Abstract
High-throughput sequencing of amplicons has been widely used to precisely and efficiently identify species compositions and analyze community structures, greatly promoting biological studies involving large amounts of complex samples, especially those involving environmental and pathogen-monitoring ones. Commercial library preparation kits for amplicon sequencing, which generally require multiple steps, including adapter ligation and indexing, are expensive and time-consuming, especially for applications at a large scale. To overcome these limitations, a “one-step PCR approach” has been previously proposed for constructions of amplicon libraries using long fusion primers. However, efficient amplifications of target genes and accurate demultiplexing of pooled sequencing data remain to be addressed. To tackle these, we present an integrative protocol for one-step PCR amplicon library construction (OSPALC). High-quality reads have been generated by this approach to reliably identify species compositions of mock bacterial communities and environmental samples. With this protocol, the amplicon library is constructed through one regular PCR with long primers, and the total cost per DNA/cDNA sample decreases to just 7% of the typical cost via the multi-step PCR approach. Empirically tested primers and optimized PCR conditions to construct OSPALC libraries for 16S rDNA V4 regions are demonstrated as a case study. Tools to design primers targeting at any genomic regions are also presented. In principle, OSPALC can be readily applied to construct amplicon libraries of any target genes using DNA or RNA samples, and will facilitate research in numerous fields.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42995-023-00182-1.
Keywords: Amplicon library preparation, Lab-made protocol, Long-primer PCR, Cost efficiency
Introduction
Amplicon sequencing of single gene fragments is widely used for studying biodiversity and community compositions, based on reference databases and relative abundance of the target genes. It can be conveniently applied to track tempo-spatial variations of population and community structures, resulting from genetic and/or environmental changes (Beser et al. 2017; Costello et al. 2009; Gous et al. 2019; McNamara et al. 2020; Pochon et al. 2013; Song et al. 2019; Xu et al. 2012). Accumulated amplicon sequences of various samples also contribute to growing databases for future research. With the development of sequencing technology and advanced analytical tools, amplicon sequencing has been adapted to various sequencing platforms, from short-reads technology, such as Illumina, Roche 454, Ion Torrent to Nanopore/PacBio long-read sequencing (Beser et al. 2017; Bokulich et al. 2018; Bolyen et al. 2019; Costello et al. 2009; Fadrosh et al. 2014; Moonsamy et al. 2013; Myer et al. 2016; Neiman et al. 2011; Robeson et al. 2020; Wu et al. 2015). Although widely applicable, costs for amplicon library preparations containing a large number of samples are much higher than those for the downstream sequencing. Even higher costs are incurred if library constructions and data analyses are outsourced to a service provider.
Illumina paired-end sequencing (PE150 and PE250) has become the most widely used amplicon sequencing technology. A series of library preparation kits are available, such as QIAseq 1Step Amplicon Lib UDI-A Kit (Cat. No.: 180419) and Vazyme VAHTS AmpSeq Library Prep Kit V2 (Cat. No.: NA201). Protocols for these kits usually contain multiple procedures, such as the adapter ligation and a further PCR for indexing (Neiman et al. 2011; Vo and Jedlicka 2014). Such protocols are always time-consuming and costly, due to the complex procedures, which require expensive reagents. In addition, the multi-step PCRs can lead to problems, such as primer dimers, sample contaminations, and biased amplifications (Bohmann et al. 2022). Although promising, recently proposed full-length amplicon sequencing by PacBio or Nanopore platforms also suffers from many problems, such as low reads accuracy, the immature analytical tools, and higher costs (Calus et al. 2018; Ciuffreda et al. 2021; Karst et al. 2018; Martijn et al. 2019; Tedersoo et al. 2021; Wagner et al. 2016).
To tackle these difficulties, a “one-step PCR approach” has been proposed for constructions of amplicon libraries using long fusion primers, which include Illumina P5/P7 adaptor sequences, indices, sequencing primer-binding sites, and target-specific primers (Parada et al. 2016). However, inefficient amplifications using long primers and cross-sample contaminations that result from index-hopped reads generated during pooled sequencing limit the application of this method (Bohmann et al. 2022; Sinha et al. 2017; van der Valk et al. 2020). To overcome these technical hurdles, we present an integrative protocol for One-Step PCR Amplicon Library Construction (OSPALC, also a letter combination which could be unscrambled into 90 words in word games), which integrates: i. the design and optimization of long primers containing sequencing adaptors and primers of the target region; ii. the selection of indices for accurate demultiplexing (Supplementary Fig. S1); iii. the library construction procedure; and iv. the sequencing strategies.
To test the efficacy of our approach, 16S rDNA V4 amplicon sequencing of two mock communities of bacteria was conducted by commercial multi-step PCR library preparation vs. OSPALC. Compositions of mock communities were reliably revealed by OSPALC. In addition, we were also able to apply the method, using RNA of time-series samples, to accurately monitor the dynamics of bacterial community compositions in a summer pond, which experienced strong disturbance from one hose-cleaning, as well as those of another undisturbed pond.
