Abstract
The elimination of seed shattering was a key step in rice (Oryza sativa) domestication. In this paper, we show that increasing the gibberellic acid (GA) content or response in the abscission region enhanced seed shattering in rice. We demonstrate that SLENDER RICE1 (SLR1), the key repressor of GA signaling, could physically interact with the rice seed shattering-related transcription factors quantitative trait locus of seed shattering on chromosome 1 (qSH1), O. sativa HOMEOBOX 15 (OSH15), and SUPERNUMERARY BRACT (SNB). Importantly, these physical interactions interfered with the direct binding of these three regulators to the lignin biosynthesis gene 4-COUMARATE: COENZYME A LIGASE 3 (4CL3), thereby derepressing its expression. Derepression of 4CL3 led to increased lignin deposition in the abscission region, causing reduced rice seed shattering. Importantly, we also show that modulating GA content could alter the degree of seed shattering to increase harvest efficiency. Our results reveal that the “Green Revolution” phytohormone GA is important for regulating rice seed shattering, and we provide an applicable breeding strategy for high-efficiency rice harvesting.
DELLA protein SLR1 interacts with 3 shattering transcription factors (qSH1, OSH15, and SNB) to alter lignin biosynthesis and regulate seed shattering in rice (Oryza sativa).
IN A NUTSHELL.
Background: Rice is one of the most important grain crops worldwide. Rice yield is determined by many factors, such as tiller number, grain weight, and number of grains. Among these factors, ideal seed shattering level and plant height reduction are 2 important biological traits. We all know that seed shattering degree and plant height are affected to natural or artificial selection during rice domestication. And, successful selection of rice semidwarf gibberellic acid (GA) biosynthesis mutants led to “Green Revolution.” But, there is very little evidence for a relationship between GA and seed shattering up to now. In this article, we found that several GA-related genes contribute to seed shattering regulation.
Question: By what way does endogenous GA affect rice seed shattering?
Findings: We showed that increased GA content or signaling in rice abscission region could enhance seed shattering. Then, we expressed GA biosynthesis gene Dwarf 18 and metabolic genes GA 2-oxidases 1 under abscission layer specific promoter Shattering Abortion 1; the shattering degree of these transgenic lines was obviously changed by influencing the lignin content in abscission region. And, core GA signaling regulator, Slender Rice 1 (SLR1), could directly interact with 3 rice shattering-related transcription factors, quantitative trait locus of seed shattering on chromosome 1 (qSH1), Oryza sativa HOMEOBOX 15 (OSH15), and SUPERNUMERARY BRACT (SNB). We also characterized that qSH1, OSH15, and SNB bind to the promoter of a lignin biosynthesis gene 4-COUMARATE: COENZYME A LIGASE 3 (4CL3) and repress its expression, which cause an easy shattering phenotype. Interaction between SLR1 and qSH1, OSH15 and SNB could interrupt their binding abilities to 4CL3 promoter, leading to more lignin deposition and causing reduced rice shattering.
Next steps: Our findings reveal a rough network on how GA regulates rice seed shattering. Based on this, we would like to characterize more about natural variation of GA-related genes in shattering differences among rice subpopulations and aim to achieve elite alleles with ideal shattering to improve yields.
Introduction
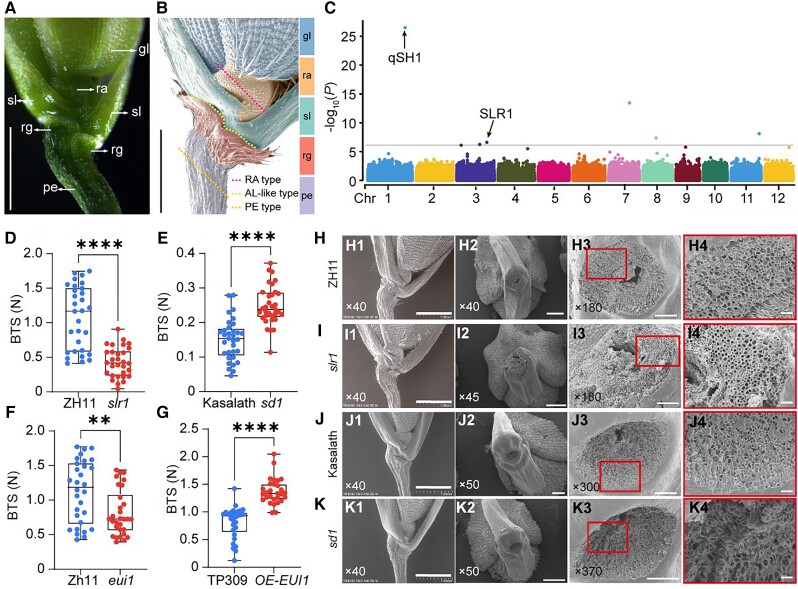
The loss of strong seed shattering was a key step in the transition of wild rice species to commonly cultivated rice during the rice domestication process. Rice shattering is correlated with abscission layer (AL) establishment with 2 or 3 layers of cells. In easy shattering rice varieties, the AL is formed between sterile lemmas and rudimentary glumes at stage 7 (Sp7) in rice spikelet development (Fig. 1, A and B), which occurs 16 to 20 d before heading when the panicles are 5 to 30 mm long (Itoh et al. 2005; Ji et al. 2010; Yu et al. 2020). As for the rice varieties without AL, seed shattering may occur in three places at the basal part of the spikelet as partially described in Yoon et al. (2014), including (i) rachilla type (RA-type), broken at rachilla, (ii) pedicel type (PE-type), broken under the AL and on the pedicel, (iii) AL-like type, broken at a position similar to AL (Fig. 1, A and B).
Figure 1.
Gibberellin contributed to rice seed-shattering regulation. A) and B) Structure diagram of rice spikelet base. Photographs A) and B) were taken by stereo microscope and SEM separately. The dashed line in B) represents three types of breaks positions in Nip. Bars = 1 mm. C) Manhattan plot demonstrating −log10 (P-values) from a genome-wide scan plotted against the position on each of the 12 chromosomes (Chr). Known genes within the 200 kb regions flanked with the associated SNPs are indicated by arrows. D to G) BTS of gibberellin (GA)-related mutants and an overexpression line. Boxplots of BTS comparing ZH11 and slr1D), Kasalath and sd1E), ZH11 and eui1F), and TP309 and EUI1 overexpression line G). Data are displayed as box and whisker plots with individual data points. Horizontal bars represent the maximum, third quantile, median, first quantile, and minimum values, respectively. **P-value ≤0.01 and ****P-value ≤0.0001 calculated from a two-tailed t-test. H to K) Characterization of rice spikelet in slr1I), sd1K), and the corresponding wild-type ZH11 H) and Kasalath J). H1) to K1) The SEM photographs of the spikelet basal part. H2) to K2) The SEM photographs of the broken area on mature seeds. H3) to K3) Close-up SEM photographs of the fracture surface corresponding to H2) to K2) separately. H4) to K4) are magnifications of the red boxes in H3) to K3), respectively. The numbers in the bottom left corner of the photos indicated the magnification. Bars = 1 mm in panels (1), 500 μm in panels (2), 100 μm in panel (3), and 20 μm in panel (4). gl, glume (blue); ra, rachillag (orange); sl, sterile lemma (green); rg, rudimentary glume (red); pe, pedicel (purple).
Rice seed shattering is also an important agronomic trait; easy shattering causes a reduction in yields, while it is difficult to harvest cultivated rice species whose seeds do not shatter (Ji et al. 2006; Wu et al. 2023). Previous studies have revealed several genetic factors controlling AL formation or seed shattering among different rice varieties. Mutations of an MYB transcription factor at a seed shattering-related quantitative trait locus (QTL) on chromosome 4 (sh4)/Shattering 1 (SHA1; an allele of sh4) lead to a nonshattering phenotype of both Asian (Oryza sativa) and African (Oryza glaberrima) cultivated rice, which were domesticated from two wild rice species, Oryza rufipogon (Li et al. 2006) and Oryza barthii (Sweeney and McCouch 2007; Wang, Yu, et al. 2014; Meyer et al. 2016), respectively.
Generally, indica-type rice (O. sativa L. subsp. indica Kato) seeds are easy to abscise because they are completely or partially AL-formed, such as Nanjing 11 (Lin et al. 2012). In contrast, japonica-type rice (O. sativa L. subsp. japonica Kato) grains are difficult to shed from the pedicel due to no obvious AL formation, like Nipponbare (Konishi et al. 2006). Some exceptions like moderate-shattering Taipei 309 belong to japonica-type rice but had incomplete AL (Li et al. 2006). A single-nucleotide polymorphism (SNP) in the 5′ regulatory region of QTL of seed shattering on chromosome 1 (qSH1), which harbors a BEL1-type homeobox gene that is a rice orthologue of REPLUMLESS (RPL) in Arabidopsis (Arabidopsis thaliana), causes loss of seed shattering owing to the absence of AL formation. This SNP explained 68.6% of the shattering difference between indica and temperate japonica cultivars (Konishi et al. 2006).
Two genes that encode APETALA2 transcription factors, namely SHATTERING ABORTION 1 (SHAT1) and SUPERNUMERARY BRACT (SNB), are required for rice seed shattering for their involvement in specifying AL development (Zhou et al. 2012; Jiang et al. 2019). Oryza sativa HOMEOBOX15 (OSH15) and SH5 form a heterodimer to enhance seed shattering by directly inhibiting lignin biosynthesis-related genes in AL (Yoon et al. 2014, 2017). Recently, Ning et al. (2023) demonstrated that African cultivated rice showed significantly reduced seed shattering by knockout of SH11 because OgSH11 represses the expression of lignin biosynthesis genes and lignin deposition by binding to the promoter of gold hull and internode-2 (GH2)/cinnamyl alcohol dehydrogenase 2 (CAD2). Therefore, rice seed shattering-related gene mutants (sh5, osh15, sh11, etc.) that present ectopic lignification in the abscission region exhibit increased tensile strength of tissues that maintain the attachment of the seeds to the plants and a reduced degree or loss of seed abscission.
Gibberellic acid (GA) is widely known as the “Green Revolution” phytohormone (Hedden 2003). The successful selection of semidwarf rice cultivars has increased rice production significantly since the 1960s. Semidwarf 1 (SD1), which is known as the “Green Revolution gene,” encodes GA20-oxidase 2 (GA20ox2), which is the key enzyme in the gibberellin biosynthesis pathway (Sasaki et al. 2002; Spielmeyer et al. 2002; Sakai et al. 2003). Dwarf 18 (D18) encodes GA3ox2, which exhibits 3β-hydroxylase activity (Itoh et al. 2001; Hu, Hu, et al. 2018). Mutations in D18 and SD1 lead to significantly decreased endogenous gibberellin contents and plant height. In contrast, GA 2-oxidases (GA2oxs) (Martins et al. 2018; Bertolotti et al. 2021) and elongated uppermost internode-1 (EUI1) products (Sakamoto et al. 2001; Sakai et al. 2003) catabolize bioactive GA into nonbioactive GA to maintain a GA balance. SLENDER RICE 1 (SLR1) encodes a DELLA protein that functions as a core repressor of GA-mediated responses in rice (Ikeda et al. 2001; Fukao and Bailey-Serres 2008). SLR1 physically interacts with different transcription factors. In the presence of GA, SLR1 proteins are degraded, which allows normal functioning of the GA response.
GA plays important roles at several developmental stages, but whether GA also regulates seed shattering is largely unknown. In this study, we evaluated the degree of seed shattering from plants composing a minicore germplasm collection, and through a genome-wide association study (GWAS), we found that GA signaling negative feedback regulator SLR1 contributes to seed shattering regulation. Using different GA mutants and several transgenic lines, we found that increased GA contents or enhanced GA signaling at the abscission region led to easy shattering. Our RNA sequencing (RNA-seq) and reverse transcription quantitative PCR (RT-qPCR) data revealed that the lignin content in the abscission region played a major role in the shattering process. Importantly, we found that SLR1, the key repressor of GA signaling, could physically interact with the rice abscission-related proteins qSH1, OSH15, and SNB. In addition, we showed that these three proteins act as transcriptional repressors that bind to the promoter of the lignin biosynthesis gene 4-Coumarate: Coenzyme A Ligase 3 (4CL3), and direct interaction between SLR1 and these rice abscission-related proteins can release the inhibition of 4CL3, which increases lignin deposition in the abscission region. As a result, the breaking tensile strength (BTS) is increased, and the degree of shattering is reduced. Our study presents a rice seed-shattering regulatory network coordinated by GA/SLR1, abscission-related proteins, and lignin content and gives examples for breeding high-yielding rice that presents ideal seed shattering and is efficiently harvested.
Results
GA regulates rice seed shattering
Rice cultivars usually have a wide range of degrees of seed shattering. To investigate the potential genes involved in the variation of rice seed shattering, we selected 134 rice accessions that experience similar growth patterns and timing from a minicore rice germplasm collection, including 45 japonica, 84 indica, and 5 intermediates (Shang et al. 2022), for a GWAS analysis (Fig. 1C; Supplemental Fig. S1B and Table S1). And, we conducted principal component analysis (PCA) and used principal component 1 (PC1) and PC2 to classify the population, which was consistent with the subpopulation classification described above (Supplemental Fig. S1A). So, the inclusion of GWAS analysis with PCA can solve the problem of population stratification. According to Fixed and Random Model Circulating Probability Unification (FarmCPU) (Liu et al. 2016), we identified a region covering a known abscission-related gene (qSH1, Chr1:36445019-36449951), and the top SNP (Chr1_36306832) was previously characterized as being strongly associated with rice seed shattering between japonica and indica (Konishi et al. 2006). These results suggested that the GWAS analysis was reliable.
