Abstract
A peptidyl prolyl cis-trans isomerase (PPIase) was purified from a thermophilic methanogen, Methanococcus thermolithotrophicus. The PPIase activity was inhibited by FK506 but not by cyclosporine. The molecular mass of the purified enzyme was estimated to be 16 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and 42 kDa by gel filtration. The enzyme was thermostable, with the half-lives of its activity at 90 and 100°C being 90 and 30 min, respectively. The catalytic efficiencies (kcat/Km) measured at 15°C for the peptidyl substrates, N-succinyl-Ala-Leu-Pro-Phe-p-nitroanilide and N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide, were 0.35 and 0.20 μM−1 s−1, respectively, in chymotrypsin-coupled assays. The purified enzyme was sensitive to FK506 and therefore was called MTFK (M. thermolithotrophicus FK506-binding protein). The MTFK gene (462 bp) was cloned from an M. thermolithotrophicus genomic library. The comparison of the amino acid sequence of MTFK with those of other FK506-binding PPIases revealed that MTFK has a 13-amino-acid insertion in the N-terminal region that is unique to thermophilic archaea. The relationship between the thermostable nature of MTFK and its structure is discussed.
Cyclosporine-binding proteins (also called cyclophilin [CyP]) and FK506-binding proteins (FKBP) are the natural targets (immunophilins) of the immunosuppressants cyclosporine and FK506 (tacrolimus), respectively (11, 32). Both of these proteins exhibit the peptidyl prolyl cis-trans isomerase (PPIase) activity that accelerates the isomerization of the peptidyl prolyl bond, a rate-limiting step in protein folding (12, 36). These two types of proteins show little sequence homology to each other (22), and the cross-inhibition of the PPIase activity by cylcosporine and FK506 was not observed. The CyP- and FKBP-type immunophilins are ubiquitous in the domains Bacteria and Eucarya (8); however, only one CyP-type immunophilin from a halophilic archaeon, Halobacterium cutirubrum, has so far been reported as a PPIase in the domain Archaea (23). While a CyP from a thermophilic bacterium, Bacillus stearothermophilus, has been reported (20), there is no available information on PPIase in thermophilic archaea.
Both CyP- and FKBP-type immunophilins accelerate the speed of the refolding of chemically denatured RNase T1 (31) and carbonic anhydrase (19) in vitro. In the refolding of chemically denatured carbonic anhydrase, a CyP homolog, human tumor recognition molecule (NK-TR) showed a chaperone-like activity that promotes correct folding of the polypeptide (30). Human FKBP52 prevents the thermal aggregation of citrate synthase in vitro in a PPIase activity-independent manner (1). It was demonstrated that the product of the ninaA gene, encoding a CyP homolog, is required for the correct folding of rhodopsin in Drosophila melanogaster in vivo (25). These in vitro and in vivo observations suggested that certain CyPs and FKBPs play important roles in protein folding and exhibit a chaperone-like activity. It has been reported that in Saccharomyces cerevisiae, heat shock induces the expression of CyP1, CyP2, and FKBP13 (27, 35). Disruption of either the CyP1 or CyP2 gene reduced the survival of this organism after the heat shock treatment (35). These results support the notion that CyPs and FKBPs contribute to the heat tolerance of yeast cells, as chaperones do.
The complete genome sequence of a hyperthermophilic archaeon, Methanococcus jannaschii, revealed that only one FKBP-like protein was encoded as PPIase in this organism (4). It was therefore interesting to investigate the roles of FKBP in the thermotolerance of thermophilic archaea. In this study, we purified an FKBP and cloned its structural gene from a thermophilic archaeon, Methanococcus thermolithotrophicus. This is the first report on the characterization of the FKBP-type PPIase in thermophilic archaea.
MATERIALS AND METHODS
Chemicals and biochemicals.
N-Succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (N-suc-A-A-P-F-pNA) was purchased from Sigma Chemical Co. (St. Louis, Mo.), and N-succinyl-Ala-Leu-Pro-Phe-p-nitroanilide (N-suc-A-L-P-F-pNA) was purchased from Peptide Institute Inc. (Osaka, Japan). Cyclosporine was purchased from Sankyo Pharmaceutical Co. (Tokyo, Japan), and FK506 was a gift from Fujisawa Pharmaceutical Co. (Osaka, Japan). They were dissolved in ethanol at 2.0 mM and stored at −20°C until use. The protein concentration was determined by the Bradford dye-binding method with a Bio-Rad protein assay kit with bovine serum albumin as the standard (3). Custom-made oligonucleotides were purchased from Nippon Bio Service Co. (Saitama, Japan). Taq DNA polymerase and a PCR kit were purchased from Nippon Gene Co. (Tokyo, Japan).
Organism and culture.
The thermophilic methanogen M. thermolithotrophicus DSM2095, whose optimum growth temperature is 65°C (16), was purchased from Deutsch Sammlung von Mikroorganismen und Zelkulturen GmbH (Braunschweig, Germany). The medium used was based on seawater and was supplemented with 2 g of yeast extract (Difco, Detroit, Mich.) per liter, 2 g of Bacto Tryptone (Difco) per liter, 1 g of sodium citrate per liter, 10 ml of DAB vitamin solution (18) per liter, 1.4 g of piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) per liter, 80 mg of resazurin per liter, and 50 mg of sodium sulfide per liter. The pH of the medium was adjusted to 6.8. The strain was grown in the seawater-based medium in a 5-liter fermentor which was gassed with H2-CO2 (4:1) at 65°C with stirring at 1,000 rpm for 16 h. The growth yield was 3 g of wet cells per 5 liters.
PPIase assay.
