Étienne Caron and colleagues discuss how immunopeptidomics and other peptide sequencing technologies could revolutionize the research for vaccine development, cancer immunotherapy, and treatment of neurodegenerative diseases and aging.
Abstract
The human immunopeptidome plays a central role in disease susceptibility and resistance. In our opinion, the development of immunopeptidomics and other peptide sequencing technologies should be prioritized during the next decade, particularly within the framework of the Human Immunopeptidome Project initiative. In this context, we present bold ideas, fresh arguments, and call upon industrial partners and funding organizations to support and champion this important initiative that we believe has the potential to save countless lives in the future.
Introduction
Immunopeptidomics is a mass spectrometry (MS)-driven discipline that investigates the plethora of antigenic peptides presented by human leukocyte antigen (HLA) molecules, collectively referred to as the human immunopeptidome. Quantifying the complexity of the human immunopeptidome is an ongoing endeavor. At the population level, over 37,000 distinct classical HLA alleles have been documented. Typically, an individual expresses up to six HLA-I and six HLA-II alleles, each of which generally presents a different set of peptides. At the cellular level, as many as 3,000,000 copies of HLA-I molecules adorn the cell surface. Each peptide can be presented in varying quantities, ranging from 1 to 10,000 copies per cell, with exceptions exceeding 100,000 copies per cell (Stutzmann et al., 2023). Furthermore, notable variations in the immunopeptidome have been observed across different organs, both within and between individuals (Kubiniok et al., 2022). Consequently, the human immunopeptidome undeniably stands as a realm of remarkable complexity and diversity.
T cells navigate through this complexity and seek for “abnormal” peptides that originate from intracellular perturbations (pathogenic sources, neoplastic transformation, etc.). The abnormal cells are then eradicated once engaged by T cells. In this context, immunopeptidomics is increasingly applied to identify tumor-specific antigens (TSAs), non-canonical HLA peptides originating from unannotated alternative reading frames or from non-coding regions of the genome, as well as post-translationally modified TSAs (Chong et al., 2022; Ouspenskaia et al., 2022; Laumont et al., 2018). Immunopeptidomics has also received much attention to help improve epitope prediction algorithms, which are highly accessible and can be rapidly tested in translational research for vaccine design or T cell-based immunotherapy (Gfeller et al., 2023).
Running parallel to the current community’s strong emphasis on the therapeutic aspects, we envision that the applications and impact of immunopeptidomics could extend far beyond the present scope. Looking at the broader perspective, we believe that uncovering hidden signatures within the intricate landscape of the immunopeptidome holds the potential to play a major role in advancing predictive and preventive medicine in the future. In our opinion, this could be achieved through the systematic profiling of the immunopeptidome in large-scale population-based association studies, an approach that we recently coined as the immunopeptidome-wide associated study (IWAS) paradigm (Vizcaíno et al., 2020). As stated by Bill Gates: “Treatment without prevention is simply unsustainable.” In this article, we put forth arguments in favor of this global effort, particularly in the context of advocating for a multi-decade NIH-funded Human Immunopeptidome Project.
The influence of the immunopeptidome in disease susceptibility and resistance: New lessons from the COVID-19 and the Black Death pandemics
Variations in HLAs and antigen presentation have been associated with the greatest number of human diseases, spanning from cancer and autoimmune diseases to neurodegenerative disorders (Wang et al., 2020; Sulzer et al., 2017; Trowsdale and Knight, 2013; Unanue et al., 2016). Furthermore, accumulating evidence indicates that the genetic susceptibility to numerous diseases finds its roots in the remarkable diversity of the human immunopeptidome (Vizcaíno et al., 2020). This being said, on a positive note, this extraordinary diversity primarily serves an evolutionary purpose—to increase the likelihood that certain individuals in the global population can effectively mobilize T cell responses against emerging infections, thus ensuring survival of our species. An illustrative case of this phenomenon can be seen in the context of the HIV epidemic, where HLA-B*57 and -B*27 have been highly associated with the ability to control viral replication and disease progression (Migueles et al., 2000; Blackwell et al., 2009). In the context of COVID-19, Augusto et al. (2023) found that HLA-B*15:01 individuals were associated with a much higher likelihood of asymptomatic infection. In addition, they demonstrated that at least one B*15:01-associated SARS-CoV-2 peptide (NQKLIANQF) was important for triggering an effective CD8+ T cell response that could enhance protection in those individuals (Augusto et al., 2023).
