Abstract
A culture-independent molecular phylogenetic survey was carried out for the bacterial community in Obsidian Pool (OP), a Yellowstone National Park hot spring previously shown to contain remarkable archaeal diversity (S. M. Barns, R. E. Fundyga, M. W. Jeffries, and N. R. Page, Proc. Natl. Acad. Sci. USA 91:1609–1613, 1994). Small-subunit rRNA genes (rDNA) were amplified directly from OP sediment DNA by PCR with universally conserved or Bacteria-specific rDNA primers and cloned. Unique rDNA types among >300 clones were identified by restriction fragment length polymorphism, and 122 representative rDNA sequences were determined. These were found to represent 54 distinct bacterial sequence types or clusters (≥98% identity) of sequences. A majority (70%) of the sequence types were affiliated with 14 previously recognized bacterial divisions (main phyla; kingdoms); 30% were unaffiliated with recognized bacterial divisions. The unaffiliated sequence types (represented by 38 sequences) nominally comprise 12 novel, division level lineages termed candidate divisions. Several OP sequences were nearly identical to those of cultivated chemolithotrophic thermophiles, including the hydrogen-oxidizing Calderobacterium and the sulfate reducers Thermodesulfovibrio and Thermodesulfobacterium, or belonged to monophyletic assemblages recognized for a particular type of metabolism, such as the hydrogen-oxidizing Aquificales and the sulfate-reducing δ-Proteobacteria. The occurrence of such organisms is consistent with the chemical composition of OP (high in reduced iron and sulfur) and suggests a lithotrophic base for primary productivity in this hot spring, through hydrogen oxidation and sulfate reduction. Unexpectedly, no archaeal sequences were encountered in OP clone libraries made with universal primers. Hybridization analysis of amplified OP DNA with domain-specific probes confirmed that the analyzed community rDNA from OP sediment was predominantly bacterial. These results expand substantially our knowledge of the extent of bacterial diversity and call into question the commonly held notion that Archaea dominate hydrothermal environments. Finally, the currently known extent of division level bacterial phylogenetic diversity is collated and summarized.
Lithotrophic microbes and the communities that they support contribute significantly to the chemistry of the biosphere. Lithotrophic metabolism, energy production from inorganic chemicals such as hydrogen, reduced iron, and reduced sulfur compounds, is phylogenetically more widely distributed among Bacteria and Archaea than is either phototrophic or organotrophic metabolism (23, 29). Environments that are expected to support primarily lithotrophs, such as the subsurface and geothermal settings, are more prevalent on Earth than are the organically rich zones of the landmasses. Considering the importance of lithotrophic metabolism to environmental processes, it is remarkable that lithotrophic microbes and the communities that they support are relatively little known. An important reason for our limited fund of information about lithotrophic organisms is that such organisms are difficult, perhaps often impossible, to cultivate in isolation from the native setting. In general, comprehensive description of microbial communities requires the use of techniques that sidestep cultivation because typically only a small fraction (<1%) of naturally occurring microorganisms is cultivatable by standard techniques (see reference 4 for a review).
One approach to the identification of the constituents of natural microbial communities is through the use of rRNA-based, molecular phylogenetic techniques. In this approach, rRNA genes are obtained directly from environmental DNA, commonly through PCR and cloning, and sequenced (4, 30). Comparative analyses of the rRNA sequences reveal the phylogenetic types of organisms that comprise the community. Some properties of otherwise unknown organisms can be inferred based on the properties of their characterized relatives, and the sequences can be used as the basis of molecular tools with which to study the respective organisms. Application of molecular methods to the characterization of natural microbial communities has significantly expanded our view of the extent of microbial diversity. Sequences obtained from the environment are seldom identical to sequences from cultured organisms. This indicates that our understanding of the phylogenetic makeup of the microbial biosphere based on cultivation studies is seriously limited.
Obsidian Pool (OP), a 75- to 95°C hot spring on the northern flank of the Yellowstone caldera, is rich in reduced iron, sulfide, CO2, and probably hydrogen. OP is a fertile ground for the discovery of novel microbial diversity in communities based on lithotrophy. A previous molecular survey of rRNA genes in DNA isolated directly from OP revealed the occurrence of a wealth of members of the Archaea (6, 7). Representatives of the phylogenetic domain Archaea are commonly associated with high-temperature geothermal settings such as OP (40). Some of the novel sequences are closely related to those of known organisms, but most of the novel archaeal rRNA genes represent organisms that are only distantly related to cultured ones. Some of these novel organisms have now been cultured, verifying that the detected rRNA genes in fact represent organisms (13, 21).
Although high-temperature environments popularly are considered a particular province of Archaea, hyperthermophilic Bacteria comprise the major constituents of some high-temperature communities (34). This proved to be the case in OP, too; we have now carried out a molecular phylogenetic survey of the bacterial constituents of a sample of OP sediment. Many novel, division level bacterial lineages have been identified, expanding significantly the known extent of diversity in the phylogenetic domain Bacteria.
MATERIALS AND METHODS
Sample collection.
Sediment samples (75 to 93°C) were collected from OP in June 1993 and stored at −80°C. The same samples used in the study of OP Archaea (7) were used in the present study. Contact slides used in the present study also were obtained in 1993; glass microscope slides in a stainless steel rack were immersed in OP for 1 week and were preserved in 4% paraformaldehyde in 1× phosphate-buffered saline at 4°C. Scanning electron micrographs were taken of the slides in 1996 as previously described (37). Samples from two additional hot pools, collected in July 1995, were used for the membrane hybridization analysis. Sample 01A was taken from a dark green microbial mat growing on the northwest side of a 72°C pool in the White Creek area, Lower Geyser Basin (near Octopus Spring at ca. 44°31′56"N and 110°47′48"W). Sample N10 was sediment taken from a small orange-colored pool (70 to 76°C, pH 4 to 5) located on the west side of the Reservoir in the Norris Geyser Basin (ca. 44°43′51"N and 110°42′33"W).
DNA extraction.
Community nucleic acids were extracted from OP sediment samples by the freeze-thaw lysis procedure of Barns et al. (7) or by physical disruption of cells by bead beating instead of freeze-thaw lysis. DNA from samples 01A and N10 was extracted by the bead-beating protocol. For the bead-beating method, 0.5 to 1.0 g of sediment was resuspended in modified 2× buffer A (200 mM Tris [pH 8.0], 50 mM EDTA, 200 mM NaCl, 2 mM sodium citrate, 10 mM CaCl2)–polyadenosine (100 μg/ml)–lysozyme (5 mg/ml) in a 2-ml screw-cap tube and incubated for 40 min at 37°C. Proteinase K (to 1 mg/ml) and sodium dodecyl sulfate (SDS) (to 0.3% [wt/vol]) were then added, and the mixture was incubated for a further 30 min at 50°C. Samples were reciprocated on a Mini-Beadbeater (Biospec Products, Inc., Bartlesville, Okla.) at low speed for 2 min in the presence of 15% (vol/vol) phenol, 2% (wt/vol) SDS, and approximately 0.5 g of acid-washed zirconium beads (0.1-mm diameter). Lysates were extracted with phenol-chloroform, then sodium acetate was added to 0.3 M, and nucleic acids were precipitated from solution by addition of 1 volume of isopropanol. Community nucleic acids were purified by electrophoresis through a 1.5% low-melting-point agarose gel (SeaPlaque GTG; FMC Bioproducts, Rockland, Maine) in the presence of 2% polyvinylpyrrolidone as described by Young et al. (48). Community DNAs ≥20 kb in size were excised from the gel and used directly as template in subsequent PCRs or purified from the gel slice with a DNA purification kit (UltraClean; Mo Bio Laboratories, Inc., Solana Beach, Calif.) and eluted in 20 μl of 10 mM Tris, pH 8.0.