The PPIase activity was determined in a two-step reaction coupled with chymotrypsin, with the oligopeptide N-suc-A-A-P-F-pNA or N-suc-A-L-P-F-pNA as the substrate (36). The reaction mixture (final volume, 2.2 ml) contained 17 μM oligopeptide and an appropriate amount of PPIase in 100 mM sodium phosphate (pH 7.8). The reaction was started by the addition of 50 μl of 1.52 mM chymotrypsin, and the increase in A390 that corresponds to the release of p-nitroanilide was monitored at 25°C for 3 min with a spectrophotometer (model UV2000; Shimadzu Co., Kyoto, Japan). The PPIase activity, Up, was calculated by the equation Up = (Kp − Kn)/Kn, where Kp and Kn are the first-order rate constants of the p-nitroanilide release in the presence and absence of PPIase, respectively. For the determination of catalytic efficiency, the reaction mixture was incubated at 15°C and the efficiency (kcat/Km at 15°C) was calculated from the relationship kcat/Km = (Kp − Kn)/E, where E is the concentration of PPIase (13).
Inhibition studies with immunosuppressants.
To measure the inhibition of the PPIase activity by cyclosporine and FK506, the enzyme was preincubated with one of the ethanol-dissolved immunosuppressants for 3 min before the addition of the substrate and chymotrypsin. The final concentration of ethanol in the assay mixture was 1% (vol/vol), which did not affect the enzyme activity. The percent inhibition of the PPIase activity was expressed as [(Up − Ui)/Up] × 100, where Up is the PPIase activity without the inhibitor and Ui is the PPIase activity with the inhibitor.
Purification of PPIase.
The cell pellet harvested by centrifugation was washed with seawater filtered through a membrane filter (pore size, 0.22 μm). The cells were disrupted by osmotic shock by suspending the 30-g (wet weight) cell pellet in 100 ml of 20 mM sodium phosphate (pH 7.0) on ice for 30 min. The supernatant was collected, and (NH4)2SO4 was added to 40% saturation on ice. After removal of the precipitate by centrifugation (13,000 × g for 20 min), 25 ml of the supernatant was applied to a Hi Trap butyl Sepharose column (5 ml; Pharmacia, Uppsala, Sweden) equilibrated with 1.8 M (NH4)2SO4 in 0.1 M sodium phosphate buffer (pH 7.0) (A buffer), and the adsorbed proteins were eluted with a linear gradient of 1.8 to 0 M (NH4)2SO4 at a flow rate of 1 ml/min. The active fractions eluted at 0.1 to 0 M (NH4)2SO4 were pooled and concentrated to 2 ml at 4°C by using an Amicon ultrafiltration device with a YM10 membrane (Millipore Corp., Bedford, Mass.). The concentrated protein solution was then applied to a Superose 12HR 10/30 gel filtration column (1.0 by 30 cm; Pharmacia) equilibrated with 0.15 M NaCl in 50 mM sodium phosphate (pH 7.0) and eluted at a flow rate of 0.2 ml/min. Active fractions at an elution volume of 10.2 to 11.0 ml were pooled, diluted with 4 volumes of 20 mM Tris-HCl (pH 7.0), and applied to a Mono Q column (0.5 by 5 cm; Pharmacia) equilibrated with 50 mM Tris-HCl (pH 7.0). After elution with a linear gradient of 0 to 0.4 M NaCl at a flow rate of 1.0 ml/min, active fractions eluted at 0.31 to 0.35 M NaCl were pooled. They were then diluted with an equal volume of 3.4 M (NH4)2SO4 in 0.1 M sodium phosphate (pH 7.0) and applied to a TSK gel Ether-5PW column (7.5 mm by 7.5 cm; Tosoh Co., Tokyo, Japan) equilibrated with A buffer. By using a linear gradient of 1.8 M to 0 M (NH4)2SO4 in 0.1 M sodium phosphate (pH 7.0) at a flow rate of 1.0 ml/min, active fractions eluted at 0.54 to 0.48 M (NH4)2SO4 were pooled. All purification procedures were carried out at a room temperature unless otherwise stated. The molecular mass of the enzyme was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or gel filtration chromatography with a TSK gel G2000 SWXL column (7.5 mm by 30 cm; Tosoh Co.) with a mobile phase of 50 mM sodium phosphate (pH 7.0) containing 0.15 M NaCl.
Sequencing of N-terminal amino acids.
To determine the N-terminal amino acid sequence of the purified PPIase, the sample was subjected to SDS-PAGE (20% polyacrylamide), electroblotted to a polyvinylidene difluoride membrane (Amersham Co., Arlington Heights, Ill.), and stained with Coomassie brilliant blue R-250. The corresponding band was cut out and subjected to automated Edman degradation with a Shimadzu PSQ-2 protein sequencer (Shimadzu Co.). To determine the amino acid sequence of the lysylendopeptidase digestion fragments of the purified enzyme, the enzyme was subjected to SDS-PAGE, blotted to a polyvinylidene difluoride membrane, and stained with a solution containing 0.1% (wt/vol) Ponceau S and 1% (vol/vol) acetic acid. The corresponding band was cut out, destained in 0.2 mM NaOH for 1 min, treated with 0.5% (wt/vol) polyvinylpyrrolidone-40 in 100 mM acetic acid at 37°C for 30 min, and then washed 10 times with distilled water. The washed membrane was sonicated for 10 min in 300 μl of 25 mM Tris-HCl (pH 8.5) containing 8% (wt/vol) CH3CN, and the enzyme was digested with 50 pmol of Achromobacter lysylendopeptidase (Wako Pure Chemical Co., Osaka, Japan) at 37°C overnight. After the digestion, the reaction mixture was sonicated for 5 min and the supernatant was recovered. The recovered peptide solution was applied to a μ-Bondasphere C18 column (particle size, 5 μm; pore size, 300 Å; 3.9 by 150 mm; Waters Co., Milford, Mass.). The column was equilibrated with a 95:5 (vol/vol) mixture of a 0.052% (vol/vol) trifluoroacetic acid solution (solution A) and the 80% (vol/vol) CH3CN solution containing 0.06% (vol/vol) trifluoroacetic acid (solution B). With a linear gradient from 95% solution A plus 5% solution B to 20% solution A plus 80% solution B, the digested peptides were separated. The three major peptides were recovered and analyzed with the protein sequencer.