Results
Primer design and the protocol for One-Step PCR Amplicon Library Construction (OSPALC)
Each long primer for the OSPALC method is about 90 nt and contains four parts, in the 5′ to 3′ direction: i. the P5 or P7 sequence for Illumina flow-cell binding; ii. the 8-nt index; iii. the Illumina sequencing primer-binding site (SP1/SP2); and iv. the forward or reverse primer targeting at the gene of interest (Fig. 1). To minimize the chance of forming primer dimers and to maximize the primer specificity, we optimized each pair of the 8-nt dual indices by eliminating palindromes or high sequence similarity. Unique dual indices were selected for each sample in a pooled library to enable exclusions of sequencing chimeras that can lead to errors in demultiplexing (Supplementary Fig. S1). The indices we used in this study and additional candidates, as well as criteria for selecting indices, are listed in Supplementary Tables S1, S2, and https://github.com/IEMB-LEG/OSPALC-1.0. The total cost for each amplicon sample starting with environmental DNA is ~ $US 2 ($US 4 if starting with total RNA), after taking long-primer synthesis, PCR for library construction, and Illumina PE250 sequencing into account. These are low compared with the charges by service providers, for example at least $US 30 for each DNA sample. Many service providers do not even accept cDNA or RNA samples.
Fig. 1.

Long primers and PCR for OSPALC. The top two horizontal stacked bars show the structures of long primers. Each long primer contains four parts: P5/P7, for the flow-cell binding; index1/index2, for sample-indexing; SP1/SP2, for Illumina sequencing primer binding; forward/reverse primer, for amplification of the target gene region. The steps below the long primers illustrate the PCR procedures for OSPLAC
The OSPALC procedures are as follows (different PCR conditions that have been tested are shown in the next section; Supplementary Table S3):
Any high-fidelity PCR kit with proof-reading DNA polymerases can be applied, and here, we used 2 × Phanta Flash Master Mix (Dye Plus) (Vazyme Inc., Cat. No.: P520-02). The 50 μL reaction system consisted of 1 μL DNA template (10 ng/μL), 25 μL master mix, 1 μL of the forward long primer (10 μmol/L), 1 μL of the reverse long primer (10 μmol/L), and 22 μL sterilized ultra-pure water, with ten regular and ten gradient PCR cycles (a miniaturized 10-μL protocol is provided in Supplementary File S1). To reduce potential biases in the yield of amplicons that PCR may introduce and reduce time costs, we suggest to minimize the number of PCR cycles (no more than 20 cycles as mentioned above), as long as the concentration of pooled amplicon library is more than 1 ng/μL. One blank control with sterilized DI water as DNA template is also preferred.
Each library (amplified gene fragments with adaptors on) of 400 to 500 bp was then size-selected by gel cutting and purified with the E.Z.N.A.® Gel Extraction Kit (OMEGA, Cat. No.: D2500–02; size selection with magnetic beads is less preferred, because this method is leaky, with many fragments smaller than the target size frequently unfiltered). If the size distribution of amplicon libraries looks uniform on the agarose gel, we recommend to perform the size selection on the pooled libraries, which could significantly reduce the workload in the gel extraction step. Illumina PE250 sequencing on the pooled amplicon libraries was then performed.
Compositions of the two mock communities are reliably revealed by OSPALC vs. multi-step PCR library preparation methods by service providers
To evaluate the data quality from OSPALC, we first prepared two mock communities by mixing purified genomic DNA of five bacterial species, including three Gram-negative species: Escherichia coli MG1655, Pseudomonas aeruginosa PAO1, Shewanella putrefaciens CGMCC1.6515, and two Gram-positive ones: Bacillus subtilis ATCC6051 and Kocuria polaris CGMCC1.8013. One mock community (Mock Equal—ME) contained genomic DNA with equal copy numbers of 16S rDNA in each species, by taking account of the specific 16S rDNA copy number of each bacterial genome. The other (Mock Gradient—MG) contained genomic DNA with gradients of 16S rDNA copy number of the five bacteria (Supplementary Table S4). We then constructed amplicon libraries of the 16S rDNA V4 region for the mock communities using OSPALC. As a comparison, we outsourced the genomic DNA samples of the same mock communities to the service providers in two batches, where amplicon libraries were constructed with kits, including end repair, adapter ligation, and indexing in three separate PCR steps, as well as multiple beads-cleaning in between (TianGen Corp.; Cat. No.: NG102). PE250 reads were generated for amplicon libraries constructed by OSPALC and the service providers. All data were analyzed with a pipeline based on QIIME2 (version: 2021.8). After the data were imported into QIIME2, the paired-end reads were joined, and chimeric feature sequences were identified and filtered out by vsearch (Rognes et al. 2016). Joined reads were then filtered by q-score and denoised by deblur (Amir et al. 2017). Through the phylogenetic inference and the classified feature sequences, we finally obtained species-abundance tables.
