Abstract
In search of novel breast cancer (BC) risk variants, we performed a whole-exome sequencing and variant analysis of 69 Finnish BC patients as well as analysed loss-of-function variants identified in DNA repair genes in the Finns from the Genome Aggregation Database. Additionally, we carried out a validation study of SERPINA3 c.918-1G>C, recently suggested for BC predisposition. We estimated the frequencies of 41 rare candidate variants in 38 genes by genotyping them in 2482–4101 BC patients and in 1273–3985 controls. We further evaluated all coding variants in the candidate genes in a dataset of 18,786 BC patients and 182,927 controls from FinnGen. None of the variants associated significantly with cancer risk in the primary BC series; however, in the FinnGen data, NTHL1 c.244C>T p.(Gln82Ter) associated with BC with a high risk for homozygous (OR = 44.7 [95% CI 6.90–290], P = 6.7 × 10–5) and a low risk for heterozygous women (OR = 1.39 [1.18–1.64], P = 7.8 × 10–5). Furthermore, the results suggested a high risk of colorectal, urinary tract, and basal-cell skin cancer for homozygous individuals, supporting NTHL1 as a recessive multi-tumour susceptibility gene. No significant association with BC risk was detected for SERPINA3 or any other evaluated gene.
Subject terms: Cancer, Genetics
Introduction
Cancer is a genetic disease in which accumulating pathogenic variants give growth advantage to malignant cells. Eukaryotic cells have specialized pathways for the repair of different mutation types and others that control the cell cycle checkpoints or initiate apoptosis. Defective DNA damage response mechanisms increase genomic instability and may lead to tumour development1.
The validated breast cancer (BC) risk genes to date function primarily in DNA double-strand break and interstrand crosslink repair via the homologous recombination and the Fanconi anaemia (FA) pathways and in DNA damage checkpoint signalling2,3. The high-penetrance BC risk genes, BRCA1 and BRCA2, encode proteins at the core of the pathways, promoting DNA repair in response to damage signalling2. The validated moderate-to-high risk BC predisposition genes, PALB2, CHEK2, ATM, BARD1, RAD51C, and RAD51D, have functions linked to BRCA1 and BRCA22,3. Studies on hereditary BC risk have most often focused on the DNA damage response genes. Other pathways may also be involved in the BC risk predisposition; for example, the syndromic cancer genes and the low-penetrance variants associated with BC risk show a wide range of affected pathways2–4.
The high- and moderate-risk variants in the established BC predisposition genes have an autosomal dominant inheritance pattern, even if with incomplete penetrance. Recessive model has also been suggested for increased risk of BC5, but to date, no recessive high- or moderate-risk BC susceptibility gene has been validated. Recently, several BC patients with pathogenic biallelic NTHL1 variants have been described6–12, indicating recessive BC predisposition. Pathogenic variants in the NTHL1 gene have been determined to cause a recessive multi-tumour syndrome, which is characterized especially by adenomatous polyposis and colorectal cancer (CRC), and with accumulating evidence, BC in women6–13.
The genes and causal variants contributing to a large proportion of the hereditary BC risk are yet to be discovered4. The genetic bottleneck events in the Finnish population have resulted in less overall variation and a higher frequency of loss-of-function (LoF) variants, including recessive disease variants, in the Finns compared to other Europeans14,15. This founder effect present in the Finns is advantageous for genetic research as it facilitates the detection of novel disease genes and variants. Only a few recurrent variants account for most of the pathogenic burden in the validated BC risk genes in Finnish BC patients16. High-risk BRCA1/2 variants have been identified in about 21% of Finnish BC families and 1.8% of unselected BC patients16–18. The combined frequency of pathogenic variants in the other validated high- and moderate-risk BC susceptibility genes is about 10% in Finnish BC families and 5% in unselected BC patients16.
With the aim of identifying novel BC risk variants, we have performed a whole-exome sequencing (WES) and variant analysis of 69 patients from Finnish BC families as well as an analysis of predicted loss-of-function (pLoF) variants in 520 DNA repair genes, detected in approximately 11,000 Finns from the Genome Aggregation Database (gnomAD), and selected candidate risk variants for a case–control study. Additionally, a recent Finnish study reported a putative novel moderate-risk BC susceptibility variant SERPINA3 c.918-1G>C19, warranting further validation. Here, we evaluated SERPINA3 c.918-1G>C alongside the other candidate variants for BC risk.
Results
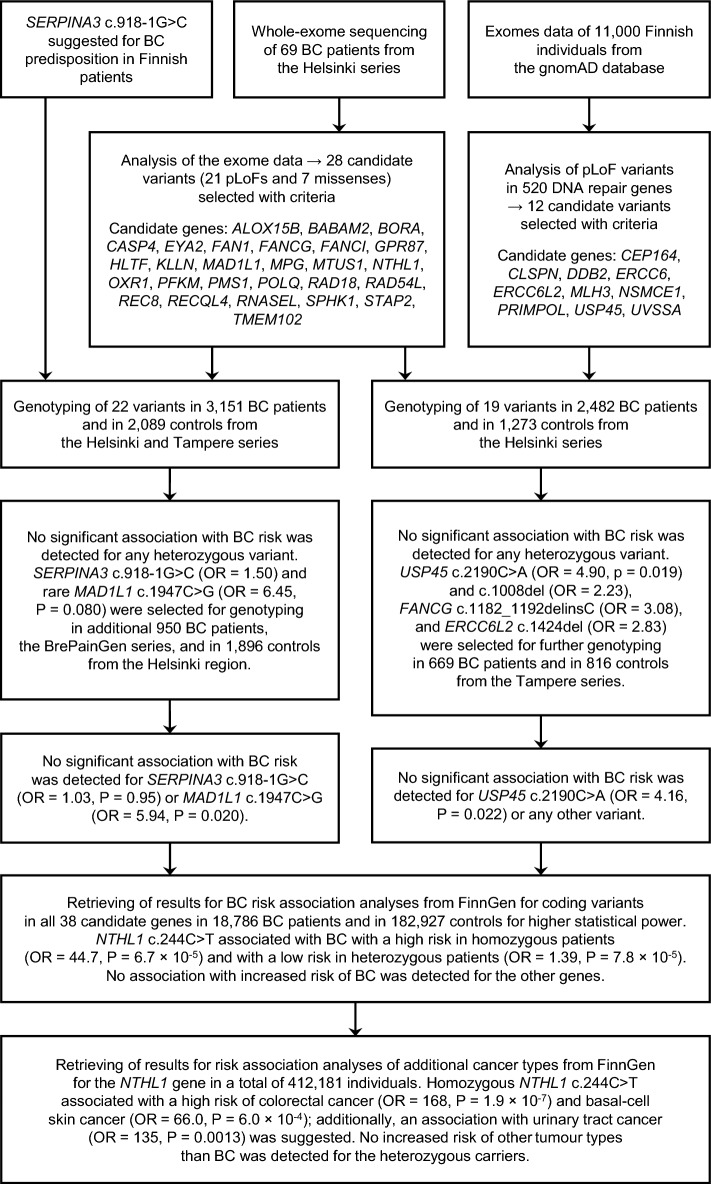
We selected altogether 41 candidate variants in 38 genes, presented in detail in the Supplementary Table S1, for genotyping in BC patients and controls from the Helsinki and Tampere regions in Southern Finland and assessed the variants for cancer risk (Fig. 1). Finally, we retrieved the data for cancer risk association analyses from the FinnGen project and examined the candidate genes and variants in this large series of cancer patients and controls.