Looking back at the historical context of the Black Death, the most devastating pandemic in human history that killed 30–50% of the Afro-Eurasia population in the mid-14th century, a recent genetic study has shed light on a fascinating discovery. Klunk et al. (2022) revealed that individuals expressing a particular variant of ERAAP (endoplasmic reticulum aminopeptidase associated with antigen processing) had a higher likelihood of surviving the bacterium Yersinia pestis, the causative agent of the Black Death. This is interesting because ERAAP is a key enzyme in antigen processing and is known to shape the composition of the HLA immunopeptidome (Serwold et al., 2002; Nagarajan et al., 2016). One could therefore hypothesize the existence of distinct immunopeptidomic signatures in individuals who succumbed to the Black Death compared to those who survived. With this in mind, several intriguing questions naturally arise: (1) What exactly are those distinctive signatures? (2) Can we establish a correlation between those immunopeptidomic signatures and factors such as T cell activity, disease progression, and the ultimate outcome of death or survival? (3) Is it possible to detect those signatures prior to any exposure to Y. pestis? And if so, (4) could we leverage those immunopeptidomic signatures as a prognostic marker for predicting disease susceptibility or resistance, before any exposure occurs?
Similarly, going back to the onset of the COVID-19 pandemic, could immunopeptidome profiling of individuals unveil distinctive “at-risk” profiles for severe infection in contrast to “resistant” profiles for asymptomatic infection? For instance, beside the B*15 peptide mentioned above, many other SARS-COV-2 peptides could be discovered and associated with asymptomatic or mild infections. Thinking forward, could we contemplate the application of robust immunopeptidomic profiling methods in future pandemics to predict disease susceptibility/resistance and personalize mitigation strategies to avoid potential disruption of societies?
To address these questions seriously, two crucial elements need to converge. First, we must access advanced technology that can swiftly and comprehensively identify and quantify the complete range of immunopeptidomic peptides, both self and non-self, in all cell and tissue types across diverse individuals. Ideally, this technology should be rapid, robust, unbiased, cost effective, and capable of offering a comprehensive view. Second, such a mapping endeavor should extend to a population-scale scope.
Currently, immunopeptidomics technologies fall relatively short in terms of the sensitivity, robustness, and accessibility essential for tackling the above questions. Yet, recognizing the profound impact of the immunopeptidome on disease susceptibility and resistance, we firmly assert that this is a matter of utmost significance. It underscores a compelling justification for mobilizing substantial funding to drive forward the necessary technological advancements. The importance of this endeavor cannot be overstated, and it is imperative that we commit the required resources to unravel the mysteries of the human immunopeptidome. As a first step, a solid scientific framework was established several years ago: the Human Immuno-Peptidome Project (HIPP).
The Human Immuno-Peptidome Project
First phase
To expedite progress in achieving robust and comprehensive immunopeptidome analysis, ultimately at population scale, leaders in the field of immunopeptidomics took a pivotal step in 2015. They established the HIPP as a pioneering initiative under the Human Proteome Organization (HUPO; https://hupo.org/Immuno-Peptidome). The HUPO-HIPP initiative was conceived with a visionary aim: to democratize access to immunopeptidomics data, robust experimental techniques, and computational tools. This accessibility would be extended to immunologists, clinical investigators, and researchers across the globe, significantly amplifying the impact of immunopeptidomics in the realm of biomedical research. As a matter of fact, the group convened in Zürich in 2017 to establish the Project’s long-term vision and framework (Caron et al., 2017). The primary objectives encompassed two core goals: first, to comprehensively map the human immunopeptidome under healthy conditions (i.e., the “baseline” immunopeptidome). Second, to bolster the technology’s robustness to the extent that it becomes readily accessible in clinical settings. The Project was also structured around four foundational pillars: (1) the advancement of methods and technology, (2) the attainment of high-quality community standards, (3) the enhancement of data-sharing efficacy, and (4) the promotion of immunopeptidomics as a field through the creation of an international-level educational program.