PCR amplification of the partial sequence of the PPIase gene.
From the N-terminal amino acid sequences of the purified enzyme, KIKVDYI, and the partial amino acid sequence of one of the three peptides described above, IPRDAFK, a forward primer, AA(AG)AT(ATC)AA(AG)GT(ATCG)GA(TC)TA(TC)AT, and a reverse primer, TT(AG)AA(ATCG)GC(AG)TC(TC)CT(ATCG)GG(ATG)AT, were designed. With these primers, PCR was carried out in a reaction mixture (100 μl) containing 250 ng of the chromosomal DNA of M. thermolithotrophicus, 0.5 U of Taq DNA polymerase, 100 μM each deoxynucleoside triphosphate, 1.0 mM MgCl2, and 2 nmol of the two primers. The mixture was preincubated for 5 min at 95°C and then subjected to 30 cycles of PCR consisting of denaturation at 95°C for 30 s, primer annealing at 52°C for 1.5 min, and primer extension at 72°C for 2 min in a model 480 DNA thermal cycler (Perkin-Elmer Co., Branchburg, N.J.). The extension reaction in the final cycle was prolonged for 10 min. The reaction mixture was frozen until use. The PCR product described above was ligated to the pT7Blue vector (Novagen Co., Madison, Wis.), and the cloned fragment was sequenced with a termination cycle-sequencing kit (Perkin-Elmer Co.) and a DNA sequencer (type ABI 373; Perkin-Elmer Co.).
Cloning and sequencing of the PPIase gene.
Genomic DNA of M. thermolithotrophicus was prepared as described previously (14). The genomic DNA was digested with BamHI, and the digested DNA fragments were ligated with BamHI-digested and bacterial alkaline phosphatase-treated pUC18. The ligated mixture was used to transform Escherichia coli JM109. A forward primer, FK-F1 (ATCAAGGTCGACTACATAGG), and a reverse primer, FK-R1 (AGAAAATACCCAGAGATGCC), which corresponded to the two ends of the partial DNA sequence of the PPIase gene described above were used to amplify the 267-bp probe for colony hybridization. PCR was carried out in 100 μl of the PCR mixture containing 100 pmol of each of these primers. Other PCR conditions were the same as those described above. The positive clones were detected with the probe labeled with a digoxigenin DNA labeling and detection kit (Boehringer, Mannheim, Germany). Prehybridization and hybridization were carried out at 63°C, and the filters (Hybond N+; Amersham, Little Chalfont, United Kingdom) were washed with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 60°C. The 5.2-kb BamHI fragments from positive clones were sequenced with a Dye termination cycle-sequencing kit (Perkin Elmer Co.).
Nucleotide sequence accession number.
The sequence determined in this study was submitted to DNA Data Bank of Japan (DDBJ) (accession no. D89881).
RESULTS
Purification and sequence analysis of PPIase from M. thermolithotrophicus.
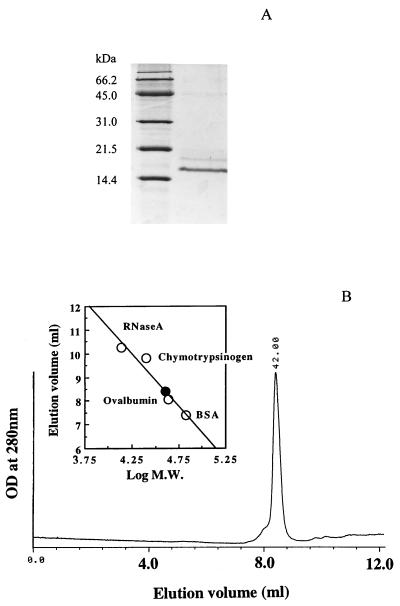
PPIase was purified from the thermophilic methanogen M. thermolithotrophicus to homogeneity by 200-fold purification with 3.6% recovery (Table 1). The molecular mass of the enzyme was estimated to be 16 kDa by SDS-PAGE (Fig. 1A) and 42 kDa by gel filtration (Fig. 1B). As shown below, the activity of this enzyme was inhibited by FK506 but not by cyclosporine. Therefore, we call this enzyme MTFK (M. thermolithotrophicus FK506-binding protein). The N-terminal amino acid sequence of the purified MTFK and those of the three peptides generated by the lysylendopeptidase digestion were determined to be VDKGVKIKVDYIGKLESGDVFDTSIEE, KDLVFTIK, KAYGNRNEMLIQK, and KIPRDAFK, respectively. The content of MTFK in cellular soluble proteins of M. thermolithotrophicus was estimated to be 0.4%.
TABLE 1.
Purification of FKBP from M. thermolithotrophicus DSM2095
| Purification step | Total vol (ml) | Protein concn (mg/ml) | Total activity (U) | Sp act (U/mg) | Recovery (%) |
|---|---|---|---|---|---|
| Crude extract | 300 | 0.86 | 1,040 | 4.03 | 100 |
| Butyl-Sepharose FF | 40 | 0.23 | 439 | 47.7 | 42 |
| Superose 12HR | 15 | 0.15 | 143 | 63.5 | 14 |
| Mono Q | 3.7 | 0.041 | 79.4 | 550 | 7.7 |
| TSK gel Ether-5PW | 1.5 | 0.027 | 37.4 | 800 | 3.6 |
FIG. 1.