Figure 1.
An overview of the work process and findings of the study.
Breast cancer risk association analyses in the Helsinki and Tampere series
We genotyped 19 of the selected candidate variants in 2482 BC patients and 1273 controls, 20 of the variants in 3151 BC patients and 2089 controls, and two of the variants in 4101 BC patients and 3985 controls from the Helsinki and Tampere regions. After the Bonferroni correction for multiple comparisons (P < 0.0012), none of the studied variants associated significantly with BC risk in this primary study (Table 1, Supplementary Table S2). We detected two variants, MAD1L1 NM_001013836.2:c.1947C>G p.(Tyr649Ter) and USP45 NM_001346022.3:c.2190C>A p.(Tyr730Ter), with a higher frequency in the patients than in the controls on a nominally significant level (P < 0.05) (Table 1); however, another pLoF in the USP45 gene, NM_001346022.3:c.1008del p.(Val337SerfsTer9), was found only slightly more often in the patients than in the controls.
Table 1.
Variant frequencies in breast cancer patients from the Helsinki and Tampere regions.
| Variant | Group | Total | Carriers | % | OR | 95% CI | P value |
|---|---|---|---|---|---|---|---|
| MAD1L1 | Controls | 3974 | 2 | 0.05 | |||
| c.1947C>G | All BC | 4083 | 12 | 0.3 | 5.94 | 1.62–38.2 | 0.020 |
| p.(Tyr649Ter) | Familial BC | 1524 | 6 | 0.4 | 8.13 | 1.86–55.7 | 0.011 |
| rs121908981 | Unselected BC | 3327 | 8 | 0.2 | 4.81 | 1.20–31.9 | 0.047 |
| ER-positive BC | 3172 | 10 | 0.3 | 6.46 | 1.70–42.1 | 0.016 | |
| ER-negative BC | 720 | 2 | 0.3 | 5.61 | 0.67–46.9 | 0.085 | |
| SERPINA3 | Controls | 3978 | 10 | 0.3 | |||
| c.918-1G>C | All BC | 4095 | 11 | 0.3 | 1.03 | 0.43–2.47 | 0.95 |
| rs199710314 | Familial BC | 1527 | 3 | 0.2 | 0.75 | 0.17–2.46 | 0.66 |
| Unselected BC | 3339 | 9 | 0.3 | 1.07 | 0.42–2.65 | 0.89 | |
| ER-positive BC | 3184 | 9 | 0.3 | 1.08 | 0.43–2.69 | 0.86 | |
| ER-negative BC | 720 | 1 | 0.1 | 0.54 | 0.03–2.83 | 0.56 | |
| ERCC6L2 | Controls | 2083 | 12 | 0.6 | |||
| c.1424del | All BC | 3142 | 12 | 0.4 | 0.76 | 0.33–1.75 | 0.51 |
| p.(Ile475ThrfsTer36) | Familial BC | 1369 | 6 | 0.4 | 1.00 | 0.34–2.68 | 1.00 |
| rs768081343 | Unselected BC | 2386 | 8 | 0.3 | 0.64 | 0.25–1.57 | 0.34 |
| ER-positive BC | 2386 | 10 | 0.4 | 0.86 | 0.35–2.03 | 0.72 | |
| ER-negative BC | 573 | 1 | 0.2 | 0.42 | 0.02–2.20 | 0.41 | |
| FANCG | Controls | 2086 | 1 | 0.05 | |||
| c.1182_1192delinsC | All BC | 3147 | 6 | 0.2 | 3.08 | 0.53–58.2 | 0.30 |
| p.(Glu395TrpfsTer5) | Familial BC | 1368 | 4 | 0.3 | 4.49 | 0.66–87.9 | 0.18 |
| rs397507559 | Unselected BC | 2391 | 3 | 0.1 | 2.21 | 0.28–44.8 | 0.49 |
| ER-positive BC | 2389 | 5 | 0.2 | 3.39 | 0.55–65.0 | 0.27 | |
| ER-negative BC | 576 | 1 | 0.2 | 2.82 | 0.11–71.4 | 0.46 | |
| NTHL1 | Controls | 2081 | 17 | 0.8 | |||
| c.244C>T | All BC | 3117 | 30 | 1.0 | 1.35 | 0.74–2.54 | 0.33 |
| p.(Gln82Ter) | Familial BC | 1357 | 16 | 1.2 | 1.77 | 0.86–3.64 | 0.12 |
| rs150766139 | Unselected BC | 2370 | 22 | 0.9 | 1.25 | 0.66–2.41 | 0.49 |
| ER-positive BC | 2366 | 17 | 0.7 | 0.99 | 0.49–1.99 | 0.98 | |
| ER-negative BC | 573 | 9 | 1.6 | 2.32 | 0.97–5.21 | 0.048 | |
| USP45 | Controls | 2088 | 3 | 0.1 | |||
| c.2190C>A | All BC | 3148 | 21 | 0.7 | 4.16 | 1.42–17.7 | 0.022 |
| p.(Tyr730Ter) | Familial BC | 1368 | 12 | 0.9 | 5.49 | 1.70–24.6 | 0.0097 |
| rs118066385 | Unselected BC | 2392 | 17 | 0.7 | 4.59 | 1.53–19.7 | 0.015 |
| ER-positive BC | 2389 | 17 | 0.7 | 4.31 | 1.43–18.6 | 0.021 | |
| ER-negative BC | 576 | 2 | 0.3 | 2.59 | 0.33–16.2 | 0.31 | |
| USP45 | Controls | 2086 | 6 | 0.3 | |||
| c.1008del | All BC | 3148 | 13 | 0.4 | 1.29 | 0.50–3.73 | 0.61 |
| p.(Val337SerfsTer9) | Familial BC | 1369 | 8 | 0.6 | 1.92 | 0.65–6.01 | 0.24 |
| rs554927779 | Unselected BC | 2392 | 9 | 0.4 | 1.23 | 0.44–3.69 | 0.70 |
| ER-positive BC | 2390 | 11 | 0.5 | 1.46 | 0.55–4.30 | 0.46 | |
| ER-negative BC | 575 | 2 | 0.3 | 1.24 | 0.18–5.53 | 0.80 |
The familial and the unselected patient groups overlap: 775 patients were included in both groups in the analyses of the MAD1L1 and SERPINA3 variants and 614 in the analyses of the other variants. Two of the NTHL1 c.244C>T carriers included in the analysis were homozygous.