Until now, HUPO-HIPP has made significant progress with regard to pillars 2 and 4. Specifically, the group has published the Minimal Information About an Immunopeptidomics Experiment (MIAIPE) to standardize the methods (Lill et al., 2018; pillar 2) and organizes a summer school every 2+ yr to expand the immunopeptidomics community (e.g., https://2022-hupo-hipp-summer-school.mailchimpsites.com; pillar 4). In addition, HUPO-HIPP has clearly contributed to the growth and visibility of the immunopeptidomics field through organization of international events, webinars, and its presence in social media (https://twitter.com/HippHupo; pillar 4). In terms of technological advancement (pillar 1) and data sharing (pillars 3), the field has also witnessed significant progress since 2015. For instance, new sample preparation and MS technologies have been developed and tested (pillar 1), and new immunopeptidomics databases have been created and are currently available online, including SysteMHC atlas (https://systemhc.sjtu.edu.cn/), caAtlas (Yi, 2021), and HLA ligand Atlas (https://hla-ligand-atlas.org/welcome; pillar 3). Notably, data accessibility has fostered the development of novel computational tools to increase peptide identification sensitivity/accuracy through machine and deep learning–based approaches (pillar 1; Zhang and Bassani-Sternberg, 2023). However, it is important to note that the technological (pillar 1) and data sharing (pillar 3) progress that we have witnessed over the last recent years primarily stemmed from individual research groups’ funds and efforts, rather than being driven by a unified, HUPO-HIPP–led initiative. This can be partly explained by the lack of funding opportunities for supporting such projects among international organizations.
Possible next phase
Based on the six lessons of the Human Genome Project (Green et al., 2015), we emphasize that technology and method development (pillar 1) should be prioritized in the upcoming phase, before embarking on population-scale association studies. Technological examples include, but are not restricted to, microfluidics, cost-effective antibody-independent techniques for rapid, efficient, and specific isolation of HLA-bound peptides, single-molecule peptide sequencing technologies, affordable methods for population-scale HLA typing, LC-MS systems, fragmentation techniques for non-tryptic peptides, and systematic absolute quantification methods for the entire spectrum of HLA-bound peptides (Caron et al., 2017; Vizcaíno et al., 2020; Stutzmann et al., 2023). This can be achieved by dedicating substantial funding to ambitious, integrated, cross-disciplinary, and milestone-driven collaborative technology projects. To this end, new funding opportunities are required and should be aligned with successful models of other major biomedical initiatives like the π-Hub Project (30-yr mission with an investment of billions of dollars; https://www.pi-hub.org.cn/) and the Human Immunome Project (https://www.humanimmunomeproject.org/). Furthermore, innovative technologies developed through the proposed next phase could be included in the curriculum of HUPO-HIPP summer courses, thus expediting their deployment.
Immunopeptidomic surveillance by 2035: A moonshot project to consider
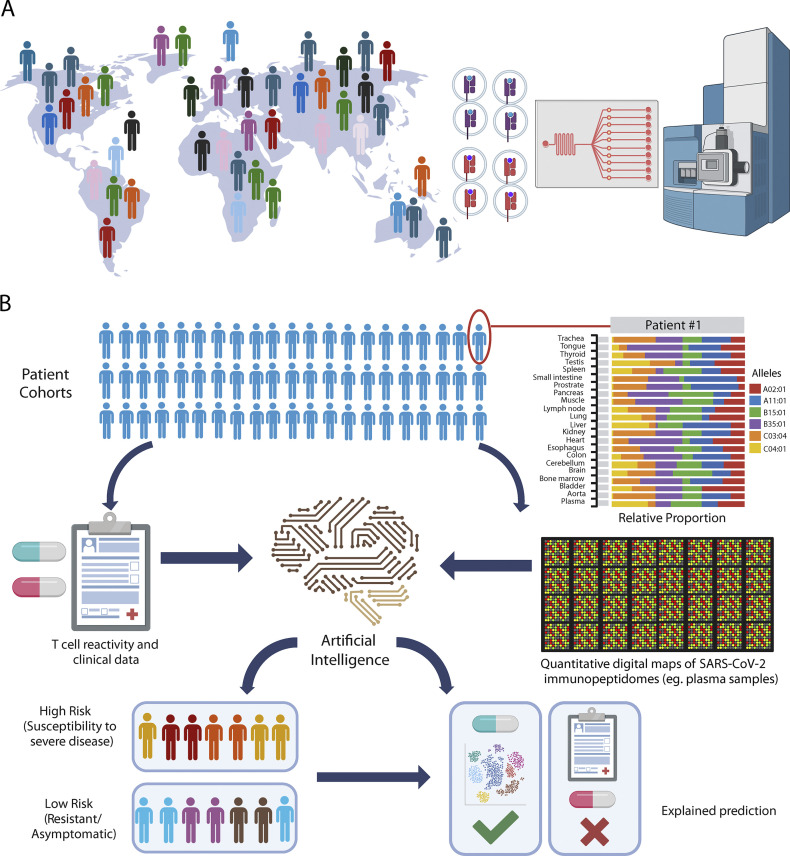
Akin to genomic surveillance and extensive sequencing efforts seen during the COVID-19 pandemic, we also recommend that a Working Group explore the concept of “immunopeptidomic surveillance” (Fig. 1). Although futuristic for now, this concept implies routine and widespread execution of robust immunopeptidome profiling studies on large patient cohorts across the globe to accelerate our understanding of T cell immunity in disease susceptibility and resistance. The team of Alessandro Sette (La Jolla Institute for Immunology) has to some extent pioneered this concept during the COVID-19 pandemic by distributing pools of synthetic SARS-CoV-2 peptides to over 200 laboratories worldwide, expediting research to enhance our global understanding of T