PCR and cloning.
Community ribosomal DNAs (rDNAs) were PCR amplified from 1 to 50 ng of bulk DNA in reactions containing (as final concentrations) 1× PCR buffer II (Perkin-Elmer, Foster City, Calif.), 2.5 mM MgCl2, 4 × 200 μM deoxynucleoside triphosphates, 300 nM each forward and reverse primer, and 0.025 U of AmpliTaq Gold or AmpliTaq LD (Perkin-Elmer) per μl. Acetamide (5% [vol/vol] final concentration) was added to most PCRs (see below) to promote amplification of templates containing high G+C content (33). Reaction mixtures were incubated in a model PT-100 thermal cycler (MJ Research Inc., Watertown, Mass.) for an initial denaturation at 94°C for 3 min followed by 30 cycles of 92°C for 1.5 min, 50°C for 1.5 min, and 72°C for 2 min.
Three clone libraries were prepared. For clone library OPS, rDNAs were amplified in the presence of acetamide with the universal oligonucleotide primers, 533FPL (5′-GCGGATCCTCTAGACTGCAGTGCCAGCAGCCGCGGTAA-3′) and 1492RPL (5′-GGCTCGAGCGGCCGCCCGGGTTACCTTGTTACGACTT-3′), which have restriction enzyme linker tails for cohesive-end cloning (24). Amplified rDNAs pooled from three reaction mixtures were cloned as NotI/PstI fragments in pBluescript KS− vector (Stratagene, La Jolla, Calif. [36]). Clone library OPT was also prepared from rDNAs that were amplified with standard universal primers, 533F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (24), in the presence of acetamide. Amplified rDNAs, pooled from three reaction mixtures, were cloned directly into the pGEM-T vector according to the manufacturer’s instructions (Promega, Madison, Wis.). Clone library OPB was prepared from rDNAs that were amplified with a Bacteria-specific primer pair, 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (24), in the presence and absence of acetamide. Amplified rDNAs, pooled from 10 reaction mixtures (four of which lacked acetamide), were cloned into the pCR2.1 vector according to the manufacturer’s instructions (Invitrogen Corp., San Diego, Calif.). Plasmid DNAs containing inserts were isolated for restriction fragment length polymorphism (RFLP) analysis and sequencing (see below), by alkaline lysis procedures, either individually (QIAprep spin columns; Qiagen, Inc., Chatsworth, Calif.) or in 96-well arrays (28).
RFLP screening of rDNA clones.
rDNA inserts from recombinant clones were reamplified by PCR in reaction mixtures containing (as final concentrations) 1× Pfu reaction buffer (Stratagene), 2.5 mM MgSO4, 4 × 100 μM deoxynucleoside triphosphates, 150 nM each forward and reverse primer, ca. 0.01 U of Pfu DNA polymerase per μl, and 50 to 100 pg of purified plasmid per μl as template. Vector primers specific to each plasmid or rDNA-specific primers were used. The cycle profile was the same as for the initial amplification of the rDNA (above). Aliquots (14.5 μl) of crude reamplified rDNA PCR products were digested with 1 U each of the 4-base-specific restriction endonucleases HinP1 I and MspI in 1× NEB buffer 2 (New England Biolabs, Beverly, Mass.)–0.01% Triton X-100 in a final volume of 20 μl, for 3 h at 37°C. Digested products were separated by agarose (4% MetaPhor; FMC Bioproducts) gel electrophoresis. Bands were visualized by staining with ethidium bromide and UV illumination. RFLP patterns for each library were grouped visually, and representatives were selected (Results) for sequencing.
Sequencing of rDNA clones.
Plasmid templates from representative clones (Table 1) were sequenced with an ABI 377 or 373 DNA sequencer (Dye-Terminator Cycle Sequencing Ready Reaction FS kit; PE Applied Biosystems, Foster City, Calif.) or a 4000L Long Read IR DNA Sequencer (infrared-labeled primers; LI-COR Inc., Lincoln, Nebr.) according to the manufacturer’s instructions. For first-pass analyses of clones, the small-subunit (SSU)-rDNA primer, 533F, in addition to the forward and reverse vector primers, was used as a sequencing primer to obtain a complete single-pass sequence of the insert (Escherichia coli positions 28 to 1491 or 534 to 1491). Second-pass analysis of selected clones to determine the sequence for both strands was performed with Applied Biosystems Inc. dye terminator chemistry and a combination of the SSU-rDNA primers; 519R (5′-GWATTACCGCGGCKGCTG-3′), 787R (5′-CTACCAGGGTATCTAAT-3′), 704F (5′-GTAGCGGTGAAATGCGTAGA-3′), 805F (5′-ATTAGATACCCTGGTAGTC-3′), and 1100R (5′-AGGGTTGCGCTCGTTG-3′) (24, 38).
TABLE 1.