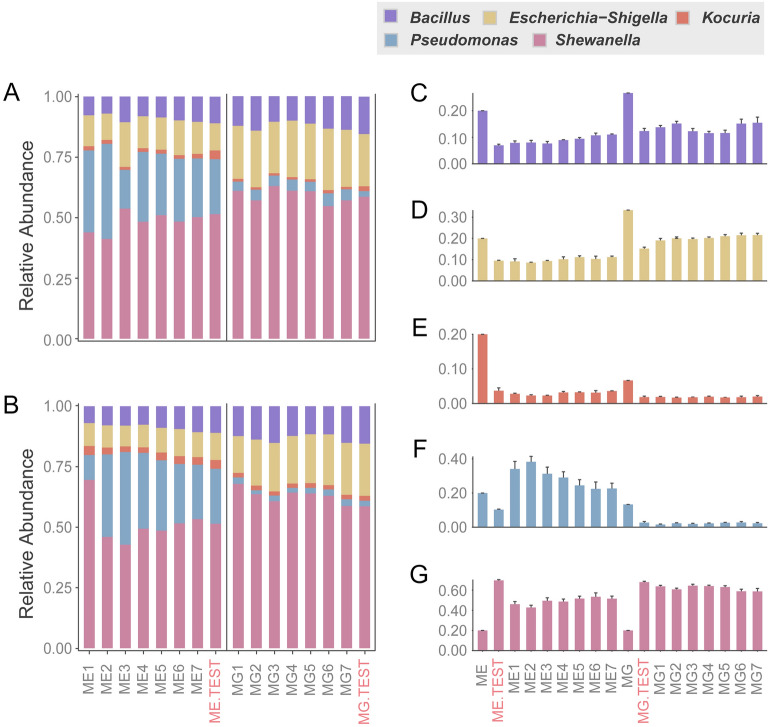
In all data, the five bacterial species of the two mock communities were detected by both OSPALC and the methods of the service providers (Fig. 2; Supplementary Fig. S2). Surprisingly, there was widespread contamination of other bacteria not belonging to the five species in the first batch of the amplicon libraries constructed by one service provider (Supplementary Fig. S2). This observation suggests that more library preparation steps increase the chance of sample contamination. Also, chimeras generated during at least two PCRs for adaptor ligation, indexing or sequencing, using multi-step amplicon library preparation methods may be identified as other species, and thus become ‘contaminating bacteria’. These demonstrate advantages of the in-lab OSPALC, since only one PCR is required, and unique indices help exclude sequencing chimeras.
Fig. 2.
Species abundance of the mock communities, based on libraries constructed by OSPALC and the service provider. ME (Mock equal, genomic DNA with equal copy numbers of 16S rDNA in each species, the specific 16S rDNA copy number of each bacterial genome was taken into account) and MG (Mock gradient, containing genomic DNA with gradients of 16S rDNA copy number of the five bacteria, details are in Supplementary Table S4) are the known preset proportions of each bacterium in the equal and the gradient mock communities. ME1-7 and MG1-7 are the equal community and the gradient community with different PCR conditions of OSPALC. Data from the service provider are labeled with ME.TEST and MG.TEST in red; A, B: mock community sequenced with PE250 and PE150, respectively. C-G: composition variation of the five bacteria detected by PE250; the standard deviations between replicates are low and both OSPALC and service provider community compositions deviated from the preset values
Sequencing details of the amplicon libraries by in-lab OSPALC and the service providers are presented in Supplementary Tables S5–S7. The service provider charged ~ $US 30 for each amplicon sample, including the library construction and the PE250 sequencing. In comparison, the total cost using the OSPALC method, from genomic DNA to Illumina reads, was ~ $US 2 per sample. After proceeding with the QIIME2 pipeline and PE250 reads, an average of 157,692 final joined reads per sample were obtained (~ 8 million raw reads in total for 56 samples; joined reads passed filters against low-quality and un-joined reads). In most cases, 50,000 joined reads per sample are sufficient for downstream analysis (corresponding sequencing fee is ~ $US 1.39 per sample). Although PE150 sequencing costs much less than PE250, we recommend PE250 sequencing. Because there is a risk that short overlaps between the PE150 forward and reverse reads result in extremely low success rate at the reads-joining step, especially if the insert size is close to 300 bp. To validate this, we also used the in-lab OSPALC libraries of the mock community samples to generate a total of 53.94 million raw Illumina PE150 reads for the 56 samples. After filtering, an average of ~ 50,000 joined reads was yielded per sample, with the sequencing cost of ~ $US 0.85 per sample. Compared with the PE250 mode, shorter read length of the PE150 mode greatly decreased the downstream join rate, i.e., only about 5% OSPALC raw reads could be joined and pass the cut-off line of 291 bp for the joined PE150 reads. By contrast, the join rate of the PE250 reads was 91.78% (Supplementary Tables S5–S7). Nonetheless, the community structures revealed by both PE150 and PE250 sequencing modes are highly similar, i.e., PE150 is still useful even if only a tiny part (~ 5%) of the reads is analyzable (Fig. 2A, B). For the 16S rDNA V4 target region (~ 264 bp) in this study, considering the cost and potential variations in the length of amplicons, PE250 is definitely a better choice. We thus recommend PE250 sequencing for OSPALC libraries, especially when the target amplification region is close to or longer than 300 bp.
