Abstract
Background
Immunotherapy has facilitated great breakthroughs in the treatment of hepatocellular carcinoma (HCC). However, the efficacy and response rate of immunotherapy are limited and vary among different patients with HCC. TP53 mutation substantially affects the expression of immune checkpoint molecules in multiple cancers. However, the regulatory relationship between programmed death ligand 1 (PD-L1) and TP53 is poorly studied in HCC. We aimed to elucidate the regulatory mechanism of PD-L1 in HCC with different TP53 statuses and to assess its role in modulating immune evasion in HCC.
Methods
HCC mouse models and cell lines with different TP53 statuses were constructed. PD-L1 levels were detected by PCR, western blotting and flow cytometry. RNA-seqencing, immunoprecipitation, chromatin immunoprecipitation and transmission electron microscopy were used to elucidate the regulatory mechanism in HCC with different TP53 status. HCC mouse models and patient with HCC samples were analyzed to demonstrate the preclinical and clinical significance of the findings.
Results
We report that loss of p53 promoted PD-L1 expression and reduced CD8+ T-cell infiltration in patient with HCC samples and mouse models. Mammalian target of rapamycin (mTOR) pathway was activated in p53-loss-of-function HCC or after knocking down TP53. The transcription factor E2F1 was found to bind to the p53 protein in TP53 wild-type HCC cells, and inhibiting mammalian target of rapamycin complex 1 (mTORC1) disrupted this binding and enhanced E2F1 translocation to the nucleus, where it bound to the PD-L1 promoter and transcriptionally upregulated PD-L1. In p53-loss-of-function HCC cells, autophagosomes were activated after mTORC1 suppression, promoting the degradation of PD-L1 protein. The combination of mTOR inhibitor and anti-PD-L1 antibody enhanced CD8+ T-cell infiltration and tumor suppression in TP53 wild-type HCC mouse models, but no benefit was observed in p53-loss-of-function HCC mouse models. In patients with TP53 wild-type HCC, PD-L1 levels were significantly higher in the high E2F1 group than in the low E2F1 group, and the low E2F1 level group had significantly superior survival.
Conclusion
We revealed the bidirectional regulatory mechanism of PD-L1 mediated by TP53/mTORC1 in HCC. The combination of mTOR inhibitor and anti-PD-L1 antibody could be a novel precise immunotherapy scheme for TP53 wild-type HCC.
Keywords: Immune Checkpoint Inhibitors, Immunotherapy, Liver Neoplasms, Tumor Microenvironment, CD8-Positive T-Lymphocytes
WHAT IS ALREADY KNOWN ON THIS TOPIC
The efficacy and response rate of anti-programmed death ligand 1 (PD-L1) immunotherapy are limited and vary among different patients with hepatocellular carcinoma (HCC).
TP53 is the most commonly mutated gene in HCC, and its mutation substantially affects the expression of immune checkpoint molecules in cancer, but the regulatory relationship between PD-L1 and p53 is poorly studied in HCC.
WHAT THIS STUDY ADDS
TP53 loss-of-function promotes PD-L1 expression and reduces CD8+ T-cell infiltration in HCC.
In TP53 wild-type HCC cells, mammalian target of rapamycin complex 1 (mTORC1) suppression upregulates PD-L1 via the transcription factor E2F1, while in TP53 loss-of-function HCC cells, mTORC1 suppression promotes PD-L1 degradation through activation of autophagy.
The combination of mTOR inhibitor and anti-PD-L1 antibody leads to significant inhibition of tumor growth and superior survival in Trp53 wild-type mouse HCC models.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Combining mTOR inhibitors and anti-PD-L1 antibodies could be a novel and promising immunotherapy strategy for TP53 wild-type HCC.
Background
Hepatocellular carcinoma (HCC) is a malignant tumor and has the fifth highest incidence rate and the second highest mortality rate among malignancies in China. The 5-year overall survival (OS) rate of HCC is less than 20%.1 Immunotherapy is a new promising treatment method that can control and eliminate tumors by reshaping the tumor immune microenvironment and restoring the normal antitumor immune response. Immunotherapies include immune checkpoint inhibitors, therapeutic antibodies, tumor vaccines, and cell therapies.2 3 In recent years, immune checkpoint inhibitors targeting the tumor microenvironment have facilitated great breakthroughs in the treatment of patients with advanced HCC. Programmed death ligand 1 (PD-L1) can be expressed on the surface of HCC cells, interact with programmed death 1 (PD-1) on the surface of cytotoxic T cells (CTLs), and inhibit the tumoricidal ability of CTLs to facilitate tumors cell escape from human immune surveillance and malignant proliferation.4 The IMbrave150 trial reported that the PD-L1 inhibitor atezolizumab combined with the antiangiogenic drug bevacizumab reduced the death risk of patients with unresectable HCC by 42% compared with the targeted drug alone.5 In general, immune checkpoint inhibitors, represented by PD-L1/PD-1-targeted drugs, have broad prospects in the treatment of HCC. Basic and translational research on PD-L1/PD-1 is currently a hot spot in HCC research.6
TP53 is one of the most widely studied tumor suppressor genes.7 The p53 protein is a monitor of cell growth. TP53 is commonly regarded as a “gene guardian” that inhibits tumor occurrence and development.8 Mutant p53 protein not only has lost antitumor function but can also inhibit the activity of wild-type p53 protein or exhibit carcinogenic activity, causing carcinogenesis.9 Functional mutation and deletion of TP53 are core driving events of hepatocarcinogenesis and have a significant negative correlation with the prognosis of patients.10–12 The mutation rate of TP53 in HCC ranges from 29.1% to 58.0%.10 13 Recent studies have also found that normal p53 protein can promote the proliferation of HCC cells, indicating that the role of TP53 in HCC cells is complex and controversial.14 The currently known TP53 mutations in HCC are mainly loss-of-function (LoF) mutations, and at the molecular level, they are mainly missense mutations, frame shift mutations and non-sense mutations.15
In recent years, with the breakthrough of tumor immunotherapy, an increasing number of researchers have begun to pay attention to the effect of TP53 mutation on the tumor microenvironment. The mutant p53 protein in tumor cells can inhibit the innate immune-related signaling pathway and promote the immune escape of tumor cells, specifically manifested by the decline in natural killer and T cells in the tumor microenvironment and the increase in tumor-associated macrophages.16 Research has also shown that TP53 mutation is associated with decreased infiltration of immune cells.17 18
In this study, we found an interesting bidirectional regulation of PD-L1 in HCC mediated by TP53/mammalian target of rapamycin complex 1 (mTORC1). In TP53 wild-type HCC, mTORC1 suppression increases the expression of the transcription factor E2F1, disrupts the binding between E2F1 and p53 in the cytoplasm, and promotes the entry of E2F1 into the nucleus, where it transcriptionally upregulates PD-L1. In TP53-mutant HCC, mTORC1 suppression promotes autophagic degradation of the PD-L1 protein. The combination of a mammalian target of rapamycin (mTOR) inhibitor and an anti-PD-L1 antibody significantly suppressed tumor growth and prolonged the survival of mice with TP53 wild-type HCC. The bidirectional regulatory mechanism of PD-L1 in tumors with different TP53 statuses may help guide precise clinical immunotherapy application to improve the prognosis of patients with HCC.
Methods
Cell culture, transfection, lentivirus infection and reagents
Human HCC cell lines (SK-HEP-1, HepG2, Hep3B, Li-7) and a murine HCC cell line (H22) were purchased from Shanghai Institute of Cell Biology, Chinese Academy of Sciences. Details regarding cell culture, siRNA, plasmids, lentivirus, and reagents are provided in the online supplemental file 1.
jitc-2023-007479supp008.pdf (215.2KB, pdf)
Patients and samples
HCC tissue samples were obtained after surgery. Details are provided in the online supplemental file 1.
RNA sequencing and gene set analysis, whole-exome sequencing, assay for transposase-accessible chromatin by sequencing and Sanger sequencing
Details are provided in the online supplemental file 1.
Co-immunoprecipitation assay, western blot analysis, chromatin immunoprecipitation, RT-qPCR analysis, immunofluorescence, luciferase reporter assay, flow cytometry staining and analysis, immunohistochemistry, transmission electron microscopy
Details are provided in the online supplemental file 1.
Animal experiments
C57BL/6J mice and BALB/c mice were obtained from the Animal Facility of Zhejiang University. This study is approved by Institutional Review Board of The First Affiliated Hospital, Zhejiang University School of Medicine (Reference number: 2018–542). Animal care and experiments were performed in strict accordance with the “Guide for the Care and Use of Laboratory Animals’’ and the ‘‘Principles for the Utilization and Care of Vertebrate Animals’’. Grouping and experimental processing details are provided in the online supplemental file 1.
Mass cytometry (CyTOF) staining, data acquisition and analysis
Details are provided in the online supplemental file 1.