Summary of the 16S rDNA sequences identified in OP sediment from three clone libraries, OPB, OPS, and OPT
| Type sequencea | Other sequenced representative(s)b | No. of clonesc | Bacterial division | Database matches (≥98% identity) | Inferred metabolism | Inferred thermophilyd |
|---|---|---|---|---|---|---|
| OPT5 | 1 | α-Proteobacteria | ||||
| OPB30 | S63, S83, S130, S140 | 14 | β-Proteobacteria | Hot spring clone OS type G | ||
| OPS122 | 2 | β-Proteobacteria | ||||
| OPB37 | S92Be | 2 | β-Proteobacteria | |||
| OPB33 | B8, B78e, S27, S105, T77 | 29 | δ-Proteobacteria | Sulfate reduction | ||
| OPB55 | S22, S86Ae | 6 | δ-Proteobacteria | Sulfate reduction? | ||
| OPT23 | T47 | 2 | δ-Proteobacteria | Sulfate reduction? | ||
| OPB16 | 1 | δ-Proteobacteria | Sulfate reduction? | |||
| OPS96 | 1 | δ-Proteobacteria | Sulfate reduction | |||
| OPT56 | 1 | δ-Proteobacteria | Sulfate reduction? | |||
| OPB67A | 1 | Nitrospira group | ||||
| OPT35 | T37 | 2 | Nitrospira group | Thermodesulfovibrio yellowstonii | Sulfate reduction | |
| OPB3 | 2 | Acidobacterium group | + | |||
| OPB35 | 1 | Verrucomicrobium group | ||||
| OPB88 | S1 | 2 | BCFf group | + | ||
| OPB73 | 1 | BCF group | ||||
| OPB56 | S133Be | 3 | BCF–green-sulfur bacteria | |||
| OPS77 | T9e | 2 | Green-sulfur bacteria | |||
| OPS185 | 1 | Green-sulfur bacteria | ||||
| OPB41 | S17, S102, T96Ae | 5 | High-G+C gram positives | + | ||
| OPT29 | 1 | High-G+C gram positives | + | |||
| OPB19 | B31, B32, S8Ae, S15, S25, T60 | 27 | Thermus-Deinococcus group | Thermus sp. strain YSPID | Organotrophy | + |
| OPS128 | 1 | OP11 | ||||
| OPS136 | S67, S68 | 5 | OP11 | |||
| OPB92 | 2 | OP11 | ||||
| OPB12 | B11, S8Be, S69Ae, S124 | 17 | Green nonsulfur bacteria | + | ||
| OPB65 | S54Be, S57, S121, T86 | 8 | Green nonsulfur bacteria | |||
| OPT90e | S143Be | 2 | Green nonsulfur bacteria | |||
| OPB9 | B34 | 2 | Green nonsulfur bacteria | |||
| OPB14 | S143Ae | 2 | OP1 | + | ||
| OPB40 | S3 | 2 | OP4 | |||
| OPS107 | B60e, S51, S54Ae, S148, S163, T69e, T76e | 16 | OP5 | |||
| OPB17 | B6, T49 | 5 | Planctomycetales | |||
| OPB95 | S35Be, S88, S92Ae, S150, S12, T3 | 12 | OP8 | |||
| OPB5 | B23, S19Ae, S37 | 7 | OP8 | |||
| OPB7 | B85e, S49Ae, S66 | 11 | Thermotogales | + | ||
| OPS196 | S69Be | 2 | Thermotogales | |||
| OPS61e | 1 | Dictyoglomus group | ||||
| OPS145 | 1 | Dictyoglomus group | Dictyoglomus thermophilum | Organotrophy | ||
| OPS152e | 1 | OP6 | ||||
| OPS133Ae | 1 | OP7 | ||||
| OPB46 | B47, S44, S118, S139Ae | 9 | OP9 | + | ||
| OPB72e | 1 | OP9 | ||||
| OPB54 | 1 | OP12 | + | |||
| OPB2 | 1 | OP3 | ||||
| OPB80 | B90, S142 | 5 | OP10 | + | ||
| OPB50 | 1 | OP10 | + | |||
| OPT4 | T53 | 2 | Thermodesulfobacterium group | Sulfate reduction | ||
| OPS7 | 1 | Thermodesulfobacterium group | Sulfate reduction | |||
| OPB45 | 1 | Thermodesulfobacterium group | Sulfate reduction | + | ||
| OPB25 | 1 | OP2 | ||||
| OPB13 | S2, S35Ae, S55, T11e, T20e | 80 | Aquificales | Hot spring clone pBLACK | Hydrogen oxidation | |
| OPS132 | 2 | Aquificales | Calderobacterium hydrogenophilum | Hydrogen oxidation | + | |
| OPS165 | T14 | 2 | Aquificales | Hot spring clone EM17 | Hydrogen oxidation | + |
| Total | 129 | 312 |
Both strands were sequenced and used as representatives in phylogenetic analyses.
Shorthand clone name which omits OP used.
With ≥98% identity to the type sequence, based on direct sequence comparison or inferred from RFLP patterns.
Positives are rDNAs with G+C content of >60%.
Short sequences ranging from 300 to 1,350 nt, due to mispriming events during PCR. 5′ and 3′ parent fragments of chimeric sequences are designated A and B, respectively.
BCF, Bacteroides-Cytophaga-Flexibacter.
Phylogenetic analyses.
Sequences were initially compared to the available databases by using the BLAST (basic local alignment search tool) network service (3) to determine their approximate phylogenetic affiliations and orientation. Partial sequences were then compiled in Sequence Navigator (PE Applied Biosystems) and aligned with rRNA sequences from the Ribosomal Database Project (SSU_rep_Prok) in the GDE multiple sequence editor (26). Chimeric sequences were identified by using the CHECK_CHIMERA program (26) and by using branching order discrepancies in phylogenetic trees inferred with independent sections of the alignment (28 to 533, 534 to 998, and 1043 to 1491; see below). Representative clone sequences used in the analyses are indicated in Table 1. Based on the bacterial mask of Lane (24), either 842 (positions 534 to 1491) or 1,249 (positions 28 to 1491) homologous nucleotide positions were included in the alignment for comparative analyses: 28 to 68, 101 to 180, 220 to 450, 480 to 837, 859 to 998, 1043 to 1126, 1147 to 1165, 1175 to 1439, and 1462 to 1491 (E. coli numbering). Evolutionary distance, maximum parsimony, and maximum likelihood analyses were performed on the alignment as described previously (7). Distance and parsimony analyses were conducted with test version 4.0d56 of PAUP*, written by David L. Swofford. Maximum likelihood analyses were calculated by fastDNAml, available through the Ribosomal Database Project (26).
Hybridization.
rDNAs for slot blot hybridizations were amplified from environmental and reference DNAs as described above with universal primers in the presence of 5% acetamide. rDNA and E. coli rRNA (100 ng) and negative controls (300 ng; 1-kb DNA ladder [New England Biolabs], HindIII digested λ DNA, pBluescript KS+) were immobilized in triplicate on a Hybond nylon membrane (Amersham, Cleveland, Ohio) with a slot blot apparatus (Minifold II; Schleicher & Schuell, Keene, N.H.) according to the manufacturers’ instructions. Oligonucleotides used as probes for hybridization were UNIV-519R, ARC/EUK-1373R (5′ AGGGGCAGGGACGTATTC 3′) and BAC-924R (5′ CCGSTTGTGCGGGCCCCCG 3′). Probes were labeled with [λ-32P]ATP (NEN Life Science Products, Boston, Mass.) as described by the supplier of T4 polynucleotide kinase (New England Biolabs). The membrane was prehybridized at room temperature for 1 h with hybridization buffer (6× SSC [0.9 M NaCl and 0.09 M sodium citrate], 5× Denhardt’s solution [36], 0.5% [wt/vol] SDS, 100 μg of salmon sperm per ml), and then approximately 106 cpm of probe UNIV-519R was added to determine the presence of DNA or RNA on the membrane. The membrane was hybridized for 5 h to overnight at 45°C and washed stepwise in 2× SSC–0.1% SDS, 1× SSC–0.1% SDS, and 0.1× SSC–0.1% SDS. Hybridization was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The membrane was stripped in boiling SDS (0.1%) for 5 min and then probed as described above, first with ARC/EUK-1373R and then with BAC-924R, to minimize loss of the archaeal signal due to successive rounds of stripping. Extents of hybridization were normalized relative to bacterial (E. coli) and archaeal-eucaryal (pJP9) controls as described previously (19).
Nucleotide sequence accession numbers.
The rDNA sequences of the 129 sequenced OP clones have the accession no. AF026978 to AF027106.
RESULTS
Analysis of rRNA-based libraries from OP.