Optimization of the PCR conditions for OSPALC
Species compositions of the mock communities, revealed by both the OSPALC and the service providers’ methods, show discrepancies from the preset ones because of the bias of amplifications. Such bias still existed after we performed qPCR on the 16S rDNA V4 region, using genomic DNA of each bacterium for the mock community and taking account of the rDNA copy number in the genome of each bacterium (Supplementary Table S4). Nonetheless, the biases among replicates of a mock population did not significantly differ (Fig. 2; Supplementary Tables S8–S12). Such bias is a widely known limitation for almost all amplicon methods (Bohmann et al. 2022; Yeh et al. 2021). It could be caused by multiple factors of different experimental steps, mainly the bias in the initial rounds of PCRs. To test whether the results of OSPALC could be optimized by changing the PCR conditions, we performed multiple PCRs with different reaction conditions, using both the ME and the MG mock community DNA as templates, each with four replicates (Supplementary Table S3). In detail, seven sets of PCR conditions were used to construct OSPALC libraries. In four of the seven PCR sets, we set up the annealing temperature of 55 °C with 35, 30, 25, and 20 amplification cycles (labeled as 1 to 4, respectively). In the other three PCR sets (5 to 7), the annealing temperature was gradually increased from 55 °C to 65 °C after the tenth cycle with a total of 20, 25, and 30 cycles. This gradient of annealing temperatures was set up to increase the primer-binding specificity, because we expect the entire long primers to fully pair with the templates in later rounds of amplifications. The results from all the above trials did not show significant differences (Fig. 2A, B) which may be explained by the simple composition of the mock communities. For complex environmental samples, minimizing amplification cycles is still recommended. The CV (coefficient of variation) of the deviations from the genuine community compositions among replicates is quite low ( 0.2%), indicating that the discrepancy from the real community compositions may result from the amplification bias generated from the original ten PCR cycles (Supplementary Tables S11, S12). Taken together, a minimized number of amplification cycles are suggested to reduce potential biases in the yield of amplicons that PCR may introduce, given that the amount of amplicons is sufficient to load on the flow cell for sequencing (usually more than 1 ng/μL). Our optimal PCR conditions for the current OSPALC protocol are: 98 °C for 5 min followed by 10 cycles of 98 °C for 30 s, 55 °C for 30 s and 72 °C for 50 s, 10 cycles of 98 °C for 30 s, gradient 55 °C to 65 °C for 30 s and 72 °C for 50 s, and one final extension step at 72 °C for 7 min.
Applications of OSPALC to RNA samples from disturbed and stable environments
Because environmental DNA is highly stable, amplicon library constructions using DNA as templates could lead to false positives due to remnant DNA from dead organisms. RNA is transient and degrades quickly in the environment, and amplicon analyses based on rRNA could thus reveal the extant biodiversity of samples. Therefore, we applied OSPALC to total RNA extracted from environmental samples to investigate the dynamics of biodiversity in a pond before and after one hose-cleaning. Such application will test the performance of the OSPALC method on natural samples. The Crescent Moon pond is located in front of the Ocean University of China (OUC) Library, Yushan Campus (samples labeled as T1–4; Supplementary Fig. S3; the pond area is around 10 m2 and 1 m deep). The pond was hose-cleaned and refilled with freshwater on June 8th, 2021. We collected samples from four spots as replicates. Each sample contains 500 mL water with sediment from the bottom of each spot, collected with zip bags on a golf-ball retriever. Samples were collected mostly every two weeks from May 21st 2021 to October 16th, 2021 (Supplementary Tables S13, S14; Supplementary Fig. S3). Before being cleaned, the pond was murky and green, full of micro-organisms that were visible even to naked eyes. After hose-cleaning, the pond was clear and started to become green a week later (Supplementary Fig. S3). For each thoroughly mixed sample, 50 mL was centrifuged at 4 °C and total RNA of the cell pellets was extracted immediately after collections and reverse-transcribed to cDNA, which was used as the template for the OSPALC PCR for the 16S-rRNA-V4 region.
The results are consistent with the previous reports on bacterial communities in summer resting freshwater, as Proteobacteria and Cyanobacteria are the dominant bacteria (Zhang et al. 2018). Interestingly, probably because of the pond’s proximity to the sea (862 m to the nearest coast), some typical marine bacteria—Pirellula, NS9_marine_group, Hyphomonas—were also detected in these freshwater samples (Fig. 3). The sea breeze or activities of other living organisms may account for the detection of marine bacteria.
Fig. 3.