Statistical analysis
SPSS V.21.0 statistical software was used for statistical analysis. The significance of the differences between groups was determined using Student’s t-test. Differences with p<0.05 were considered significantly different. The χ2 test was used to analyze the correlation between quantitative data. For survival analysis, OS was estimated using the Kaplan-Meier method.
Data availability
The data of RNA sequencing, whole-exome sequencing, assay for transposase-accessible chromatin by sequencing (ATAC-seq) can be viewed in SRA database and GEO database. The accession IDs are PRJNA999626 and GSE240197 and GSE239926. The persistent URL are http://www.ncbi.nlm.nih.gov/bioproject/999626, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE240197, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE239926, retrospectively.
Results
Loss of p53 promotes PD-L1 expression and modulates immune evasion in HCC
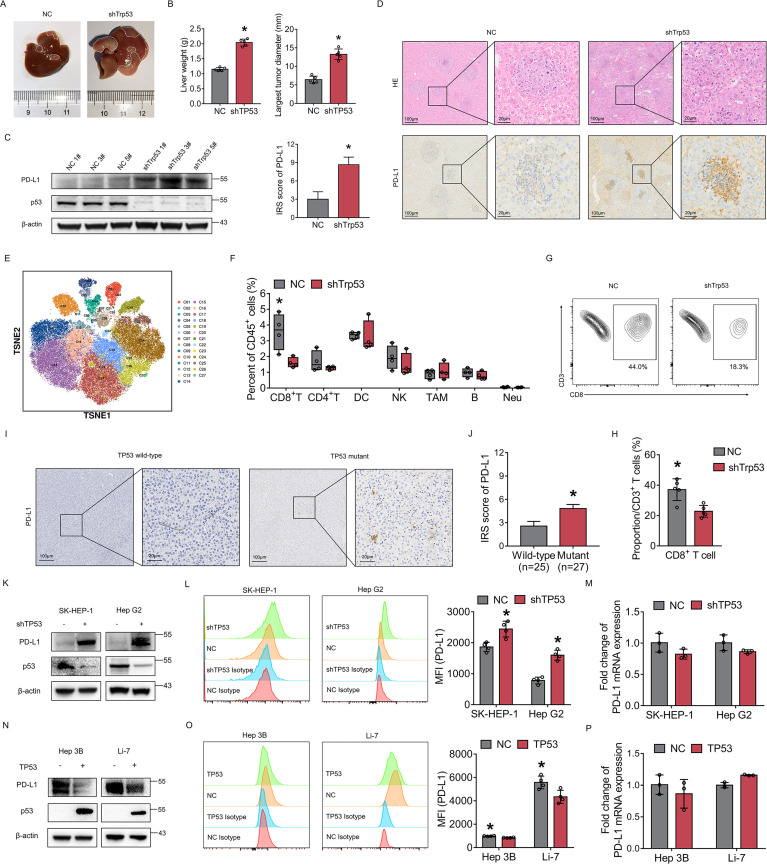
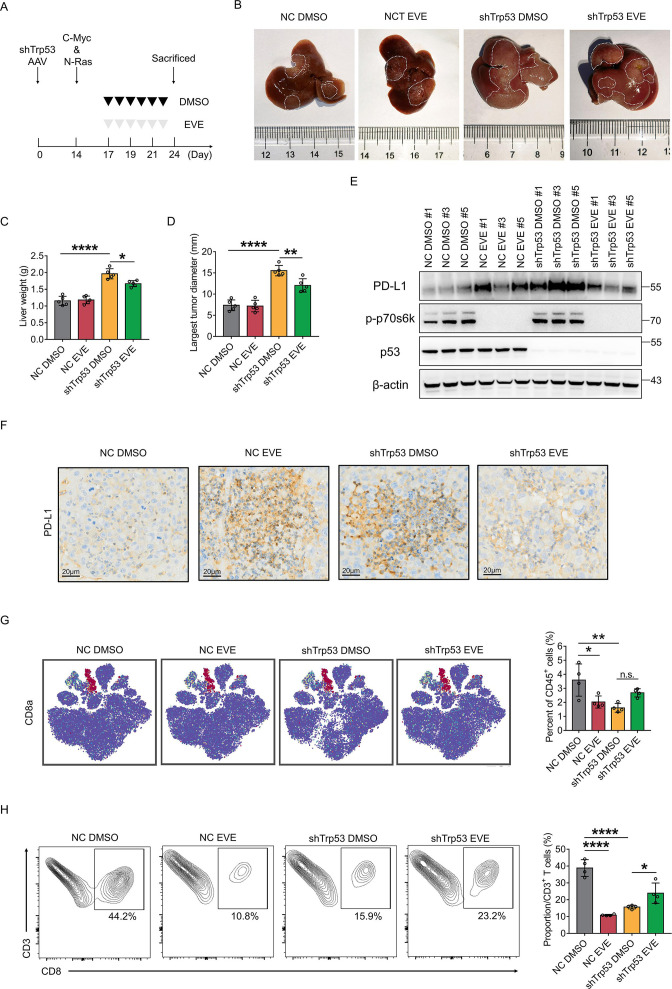
To explore the significance of p53 protein on PD-L1 expression, we first constructed Trp53 knockdown mice through injection with adreno-associated virus (AAV) and then established an HCC mouse model by hydrodynamic gene delivery of c-Myc and NRAS plasmids. Results showed that Trp53 knockdown mice had wider tumor areas (macroscopically and microscopically) and had heavier livers (figure 1A, B and D). PD-L1 protein expression was increased in Trp53 knockdown mouse tumors compared with Trp53 wild-type mouse tumors (figure 1C,D). To elucidate the effect of p53 on the immune microenvironment, we performed Mass Cytometry (CyTOF) analysis on mouse tumors with and without Trp53 knockdown. The results revealed that infiltration of CD8+ T cells was significantly decreased in Trp53 knockdown mouse tumors (figure 1E,F). Flow cytometry also confirmed that CD8+ T-cell infiltration was significantly decreased in Trp53 knockdown mouse tumors (figure 1G,H). The CD8 T-cell proportion was decreased from approximately 44.0% in Trp53 wild-type mouse tumors to approximately 18.3% in Trp53 knockdown mouse tumors. Furthermore, according to TP53 mutation status, immunohistochemistry (IHC) was performed on HCC tissues from patients. The results also suggested that PD-L1 expression was increased in TP53-mutant HCC (figure 1I,J).
Figure 1.
Loss of p53 promotes PD-L1 expression and reshapes immune microenvironment in HCC. (A) The plasmid-induced mouse HCC model was constructed. Representative images of liver tumors in NC and shTrp53 group. The tumor area of shTrp53 group was larger than that of NC group. (B) The mice liver weight and the largest tumor diameter were measured in NC group and shTrp53 group. (C) PD-L1 protein was detected by western blotting in mouse tumors of each group. β-actin was used as the loading control for western blotting. (D) H&E staining and IHC staining of PD-L1 expression in mouse tumor tissues. Comparison of IRS score of PD-L1 between Trp53 wild-type and Trp53 knockdown groups. (E) t-SNE plots of mouse tumor CD45+ cell clusters. (F) Percentages of immune cell subtypes in mouse tumors of each group. (G) CD45+, CD3+ and CD8+ T cells extracted from mouse tumors were analyzed by flow cytometry. (H) Percentages of CD8+ T cells in NC group and shTrp53 group. (I) IHC staining of PD-L1 expression of tumor tissues from patients with TP53 wild-type and TP53 mutant HCC. (J) IRS of PD-L1 in TP53 wild-type and TP53 mutant HCC tissues. (K–M) The expression of PD-L1 in SK-HEP-1 and HepG2 cells with or without TP53 knockdown were detected by western blotting (K) flow cytometry (L) and qRT-PCR (M). (N–P) The expression of PD-L1 in Hep 3B and Li-7 cells with or without TP53 overexpression were detected by western blot (N) flow cytometry (O) and RT-qPCR (P). The bars were compared by Student’s t-test. *p<0.05, compared with the NC (B, D, G, H, L, M,O, P) or p53 wild-type (J) groups. DC, dendritic cell; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; IRS, IRS, Immune Reactive Score; MFI, mean fluorescent intensity; mRNA, messenger RNA; Neu, neutrophils; NK, natural killer cell; PD-L1, programmed death ligand 1; RT-qPCR, Real Time Quantitative Polymerase Chain Reaction; TAM, tumor associated macrophage; t-SNE, t-Distributed Stochastic Neighbor Embedding.
In addition, human HCC cell lines were used to further prove the above results. In SK-HEP-1 and HepG2 cells, which are p53 wild-type HCC cells, PD-L1 protein expression was increased after TP53 knockdown (figure 1K,L). In Hep3B and Li-7 cells, which exhibit p53 LoF, PD-L1 protein expression was decreased after TP53 overexpression (figure 1N,O). At the transcriptional level, there was no significant difference in PD-L1 messenger RNA expression after TP53 knockdown or overexpression (figure 1M,P). The above results indicated that p53 plays an important role in regulating PD-L1 expression and modulating immune evasion in HCC.