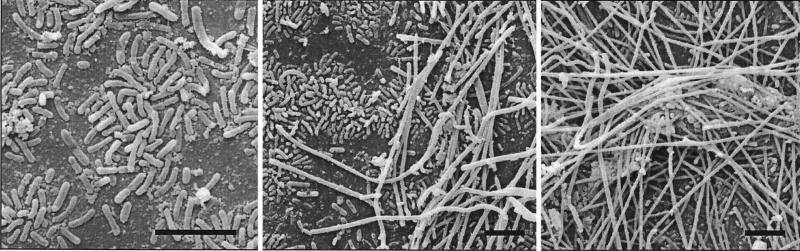
Glass contact slides immersed in OP were rapidly and prolifically colonized by microorganisms, indicating that a thriving microbial community exists in this hot spring. Typically, a variety of rods and filaments colonized glass surfaces, apparently with a succession as the biofilm accumulated (Fig. 1). Most microbial biomass in OP sediment was observed by microscopy to be associated with dense stromatolite-like objects on the bottom of the shallow pool. Samples obtained from the objects are fine and slimy in texture, suggesting the presence of exopolysaccharides or other biological materials. In contrast, coarse obsidian sand that fills the channels between the stromatolites seems relatively poor in biomass as judged by microscopy and low recovery of DNA. DNA extracted from the stromatolite sediment samples was used in the present study.
FIG. 1.
Scanning electron micrographs of glass slides colonized by microbial cells after immersion in OP for 1 week. Slides were fixed and prepared as described previously (37). Note the apparent succession of the primary colonizing rod morphotypes (left panel) by overlying filament morphotypes (center and right panels). Bar = 5 μm in all panels.
In order to determine the nature of the microbial constituents of the OP sediment samples, we analyzed the sequences of SSU-rRNA genes obtained by PCR with OP sediment DNA and SSU-rDNA primers and with cloning. Since different DNA extraction procedures and PCR conditions can result in differential recovery of rRNA genes, we prepared and analyzed three types of clone libraries, as detailed in Materials and Methods. Library OPS was prepared from community DNA extracted by a freeze-thaw lysis procedure (7) and amplified with universal primers (533F and 1492R), and the PCR products were cloned into pBluescript. Community DNA for library OPT was prepared by the above-described bead-beating protocol, and rRNA genes were obtained by amplification with universal primers and cloning into a T-tailed vector (Materials and Methods). DNA for library OPB also was prepared by the bead-beating protocol and with a T-tailed vector, but amplification was carried out with a Bacteria-specific primer set (27F and 1492R). In general, the bead-beating protocol resulted in significantly higher yields of DNA than did the freeze-thaw method. A total of about 300 clones containing inserts of the expected sizes (ca. 1 or 1.5 kb) were selected from the three libraries for further analysis.
rRNA gene-containing clones were screened by RFLP to identify unique types for sequence determination. As detailed in Materials and Methods, reamplified rDNA inserts were digested with the 4-base-specific restriction endonucleases HinP1 I and MspI and the products were analyzed by gel electrophoresis. Typically, 5 to 15 bands resulted from each rDNA digest in the discernible fragment size range of 50 to 400 bp (data not shown). In the three libraries taken together, 95 RFLP types were distinguished visually and 16S rDNA inserts from 122 clones representing the unique RFLP types were sequenced: 1.5 kb for clones obtained with the Bacteria-specific primer set (library OPB; nucleotide [nt] positions 28 to 1491, E. coli numbering) and 1 kb for clones obtained with the universal primer set (libraries OPS and OPT; nt 534 to 1491). Sequences differing only slightly (≤2%) were considered as a single relatedness group, and one representative of each group was fully sequenced on both strands with additional rDNA sequencing primers (Materials and Methods).
The collection of OP sequences was inspected for the occurrence of chimeric sequences, artifacts resulting from PCR-mediated recombination of different rRNA genes, by three different methods. In one method, independent phylogenetic trees were constructed with the 5′ and alternatively the 3′ halves of each sequence, and the trees were compared to identify branching discrepancies indicative of different sources for the two halves. In a second approach, each sequence was checked for the maintenance of long-range complementarities that may be disrupted in chimeric sequences. In the third method, each sequence was subjected to analysis by the program CHECK_CHIMERA (26), which tests for deviation along the sequence length in the extent of similarity of the test sequence to its closest relative in a set of reference sequences. We note that CHECK_CHIMERA is most useful if sequences closely related to the parent molecules of the chimera are available in the reference set for matching, because the program relies on the detection of abrupt changes in match with a parent sequence to indicate the presence of a chimera. If no close matches occur in the database, then any transition in similarity to the reference sequence cannot be detected above the background of random mismatches to other distantly related sequences. Since many of the OP sequences are only distantly related to known ones, in these cases CHECK_CHI MERA with known reference sequences was of limited use. Consequently, we compared the OP sequences to one another to increase the chance of a close match for comparison. Thirteen chimeric sequences were detected in the libraries, generally produced from phylogenetically disparate parent molecules and with recombination sites in highly conserved regions of the gene, such as nt 920 to 960 and around nt 1400. In all cases, recombination sites occurred in regions of near identity between parent sequences, which, therefore, could act as priming sites to produce the chimeric sequence. Fifty-four representative OP rDNA sequences, sequenced on both strands and determined to be nonchimeric, were used in subsequent phylogenetic analyses.
Phylogenetic distribution of OP sequences.
The OP sequences analyzed, the frequencies with which they were obtained in the libraries, their phylogenetic positions, and some inferred properties (Discussion) of the organisms represented by the rRNA sequences are summarized in Table 1. Figure 2 shows an evolutionary distance dendrogram of OP type sequences in the context of bacterial divisions currently recognized in the Ribosomal Database Project (26), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Taxonomy/tax.html), and the ARB database (http://pop.mikro.biologie.tu-muenchen.de/pub/ARB/) taxonomic listings. The analysis of the sequences showed that all were bacterial in nature. Even in the case of clone libraries developed with the universal rDNA primers, no archaeal or eucaryal sequences were detected. Only 70% (38 of 54) of the OP sequences could be assigned reproducibly (albeit often deeply) to recognized bacterial divisions. All of these specific affiliations were confirmed independently with subsets of taxa by a variety of tree inference methods (Materials and Methods; see supported nodes in Fig. 2). The remaining 16 sequence types appear to represent 12 novel, division level, bacterial lineages. Thus, this study substantially expands the known breadth of bacterial diversity (Discussion).
FIG. 2.
Evolutionary distance dendrogram of the bacterial domain and bacterial 16S rDNA sequence types obtained from OP sediment. Reference sequences were chosen to represent the broadest diversity of Bacteria. Sulfolobus acidocaldarius and Methanococcus vannielii were used as outgroups for the analysis. Division level groupings are bracketed at the right of the figure. Twelve novel candidate divisions determined in the present study are indicated as OP1 to OP12. Branch points supported (bootstrap values, ≥75%) by most or all phylogenetic analyses (see Materials and Methods) are indicated by filled circles; open circles indicate branch points supported by some analyses but only marginally supported (bootstrap, 50 to 74%), or not supported (bootstrap, <50%) by others. Branch points without circles are not resolved (bootstrap, <50%) as specific groups in different analyses.
