Abstract
Background
An accumulation of somatic mutations in tumors leads to increased neoantigen levels and antitumor immune response. Tumor mutational burden (TMB) reflects the rate of somatic mutations in the tumor genome, as determined from tumor tissue (tTMB) or blood (bTMB). While high tTMB is a biomarker of immune checkpoint inhibitor (ICI) treatment efficacy, few studies have explored the clinical utility of bTMB, a less invasive alternative for TMB assessment. Establishing the correlation between tTMB and bTMB would provide insight into whether bTMB is a potential substitute for tTMB. We explored the tumor genomes of patients enrolled in CheckMate 848 with measurable TMB. The correlation between tTMB and bTMB, and the factors affecting it, were evaluated.
Methods
In the phase 2 CheckMate 848 (NCT03668119) study, immuno-oncology-naïve patients with advanced, metastatic, or unresectable solid tumors and tTMB-high or bTMB-high (≥10 mut/Mb) were prospectively randomized 2:1 to receive nivolumab plus ipilimumab or nivolumab monotherapy. Tissue and plasma DNA sequencing was performed using the Foundation Medicine FoundationOne CDx and bTMB Clinical Trial Assays, respectively. tTMB was quantified from coding variants, insertions, and deletions, and bTMB from somatic base substitutions. Correlations between tTMB and bTMB were determined across samples and with respect to maximum somatic allele frequency (MSAF). Assay agreement and variant composition were also evaluated.
Results
A total of 1,438 and 1,720 unique tissue and blood samples, respectively, were obtained from 1,954 patients and included >100 screened disease ontologies, with 1,017 unique pairs of tTMB and bTMB measurements available for assessment. Median tTMB and bTMB were 3.8 and 3.5 mut/Mb, respectively. A significant correlation between tTMB and bTMB (r=0.48, p<0.0001) was observed across all sample pairs, which increased to r=0.54 (p<0.0001) for samples with MSAF≥1%. Assay concordance was highest for samples with MSAF≥10% across multiple disease ontologies and observed for both responders and non-responders to ICI therapy. The variants contributing to tTMB and bTMB were similar.
Conclusions
We observed that tTMB and bTMB had a statistically significant correlation, particularly for samples with high MSAF, and that this correlation applied across disease ontologies. Further investigation into the clinical utility of bTMB is warranted.
Keywords: Biomarkers, Tumor; Immunotherapy; Ipilimumab; Nivolumab
WHAT IS ALREADY KNOWN ON THIS TOPIC
Tumor mutational burden (TMB) represents the number of somatic mutations in the tumor genome, and when assessed from tissue (tTMB), serves as a biomarker of immune checkpoint inhibitor (ICI) treatment efficacy. Assessment of TMB from blood (bTMB) involves a less invasive approach and captures a more representative sample of tumor DNA compared with tTMB assessment, but its clinical utility has not been widely studied.
WHAT THIS STUDY ADDS
We tested the correlation between tTMB and bTMB assays in a cohort of nearly 2000 patients enrolled in CheckMate 848 who received ICI therapy. This cohort is one of the largest evaluated for a correlation between tTMB and bTMB, and comprised many types of solid tumors. Our results suggest that tTMB and bTMB are generally correlated. Moreover, we observed strong concordance between tTMB and bTMB assays for both responders and non-responders to ICI therapy, and similarity in the variant pool detected by the two types of assays.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study supports the ongoing use of tTMB for TMB assessment, suggests that bTMB may have clinical utility, and lays the groundwork for further research in this area. If validated for clinical use, bTMB would offer a less invasive approach to TMB assessment compared with tTMB, thereby reducing patient risk.
Background
As tumors grow and evolve, they can accumulate somatic mutations, which may lead to increased levels of neoantigens and enhanced antitumor immune responses.1–3 Tumor mutational burden (TMB) is an estimate of the number of somatic mutations per megabase (mut/Mb) in the sequenced portion of the tumor genome.4 TMB can be determined using DNA from tumor tissue biopsies (tissue TMB (tTMB)) or blood samples (blood TMB (bTMB)). tTMB is increasingly recognized as an important biomarker for the efficacy of immune checkpoint inhibitor (ICI) treatment in patients with advanced solid tumors.5–12 Evidence suggests that across a range of solid tumor types, high tTMB (tTMB-H; ≥10 mut/Mb5 6 13 14 can be used to identify subgroups of patients who are likely to respond to treatment with programmed death-1 (PD-1) inhibitors.5 6 14 In addition, high tTMB is associated with improved overall survival (OS) following ICI treatment for patients with many types of cancer.8
Assessment of bTMB from circulating tumor DNA (ctDNA) may offer a less invasive approach for genomic profiling than tissue biopsy-based methods.15 16 In addition, bTMB has the potential to overcome the challenge of accurately determining TMB from a single region of a heterogeneous tumor.17 High bTMB may be associated with improved clinical benefit in patients with non-small cell lung cancer (NSCLC) treated with ICIs, but the clinical utility of bTMB has not been as widely studied as that of tTMB.14 15 17–21
Both biological and analytical factors, such as disease ontology, disease stage, intratumor heterogeneity, intertumor heterogeneity within a patient,22 tumor clonality, sample type, sample purity, and computational methodologies, can contribute to differences between tissue- and blood-derived genomic assessments.17 23 24 Notably, increased rates of tumor shedding may contribute to higher levels of ctDNA available for analysis in plasma, thus increasing the correlation between tissue- and blood-based assessments.25
Correlation between tTMB and bTMB has been studied previously in a small number of tumor types, including NSCLC.17 24 26–30 Evidence suggests that lower maximum somatic allele frequency (MSAF), which correlates with ctDNA levels in the blood, contributes to decreased concordance between tTMB and bTMB.17 However, further investigation is needed to fully understand the influence of biological and analytical factors on tTMB versus bTMB concordance. This will enable the successful adoption of non-invasive tumor profiling in a clinical setting, ultimately helping to guide the selection of patients who are most likely to respond to treatment with ICIs.