Bidirectional regulation of PD-L1 by mTORC1 suppression in HCC cells with different TP53 statuses
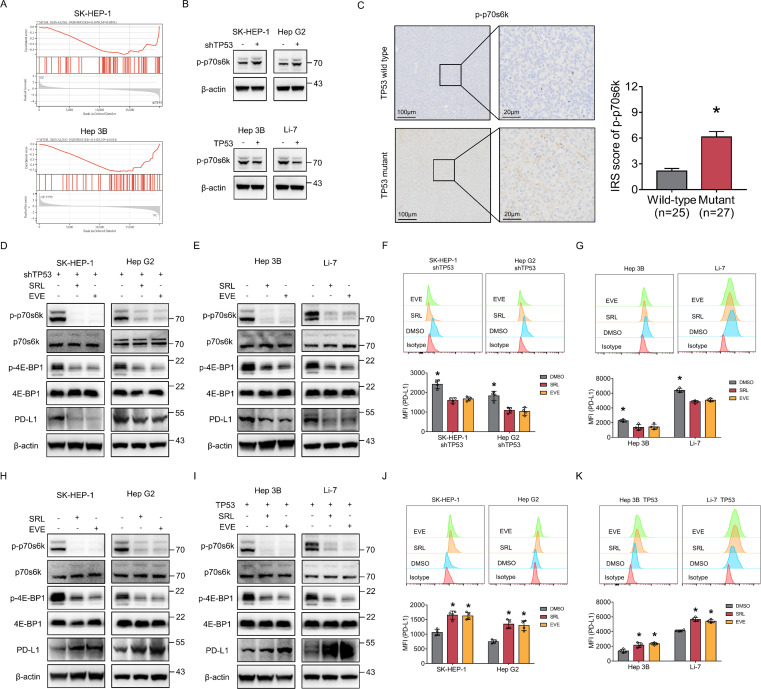
Gene set enrichment analysis revealed that loss of p53 protein expression led to significant activation of the mTOR pathway in HCC cells (figure 2A), which was further confirmed by western blotting (figure 2B). IHC results of HCC tissues from patients also suggested that p-p70s6k expression was increased in TP53 mutant HCC, indicating activation of the mTOR pathway (figure 2C). Therefore, we investigated whether mTORC1 suppression can affect the expression of PD-L1 in HCC cells with p53 LoF. We applied two mTOR pathway inhibitors, sirolimus and everolimus, and western blotting and flow cytometry showed that PD-L1 protein expression was decreased in four HCC cell lines with p53 LoF (figure 2D–G). The presence of TP53 knockdown in SK-HEP-1 and Hep G2 was presented in online supplemental figure 1.
Figure 2.
TP53/mammalian target of rapamycin complex 1-mediated bidirectional regulation of PD-L1 in HCC cells. (A) Pathway enriched in the mammalian target of rapamycin signaling for SK-HEP-1 with or without TP53 knockdown and Hep3B with or without TP53 overexpression by gene set enrichment analysis. (B) The expression level of p-p70s6k was detected by western blotting in TP53 wild-type cells (SK-HEP-1 and HepG2) with or without TP53 knockdown and in TP53 loss-of-function cells (Hep3B and Li-7) with or without TP53 overexpression. (C) p-p70s6k expression of TP53 wild-type and TP53 mutant tumor tissues from patients with HCC was detected by immunohistochemistry staining. Comparison of IRS score of p-p70s6k between TP53 wild-type and TP53 mutant groups. (D–G) TP53 wild-type cells (SK-HEP-1 and HepG2) with siTP53 plasmid transfection and TP53 loss-of-function cells (Hep3B and Li-7) were treated with 50 nM everolimus or sirolimus. The protein level of p70s6k, p-p70s6k, 4E-BP1, p-4E-BP1 and PD-L1 were detected by western blotting (D, E) and flow cytometry (F, G). (H–K) TP53 wild-type cells (SK-HEP-1 and HepG2) and TP53 loss-of-function cells (Hep3B and Li-7) with TP53 overexpression were treated with 50 nM everolimus or sirolimus. The protein level of p70s6k, p-p70s6k, 4E-BP1, p-4E-BP1and PD-L1 were detected by western blotting (H, I) and flow cytometry (J, K). β-actin was used as the loading control for western blotting. *p<0.05, compared with the DMSO (F, G, J, K) and TP53 wild-type (C) groups. EVE, everolimus; HCC, hepatocellular carcinoma; IRS, Immune Reactive Score; MFI, mean fluorescent intensity; PD-L1, programmed death ligand 1; SRL, sirolimus.
jitc-2023-007479supp001.pdf (461.5KB, pdf)
In addition, we investigated the effect of mTORC1 suppression on PD-L1 expression in HCC cells with wild-type p53. Interestingly, western blotting and flow cytometry showed that PD-L1 protein expression was increased in four HCC cell lines with wild-type p53 after sirolimus and everolimus treatment (figure 2H–K), which was completely opposite to the results in HCC cells with p53 LoF. The presence of TP53 overexpression in Hep 3B and Li-7 was presented in online supplemental figure 1.
The opposite results of PD-L1 indicated an intriguing bidirectional regulation by mTORC1 suppression in HCC cells, suggesting the importance of different TP53 statuses. Given the high mutation rate of TP53 in HCC, it is highly possible that this bidirectional regulatory mechanism may play an important role in HCC immune escape.
We also performed TSC1 knockdown to activate mTORC1 in four HCC cell lines with wild-type p53 or p53 LoF. The effect on regulating PD-L1 expression was opposite to that of mTORC1 suppression in corresponding HCC cells (online supplemental figure 2). The above results indicate that PD-L1 expression can be bidirectionally regulated by mTORC1 suppression in HCC cells with different p53 statuses.
jitc-2023-007479supp002.pdf (2.5MB, pdf)
E2F1 promotes PD-L1 transcription after mTORC1 suppression in p53 wild-type HCC cells
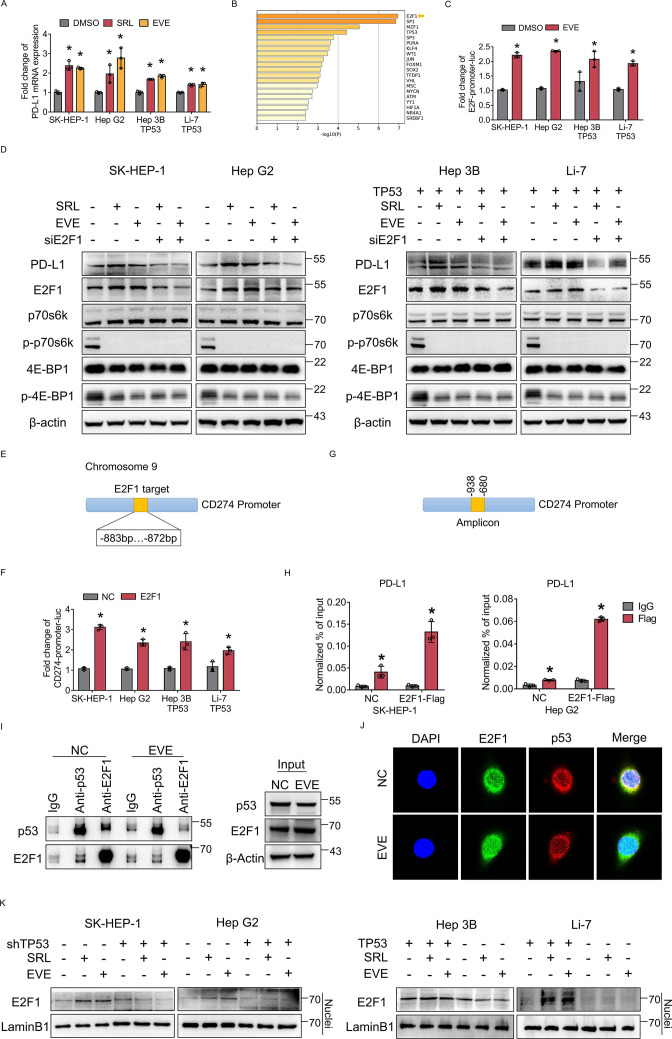
In p53 wild-type HCC cells, PD-L1 transcription was activated after mTORC1 suppression (figure 3A), which suggested that PD-L1 expression might be regulated by transcription. Therefore, ATAC-seq was performed. ATAC-seq analysis revealed several transcription factors that were upregulated in p53 wild-type HCC cells after mTORC1 suppression; among these transcription factors, E2F1 was the most obviously upregulated (figure 3B). According to the ATAC-seq analysis and prediction of PD-L1 transcription factors, we performed a luciferase reporter assay. The results showed that transcriptional activity was only upregulated in E2F1 after mTORC1 suppression (figure 3C and online supplemental figure 3). Thus, mTORC1 suppression may promote PD-L1 expression by transcriptionally promoting E2F1 expression.
Figure 3.