Relative bacterial abundance of the environmental samples from the two collection sites. The sequences of the four samples from each site on the same day were merged for analysis. T stands for the Crescent Moon pond (light blue background on the left panel) in front of the library of Ocean University of China (Yushan Campus), Z stands for the Xiaoxihu pond (light purple background on the right panel) in the Zhongshan Park. The four digits after “T” or “Z” stand for the date (month-day) in 2021. Crescent Moon pond was hose-cleaned on June 8th, 2021
The community composition, revealed by the amplicon sequencing, also indicated clear-cut community successions, particularly for the samples collected before and after the hose-cleaning (June 8th, 2021; Fig. 3; Supplementary Table S13). There was a substantial increase of the bacteria Zoogloea (from 0.08% to 21.44%) and Hydrogenophaga (from 0% to 31.23%) right after the environmental disturbance, but they almost disappeared in subsequent samples. Zoogloea are frequently reported in sewage or from sewage-treatment systems and Hydrogenophaga are able to perform dissimilatory nitrate reduction to ammonium (Dugan 1981; Tang et al. 2020). These bacteria may be opportunistic growers, particularly when competitors/predators are reduced after the hose-cleaning. Several species such as Brevundimonas and Rhodococcus, which were not detected before the hose-cleaning, also appeared afterward, with low abundance. Several genera, such as Crenothrix and Margulisbacteria, gradually disappeared after the cleaning. Crenothrix are important methane consumers (Oswald et al. 2017). These bacteria can be greatly affected by environmental changes such as the removal of leaves and sediments. At the phylum level, four days after the hose-cleaning, the proportion of Proteobacteria rose from 17.95% to 73.10%, and Fusobacteriota decreased from 43.30% to 3.18% (Fig. 3).
Besides the samples from an environment with perturbations, we also tested OSPALC on samples collected from a stable environment, Xiaoxihu pond, at Zhongshan Park of Qingdao, China (700 m to the above Crescent Moon pond; area ~ 8000 m2, depth ~ two meters, labeled as Z5–8). This pond has a history of 99 years and is rarely disturbed. We expected no extreme community successions around the hose-cleaning time of the Crescent Moon pond and performed parallel sample collections with similar procedures as those for the Crescent Moon pond (Supplementary Tables S13, S14). We then prepared OSPALC libraries using the total RNA extracted from the Xiaoxihu pond samples. To note, the PCR success rate of OSPALC could be elevated by diluting cDNA to ~ 1 ng/μL (data not shown), especially for the repeatedly-PCR-failed environmental samples. As expected, the community structure of the Xiaoxihu pond was stable across all sampling time points (Fig. 3). Taken together, applications of OSPALC to RNA samples from disturbed and stable environments suggest that it can be reliably used to reveal community structure and biodiversity for environmental samples.
Discussion
Compared with metagenomic sequencing, short-reads-based amplicon sequencing is not able to fully reveal community function, since only part of the target gene is investigated (Myer et al. 2016). However, even the biodiversity indices and community compositions revealed from amplicon sequencing can be tremendously useful, compared with the traditional methods using microscopy and staining (Eisenstein 2018; Gupta et al. 2019; Pochon et al. 2013). The protocol in this study can achieve this goal in a fast and cost-efficient way, compared with most amplicon library construction methods, especially those with the adapter ligation and indexing steps. Besides 16S rDNA V4 regions, OSPALC can in principle be applied to any genomic regions. We have recently applied this protocol to 18S rDNA V8-V9 regions to detect community structures in coastal waters, stomach content of fish with success (unpublished data). The strategy of OSPALC protocol is different from the miniaturization to reduce costs for the whole-genome library construction (Li et al. 2019).
To overcome limitations of insufficient amplifications using long fusion primers and cross-sample contaminations during pooled sequencing, we provided a bioinformatic tool to design primers with high specificities and assign unique dual indices for pooled samples. Including unique molecular identifiers (UMIs) enables quantitative analyses on community compositions, but degenerate nucleotides may also create reverse complementary regions within a pair of primers or between primers and nontargeted templates (Yeh et al. 2021). In the present protocol, UMIs are excluded from the long primers to potentially increase the efficiency of amplifications.
In principle, OSPALC could be applied to any conservative genes of most organisms, not limited to bacteria, contrastingly different from the service providers, which usually provide amplicon sequencing for 16S/18S rDNA only. The OSPALC method has been used to produce reliable results with no contaminating species and fewer chimeras when analyzing mock communities (Supplementary Tables S5–S7). With its low cost and efficient workflow, OSPALC shows great potential for applications in biodiversity and evolution. As it continues to be explored, OSPALC has the potential to bring about significant advancements and discoveries in these fields (Zhao et al. 2021). OSPALC could not get full-length sequencing for 16S/18S rDNA. Long-read sequencing remains costly and prone to errors when compared to short-read sequencing. Additionally, being in the same "genome fragment" status as partial gene amplicons does not provide a significant advantage over short-read amplicons in terms of improved resolution (Myer et al. 2016). However, we hope that with the continuous development of sequencing technology, full-length amplicons may become the mainstream method for studying community structure in the future. Future advancement may also facilitate the exploration of genetic diversity in target sites even in organisms with extremely low mutation rates (Pan et al. 2021).
We have shown that the deviation between genuine community composition and that from the amplicon analysis may arise during the initial cycles of PCR because of biases in GC content and copy number of target regions (Laursen et al. 2017). To solve these problems, we are developing a model that could hopefully calibrate the community composition deviation of OSPALC. And as reference databases (such as taxonomic and functional databases) continue to improve and become more comprehensive, OSPALC would become more reliable and more widely used in predictions based on Operational Taxonomic Units (OTUs), e.g., PICRUSt2 pathway prediction (Douglas et al. 2020). The next version of OSPALC will thus integrate cutting-edge sequencing technology, sophisticated algorithms, and target much larger genomic regions, from sequencing to data analysis.
