) 2021;60:4245–51. 10.1093/rheumatology/keaa921 [DOI] [PubMed] [Google Scholar]
- 25. Komatsu H, Fujimoto S, Maruyama S et al. Distinct characteristics and outcomes in elderly-onset IgA vasculitis (Henoch-Schonlein purpura) with nephritis: nationwide cohort study of data from the Japan Renal Biopsy Registry (J-RBR). PLoS One 2018;13:e0196955. 10.1371/journal.pone.0196955 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.