CheckMate 848 (NCT03668119) is a prospective phase 2 study of the anti-PD-1 antibody nivolumab in combination with the anti-cytotoxic T-lymphocyte-associated protein 4 antibody ipilimumab versus nivolumab monotherapy in patients with advanced, metastatic, or unresectable solid tumors refractory to standard therapies, and with tTMB-H and/or high bTMB (bTMB-H; ≥10 mut/Mb).31 32 We explored the genomic landscape of tumor samples from 1,954 patients with measurable TMB who were pre-screened for enrollment into CheckMate 848. With over 100 disease ontologies included in this study, this genomic data set represents one of the largest to date for a study of this kind. The correlation between tTMB and bTMB was evaluated, and biological factors that may influence the correlation, including disease ontology, variant clonality, and MSAF, were investigated.
Methods
Trial design
CheckMate 848 (NCT03668119) is a prospective, open-label phase 2 study in which immuno-oncology-naïve patients with advanced, metastatic, or unresectable solid tumors refractory to standard therapies or who lacked an available standard treatment and were tTMB-H or bTMB-H were randomized 2:1 to receive nivolumab plus ipilimumab or nivolumab monotherapy.32 These patients may have received multiple lines of non-immunotherapy prior to screening. Patients with melanoma, NSCLC, renal cell carcinoma, or hematological malignancy as the primary site of disease were excluded from the study. In addition, patients who had received prior treatment with ICIs were excluded, as were patients who had received chemotherapy, radiation therapy, biologics for cancer, or investigational therapy within 28 days of first administration of study treatment. Patients in the nivolumab plus ipilimumab arm received 240 mg nivolumab every 2 weeks (Q2W) plus 1 mg/kg of ipilimumab every 6 weeks (Q6W). Those in the nivolumab monotherapy arm received 480 mg of nivolumab every 4 weeks; however, on progression, patients could opt for nivolumab 240 mg Q2W plus ipilimumab 1 mg/kg Q6W. Response was evaluated according to Response Evaluation Criteria in Solid Tumors (RECIST) v.1.1. Best overall response (BOR) was classified as complete response (CR), partial response (PR), stable disease (SD), progressive disease (PD), or not evaluable for response (NE). Clinical endpoints from CheckMate 848, including the primary endpoint of objective response rate, and secondary endpoints of OS, and progression-free survival, have been presented elsewhere.32 Data from 1,954 pre-screened patients from CheckMate 848, including over 100 screened disease ontologies (taken from the tissue diagnosis), were included in this analysis.
Genomic profiling from tumor and blood samples
Both fresh and archival (<9 months) formalin-fixed, paraffin-embedded (FFPE) tissue samples were included in the study. Tissue-based analyses were performed using the FoundationOne CDx-based Clinical Trial Assay (Foundation Medicine, Cambridge, Massachusetts, USA), which features the hybridization-based capture and next-generation sequencing (NGS) of all exons in 324 cancer-related genes. tTMB was determined by quantification of all coding single nucleotide variants (SNVs) and small insertions and deletions (indels) present at an allele frequency of ≥5%. Blood-based analyses were performed on plasma samples using the Foundation Medicine bTMB Clinical Trial Assay (Foundation Medicine)17 which uses hybridization-based capture technology to report tumor-derived SNVs of 394 cancer-related genes. bTMB was determined by quantification of somatic base substitutions down to an allele frequency of 0.5%.17
Error-corrected reads were aligned to the hg19 reference genome,33 and base substitutions were called, while germline variants and oncogenic driver mutations were removed. The final outputs are the bTMB score and the MSAF, an estimate of the tumor fraction. Detailed methods are described in Gandara et al.17
To determine microsatellite instability (MSI) status, a fraction-based (FB) MSI algorithm was used to categorize a tumor specimen as MSI-high (MSI-H) or microsatellite stable (MSS) based on a genome-wide analysis across >2,000 microsatellite loci using the FoundationOne CDx assay. MSI status was considered as MSI-H for samples with FB-MSI scores ≥0.0124, and MSS for samples with FB-MSI scores ≤0.0041. For samples with FB-MSI scores >0.0041 and <0.0124, an MSI result of “Cannot be determined” was reported.
Quality control criteria
Pre- and post-sequencing quality control (QC) steps were performed for the analysis of tTMB and bTMB. Prior to conducting the analysis using the FoundationOne CDx-based Clinical Trial Assay, tissue samples with sufficient tumor content (20% tumor nuclei) and 0.6 mm3 volume were identified, and extracted DNA was quantified. Post-sequencing QC steps were used to identify samples with sufficient sequencing coverage, low error rates, and low levels of contamination.