FANCG NM_004629.2:c.1182_1192delinsC p.(Glu395TrpfsTer5), NTHL1 NM_002528.7:c.244C > T p.(Gln82Ter) (also known as NM_002528.6:c.268C > T p.(Gln90Ter) in reference to the previous transcript version), and ERCC6L2 NM_020207.7:c.1424del p.(Ile475ThrfsTer36) (previously denoted as NM_020207.5:c.1457del p.(Ile486ThrfsTer36)) have been identified to cause recessive hereditary diseases with increased risk of cancer6,9,20–23. Here, we detected no significant association between the heterozygous pLoFs and BC risk. Of note, FANCG c.1182_1192delinsC was very rare in our patient series and only detected in 0.2% (6/3147) of the patients and in 0.05% (1/2086) of the controls. Only two patients were homozygous for the NTHL1 c.244C > T variant, and we were unable to study any recessive BC risk associated with NTHL1 in our patient series. No study subject was homozygous for ERCC6L2 c.1424del.
We found SERPINA3 NM_001085.5:c.918-1G>C with a similar frequency in the patients and in the controls and detected no association between the variant and BC risk. Previously, the c.918-1G>C carriers were reported to have a medullary breast tumour type more often than noncarriers19. Here, no c.918-1G>C carrier had medullary BC: eight patients had ductal, one patient had lobular, and two patients had carcinomas of mixed type.
The other studied variants were either detected only in a few patients or the analyses did not suggest an increased risk of BC (Supplementary Table S2).
Breast cancer risk association analyses from FinnGen
To further evaluate the candidate genes and variants in a dataset with higher statistical power, we retrieved the results for BC risk association analyses from the FinnGen study, data release 10, for all coding variants in the studied genes in 18,786 Finnish BC patients and in 182,927 controls15,24. The FinnGen data also provided recessive association analyses for NTHL1 c.244C>T and ERCC6L2 c.1424del, which we were unable to perform in the Helsinki and Tampere BC series.
The genotype data suggested a low increased risk of BC for heterozygous NTHL1 c.244C>T carriers in the additive model (odds ratio (OR) = 1.39 [95% confidence interval (CI) 1.18–1.64], P = 7.8 × 10–5) (Tables 2, 3). Carriers were detected with a similar frequency in the oestrogen receptor (ER)-positive patient group (OR = 1.41 [1.14–1.73], P = 0.0012) and in the ER-negative patient group (OR = 1.44 [1.06–1.95], P = 0.020) (Table 3). The recessive model suggested a notable risk of BC for homozygous individuals (OR = 44.7 [6.90–290], P = 6.7 × 10–5), both in the ER-positive patient group (OR = 82.1 [10.2–660], P = 3.4 × 10–5) and in the ER-negative patient group (OR = 86.3 [4.89–1523], P = 0.0023) (Table 3). Another, a much rarer pLoF in the NTHL1 gene, c.674dup p.(Ser226ValfsTer39), was found only in heterozygous state (OR = 3.01 [0.67–13.6], P = 0.15) (Table 2); therefore, recessive analysis was not available for this variant.
Table 2.
Breast cancer risk association analyses from FinnGen for heterozygous pLoF variants in the candidate genes.
| Gene | Variant | Effect allele frequecy | OR | 95% CI | P value |
|---|---|---|---|---|---|
| ERCC6L2 | c.123dup p.(Ile42TyrfsTer5) | 8.08 × 10–5 | 5.08 | 1.56–16.5 | 0.0070 |
| ERCC6L2 | c.1125dup p.(Ile376TyrfsTer7) | 4.30 × 10–5 | 1.07 | 0.19–6.06 | 0.94 |
| ERCC6L2 | c.1424del p.(Ile475ThrfsTer36)a | 3.78 × 10–3 | 1.09 | 0.89–1.33 | 0.42 |
| ERCC6L2 | c.1930C>T p.(Arg644Ter) | 1.55 × 10–4 | 0.69 | 0.29–1.65 | 0.40 |
| FANCG | c.832dup p.(Ala278GlyfsTer11) | 1.10 × 10–4 | 0.84 | 0.26–2.74 | 0.78 |
| FANCG | c.1076+1G>A | 4.00 × 10–5 | 1.45 | 0.23–9.18 | 0.69 |
| MAD1L1 | c.150+1G>T | 4.61 × 10–5 | 0.33 | 0.04–2.57 | 0.29 |
| MAD1L1 | c.538dup p.(Val180GlyfsTer47) | 5.91 × 10–5 | 0.48 | 0.11–2.12 | 0.33 |
| MAD1L1 | c.1396C>T p.(Gln466Ter) | 6.15 × 10–5 | 1.09 | 0.21–5.53 | 0.92 |
| MAD1L1 | c.1505+2T>A | 1.54 × 10–5 | 3.73 | 0.40–34.8 | 0.25 |
| MAD1L1 | c.1947C>G p.(Tyr649Ter)a | 9.47 × 10–4 | 0.87 | 0.59–1.27 | 0.47 |
| NTHL1 | c.244C>T p.(Gln82Ter)a | 4.65 × 10–3 | 1.39 | 1.18–1.64 | 7.8 × 10–5 |
| NTHL1 | c.674dup p.(Ser226ValfsTer39) | 6.15 × 10–5 | 3.01 | 0.67–13.6 | 0.15 |
| SERPINA3 | c.511C>T p.(Gln171Ter) | 2.49 × 10–4 | 1.03 | 0.49–2.14 | 0.95 |
| SERPINA3 | c.918-1G>Ca | 1.96 × 10–3 | 1.15 | 0.86–1.54 | 0.35 |
| USP45 | c.7del p.(Val3Ter) | 1.51 × 10–4 | 0.62 | 0.23–1.66 | 0.34 |
| USP45 | c.658G>T p.(Glu220Ter) | 8.92 × 10–5 | 1.97 | 0.67–5.77 | 0.22 |
| USP45 | c.845+2T>C | 4.28 × 10–3 | 0.86 | 0.71–1.03 | 0.10 |
| USP45 | c.1008del p.(Val337SerfsTer9)a | 4.43 × 10–4 | 0.78 | 0.41–1.46 | 0.43 |
| USP45 | c.2190C>A p.(Tyr730Ter)a | 1.70 × 10–3 | 0.90 | 0.67–1.21 | 0.48 |
The variants denoted with a were genotyped in the Helsinki and Tampere BC series. Reference transcripts: ERCC6L2 NM_020207.7, FANCG NM_004629.2, MAD1L1 NM_001013836.2, NTHL1 NM_002528.7, SERPINA3 NM_001085.5, and USP45 NM_001346022.3.
Table 3.