In addition to the peak of this key SNP, we also found several peaks that surpassed the inspection threshold. Among them, we found an SNP in chromosome 3, which was related to the gibberellin signal transduction gene SLR1 (Chr3:28512754-28515086) (Fig. 1C). And in the genome region of SLR1, we identified 6 haplotypes and 3 haplotypes based on CDS and promoter SNPs, respectively (Supplemental Fig. S2, A to C). The mutation of ACG in japonica (43 varieties) to ATG in indica (78 varieties) resulted in the substitution of threonine (T) for methionine (M). The decrease in phenotype values of Hap.4 and Hap.5 may be due to this difference (Supplemental Fig. S2F). The mutation of CAC in japonica (42 varieties) to CGC in indica (83 varieties) resulted in the substitution of histidine (H) for arginine (R), and the reduced phenotypic values of Hap.4, Hap.5, and Hap.6 may be due to this difference (Supplemental Fig. S2H). These two SLR1 mutation types may be associated with the seed-shattering degree between japonica and indica. The other haplotypes were not significant (Supplemental Fig. S2, D, E, and G). And the phenotypic value of Hap.a differs significantly only from that of Hap.b (Supplemental Fig. S2F). Thus, natural variations that exist in the coding regions and promoters in SLR1 may contribute to seed shattering among indica and japonica (Supplemental Fig. S2 and Table S2).
Then, we examined whether SLR1 was associated with seed shattering. In the slr1 mutant, the pedicel BTS level of slr1 (Fig. 1D) was significantly decreased compared with that of wild-type Zhonghua 11 (ZH11). To precisely distinguish the anatomical differences in the abscission region, we used scanning electron microscopy (SEM) to examine the broken interface from the abscission region in the spikelet basal part of slr1. The SEM photos of the broken interface showed that there was a smooth fracture surface of the mutant slr1 (Fig. 1, I3 and I4) compared with the wild-type ZH11 (Fig. 1, H3 and H4). These results suggested that the negative feedback regulator of gibberellin, SLR1, is indeed related to seed shattering.
As we mentioned above, SLR1 is a core functional protein in gibberellin signal transduction. SLR1 protein level changes in several gibberellin biosynthesis or catabolism mutants or transgenic lines. So, we hypothesized that the seed-shattering degree might also change in other gibberellin-related mutants. As shown in Fig. 1, E to G, the BTSs of sd1 (Fig. 1E) and OE-EUI1 (Fig. 1G) were 1.65 and 1.4 times higher than those of their corresponding wild-types, and the pedicel BTS of eui1 (Fig. 1E) was significantly decreased compared with that of Zhonghua 11 (ZH11). Consistent with the BTS testing results, compared with Kasalath (Fig. 1, J3 and J4), the sd1 mutant (Fig. 1, K3 and K4) exhibited a rough and broken cross-section. The fracture surface of eui1 was smooth, but the corresponding wild-type ZH11 was rough with spring-like broken vascular bundles (Supplemental Fig. S3, A and B). And, the opposite result was shown between TP309 and OE-EUI1. The transverse fracture plane of OE-EUI1 was uneven, and TP309 was smooth (Supplemental Fig. S3, C and D).
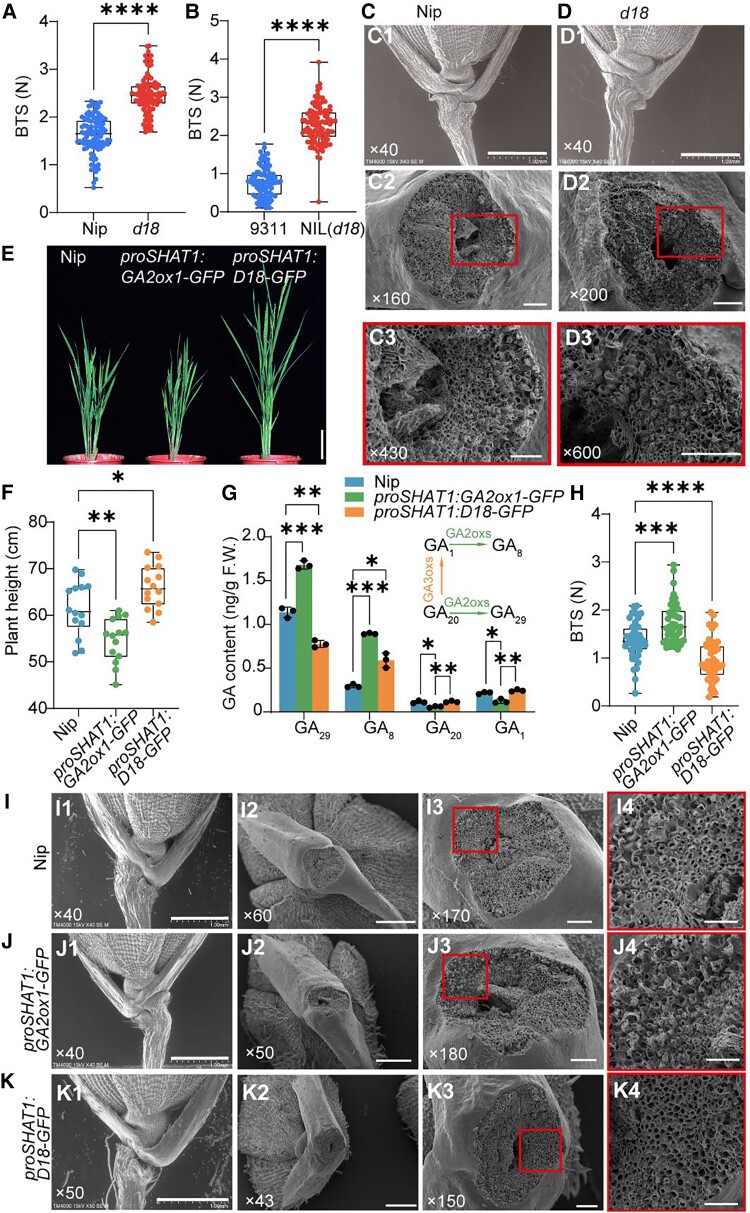
To further verify that GA regulates seed shattering, we checked another GA biosynthesis mutant, d18. The BTSs of the d18 mutant and d18-NIL-9311 were much higher than those of Nip and 9311, respectively (Fig. 2, A and B). In addition, compared with Nip (Fig. 2, C2 and C3), d18 (Fig. 2, D2 and D3) exhibited a rough and broken cross-section (Fig. 2, C and D). The 9311 belongs to indica-type rice and has an AL structure. So, a smooth fracture surface structure was observed in 9311 and NIL (d18). Even so, a rough region was observed in the area near the edge of NIL (d18) (Supplemental Fig. S3, E and F). Together, these results further confirmed that GA content or signaling is negatively associated with the degree of seed shattering.
Figure 2.
GA content alteration in abscission region influenced seed-shattering degree in Nipponbare. A) and B) Boxplots of BTS comparing Nipponbare (Nip) and d18 mutant in Nip A) and 9311 background B). C) and D) Characterization of rice spikelet in Nip C) and d18D). C1) to D1) The SEM photographs of the spikelet basal part. C2) to D2) The SEM photographs of the fracture surface. C3) to D3) are magnifications of the red boxes in C2) to D2), respectively. Bars = 1 mm in panels (1), 100 μm in panels (2), 50 μm in panel (3). The numbers in the bottom left corner of the photos indicated the magnification. E) Appearance of the proSHAT1:GA2ox1-GFP, proSHAT1:D18-GFP transgenic lines, and the wild-type Nip plants. Bar = 10 cm. F) The plant height of wild-type and transgenic lines. G) Endogenous GA levels in young fluorescence as described above. The upper right corner diagram indicated the partial gibberellin metabolism pathway involved in GA3oxs and GA2oxs. The data are the mean ± Sd of 3 biological repeats. H) Boxplots of BTS comparing Nip, proSHAT1:D18-GFP, and proSHAT1:GA2ox1-GFP. Data in A), B), F), and H) are displayed as box and whisker plots with individual data points. Horizontal bars represent the maximum, third quantile, median, first quantile, and minimum values respectively. *P-value ≤0.05, **P-value ≤0.01,***P-value ≤0.001, and ****P-value ≤0.0001 calculated from two-tailed t-test A) and B) and one-way ANOVA test F) to H). The data are the mean ± Sd. I) to K) Morphological characteristics of abscission regions. The 3 rows from top to bottom represent morphological analyses of the Nip I), proSHAT1:GA2ox1-GFPJ), and proSHAT1:D18-GFPK), respectively. I1) to K1) The SEM photographs of the spikelet basal part. I2) to K2) The SEM photographs of the broken area on mature seeds. I3) to K3) Close-up SEM photographs of the fracture surface corresponding to I2) to K2). I4) to K4) are magnifications of the red boxes in I3) to K3), respectively. The numbers in the bottom left corner of the photos indicated the magnification. Bars = 1 mm in panels (1), 500 μm in panels (2), 100 μm in panel (3), and 50 μm in panel (4). F.W., fresh weight.
qSH1 is important for AL-like region development in the nonshattering rice cultivar Nipponbare
Konishi et al. (2006) demonstrated that the reduction of qSH1 expression level caused loss of seed shattering owing to the absence of AL formation among japonica subspecies of rice, like Nip (O. sativa L. ssp. japonica cv. Nipponbare). But even though there is no AL formed in Nip, seed shattering can still happen in the basal portion of the spikelet (Fig. 1B; Supplemental Fig. S4A). And, the proportion of break position in Nip was calculated. About 60% of shattering occurs in the AL-like region, which is significantly higher than the rachilla (6%) and pedicel (34%) (Supplemental Fig. S4B). These results suggested that an AL-like region was formed, which may fulfill a real AL function even if no visible AL is established.
So, we measured the BTS and broken surface observation only in AL-like-type to ensure consistency and validity of the data in the following experiments. qSH1 is known as an important regulator for AL formation in the indica subpopulation, and we found that the nonshattering japonica accession Nip also has a similar AL region. These results suggested that qSH1 may also perform its function in AL-like region formation in Nip, at least partially. We investigated qSH1 expression by RNA in situ hybridization in Nipponbare and Kasalath (Supplemental Fig. S5). In Nipponbare, qSH1 was highly expressed in the boundary region between the floral meristem and sterile lemma primordium in spikelet development Sp2. And, the transcripts were also detected in a small area above the rudimentary glume (Supplemental Fig. S5A). Later, strong signals were observed in the floral meristem in Sp4. And, a clear ribbon-like expression pattern appeared between the sterile lemma and rudimentary glume, where the AL generally initiated even though there is no obvious AL in Nip (Supplemental Fig. S5B). Subsequently, the expression of qSH1 was primarily restricted to the floral meristem and abscission region in Sp6 (Supplemental Fig. S5C). During Sp7, the transcripts of qSH1 became a bit weaker but still expressed clearly in the pistil, abscission region, and rachilla (Supplemental Fig. S5D). In Kasalath, the qSH1 signal appeared in Sp4 in the floral meristem. And, the qSH1 signal became increasingly restricted to ALs in Sp8 (Supplemental Fig. S5, E to L).
To verify the function of qSH1 in seed shattering in Nipponbare, we generated a qSH1 mutant via Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated 9 (Cas9) genome editing. A 13-bp deletion from position 8 to 20 led to disorderly protein coding and premature termination (Supplemental Fig. S6, A and B). And, calculations of BTS showed that the CRISPR-qSH1 grains were more difficult to shatter compared with wild-type Nip (Supplemental Fig. S6C). And according to the results of SEM, the fracture surface at the AL-like region of the CRISPR-qSH1 line was rougher than Nip (Supplemental Fig. S6, D and E). These data demonstrated that qSH1 plays an important role in determining the degree of seed shattering in Nip, even if real AL is not visibly formed.
Ectopic expression of D18 and GA2ox1 in the abscission region alters the degree of shattering
As several GA-related mutants have different genetic backgrounds and usually exhibit strong developmental defects, it is necessary to generate specific transgenic lines with minor defects. Previous studies have reported that SHAT1 is fairly expressed in the AL-like region of Nip (Zhou et al. 2012). We also generated similar GUS lines to check the promoter activity. GUS staining of proSHAT1:GUS transgenic lines (#1 and #2) showed that SHAT1 exhibited an intense signal in the spike basal part (Supplemental Fig. S7, A to D). Similarly, according to RNA in situ hybridization, SHAT1 is expressed between sl and rg in Sp6 in the Nip AL-like region as a clear curved and broader banded-like pattern (Supplemental Fig. S7, E to G). Therefore, we selected the SHAT1 gene promoter to ectopically express the GA biosynthesis gene D18 or the catabolism gene GA2ox1 in the abscission region.
The plant height of these two transgenic lines was altered (Fig. 2, E and F), and agronomic traits were not changed (Supplemental Fig. S8, A to C), except proSHAT1:D18-GFP lines had more grains on each spike (Supplemental Fig. S8, D and F); however, compared with wild-type Nip, neither line presented any significant yield loss (Supplemental Fig. S8E). On the contrary, both transgenic lines exhibited increased grain length and similar grain width compared with Nip (Supplemental Fig. S8, G to K). As expected, endogenous bio-active GA1 was significantly higher in proSHAT1:D18-GFP than proSHAT1:GA2ox1-GFP due to ectopically expressed d18 (GA3ox2) and slightly higher than Nip (Fig. 2G), probably because of dynamic equilibrium between the contents of different forms (free-form or bound-form) of GAs.