cell immunity in COVID-19. Moving forward, establishing a sustainable international infrastructure for global, robust, and unbiased physical measurements of immunopeptidomes could represent an important milestone in immunology (Fig. 1 A). This infrastructure would empower the generation of quantitative digital maps of HLA-bound peptides from large cohorts of patient samples, including identification and quantification of all self and non-self peptides, setting up the stage for a global T cell digital surveillance network. Once the technology has reached the robustness and sensitivity required, this concept could be explored in pilot studies using plasma samples from large populations of COVID-19 patients. Indeed, a proof-of-concept study has already been reported by comparing soluble immunopeptidomes of COVID-19 patients versus those of convalescent or healthy individuals (Nelde et al., 2022; Fig. 1 B). Simultaneously, T cell experiments could be performed from the same samples to assess T cell-epitope reactivity. Subsequently, data could be digitized and processed through artificial intelligence systems, enabling the discovery of immunopeptidomic-T-cell activity patterns across human populations, which could then be linked to disease severity/resistance and other clinical phenotypes to personalize mitigation strategies and treatments. Findings derived from such association studies could subsequently serve as the foundation for more in-depth investigations into the precise mechanisms of action using conventional model systems.
Figure 1.
Moving from genomics to immunopeptidomics surveillance. (A) World map illustrating the ambition of profiling immunopeptidomes across human populations using advanced immunopeptidomics technologies. A microfluidic device for automated sample preparation and a mass spectrometer for data acquisition are shown as representative examples of technologies that could be further developed. (B) Schematic of the workflow to identify immunopeptidomic signatures of disease susceptibility/resistance to stratify patients and help personalize mitigation strategies in future pandemics. SARS-CoV-2 is used for illustrating the concept. In this example, viral peptides are isolated at populations-scale from plasma samples (patient cohorts). Fictive bar plot of HLA-I-allele-specific self peptides in different organs (patient #1) and population-scale quantitative digital maps of soluble HLA-associated SARS-CoV-2 peptides from plasma samples are illustrated.
We wish to emphasize that the establishment of an immunopeptidomic surveillance approach is ambitious, but not beyond reach, as it can be expedited by prioritizing the development of innovative peptide sequencing and immunopeptidomics technologies (pillar 1), especially within the framework of a large, multi-decade NIH-funded Human Immunopeptidome Project.
Final perspective: Going global for big impact
The Human Genome Project has transformed biology through its integrated big science approach. This ambitious endeavor led to the development of novel technologies and analytical tools, and brought the expertise of engineers, computer scientists, and mathematicians together with that of biologists (Hood and Rowen, 2013). Inspired by the success of the Human Genome Project, we advocate herein for a visionary, multi-decade NIH-funded Human Immunopeptidome Project. This Project should aim at elevating the field of immunopeptidomics to catalyze discoveries across disciplines, offering tangible benefits in predictive, preventive, and personalized medicine. With this in mind, we are proactively initiating collaborative discussions with synergistic communities, including individuals affiliated with the Yale Center for Infection and Immunity, the Yale Center for Immuno-Oncology, and the Yale Center for Systems and Engineering Immunology, along with academic and industrial partners. Most significantly, we aim to engage the HUPO-HIPP community in a collective effort to present a compelling proposal for the launch of this game-changing scientific endeavor.