Materials and methods
Preparation of the two mock communities
Each mock community was a mixture of genomic DNA of five bacteria: two Gram-positive species—Bacillus subtilis ATCC6051, Kocuria polaris CGMCC1.8013; three Gram-negative species—Escherichia coli MG1655, Pseudomonas aeruginosa PAO1 and Shewanella putrefaciens CGMCC1.6515. All bacteria were cultured with conditions shown in Supplementary Table S4. The Marine LB Broth was prepared with a lab-developed recipe (Strauss et al. 2017): 1000 mL natural seawater with PSU of ~ 30, 5 g Bacto™ peptone (BD, Cat. No.: 9030688), 1 g Bacto™ yeast extract (BD, Cat. No.: 8344948), and 0.18 g ferric-EDTA (SIGMA, Cat. No.: SLBV7746). DNA was extracted with the MasterPure™ Complete DNA and RNA Purification Kit (Epicentre, Cat. No.: MC85200). The Qubit 3.0 fluorometer and Nano-300 (Allsheng™) were used to measure the concentration and the purity of DNA, respectively. Four replicates were prepared for each mock community (Mock-Equal replicate 1 was discarded due to operation errors during mixing). Kocuria polaris CGMCC1.8013 genome was assembled by Unicycler 0.4.8 (Wick et al. 2017).
Sample collection
Samples were collected from two sites in Qingdao, China: the Crescent Moon pond in front of the library of Ocean University of China (Yushan Campus) and the Xiaoxihu pond at Zhongshan Park. About every two weeks (May 21st – October 16th, 2021), we collected 500 mL bottom-water samples at each of the four spots of each site (Supplementary Fig. S3), using a golf-ball retriever with a zip bag. The Crescent Moon pond experienced draining and hose-cleaning (without using any disinfectant) on June 8th, 2021. The Xiaoxihu pond of Zhongshan Park was investigated as an undisturbed comparison, and sampled similarly (Supplementary Fig. S3), which was much less human-disturbed as a site of view. After collection, each sample was immediately shipped to the lab and completely mixed, and 50 mL sub-sample was centrifuged at 4000 g and 4 °C. Temperature, dissolved oxygen, pH, salinity, and conductivity were measured on site using one YSI Professional Plus Multiparameter Instrument (Supplementary Table S14).
Nucleic acids extraction, amplicon library construction, and Illumina sequencing of environmental samples
After centrifugation of 50 mL of each sample, we discarded the supernatant and transferred the pellets to 1.5 mL Eppendorf tubes. We then centrifuged again at 15,000 g and 4 °C, for 3 min. RNA and DNA were extracted with MasterPure™ Complete DNA and RNA Purification Kit (Epicentre, Cat. No.: MC85200). For reverse transcription of RNA to cDNA, we used Vazyme HiScript III 1st Strand cDNA Synthesis Kit (+ gDNA wiper) (Cat. No.: R312). Details for all DNA or cDNA OSPALC library construction are in Supplementary File S1, and no contamination was detected after gel electrophoresis. Illumina Novaseq6000 sequencing was performed at Berry Genomics, Beijing and Novogene, Tianjin. All raw sequences were submitted to NCBI SRA (BioProject No.: PRJNA906703).
qPCR
We diluted genomic DNA of the five bacteria to 10 ng/μL, each with three replicates. With four serial tenfold dilutions, five different concentrations of DNA for each bacterium were used for qPCR with one ABIStepOnePlus™ Real-Time PCR instrument. The 20 μL reaction system consisted of 2 μL DNA template, 10 μL AceQ® Universal SYBR qPCR Master Mix (Cat. No.: Q511), 0.4 μL 515F primer (10 μM), 0.4 μL 806Y primer (10 μM), and 7.2 μL sterilized ultra-pure water. The amplification conditions were: 95 °C for 5 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 25 s.
Amplicon analysis
The amplicon analysis was based on QIIME2 (version: 2021.8) (Bolyen et al. 2019). Raw reads were joined with qiime vsearch (Rognes et al. 2016) and filtered by qiime q-score. The 16S rDNA V4 region database Silva was used—Silva 138 99% OTUs based on the 515F/806R sequences (Bokulich et al. 2018; Quast et al. 2012; Robeson et al. 2021). We performed feature classification with qiime deblur (Amir et al. 2017). We ran qiime alignment mafft for feature reads alignment and performed phylogenetic analysis with qiime phylogeny fasttree (Price et al. 2010). Overlapping bar charts were plotted with ggplot2 (Wickham 2016).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31961123002, 31872228), the Fundamental Research Funds for the Central Universities of China (202041001), the Young Taishan Scholars Program of Shandong Province (tsqn201812024), and the National Science Foundation (DEB-1927159). We appreciate the technical help from Vazyme Biotech Co., Ltd., Nanjing and Wei Yang, OUC. All bioinformatic analyses were performed with IEMB-1 computation clusters at OUC. We also thank Haoyu Li, Jingjing Baoli and Zhirong Zhang for technical help.
Author contributions
JN, JP, HL, and WL designed this study; JN, TC, XF, and YL performed experiments; JN, YW, and TL analyzed data; JN, JP, YW, HL, ML, and WL wrote the manuscript. All authors read and approved the submitted manuscript.