GA1 and GA20 are catalytically decomposed into inactive GA8 and GA29 by GA2oxs. As a result, GA1 and GA20 content decreased while GA8 and GA29 content increased in proSHAT1:GA2ox1-GFP (Fig. 2G). Importantly, compared with Nip, the proSHAT1:GA2ox1-GFP transgenic lines showed a significantly increased BTS level, and the proSHAT1:D18-GFP transgenic lines exhibited the opposite phenotype (Fig. 2H). And, the breaking position ratio at the AL-like region in proSHAT1:GA2ox1-GFP (32.6%) was significantly decreased compared with Nip (56.2%), while it increased in proSHAT1:D18-GFP (64.7%) (Supplemental Fig. S9). The SEM results showed that, compared with the AL-like region (Fig. 2, I2 to I4) in the Nip spikelet, proSHAT1:GA2ox1-GFP had an uneven and rough fracture surface, filled with many cracked spring-shaped vascular bundles (Fig. 2, J2 to J4). In contrast, the proSHAT1:D18-GFP transgenic lines showed a smooth and flat section surface (Fig. 2, K2 to K4), which is consistent with the BTS results (Fig. 2H). And, there is no difference between them in the basal part of the spikelet (Fig. 2, I1 to K1).
Moreover, we also detected the SHAT1 expression pattern in the spikelet development of Kasalath, an indica rice variety with ALs. In Sp4, SHAT1 signals were mainly enriched in rg and detected in st and rg in Sp6 (Supplemental Fig. S7, H and I). In Sp7, the SHAT1 transcript appeared in lo and was also found in rg and sl (Supplemental Fig. S7J). Afterward, during the Sp8 stage, SHAT1 expression accumulated to higher levels in the AL (Supplemental Fig. S7, K to M). Then, we used Kasalath as a transgene background to explore if the same mechanism is present in the indica rice variety with an obvious AL. As shown in Supplemental Fig. S10A, plant height in the elongation stage is also changed when expressed proSHAT1:GA2ox1-GFP and proSHAT1:D18-GFP. The proSHAT1:GA2ox1-GFP line is shorter and the proSHAT1:D18-GFP is taller than the wild-type in the flowering stage (Supplemental Fig. S10B).
And, the bio-active GA1 content was increased in the young inflorescence of proSHAT1:D18-GFP and decreased in proSHAT1:GA2ox1-GFP (Supplemental Fig. S10C). Then, the tendency of BTS levels was similar to those in the Nip background. The proSHAT1:GA2ox1-GFP transgenic lines showed a significantly increased BTS level, and the proSHAT1:D18-GFP transgenic lines were easier to shed compared with wild-type Kasalath (Supplemental Fig. S10D). The two transgenic lines and the wild-type all have a clear circular fracture edge (Supplemental Fig. S10, E to G). But, there is a difference near the central vascular bundle between them. The wild-type Kasalath fracture surface is smooth away from the vascular bundle and relatively rough around it (Supplemental Fig. S10, E2 and E3). But, the fracture surface of the proSHAT1:D18-GFP transgenic line is flat from the edge to the center (Supplemental Fig. S10, F2 and F3). And, the proSHAT1:GA2ox1-GFP transgenic lines have rough spots even around the edges (Supplemental Fig. S10, G2 and G3). These results indicated that the level of gibberellin content around the abscission region will affect the seed-shattering degree.
GA affects seed shattering by altering the lignin content in the abscission region
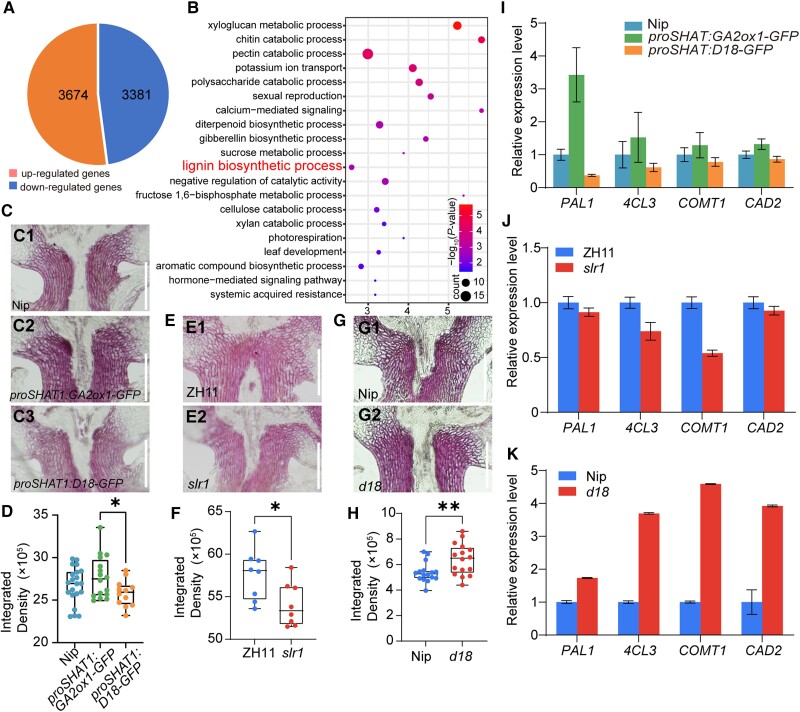
We demonstrated that ectopically expressed GA-related genes could alter the degree of rice seed shattering (Fig. 2H; Supplemental Fig. S10). To further explore this conclusion and evaluate the signaling pathway involved, RNA-seq analysis of rice panicles from Nip and proSHAT1:D18-GFP plants at the booting stage was conducted. A total of 7,055 differentially expressed genes (DEGs) was detected (false discovery rate set to P < 0.05), of which 3,674 genes were upregulated and 3,381 genes were downregulated in the proSHAT1:D18-GFP plants compared with the wild-type (Fig. 3A).
Figure 3.
GA affects seed shattering by altering the lignin content in the abscission region. A) DEGs detected by RNA-seq in inflorescence meristem of proSHAT1:D18-GFP lines with 3 biological replicates (P < 0.05). B) BP enrichment analysis of DEGs. The letters in red indicate lignin biosynthetic progress. The X-axis represents the enrichment factor −log10 (P-value) ranging is from 1.45 to 5.70. The color and size of the dots represent the range of the P-value and the number of genes. C) to H) Analysis of lignin deposition. Longitudinal sections across the abscission region of Nip C1), proSHAT1:GA2ox1-GFPC2), proSHAT1:D18-GFPC3); ZH11 E1) and slr1E2); Nip G1) and d18G2). Sections were stained with phloroglucinol-HCl. Scale bars = 100 μm. D), F), and H) Quantitative results of lignin stained with phloroglucinol-HCl according to C), E), and G). Different sections were used to estimate the lignin content by Image J. Data in D), F), and H) are displayed as box and whisker plots with individual data points. Horizontal bars represent the maximum, third quantile, median, first quantile, and minimum values respectively. *P-value ≤0.05 and **P-value ≤0.01, calculated from a two-tailed t-test F) and H) and one-way ANOVA test D). I) to K) Expression analysis of lignin biosynthesis genes in transgenic lines I), slr1J), d18K), and their corresponding wild-type as revealed by RT-qPCR. Ubiquitin was used as a loading control. The data are the mean ± Sd of 4 biological repeats.
Interestingly, the expression of previously known shattering-related genes was not significantly changed (Supplemental Fig. S11), suggesting that the shifted degree of seed shattering for the transgenic line was probably not due to changes in the expression of abscission-related genes. Afterward, Gene Ontology (GO) analysis of the DEGs was performed to identify the biological process (BP) (Fig. 3B), cellular component (Supplemental Fig. S12A), and molecular function (Supplemental Fig. S12B) terms, followed by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Supplemental Fig. S12C). Among the BP (Fig. 3B), several DEGs were found to be associated with lignin biosynthetic processes, as reported previously (Zhou et al. 2012; Yoon et al. 2014, 2017).
To test the hypothesis that GA may affect the lignin content in the abscission region, we measured lignin deposition via phloroglucinol staining in transgenic lines and wild-type at the spikelet developmental stage Sp8. The lignin content in Nip (Fig. 3, C1) was higher than that in proSHAT1:D18-GFP (Fig. 3, C3) and lower than that in proSHAT1:GA2ox1-GFP (Fig. 3, C2). According to the quantification of the staining degree, the lignin integrated density in proSHAT1:GA2ox1-GFP was significantly higher than in proSHAT1:D18-GFP. The content in Nip was between them but not statistically significant (Fig. 3D). Similarly, lignin deposition was lower in the slr1 (Fig. 3, E2) mutant than in ZH11 (Fig. 3, E1) and higher in d18 (Fig. 3, G2) than in Nip (Fig. 3, G1). The quantification of the staining results showed that the value for slr1 was much lower than that for ZH11 and that the value for d18 was higher than that for the Nip (Fig. 3, F and H).
Then, we selected 4 genes (Fig. 3, I to K) with higher expression levels in rice panicles from 10 genes (Supplemental Fig. S13B and Table S3) that have been reported previously (Kawasaki et al. 2006; Gui et al. 2011; Hirano et al. 2012; Zhou et al. 2018; He et al. 2020; Bang et al. 2022; Huangfu et al. 2022) according to lignin biosynthesis pathway (Whetten and Sederoff 1995; Weng and Chapple 2010) (Supplemental Fig. S13, A and B) to analyze their transcripts in Nip, proSHAT1:D18-GFP, and proSHAT1:GA2ox1-GFP. We found that PHENYLALANINE AMMONIA-LYASE 1 (PAL1), 4CL3, CAFFEIC ACID O-METHYLTRANSFERASE 1 (COMT1), and CAD2 expression levels in the junction region of developing panicles in Nip were reduced in proSHAT1:D18-GFP and increased in proSHAT1:GA2ox1-GFP compared with Nip (Fig. 3I). These 4 genes’ transcripts were slightly decreased in slr1 and increased in d18 compared with their corresponding wild-types (Fig. 3, J and K), which are consistent with the lignin content.
Moreover, we also measured the lignin content in these transgenic lines in Kasalath background. The lignin deposition in Kasalath (Supplemental Fig. S10, E4) was higher than that in proSHAT1:D18-GFP (Supplemental Fig. S10, F4) and lower than that in proSHAT1:GA2ox1-GFP (Supplemental Fig. S10, G4). The quantification of the staining degree also validated this result (Supplemental Fig. S10H). These results demonstrated that in both rice varieties without and with ALs, changing the gibberellin content in the abscission region affected the lignin deposition.
SLR1 interacts with qSH1, OSH15, and SNB
SLR1 is a core component of GA signaling and mediates downstream responses through protein–protein interactions (Arnaud et al. 2010; Lim et al. 2013; Huang et al. 2015; Fukazawa et al. 2017; Hu, Israeli, et al. 2018; Liao et al. 2019). We hypothesize that GA regulates seed shattering similarly. Previous studies have reported that 8 genes (qSH1, SHATTERING 1 [Sh1], sh4, SH5, CTD PHOSPHATASE-LIKE [CPL1], SHATTERING ABORTION1 [SHAT1], OSH15, and SNB) are important for AL formation or seed shattering (Konishi et al. 2006; Li et al. 2006; Lin et al. 2007, 2012; Ji et al. 2010; Zhou et al. 2012; Yoon et al. 2014, 2017; Jiang et al. 2019).