Many of the currently recognized taxonomic divisions of Bacteria are represented by OP sequences (14 of 24 divisions). These include the Aquificales, Thermotogales, Dictyoglomus group, Thermodesulfobacterium group, Thermus-Deinococcus group, and the green nonsulfur bacteria (Table 1; Fig. 2). Organisms belonging to these divisions might be expected to be found in OP, since many are thermophilic and often are cultured from geothermal habitats. Representatives of the Aquificales, in particular, are abundant (84 of 312 clones), suggesting the importance of hydrogen metabolism in this ecosystem (Discussion). However, several other bacterial divisions characterized mainly by mesophilic cultivars also have OP representatives, including the Proteobacteria, high-G+C Gram-positive bacteria, the Bacteriodes-Cytophaga-Flexibacter group, green-sulfur bacteria, the Nitrospira group, and the Planctomycetales. Figure 3 shows the distribution of OP sequences associated with the δ subdivision of the Proteobacteria, which are abundant (40 of 312 clones) in the clone libraries and implicate sulfate reduction as an important component of the OP metabolic spectrum (Discussion). Two OP sequence types, OPB33 and OPS96, of the six associated with the δ-Proteobacteria could be assigned reproducibly to a monophyletic group comprised of sulfate-reducing bacteria (SRB) within the δ-Proteobacteria. The remaining OP sequence types which associate with the SRB, OPB16, OPB55, OPT23, and OPT56, appear to constitute independent lines of descent within the δ-Proteobacteria; however, by bootstrap analysis this is not certain. In particular, OPT56 may constitute a novel bacterial line of descent (Fig. 3).
FIG. 3.
Evolutionary distance dendrogram of the δ-Proteobacteria and associated OP sequence types. Rhodocyclus purpureus (β-Proteobacteria) and E. coli (γ-Proteobacteria) were used as outgroups for the analysis. Branch points supported (bootstrap values, ≥75%) by rate-corrected maximum likelihood, parsimony, and distance analyses are indicated by filled circles. Branch points without circles are not resolved (bootstrap, <50%) as specific groups in different analyses. SRB are indicated with asterisks.
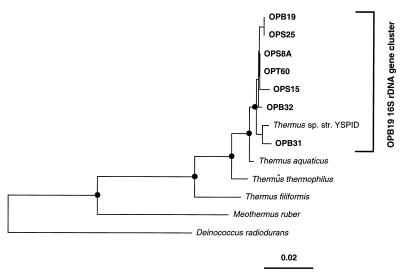
Several of the OP sequences are closely related to those of cultivated organisms or clone sequences determined in previous studies. Figure 4, for instance, shows the close relationships (>99% sequence identity) of representative sequences of the abundant OPB19 relatedness group (27 of 312 clones) to the cultivated Thermus species strain YSPID. The OPB19 group, together with Thermus sp. strain YSPID, comprises a gene cluster. Phylogenetic clusters of closely related but distinct SSU-rDNA sequences (>98% identity) have repeatedly been observed in culture-independent molecular phylogenetic environmental analyses (18). The occurrence of such gene clusters was the main reason for reducing groups of sequences with >98% identity to one representative sequence type (Table 1), to simplify the presentation of phylogenetic trees. Gene clusters are thought to be biologically relevant, the result of intercistronic variation in the same organism or sets of closely related cellular lineages of organisms (18); however, artifacts of PCR cannot be ruled out.
FIG. 4.
Evolutionary distance dendrogram of the Thermus-Deinococcus division showing the OPB19 sequence type rDNA gene cluster. Branch points supported (bootstrap values, ≥75%) by maximum likelihood, parsimony, and distance analyses are indicated by filled circles.
Other examples of OP sequences that are closely related to previously described cultivar or environmental clone sequences, all from Yellowstone geothermal sites, include OPT35 (Thermodesulfovibrio yellowstonii; 98.4%), OPS165 (hot spring clone EM17; 98.4%), OPB30 (hot spring clone OS type G; 99.1%), and OPB45 (Thermodesulfobacterium commune; 95.2%). Most of the OP sequences, however, are not closely related to known rRNA sequences (<90% identity). Some of the OP sequence types constitute new deepest branches within recognized bacterial divisions, such as OPB13 in the Aquificales, OPB7 (together with clone EM3) in the Thermotogales, and OPS77 and OPS185 in the green-sulfur bacteria (Fig. 2).
A number of the OP sequence types could not be phylogenetically placed within currently recognized bacterial divisions, and the majority are only modestly related (<85% sequence identity) to known rDNA sequences. These divergent sequences seem to represent new divisions, termed here candidate divisions, designated as OP1 to OP12 in Table 1 and Fig. 2. We establish a candidate division as an unaffiliated lineage in multiple analyses of data sets with varying types and number of taxa and having <85% identity to reported sequences, indicating its potential to represent a new bacterial division. Since the depth of a lineage cannot be estimated based on only one or a few examples, analysis of further sequences will be necessary to confirm or refute the status of a candidate division. The net effect of adding novel unaffiliated sequences to the bacterial tree has been the loss of resolution (collapse) of the main backbone of the tree, with the near-simultaneous radiation of multiple distinct divisions (Discussion).
Abundance of bacterial versus archaeal rDNAs in selected hot springs.
A broad diversity of archaeal rRNA genes previously had been detected in DNA isolated from OP and amplified with Archaea-specific PCR primers (6, 7). Moreover, the cultivation of many hyperthermophilic Archaea isolates over the past decade has resulted in a general perception that high-temperature environments may be a selective province for Archaea. We were surprised, therefore, that only bacterial rRNA genes were obtained upon amplification with universal primers as described above. In order to gain a rough assessment of the relative abundances of Archaea relative to Bacteria in OP sediments, we carried out domain-specific oligonucleotide hybridization analyses with amplified rDNA from OP as well as samples from two other Yellowstone hot springs, a microbial mat sample, O1A, and sediment sample N10, as detailed in Materials and Methods. Although not optimal for assessments of abundance, we found it necessary to use PCR products as hybridization targets because we were unable to obtain sufficient community RNA or DNA from the sediments for direct analysis.