The implications of scaling up robust immunopeptidomics technologies for the establishment of a global T cell surveillance digital network are profound. It will not only advance our fundamental understanding of the immune system but will also have practical applications in diverse fields, well beyond pandemic preparedness. In vaccine development, this approach can aid in the design of more effective vaccines by identifying immunogenic peptides. In autoimmune, neurodegenerative, and aging-related disease research, it can help uncover the triggers of aberrant immune responses. Additionally, in cancer immunotherapy, it can guide the development of personalized treatments by identifying TSAs. In conclusion, the endeavor of expediting the scalability of robust immunopeptidomics technologies represents a pivotal milestone in immunology and healthcare. It is an effort that will demonstrate our collective commitment to harnessing the power of technology and knowledge to improve human health on a global scale.
Acknowledgments
This work was supported by the National Sciences and Engineering Research Council (grant #RGPIN-2020-05232), the Canadian Institutes of Health Research (CIHR; grant #174924), and the CIHR Coronavirus Variants Rapid Response Network (CoVaRR-Net).
References
- Augusto, D.G., et al. 2023. Nature. 10.1038/s41586-023-06331-x [DOI] [Google Scholar]
- Blackwell, J.M., et al. 2009. Clin. Microbiol. Rev. 10.1128/CMR.00048-08 [DOI] [Google Scholar]
- Caron, E., et al. 2017. Immunity. 10.1016/j.immuni.2017.07.010 [DOI] [Google Scholar]
- Chong, C., et al. 2022. Nat. Biotechnol. 10.1038/s41587-021-01038-8 [DOI] [PubMed] [Google Scholar]
- Gfeller, D., et al. 2023. Semin. Immunol. 10.1016/j.smim.2022.101708 [DOI] [PubMed] [Google Scholar]
- Green, E.D., et al. 2015. Nature. 10.1038/526029a [DOI] [Google Scholar]
- Hood, L., and Rowen L.. 2013. Genome Med. 10.1186/gm483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk, J., et al. 2022. Nature. 10.1038/s41586-022-05349-x [DOI] [Google Scholar]
- Kubiniok, P., et al. 2022. iScience. 10.1016/j.isci.2022.103768 [DOI] [Google Scholar]
- Laumont, C.M., et al. 2018. Sci. Transl. Med. 10.1126/scitranslmed.aau5516 [DOI] [PubMed] [Google Scholar]
- Lill, J.R., et al. 2018. Proteomics. 10.1002/pmic.201800110 [DOI] [Google Scholar]
- Migueles, S.A., et al. 2000. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.050567397 [DOI] [Google Scholar]
- Nagarajan, N.A., et al. 2016. J. Immunol. 10.4049/jimmunol.1500654 [DOI] [Google Scholar]
- Nelde, A., et al. 2022. iScience. 10.1016/j.isci.2022.105643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouspenskaia, T., et al. 2022. Nat. Biotechnol. 10.1038/s41587-021-01021-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwold, T., et al. 2002. Nature. 10.1038/nature01074 [DOI] [Google Scholar]
- Stutzmann, C., et al. 2023. Cell Rep. Methods. 10.1016/j.crmeth.2023.100511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer, D., et al. 2017. Nature. 10.1038/nature22815 [DOI] [Google Scholar]
- Trowsdale, J., and Knight J.C.. 2013. Annu. Rev. Genom Hum. G. 10.1146/annurev-genom-091212-153455 [DOI] [Google Scholar]
- Unanue, E.R., et al. 2016. Annu. Rev. Immunol. 10.1146/annurev-immunol-041015-055420 [DOI] [PubMed] [Google Scholar]
- Vizcaíno, J.A., et al. 2020. Mol. Cell. Proteomics. 10.1074/mcp.R119.001743 [DOI] [Google Scholar]
- Wang, J., et al. 2020. Cell. 10.1016/j.cell.2020.09.054 [DOI] [Google Scholar]
- Yi, X., et al. 2021. iScience. 10.1016/j.isci.2021.103107 [DOI] [Google Scholar]
- Zhang, B., and Bassani-Sternberg M.. 2023. J. Immunother. Cancer. 10.1136/jitc-2023-007073 [DOI] [PMC free article] [PubMed] [Google Scholar]