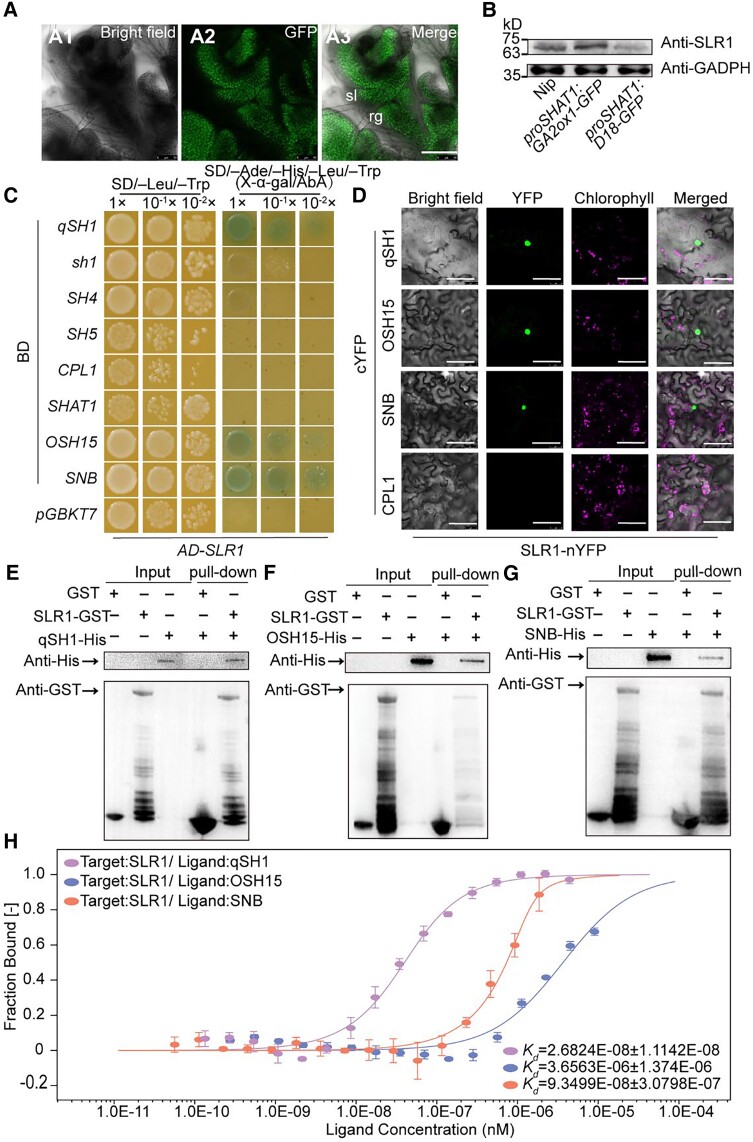
As such, we first characterized the SLR1 expression pattern in Nip by RNA in situ hybridization. SLR1 was expressed mostly throughout the whole early-reproductive stage (Supplemental Fig. S14, A to K). Especially during later spikelet development Sp7, SLR1was mainly expressed in the junction between sl and rg (Supplemental Fig. S14J). Taken together, the results of in situ hybridization suggest that the expression of SLR1 covered the abscission region during spikelet development. Moreover, we generated a proSLR1:SLR1-GFP transgenic line to determine protein accumulation. The GFP signal was detected in the rg, sl, and the position between them during spikelet development Sp8 (Fig. 4A). In addition, the increased BTS level of proSLR1:SLR1-GFP indicated that SLR1 plays a role in seed shattering (Supplemental Fig. S14L). Taken together, these results indicated that SLR1 is coexpressed with shattering-related transcription factors in the abscission region, thus enabling direct interaction between SLR1 and shattering-related transcription factors. Furthermore, the protein level of SLR1 in the wild-type was higher than proSHAT1:D18-GFP lines but lower than proSHAT1:GA2ox1-GFP lines (Fig. 4B), indicating that expressing D18 and GA2ox1 in the abscission region altered the endogenous SLR1 protein level in rice inflorescence.
Figure 4.
SLR1 interacts with qSH1, OSH15, and SNB. A) GFP signals detected in Sp8 in proSLR1:SLR1-GFP. Bar = 100 μm. B) The levels of SLR1 protein in young panicles of Nip, proSHAT1:GA2ox1-GFP, and proSHAT1:D18-GFP. Proteins extracted from young panicles were subjected to immunoblot analysis using the anti-SLR1 antibody. The anti-GADPH antibody was used as the loading control. C) Yeast two-hybrid assay revealing the interaction of SLR with qSH1, OSH15, and SNB. CPL1 was used as a negative control. The transformed yeast cells were grown on minimal, synthetically defined (SD) medium: SD-Leu/-Trp and SD-Ade/-His/-Leu/-Trp/+AbA (aureobasidin A). Yeast growth is presented at three dilutions. pGADT7 (AD), pGBKT7 (BD). D) BiFC analysis of the interaction between SLR1 and qSH1, OSH15, and SNB in N. benthamiana. Merge indicates merged images of enhanced yellow fluorescent protein. In each experiment, at least five independent N. benthamiana leaves were infiltrated and evaluated. Bars = 50 μm. E) to G) Pull-down assay demonstrating the interaction between SLR1 interacts with qSH1 E), OSH15 F), and SNB G). H) Determination of the binding affinity of SLR1 to qSH1-GST, OSH15-GST, and SNB-GST by MST. The curve is fit by the standard Kd-fit function. Kd, dissociation constant. Bars represent ± Sd (n = 3 biological replicates). sl, sterile lemma; rg, rudimentary glume.
We used multiple methods to determine the possible interactions. The results of our yeast two-hybrid assays indicated that SLR1 directly interact with qSH1, OSH15, and SNB, and no direct interactions were observed with the other 5 proteins (Fig. 4C). Bimolecular fluorescence complementation (BiFC) analysis in Nicotiana benthamiana also revealed a direct interaction between SLR1 and qSH1, OSH15, and SNB in the nuclei of leaf pavement cells (Fig. 4D), and yellow fluorescent protein (YFP) signal was not observed between SLR1 and CPL1. These interactions were also confirmed by the results of an in vitro pull-down assay in which recombinant SLR1 fused to glutathione-S-transferase (GST) (SLR1-GST), and His-tagged qSH1 (qSH1-His), OSH15 (OSH15-His), and SNB (SNB-His) were expressed in Escherichia coli. SLR1-GST but not GST alone was able to pull down qSH1-His (Fig. 4E), OSH15-His (Fig. 4F), and SNB-His (Fig. 4G). Here, to verify the affinity of SLR1 to qSH1, OSH15, and SNB, microscale thermophoresis (MST) was performed. The results showed that SLR1 bound to qSH1, OSH15, and SNB when the titrant was at a low micromolar concentration; the dissociation constant (Kd) values were 0.27 ± 0.11 nm for qSH1, 3.66 ± 1.37 µm for OSH15, and 0.93 ± 0.3 nm for SNB (Fig. 4H). These results suggest that there is a relatively strong interaction between the tested proteins, and the affinity of qSH1 is much stronger than that of OSH15 or SNB concerning binding to SLR1. Therefore, GA may modulate seed shattering via interactions between SLR1 with qSH1, OSH15, and SNB.
Identification of qSH1, OSH15, and SNB cotarget genes
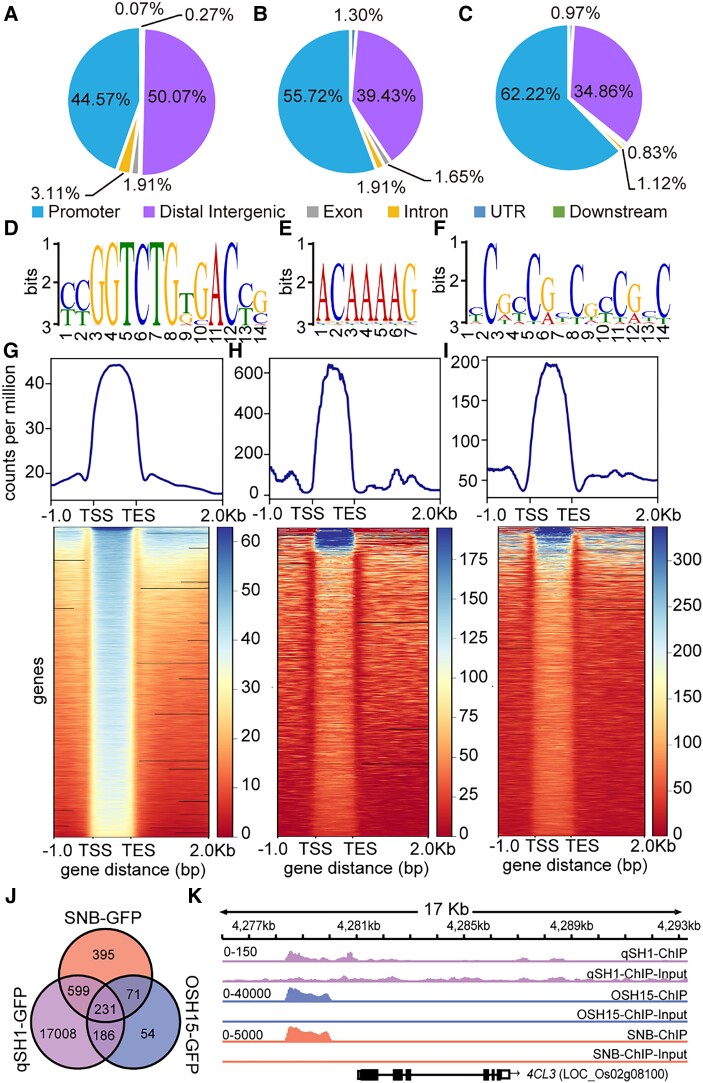
It is known that OSH15 can inhibit lignin biosynthesis to enhance seed shattering, and our RNA-seq analysis also revealed that GA may regulate seed shattering through lignin biosynthesis. Therefore, to further understand how the interaction of SLR1 with qSH1, OSH15, and SNB regulates seed shattering, we performed chromatin immunoprecipitation sequencing (ChIP-seq) separately with pro35S:qSH1-GFP, pro35S:OSH15-GFP, and pro35S:SNB-GFP transgenic lines. Abscission-related gene binding sites were highly enriched in the promoter regions, accounting for 44.57% (Fig. 5A), 55.72% (Fig. 5B), and 62.22% (Fig. 5C), respectively, of all the peak data. The binding sites were also present in distal intergenic regions, UTRs, exons, and introns.
Figure 5.
Chromatin profiling analysis of qSH1, OSH15, and SNB-regulated genes. A) to C) Peak distribution of each mark surrounding various genomic features in qSH1-GFP A), OSH15-GFP B), and SNB-GFP C) target genes. D) to F) DNA-sequence of the motif enriched in their binding sites and positional distribution of the YCGGTCTGTGACYG motif in qSH1 binding peaks D), ACAAAAG motif in OSH15 binding peaks E), and YCGCCGYCGYCGYC motif in SNB binding peaks F). G) to I) Genes marked by different combinations and positions of qSH1 G), OSH15 H), and SNB I). The normalized intensity of each mark in the surrounding genes was recorded for k-means clustering. J) Venn diagram showing the number of gene regions bound by qSH1, OSH15, and SNB, as detected by ChIP-seq. K) Genome-browser view of qSH1, OSH15, and SNB binding at 4CL3 loci. The number range in the top left corner of each genome indicated the data scale. TSS, transcriptional start site; TES, transcriptional end site.
To identify their binding motifs, the flanking sequences ±150 bp around the regions corresponding to peak summits were analyzed using MEME-ChIP. The DNA sequences YCGGTCTGTGACYG, ACAAAAG, and YCGCCGYCGYCGYC (Y represents C or T) were identified as being the most significantly enriched motifs separately in qSH1 (Fig. 5D), OSH15 (Fig. 5E), and SNB (Fig. 5F). These binding sites were similar to those in previous studies showing that qSH1 and SNB, respectively, bind to TGAC (Sharma et al. 2014) and GCCGCC (Sharoni et al. 2011) cis-acting elements in the target genes. Genome-wide distribution analysis revealed that the qSH1-GFP, OSH15-GFP, and SNB-GFP binding sites were highly enriched between the transcriptional start site and transcriptional end site (Fig. 5, G to I). To determine whether genes with specific functions are enriched in qSH1, OSH15, and SNB binding regions, we performed a GO analysis. The results showed that qSH1 target genes were mainly involved in cell wall organization, the auxin signaling pathway, lignin metabolism, and flower development (Supplemental Fig. S15A). The OSH15 target genes were enriched in cell fate commitment and stress responses (Supplemental Fig. S15B), and those of SNB were involved in the plant hormone signaling pathway and flower development (Supplemental Fig. S15C).
We identified 18,024, 542, and 1,296 target genes that were bound by qSH1, OSH15, and SNB, respectively, and 231 of these targets were shared by these proteins (Fig. 5J). Among these genes, we found that 4CL3 presented a strong TF-binding signal (Fig. 5K). According to the results of the genome browser view of the abscission-related gene binding site, all 3 genes could individually bind to the promoter of 4CL3, which contains one copy of the identified binding motif (Supplemental Fig. S16).
4CL3 is directly suppressed by qSH1, OSH15, and SNB
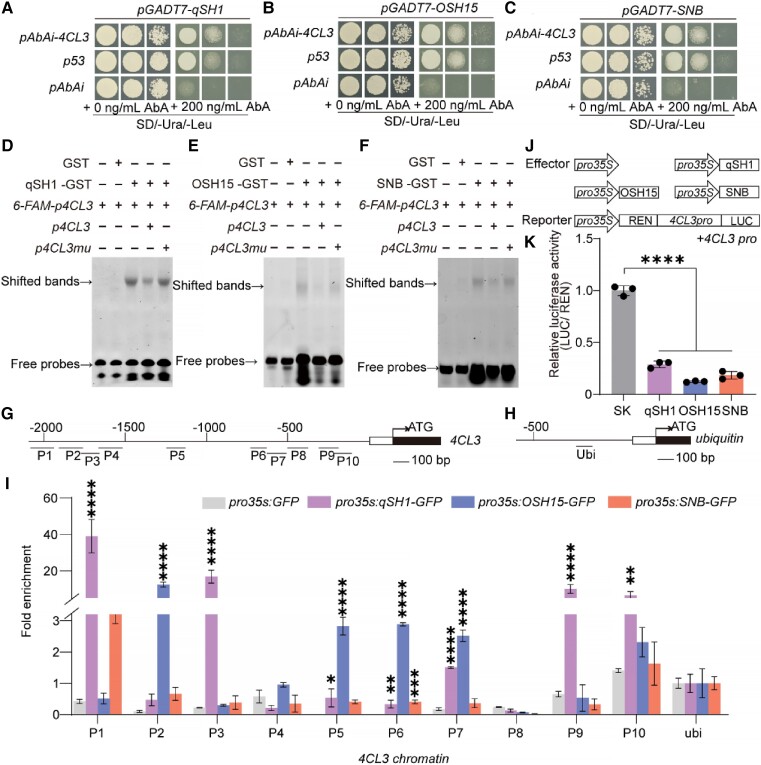
To further confirm that qSH1, OSH15, and SNB directly bind to the 4CL3 promoter, we first performed a yeast one-hybrid analysis. The yeast (Saccharomyces cerevisiae) grew on selective SD/-Ura/-Leu media supplemented with 200 ng/mL aureobasidin A (AbA) when the promoter of 4CL3 was cotransformed with qSH1 (Fig. 6A), OSH15 (Fig. 6B), and SNB (Fig. 6C), suggesting that qSH1, OSH15, and SNB indeed directly bind to the 4CL3 promoter. Next, 4CL3 promoter fragments containing each cis-acting element were examined through an electrophoretic mobility shift assay (EMSA). A 30-bp probe from the promoter of 4CL3 (Supplemental Fig. S16) was bound by the recombinant qSH1 (Fig. 6D), OSH15 (Fig. 6E), and SNB (Fig. 6F) proteins fused to GST, which resulted in a mobility shift (Lane 3); as a negative control, GST alone (Lane 2) did not cause a mobility shift. The binding ability to 4CL3 was suppressed by the addition of increasing amounts of unlabeled probes (Lane 4), and the competition was abolished when the unlabeled probes were mutated (Lane 5).