E2F1 promotes PD-L1 transcription after mammalian target of rapamycin complex 1 suppression in p53 wild-type HCC cells. (A) TP53 wild-type cells (SK-HEP-1, HepG2) and TP53 loss-of-function cells (Hep3B, Li-7) with TP53 overexpression were treated with everolimus or sirolimus. PD-L1 mRNA expression was detected by qRT-PCR. (B) ATAC-seq analysis of TP53 wild-type cells treated with SRL. (C) The fold change in the relative luciferase activity was examined in HCC cells treated with everolimus. (D) SK-HEP-1, HepG2, Hep3B (TP53 overexpression), Li-7 (TP53 overexpression) with or without E2F1 knockdown were treated with everolimus or sirolimus. The protein level of PD-L1, E2F1, p70s6k, p-p70s6k, 4E-BP1 and p-4E-BP1 were detected using western blotting. β-actin was used as the loading control. (E) Predicted E2F1 binding site sequences in the PD-L1 promoter. (F) The fold change in the relative luciferase activity was examined in HCC cells transfected with E2F1 plasmids (E2F1) or empty plasmids (NC). (G) Amplicons in the CD274 promoter were used for ChIP assay (H) which was performed to determine the binding of E2F1 to the binding site sequence of the CD274 promoter in SK-HEP-1 and HepG2 cells transfected with E2F1 plasmids (E2F1) or empty plasmids (NC). (I) The interaction of endogenous p53 and E2F1 was tested in HCC cells treated with or without everolimus. Normal rabbit IgG was used as a control. (J) Colocalization (yellow) of E2F1 (green) with p53 (red) by confocal microscopy. The nuclei were stained with DAPI. (K) Nuclear proteins were isolated and subjected to western blotting for expression of E2F1. LaminB1 was used as the loading control. *p<0.05, compared with the DMSO (A, C) or NC (F) or IgG (H) groups. ACTC-seq, assay for transposase accessible chromatin; CD274, cluster of differentiation 274; ChIP, chromatin immunoprecipitation; DAPI, 4,6-diamino-2-phenyl indole; DMSO, Dimethylsulfoxide; EVE, everolimus; HCC, hepatocellular carcinoma; mRNA, messenger RNA; PD-L1, programmed death ligand 1; RT-qPCR, Real Time Quantitative Polymerase Chain Reaction; SRL, sirolimus.
jitc-2023-007479supp003.pdf (785.3KB, pdf)
Therefore, we further knocked down E2F1 and found that the upregulation of PD-L1 after mTORC1 suppression in p53 wild-type HCC cells was attenuated by E2F1 knockdown (figure 3D). Luciferase reporter and chromatin immunoprecipitation assays were used and demonstrated that E2F1 can bind to the TTGGCGGATCAC sequence at −883 to −872 of the PD-L1 promoter (figure 3E–H and online supplemental figure 4). According to our previous results, E2F1 can transcriptionally upregulate PD-L1.
jitc-2023-007479supp004.pdf (1.3MB, pdf)
To clarify the interaction between p53 and E2F1 under different mTORC1 suppression states, we performed Co-immunoprecipitation assay (Co-IP) in p53 wild-type HCC cells. The results showed that the interaction between p53 and E2F1 was weakened after mTORC1 suppression (figure 3I and online supplemental figure 5). Moreover, the immunofluorescence results showed that colocalization of E2F1 and p53 was decreased and E2F1 nuclear entry was increased after mTORC1 suppression (figure 3J). Furthermore, nuclear proteins were isolated and subjected to western blotting to assess the expression of E2F1. In p53 wild-type HCC cells, the nuclear protein level of E2F1 was increased after mTORC1 suppression, while in HCC cells with p53 LoF, E2F1 protein was expressed at a low level and did not change obviously after mTORC1 suppression (figure 3K). The above results indicated that mTORC1 suppression promoted the expression and nuclear entry of E2F1, which may further promote the transcription of PD-L1.
jitc-2023-007479supp005.pdf (1.1MB, pdf)
mTORC1 suppression induces autophagic degradation of PD-L1 in HCC cells with p53 loss-of-function
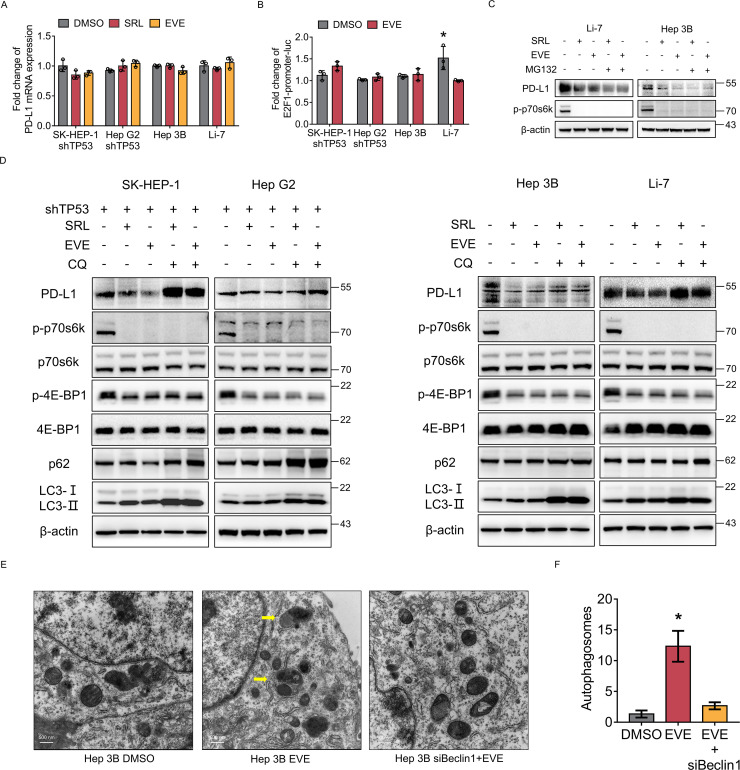
In HCC cells with p53 LoF, PD-L1 transcription was not activated after mTORC1 suppression (figure 4A). In addition, the results of the luciferase reporter assay showed that the transcriptional activity of E2F1 presented no difference after mTORC1 suppression (figure 4B). These results suggest that PD-L1 expression may not be regulated by transcription in HCC cells with p53 LoF.
Figure 4.
Mammalian target of rapamycin complex 1 suppression induces autophagic degradation of PD-L1 in HCC cells with p53 loss-of-function. (A) TP53 wild-type cells (SK-HEP-1, HepG2) with TP53 knockdown and TP53 loss-of-function cells (Hep3B, Li-7) were treated by everolimus or sirolimus. PD-L1 messenger RNA expression of each group was detected by qRT-PCR. (B) The fold change in the relative luciferase activity was examined in HCC cells treated with everolimus. (C) TP53 loss-of-function cells (Hep3B, Li-7) were treated with everolimus or sirolimus. Then, the cells were treated with or without MG132 (10 µM) for 2 hours. The protein levels of p-p70s6k and PD-L1 were analyzed by western blotting. β-actin was used as the loading control. (D) SK-HEP-1, HepG2 with TP53 knockdown and Hep3B, Li-7 were treated with everolimus, sirolimus and CQ alone or in combination for the indicated time. The protein levels of PD-L1, E2F1, p70s6k, p-p70s6k, 4E-BP1, p-4E-BP1, p62 and LC3 were analyzed by western blot. β-actin was used as the loading control. (E) Autophagosomes as revealed by transmission electron microscopy in Hep3B cells treated without (DMSO) or with everolimus alone (EVE) or with Beclin1 knockdown and everolimus (siBeclin 1+EVE). Yellow arrows, autophagosomes. (F) The numbers of autophagosomes in each group were determined. *p<0.05, compared with the DMSO groups (A, B, G). CQ, autophagy inhibitor chloroquine; DMSO, Dimethylsulfoxide; HCC, hepatocellular carcinoma; MG132, proteasome inhibitor; PD-L1, programmed death ligand 1; RT-qPCR, Real Time Quantitative Polymerase Chain Reaction; SRL, sirolimus.