Cancer risk association analyses from FinnGen for the NTHL1 c.244C>T variant.
| Cancer type | Total number of individuals | Recessive model | Additive model | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients | Controls | OR | 95% CI | P value | OR | 95% CI | P value | ||
| Breast | Breast cancer | 18,786 | 182,927 | 44.7 | 6.90–290 | 6.7 × 10–5 | 1.39 | 1.18–1.64 | 7.8 × 10–5 |
| ER-positive breast cancer | 10,404 | 182,678 | 82.1 | 10.2–660 | 3.4 × 10–5 | 1.41 | 1.14–1.73 | 0.0012 | |
| ER-negative breast cancer | 6188 | 182,678 | 86.3 | 4.89–1523 | 0.0023 | 1.44 | 1.06–1.95 | 0.020 | |
| Colon | Colorectal cancer | 6847 | 314,193 | 168 | 24.4–1152 | 1.9 × 10–7 | 1.14 | 0.86–1.52 | 0.35 |
| Colorectal adenocarcinoma | 5610 | 314,193 | 204 | 22.7–1837 | 2.1 × 10–6 | 0.99 | 0.73–1.36 | 0.96 | |
| Colon cancer | 4143 | 314,193 | 166 | 14.8–1856 | 3.4 × 10–5 | 1.19 | 0.83–1.70 | 0.35 | |
| Colon adenocarcinoma | 3212 | 314,193 | 224 | 10.8–4643 | 4.7 × 10–4 | 0.99 | 0.65–1.49 | 0.95 | |
| Rectal cancer | 2490 | 314,193 | 447 | 49.7–4023 | 5.2 × 10–8 | 1.04 | 0.67–1.63 | 0.85 | |
| Adenocarcinoma, papilloma adenocarcinoma, and mucinous carcinoma of rectum | 2545 | 314,193 | 472 | 52.1–4279 | 4.4 × 10–8 | 1.13 | 0.72–1.75 | 0.60 | |
| Urinary tract | Cancer of the urinary organs | 2619 | 314,193 | 135 | 6.73–2713 | 0.0013 | 0.94 | 0.60–1.47 | 0.79 |
| Cancer of the renal pelvis | 138 | 314,193 | 146 | 3.55–5985 | 0.0086 | 3.14 | 0.41–24.0 | 0.27 | |
| Bladder cancer | 2193 | 314,193 | 238 | 8.92–6334 | 0.0011 | 1.45 | 0.87–2.39 | 0.15 | |
| Other | Basal-cell carcinoma of the skin | 20,506 | 314,193 | 66.0 | 6.02–723 | 6.0 × 10–4 | 1.16 | 0.97–1.38 | 0.11 |
| Prostate cancer | 15,199 | 131,266 | 365 | 1.97–67,342 | 0.027 | 1.04 | 0.84–1.29 | 0.73 | |
The controls included only women for BC and only men for prostate cancer. The risk association analyses of other cancer types for heterozygous NTHL1 c.244C>T carriers are presented in the Supplementary Table S4.
No variant significantly associated with BC risk (P < 0.0012) in the other candidate genes (Table 2, Supplementary Table S3). In more detail, no risk association was detected for MAD1L1 c.1947C>G (OR = 0.87 [0.59–1.27], P = 0.47), SERPINA3 c.918-1G>C (OR = 1.15 [0.86–1.54], P = 0.35), or USP45 c.2190C>A (OR = 0.90 [0.67–1.21, P = 0.48). FANCG c.1182_1192delinsC was not included in the FinnGen data, but two other, albeit very rare FANCG pLoFs, c.832dup p.(Ala278GlyfsTer11) and c.1076+1G>A, were detected in the study subjects. ERCC6L2 c.1424del was found with a similar frequency in the patients as in the controls (OR = 1.09 [0.89–1.33], P = 0.42); however, another pLoF in ERCC6L2, c.123dup p.(Ile42TyrfsTer5), was more frequent in the patients compared with the controls (OR = 5.08 [1.56–16.5], P = 0.0070). Of the ERCC6L2 variants, recessive analysis was available only for c.1424del (recessive OR = 20.6 [1.40–303], P = 0.027).
Breast tumour characteristics of the patients with the NTHL1 c.244C>T variant
We were able to evaluate the breast tumours of the patients with the NTHL1 c.244C>T variant further in the Helsinki and Tampere BC series. Two patients from Helsinki were homozygous for the variant. One homozygous patient had been diagnosed with BC at the age of 41 years and with rectal and cecum cancers at the age of 47 years. The breast tumour of this patient was ER-positive and progesterone receptor (PR)-positive ductal carcinoma with grade 3. The other homozygous patient had BC at the age of 47 years and cancer of the sigmoid colon at the age of 51 years. This patient had an ER-positive, PR-positive, and HER2-negative ductal breast carcinoma with grade 2. Neither of the homozygous patients had a family history of BC or OC.
The average age of BC diagnosis among the 28 heterozygous carriers was 58.3 years (range 39–88 years), which was higher than the average age of 56.5 years (range 21–95) for all patients in the Helsinki and Tampere series. Of the heterozygous carriers, 75.0% (21/28) had ductal, 17.9% (5/28) had lobular, and 7.1% (2/28) had other invasive breast tumour type. Additionally, 65.4% (17/26) of the patients had ER-positive and 34.6% (9/26) had ER-negative BC, including three patients with triple-negative BC, and 78.3% (18/23) of the patients had a breast tumour with a grade 2 or 3. Additional cancer diagnoses were available only for the patients from the Helsinki BC series: of the 18 heterozygous carriers, two patients had bilateral BC, one had BC and uterus cancer, and one had BC and pancreatic cancer. One patient with bilateral BC and one other heterozygous BC patient also carried a pathogenic CHEK2 c.1100del variant; no other high- or moderate-risk BC predisposition variants had been found in the NTHL1 c.244C>T carriers from Helsinki.
Association of NTHL1 c.244C>T with increased risk of other cancer types than breast cancer
We obtained the data for recessive risk association analyses from FinnGen for all malignant tumour types diagnosed in the individuals homozygous for the NTHL1 c.244C>T variant. Besides BC, homozygous NTHL1 c.244C>T significantly associated with a high risk of CRC (OR = 168 [24.4–1152], P = 1.9 × 10–7) and basal-cell skin cancer (OR = 66.0 [6.02–723], P = 6.0 × 10–4) (Table 3). Additionally, the results suggested an increased risk of urinary tract cancers (OR = 135 [6.73–2713], P = 0.0013).
Ten individuals with the homozygous NTHL1 c.244C>T variant were identified in the FinnGen study: nine of them had been diagnosed with one or multiple tumour types as verified by the Finnish Cancer Registry, and one had no cancer diagnosis. The diagnosed malignant tumour types were rectal, colon, breast, bladder, renal pelvis, basal-cell skin, and prostate cancer, and the non-invasive tumour types were rectal, bladder, and meningeal tumour. Altogether, the nine patients had 19 tumour diagnoses.
To examine the cancer risks for the heterozygous carriers, we retrieved the results for additive risk association analyses from FinnGen for the available malignant tumour types, which have been diagnosed in the patients with biallelic NTHL1 variants in the FinnGen data or reported previously6–11,13. No increased risk of cancer was suggested for the heterozygous carriers for other cancer types than BC (Table 3, Supplementary Table S4).
Discussion
We have performed a WES study of BC patients and a gnomAD database analysis of pLoF variants, with the aim of identifying novel BC risk variants. Furthermore, a recent exome-sequencing study of Finnish patients identified SERPINA3 as a novel candidate gene for moderate-risk BC predisposition19. We assessed the cancer risk associated with the candidate variants by evaluating them in series of BC patients and controls from the Helsinki and Tampere regions and from the FinnGen project.