Prior to performing analysis using the Foundation Medicine bTMB Clinical Trial Assay, plasma samples were screened for sufficient volume, and ctDNA was quantified. Post sequencing, samples were screened for adequate sequencing coverage.
Analysis of programmed death ligand 1 expression
Programmed death ligand 1 (PD-L1) expression was assessed manually as the percentage of PD-L1-positive tumor cells (%) using the Dako/Agilent PD-L1 immunohistochemistry 28–8 pharmDx assay (Agilent, Santa Clara, California, USA). Tissue processing and PD-L1 staining were performed following protocols optimized and approved for FFPE non-squamous NSCLC samples.
Statistical analyses
Correlations between tTMB and bTMB were assessed using Spearman correlation coefficients (r). Additional correlations were performed in which sample pairs were stratified by MSAF, using various cut-offs, including ≥10%, a cut-off that was previously associated with greater sensitivity of liquid biopsies to detect actionable variants in several malignancies.34 Agreement in assessment of TMB status between tTMB and bTMB assays was evaluated using the prespecified cut-off of ≥10 mut/Mb to define high tumor mutational burden (TMB-H) and <10 mut/Mb to define low TMB. Concordance between tTMB and bTMB assays was calculated as positive percentage agreement (PPA), negative percentage agreement (NPA), and overall percentage agreement (OPA). Further details on the Methods and Statistical Analyses can be found in the protocol (online supplemental file 2)
jitc-2023-007339supp002.pdf (10.1MB, pdf)
Results
Tumor and blood sample characteristics
A total of 1,438 and 1,720 unique tissue and blood samples, respectively, were obtained from 1,954 patients pre-screened for enrollment into CheckMate 848 (online supplemental figure S1). For each patient, only one tissue and one blood sample were included in the study. A total of 1,279 tissue and 1,606 blood samples passed QC steps for sequencing. Reportable data were obtained from 1,141 tissue and 1,573 plasma samples, resulting in ascertainment rates of 89.2% for tTMB and 97.9% for bTMB. The most common reasons for tissue sample failure were low percentage of tumor nuclei, insufficient sample volume, low DNA yield, and failure to meet NGS quality control metrics. Low DNA yield and failure to meet NGS QC metrics were the most common reasons for blood sample failure.
jitc-2023-007339supp001.pdf (192.7KB, pdf)
The analysis cohort comprised 1,017 patients who had available paired tTMB and bTMB data. Of over 100 screened disease ontologies in this cohort, the most prevalent ontologies were pancreatic (9.7%), breast (8.8%), and ovarian (6.1%) (online supplemental table S1). Among the 1,017 evaluable samples, the median tTMB was 3.8 mut/Mb (IQR, 2.5–6.3), and the median bTMB was 3.5 mut/Mb (IQR, 0.9–7.9) (figure 1).
Figure 1.
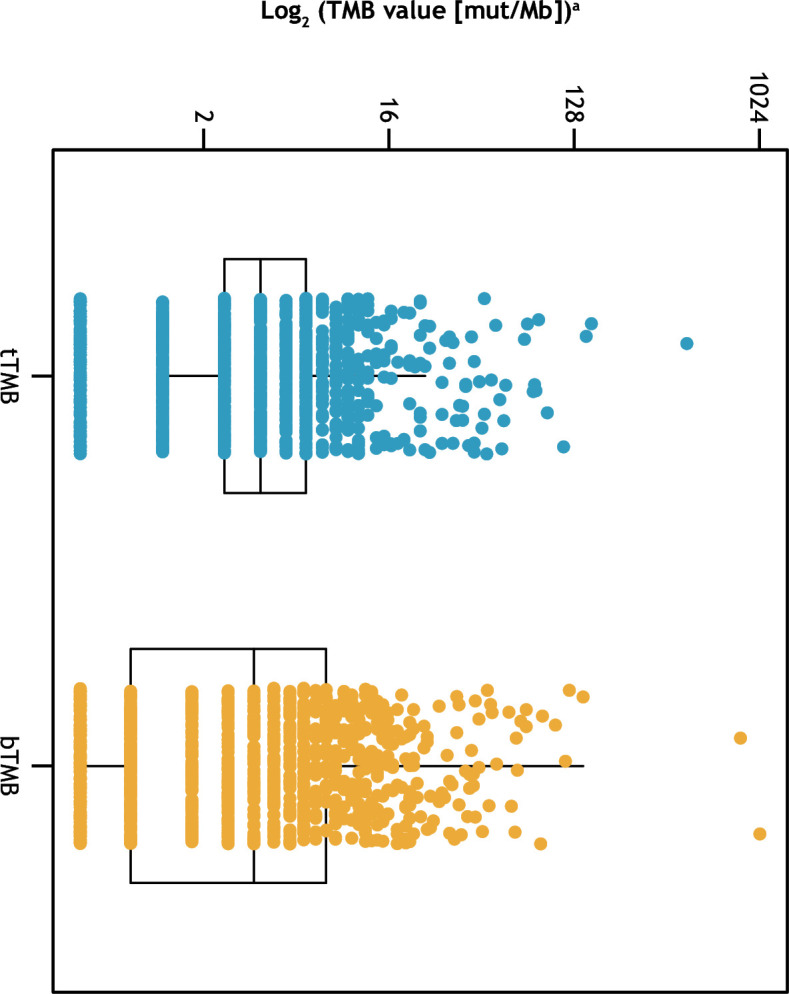
Distribution of tTMB and bTMB. Each box shows the IQR, and horizontal lines represent median values. Whiskers represent values 1.5× the upper and lower limits of the IQR. aTo accommodate TMB values of 0 for taking logarithms, 0.5 was added to all TMB values prior to taking the logarithm, for plotting purposes. bTMB, blood tumor mutational burden; mut/Mb, mutations/megabase; TMB, tumor mutational burden; tTMB, tissue tumor mutational burden.