(A) SDS-PAGE analysis of the purified MTFK. Molecular mass markers are shown in the left lane. The active fraction of the TSK gel Ether-5PW column is shown in the right lane. (B) Elution profile of the purified MTFK on TSK gel G2000 SWxl. The data above the chromatogram gives the estimation of the molecular mass (M.W.) of MTFK. The molecular mass standards (open circles) are 67, 43, 25, and 13.7 kDa, respectively. MTFK is shown as the solid circle. BSA, bovine serum albumin.
Catalytic efficiency and inhibition by immunosuppressants.
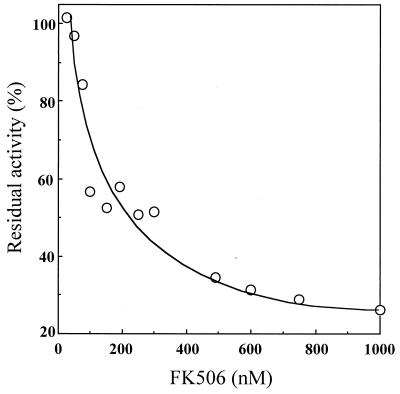
The catalytic efficiency (kcat/Km) of MTFK at 15°C for N-suc-A-L-P-F-pNA (0.35 μM−1 s−1) was higher than that for N-suc-A-A-P-F-pNA (0.20 μM−1 s−1) (Table 2). This specificity was similar to that of Escherichia coli trigger factor but much lower than those of Legionella pneumophila MIP and bovine FKBP, which exhibit greater specificity for N-suc-A-L-P-F-pNA than for N-suc-A-A-P-F-pNA (Table 2). The activity of MTFK was inhibited by FK506, with a 50% inhibitory concentration (IC50) of 250 nM (Fig. 2), but not by cyclosporine, even at a concentration of 10 μM.
TABLE 2.
Catalytic efficiencies of MTFK and other FKBPs to N-suc-Ala-Xaa-Pro-Phe-pNA
FIG. 2.
Inhibition of the PPIase activity of MTFK by FK506. The PPIase assay mixture (25°C) contained 0.1 M sodium phosphate buffer (pH 7.8), 17.0 μM N-suc-A-L-P-F-pNA, 53 nM MTFK, and 34.5 μM chymotrypsin.
Effects of temperature on stability and activity of MTFK.
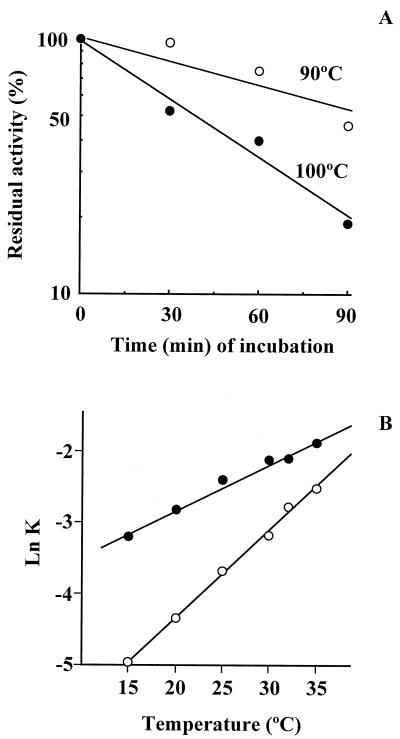
The thermostability of MTFK between 30 and 100°C was investigated. The activity of MTFK was unchanged after incubation for 30 min at 90°C or below. The half-lives of the activity at 90 and 100°C were 90 and 30 min, respectively (Fig. 3A). The PPIase activity was measured at temperatures between 15 and 35°C. The first-order rate constants of the p-nitroanilide release in the absence and presence of MTFK increased as the temperature increased (Fig. 3B). The slope of the graph in Fig. 3B, representing the rate of the increase (Δln K/ΔT), was more steep in the spontaneous reaction than in the reaction in the presence of MTFK. Measurement of the PPIase activity of MTFK was difficult above 35°C, because the spontaneous isomerization of the substrates ended less than 20 s after the addition of chymotrypsin at these temperatures.
FIG. 3.
(A) Thermostability of MTFK. The PPIase assay was used as the indicator of the thermostability of MTFK. The purified MTFK was incubated at the indicated temperature. The PPIase activity was then assayed after a 15-min incubation at 50°C. The PPIase activity was expressed as a percentage of the original activity before heat treatment and is plotted on a semilogarithmic scale. (B) The first-order rate constants of pNA release in the presence and absence of MTFK were determined by the chymotrypsin-coupled method (see Materials and Methods) at the indicated temperatures. The reactions were monitored for 250 s (15 and 20°C), 200 s (25°C), 100 s (30°C), and 50 s (35°C). ○, absence of MTFK; •, presence of MTFK.
Cloning and sequencing of the FKBP gene in M. thermolithotrophicus.
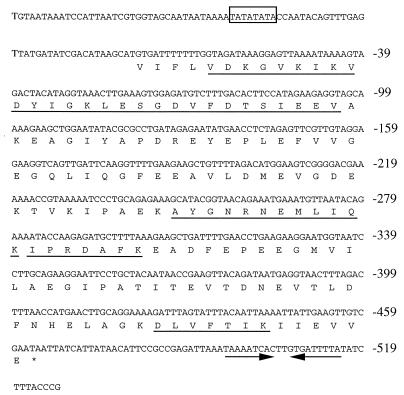
With the probe which had been amplified from the genomic DNA of M. thermolithotrophicus by PCR with the FK-F1 and FK-R1 primers, three positive clones were isolated from a genomic library of M. thermolithotrophicus. All the positive clones contained a 5.2-kb BamHI fragment. An open reading frame of 462 bp encoding a protein of 154 amino acids (Fig. 4) was found. The amino acid sequences deduced from the nucleotide sequence contained the N-terminal sequences of MTFK and the lysylendopeptidase fragments. From the deduced amino acid sequence of MTFK, the molecular mass of this enzyme was calculated to be 16.8 kDa. The open reading frame started at the codon GTG, which is frequently used as the translation initiation codon in methanogenic archaea (29). The putative (T/A)(T/A)TATATA box (37) was found at approximately 40 bp upstream from the initiation codon GTG. An inverted repeat sequence was found downstream of the stop codon (TAA).