Data availability
Data are uploaded to https://submit.ncbi.nlm.nih.gov/subs/sra/SUB12307788.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest. Hongan Long is one of the Editorial Board Members, but he was not involved in the journal’s review of, or decision related to, this manuscript.
Animal and human rights statement
This article does not contain any studies performed with human and animals.
Footnotes
Special Topic: EvoDevo.
Edited by Jiamei Li.
Jiahao Ni and Jiao Pan have contributed equally to this work.
References
- Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech XZ, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2:e00191–16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beser J, Hallström BM, Advani A, Andersson S, Östlund G, Winiecka-Krusnell J, Lebbad M, Alm E, Troell K, Arrighi RBG. Improving the genotyping resolution of Cryptosporidium hominis subtype IbA10G2 using one step PCR-based amplicon sequencing. Infect Genet Evol. 2017;55:297–304. doi: 10.1016/j.meegid.2017.08.035. [DOI] [PubMed] [Google Scholar]
- Bohmann K, Elbrecht V, Carøe C, Bista I, Leese F, Bunce M, Yu DW, Seymour M, Dumbrell AJ, Creer S. Strategies for sample labelling and library preparation in DNA metabarcoding studies. Mol Ecol Resour. 2022;22:1231–1246. doi: 10.1111/1755-0998.13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calus ST, Ijaz UZ, Pinto AJ. NanoAmpli-Seq: a workflow for amplicon sequencing for mixed microbial communities on the nanopore sequencing platform. GigaScience. 2018;7:giy140. doi: 10.1093/gigascience/giy140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffreda L, Rodríguez-Pérez H, Flores C. Nanopore sequencing and its application to the study of microbial communities. Comput Struct Biotechnol J. 2021;19:1497–1511. doi: 10.1016/j.csbj.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan PR. The Genus Zoogloea. In: Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG, editors. The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Berlin: Springer; 1981. pp. 764–770. [Google Scholar]
- Eisenstein M. Microbiology: making the best of PCR bias. Nat Methods. 2018;15:317–320. doi: 10.1038/nmeth.4683. [DOI] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gous A, Swanevelder DZH, Eardley CD, Willows-Munro S. Plant-pollinator interactions over time: Pollen metabarcoding from bees in a historic collection. Evol Appl. 2019;12:187–197. doi: 10.1111/eva.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Mortensen MS, Schjørring S, Trivedi U, Vestergaard G, Stokholm J, Bisgaard H, Krogfelt KA, Sørensen SJ. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun Biol. 2019;2:291. doi: 10.1038/s42003-019-0540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Dueholm MS, McIlroy SJ, Kirkegaard RH, Nielsen PH, Albertsen M. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat Biotechnol. 2018;36:190–195. doi: 10.1038/nbt.4045. [DOI] [PubMed] [Google Scholar]
- Laursen MF, Dalgaard MD, Bahl MI. Genomic GC-content affects the accuracy of 16S rRNA gene sequencing based microbial profiling due to PCR bias. Front Microbiol. 2017;8:1934. doi: 10.3389/fmicb.2017.01934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wu K, Ruan C, Pan J, Wang Y, Long H. Cost-reduction strategies in massive genomics experiments. Mar Life Sci Technol. 2019;1:15–21. doi: 10.1007/s42995-019-00013-2. [DOI] [Google Scholar]
- Martijn J, Lind AE, Schön ME, Spiertz I, Juzokaite L, Bunikis I, Pettersson OV, Ettema TJG. Confident phylogenetic identification of uncultured prokaryotes through long read amplicon sequencing of the 16S-ITS-23S rRNA operon. Environ Microbiol. 2019;21:2485–2498. doi: 10.1111/1462-2920.14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara RP, Caro-Vegas C, Landis JT, Moorad R, Pluta LJ, Eason AB, Thompson C, Bailey A, Villamor FCS, Lange PT, Wong JP, Seltzer T, Seltzer J, Zhou Y, Vahrson W, Juarez A, Meyo JO, Calabre T, Broussard G, Rivera-Soto R, et al. High-density amplicon sequencing identifies community spread and ongoing evolution of SARS-CoV-2 in the Southern United States. Cell Rep. 2020;33:108352. doi: 10.1016/j.celrep.2020.108352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonsamy PV, Williams T, Bonella P, Holcomb CL, Höglund BN, Hillman G, Goodridge D, Turenchalk GS, Blake LA, Daigle DA, Simen BB, Hamilton A, May AP, Erlich HA. High throughput HLA genotyping using 454 sequencing and the Fluidigm Access Array™ system for simplified amplicon library preparation. Tissue Antigens. 2013;81:141–149. doi: 10.1111/tan.12071. [DOI] [PubMed] [Google Scholar]