Figure 6.
4CL3 was a direct cotarget of qSH1, OSH15, and SNB. A) to C) Yeast one-hybrid assays testing the binding of qSH1 A), OSH15 B), and SNB C) to the 4CL3 promoter. D) to F) Electrophoresis mobility shift assay of qSH1 D), OSH15 E), SNB F), and 6-Fam-labeled probes containing different cis-acting elements in the 4CL3 promoter region. The upper and lower arrows indicate the shift bands and free probes, respectively. G) to I) ChIP-qPCR assays of 4CL3 using ChIP-DNA complexes isolated from 0 to 4 cm young panicles of the pro35S:qSH1-GFP, pro35S:OSH15-GFP, and pro35S:SNB-GFP transgenic plants. G) and H) The genomic structures of 4CL3 and ubiquitin, respectively. The numbers (P1 to P10) and ubi indicated the tested regions. I) The enrichment of the 4CL3 chromatin. For enrichment of qSH1, OSH15, and SNB-GFP on the indicated fragments was calculated as the ratio of anti-GFP IP to control beads immunoprecipitation of each independent replicate. Ubiquitin was used as a negative control. Values are mean ± Sd (n = 3 pooled tissues, 10 plants per pool). J) Schematic diagrams of the effector and reporter plasmids used in the transient assay. K) Relative LUC activity in N. benthamiana leaves cotransformed with the indicated reporter and effector plasmids. LUC (Firefly Luciferase), REN (Rellina Luciferase), and SK (pGreenII 62-SK). The LUC activity in control was set as “1.” Error bars indicate means ± Sd of 3 biological repeats. *P-value ≤0.05, **P-value ≤0.01, ***P-value ≤0.001, and ****P-value ≤0.0001 calculated from the two-way ANOVA test I) and one-way ANOVA test K).
Furthermore, we performed a chromatin immunoprecipitation qPCR (ChIP-qPCR) assay involving 0 to 4 cm long young panicles from pro35S:qSH1-GFP, pro35S:OSH15-GFP, and pro35S:SNB-GFP transgenic plants. Our data showed that the qSH1-bound (P1, P3, P9, and P10), OSH15-bound (P2, P5, P6, P7, and P10), and SNB-bound (P1 and P10) DNA fragments (Fig. 6G) were enriched at the different promoter regions of the 4CL3 gene (Fig. 6I) compared with ubiquitin (Fig. 6H). Moreover, a transactivation analysis was performed, and the results revealed reduced luciferase activity when the cauliflower mosaic virus (CaMV) 35S promoter driving qSH1, OSH15, and SNB was coexpressed with the 4CL3 promoter driving the luciferase reporter in rice protoplasts (Fig. 6J), demonstrating that qSH1, OSH15, and SNB suppressed 4CL3 expression (Fig. 6K). Thus, qSH1, OSH15, and SNB bind to the promoter of 4CL3 directly and negatively regulate its expression.
The interaction between SLR1 and qSH1, OSH15, and SNB interferes with the qSH1, OSH15, and SNB-4CL3 regulatory pathway
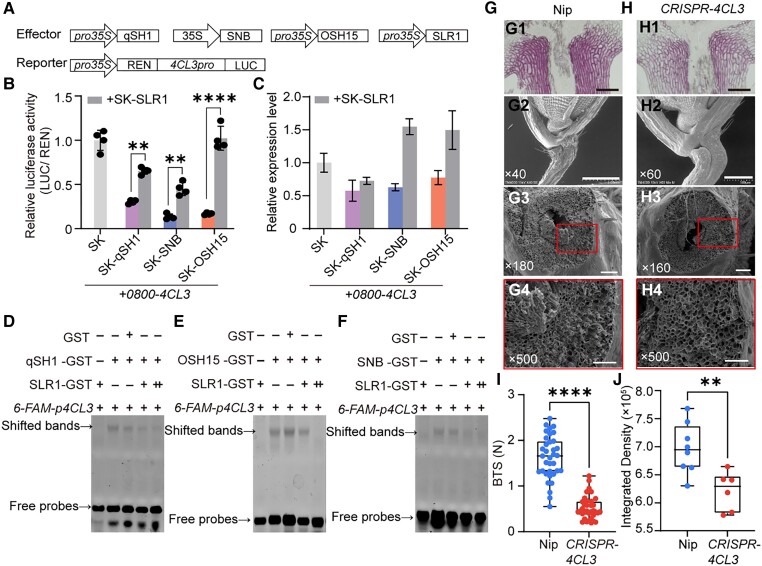
As SLR1 interacts with qSH1, OSH15, and SNB, we hypothesize that this interaction may influence their ability to bind to the 4CL3 promoter. Transactivation analysis showed that luciferase activity in the cells coexpressing a reporter containing the 4CL3 promoter driving luciferase and effectors containing qSH1, OSH15, and SNB separately was significantly increased by the additional coexpression of SLR1 (Fig. 7, A and B). In consideration of these three TFs, there were transcription inhibitors of 4CL3, demonstrating that SLR1 inhibits the qSH1, OSH15, and SNB-mediated regulatory pathways.
Figure 7.
Effect of SLR1-qSH1, OSH15, and SNB interaction on the qSH1, OSH15, and SNB-4CL3 signaling cascade. A) Schematic diagrams of the effector and reporter plasmids used in the transient assay in rice protoplasts. B) Luciferase activities in protoplasts con-transfected with the reporter and different combinations of effectors. The transactivation activity was monitored by assaying the luciferase activities. **P-value ≤0.01 and ****P-value ≤0.0001 calculated from a two-way ANOVA test. Error bars indicate ± Sd (n = 4). C) RT-qPCR examination of the transcripts of 4CL3 in rice protoplasts expression of the different combinations of effectors shown in A). Actin1 was used as an internal control. Error bars indicate the SD of three biological repeats. The plasmids combination represented by purple, blue and orange columns are the same as in Fig. 6K. D) to F) EMSA showing that SLR1 attenuates or inhibits the binding of qSH1 D), OSH15 E), and SNB F) to the 4CL3 promoter. A gradient concentration of SLR1-GST was applied (+, 1.0 μg; ++, 2.0 μg). G) and H) Characterization of rice spikelet in Nip G), CRISPR-4CL3H). G1) and H1) Longitudinal sections across the abscission region of Nip G1) and CRISPR-4CL3H1). Sections were stained with phloroglucinol-HCl. G2) and H2) SEM photographs of the spikelet basal part. G3) to H3) The SEM photographs of the fracture surface. G4) to H4) are magnifications of the red boxes in G3) to H3), respectively. Bars = 100 μm in panels (1) and (3), 1 mm in (G2), 500 μm in H2) and 50 μm in panels (4). The numbers in the bottom left corner of the photos indicated the magnification. I) Boxplots of BTS comparing Nip and CRISPR-4CL3 line. J) Quantitative results of lignin stained with phloroglucinol-HCl according to G1) and H1). Data in I) and J) are displayed as box and whisker plots with individual data points. Horizontal bars represent the maximum, third quantile, median, first quantile, and minimum values, respectively. ****P-value ≤0.0001 and **P-value ≤0.01 calculated from a two-tailed t-test.
The effect of this interaction on 4CL3 was further determined by measuring their transcripts in rice protoplasts. The decreased 4CL3 levels in cells expressing qSH1, OSH15, and SNB increased in response to the additional coexpression of SLR1 (Fig. 7C). Another proof of this sequestration role was derived from EMSAs (Fig. 7, D to F). Additional SLR1-GST proteins (added in line with a gradient) in the reaction substantially reduced the binding of rice qSH1, OSH15, and SNB to the 4CL3 promoter (Lanes 4 and 5). Concerning the negative control, there was no reducing effect when GST alone was added (Lane 3). Moreover, SLR1 could not bind to the 4CL3 promoter (Lane 1). The in vitro evidence indicated that SLR1 directly interacts with qSH1, OSH15, and SNB to sequester these factors and inhibit their binding to 4CL3.
Within the lignin biosynthesis pathway, 4CL3 mediates the activation of several hydroxycinnamic acids for the biosynthesis of monolignols and other phenolic secondary metabolites in vascular plants (Hu et al. 1998; Ehlting et al. 1999; Hamberger and Hahlbrock 2004). To verify the function of 4CL3 in lignin biosynthesis and its possible function in seed shattering, we generated a 4CL3 mutant via CRISPR/Cas9. A single nucleotide (A) insertion at position 34 led to disorderly protein coding and premature termination (Supplemental Fig. S17). Calculations of BTS showed that the CRISPR-transformed line displayed an easy seed shattering phenotype (Fig. 7I), and lignin deposition was reduced significantly (Fig. 7, G1, H1, and J). There was a smooth fracture at the broken position of the CRISPR-4CL3 line (Fig. 7, H3 and H4) compared with wild-type Nip, which presented a rough and broken cross-section (Fig. 7, G3 and G4). These observations supported our conclusion that lignin deposition in the abscission region plays an important role in determining the degree of seed shattering.
Discussion
GA is important for rice seed shattering through lignin biosynthesis regulation
Seed shattering is an important agronomic trait that strongly influences harvest efficiency and is closely associated with rice yield. In addition, the elimination of seed shattering also provides evolutionary evidence for the domestication process from wild rice to cultivated rice. In recent years, several genes involved in rice abscission region formation or development in both wild and cultivated rice have been characterized. However, the fundamental mechanisms, especially the complete signal transduction pathway for seed-shattering regulation, are still unclear. In this study, we revealed the role of GA in the control of rice seed shattering (Fig. 8). GA signals are transduced by interactions between SLR1, the key component of GA signaling, and three AL development-related proteins, qSH1, SNB, and OSH15. In the presence of GA, SLR1 is degraded and releases qSH1, SNB, and OSH15, enabling these transcription factors to bind to the promoter of the lignin biosynthesis-related gene 4CL3 and repress its expression. OSH15 and SNB are known as lignin biosynthesis repressors during AL formation, and we showed here that qSH1 also represses lignin biosynthesis via direct binding to the 4CL3 promoter. Downregulation of 4CL3 expression results in low lignin deposition in the abscission region and causes easy seed shattering.
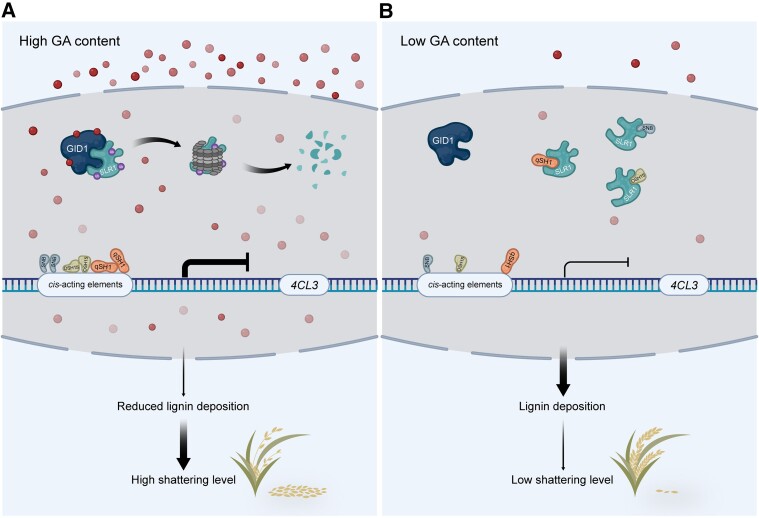
Figure 8.
Proposed working model of the role of SLR1. SLR1 interacts with abscission genes qSH1, OSH15, and SNB to repress their DNA binding activity. In the presence of GA, GA triggers the degradation of SLR1 and frees the three abscission genes to deactivate the expression of common downstream lignin biosynthesis gene, 4CL3, which consequently decreases lignin content in the abscission region, leading to an easy shattering phenotype in the rice (Fig. 8A). When endogenous GA content is low, undegraded SLR1 protein interacts with qSH1, OSH15 and SNB, which prevents them from binding to the 4CL3 promoter. Lignin can be deposited on abscission region normally, exhibiting a low shattering level phenotype in rice (Fig. 8B). The red spheres represent gibberellin. The thickness of the lines with different arrow types represents the degree of effects. Flat head means inhibition and pointed head means activation.