FIG. 4.
DNA sequence and deduced amino acid sequence of MTFK. The putative archaeal promoter is boxed. The underlined sequences indicate the N terminus of the purified MTFK and the peptides whose sequences were determined after lysylendopeptidase digestion of the purified MTFK. Two arrows after the stop codon (TAA) show the inverted-repeat sequence.
Comparison of the MTFK sequences with those of other FKBPs.
The amino acid sequence of MTFK was compared with those of other FKBPs in the SWISS-PROT database. Protein sequences similar to that of MTFK were also searched for in the genome database of Methanococcus jannaschii (http://www.tigr.org/tdb/mdb/mjdb/html) (Fig. 5). Two genes encoding identical FKBP-like proteins (genes 0278 and 0825) were found. The FKBP homolog in M. jannaschii is called MJFK.
FIG. 5.
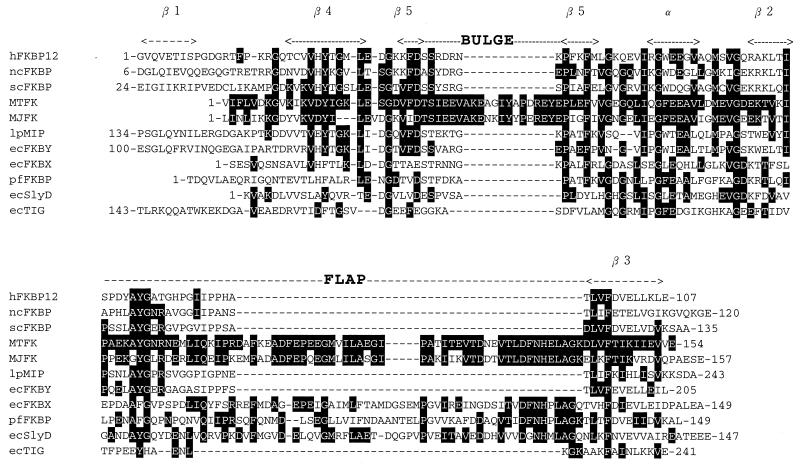
Alignment of the amino acid sequence of MTFK with those of other FKBPs. The secondary structure of hFKBP12 is given above its sequence. The bulge and flap regions of hFKBP12 (6) are shown above its sequences. The residues of MTFK identical to other FKBPs are shown in white letters on a dark background. The first and last amino acids of each sequence are indicated. hFKBP12, human FKBP12; ncFKBP, N. crassa FKBP (38); scFKBP, S. cerevisiae FKBP (24); MTFK, M. thermolithotrophicus FKBP (this study); MJFK, M. jannaschii FKBP (4); lpMIP, L. pneumophila FKBP (9); ecFKBX, E. coli FKBP homolog (2); ecFKBY, E. coli 22-kDa FKBP (28); pfFKBP, P. fluorescens FKBP homolog (17); ecSlyD, E. coli FKBP homolog (15); ecTIG, E. coli trigger factor (6).
The three-dimensional structure of a human 12-kDa FKBP (hFKBP12) has been resolved (6). It consists of five β sheets, one α helix, and loops connecting them. These secondary structures in hFKBP12 were arranged in the order (N terminus)-β1-β4-β5-α-β2-β3-(C terminus). Between β2 and β3, a surface loop called a “flap” exits, and in the middle of β5, an intervening sequence splits β5 into two. The intervening sequence, called the bulge, is of variable length in members of the FKBP family, being between 2 and 14 amino acid residues (6) (Fig. 5).
The amino acid sequence corresponding to the β1 strand was missing in MTFK and some other FKBPs, namely, MJFK, ecFKBX (the FKBP homolog in E. coli) (2), pfFKBX (the FKBP homolog in Pseudomonas fluorescens) (17), and ecSlyD (another FKBP homolog in E. coli) (15) (Fig. 5). The nomenclature of various FKBPs and their characteristics are summarized in Table 3. It was notable that MTFK and MJFK, FKBPs from thermophilic methanogens, have long bulge and flap regions. While the long flap region was also found in some bacterial FKBP homologs, ecFKBX, ecSlyD, and pfFKBX, the long bulge region was found only in FKBPs of archaea (MTFK and MJFK). The amino acid sequence of MTFK shows 66, 24, 34, and 27% identity to those of MJFK, ecFKBX, ecSlyD, and pfFKBX, respectively.
TABLE 3.
Characteristics of FKBPs
| FKBP | Organisms | Size (aa)b of:
|
PPIase activitya | FK506 sensitivitya | Molecular mass (kDa) | Reference | Swiss-Prot accession no. | |
|---|---|---|---|---|---|---|---|---|
| Bulge | Flap | |||||||
| Eucarya | ||||||||
| hFKBP12 | Human | 7 | 20 | + | + | 12 | 12 | P20071 |
| scFKBP | S. cerevisiae | 7 | 20 | + | + | 12 | 24 | P32472 |
| ncFKBP | N. crassa | 7 | 20 | + | + | 13 | 38 | P20080 |
| Bacteria | ||||||||
| lpMIP | L. pneumophila | 7 | 20 | + | + | 26 | 9 | P20380 |
| ecFKBY | E. coli | 7 | 20 | + | + | 22 | 28 | P39311 |
| ecFKBX | E. coli | 7 | 68 | + | − | 16 | 15 | P22563 |
| pfFKBP | P. fluorescens | 6 | 67 | ND | ND | 16 | 17 | P21863 |
| ecSlyD | E. coli | 6 | 67 | + | − | 21 | 15 | P03856 |
| ecTIG | E. coli | 5 | 15 | + | − | 48 | 34 | P22257 |
| Archaea | ||||||||
| MTFK | M. thermolithotrophicus | 20 | 64 | + | + | 16 | This study | |
| MJFK | M. jannaschii | 20 | 64 | ND | ND | 16 | 4 | Q58235 |
ND, not determined; +, positive in activity or sensitivity; −, negative in sensitivity.
aa, amino acids.