Even though we did not detect a significant association between NTHL1 c.244C>T p.(Gln82Ter) and BC risk in our patient series, a much larger genotype dataset from FinnGen showed a high increased risk of BC for homozygous (OR = 44.7, P = 6.7 × 10–5) and a low increased risk for heterozygous women (OR = 1.39, P = 7.8 × 10–5). Different cancer studies have reported a high frequency of BC (55%) among women with biallelic pathogenic NTHL1 variants, as reviewed by Beck et al.6–13. The association of NTHL1 variants with BC predisposition has previously been evaluated in a large international case–control study; however, just one biallelic patient was identified and the BC risk remained unclear also for the heterozygous carriers25. In that study, the carrier frequencies and associated BC risk for the c.244C>T variant varied between patient series, but the results for other, rarer heterozygous pLoF and pathogenic missense variants suggested a low increased risk of BC25. The c.244C>T variant (previously reported as c.268C>T p.(Gln90Ter)) is the most frequent LoF variant identified in the patients with NTHL1 tumour syndrome as well as in the NTHL1 gene in the gnomAD database13,26. The variant is enriched in the uniform Finnish population—it was found with a minor allele frequency (MAF) of 0.0044 in the controls from the FinnGen study—which facilitates the detection of increased risk.
Biallelic pathogenic variants in the NTHL1 gene cause a high-penetrance multi-tumour syndrome, which is especially manifested with colorectal, breast, endometrial, urothelial, and basal-cell skin cancer, as well as meningeal tumours6–13. Of the previously reported homozygous and compound heterozygous individuals, 49% had CRC, and of the individuals who had undergone a colonoscopy, even 93% had colonic adenomas13. The FinnGen results support the previous findings on high risk of CRC for the individuals with biallelic variants6,9–11,13. The present study also indicates a high recessive risk of BC; furthermore, high risks of basal-cell skin carcinoma and urinary tract cancer are suggested. Combining the FinnGen and the Helsinki patient series, 11 out of the identified 12 homozygous individuals had a total of 24 tumour diagnoses, further supporting high-penetrance cancer risk. Other cancer types, which have been reported in more than one biallelic case, include hematologic malignancies, squamous cell carcinomas of the head and neck, thyroid, pancreatic, and prostate cancer, and melanoma6,7,9–11,13.
Monoallelic NTHL1 variants are unlikely to cause a substantially increased risk of cancer if any8,12,25,27. In the current study, we examined the risks for the heterozygous carriers to malignant tumours, which have been detected in the patients with biallelic NTHL1 variants6–13. We observed no increased risk of any other cancer type than BC; however, for some tumour types, the case groups were small. In addition to BC, the risk associated with monoallelic NTHL1 variants has previously been investigated in CRC, polyposis, and in a pan-cancer patient population8,12,27. In line with our results, no increased risk of other cancer types was detected.
The premature stop codon caused by the NTHL1 c.244C>T variant has been reported to activate the nonsense-mediated mRNA decay surveillance mechanism6, resulting in loss of the NTHL1 gene product in homozygous individuals. Consistently, reduced NTHL1 protein expression has been observed in heterozygous carriers25. The NTHL1 protein is a bifunctional DNA glycosylase, which catalyses the initial step of base excision-repair pathway to remove oxidative DNA damage28–30. NTHL1 has glycosylase activity on damaged bases, with a preference for oxidized pyrimidines as the substrate, and apurinic/apyrimidinic lyase activity on the DNA phosphate backbone28,29. Disruption of the NTHL1 function may lead to mispairing of damaged bases in replication and accumulation of sequence-specific mutations30. Biallelic LoF variants in the NTHL1 gene have been shown to drive a mutational process causing the COSMIC signature SBS30, which is characterized by somatic C>T transitions at non-CpG sites over different tumour types, including BC6,9,12,25,31,32. Although there is some contradiction, the mutational signature 30, somatic loss of a second allele, or promoter methylation have typically not been observed in heterozygous NTHL1 variant carriers12,25,27,32,33—in these individuals, the possible increased risk of cancer has been suggested to be caused by haploinsufficiency25.
The current study is a comprehensive cancer risk analysis for NTHL1 in an extensive case–control material. Previous studies have been unable to estimate the associated risks for the biallelic individuals in a case–control setting. In the FinnGen data, the prevalence of individuals homozygous for the NTHL1 c.244C>T variant was 1 in every 41,200. This is higher than the estimate of 1 in 114,770 Europeans30. Still, due to the rarity of homozygous individuals, the observed effect sizes for the increased recessive risk associated with the c.244C>T variant, here, are uncertain and the CIs are wide, and the NTHL1 gene warrants further evaluation for more precise risk estimates for different cancer types. Nevertheless, because of the high cancer risk, we suggest that NTHL1 should be included in cancer gene panels in clinical diagnostics, at least for the most common tumour types reported in the patients with pathogenic biallelic NTHL1 variants. Additionally, the susceptibility to multiple tumour types should be considered in surveillance and cancer-prevention strategies for the individuals with biallelic variants, and clinical practice guidelines should be developed for the NTHL1 gene.
FANCG c.1182_1192delinsC p.(Glu395TrpfsTer5) was rare in our patient series, and it was not included in the FinnGen dataset; hence, we were unable to statistically assess any BC risk associated with it. FANCG is an established FA risk gene, with p.(Glu395fs) among the first described causative FANCG mutations for the syndrome20,21. Monoallelic variants in several FA genes are known to predispose to BC3. Two other FANCG pLoF variants, c.832insG p.(Ala278GlyfsTer11) and c.1076+1G>A, identified in the BC patients in the FinnGen study, have been discovered also in Finnish FA patients34. No association with increased risk of BC was detected for these two variants in the FinnGen data; however, both variants were very rare in the study subjects. We did not find heterozygous ERCC6L2 variants associated with BC risk. The additive ORs were inconsistent between the different ERCC6L2 variants in the FinnGen data, which may have been influenced by the rarity of the variants. Biallelic LoF variants in the ERCC6L2 gene, including homozygous c.1424del p.(Ile475ThrfsTer36) (previously known as c.1457del), have been described in patients with inherited bone marrow failure and acute myeloid leukaemia22,23. Additionally, a BC patient with biallelic variants has been reported23. The homozygous c.1424del variant was detected among the BC patients also in the current study, and the contribution of ERCC6L2 to BC remains unclear.
We identified MAD1L1 c.1947C>G p.(Tyr649Ter) and USP45 c.2190C>A p.(Tyr730Ter) in about four- to fivefold higher frequency in the unselected patient group compared with the controls from the Helsinki and Tampere regions. A recent copy number variant analysis reported a twofold increased frequency of MAD1L1 gene deletions among patients in a large BC dataset35; additionally, p.(Tyr649Ter) has been suggested to have a dominant-negative effect on the MAD1L1 protein function and impair the mitotic spindle-assembly checkpoint36. Other studies have connected USP45 to hypersensitivity to mitomycin C -induced interstrand crosslinks and as a candidate gene to multiple myeloma37,38. Our results did not remain significant after adjusting the P value threshold for multiple comparisons and no association with BC risk was detected for the MAD1L1 and USP45 genes in the FinnGen data. We found the SERPINA3 c.918-1G>C variant with a similar frequency in the BC patients and in the controls both in the Helsinki and Tampere BC series and in the FinnGen data; therefore, in the current study, no association with increased BC risk was detected.