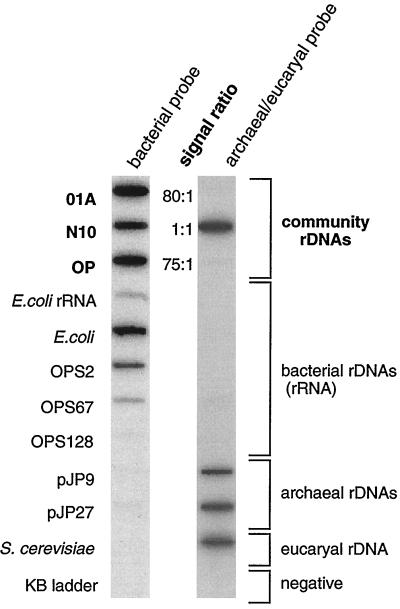
PCR products amplified from natural DNAs or control rDNAs were applied in triplicate to a nylon membrane, and sequential hybridizations were carried out with 32P-labeled oligonucleotides specific for archaeal and eucaryal rDNAs and, alternatively, bacterial rDNAs, all as detailed in Materials and Methods. Autoradiograms of the membrane are shown in Fig. 5. The oligonucleotide specific for the archaeal and eucaryal domains specifically bound to the archaeal and eucaryal controls and did not bind to the bacterial controls or to negative controls. Conversely, the bacterium-specific probe hybridized to the bacterial controls and not to the archaeal and eucaryal controls, with the exception of the atypical bacterial sequence (OPS128) belonging to candidate division OP11 (Table 1; Discussion), which has five internal mismatches to the BAC-924R probe. The universally conserved oligonucleotide hybridized to all rDNAs and not to the negative controls (data not shown). The specificity of the probes having been confirmed, the ratio of bacterial and archaeal-eucaryal probes bound to the community rDNAs was estimated with a phosphorimager (Materials and Methods). Community rDNAs from OP sediment and the 01A microbial mat had a ratio of Bacteria to Archaea-Eucarya of 75:1 and 80:1, respectively, consistent with the cloning results showing that Bacteria dominated Archaea in the OP sediment sample. Bacterial dominance is not general, however; the N10 sediment had a 1:1 ratio of Bacteria to Archaea-Eucarya. Thus, although further survey is necessary, these preliminary results suggest that Archaea members do not dominate, and indeed are exceptional, in some high-temperature environments.
FIG. 5.
Representative membrane hybridization of rDNAs PCR amplified with universal primers and E. coli rRNA, hybridized with a Bacteria-specific and an Archaea-Eucarya-specific probe. rDNAs were amplified from three hot spring communities including OP and representative organisms or clones. Reference archaeal pJP clone rDNAs were described in a previous study (7).
DISCUSSION
Inference of physiology from phylogeny of OP organisms.
Relatively few organisms in the environment are cultivatable by standard techniques. If we are to understand the roles of uncultivated microbes in an ecosystem, it is a challenge to us to infer physiological properties of organisms based on molecular data. An rRNA sequence in the absence of additional information provides little insight into the physiological nature of the organism that contributed the sequence. If, however, phylogenetic analysis groups the sequence with those of organisms that all possess a particular trait, then that trait is likely to occur in the organism which is otherwise known only by a sequence. Many of the OP sequences group robustly with assemblages of organisms that display uniform physiological properties. Consequently, presuming that the distribution of sequence types approximately reflects the in situ distribution of organisms, we can with some confidence infer the nature of energy metabolism employed by the organisms that constitute much of the OP community and serve as the main source of primary productivity.
Approximately 27% of the rRNA sequences obtained from OP sediment are associated phylogenetically with the bacterial division Aquificales (Table 1; Fig. 2). Cloned rRNA sequences representative of Aquificales also have been identified as major constituents of communities in other Yellowstone hot springs, including alkaline Octopus Spring (34) and hydrocarbon-rich Calcite Spring (35). All cultivated representatives of this division thrive obligatorily by microaerophilic oxidation of molecular hydrogen (31). Consequently, we infer that these dominant members of the OP community also likely are supported by hydrogen metabolism. It is probable that OP organisms other than the Aquificales group also utilize hydrogen metabolism, since that property is widely distributed in the bacterial domain (5). Other divisions with representatives that oxidize hydrogen are not restricted to that type of metabolism (44), however, and so the trait is not necessarily predictable from 16S rRNA phylogeny alone. Although hydrogen has not been directly measured in OP, the OP sediment is very rich in reduced iron (>15 g/kg), which interacts with water to generate hydrogen (5), and so this metabolic energy source is expected to be abundant in this environment.
OP also is a sulfide-rich hot spring, and so organisms engaged in sulfur metabolism are expected to abound in this habitat. Sequences representing almost all groups of currently recognized (cultivated) dissimilatory sulfate reducers (39) are present in the rRNA gene libraries (Table 1; Fig. 2). Examples are sequences closely related to those of the thermophilic sulfate-reducing genera Thermodesulfovibrio (Nitrospira group) and Thermodesulfobacterium and the SRB belonging to the δ-Proteobacteria. After the Aquificales, the sequences representing δ-Proteobacteria are the most abundant in the clone libraries. Most of the OP δ-Proteobacteria sequence types branch deeply within that group (Fig. 3) and so are not specifically associated with any named organisms. Nonetheless, the wide distribution of sulfate reduction in the δ-Proteobacteria indicates that this physiological trait is likely to be predictable from phylogenetic assignment to that group. Moreover, the most abundant OP δ-proteobacterial sequence type, represented by OPB33, falls convincingly within the radiation of a subgroup of the SRB (Fig. 3) and is 95% identical to the rDNA sequence of a cultivated thermophilic sulfate reducer, Thermodesulforhabdus norvegicus (8). We can reasonably infer, then, that sulfate reduction, possibly with hydrogen as electron donor, is another major metabolic theme in the OP community. The archaeal constituents of the OP community (6, 7) also include at least one sulfate reducer, a close phylogenetic relative of Archaeoglobus, currently the only recognized sulfate-reducing archaeal genus (39).
Organotrophic organisms probably are common in the OP community, supported by the primary productivity of the hydrogen metabolizers. However, only a few of the OP sequences are sufficiently closely related to those of known organotrophs to assign that trait to the OP organisms. One instance is the abundant group of OP sequences associated with the Thermus-Deinococcus group that are nearly identical to those of the cultured organotroph Thermus sp. strain YSPID (Table 1; Fig. 4). Because of this close relationship, the organotrophic nature of organisms that correspond to those OP sequences is predictable from the properties of the cultured organism.
OP is a high-temperature environment, and so it is expected that sequences derived from the OP sediment are from thermophilic organisms. Nonetheless, contamination of the hot spring with low-temperature organisms is possible, for instance, from groundwater flow into the hot spring. Therefore, we sought independent verification of the thermophilic nature of the organisms from which the rRNA genes were obtained. Thermophily of Bacteria and Archaea has been positively correlated with the G+C content of their rDNAs; thermophiles often (but not always) have rDNA G+C contents of >60%, whereas mesophiles generally have rDNA G+C contents of 55% or less (14, 47). Only 15 of the 54 different types of OP sequences have G+C contents of >60% and so by this criterion are indicated to be thermophilic (Table 1). This includes sequences belonging to divisions characterized (thus far) exclusively by thermophilic organisms, such as members of the Aquificales, the Thermotogales, and the Thermodesulfobacterium group, as well as to divisions not generally recognized for thermophily, such as the Acidobacterium group and the Planctomycetales (Table 1). Most of the other types of OP sequences (28 of 54) had rDNA G+C contents between 57 and 60%, and a few were below 55%, including proteobacterial, green-sulfur, and Bacteroides-Cytophaga-Flexibacter group sequences. Correlation of mesophily with low G+C content is poor, however (19a), and so cannot be used as a specific indicator of mesophilic bacteria. We believe, however, that all of the organisms indicated by OP sequences probably are thermophilic because nonviable low-temperature contaminants in the hot spring would be sufficiently rare that it is unlikely that they would be detected in the analysis of only 300 clones from a robust indigenous community (Fig. 1).
Bacterial dominance of OP rRNA genes.