Therefore, we hypothesized that mTORC1 suppression promotes PD-L1 degradation. Thus, we first used the inhibitor MG-132 and found that inhibition of proteasome degradation did not restore PD-L1 expression (figure 4C). Then, we tried to use the autophagy inhibitor chloroquine (CQ). Surprisingly, the results showed that the downregulation of PD-L1 induced by mTORC1 suppression in HCC cells with p53 LoF was reversed after CQ treatment (figure 4D). In addition, the electron microscopy results suggested that the number of autophagosomes increased significantly after mTORC1 suppression but decreased when Beclin1 was knocked down (figure 4E). To further exclude the influence of CQ, the key molecule Beclin1 in the autophagy pathway was knocked down, and the results showed that the downregulation of PD-L1 on mTORC1 suppression was reversed after knocking down Beclin1 (online supplemental figure 6). These results suggested that mTORC1 suppression induces the autophagic degradation of PD-L1 in HCC cells with p53 LoF.
jitc-2023-007479supp006.pdf (2.1MB, pdf)
Bidirectional regulation of PD-L1 by mTORC1 suppression in HCC with different p53 statuses in vivo
To verify the bidirectional regulation of PD-L1 by mTORC1 suppression in HCC with different p53 statuses in vivo, we constructed a mouse model by injecting Trp53-knockdown AAV followed by delivery of c-Myc and NRas oncogenic plasmids (figure 5A). Trp53 knockdown mice had larger tumor areas and heavier livers, while the tumor burden was reduced in Trp53 knockdown mice administered everolimus (figure 5B–D). In Trp53 wild-type mice, PD-L1 was upregulated with everolimus administration. In Trp53 knockdown mice, PD-L1 was downregulated with everolimus administration (figure 5E,F). CyTOF analysis and flow cytometry indicated that infiltration of CD8+ T cells was significantly decreased in Trp53 wild-type tumors after everolimus administration but slightly increased in Trp53 knockdown tumors after everolimus administration (figure 5G,H), which was consistent with the PD-L1 expression results. The above results validated the bidirectional regulation of PD-L1 by mTORC1 suppression in HCC with different p53 statuses in vivo.
Figure 5.
Bidirectional regulation of PD-L1 by mammalian target of rapamycin complex 1 suppression in HCC with different p53 statuses in vivo. (A) Process of plasmid-induced mouse HCC model construction and drug administration. Mice were divided into four groups, including Trp53 wild-type mice administrated with (WT EVE) or without (WT DMSO) everolimus, Trp53 knockdown mice administrated with (shTrp53 EVE) or without (shTrp53 DMSO) everolimus. (B) Representative images of liver tumors of the four groups. (C–D) Liver weight and the largest tumor diameters of the four groups were measured. (E) The protein level of PD-L1 and p-p70s6k in mouse tumors was detected by western blotting. β-actin was used as the loading control for western blotting. (F) IHC staining of PD-L1 expression in mouse tumor tissues. (G) t-SNE maps of CD8+ T-cell clusters distribution in mouse tumors of the four groups. Percentages of CD8+ T cells in mouse tumors of each group. (H) CD45+, CD3+ and CD8+ T cells extracted from mouse tumors were analyzed by flow cytometry. Percentages of CD8+ T cells in the four groups. *p<0.05, compared with the WT DMSO (C, D, E, F) or shTrp53 DMSO (E, F) groups. AAV, adreno-associated virus; EVE, everolimus; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; PD-L1, programmed death ligand 1; RT-qPCR, Real Time Quantitative Polymerase Chain; t-SNE, t-Distributed Stochastic Neighbor Embedding; WT, wild-type.
Treatment efficacy of the mTOR inhibitor and αPD-L1 antibody combination depends on TP53 status in HCC
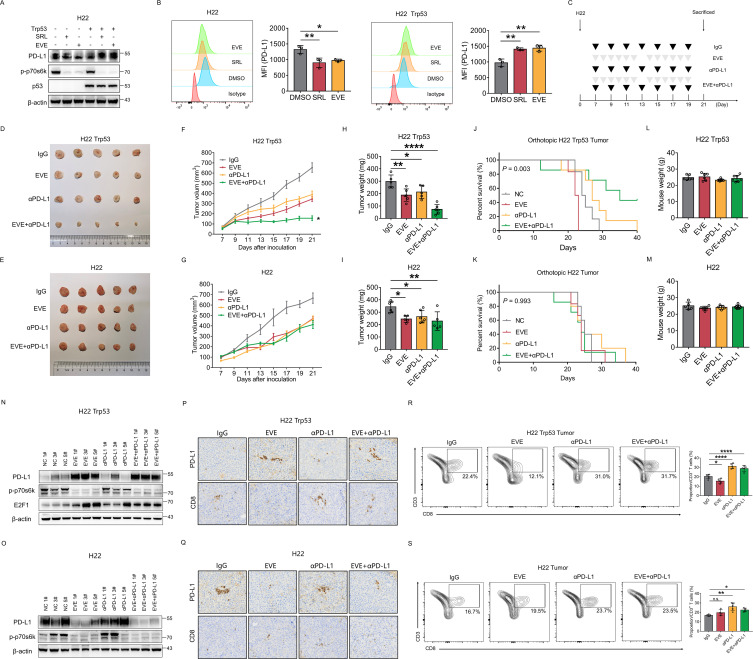
The mouse HCC cell lines H22 and Hepa1-6 were assessed by DNA exon sequencing. The results indicated that H22 was genetically Trp53 wild type and Hepa1-6 was genetically Trp53 mutant. However, western blotting showed that H22 cell did not express p53 protein (online supplemental figure 7). Thus, we defined H22 as a mouse HCC cell line with p53 LoF. Similar to the results in human HCC cells, PD-L1 was downregulated by mTORC1 suppression in H22 cell with p53 LoF, while PD-L1 was upregulated by mTORC1 suppression in H22 cell overexpressing p53 (figure 6A,B). We used H22 cells to establish the subcutaneous tumor model and the orthotopic tumor model (figure 6C). Mice were intraperitoneally injected with αPD-L1 and/or everolimus by gavage. In Trp53-overexpressing H22 subcutaneous tumors, compared with the control or single drug treatments, the combination of αPD-L1 and everolimus led to significant inhibition of tumor growth (figure 6D, F and H). The tumor volume of the combination group was reduced by 76.1%, and the tumor weight was reduced by 75.5% compared with that in the control group. In addition, the combination group also had significantly superior survival compared with the control or single drug groups in Trp53-overexpressing H22 orthotopic tumor model (figure 6J). In H22 subcutaneous tumors with p53 LoF, the combination of αPD-L1 and everolimus did not effectively inhibit tumor growth compared with that in the control or single drug groups (figure 6E,G,I). No survival difference was observed among the four groups in H22 orthotopic tumor model with p53 LoF (figure 6K). In addition, there was no difference in the weight of mice between different groups, suggesting that the drug had little adverse effect (figure 6L,M).
Figure 6.
Treatment efficacy of mammalian target of rapamycin complex 1 inhibitor and αPD-L1 antibody combination depends on TP53 statuses in HCC. (A–B) H22, the mouse HCC cell, with or without TP53 overexpression were treated with everolimus or sirolimus. The level of PD-L1, p-p70s6k, p53 were detected by western blot (A) and flow cytometry (B). β-actin was used as the loading control for western blot. (C) Process of subcutaneous tumor xenograft model and orthotopic tumor model construction and drug administration. Mice were administered with anti-PD-L1 antibody and everolimus alone or in combination. (D–I) Comparison of H22 tumors (D, E) tumor volume (F, G) and tumor weight (H, I) in different groups were shown. (J–O) Survival analysis was performed among different groups (J, K). Mouse weight was measured before sacrifice (L, M). The level of PD-L1, p-p70s6k, E2F1 protein were detected by western blot (N, O). β-actin was used as the loading control for western blotting. (P–Q) Immunohistochemistry staining of PD-L1 and CD8 expression in the tumor tissues of each group. (R–S) CD45+, CD3+ and CD8+ T cells extracted from mouse tumors were analyzed by flow cytometry. Percentages of CD8+ T cells in each group. *p<0.05, compared with the IgG group. DMSO, Dimethylsulfoxide; EVE, everolimus; HCC, hepatocellular carcinoma; MFI, mean fluorescent intensity; PD-L1, programmed death ligand 1; SRL, sirolimus.
jitc-2023-007479supp007.pdf (1.6MB, pdf)
In Trp53-overexpressing H22 tumors, PD-L1 was significantly upregulated in groups administered everolimus (figure 6N,P). E2F1 was also upregulated in mice treated with everolimus. In addition, the levels of tumor-infiltrating CD8+ T cells were significantly decreased in the everolimus group but increased in the αPD-L1 group and combination group compared with the control group (figure 6P,R). In p53 LoF H22 tumors, PD-L1 was downregulated in groups administered everolimus (figure 6O,Q). The level of tumor-infiltrating CD8+ T cells was slightly increased in the everolimus group and significantly increased in the αPD-L1 group and combination group compared with the control group (figure 6Q,S). The above results proved the bidirectional regulation of PD-L1 by mTORC1 suppression in HCC with different p53 status in vivo and indicated that combination therapy with an mTOR inhibitor and αPD-L1 may significantly improve the prognosis of p53 wild-type HCC.