In conclusion, our results indicate that biallelic LoF variants in the NTHL1 gene cause a high risk of multiple cancer types, including BC. We also suggest NTHL1 as a low-risk gene for BC predisposition in heterozygous women. However, further studies are required to estimate the effect sizes for the increased risk of different cancer types more precisely. Finally, we propose that NTHL1 should be included in cancer gene panels in clinical diagnostics and clinical practice guidelines should be developed for cancer screening strategies for individuals with pathogenic biallelic NTHL1 variants.
Materials and methods
Whole-exome sequencing and variant calling
We included 69 BC patients from 44 families in the WES. Of the families, 39 had at least three patients with BC or OC among first- and second-degree relatives and 4 had two affected first-degree relatives. Furthermore, 10 of the families included male BC patients, 19 families had uterine cancer cases, and 8 families were suspected of Li-Fraumeni-like syndrome. None of the exome-sequenced patients had a pathogenic BRCA1/2 or TP53 variant. The index patients and their family members were collected among the Helsinki BC series as described below. The WES was carried out using genomic DNA extracted from peripheral blood samples.
The sequencing and variant calling was performed at the McGill University and Génome Québec Innovation Centre, Montreal, Canada. Exome libraries were created with Roche Nimblegen SeqCap EZ Exome + UTR capture kit for 39 of the samples and Roche Nimblegen SeqCap EZ Exome v3 kit for 30 of the samples. Sequencing of the libraries was performed with Illumina HiSeq 2000 platform with 100 bp paired-end reads. The read quality trimming of FASTQ files was executed with FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The reads were aligned to the human reference genome GRCh37/hg19 with Burrows-Wheeler Aligner39. Insertion and deletion variants (indels) were realigned and duplicates were marked with Picard (https://broadinstitute.github.io/picard/). The metrics were computed with Integrative Genomics Viewer40, and the variant calling was performed with SAMtools and BCFtools41,42.
Variant selection from the whole-exome sequencing data
The candidate variants were selected for genotyping based on MAF, pathogenicity of the variant, and relevant gene function. We annotated the variants with Annovar43 and retrieved gene ontology (GO) terms from the AmiGO2 website by the Gene Ontology Consortium44,45. We excluded variants with a raw read depth of < 30 and a phred-scaled quality probability of < 10. Common variants with a MAF of > 0.03 were excluded using the Exome Aggregation Consortium (in any population) and the 1000Genomes variant databases46,47. This selection stage yielded 22,531 variants, which were predicted to alter the protein sequence. We included pLoF variants, defined as stop-gain, frameshift, and essential splice site variants, involved in DNA repair (GO:0006281), cell cycle (GO:0007049), or apoptotic pathways (GO:0006915), totalling in 178 variants in 160 genes. PLoF variants outside of these pathways were considered based on relevance in tumorigenesis. Missense variants involved in DNA repair or cell cycle pathways were considered if predicted to be pathogenic by CADD48 (phred ≥ 20) and by the majority of the other pathogenicity prediction tools included in the LJB (dbNSFP) database in Annovar43 (201 variants in 174 genes). Finally, we focused on plausible candidate genes based on gene function, queried from the UniProt and the NCBI Gene databases49,50, and selected 28 variants in well-supported transcripts51 for genotyping, including 21 pLoFs and seven missenses (Supplementary Table S1). All selected variants had a raw read depth of ≥ 600 and a phred-scaled quality probability of ≥ 150 in the WES data. We further confirmed the indel variants with Sanger sequencing. The variant descriptions were confirmed with Mutalyzer 3 and comply with the current HGVS nomenclature52,53.
Variant selection from the gnomAD database
We downloaded the exomes data of approximately 11,000 Finns from the gnomAD database, release 2.0.1, for about 520 DNA repair genes (GO:0006281, release 2017-07-01)26,44,45. We selected only high-confidence pLoF variants with a MAF of 0.0001–0.03 in the Finnish population; furthermore, we excluded the variants with a MAF of > 0.03 in any other population. We excluded the variants in the validated BC risk genes and in the candidate risk genes previously published from the Helsinki BC series3,54,55. This selection stage yielded 124 pLoF variants in 92 genes in well-supported transcripts (transcript support level 1 and 2), annotated with transcript flags from the Ensembl database through BioMart51,56. We prioritized the candidate variants based on gene function49,50, similarly as for variants chosen from the WES data, and selected twelve pLoF variants in ten candidate genes for genotyping (Supplementary Table S1).
Patient and control series
The case–control series included a total of 4101 BC patients and 3985 population controls from the Helsinki and Tampere regions. All study subjects from Helsinki were women, whereas the Tampere control group also included men. The genomic DNA used in genotyping had been extracted from peripheral blood samples.
Breast cancer patients
The unselected Helsinki BC series consisted of 1726 patients who had been diagnosed with their first primary invasive BC. The patients were recruited consecutively in the Helsinki University Hospital at the Department of Oncology in 1997–1998 and 2000 (n = 847) and at the Department of Surgery in 2001–2004 (n = 879)18,57,58 without any selection criteria for family history of BC or age of diagnosis. The familial Helsinki BC series was combined from 380 index patients with a family history of BC or OC from the unselected series and from 756 additional index patients who were recruited at the Department of Oncology and at the Department of Clinical Genetics until 201558–60. Of these 1136 familial patients, 606 had a family history of at least three BC or OC patients among first- or second-degree relatives (including the proband) and 530 had one affected first-degree relative. The familial patients had been tested at least for BRCA1/2 founder mutations in Finland and the carriers had been excluded from the series. The cancer diagnoses of the patients and their family members were confirmed from hospital records and/or the Finnish Cancer Registry. Altogether, the Helsinki BC series included a total of 2482 patients.
Additional unselected BC patients from the Helsinki region, the BrePainGen series, had been collected in the Helsinki University Hospital at the Breast Surgery Unit in 2006–201061. The series consisted of 950 patients with invasive breast tumour, which had been unilateral and non-metastasised at the time of recruitment; however, no selection for family history of the disease or age of diagnosis had been performed. Of the patients, 161 had at least one first- or second-degree relative diagnosed with BC or OC and were classified as familial.
The unselected Tampere BC series consisted of 669 patients who had been recruited at the Tampere University Hospital consecutively in 1997–1999 and additionally in 1996–200418,58. All patients had been newly diagnosed with invasive BC. Altogether 234 patients had at least one first- or second-degree relative diagnosed with BC or OC and were defined familial.
Population controls
The geographically matched population controls from the Helsinki region consisted of 1273 anonymous blood donors, collected in 2002–2003, and 1896 additional controls with no cancer diagnosis from the Helsinki Biobank. The population controls from the Tampere region consisted of 816 blood donors.