Correlations between tTMB and bTMB
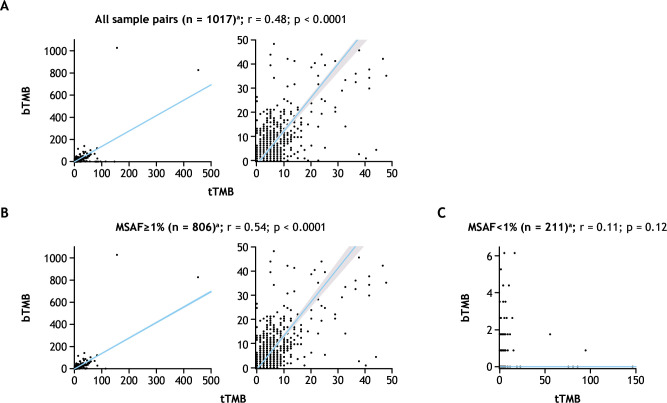
A Spearman correlation coefficient (r) of 0.48 was observed for tTMB versus bTMB scores across all 1,017 unique tissue and plasma sample pairs (p<0.0001) (figure 2A). To understand the impact of factors such as DNA shedding, we used MSAF, which has previously been demonstrated to correlate with rates of ctDNA shedding.17 MSAF metrics were derived from somatic mutations, including driver mutations detected from blood samples. Correlations were determined using various cut-offs for MSAF. Samples with MSAF<1%, ≥1% to <10%, and ≥10% had correlations of 0.11, 0.42 and 0.63, respectively (online supplemental figure S2A). A continuous increase in the correlation coefficient between tTMB and bTMB was observed with increasing MSAF with incremental cut-offs between 1% and 15% (online supplemental figure S2B). Among the 806 samples with MSAF≥1% (79.3% of 1,017 evaluable sample pairs), the correlation coefficient was 0.54 (p<0.0001), while no correlation was observed among the 211 samples with MSAF<1% (r=0.11; p=0.12; figure 2B).
Figure 2.
Correlation of bTMB and tTMB for (A) all sample pairs; (B) sample pairs with MSAF≥1%; or (C) sample pairs with MSAF<1%. aRight-hand panel shows enlargement of the left-hand panel. Shaded area around the regression line in the right-hand panel shows the 95% CIs for Passing–Bablok regression analysis. bTMB, blood tumor mutational burden; MSAF, maximum somatic allele frequency; r, Spearman’s correlation coefficient; tTMB, tissue tumor mutational burden.
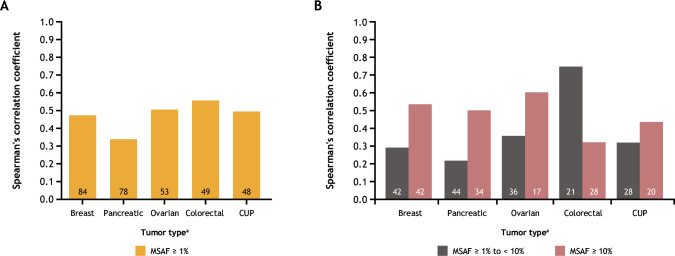
The effect of increasing MSAF on correlation between tTMB and bTMB was examined in several disease ontologies. Correlations between tTMB and bTMB varied across a range of MSAF cut-offs (≥1%, ≥1% to <10%, and ≥10%) (figure 3). For breast, pancreatic, ovarian, and colorectal tumors and for carcinoma of unknown primary site of disease (CUP) with MSAF≥1%, correlation coefficients ranged from 0.34 to 0.55 (figure 3A). Correlation coefficients were higher for samples with MSAF≥10% than for those with MSAF≥1% to <10% among breast (r=0.53 vs 0.29), pancreatic (r=0.50 vs 0.21), and ovarian (r=0.60 vs 0.36) cancers, and in CUP (r=0.43 vs 0.32) (figure 3B). However, for colorectal cancer samples, the correlation was highest (r=0.75) in samples with MSAF≥1% to <10%.
Figure 3.
Correlations between tTMB and bTMB by disease ontology for (A) samples with MSAF≥1% and for (B) samples with MSAF≥1% to <10% versus ≥10%. Number of sample pairs is shown inside each bar. aData are shown for the top five most frequent disease ontologies among samples with MSAF≥1%. bTMB, blood tumor mutational burden; CUP, carcinoma of unknown primary; MSAF, maximum somatic allele frequency; tTMB, tissue tumor mutational burden.