The complete absence of archaeal sequences among 217 clones screened from libraries developed with universal PCR primers was unexpected, considering the great diversity of Archaea previously detected in OP with Archaea-specific primers (6, 7) and considering that Archaea members are commonly thought to be the dominating microflora in hot springs (40). A similar absence of archaeal sequences from a universal clone library of another Yellowstone hot spring (Octopus Spring) has been noted previously (34). To test whether these findings stem from biases introduced in cloning, we hybridized labeled domain-specific probes against PCR-amplified community rDNAs from OP and two other Yellowstone hot springs to estimate the relative abundance of bacterial and archaeal rRNA genes. The results confirmed that bacterial rRNA genes heavily dominate the PCR-amplified community rDNAs of OP, by 75:1. Of course, the abundance of rRNA genes does not necessarily reflect the relative abundance of organisms, since the number of rRNA genes varies in different organisms (17). Additionally, PCR potentially skews relative proportions of different sequence types during the amplification cycles (17, 33, 41). However, a number of lines of evidence suggest that the relative abundance of PCR rDNAs reasonably reflects the relative in situ abundance of bacterial and archaeal rRNA genes in the samples taken. First, PCRs for the hybridizations were carried out in the presence of acetamide, which relieves bias against some thermophilic rDNAs, particularly thermophilic archaeal rDNAs (33). Second, good correlation has been noted between bacterial sequence abundance in the water column based on probing against PCR-amplified rDNAs and abundance based on probing against total RNA (18, 19). Finally, the hot pool sediment sample, N10, had a bacterium-to-archaeon ratio of 1:1 (Fig. 5), suggesting that there is no systematic skewing of relative abundances away from Archaea, by PCR amplification. We conclude, the detail of the numbers aside, that Bacteria members dominate Archaea members in the OP sediment analyzed.
Increasingly, perceived ecological boundaries between bacterial habitats (e.g., temperate soils and waters) and archaeal habitats (extreme environments such as hot springs and hypersaline waters) are becoming blurred. Archaea members of the types previously thought restricted to high temperatures (Crenarchaeota) are abundant in many temperate environments (e.g., references 9, 15, and 20), and Bacteria members evidently play a more important role in extreme environments, such as hot springs, popularly thought to be the province of Archaea.
Novel candidate divisions and the organization of the bacterial phylogenetic tree.
The analyses of OP sequences in comparison with representatives of currently recognized divisions of the bacterial domain (Fig. 2) show that the OP sediment contains a great bacterial diversity, spanning the entire domain. Only 70% of the new sequences fall into taxonomic divisions previously characterized by molecular criteria; the rest comprise novel, candidate divisions. It is now clear from this culture-independent study of OP and similar analyses of other habitats (see reference 4 for a review; also see references 10–12, 34, 42, and 45), as well as from molecular characterization of novel isolates (e.g., references 1, 2, 16, 25, 27, 32), that the phylogenetic description of bacterial diversity is probably incomplete.
Twelve novel, division level lineages were encountered in the present study (Table 1; Fig. 2). Of particular note are lineages with two or more representatives (OP8 to OP11), which gives some notion of the phylogenetic depth of these candidate divisions. The candidate division designated OP11, represented by several sequences, is a particularly interesting group for a number of reasons. OP11 sequences have very low sequence identity to existing rDNA sequences (≤80%) and have highly atypical sequence signatures for the domain Bacteria (30 to 40% mismatch with signatures used to define the Bacteria, compared to Aquifex with 20% nonbacterial signatures [46]), which is reflected in their long branch lengths in Fig. 2. If not corrected for rate variation, OP11 sequences branch falsely deep in the bacterial domain. A reverse primer specific for the OP11 lineage was designed and used in concert with a universal or bacterial forward primer to selectively amplify representatives of this phylogenetic group from various habitats, including other Yellowstone hot springs and low-temperature, freshwater sediment. A wide diversity of sequences belonging to the group were successfully obtained from freshwater sediment, indicating that there are mesophilic representatives of the group (22). Additional representatives of the OP11 group have been detected independently by rRNA sequence analysis in other environments, including Carolina Bay sediment (clone RB39 [45]), Amazonian soil (clone P36 [12]), hydrocarbon-contaminated soils under methanogenic and sulfate-reducing conditions (15a), and Australian deep-subsurface water (15b), indicating that the group is widespread in nature. Additional sequences belonging to candidate divisions OP5, OP8, and OP10 also have been obtained from the hydrocarbon-contaminated soil under methanogenic conditions (15a), suggesting that these groups may represent ecologically significant bacterial divisions. As yet, little beyond the general properties of Bacteria can be inferred about the physiology of the organisms that these novel sequence types represent and what role each may be playing in their respective environments.
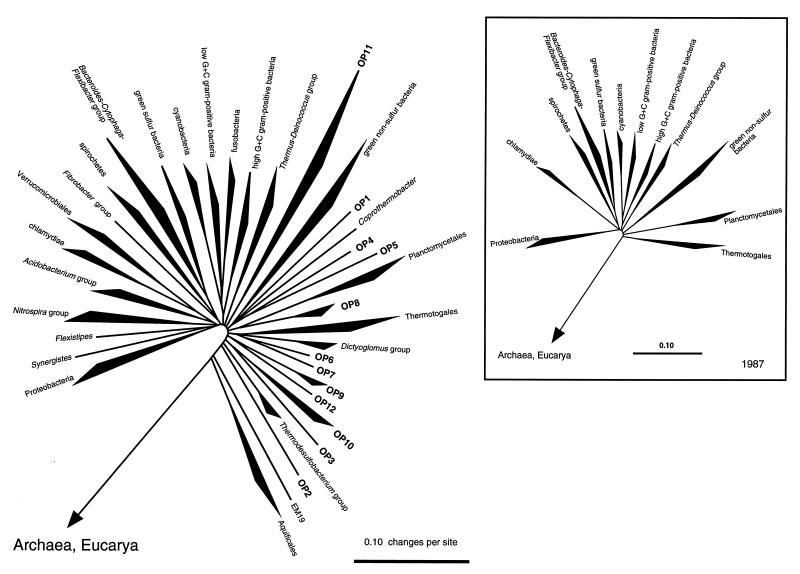
In his landmark 1987 paper that outlined the pattern of bacterial evolution, Carl Woese defined a dozen main bacterial lines of descent based primarily on signature sequences of rRNAs from cultivated organisms (46). As additional rRNA sequences have accumulated, the depth of branching within many of these divisions has increased and many more division level branches in the bacterial tree have been recognized. As summarized in the radial tree in Fig. 6, at least 36 putative division level lineages are now identifiable, comprising the 12 lines of descent described in 1987 (inset) (46), 12 additional lineages collated from various studies since that time (26), and 12 clonal OP lineages from the present study. It has long been suggested (23, 29, 43, 46) that the phylogenetic diversity of the bacterial domain did not result from an ordered progression of bifurcating divergences from a main line of bacterial descent but rather arose as an explosion of diversity from common ancestry. This effect is even more striking with the addition of new division level lineages (Fig. 6), to the point that even the deeply divergent bacterial divisions, Aquificales and Thermotogales, which at this time seem to have diverged prior to the massive radiation, may be subsumed into the main radiation. If this near-simultaneous radiation of most of the modern bacterial divisions in fact occurred and is not the result of some limitation in the resolution afforded by the SSU-rRNA molecule and/or current phylogenetic inference methods, then one or more important developments in bacterial evolution must have occurred to trigger the massive increase in biodiversity. Two possibilities which have been suggested include the advent of a rigid murein saculus and photoautotrophic growth (23), two properties which undoubtedly made Earth’s niches more accessible to the microbial world. It remains to be seen how many division level taxa will constitute the bacterial domain.