DISCUSSION
While many organisms have both CyPs and FKBPs, only one CyP has so far been purified from a halophilic archaeon, H. cutirubrum (23). In the present study, we purified an FKBP from M. thermolithotrophicus and cloned the structural gene for this protein in E. coli. We did not find any evidence for the CyP activity in this organism: the PPIase activity in crude extract of M. thermolithotrophicus was completely inhibited by FK506, and no PPIase activity other than that of MTFK was detected in the purification steps (data not shown); furthermore, attempts to detect genes for CyP homologs in M. thermolithotrophicus by PCR techniques have been unsuccessful (data not shown). Thus, it is likely that M. thermolithotrophicus expresses only one type of PPIase, MTFK. In this context, it is worthwhile to mention that two FKBP homologs, but no CyP homolog, are encoded in the genome of a hyperthermophilic archaeon, M. jannaschii (4).
MTFK is abundant in the cytosol of M. thermolithotrophicus, accounting for about 0.4% of the soluble proteins. This situation is similar to that of other CyP proteins (23) and FKBPs (33). The obvious question is the function of this abundant MTFK. Chaperone-like activities have been demonstrated in vitro in a CyP homolog, human tumor recognition molecule (NK-TR) (30), and in human FKBP52 (1). In canine kidney cells, the accumulation of unfolded or misfolded proteins in the endoplasmic reticulum enhanced the expression of mRNA for FKBP (5). The involvement of PPIases in thermotolerance in S. cerevisiae has been reported (27, 35). Therefore, an interesting possibility is that the only FKBP found in M. thermolithotrophicus, MTFK, exhibits a chaperone-like activity. We are investigating the chaperone activity of MTFK at various temperatures.
The comparison of the catalytic efficiencies (kcat/Km) of MTFK and other PPIases revealed that the kcat/Km of MTFK is similar to those of other FKBPs but much smaller than those of CyPs (Table 2). However, since most experiments for the determination of these catalytic parameters were performed at lower than physiological temperatures with artificial substrates, the kcat/Km values at low temperatures, e.g., 10 to 15°C, would not necessarily indicate the physiological properties of these PPIases. Further biochemical and molecular biological studies would be required to link the catalytic properties of these PPIases and their physiological functions.
The PPIase activity of MTFK was inhibited by FK506 with an IC50 of 250 nM (Fig. 2). This was higher than the IC50s of most FKBPs from Eucarya (8). The IC50s for FK506 of FKBPs in bacteria are diverse. Those of Legionella pneumophila (9) and Streptomyces chrysomallus (26) are quite low, approximately 50 nM. On the other hand, the FKBP homolog in E. coli, trigger factor, is not sensitive to FK506 (this protein is grouped to the FKBP family because of its sequence similarity to FKBPs) (34). The amino acid residues involved in the FK506 binding pocket of human FKBP12 were investigated (39). The difference in the sensitivity to FK506 among FKBPs may be explained by their primary structures. Of 15 amino acid residues corresponding to the FK506-binding pocket of hFKBP12, 7 were conserved in MTFK (Table 4). This is a larger number than those of FK506-insensitive FKBPs (5 of 15 for ecSlyD and 6 of 15 for ecTIG) but smaller than those of highly FK506-sensitive FKBPs (11 of 15 for ecFKBY, 11 of 15 for stcFKBP, and 10 of 15 for lpMIP). Four amino acid residues corresponding to Y26, G28, F36, and D37 of hFKBP12 are conserved in all FK506-sensitive FKBPs, including MTFK (Table 4). F and E were substituted for Y26 and D37 (numbering according to the hFKBP12 sequence), respectively, in FK506-insensitive ecTIG. The substitution of V for D37 in hFKBP12 resulted in a substantial increase in the Ki value of FK506 from 0.6 to 350 nM (10). The six residues (V55, I56, W59, Y82, I91, and F99) are conserved in highly FK506-sensitive FKBPs (Table 4). Thus, one or several of the substitutions of L, F, and Q for V55, W59, and I91, respectively, in MTFK may be responsible for its moderate sensitivity to FK506.
TABLE 4.
Conservation of residues involved in the FK506-binding pocket of human FKBP12
| FKBP | Residuea at:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β4 (Y26) | β4 (G28) | β5 (F36) | β5 (D37) | Bulge (R42) | β5 (F46) | Q53 | E54 | V55 | α (I56) | α (W59) | Flap (Y82) | Flap (H87) | β3 (I91) | β3 (F99) | |
| ecSlyDb (—)c | Y | V | V | D | S | L | G | S | L | I | L | Y | E | Q | F |
| ecTIGd (—) | F | G | F | E | F | G | R | M | I | F | Y | E | F | ||
| MTFK (250) | Y | G | F | D | E | L | G | Q | L | I | F | Y | E | Q | F |
| ecFKBYe (Ki = 25) | Y | G | F | D | R | A | G | V | I | W | Y | A | I | F | |
| lpMIPf (45) | Y | G | F | D | T | A | V | I | W | Y | V | I | F | ||
| ncFKBPg (20) | Y | G | F | D | R | L | G | R | V | I | W | Y | V | I | F |
| stcFKBPh (30–60) | Y | G | F | D | R | L | G | Q | V | I | W | Y | A | I | F |
The structure position (β, bulge, flap, α) indicates the positions in human FKBP12. The conserved amino acid residues compared to the residues of the human FKBP12-binding pocket are shown in boldface type.