Interactions between DELLAs and different transcription factors constitute a robust module for seed-shattering regulation
An increasing number of studies have revealed that GA regulates the plant development process via physical interactions between DELLAs and different types of transcription factors, which together form a central regulatory system by integrating various development signals. Previous studies have shown that several transcription factors regulate AL formation at different stages (Yoon et al. 2014, 2017; Jiang et al. 2019). AL formation in rice starts at Sp6, and qSH1 is important at this stage for AL differentiation and maintenance. OSH15 and SNB are required for lignin deposition in the AL beginning at Sp7. Here, we show that SLR1 could physically interact with qSH1, OSH15, and SNB and especially exhibited a stronger interaction with qSH1, suggesting that GA is involved in almost the whole AL development process, beginning at the early stage.
Importantly, qSH1, OSH15, and SNB encode different types of transcription factors, but all of them could interact with SLR1. These results further confirmed that the GA/SLR1 transcription factor signaling cascade works as a central command for AL development. Interestingly, in Arabidopsis, the silique dehiscence process also requires the GA/DELLAs transcription factor signaling cascade. The valve margin-specific gene INDEHISCENT (IND) activates GA3ox1 expression and increases the GA content. High GA levels trigger the degradation of DELLA proteins and release the DELLA interaction transcription factor ALCATRAZ (ALC), resulting in valve margin development and promoting silique dehiscence (Arnaud et al. 2010). The GA/DELLA transcription factor signaling cascade might be a conserved regulatory module for seed dispersal among different plant species.
GA response output may orchestrate with shattering genes which leads to rice seed-shattering variation
GWAS analysis showed several natural variations of SLR1 contributed to seed-shattering regulation (Supplemental Fig. S2). And, these variations exist in the form of single nucleobase substitutions in the promoter region, or CDS, but do not cause severe defects. GA is very important for plant development, and strong mutations in the SLR1 gene may result in severe defects. The abscission process normally occurs in a narrow region, so GA regulation in shattering might be mild. Similarly, the qSH1 gene regulation difference in japonica and indica is due to a long-distance SNP, which also suggests that qSH1 regulates shattering in a mildly regulated way (Konishi et al. 2006). And according to the haplotype analysis of SLR1, an SNP (A in japonica and G in indica) may contribute to the reduced seed-shattering degrees from japonica to indica (Supplemental Fig. S2H). So, in a general way, natural variation of SLR1 may influence its protein stability, leading to different GA response outputs, which may orchestrate with shattering genes to regulate the abscission process.
GA may play similar roles in shattering and nonshattering cultivars through lignin deposition
It is known that indica subpopulations have at least partially formed ALs, and japonica subpopulations usually have less developed or invisible ALs. Different AL formations cause seed-shattering variation, and some japonica cultivars are known as nonshattering, like Nip used in this study. In nonshattering cultivars, seeds are usually dropped at a place that may have less mechanical support, as we have shown here that broken positions in nonshattering cultivars could occur at 3 different places: (i) rachilla type (RA-type), broken at rachilla, (ii) PE-type, broken under the AL and on the pedicel, (iii) AL-like-type, broken at a position similar to AL (Fig. 1, A and B). And most broken positions in Nip were AL-like-type. In situ hybridization showed that AL-specific genes qSH1 and SHAT1 have a fair expression at the AL-like region in nonshattering Nip (Supplemental Figs. S5, A to D and S7, E to G), which was different from the easy shattering cultivar Kasalath, where qSH1 and SHAT1 are expressed in a sharp band layer (Supplemental Figs. S5, E to L and S7, H to M). This fair expression of qSH1 and SHAT1 may be important for an AL-like region's establishment in nonshattering cultivars.
We showed here that GA regulates shattering in both shattering and nonshattering cultivars, and this regulation may be through lignin deposition. Several previous studies showed that lignin is very important for seed shattering in both wild rice and cultivated rice varieties (Yoon et al. 2014, 2017; Ning et al. 2023). As presented in Fig. 3, lignin was deposited in a broad region rather than being limited to AL. It could be that different lignin deposition regions could influence the seed dropping-off position. In easy shattering cultivars, like Kasalath, where lignin is almost absent from AL, modulation of GA content could change the lignin deposition level, which will influence the seed shattering level (Supplemental Fig. S10). While in nonshattering Nip, modulation of GA content could influence lignin, which may influence both shattering position and shattering degree, as we could see that detached position in the proSHAT1:GA2ox1-GFP transgenic line of Nip has more at PE-type, and shattering degree also changed (Fig. 2H; Supplemental Fig. S9). It will be interesting to characterize the regulatory network among GA/SLR1, shattering-related transcription factors, and lignin biosynthesis, which may regulate lignin deposition site and level and result in various shattering levels in both shattering and nonshattering rice cultivars or species.
Specific modulation of GA response output may help ideal shattering rice breeding
In recent years, more and more mechanized facilities have been used in agricultural production globally. Rice harvesting methods are also changing from manual to mechanical, step by step. Height reduction and ideal seed-shattering level are two important traits that could deeply influence harvest efficiency. GA is well known for its role in plant height regulation, and the selection of GA biosynthesis-related sd1 mutants led to the “Green Revolution” in the 1960s.
Here, we showed that knockout of SD1 in the cultivar Kasalath, which has clear AL formation, leads to reduced plant height and seeds that are less likely to fall off (Fig. 1, E, J, and K). Importantly, Kasalath sd1 mutants do not cause yield losses (Hu et al. 2019), and the reduced plant height and difficulty of the seeds to shatter could dramatically improve harvest efficiency. Decreasing GA content via proSHAT1:GA2ox-GFP in Kasalath could reduce seed shattering levels, which may result in fewer seeds dropping off during the harvesting process. Similarly, increasing the GA content specific to the abscission region (proSHAT1:D18-GFP) in “nonshattering” Nip reduced the degree of seed shattering without yield loss and may increase harvest efficiency. In summary, our study highlights the role of GA in rice seed-shattering regulation and provides an important and applicable breeding strategy for rice with ideal seed shattering that could fulfill the needs of modern agricultural facilities.
Materials and methods
Plant materials, growth conditions, and agronomic analysis
The rice plants (O. sativa) used in this study, including the wild-type plants, Nipponbare (Nip), Taipei 309 (TP309), ZhongHua11 (ZH11), 9311, Kasalath, the GA-related mutants, d18 (Nip) (Hu, Hu, et al. 2018), NIL (d18), sd1(Hu et al. 2019), slr1 (Ikeda et al. 2001), and eui1 (Zhu et al. 2006), and the relevant transgenic plants were grown in the experimental fields with suitable planting conditions at the Agricultural Genomics Institute in Shenzhen (114°30′E, 22°36′N) and Lingshui (110°0′E, 18°32′N) in Hainan province to speed up the breeding process. Nicotiana benthamiana seedlings were grown in soil pots at 22°C to 24°C under long-day conditions of 3,000 lx light intensity (16 h of light and 8 h of darkness) in a greenhouse with additional cool-blue fluorescent lights (450 to 470 nm). For agronomic analysis, each plot was planted with rice seedlings in six rows, six seedlings per row. At the maturity stage, at least five consecutive plants were selected from the middle row of each plot. Agronomic traits such as plant height, spikelet fertility, productive tiller, 1,000-grain weight, grains per spikelet, grain length, and grain width were measured. The theoretical yield is calculated by the following formula: theoretical yield (kg/mu) = productive tiller (10,000/mu) × grains per spikelet × spikelet fertility (%) × 1,000-grain weight (g) × 10−6 (mu, a Chinese unit of land measurement that is commonly 666.7 m2). Photos were taken by a scanner (Perfection V800 Photo, EPSON).
GWAS analysis
The SNP information of 134 materials completed by the research group in the early stages and the seed-shattering degree phenotype data measured in Ling Shui, Hainan province, were selected for GWAS analysis. Use vcftools (version 0.1.16) (Danecek et al. 2011) to filter SNPs, retain sites with integrity >0.9, minimum allele frequency >0.05, no multiple alleles (parameter: -maf 0.05 -max-missing 0.9). GWAS was performed using the FarmCPU method (Liu et al. 2016) from the rMVP r-package (Yin et al. 2021). Incorporate the five PCA and kinship matrices into the GWAS analysis within the r-package. The threshold for GWAS was calculated using a genetic Type I error calculator (GEC) (Li et al. 2012).
Transformation vector construction
For transgenic rice plant generation, the GreenGate cloning system (Lampropoulos et al. 2013) was used to construct the vector, namely proSLR1:SLR1-GFP, proSHAT1:D18-GFP, proSHAT1:GA2ox1-GFP, pro35s:qSH1-GFP, pro35s:OSH15-GFP, and pro35s:SNB-GFP. The full-length cDNA sequences, promoters, and terminators (internal BsaI restriction enzyme cutting site removed) of these genes were amplified by PCR using the primers listed in Supplemental Table S4 and inserted into the corresponding entry vectors, pGGC000, pGGA000, and pGGE000 (Lampropoulos et al. 2013). Then, these modules were assembled with the destination vector, GFP module, plant resistance cassette, and other modules provided by this system using the PCR thermocycler instrument, followed by the GreenGate reaction instructions.
For qSH1 and 4CL3 CRISPR lines generation, the VK005-01 (Zhang et al. 2022) vector was used provided by Beijing view solid biotechnology company. Briefly, the 20-bp gRNA target sequence was synthesized and inserted into the VK005-01 vector digested by BspQI using T4 DNA ligase (New England Biolabs). The sequences of gRNA targets were shown in Supplemental Table S4.
Shattering degree tests and breaking position ratio calculation
The BTS upon detachment of seeds from the pedicels by pulling was measured by a digital force gauge (ELECALL, ELK-5). Measurements were made at the rice's full ripening stage. BTS values were recorded for at least 30 seeds. For breaking position ratio calculations, the experiment was performed three times. Photos were taken with a stereo microscope (M165FC, Leica).
Scanning electron microscopy
The bases of the spikelet were observed by a TM4000PLUS (Hitachi) SEM. The fracture surface was gold-plated with an ion sputter coater (JFC-1600, JEOL) and observed using an SEM (JSM-6390LV, JEOL) at 15 kV.
RT-qPCR assays
The panicle was harvested from booting stage wild-type, GA-related mutants, and transgenic lines subjected to total RNA isolation. The concert plant RNA reagent (RNeasy Plus Mini Kit, Qiagen) was used to isolate the total RNA. And, total RNA was transcribed with oligo (dT)18 primers using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc.). RT-qPCR was performed on a cycler apparatus (Bio-Rad CFX connect/384) with the ChamQ Universal SYBR qPCR Master Mix (Vazyme, 11184ES08) followed by the reagent specification. Primers for the expression analysis are summarized in Supplemental Table S4.
Immunoblotting
For total protein isolation, young inflorescences of Nip, proSHAT1:D18-GFP, and proSHAT1:GA2ox1-GFP were collected and ground into powder in liquid nitrogen and then suspended in protein extraction buffer (62.5 mm Tris-HCl [pH 7.4], 10% [v/v] glycerol, 2% [w/v] SDS, 2 mm EDTA [pH 7.4], 1 mm PMSF, and 5% [v/v] β-mercaptoethanol). The samples were boiled, and the supernatants were resolved on a 12% (w/v) SDS-PAGE gel. The separated proteins were transferred to a PVDF membrane (Millipore, IPVH00010) and detected by immunoblotting with the SLR1 mouse monoclonal antibodies (generated by Dia-An Biotechnology, 1:1,000) and GAPDH mouse monoclonal antibody (Beyotime Biotechnology, AF0006, 1:1,000), and then HRP-conjugated affinity goat anti-mouse IgG (H + L) (Proteintech, SA00001-1, 1:20,000) was used. Enhanced ECL Chemiluminescent Substrate Kit was used for chemiluminescence immunoassay (Yeasen Biotechnology, BE6706-100). Images were captured by a chemiluminescence imaging system (Bio-Rad ChemiDoc).
Yeast two-hybrid
The full lengths of eight abscission genes were cloned into pGBKT7 and SLR1 was cloned into pGADT7 using ClonExpress Ultra One-Step Cloning Kit (Vazyme), respectively (Zheng et al. 2020). Interactions in Y2H Gold yeast strain (S. cerevisiae) were tested on SD/-Trp/-Leu/-His/-Ade/+AbA medium. The detailed experimental procedures were followed by the yeast instructions (Yeastmaker Yeast Transformation System 2 User Manual). The PCR primers are listed in Supplemental Table S4.
BiFC
The CDS of SLR1 and qSH1, OSH15, and SNB were amplified (Supplemental Table S4) and cloned into the YNE and YCE vectors separately, which contained either N- or C-terminal enhanced yellow fluorescent protein fragments. The resulting constructs were then introduced into Agrobacterium (Agrobacterium tumefaciens) strain GV3101 and coinfiltrated onto the abaxial surface of the leaves of 4-wk-old N. benthamiana plants according to Chen et al. (2008). Fluorescence was observed with a confocal laser-scanning microscope (TCS SP8, Leica) using the preset settings for YFP (Ex: 488 nm, Em: 510 to 550 nm) and chlorophyll (Ex: 561 nm, Em: 650 to 680 nm). Lasers: 488 and 561 nm, intensity: 3.0% to 4.9%, collection bandwidth: hybrid detector (HyD, 500 to 550 nm) and photomultiplier (PMT, 650 to 750 nm), gains (37 to 834). The PCR primers are listed in Supplemental Table S4.