Evaluation of assay agreement
Among 1,017 evaluable pairs, 161 (15.8%) and 196 (19.3%) were tTMB-H and bTMB-H, respectively. PPA, NPA, and OPA between assays were 60%, 88%, and 84%, respectively. Among the 806 sample pairs with MSAF≥1%, PPA was improved to 66% (table 1); however, there was a slight decrease in NPA and OPA to 85% and 81%, respectively.
Table 1.
Assessment of agreement on TMB status between tTMB and bTMB assays
| All evaluable pairs (n=1,017) | MSAF≥1% (n=806) | |||
| tTMB≥10 mut/Mb | tTMB<10 mut/Mb | tTMB≥10 mut/Mb | tTMB<10 mut/MB | |
| bTMB≥10 mut/Mb | 96 | 100 | 96 | 100 |
| bTMB<10 mut/Mb | 65 | 756 | 50 | 560 |
| PPA, % | 60 | 66 | ||
bTMB, blood tumor mutational burden; MSAF, maximum somatic allele frequency; mut/Mb, mutations/megabase; PPA, positive percentage agreement; TMB, tumor mutational burden; tTMB, tissue tumor mutational burden.
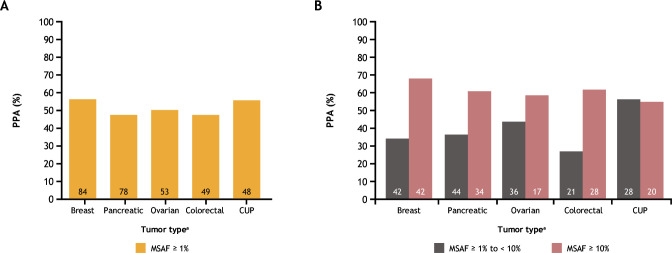
Agreement in TMB status determined by tissue-based and blood-based assays varied by disease ontology and by MSAF. For breast, pancreatic, ovarian, and colorectal cancer sample pairs with MSAF≥1%, PPA values ranged from approximately 48% to 56% (figure 4A). For sample pairs with MSAF≥1% to <10%, PPA varied from approximately 27% to 55%, and the highest concordance among these disease ontologies was observed when MSAF was ≥10% (PPA ranging from approximately 55% to 68%) (figure 4B).
Figure 4.
Assessment of TMB status between tissue-based and blood-based assays by disease ontology, for (A) samples with MSAF≥1% and (B) samples with MSAF≥1% to <10% versus ≥10%. Number of sample pairs is shown inside each bar. aData are shown for the top five most frequent disease ontologies among samples with MSAF≥1%. CUP, carcinoma of unknown primary; MSAF, maximum somatic allele frequency; PPA, positive percentage agreement; TMB, tumor mutational burden.
Comparison of assays for genomic variant detection
To examine how genetic variants contribute to tTMB and bTMB agreement, genomic variant detection was evaluated across 43 sample pairs that had tTMB and bTMB≥20 mut/Mb, as well as MSAF≥1%. These cutoffs were selected to ensure a sufficient number of somatic variants per sample pair for accurate comparison by tissue-based and blood-based assays. tTMB and bTMB assays detected overlapping, but not completely identical, sets of variants (online supplemental figure S3A). In most samples, the variants that contributed to tTMB and bTMB were highly concordant; however, there was a small subset of paired samples showing greater unique variants. In general, more unique variants were identified using the bTMB assay. To understand this further, the clonality of bTMB variants was investigated according to whether the variant was shared with tTMB versus being unique to bTMB across blood samples with MSAF≥1%. The majority of unique bTMB variants in blood samples were subclonal (online supplemental figure S3B, table S2).
The prevalence of MSI-H samples was low, with MSI-H detected in only 25 (2.5%) of 996 MSI-evaluable tissue samples. Among samples with MSI-H, the median tTMB was 25.2 mut/Mb.
Evaluation of assay agreement by treatment response
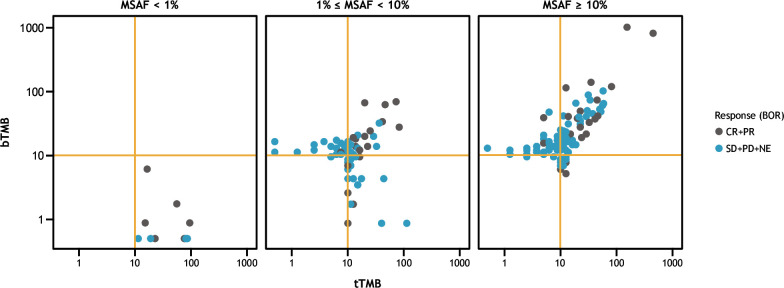
Of 1,954 patients screened for enrollment in CheckMate 848, 212 with tTMB-H and/or bTMB-H status were randomized to receive nivolumab plus ipilimumab or nivolumab monotherapy, with a cap of 15% per disease ontology. Samples from both treatment arms were pooled for analysis. Concordance between tTMB and bTMB was observed in both responders (patients with BOR of CR+PR) and nonresponders (BOR of SD+PD+NE) to ICI therapy (figure 5). Among both responders and nonresponders, concordance was improved in patients with MSAF≥10% compared with other MSAF subgroups (<1% and ≥1% to <10%).
Figure 5.