Clinical significance of TP53 status and E2F1 in patients with HCC
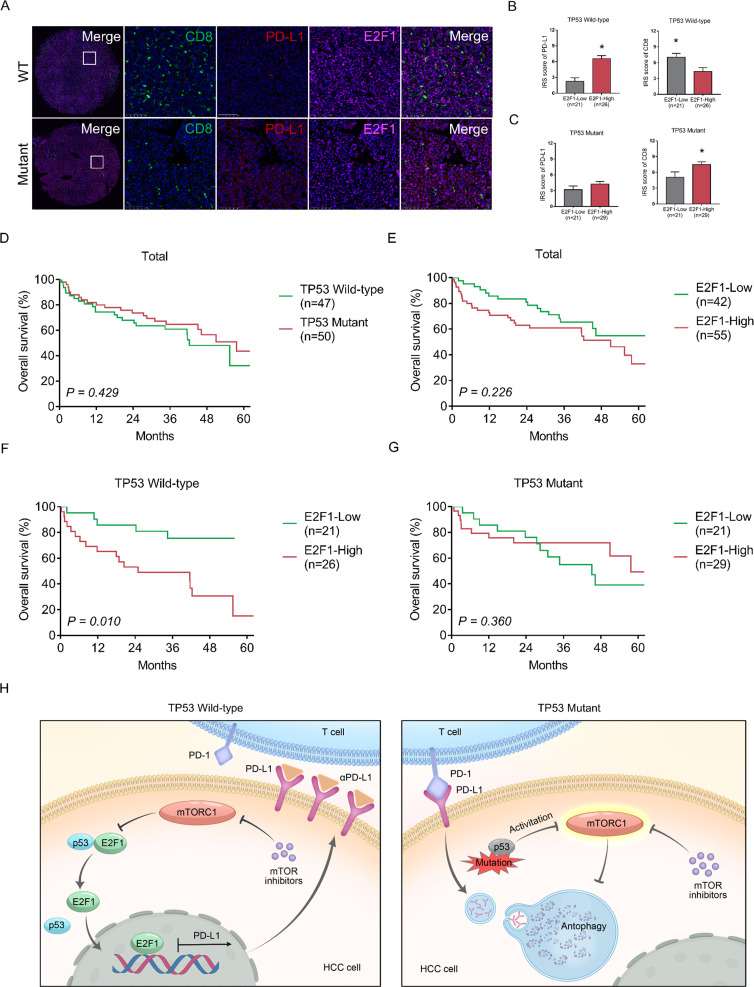
TP53 mutations in 97 patient with HCC samples after surgery were detected by Sanger sequencing. Forty-seven (48.5%) samples were TP53 wild-type, and 50 (51.5%) samples were TP53 mutant. Clinical characteristics are shown in online supplemental table 1. We found that TP53 mutations in these HCC tissues are mainly p53 LoF mutations, including missense mutations, frame shift mutations and non-sense mutations. Thus, TP53 mutations in patients with HCC in this study are actually p53 LoF mutations. HCC samples underwent multicolor fluorescence staining. The results indicated that TP53-mutant HCC has higher PD-L1 expression and less CD8+ T-cell infiltration (figure 7A) than TP53 wild-type HCC. However, E2F1 expression was not correlated with TP53 mutation status (TP53 wild-type vs TP53 mutant) in HCC.
Figure 7.
Clinical significance of TP53 status and E2F1 in patients with HCC. (A) Tumor tissues from patients with HCC were isolated and infiltrated CD8 (green), PD-L1 (red) and E2F1 (pink) were stained. (B–C) Comparison of IRS score of PD-L1 and CD8 between E2F1 low expression group and high expression groups in different TP53 statuses were shown. (D–G) OS of patients with HCC after surgery based on the TP53 statuses and level of E2F1 in HCC was evaluated by the Kaplan-Meier method. (H) Graphical summary of the bidirectional regulatory mechanisms of PD-L1 in HCC. HCC, hepatocellular carcinoma; IRS, Immune Reactive Score; mTOR, mammalian target of rapamycin; mTORC1, mammalian target of rapamycin complex 1; PD-1, programmed death 1; PD-L1, programmed death ligand 1; OS, overall survival; WT, wild-type.
jitc-2023-007479supp009.pdf (50.9KB, pdf)
Patients were grouped according to the immunoreactive scores of E2F1 and TP53 status. Clinical characteristics are shown in online supplemental tables 2-4. In TP53 wild-type HCC, PD-L1 levels were significantly higher in patients with high E2F1 levels than in patients with low E2F1 levels; CD8 levels were significantly lower in patients with high E2F1 levels than in patients with low E2F1 levels (figure 7B). In TP53-mutant HCC, no difference was observed in PD-L1 levels between the two groups, but CD8 levels were significantly higher in patients with high E2F1 levels than in patients with low E2F1 levels (figure 7C). These results suggest that the regulatory mechanism among E2F1, PD-L1 and CD8 in TP53 wild-type HCC was consistent with that in HCC cell lines and mouse models.
We further investigated the clinical significance of TP53 status and E2F1 in predicting the survival of patients with HCC after surgery. Patients were grouped by TP53 status and E2F1 expression level, and their clinical characteristics are shown in online supplemental table. No difference in OS was observed between TP53 wild-type patients and TP53 mutant patients (figure 7D) or between patients with low and high E2F1 levels (figure 7E). However, in patients with TP53 wild-type HCC, patients with low E2F1 levels had significantly superior OS compared with patients with high E2F1 levels (figure 7F). High E2F1 level was an independent risk factor for OS (HR=2.906, p=0.042) among patients with TP53 wild-type HCC (online supplemental table 6). In patients with TP53-mutant HCC, no difference in OS was observed between the two groups (figure 7G, online supplemental table 7).
In this study, our results revealed the bidirectional regulatory mechanism of PD-L1. To summarize, in TP53 wild-type HCC cells, mTORC1 suppression promotes PD-L1 expression, while in HCC cells with p53 LoF, mTORC1 suppression promotes PD-L1 degradation. Mechanistically, mTORC1 suppression weakens the interaction between p53 and E2F1 and leads to increased E2F1 entry into the nucleus, further promoting the transcription of PD-L1 in TP53 wild-type HCC cells. On the other hand, mTORC1 suppression induces the autophagic degradation of PD-L1 in HCC cells with p53 LoF (figure 7H).
Discussion
Molecular targeted drugs and immunotherapy have facilitated great breakthroughs in the treatment of patients with HCC.5 19 20 However, the efficiency and response rate of these treatments alone are limited and vary among different patients with HCC due to tumor heterogeneity.4 Thus, the combined use of multiple therapies has received more attention clinically.21 Nevertheless, using combination therapies blindly does not bring benefit to all patients. Clinicians are facing challenges regarding the development of individualized and precise treatment for patients with HCC. Therefore, research on treatment efficacy-oriented molecular characteristics or molecular subtyping of HCC is urgently needed to find effective drugs or therapies in clinical practice and to develop new treatment strategies for treating HCC precisely in the future.
Research has demonstrated that mutations in key genes can lead to changes in the tumor microenvironment and cause immune evasion, which can greatly influence the efficacy of immunotherapy.16–18 Dong et al reported that KRAS/TP53 mutation in human lung adenocarcinoma is significantly associated with increased PD-L1 expression and CD8+ T-cell infiltration, and patients with KRAS/TP53-mutant lung adenocarcinoma had a better response to PD-1 blockade immunotherapy and significantly better prognosis than those with wild-type cancer.22 Based on the largest original multiomics data set on triple-negative breast cancer, Xiao et al found an “innate immune-inactivated” cluster with resting innate immune cell and non-immune stromal cell infiltration. This cluster is characterized by inactivation of innate immunity and low tumor antigen burden contributing to tumor immune escape, and inclusion in this cluster is directly correlated with mutations in the PI3K-AKT pathway.23 Similarly, HCC is a tumor with a high incidence of mutations. TP53 gene mutation is the most common mutation in HCC. Genome-wide analysis of large-scale public databases showed that the mutation rate of TP53 in HCC ranges from 29.1% to 58.0%.10 13 In our study, 63 of 114 patients with HCC (55.3%) had p53 LoF mutations. Therefore, the changes in the tumor microenvironment caused by TP53 mutation deserve great attention. Our findings suggest that TP53-mutant HCC had a lower level of CD8+ T-cell infiltration and higher PD-L1 expression levels than TP53 wild-type HCC, indicating that TP53 mutation may represent a state of adaptive immune resistance and high immunogenicity. These results are helpful to address the association among TP53 mutation in tumors and PD-L1 expression levels as well as with the CD8+ T-cell infiltration. In addition, due to the widespread mutation of TP53 in cancer, similar regulatory mechanisms may also exist in other tumors, which deserves further basic, preclinical and clinical research. However, other gene mutations can also coexist with TP53 mutations; these include CTNNB1 and TERT mutations, which may also cause changes in the tumor microenvironment.24 25 Therefore, more comprehensive analysis of the HCC microenvironment based on key gene mutations is needed.
mTOR is a serine/threonine protein kinase that regulates cell growth and metabolism, of which mTORC1 is the main form.26 Previous research has reported activation of the mTOR pathway after TP53 mutation in tumor cells and the important role of the mTOR pathway in regulating the tumor microenvironment.27 28 However, several studies have reported the regulatory relationship between PD-L1 and mTOR pathway in renal cell carcinoma, lung cancer, ovarian cancer and melanoma.29–31 Our study is the first to explore the mechanism of PD-L1 regulation via the mTOR pathway in the context of different gene mutation backgrounds in HCC. The results of our study indicate that it is important to consider different gene backgrounds and key signaling pathways together to comprehensively explore tumor behavior and the microenvironment due to cancer heterogeneity, especially in HCC.