FIG. 6.
Diagrammatic radial representation of the known phylogenetic span of Bacteria in 1987 (inset [46]) and today. The figure is based directly on Fig. 2. Filled sectors indicate that two or more representative sequences fall within the indicated depth of branching. Twelve novel candidate divisions determined in the present study are indicated as OP1 to OP12.
ACKNOWLEDGMENTS
We thank Brett Goebel and Dan Frank for providing valuable comments on the manuscript, and Susan Barns for sample collection, design of the ARC/EUK probe, and advice on phylogenetic analyses. We thank Rudi Turner for preparation of the scanning electron microscopy samples, Yen Shu for operating the 373 ABI sequencer, and Lawrence Washington and Georgia Zeigler for operating the LI-COR sequencer.
This work was supported by grants from the U.S. Department of Energy and NIH.
REFERENCES
- 1.Albrecht W, Fischer A, Smida J, Stackebrandt E. Verrucomicrobium spinosum, a eubacterium representing an ancient line of descent. Syst Appl Microbiol. 1987;10:57–62. [Google Scholar]
- 2.Allison M J, Mayberry W R, McSweeney C S, Stahl D A. Synergistes jonesii, gen. nov., sp. nov.: a rumen bacterium that degrades toxic pyridinediols. Syst Appl Microbiol. 1992;15:522–529. [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aragno M. Thermophilic, aerobic, hydrogen-oxidizing (knallgas) bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. IV. New York, N.Y: Springer-Verlag; 1992. pp. 3917–3933. [Google Scholar]
- 6.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beeder J, Torsvik T, Lien T. Thermodesulforhabdus norvegicus gen. nov., sp. nov., a novel thermophilic sulfate-reducing bacterium from oil field water. Arch Microbiol. 1995;164:331–336. [PubMed] [Google Scholar]
- 9.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burggraf S, Heyder P, Eis N. A pivotal Archaea group. Nature. 1997;385:780. doi: 10.1038/385780a0. [DOI] [PubMed] [Google Scholar]
- 14.Dalgaard J Z, Garrett R A. Archaeal hyperthermophilic genes. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of Archaea (Archaebacteria) Vol. 26. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1993. pp. 535–563. [Google Scholar]
- 15.DeLong E F, Wu K Y, Prezelin B B, Jovine R V M. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 15a.Dojka, M. A., P. Hugenholtz, and N. R. Pace. Unpublished data.
- 15b.Durand, P. Unpublished data.
- 16.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrate-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 17.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field K G, Gordon D, Wright T, Rappe M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter in the Atlantic and Pacific oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Gutell, R. Personal communication.
- 20.Hershberger K L, Barns S M, Reysenbach A-L, Dawson S C, Pace N R. Wide diversity of Crenarchaeota. Nature. 1996;384:420. doi: 10.1038/384420a0. [DOI] [PubMed] [Google Scholar]
- 21.Huber R, Burggraf S, Mayer T, Barns S M, Rossnagel P, Stetter K O. Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature. 1995;376:57–58. doi: 10.1038/376057a0. [DOI] [PubMed] [Google Scholar]
- 22.Hugenholtz P, Hershberger K L, Flanagan J L, Kimmel B, Pace N R. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Widespread distribution of a novel phylum-depth bacterial lineage in nature, abstr. N-23; p. 385. [Google Scholar]
- 23.Kandler O. The early diversification of life. Nobel Symp. 1994;84:152–160. [Google Scholar]
- 24.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 25.Love C A, Patel B K C, Ludwig W, Stackebrandt E. The phylogenetic position of Dictyoglomus thermophilum based on 16S rRNA sequence analysis. FEMS Microbiol Lett. 1993;107:317–320. [Google Scholar]
- 26.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maymo-Gatell X, Chien Y-T, Gossett J, Zinder S. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 28.Ng W-L, Schummer M, Cirisano F D, Baldwin R L, Karlan B Y, Hood L. High-throughput plasmid mini preparations facilitated by micro-mixing. Nucleic Acids Res. 1996;24:5045–5047. doi: 10.1093/nar/24.24.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 30.Pace N R, Stahl D A, Lane D J, Olsen G J. Analyzing natural microbial populations by rRNA sequences. ASM News. 1985;51:4–12. [Google Scholar]
- 31.Pitulle C, Yang Y, Marchiani M, Moore E R B, Siefert J L, Aragno M, Jurtshuk P, Fox G E. Phylogenetic position of the genus Hydrogenobacter. Int J Syst Bacteriol. 1994;44:620–626. doi: 10.1099/00207713-44-4-620. [DOI] [PubMed] [Google Scholar]
- 32.Rainey F A, Stackebrandt E. Phylogenetic analysis of the bacterial genus Thermobacteroides indicates an ancient origin of Thermobacteroides proteolyticus. Lett Appl Microbiol. 1993;16:282–286. [Google Scholar]
- 33.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reysenbach A-L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reysenbach, A.-L., M. A. Ehringer, K. L. Hershberger, and N. R. Pace. Unpublished data.
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stackebrandt E, Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol. 1990;136:37–43. doi: 10.1099/00221287-136-1-37. [DOI] [PubMed] [Google Scholar]
- 39.Stackebrandt E, Stahl D A, Devereux R. Taxonomic relationships. In: Barton L L, editor. Sulfate-reducing bacteria. New York, N.Y: Plenum Press; 1995. pp. 49–87. [Google Scholar]
- 40.Stetter K O, Fiala G, Huber G, Huber R, Segerer A. Hyperthermophilic microorganisms. FEMS Microbiol Rev. 1990;75:117–124. [Google Scholar]
- 41.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 43.Wais A C. Archaebacteria: the road to the universal ancestor. Bioessays. 1986;5:75–78. doi: 10.1002/bies.950050207. [DOI] [PubMed] [Google Scholar]
- 44.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. IV. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 45.Wise M G, McArthur J V, Shimkets L J. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl Environ Microbiol. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woese C R, Achenbach L, Rouviere P, Mandelco L. Archaeal phylogeny: reexamination of the phylogenetic position of Archaeoglobus fulgidus in light of certain compostion-induced artifacts. Syst Appl Microbiol. 1991;14:364–371. doi: 10.1016/s0723-2020(11)80311-5. [DOI] [PubMed] [Google Scholar]
- 48.Young C C, Burghoff R L, Keim L G, Minak-Bernero V, Lute J R, Hinton S M. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA from soils. Appl Environ Microbiol. 1993;59:1972–1974. doi: 10.1128/aem.59.6.1972-1974.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]