Concordance between tTMB and bTMB status among responders and non-responders to ICI therapy.a aPatients with either tTMB-H or bTMB-H status who were randomized in the CheckMate 848 trial. Samples were pooled across the nivolumab+ipilimumab and nivolumab monotherapy treatment arms. BOR, best overall response; bTMB, blood tumor mutational burden; bTMB-H, high blood tumor mutational burden; CR, complete response; ICI, immune checkpoint inhibitor; MSAF, maximum somatic allele frequency; NE, not evaluable (for response); PD, progressive disease; PR, partial response; SD, stable disease; tTMB, tissue tumor mutational burden; tTMB-H, high tissue tumor mutational burden.
Discussion
In CheckMate 848, we found that the distributions of tTMB and bTMB scores were comparable and moderately correlated, and that the correlation increased with increasing ctDNA levels inferred from MSAF. In addition, a correlation between tTMB and bTMB was observed for both responders and non-responders to ICI therapy. These findings suggest that bTMB is a pragmatic, minimally invasive tool for assessing tTMB levels in patients with adequate levels of ctDNA shedding. Although analyses performed in this study were limited by low numbers of patients for each individual disease ontology, correlations between tTMB and bTMB were greatest among the most common disease ontologies, particularly when MSAF was elevated. These results are particularly interesting given that NSCLC, for which previous studies have shown a correlation between bTMB and tTMB and an association between bTMB and ICI efficacy,21 was excluded from this study, lending further evidence for the correlation of bTMB and tTMB across cancer types. Longitudinal monitoring and quantification of ctDNA levels in samples used for bTMB assessment could potentially be used to guide adaptive disease-specific treatment decisions.
Developing bTMB in correlation with tTMB offers several advantages, including (1) the ability to compare bTMB performance against the established clinical utility of tTMB for ICI treatment in the pan-tumor setting; and (2) the availability of analytically robust algorithms for tTMB assessment that take important factors into account, such as driver mutations, germline variants, and the size of the target region. The correlations between bTMB and tTMB observed in this study were moderately strong and comparable with those exhibited by other commercially available or investigational assays.24 30 35 A previous study found that the most important factors affecting concordance between tTMB and bTMB were low MSAF and longer time between sample collections.17 Additionally, discordance between bTMB and tTMB is reportedly more common for the combination of bTMB-H/low tTMB than for low bTMB/tTMB-H.30 Taken together, these results suggest that identification of the key biological and analytical factors in the bTMB algorithm will be crucial for enabling the use of this less invasive, blood-based approach to TMB assessment.
In addition to characterizing the impact of tumor shedding on bTMB across different disease ontologies, our study identified additional, biologically driven factors affecting bTMB assessment. For example, we observed tumor heterogeneity-introduced discordance when comparing DNA from a single metastatic tissue site with ctDNA samples comprising cell-free DNA shed from multiple tissue sites, and exhibiting subclonal variants. These subclonal variants may have accounted for some of the observed discordance between the tTMB assays and bTMB assays. Of importance, the bTMB assay used in this study was an early version developed for clinical trial use. Recent studies have described the prevalence of mutations associated with clonal hematopoiesis (CH), which are found in the general population and detectable in cell-free DNA.36 There is as yet no classification of CH mutations, and their impact on bTMB assessment is unknown.36 Detailed investigation is needed to characterize these CH variants and their role in the discordance between tTMB and bTMB assessment, and to improve the prediction of ICI treatment efficacy using bTMB.
A limitation of this study was the use of MSAF as a surrogate for plasma ctDNA levels. As MSAF is estimated from the variant with the highest allele frequency, which could potentially introduce bias into the analysis, there are opportunities to optimize quantification of the tumor fraction in plasma ctDNA in future studies. For example, improvement of clonal hematopoiesis of indeterminate potential (CHIP) variant filtering may enhance tumor purity estimation based on robust copy number modeling. An additional limitation is that the study did not evaluate whether a higher rate of response among patients with low MSAF resulted from lower tumor shedding rates.
Together, our findings suggest that distinct biological and analytical factors impact TMB measurement in plasma ctDNA samples. Failure to address these confounding factors may result in inflated estimation of bTMB and higher discordance between tTMB and bTMB assays.30 In future investigations, these biological and analytical factors should be integrated into the calculation of bTMB in order to define robust clinical cut-offs. Moreover, other biological factors affecting bTMB assessment, along with subsequent patient response to nivolumab with or without ipilimumab therapy, require further investigation.
In conclusion, tTMB and bTMB are strongly correlated in patients with advanced, metastatic, or unresectable solid tumors that are refractory to standard therapies, particularly when ctDNA is elevated. In addition, the correlation between tTMB and bTMB appears to span many disease ontologies. Further interrogation of the biological and analytical factors affecting tumor-derived and blood-derived genomic profiling is warranted to support implementation of this technique in clinical settings.
Acknowledgments
The authors wish to thank the patients and families who made this study possible and the clinical teams who participated. We also thank Bristol Myers Squibb (Princeton, New Jersey, USA) and Ono Pharmaceutical Company Ltd. (Osaka, Japan). Medical writing support and editorial assistance were provided by Sandra J Page, PhD, and Agata Shodeke, PhD, of Spark Medica, according to Good Publication Practice guidelines, funded by Bristol Myers Squibb.