mTOR inhibitors have demonstrated antitumor effects and are widely used for cancer treatment; for example, they are used for the treatment of renal cell carcinoma and for immunoregulation after transplantation.29 32 Several clinical trials have also assessed its antitumor effects for advanced HCC. However, due to unsatisfactory results, they have not yet been approved as a treatment for HCC.33 The international, randomized, phase 3 study, EVOLVE-1, was intended to assess the efficacy of everolimus in patients with advanced HCC after sorafenib treatment failure.34 This study totally enrolled 546 patients with HCC whose disease progressed during or after sorafenib or who were intolerant of sorafenib. Among them, 362 patients were randomized to the everolimus group and 184 patients to the placebo group. No benefit was observed in the everolimus group. The median OS was 7.6 months in the everolimus group compared with 7.3 months in the placebo group, and median time to progression were 3.0 months and 2.6 months, respectively. Another randomized multicenter, multinational phase 2 trial, SAKK 77/08 and SASL 29, was to investigate the efficacy of the combination of sorafenib plus everolimus compared with sorafenib alone in treating advanced HCC.35 A total of 106 patients were randomly grouped to the sorafenib group (46 patients) and the sorafenib plus everolimus group (60 patients). Median progression-free survival (6.6 vs 5.7 months), time to progression (7.6 vs 6.3 months), duration of disease stabilization (6.7 vs 6.7 months), and OS (10 vs 12 months) were similar between the two groups, respectively. Thus, no evidence was found that the combination of sorafenib and everolimus improves the efficacy compared with sorafenib alone. Previous clinical trials have focused on the efficacy of mTOR inhibitors as monotherapy or in combination with traditional molecular targeted drugs such as sorafenib. As anti-PD-1 and anti-PD-L1 antibodies show better efficacy in treating HCC than molecular targeted drugs, researchers are paying more attention to immunotherapy. The IMbrave150 trial reported that the PD-L1 inhibitor atezolizumab combined with the antiangiogenic drug bevacizumab significantly prolonged the overall survival and reduced the death risk of patients with unresectable HCC by 42% compared with sorafenib alone.5 However, only about one-third of patients present a response to immunotherapy.36 How to improve the response rate, efficacy and precision of immunotherapy is currently an urgent problem to be solved. A variety of combined use schemes are currently being studied in basic research. But, there is no clinical trial conducted on the combination of everolimus and immunotherapy for advanced HCC at present. Our findings demonstrate that the gene mutation status of TP53 can guide the combined use of mTOR inhibitors and anti-PD-L1 antibodies. This study may support the reconsideration of mTOR inhibitors for treating HCC and represents a new step toward the application of precision immunotherapy for HCC. In this study, the combination of an mTOR inhibitor and an anti-PD-L1 antibody significantly suppressed tumor growth and prolonged the survival of mice with TP53 wild-type HCC. Based on our results, mTOR inhibitors represent a new opportunity for treating TP53 wild-type HCC and can enhance the effects of anti-PD-L1 antibody treatment.
Research has revealed that the proportion of patients with HCC receiving mTOR inhibitors after liver transplant ranges from 37.2% to 42.2%.37 38 The bidirectional regulatory mechanism of PD-L1 mediated by TP53/mTORC1 in this study has guiding significance for clinical practice and is helpful in preventing tumor recurrence after liver transplantation for HCC. However, the use of PD-L1 antibody after transplantation has a risk of inducing rejection.39 Therefore, further research is still needed.
E2F1, as a transcriptional activator, is a member of the transcription factor E2F family that regulates cell progression, division and genome replication.40 The most classic axis is the cyclin-dependent kinase/RB/E2F axis. In the present study, we found an interaction between E2F1 and p53 in HCC, which is not involved in the classic pathway. Previous studies reported that the interaction of E2F1 and p53 can influence their respective functions.41–43 Fogal et al reported that the amino terminal domain in E2F1 binds to amino acid residues of p53, enhancing nuclear retention of p-p53.42 In another case, the E2F1-p53 complex bound to the E2F target. Zhou et al reported that p53 interacts with E2F1 to form the p53-E2F1-DNA complex, which suppresses E2F1-dependent PLK1 expression, leading to apoptosis in response to DNA damage.43 Although some studies have reported the cancer-promoting function of E2F1 and its binding with p53, its regulatory effects on PD-L1 expression and immune evasion have not been reported.44 45 In our study, we found that p53 can bind with E2F1 in the cytoplasm, obstruct its entry into the nucleus and suppress its transcriptional activation in TP53 wild-type HCC cells. mTORC1 suppression disrupts the binding and promotes the nuclear translocation of E2F1. Thus, E2F1 binds to the PD-L1 promoter, facilitating its transcription and modulating immune evasion. Therefore, E2F1 might be a potential target for immunotherapy. However, our study did not determine how mTORC1 suppression disrupts the binding of p53 to E2F1, which deserves further exploration.
In addition, E2F1 does not play a regulatory role in TP53-mutant HCC. mTORC1-mediated autophagic degradation of PD-L1 plays a major role in this circumstance. For TP53-mutant HCC, compared with monotherapy, the combination of mTOR inhibitor and anti-PD-L1 antibody did not show a superior benefit. Therefore, other strategies to improve treatment efficacy for these patients remain to be explored.
In summary, we revealed the mechanism of bidirectional regulation of PD-L1 mediated by TP53/mTORC1 in HCC and its effect on modulating immune evasion. These data provide strong evidence for the combined use of mTOR inhibitors and anti-PD-L1 antibodies in the clinic, which can be a novel and promising immunotherapy strategy for TP53 wild-type HCC. Further clinical trials are needed due to the complexity and heterogeneity of HCC in humans.
Footnotes
Contributors: JY, SL, XX conceived and designed the study. JY, JH, LZ, WZ, LY performed the experiments. JY, JH, YW, QZ, QY, JB, NX, YL, KC, XW, ZL, TF, SL, JL prepared the manuscript. JY, JH, SX, QZ, SW analyzed the data. JY, LZ, HX, SZ, SL, XX collected the clinical samples. XX is responsible for the overall content as guarantor.
Funding: This study was supported by the National Natural Science Foundation of China (No.92159202, 82273270, 82203070, 82200726, 82200727) and the National Key Research and Development Program of China (No.2022YFA1106800).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Not applicable.