Variant genotyping
Twenty-one variants selected from the WES data were genotyped in 3143 BC patients and 2089 controls from the Helsinki and Tampere BC series with the Sequenom MassARRAY. Seven indel variants from the WES data were genotyped outside of the array for technical reasons. Changes of ≤ 6 base pairs were genotyped with TaqMan real-time PCR and larger indels with 3% agarose gel electrophoresis in 2482 BC patients and 1273 controls from Helsinki. Positive control samples were included in all analyses and the carriers detected with 3% agarose gel electrophoresis were confirmed with Sanger sequencing. Twelve variants selected from the gnomAD data were genotyped in 2482 BC patients and 1273 controls from Helsinki with the Sequenom MassARRAY.
The genotyping of four variants, which had been analysed in the Helsinki BC series, was continued to the 669 BC patients and 816 controls of the Tampere BC series. We genotyped ERCC6L2 c.1424del and USP45 c.2190C>A with TaqMan real-time PCR, USP45 c.1008del with Sanger sequencing, and FANCG c.1182_1192delinsC with 3% agarose gel electrophoresis. The genotyping of MAD1L1 c.1947C>G was further continued to additional 950 BC patients from the BrePainGen series and 1896 controls from the Helsinki Biobank with TaqMan real-time PCR. SERPINA3 c.918-1G>C, selected for genotyping outside of the WES or the gnomAD variant data, was genotyped in all 4101 BC patients and 3985 controls with TaqMan real-time PCR. We confirmed the detected carriers for the ERCC6L2 c.1424del, FANCG c.1182_1192delinsC, MAD1L1 c.1947C>G, NTHL1 c.244C>T, SERPINA3 c.918-1G>C, and USP45 c.2190C>A and c.1008del variants with Sanger sequencing. Further details on genotyping are given in the Supplementary Information Methods.
Statistical analyses
We performed the statistical analyses using the R environment for statistical computing (version 4.2.2)62. We used region-adjusted logistic regression for the combined analyses including patients from Helsinki and Tampere BC series and Fisher’s exact test for the Helsinki BC series, with two-sided P values. After the Bonferroni correction for multiple comparisons, P values < 0.0012 were considered statistically significant.
FinnGen data
To further evaluate the candidate genes, we obtained the data for cancer risk association analyses for a total of 412,181 individuals (230,310 women and 181,871 men) from the FinnGen research project (https://www.finngen.fi/en), which produces genotype data from samples of Finnish biobank participants and combines it with longitudinal data from Finnish health registries24. The biobank sample and data accession numbers for the FinnGen data release 10 are presented in the Supplementary Information Materials.
We retrieved the results for BC risk association analyses for all 38 candidate genes with the endpoint C3_BREAST_EXALLC, which included 18,786 female BC patients and 182,927 female controls with no cancer diagnosis. We annotated the variants with Annovar43; from these results, we included pLoF, missense, and in-frame indel variants with a MAF of ≤ 0.03 in the controls. Additionally, we retrieved the data for risk association analyses for all available tumour types, which had been detected in cancer patients with biallelic pathogenic variants in the NTHL1 gene in the FinnGen study and in previous reports6–13. We excluded the endpoints for benign and in situ tumours (ICD-10 D-coded tumours), as the registry entries may be incomplete for them, except for the endpoint C3_BREAST_EXALLC, which included both malignant and in situ tumours (ICD-O-3 behaviour codes 3 and 2). We used the analyses in which the controls with any cancer diagnosis had been excluded. All included cancer endpoints are given in the Supplementary Table S5 and the endpoint definitions are available at https://risteys.finregistry.fi.
The cancer risk associated with heterozygous variants was detected with the additive model in the FinnGen data; homozygous and compound heterozygotes had been excluded from the analyses as described in15. The recessive model compared homozygous individuals against heterozygotes and noncarriers15. Of the additive analyses, we included only variants which had been genotyped on array, whereas the recessive analyses for NTHL1 c.244C>T and ERCC6L2 c.1424del included also imputed genotypes. The imputation quality scores were 0.9974 for NTHL1 c.244C>T and 0.9951 for ERCC6L2 c.1424del. The association analyses in the FinnGen data had been performed with the REGENIE software (version 2.2.4)63. The genotyping and production of the FinnGen dataset has been described in24 and at https://finngen.gitbook.io/documentation.
Ethics declarations
The study was conducted in accordance with the Declaration of Helsinki and with approval by the Ethics Committee of the Helsinki University Hospital (Dnro207/E9/07 and HUS71597/2016). The Tampere study protocol was approved by the Ethics Committee of the Pirkanmaa Hospital District (97247) and the BrePainGen study protocol by the Coordinating Ethics Committee (136/E0/2006) and the Ethics Committee of the Department of Surgery (Dnro 148/E6/05) of the Hospital District of Helsinki and Uusimaa. The ethics statement for FinnGen is given in the Supplementary Information Materials. Informed consent was obtained from all patients.
Supplementary Information
Acknowledgements
We thank the study participants for their valuable participation. We thank research nurses Outi Malkavaara and Irja Erkkilä for their help with the patient data, M.Sc. Erja Nynäs with bioinformatics and with the FinnGen data, and M.Sc. Lotta Mielikäinen with the genotyping, Dr. Taru A. Muranen for valuable advice, and Prof. Katri Pylkäs for providing a positive control sample for genotyping. We gratefully acknowledge the participants and investigators of the FinnGen study and thank Prof. Aarno Palotie and Dr. Risto Kajanne for their kind help. The FinnGen consortium members are presented in the Supplementary Table S6. The following biobanks are acknowledged for delivering biobank samples to FinnGen: Auria Biobank (https://www.auria.fi/biopankki), THL Biobank (https://www.thl.fi/biobank), Helsinki Biobank (https://www.helsinginbiopankki.fi), Biobank Borealis of Northern Finland (https://www.ppshp.fi/Tutkimus-ja-opetus/Biopankki/Pages/Biobank-Borealis-briefly-in-English.aspx), Finnish Clinical Biobank Tampere (https://www.tays.fi/en-US/Research_and_development/Finnish_Clinical_Biobank_Tampere), Biobank of Eastern Finland (https://www.ita-suomenbiopankki.fi/en), Central Finland Biobank (https://www.ksshp.fi/fi-FI/Potilaalle/Biopankki), Finnish Red Cross Blood Service Biobank (https://www.veripalvelu.fi/verenluovutus/biopankkitoiminta), Terveystalo Biobank (https://www.terveystalo.com/fi/Yritystietoa/Terveystalo-Biopankki/Biopankki/), and Arctic Biobank (https://www.oulu.fi/en/university/faculties-and-units/faculty-medicine/northern-finland-birth-cohorts-and-arctic-biobank). All Finnish Biobanks are members of BBMRI.fi infrastructure (https://www.bbmri.fi). Finnish Biobank Cooperative -FINBB (https://finbb.fi/) is the coordinator of BBMRI-ERIC operations in Finland. The Finnish biobank data can be accessed through the Fingenious® services (https://site.fingenious.fi/en/) managed by FINBB. We thank the Helsinki Biobank also for providing control samples for genotyping in the current study, the staff at the McGill University and Génome Québec Innovation Centre for exome-sequencing services, the staff at the Genotyping laboratory of Institute for Molecular Medicine Finland (FIMM) Technology Centre, University of Helsinki, for variant genotyping services, and the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to the database. A full list of contributing groups for the gnomAD database can be found at https://gnomad.broadinstitute.org/about.