Footnotes
Contributors: JH, DF, and JB contributed to the design of the study. NK contributed to the acquisition of the data set. JH, NK, DCP, EME, JP, and JB contributed to the analysis of the data, and JH, NK, PD, LAA, EME, GF, GG, JP, and JB contributed to their interpretation. All authors reviewed and revised the work and approved the final draft for submission. JH and JB are responsible for the overall content as guarantors.
Funding: The study was supported by Bristol Myers Squibb.
Competing interests: DF, DCP, EME, GRO, HT, JH, and LAA are employees of Foundation Medicine, Inc. and hold stock in Bristol Myers Squibb. GF, GG, and JP are employees of and hold stock in Bristol Myers Squibb. JB is an employee of and holds stock in Bristol Myers Squibb, and holds stock in Johnson and Johnson. NK holds stock in Bristol Myers Squibb. PD is a former employee of Bristol Myers Squibb.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data may be obtained from a third party and are not publicly available. BMS will honor legitimate requests for clinical trial data from qualified researchers with a clearly defined scientific objective. Data sharing requests will be considered for Phase II–IV interventional clinical trials that completed on or after January 1, 2008. In addition, primary results must have been published in peer-reviewed journals and the medicines or indications approved in the USA, EU, and other designated markets. Sharing is also subject to protection of patient privacy and respect for the patient’s informed consent. Data considered for sharing may include non-identifiable patient-level and study-level clinical trial data, full clinical study reports, and protocols. Requests to access clinical trial data may be submitted using the enquiry form at https://vivli.org/ourmember/bristol-myers-squibb/.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This was an international, Bristol Myers Squibb-led clinical trial with 60 study sites worldwide. Listing every international review board that approved the study at each location would therefore not be feasible. We currently have the following language in the manuscript: ‘This trial was approved by the institutional review board or ethics committee at each site, and was conducted according to Good Clinical Practice guidelines, defined by the International Conference on Harmonisation. The study was conducted in compliance with the protocol. The protocol and any amendments and the participant informed consent received approval/favorable opinion by the Institutional Review Board/Independent Ethics Committee (IRB/IEC), and regulatory authorities according to applicable local regulations prior to initiation of the study. Study locations can be found listed on ClinicalTrials.gov (https://classic.clinicaltrials.gov/ct2/show/study/NCT03668119). All the patients provided written informed consent that was based on the Declaration of Helsinki principles.’
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719–24. 10.1038/nature07943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chabanon RM, Pedrero M, Lefebvre C, et al. Mutational landscape and sensitivity to immune checkpoint blockers. Clin Cancer Res 2016;22:4309–21. 10.1158/1078-0432.CCR-16-0903 [DOI] [PubMed] [Google Scholar]
- 4.Merino DM, McShane LM, Fabrizio D, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer 2020;8:e000147. 10.1136/jitc-2019-000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353–65. 10.1016/S1470-2045(20)30445-9 [DOI] [PubMed] [Google Scholar]
- 6.Ready N, Hellmann MD, Awad MM, et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (Checkmate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol 2019;37:992–1000. 10.1200/JCO.18.01042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samstein RM, Lee C-H, Shoushtari AN, et al. Tumor mutational load predicts survival after Immunotherapy across multiple cancer types. Nat Genet 2019;51:202–6. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valero C, Lee M, Hoen D, et al. The association between tumor mutational burden and prognosis is dependent on treatment context. Nat Genet 2021;53:11–5. 10.1038/s41588-020-00752-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 2017;376:2415–26. 10.1056/NEJMoa1613493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodi FS, Wolchok JD, Schadendorf D, et al. TMB and inflammatory gene expression associated with clinical outcomes following immunotherapy in advanced melanoma. Cancer Immunol Res 2021;9:1202–13. 10.1158/2326-6066.CIR-20-0983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reck M, Schenker M, Lee KH, et al. Nivolumab plus ipilimumab versus chemotherapy as first-line treatment in advanced non-small-cell lung cancer with high tumour mutational burden: patient-reported outcomes results from the randomised, open-label, phase III Checkmate 227 trial. Eur J Cancer 2019;116:137–47. 10.1016/j.ejca.2019.05.008 [DOI] [PubMed] [Google Scholar]
- 12.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.US Food and Drug Administration . FDA approves pembrolizumab for adults and children with TMB-H solid tumors.[Online]. 2020. Available: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors [Accessed 7 Nov 2022].