GST pull-down
The experiment was conducted following modified protocols (He et al. 2018). The full-length coding region of SLR1 in the pEasy blunt vector was subcloned into the expression vector pGEX 6p-1 to generate SLR1-GST. The coding regions of qSH1, OSH15, and SNB were introduced into the pET30a vector to generate qSH1-His, OSH15-His, and SNB-His. These constructs were expressed in E. coli (Strain BL21), and the fusion proteins were purified using corresponding affinity chromatography. GST or SLR1-GST coupled GST beads were respectively incubated with qSH1-His, OSH15-His, and SNB-His for 2 h at 4°C and then washed thoroughly, boiled in 1× SDS-PAGE sample buffer (Beyotime Biotechnology), and analyzed by immunoblot using anti-His (HRP-conjugated) mouse monoclonal antibody (EasyBio, BE2062-100, 1:1,000) and anti-GST (HRP-conjugated) mouse monoclonal antibody (EasyBio, BE2065-100, 1:1,000). The PCR primers are listed in Supplemental Table S4.
MST
All MST measurements were performed using Monolith NT standard Capillaries and the Monolith NT.015T device (NanoTemper Technologies) with the laser on for 40 s, resulting in a temperature increase of 6 K. Affinity measurements were also performed using MST buffer (1×PBS with 0.05% [v/v] Tween 20, and pH = 7.4). Measurements were performed at 23°C, 1% to 40% IR laser power, and at a constant concentration of 30 and 0.175 nm of RED-NHS labeled protein with increasing concentrations of purified proteins. The concentration of the target protein SLR1 is kept constant at 0.175 or 30 nm, while the ligand concentration varies from 4.4 to 0.000134 µm for qSH1-GST, 9 to 0.000275 µm for OSH15-GST, and 1.85 µm to 5.65E−05 µm for SNB-GST.
Yeast one-hybrid
The experimental procedures were conducted as described by Wu et al. (2016). The full-length qSH1, OSH15, and SNB ORFs were amplified with gene-specific primers and fused to the GAL4 activation domain of the pGADT7 vector. And, the promoter fragment of 4CL3 was amplified from O. sativa genomic DNA according to the primers and fused separately to the pAbAi vector. The resulting constructs were linearized with BstBI and transformed into the Y1H Gold yeast strain (S. cerevisiae). The plasmid containing AD-qSH1, OSH15, and SNB was subsequently transformed into the Y1H Gold strain containing the pABAi-4CL3 constructs, separately. The p53-AbAi was used as a positive control, and the empty pGADT7 was used as a negative control. The DNA–protein interaction in yeast was selected by SD/-Ura/-Leu agar plates containing 150 ng/mL aureobasidin A (AbA). The PCR primers are listed in Supplemental Table S4.
EMSA
The CDS of qSH1, OSH15, and SNB was amplified and inserted into the pGEX 6p-1 vector to generate the recombinant proteins. The plasmid was introduced into E. coli (BL21) strains. The recombinant proteins were purified according to the manufacturer's instructions. A 30-bp single-strand fragment containing the specific cis-acting element was synthesized based on their promoter sequences and labeled with the fluorescent probe 6-FAM (Sangon Biotech). The same fragment without 6-FAM labeling was used as a competitor. The probes were incubated with the fusion protein in a 20μL reaction solution with or without the 200-fold competitor for 30 min at room temperature. The reaction products were then separated by electrophoresis on a 6% native polyacrylamide gel, and migration of the 6-FAM-labeled probe was visualized using the multispectral laser imager GE Amersham Typhoon. The probes are listed in Supplemental Table S4.
Transient expression in rice protoplast
For the construction of the effector expression vector, the full-length qSH1, OSH15, and SNB cDNA were amplified and cloned into the pGreenII 62-SK vector under the control of the CaMV 35S promoter. Intact (1,817 bp) 4CL3 promoters were amplified and constructed into the pGreenII 0800-LUC reporter vector (Hellens et al. 2005). Isolation of rice sheath protoplasts and PEG-mediated transfection were performed as previously described. Plasmids (5 μm) were transformed into protoplasts. The firefly and Renilla luciferase activity ratios were measured using a Dual-Luciferase Assay Kit (Promega) followed by its instructions. The PCR primers are listed in Supplemental Table S4.
ChIP-qPCR and ChIP-seq analysis
ChIP was conducted following modified protocols from Abcam. Three biological replicates of pro35S:GFP as a control. A biological replicate refers to an assay from an independent sample collection. Formaldehyde cross-linked chromatin DNA was isolated from young inflorescences of wild-type and qSH1, OSH15, and SNB overexpression plants with a GFP tag. Immunoprecipitation was performed with an anti-GFP (Invitrogen, A-11122) antibody with a 1:100 dilution (Invitrogen). The immunoprecipitated DNA was purified by the Qiagen DNA Purification Kit (Qiagen) and used as a template for library construction and sequencing (Novogene) and for PCR amplification (Supplemental Table S4) using a cycler apparatus (Bio-Rad CFX384) with the ChamQ Universal SYBR qPCR Master Mix (Vazyme). Enrichment folds of qSH1, OSH15, and SNB-bound DNA fragments were obtained as the ratio of anti-GFP IP to control beads IP of each independent replicate. The data are presented as means ± Sd of three biological repeats.
For ChIP-seq data analysis, with a 6 Gb (Gbase) sequencing depth, the output reads were trimmed with fastp to obtain clean data at first (Chen et al. 2018). Then, these filtered reads were aligned to Rice Genome build MSU 7.0 by BWA (Li 2013) using default parameters. Duplicated reads and reads with low mapping quality were further identified and removed with SAMtools (Li et al. 2009). Enriched intervals were identified by MACS2 (Zhang et al. 2008) with default parameters, and deepTools was utilized to plot (Ramírez et al. 2014).
RNA-seq analysis
Total RNA was isolated from the wild-type's young panicles (0 to 4 cm) and the proSHAT1:D18-GFP transgenic lines with three biological replicates each containing five plants. The total RNA was sent to the Novogene company for library construction and sequencing. The raw reads were mapped to the reference genome (Os-Nipponbare-Reference-IRGSP-1.0, MSU7) using HISAT2 with the default parameters (Kawahara et al. 2013; Kim et al. 2019). StringTie (Pertea et al. 2015) was used to calculate each gene's transcripts per kilobase of exon model per million mapped reads (TPM). Then, the DEGs (fold change ≥ 1.5, FDR < 0.001) were identified with DESeq2 (Love et al. 2014) between the transgenic lines and the wild-type. The functional category analysis (GO and KEGG analysis) of the DEGs was performed using DAVID (the Database for Annotation, Visualization, and Integrated Discovery) (Huang et al. 2009).
Examination of endogenous GAs
The young inflorescences were collected from Nip, proSHAT1:D18-GFP, and proSHAT1:GA2ox1-GFP and frozen rapidly in liquid nitrogen for endogenous GA measurement. GA content was detected by Nanjing Convinced-test Technology Co., Ltd based on the High-Performance Liquid Chromatography (Agilent1290) and tandem MS (MS/MS, Applied Biosystems 6500 Quadrupole Trap).
In situ hybridization
Fresh young panicles were collected and fixed in a 4% polyformaldehyde solution overnight at 4°C and then dehydrated through a graded series of ethanol from 30% to 85% (Coen et al. 1990; Jackson 1991). Tissues were dried and embedded using Leica HistoCore PEARL and Leica HistoCore Acradia H. And, wax blocks were sectioned using a microtome (Leica RM2235). Probes were labeled with digoxigenin using the DIG RNA Labeling Kit (SP6/T7, Roche) and primers in Supplemental Table S4. Pretreatment of sections, hybridization, and digoxigenin signal detection were performed following Wang, Kohlen, et al. (2014). Images were obtained using an Olympus microscope (BX53).
Confocal imaging
To observe the SLR1-GFP signal in rice inflorescences, the leaf sheath was artificially removed, and the spikelet meristem was exposed. The rice spikelet in the booting stage was embedded in tissue freezing medium (Richard-Allan Scientific Neg-50) and sliced into 10 μm thick sections using a freezing microtome (Thermo Scientific CryoStar NX50 OP). Fluorescent and bright field images were taken on a confocal laser-scanning microscope (TCS SP8, Leica). The GFP signal was imaged using 488 nm excitation and 505 to 530 nm emission. Set other parameters to: Intensity, 2.0%, Hyd (500 to 550 nm, gain 150.7), PMT (640 to 720 nm, gain 553.5), PMT Transmission Channel (gain, 265.2).
Lignin deposition analysis
The spikelet embedded in the paraplast was longitudinally sectioned, and these slides were placed in a container of histoclear to remove the paraffin. Then place the slides in two changes of absolute ethanol for a few minutes to remove the histoclear. Stain the slides with a 2% (w/v) phloroglucinol solution (200 mg phloroglucinol, 10 mL 95% ethanol) for 2 min before soaking them in 20% (v/v) HCL. Examine the slides immediately using an Olympus microscope (BX53) (Liljegren 2010). The quantitative analysis of the staining degree is calculated by ImageJ (version 1.53t).
Statistical analysis
The two-tailed Student's t-tests were used to compare data from two groups, and the one-way/two-way ANOVA test used to compare multiple groups was performed using GraphPad Prism 9. Statistical data are provided in Supplemental Data Set 1.
Accession numbers
All gene information in this study was obtained from the Rice Genome Annotation Project (MSU-RGAP) according to accession numbers as follows: D18 (LOC_Os01g08220), SLR1 (LOC_Os03g49990), GA2ox1 (LOC_Os05g06670), EUI1 (LOC_Os05g40384), SD1 (LOC_Os01g66100), qSH1 (LOC_Os01g62920), sh4(LOC_Os04g57530), CPL1 (LOC_Os07g10690), SHAT1 (LOC_Os04g55560), Sh1 (LOC_Os03g44710), SH5 (LOC_Os05g38120), OSH15 (LOC_Os07g03770), SNB (LOC_Os07g13170), 4CL3 (LOC_Os02g08100), PAL1 (LOC_Os02g41630), CCoAoMT (LOC_Os08g38910), CCR1 (LOC_Os02g56460), COMT1 (LOC_Os08g06100), CAD2 (LOC_Os02g09490), 4CL4 (LOC_Os06g44620), 4CL5 (LOC_Os06g44620), PAL6 (LOC_Os04g43800), PAL8 (LOC_Os11g48110), CCR10 (LOC_Os02g56700), and ubiquitin (LOC_Os03g13170).
Supplementary Material
Acknowledgments
We thank Prof. He Zuhua (CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for kindly sharing materials.
Contributor Information
Hao Wu, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Qi He, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Bing He, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Shuyi He, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China; State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng 475001, China; Shenzhen Research Institute of Henan University, Shenzhen 518000, China.
Longjun Zeng, Yichun Academy of Science, Yichun 336000, China.
Longbo Yang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Hong Zhang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Zhaoran Wei, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Xingming Hu, College of Agronomy, Anhui Agricultural University, Heifei 230026, China.
Jiang Hu, State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou 311401, China.
Yong Zhang, Department of Biotechnology, School of Life Sciences and Technology, Center of Informational Biology, University of Electronic Science and Technology of China, Chengdu 611731, China.
Lianguang Shang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Suikang Wang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Peng Cui, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.
Guosheng Xiong, Academy for Advanced Interdisciplinary Studies, Plant Phenomics Research Center, Nanjing Agricultural University, Nanjing 210095, China.
Qian Qian, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China; State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou 311401, China.
Quan Wang, Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China; College of Agricultural Sciences, Nankai University, Tianjin 300071, China.
Author contributions
Q.W. conceptualized and supervised the project. H.W. and Q.H. performed most of the experiments. B.H. and P.C. participated in ChIP-seq data analysis. L.Y., H.Z., Z.W., and L.S. participated in GWAS data analysis. S.H. and L.Z. participated in rice cultivation. X.H., J.H., Y.Z., S.W., G.X., and Q.Q. provided modification suggestions. H.W., Q.H., and Q.W. drafted the manuscript. All authors contributed to review and editing the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. PCA and (Q-Q) plot for GWAS analysis.
Supplemental Figure S2. Haplotype analysis of SLR1 across the promoter and ORF regions.
Supplemental Figure S3. Morphological characteristics of fracture surfaces in GA-related mutants and their corresponding wild-type.
Supplemental Figure S4. The breaking position is calculated in Nip.
Supplemental Figure S5. Expression analysis of qSH1 in the different spikelet development stages of the Nip and Kasalath using in situ hybridization.
Supplemental Figure S6. qSH1 is also functional in Nip.
Supplemental Figure S7. Expression pattern of SHAT1 in rice spikelet development in Nipponbare and Kasalath backgrounds.
Supplemental Figure S8. Agronomic trait of proSHAT1:D18-GFP and proSHAT1:GA2ox1-GFP transgenic lines.
Supplemental Figure S9. The ratio of breaking position in Nip, proSHAT1:GA2ox1-GFP, and proSHAT1:D18-GFP.
Supplemental Figure S10. GA content alteration in the abscission region-influenced seed-shattering degree in Kasalath.
Supplemental Figure S11. TPMs of 8 abscission genes in inflorescence meristem of transgenic line proSHAT1:D18-GFP and Nip in RNA-seq data with three biological replicates (P < 0.05).
Supplemental Figure S12. GO enrichment and KEGG pathway analysis of DEGs.
Supplemental Figure S13. The monolignol biosynthetic pathway and tissue-specific expressions.
Supplemental Figure S14. Expression pattern of SLR1 in rice inflorescence development.
Supplemental Figure S15. GO enrichment of qSH1, OSH15, and SNB target genes.
Supplemental Figure S16. The distribution of qSH1, OSH15, and SNB binding elements in 4CL3.
Supplemental Figure S17. Sequences alignment of wild-type and 4CL3 CRISPR line.
Supplemental Table S1. Rice germplasm used in GWAS.