E. coli FKBP homolog (15).
Values in parentheses show the IC50 for FK506 in nanomolar; — means that it is insensitive to FK506. In the case of ecFKBY, sensitivity to FK506 is expressed by a Ki value.
E. coli trigger factor (34).
E. coli 22-kDa FKBP (28).
L. pneumophila FKBP (9).
N. crassa FKBP (38).
Streptomyces chrysomallus FKBP (26).
Alignment of the deduced amino acid sequence of MTFK with other reported FKBPs (Fig. 5) revealed the absence of the β1 sheet and the presence of the long insertion (44 amino acids) and the other insertion (13 amino acids) in the flap and bulge regions, respectively. The β1 sheet is lacking not only in MTFK but also in other FKBPs, as described in Results. Therefore, the β1 sheet is not important for the PPIase activity or for the FK506 binding. The long flap sequence is also found in MJFK and some bacterial FKBP homologs (Fig. 5), although it has not yet been reported in eukaryotic FKBPs. The long flap sequence of MTFK is 64 amino acid residues, corresponding to 40% of the whole sequence. This region may have another function than PPIase activity and FK506 binding. Since the MTFK gene has been cloned in the present study, subsequent site-directed mutagenesis followed by the introduction of the mutated MTFK gene would be required to examine this possibility.
The insertion in the bulge region is unique in thermophilic archaeal FKBPs, MTFK and MJFK, and an interesting possibility is that the insertion is responsible for the thermostability. Site-directed mutagenesis experiments would answer this question.
ACKNOWLEDGMENTS
We express thanks to N. Yano and M. Uematsu for technical assistance, S. Suzuki for DNA sequence determination, and T. Hoaki for discussions. S. Harayama and J. H. Waite are acknowledged for critical reading of the manuscript. We thank Fujisawa Pharmaceutical Co., Ltd., for providing FK506.
This work was performed as a part of the Industrial Science and Technology Frontier Program supported by the New Energy and Industrial Technology Development Organization.
REFERENCES
- 1.Bose S, Weikl T, Bügel H, Buchner J. Chaperone function of hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 2.Bouvier J, Stragier P. Nucleotide sequence of the lsp-dapB interval in Escherichia coli. Nucleic Acids Res. 1990;19:180. doi: 10.1093/nar/19.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelly J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Bush K T, Hendrickson B A, Nigam S K. Induction of the FK506-binding protein, FKBP13, under conditions which misfold proteins in the endoplasmic reticulum. Biochem J. 1994;303:705–708. doi: 10.1042/bj3030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callebaut I, Mornon J-P. Trigger factor, one of the Escherichia coli chaperone proteins, is an original member of the FKBP family. FEBS Lett. 1995;374:211–215. doi: 10.1016/0014-5793(95)01109-r. [DOI] [PubMed] [Google Scholar]
- 7.Compton L A, Davis J M, Macdonald J R, Bächinger H P. Structural and functional characterization of Escherichia coli peptidyl-prolyl cis-trans isomerases. Eur J Biochem. 1992;206:927–934. doi: 10.1111/j.1432-1033.1992.tb17002.x. [DOI] [PubMed] [Google Scholar]
- 8.Fischer G. Peptidyl-prolyl cis/trans isomerases and their effectors. Angew Chem Int Ed Engl. 1994;33:1415–1436. [Google Scholar]
- 9.Fischer G, Bang H, Ludwig B, Mann K, Hacker J. Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPIase) activity. Mol Microbiol. 1992;6:1375–1383. doi: 10.1111/j.1365-2958.1992.tb00858.x. [DOI] [PubMed] [Google Scholar]
- 10.Futer O, DeCenzo M T, Aldape R A, Livingston D J. FK506 binding protein mutational analysis. J Biol Chem. 1995;270:18935–18940. doi: 10.1074/jbc.270.32.18935. [DOI] [PubMed] [Google Scholar]
- 11.Handschumacher R E, Harding M W, Rice J, Drugge R J. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 12.Harding M W, Galat A, Uehling D E, Schreiber S L. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature (London) 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 13.Harrison R K, Stein R L. Substrate specificities of the peptidyl prolyl cis-trans isomerase activities of cyclophilin and FK506 binding protein: evidence for the existence of a family of distinct enzymes. Biochemistry. 1990;29:3813–3816. doi: 10.1021/bi00468a001. [DOI] [PubMed] [Google Scholar]
- 14.Hoaki T, Nishijima M, Kato M, Adachi K, Mizobuchi S, Hanzawa N, Maruyama T. Growth requirements of hyperthermophilic sulfur-dependent heterotrophic archaea isolated from a shallow submarine geothermal system with reference to their essential amino acids. Appl Environ Microbiol. 1994;60:2898–2904. doi: 10.1128/aem.60.8.2898-2904.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hottenrott S, Schumann T, Plückthun A, Fischer G, Rahfeld J-U. The Escherichia coli slyD is a metal ion-regulated peptidyl-prolyl cis/trans isomerase. J Biol Chem. 1997;272:15697–15701. doi: 10.1074/jbc.272.25.15697. [DOI] [PubMed] [Google Scholar]
- 16.Huber H, Thomm M, König H, Thies G, Stetter K O. Methanococcus thermolithotrophicus, a novel thermophilic lithotrophic methanogen. Arch Microbiol. 1982;132:47–50. [Google Scholar]