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Tagliamonte M, Mauriello A, Cavalluzzo B, et al. Tackling hepatocellular carcinoma with individual or Combinatorial Immunotherapy approaches. Cancer Lett 2020;473:25–32. 10.1016/j.canlet.2019.12.029 [DOI] [PubMed] [Google Scholar]
- 3.Tian Y, Hu D, Li Y, et al. Development of therapeutic vaccines for the treatment of diseases. Mol Biomed 2022;3:40. 10.1186/s43556-022-00098-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol 2022;19:151–72. 10.1038/s41571-021-00573-2 [DOI] [PubMed] [Google Scholar]
- 5.Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus Bevacizumab in Unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894–905. 10.1056/NEJMoa1915745 [DOI] [PubMed] [Google Scholar]
- 6.Khan AA, Liu Z-K, Xu X. Recent advances in Immunotherapy for hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int 2021;20:511–20. 10.1016/j.hbpd.2021.06.010 [DOI] [PubMed] [Google Scholar]
- 7.Meek DW. Tumour suppression by P53: a role for the DNA damage response Nat Rev Cancer 2009;9:714–23. 10.1038/nrc2716 [DOI] [PubMed] [Google Scholar]
- 8.Aylon Y, Oren M. Living with P53, dying of P53. Cell 2007;130:597–600. 10.1016/j.cell.2007.08.005 [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, Liu J, Xu D, et al. Gain-of-function mutant P53 in cancer progression and therapy. J Mol Cell Biol 2020;12:674–87. 10.1093/jmcb/mjaa040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata T. Genomic landscape of Hepatocarcinogenesis. J Hum Genet 2021;66:845–51. 10.1038/s10038-021-00928-8 [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Yan K, He X, et al. Lrp1B or Tp53 mutations are associated with higher tumor mutational burden and worse survival in hepatocellular carcinoma. J Cancer 2021;12:217–23. 10.7150/jca.48983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling S, Shan Q, Zhan Q, et al. Usp22 promotes hypoxia-induced hepatocellular carcinoma Stemness by a Hif1Α/Usp22 positive feedback loop upon Tp53 inactivation. Gut 2020;69:1322–34. 10.1136/gutjnl-2019-319616 [DOI] [PubMed] [Google Scholar]
- 13.Gao Q, Zhu H, Dong L, et al. Integrated Proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell 2019;179. 10.1016/j.cell.2019.10.038 [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Yu L, Chen W, et al. Wild-type P53 promotes cancer metabolic switch by inducing PUMA-dependent suppression of oxidative Phosphorylation. Cancer Cell 2019;35:191–203. 10.1016/j.ccell.2018.12.012 [DOI] [PubMed] [Google Scholar]
- 15.Lam YK, Yu J, Huang H, et al. Tp53 R249S Mutation in hepatic Organoids captures the predisposing cancer risk. Hepatology 2023;78:727–40. 10.1002/hep.32802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh M, Saha S, Bettke J, et al. Mutant P53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell 2021;39:494–508. 10.1016/j.ccell.2021.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyu H, Li M, Jiang Z, et al. Correlate the Tp53 Mutation and the HRAS Mutation with immune signatures in head and neck squamous cell cancer. Computational and Structural Biotechnology Journal 2019;17:1020–30. 10.1016/j.csbj.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Z, Liu Z, Li M, et al. Immunogenomics analysis reveals that Tp53 mutations inhibit tumor immunity in gastric cancer. Transl Oncol 2018;11:1171–87. 10.1016/j.tranon.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel A, Qin S, Kudo M, et al. Lenvatinib versus sorafenib for first-line treatment of Unresectable hepatocellular carcinoma: patient-reported outcomes from a randomised, open-label, non-inferiority, phase 3 trial. Lancet Gastroenterol Hepatol 2021;6:649–58. 10.1016/S2468-1253(21)00110-2 [DOI] [PubMed] [Google Scholar]
- 20.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–90. 10.1056/NEJMoa0708857 [DOI] [PubMed] [Google Scholar]
- 21.Shang R, Song X, Wang P, et al. Cabozantinib-based combination therapy for the treatment of hepatocellular carcinoma. Gut 2021;70:1746–57. 10.1136/gutjnl-2020-320716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Z-Y, Zhong W-Z, Zhang X-C, et al. Potential predictive value of Tp53 and KRAS Mutation status for response to PD-1 blockade Immunotherapy in lung adenocarcinoma. Clin Cancer Res 2017;23:3012–24. 10.1158/1078-0432.CCR-16-2554 [DOI] [PubMed] [Google Scholar]
- 23.Xiao Y, Ma D, Zhao S, et al. Multi-Omics profiling reveals distinct Microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin Cancer Res 2019;25:5002–14. 10.1158/1078-0432.CCR-18-3524 [DOI] [PubMed] [Google Scholar]
- 24.Ambrozkiewicz F, Trailin A, Červenková L, et al. Ctnnb1 mutations, TERT polymorphism and Cd8+ cell densities in Resected hepatocellular carcinoma are associated with longer time to recurrence. BMC Cancer 2022;22:884. 10.1186/s12885-022-09989-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montironi C, Castet F, Haber PK, et al. Inflamed and non-inflamed classes of HCC: a revised Immunogenomic classification. Gut 2023;72:129–40. 10.1136/gutjnl-2021-325918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Liu Y, Wang D, et al. The upstream pathway of mTOR-mediated Autophagy in liver diseases. Cells 2019;8:1597. 10.3390/cells8121597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montal R, Sia D, Montironi C, et al. Molecular classification and therapeutic targets in extrahepatic Cholangiocarcinoma. J Hepatol 2020;73:315–27. 10.1016/j.jhep.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumas AA, Pomella N, Rosser G, et al. Microglia promote glioblastoma via mTOR-mediated immunosuppression of the tumour Microenvironment. EMBO J 2020;39:e103790. 10.15252/embj.2019103790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C, Duan Y, Xia M, et al. TFEB mediates immune evasion and resistance to mTOR inhibition of renal cell carcinoma via induction of PD-L1. Clin Cancer Res 2019;25:6827–38. 10.1158/1078-0432.CCR-19-0733 [DOI] [PubMed] [Google Scholar]
- 30.Clark CA, Gupta HB, Sareddy G, et al. Tumor-intrinsic PD-L1 signals regulate cell growth, pathogenesis, and Autophagy in ovarian cancer and Melanoma. Cancer Res 2016;76:6964–74. 10.1158/0008-5472.CAN-16-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lastwika KJ, Wilson W, Li QK, et al. Control of PD-L1 expression by Oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res 2016;76:227–38. 10.1158/0008-5472.CAN-14-3362 [DOI] [PubMed] [Google Scholar]
- 32.Chen J, Shen T, Li J, et al. Clinical practice guideline on liver transplantation for hepatocellular carcinoma in China. Chinese Medical Journal 2022;135:2911–3. 10.1097/CM9.0000000000002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X, Paliogiannis P, Calvisi DF, et al. Role of the mammalian target of rapamycin pathway in liver cancer: from molecular Genetics to targeted therapies. Hepatology 2021;73 Suppl 1(Suppl 1):49–61. 10.1002/hep.31310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu AX, Kudo M, Assenat E, et al. Effect of everolimus on survival in advanced hepatocellular carcinoma after failure of sorafenib: the EVOLVE-1 randomized clinical trial. JAMA 2014;312:57–67. 10.1001/jama.2014.7189 [DOI] [PubMed] [Google Scholar]
- 35.Koeberle D, Dufour J-F, Demeter G, et al. Sorafenib with or without everolimus in patients with advanced hepatocellular carcinoma (HCC): a randomized multicenter, multinational phase II trial (SAKK 77/08 and SASL 29). Ann Oncol 2016;27:856–61. 10.1093/annonc/mdw054 [DOI] [PubMed] [Google Scholar]
- 36.Greten TF, Abou-Alfa GK, Cheng A-L, et al. Society for Immunotherapy of cancer (SITC) clinical practice guideline on Immunotherapy for the treatment of hepatocellular carcinoma. J Immunother Cancer 2021;9:e002794. 10.1136/jitc-2021-002794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Q, Ling S, Jiang G, et al. Sirolimus-based immunosuppression improves the prognosis of liver transplantation recipients with low Tsc1/2 expression in hepatocellular carcinoma beyond the Milan criteria. Eur J Surg Oncol 2021;47:2533–42. 10.1016/j.ejso.2021.04.001 [DOI] [PubMed] [Google Scholar]
- 38.Ling S, Feng T, Zhan Q, et al. Sirolimus-based immunosuppression improves outcomes in liver transplantation recipients with hepatocellular carcinoma beyond the Hangzhou criteria. Ann Transl Med 2020;8:80. 10.21037/atm.2020.01.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X. State of the art and perspectives in liver transplantation. Hepatobiliary Pancreat Dis Int 2023;22:1–3. 10.1016/j.hbpd.2022.12.001 [DOI] [PubMed] [Google Scholar]
- 40.Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer 2019;19:326–38. 10.1038/s41568-019-0143-7 [DOI] [PubMed] [Google Scholar]
- 41.Nip J, Strom DK, Eischen CM, et al. E2F-1 induces the stabilization of P53 but blocks P53-mediated Transactivation. Oncogene 2001;20:910–20. 10.1038/sj.onc.1204171 [DOI] [PubMed] [Google Scholar]
- 42.Fogal V, Hsieh JK, Royer C, et al. Cell cycle-dependent nuclear retention of P53 by E2F1 requires Phosphorylation of P53 at Ser315. EMBO J 2005;24:2768–82. 10.1038/sj.emboj.7600735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z, Cao J-X, Li S-Y, et al. P53 suppresses E2F1-dependent Plk1 expression upon DNA damage by forming P53-E2F1-DNA complex. Exp Cell Res 2013;319:3104–15. 10.1016/j.yexcr.2013.09.012 [DOI] [PubMed] [Google Scholar]
- 44.González-Romero F, Mestre D, Aurrekoetxea I, et al. E2F1 and E2F2-mediated repression of Cpt2 establishes a lipid-rich tumor-promoting environment. Cancer Res 2021;81:2874–87. 10.1158/0008-5472.CAN-20-2052 [DOI] [PubMed] [Google Scholar]
- 45.Zhang T, Guan G, Zhang J, et al. E2F1-mediated Auf1 upregulation promotes HCC development and enhances drug resistance via stabilization of Akr1B10. Cancer Sci 2022;113:1154–67. 10.1111/cas.15272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007479supp008.pdf (215.2KB, pdf)
jitc-2023-007479supp001.pdf (461.5KB, pdf)
jitc-2023-007479supp002.pdf (2.5MB, pdf)
jitc-2023-007479supp003.pdf (785.3KB, pdf)
jitc-2023-007479supp004.pdf (1.3MB, pdf)
jitc-2023-007479supp005.pdf (1.1MB, pdf)
jitc-2023-007479supp006.pdf (2.1MB, pdf)
jitc-2023-007479supp007.pdf (1.6MB, pdf)
jitc-2023-007479supp009.pdf (50.9KB, pdf)
Data Availability Statement
The data of RNA sequencing, whole-exome sequencing, assay for transposase-accessible chromatin by sequencing (ATAC-seq) can be viewed in SRA database and GEO database. The accession IDs are PRJNA999626 and GSE240197 and GSE239926. The persistent URL are http://www.ncbi.nlm.nih.gov/bioproject/999626, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE240197, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE239926, retrospectively.
Data are available in a public, open access repository.