- 14.Paz-Ares L, Ciuleanu T-E, Cobo M, et al. 98O first-line nivolumab (nivo) + ipilimumab (ipi) + 2 cycles chemotherapy (chemo) vs 4 cycles chemo in advanced non-small cell lung cancer (aNSCLC): association of blood and tissue tumor mutational burden (TMB) with efficacy in checkmate 9la. J Thorac Oncol 2021;16:S750–1. 10.1016/S1556-0864(21)01940-7 [DOI] [Google Scholar]
- 15.Fenizia F, Pasquale R, Roma C, et al. Measuring tumor mutation burden in non-small cell lung cancer: tissue versus liquid biopsy. Transl Lung Cancer Res 2018;7:668–77. 10.21037/tlcr.2018.09.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ossandon MR, Agrawal L, Bernhard EJ, et al. Circulating tumor DNA assays in clinical cancer research. J Natl Cancer Inst 2018;110:929–34. 10.1093/jnci/djy105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441–8. 10.1038/s41591-018-0134-3 [DOI] [PubMed] [Google Scholar]
- 18.Si H, Kuziora M, Quinn KJ, et al. A blood-based assay for assessment of tumor mutational burden in first-line metastatic NSCLC treatment: results from the MYSTIC study. Clin Cancer Res 2021;27:1631–40. 10.1158/1078-0432.CCR-20-3771 [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Duan J, Cai S, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol 2019;5:696–702. 10.1001/jamaoncol.2018.7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Li Q, Du Y, et al. Blood tumor mutational burden as a predictive biomarker in patients with advanced non-small cell lung cancer (NSCLC). Front Oncol 2021;11:640761. 10.3389/fonc.2021.640761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Fang L, Zhu Y, et al. Blood tumor mutation burden can predict the clinical response to immune checkpoint inhibitors in advanced non-small cell lung cancer patients. Cancer Immunol Immunother 2021;70:3513–24. 10.1007/s00262-021-02943-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen B, Fong C, Luthra A, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 2022;185:563–75.e11. 10.1016/j.cell.2022.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer. American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631–41. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Chang L, Yang Y, et al. The correlations of tumor mutational burden among single-region tissue, multi-region tissues and blood in non-small cell lung cancer. J Immunother Cancer 2019;7:98. 10.1186/s40425-019-0581-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baden J, Chang H, Greenawalt DM, et al. Comparison of platforms for determining tumor mutational burden (TMB) from blood samples in patients with non-small cell lung cancer (NSCLC). Ann Oncol 2019;30:v28. 10.1093/annonc/mdz239.010 [DOI] [Google Scholar]
- 26.Rizvi NA, Cho BC, Reinmuth N, et al. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the MYSTIC phase 3 randomized clinical trial. JAMA Oncol 2020;6:661–74. 10.1001/jamaoncol.2020.0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinn K, Helman E, Nance T, et al. Development and analytical validation of a plasma-based tumor mutational burden (TMB) score from next-generation sequencing panels. Ann Oncol 2018;29:viii41. 10.1093/annonc/mdy269.129 [DOI] [Google Scholar]
- 28.Jiang T, Zhang S, Jager A, et al. Accurate measurement of tumor mutation burden in liquid biopsy (bTMB) using a 500 gene panel. Ann Oncol 2018;29:viii51. 10.1093/annonc/mdy269.161 [DOI] [Google Scholar]
- 29.Baden JF, Sausen M, Kalinava N, et al. Concordance of tissue- and plasma-derived genomic profiling in Checkmate 9LA, using the FoundationOne® Cdx and GuardantOMNI® assays. J Clin Oncol 2021;39:9010. 10.1200/JCO.2021.39.15_suppl.9010 [DOI] [Google Scholar]
- 30.Sturgill EG, Misch A, Jones CC, et al. Discordance in tumor mutation burden from blood and tissue affects association with response to immune checkpoint inhibition in real-world settings. Oncologist 2022;27:175–82. 10.1093/oncolo/oyab064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J, Kalinava N, Doshi P, et al. Abstract 2139: evaluation of Tissue- and plasma-derived tumor mutational burden and Genomic alterations of interest from the Checkmate 848 clinical trial. Cancer Res 2022;82:2139. 10.1158/1538-7445.AM2022-2139 [DOI] [Google Scholar]
- 32.Schenker M, Burotto M, Richardet M, et al. Checkmate 848: a randomized, open-label, phase 2 study of Nivolumab in combination with Ipilimumab or Nivolumab monotherapy in patients with advanced or metastatic solid tumors of high tumor mutational burden. Cancer Res 2022;82:CT022. 10.1158/1538-7445.AM2022-CT022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genome Reference Consortium . Genome Reference Consortium Human Build 37 (GRCh37) [online]. 2009: GRCh37. [Google Scholar]
- 34.Husain H, Pavlick DC, Fendler BJ, et al. Tumor fraction correlates with detection of actionable variants across > 23,000 circulating tumor DNA samples. JCO Precis Oncol 2022;6:e2200261. 10.1200/PO.22.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuurbiers M, Huang Z, Saelee S, et al. Biological and technical factors in the assessment of blood-based tumor mutational burden (bTMB) in patients with NSCLC. J Immunother Cancer 2022;10:e004064. 10.1136/jitc-2021-004064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan HT, Chin YM, Nakamura Y, et al. Clonal hematopoiesis in liquid biopsy: from biological noise to valuable clinical implications. Cancers (Basel) 2020;12:2277. 10.3390/cancers12082277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007339supp002.pdf (10.1MB, pdf)
jitc-2023-007339supp001.pdf (192.7KB, pdf)
Data Availability Statement
Data may be obtained from a third party and are not publicly available. BMS will honor legitimate requests for clinical trial data from qualified researchers with a clearly defined scientific objective. Data sharing requests will be considered for Phase II–IV interventional clinical trials that completed on or after January 1, 2008. In addition, primary results must have been published in peer-reviewed journals and the medicines or indications approved in the USA, EU, and other designated markets. Sharing is also subject to protection of patient privacy and respect for the patient’s informed consent. Data considered for sharing may include non-identifiable patient-level and study-level clinical trial data, full clinical study reports, and protocols. Requests to access clinical trial data may be submitted using the enquiry form at https://vivli.org/ourmember/bristol-myers-squibb/.