- Myer PR, Kim M, Freetly HC, Smith TPL. Evaluation of 16S rRNA amplicon sequencing using two next-generation sequencing technologies for phylogenetic analysis of the rumen bacterial community in steers. J Microbiol Methods. 2016;127:132–140. doi: 10.1016/j.mimet.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Neiman M, Lundin S, Savolainen P, Ahmadian A. Decoding a substantial set of samples in parallel by massive sequencing. PLoS ONE. 2011;6:e17785. doi: 10.1371/journal.pone.0017785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald K, Graf JS, Littmann S, Tienken D, Brand A, Wehrli B, Albertsen M, Daims H, Wagner M, Kuypers MMM, Schubert CJ, Milucka J. Crenothrix are major methane consumers in stratified lakes. ISME J. 2017;11:2124–2140. doi: 10.1038/ismej.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Williams E, Sung W, Lynch M, Long H. The insect-killing bacterium Photorhabdus luminescens has the lowest mutation rate among bacteria. Mar Life Sci Technol. 2021;3:20–27. doi: 10.1007/s42995-020-00060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- Pochon X, Bott NJ, Smith KF, Wood SA. Evaluating detection limits of next-generation sequencing for the surveillance and monitoring of international marine pests. PLoS ONE. 2013;8:e73935. doi: 10.1371/journal.pone.0073935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2 — approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robeson MS, O’Rourke DR, Kaehler BD, Ziemski M, Dillon MR, Foster JT, Bokulich NA. RESCRIPt: reproducible sequence taxonomy reference database management. PLoS Comput Biol. 2021;17:e1009581. doi: 10.1371/journal.pcbi.1009581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Stanley G, Gulati GS, Ezran C, Travaglini KJ, Wei E, Chan CKF, Nabhan AN, Su T, Morganti RM, Conley SD, Chaib H, Red-Horse K, Longaker MT, Snyder MP, Krasnow MA, Weissman IL. Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. bioRxiv. 2017 doi: 10.1101/125724. [DOI] [Google Scholar]
- Song C, Zhong L, Liu Y, Yin Y, Zhang C, Zhang X, Chen J. Spatial and temporal succession of bacterial communities in three artificial fishponds. Aquac Res. 2019;50:2793–2801. doi: 10.1111/are.14231. [DOI] [Google Scholar]
- Strauss C, Long H, Patterson CE, Te R, Lynch M, Moran NA. Genome-wide mutation rate response to pH change in the coral reef pathogen Vibrio shilonii AK1. MBio. 2017;8:e01021–01017. doi: 10.1128/mBio.01021-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Liao Y, Xu Y, Dang Z, Zhu X, Ji G. Microbial coupling mechanisms of nitrogen removal in constructed wetlands: a review. Bioresour Technol. 2020;314:123759. doi: 10.1016/j.biortech.2020.123759. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Albertsen M, Anslan S, Callahan B. Perspectives and benefits of high-throughput long-read sequencing in microbial ecology. Appl Environ Microbiol. 2021;87:e0062621. doi: 10.1128/AEM.00626-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk T, Vezzi F, Ormestad M, Dalén L, Guschanski K. Index hopping on the Illumina HiseqX platform and its consequences for ancient DNA studies. Mol Ecol Resour. 2020;20:1171–1181. doi: 10.1111/1755-0998.13009. [DOI] [PubMed] [Google Scholar]
- Vo A-TE, Jedlicka JA. Protocols for metagenomic DNA extraction and Illumina amplicon library preparation for faecal and swab samples. Mol Ecol Resour. 2014;14:1183–1197. doi: 10.1111/1755-0998.12269. [DOI] [PubMed] [Google Scholar]
- Wagner J, Coupland P, Browne HP, Lawley TD, Francis SC, Parkhill J. Evaluation of PacBio sequencing for full-length bacterial 16S rRNA gene classification. BMC Microbiol. 2016;16:274. doi: 10.1186/s12866-016-0891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Cham: Springer International Publishing; 2016. pp. 189–201. [Google Scholar]
- Wu L, Wen C, Qin Y, Yin H, Tu Q, Van Nostrand JD, Yuan T, Yuan M, Deng Y, Zhou J. Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol. 2015;15:125. doi: 10.1186/s12866-015-0450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ravnskov S, Larsen J, Nilsson RH, Nicolaisen M. Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol Biochem. 2012;46:26–32. doi: 10.1016/j.soilbio.2011.11.010. [DOI] [Google Scholar]
- Yeh Y-C, McNichol J, Needham DM, Fichot EB, Berdjeb L, Fuhrman JA. Comprehensive single-PCR 16S and 18S rRNA community analysis validated with mock communities, and estimation of sequencing bias against 18S. Environ Microbiol. 2021;23:3240–3250. doi: 10.1111/1462-2920.15553. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Chen S, Zhao Z, Feng J, Zhang Z, Lu K, Jia J. Water bacterial and fungal community compositions associated with urban lakes, Xi’an, China. Int J Environ Res Public Health. 2018;15:469. doi: 10.3390/ijerph15030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Gao F, Gao S, Liang Y, Long H, Lv Z, Su Y, Ye N, Zhang L, Zhao C, Wang X, Song W, Zhang S, Dong B. Biodiversity-based development and evolution: the emerging research systems in model and non-model organisms. Sci China Life Sci. 2021;64:1236–1280. doi: 10.1007/s11427-020-1915-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are uploaded to https://submit.ncbi.nlm.nih.gov/subs/sra/SUB12307788.