Supplemental Table S2. Haplotypes of SLR1 loci and BTS value for GWAS.
Supplemental Table S3. Rice tissue-specific expression derived from RNA-Seq data.
Supplemental Table S4. Primers used in this study.
Supplemental Data Set 1. Statistical analysis.
Funding
This research was supported by the National Natural Science Foundation of China (31901527 and 32070202), the China Postdoctoral Science Foundation (2018M641555), the Science Technology and Innovation Commission of Shenzhen Municipality (JCYJ20170303154319837 and JCYJ20170412155447658), the Guangdong Basic and Applied Basic Research Foundation (2020B1515120086), the National Key R&D Program (2020YFE0202300), the Key-Area Research and Development Program of Guangdong Province (2021B0707010006), and the Science Technology Innovation and Industrial Development of Shenzhen Dapeng New District (PT201901-18).
Data availability
All study data are included in the article and the supplementary information. The data of this study are available from the corresponding author upon reasonable request.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Arnaud N, Girin T, Sorefan K, Fuentes S, Wood TA, Lawrenson T, Sablowski R, Ostergaard L. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010:24(19):2127–2132. 10.1101/gad.593410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang SW, Choi S, Jin X, Jung SE, Choi JW, Seo JS, Kim J-K. Transcriptional activation of rice CINNAMOYL-CoA REDUCTASE 10 by OsNAC5, contributes to drought tolerance by modulating lignin accumulation in roots. Plant Biotechnol J. 2022:20(4):736–747. 10.1111/pbi.13752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti G, Unterholzner SJ, Scintu D, Salvi E, Svolacchia N, Di Mambro R, Ruta V, Linhares Scaglia F, Vittorioso P, Sabatini S, et al. A PHABULOSA-controlled genetic pathway regulates ground tissue patterning in the Arabidopsis root. Curr Biol. 2021:31(2):420–426.e6. 10.1016/j.cub.2020.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou J-M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008:146(2):368–376. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018:34(17):i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Romero J, Doyle S, Elliott R, Murphy G, Carpenter R. floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell. 1990:63(6):1311–1322. 10.1016/0092-8674(90)90426-F [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011:27(15):2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999:19(1):9–20. 10.1046/j.1365-313X.1999.00491.x [DOI] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci U S A. 2008:105(43):16814–16819. 10.1073/pnas.0807821105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Mori M, Watanabe S, Miyamoto C, Ito T, Takahashi Y. DELLA-GAF1 complex is a main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol. 2017:175(3):1395–1406. 10.1104/pp.17.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Shen J, Li L. Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol. 2011:157(2):574–586. 10.1104/pp.111.178301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Hahlbrock K. The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci U S A. 2004:101(7):2209–2214. 10.1073/pnas.0307307101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Liu Y, Yuan D, Duan M, Liu Y, Shen Z, Yang C, Qiu Z, Liu D, Wen P, et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc Natl Acad Sci U S A. 2020:117(1):271–277. 10.1073/pnas.1902771116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Tang Q-Y, Sun Y-F, Zhou M, Gartner W, Zhao K-H. Chromophorylation of cyanobacteriochrome Slr1393 from Synechocystis sp. PCC 6803 is regulated by protein Slr2111 through allosteric interaction. J Biol Chem. 2018:293(46):17705–17715. 10.1074/jbc.RA118.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. The genes of the green revolution. Trends Genet. 2003:19(1):5–9. 10.1016/S0168-9525(02)00009-4 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005:1(1):13. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Aya K, Kondo M, Okuno A, Morinaka Y, Matsuoka M. OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep. 2012:31(1):91–101. 10.1007/s00299-011-1142-7 [DOI] [PubMed] [Google Scholar]
- Hu J, Israeli A, Ori N, Sun T-P. The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell. 2018:30(8):1710–1728. 10.1105/tpc.18.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Hu X, Hu J, Shang L, Dong G, Zeng D, Guo L, Qian Q. Xiaowei, a new rice germplasm for large-scale indoor research. Mol Plant. 2018:11(11):1418–1420. 10.1016/j.molp.2018.08.003 [DOI] [PubMed] [Google Scholar]
- Hu W-J, Kawaoka A, Tsai C-J, Lung J, Osakabe K, Ebinuma H, Chiang VL. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc Natl Acad Sci U S A. 1998:95(9):5407–5412. 10.1073/pnas.95.9.5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Cui Y, Dong G, Feng A, Wang D, Zhao C, Zhang Y, Hu J, Zeng D, Guo L, et al. Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces. Sci Rep. 2019:9(1):19096. 10.1038/s41598-019-55757-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wang S, Zhang B, Shang-Guan K, Shi Y, Zhang D, Liu X, Wu K, Xu Z, Fu X, et al. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell. 2015:27(6):1681–1696. 10.1105/tpc.15.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009:4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huangfu L, Chen R, Lu Y, Zhang E, Miao J, Zuo Z, Zhao Y, Zhu M, Zhang Z, Li P, et al. OsCOMT, encoding a caffeic acid O-methyltransferase in melatonin biosynthesis, increases rice grain yield through dual regulation of leaf senescence and vascular development. Plant Biotechnol J. 2022:20(6):1122–1139. 10.1111/pbi.13794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001:13(5):999–1010. 10.1105/tpc.13.5.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. Cloning and functional analysis of two gibberellin 3 beta -hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci U S A. 2001:98(15):8909–8914. 10.1073/pnas.141239398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005:46(1):23–47. 10.1093/pcp/pci501 [DOI] [PubMed] [Google Scholar]
- Jackson D. In-situ hybridisation in plants. In: Bowles DJ, Gurr SJ, McPhereson M, editors. Molecular plant pathology: a practical approach. UK: Oxford University Press; 1991. p. 17–25. [Google Scholar]
- Ji H, Kim S-R, Kim Y-H, Kim H, Eun M-Y, Jin I-D, Cha Y-S, Yun D-W, Ahn B-O, Lee MC, et al. Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J. 2010:61(1):96–106. 10.1111/j.1365-313X.2009.04039.x [DOI] [PubMed] [Google Scholar]
- Ji H-S, Chu S-H, Jiang W, Cho Y-I, Hahn J-H, Eun M-Y, McCouch SR, Koh H-J. Characterization and mapping of a shattering mutant in rice that corresponds to a block of domestication genes. Genetics. 2006:173(2):995–1005. 10.1534/genetics.105.054031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Ma X, Zhao S, Tang Y, Liu F, Gu P, Fu Y, Zhu Z, Cai H, Sun C, et al. The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size. Plant Cell. 2019:31(1):17–36. 10.1105/tpc.18.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013:6(1):4. 10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K, Takahashi H, Umemura K, Umezawa T, Shimamoto K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc Natl Acad Sci U S A. 2006:103(1):230–235. 10.1073/pnas.0509875103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37(8):907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M. An SNP caused loss of seed shattering during rice domestication. Science (80-). 2006:312(5778):1392–1396. 10.1126/science.1126410 [DOI] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J. GreenGate - a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One. 2013:8(12):e83043. 10.1371/journal.pone.0083043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science (80-). 2006:311(5769):1936–1939. 10.1126/science.1123604 [DOI] [PubMed] [Google Scholar]
- Li, H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, arXiv:1303.3997, preprint: not peer reviewed.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009:25(16):2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M-X, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012:131(5):747–756. 10.1007/s00439-011-1118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z, Yu H, Duan J, Yuan K, Yu C, Meng X, Kou L, Chen M, Jing Y, Liu G, et al. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat Commun. 2019:10(1):2738. 10.1038/s41467-019-10667-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S. Phloroglucinol stain for lignin. Cold Spring Harb Protoc. 2010;1:pdb.prot4954. 10.1101/pdb.prot4954 [DOI] [PubMed] [Google Scholar]
- Lim S, Park J, Lee N, Jeong J, Toh S, Watanabe A, Kim J, Kang H, Kim DH, Kawakami N, et al. ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell. 2013:25(12):4863–4878. 10.1105/tpc.113.118604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Griffith ME, Li X, Zhu Z, Tan L, Fu Y, Zhang W, Wang X, Xie D, Sun C. Origin of seed shattering in rice (Oryza sativa L.). Planta. 2007:226(1):11–20. 10.1007/s00425-006-0460-4 [DOI] [PubMed] [Google Scholar]
- Lin Z, Li X, Shannon LM, Yeh C-T, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE, et al. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 2012:44(6):720–724. 10.1038/ng.2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, Buckler ES, Zhang Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016:12(2):e1005767. 10.1371/journal.pgen.1005767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins AO, Nunes-Nesi A, Araujo WL, Fernie AR. To bring flowers or do a runner - GAs make the decision. Mol Plant. 2018:11(1):4–6. 10.1016/j.molp.2017.12.005 [DOI] [PubMed] [Google Scholar]
- Meyer RS, Choi JY, Sanches M, Plessis A, Flowers JM, Amas J, Dorph K, Barretto A, Gross B, Fuller DQ, et al. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat Genet. 2016:48(9):1083–1088. 10.1038/ng.3633 [DOI] [PubMed] [Google Scholar]
- Ning J, He W, Wu L, Chang L, Hu M, Fu Y, Liu F, Sun H, Gu P, Ndjiondjop M-N, et al. The MYB transcription factor Seed Shattering 11 controls seed shattering by repressing lignin synthesis in African rice. Plant Biotechnol J. 2023;21(5):931–942. 10.1111/pbi.14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015:33(3):290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014:42(W1):W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M, Sakamoto T, Saito T, Matsuoka M, Tanaka H, Kobayashi M. Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. J Plant Res. 2003:116(2):161–164. 10.1007/s10265-003-0080-z [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001:125(3):1508–1516. 10.1104/pp.125.3.1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. A mutant gibberellin-synthesis gene in rice. Nature. 2002:416(6882):701–702. 10.1038/416701a [DOI] [PubMed] [Google Scholar]
- Shang L, Li X, He H, Yuan Q, Song Y, Wei Z, Lin H, Hu M, Zhao F, Zhang C, et al. A super pan-genomic landscape of rice. Cell Res. 2022:32(10):878–896. 10.1038/s41422-022-00685-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Lin T, Grandellis C, Yu M, Hannapel DJ. The BEL1-like family of transcription factors in potato. J Exp Bot. 2014:65(2):709–723. 10.1093/jxb/ert432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, Choi IR, Omura T, Kikuchi S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011:52(2):344–360. 10.1093/pcp/pcq196 [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A. 2002:99(13):9043–9048. 10.1073/pnas.132266399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M, McCouch S. The complex history of the domestication of rice. Ann Bot. 2007:100(5):951–957. 10.1093/aob/mcm128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yu Y, Haberer G, Marri PR, Fan C, Goicoechea JL, Zuccolo A, Song X, Kudrna D, Ammiraju JSS, et al. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat Genet. 2014:46(9):982–988. 10.1038/ng.3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Kohlen W, Rossmann S, Vernoux T, Theres K. Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell. 2014:26(5):2068–2079. 10.1105/tpc.114.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J-K, Chapple C. The origin and evolution of lignin biosynthesis. New Phytol. 2010:187(2):273–285. 10.1111/j.1469-8137.2010.03327.x [DOI] [PubMed] [Google Scholar]
- Whetten R, Sederoff R. Lignin biosynthesis. Plant Cell. 1995:7(7):1001–1013. 10.2307/3870053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Fu B, Sun P, Xiao C, Liu J-H. A NAC transcription factor represses putrescine biosynthesis and affects drought tolerance. Plant Physiol. 2016:172(3):1532–1547. 10.1104/pp.16.01096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, He Q, Wang Q. Advances in rice seed shattering. Int J Mol Sci. 2023:24(10):8889. 10.3390/ijms24108889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Zhang H, Tang Z, Xu J, Yin D, Zhang Z, Yuan X, Zhu M, Zhao S, Li X, et al. rMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for Genome-Wide Association Study. Genomics Proteomics Bioinformatics. 2021:19(4):619–628. 10.1016/j.gpb.2020.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Cho L-H, Antt HW, Koh H-J, An G. KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 2017:174(1):312–325. 10.1104/pp.17.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Cho L-H, Kim SL, Choi H, Koh H-J, An G. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 2014:79(5):717–728. 10.1111/tpj.12581 [DOI] [PubMed] [Google Scholar]
- Yu Y, Leyva P, Tavares RL, Kellogg EA. The anatomy of abscission zones is diverse among grass species. Am J Bot. 2020:107(4):549–561. 10.1002/ajb2.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zeng P, Yu H, Meng X, Li J. Protocol for genome editing in wild allotetraploid rice Oryza alta. STAR Protocols. 2022:3(4):101789. 10.1016/j.xpro.2022.101789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008:9(9):R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Hong K, Zeng L, Wang L, Kang S, Qu M, Dai J, Zou L, Zhu L, Tang Z, et al. Karrikin signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell. 2020:32(9):2780–2805. 10.1105/tpc.20.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao H, Chern M, Yin J, Chen Y, Wang J, Zhu X, Chen Z, Yuan C, Zhao W, et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc Natl Acad Sci U S A. 2018:115(12):3174–3179. 10.1073/pnas.1705927115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lu D, Li C, Luo J, Zhu BF, Zhu J, Shangguan Y, Wang Z, Sang T, Zhou B, et al. Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell. 2012:24(3):1034–1048. 10.1105/tpc.111.094383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006:18(2):442–456. 10.1105/tpc.105.038455 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and the supplementary information. The data of this study are available from the corresponding author upon reasonable request.