- 17.Isaki L, Beers R, Wu H C. Nucleotide sequence of the Pseudomonas fluorescens signal peptidase II gene (lsp) and flanking genes. J Bacteriol. 1990;172:6512–6517. doi: 10.1128/jb.172.11.6512-6517.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jannasch H W, Wirsen C O, Molyneaux S J, Langworthy T A. Extremely thermophilic fermentative archaebacteria of the genus Desulfurococcus from deep-sea hydrothermal vents. Appl Environ Microbiol. 1988;54:1203–1209. doi: 10.1128/aem.54.5.1203-1209.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kern G, Kern D, Schmid F X, Fischer G. Reassessment of the putative chaperone function of prolyl-cis/trans-isomerases. FEBS Lett. 1994;348:145–148. doi: 10.1016/0014-5793(94)00591-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim D-J, Morikawa M, Takagi M, Imanaka T. Gene cloning and characterization of thermostable peptidyl prolyl cis-trans isomerase (PPIase) from Bacillus stearothermophilus SIC1. J Ferment Bioeng. 1995;79:87–94. [Google Scholar]
- 21.Ludwig B, Rahfeld J, Schmidt B, Mann K, Wintermeyer E, Fischer G, Hacker J. Characterization of Mip proteins of Legionella pneumophila. FEMS Microbiol Lett. 1994;118:23–30. doi: 10.1111/j.1574-6968.1994.tb06798.x. [DOI] [PubMed] [Google Scholar]
- 22.Maki N, Sekiguchi F, Nishimaki J, Miwa K, Hayano T, Takahashi N, Suzuki M. Complementary DNA encoding the human T-cell FK506-binding protein, a peptidylprolyl cis-trans isomerase distinct from cyclophilin. Proc Natl Acad Sci USA. 1990;87:5440–5443. doi: 10.1073/pnas.87.14.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagashima K, Mitsuhashi S, Kamino K, Maruyama T. Cyclosporin A sensitive peptidyl prolyl cis-trans isomerase in a halophilic archaeum, Halobacterium cutirubrum. Biochem Biophys Res Commun. 1994;198:466–472. doi: 10.1006/bbrc.1994.1068. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen J B, Foor F, Siekierka J J, Hsu M J, Ramadan N, Morin N, Shafiee A, Dahl A M, Brizuela L, Chrebet G, Bostian K A, Parent S A. Yeast FKBP-13 is a membrane-associated FK506-binding protein encoded by the nonessential gene FKB2. Proc Natl Acad Sci USA. 1992;89:7471–7475. doi: 10.1073/pnas.89.16.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ondek B, Hardy R W, Baker E K, Stamnes M A, Shieh B-H, Zuker C S. Genetic dissection of cyclophilin function. J Biol Chem. 1992;267:16460–16466. [PubMed] [Google Scholar]
- 26.Pahl A, Keller U. FK-506-binding proteins from streptomycetes producing immunosuppressive macrolactones of the FK506 type. J Bacteriol. 1992;174:5888–5894. doi: 10.1128/jb.174.18.5888-5894.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Partaledis J A, Berlin V. The FKB2 gene of Saccharomyces cerevisiae, encoding the immunosuppressant-binding protein FKBP-13, is regulated in response to accumulation of unfolded proteins in the endoplasmic reticulum. Proc Natl Acad Sci USA. 1993;90:5450–5454. doi: 10.1073/pnas.90.12.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahfeld J-U, Rücknagel K P, Stoller G, Horne S M, Schierhorn A, Young K D, Fischer G. Isolation and amino acid sequence of a new 22-kDa FKBP-like peptidyl-prolyl cis/trans-isomerase of Escherichia coli. J Biol Chem. 1996;271:22130–22138. doi: 10.1074/jbc.271.36.22130. [DOI] [PubMed] [Google Scholar]
- 29.Reeve J N. Molecular biology of methanogens. Annu Rev Microbiol. 1992;46:165–191. doi: 10.1146/annurev.mi.46.100192.001121. [DOI] [PubMed] [Google Scholar]
- 30.Rinfret A, Collins C, Menard R, Anderson S K. The N-terminal cyclophilin-homologous domain of a 150-kilodalton tumor recognition molecule exhibits both peptidylprolyl cis-trans-isomerase and chaperone activities. Biochemistry. 1994;33:1668–1673. doi: 10.1021/bi00173a008. [DOI] [PubMed] [Google Scholar]
- 31.Schönbrunner E R, Mayer S, Tropschug M, Fischer G, Takahashi N, Schmid F X. Catalysis of protein folding by cyclophilins from different species. J Biol Chem. 1991;266:3630–3635. [PubMed] [Google Scholar]
- 32.Siekierka J J, Staruch M J, Hung S H Y, Sigal N H. FK-506, a potent novel immunosuppressive agent, binds to a cytosolic protein which is distinct from the cyclosporin A-binding protein, cyclosporin A. J Immunol. 1989;143:1580–1583. [PubMed] [Google Scholar]
- 33.Siekierka J J, Hung S H Y, Poe M, Lin C S, Sigal N H. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature (London) 1989;341:755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 34.Stoller G, Rücknagel K P, Nierhaus K H, Schmid F X, Fischer G, Rahfeld J-U. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 1995;14:4939–4948. doi: 10.1002/j.1460-2075.1995.tb00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sykes K, Gething M-J, Sambrook J. Proline isomerases function during heat shock. Proc Natl Acad Sci USA. 1993;90:5853–5857. doi: 10.1073/pnas.90.12.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi N, Hayano T, Suzuki M. Peptidyl prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature (London) 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 37.Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 38.Tropschug M, Wachter E, Mayer S, Schönbrunner E R, Schmid F X. Isolation and sequence of an FK506-binding protein from N. crassa which catalyses protein folding. Nature (London) 1990;346:674–677. doi: 10.1038/346674a0. [DOI] [PubMed] [Google Scholar]
- 39.Van Duyne G D, Standaert R F, Karplus P A, Schreiber S L, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]