Author contributions
H.N., A.K.N., and L.M.P. designed the study. S.K. annotated and pre-processed the WES data. M.N. pre-processed the gnomAD data. A.K.N., J.I.K., L.M.P., and H.N. performed the variant selection. A.K.N., M.S., N.A., L.M.P., and J.I.K. carried out the variant genotyping. A.K.N. did the statistical analyses and processed the FinnGen data. K.A., C.B., P.H., J.S., A.K., E.K., and T.T. contributed samples and patient information. A.K.N. and H.N. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Helsinki University Hospital Research Fund, the Sigrid Jusélius Foundation, and the Cancer Foundation Finland. The FinnGen project is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and the following industry partners: AbbVie Inc., AstraZeneca UK Ltd, Biogen MA Inc., Bristol Myers Squibb (and Celgene Corporation & Celgene International II Sàrl), Genentech Inc., Merck Sharp & Dohme LCC, Pfizer Inc., GlaxoSmithKline Intellectual Property Development Ltd., Sanofi US Services Inc., Maze Therapeutics Inc., Janssen Biotech Inc., Novartis AG, and Boehringer Ingelheim International GmbH.
Data availability
For the Helsinki and Tampere BC series, the data that support the findings of our study are available on reasonable request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions. Instructions on accessing the FinnGen data are available at https://www.finngen.fi/en/access_results.
Competing interests
L.M.P. and M.S. are currently employed by Blueprint Genetics. The other authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Heli Nevanlinna, Email: heli.nevanlinna@hus.fi.
FinnGen:
Aarno Palotie, Mark Daly, Bridget Riley-Gillis, Howard Jacob, Dirk Paul, Slavé Petrovski, Heiko Runz, Sally John, George Okafo, Nathan Lawless, Heli Salminen-Mankonen, Robert Plenge, Joseph Maranville, Mark McCarthy, Margaret G. Ehm, Kirsi Auro, Simonne Longerich, Anders Mälarstig, Katherine Klinger, Clement Chatelain, Matthias Gossel, Karol Estrada, Robert Graham, Robert Yang, Chris O’Donnell, Tomi P. Mäkelä, Jaakko Kaprio, Petri Virolainen, Antti Hakanen, Terhi Kilpi, Markus Perola, Jukka Partanen, Anne Pitkäranta, Taneli Raivio, Jani Tikkanen, Raisa Serpi, Tarja Laitinen, Veli-Matti Kosma, Jari Laukkanen, Marco Hautalahti, Outi Tuovila, Raimo Pakkanen, Jeffrey Waring, Fedik Rahimov, Ioanna Tachmazidou, Chia-Yen Chen, Zhihao Ding, Marc Jung, Shameek Biswas, Rion Pendergrass, David Pulford, Neha Raghavan, Adriana Huertas-Vazquez, Jae-Hoon Sul, Xinli Hu, Åsa Hedman, Manuel Rivas, Dawn Waterworth, Nicole Renaud, Ma’en Obeidat, Samuli Ripatti, Johanna Schleutker, Mikko Arvas, Olli Carpén, Reetta Hinttala, Johannes Kettunen, Arto Mannermaa, Katriina Aalto-Setälä, Mika Kähönen, Johanna Mäkelä, Reetta Kälviäinen, Valtteri Julkunen, Hilkka Soininen, Anne Remes, Mikko Hiltunen, Jukka Peltola, Minna Raivio, Pentti Tienari, Juha Rinne, Roosa Kallionpää, Juulia Partanen, Ali Abbasi, Adam Ziemann, Nizar Smaoui, Anne Lehtonen, Susan Eaton, Sanni Lahdenperä, Natalie Bowers, Edmond Teng, Fanli Xu, Laura Addis, John Eicher, Qingqin S. Li, Karen He, Ekaterina Khramtsova, Martti Färkkilä, Jukka Koskela, Sampsa Pikkarainen, Airi Jussila, Katri Kaukinen, Timo Blomster, Mikko Kiviniemi, Markku Voutilainen, Tim Lu, Linda McCarthy, Amy Hart, Meijian Guan, Jason Miller, Kirsi Kalpala, Melissa Miller, Kari Eklund, Antti Palomäki, Pia Isomäki, Laura Pirilä, Oili Kaipiainen-Seppänen, Johanna Huhtakangas, Nina Mars, Apinya Lertratanakul, Coralie Viollet, Marla Hochfeld, Jorge Esparza Gordillo, Fabiana Farias, Nan Bing, Margit Pelkonen, Paula Kauppi, Hannu Kankaanranta, Terttu Harju, Riitta Lahesmaa, Hubert Chen, Joanna Betts, Rajashree Mishra, Majd Mouded, Debby Ngo, Teemu Niiranen, Felix Vaura, Veikko Salomaa, Kaj Metsärinne, Jenni Aittokallio, Jussi Hernesniemi, Daniel Gordin, Juha Sinisalo, Marja-Riitta Taskinen, Tiinamaija Tuomi, Timo Hiltunen, Amanda Elliott, Mary Pat Reeve, Sanni Ruotsalainen, Audrey Chu, Dermot Reilly, Mike Mendelson, Jaakko Parkkinen, Tuomo Meretoja, Heikki Joensuu, Johanna Mattson, Eveliina Salminen, Annika Auranen, Peeter Karihtala, Päivi Auvinen, Klaus Elenius, Esa Pitkänen, Relja Popovic, Margarete Fabre, Jennifer Schutzman, Diptee Kulkarni, Alessandro Porello, Andrey Loboda, Heli Lehtonen, Stefan McDonough, Sauli Vuoti, Kai Kaarniranta, Joni A. Turunen, Terhi Ollila, Hannu Uusitalo, Juha Karjalainen, Mengzhen Liu, Stephanie Loomis, Erich Strauss, Hao Chen, Kaisa Tasanen, Laura Huilaja, Katariina Hannula-Jouppi, Teea Salmi, Sirkku Peltonen, Leena Koulu, David Choy, Ying Wu, Pirkko Pussinen, Aino Salminen, Tuula Salo, David Rice, Pekka Nieminen, Ulla Palotie, Maria Siponen, Liisa Suominen, Päivi Mäntylä, Ulvi Gursoy, Vuokko Anttonen, Kirsi Sipilä, Hannele Laivuori, Venla Kurra, Laura Kotaniemi-Talonen, Oskari Heikinheimo, Ilkka Kalliala, Lauri Aaltonen, Varpu Jokimaa, Marja Vääräsmäki, Outi Uimari, Laure Morin-Papunen, Maarit Niinimäki, Terhi Piltonen, Katja Kivinen, Elisabeth Widen, Taru Tukiainen, Niko Välimäki, Eija Laakkonen, Jaakko Tyrmi, Heidi Silven, Eeva Sliz, Riikka Arffman, Susanna Savukoski, Triin Laisk, Natalia Pujol, Janet Kumar, Iiris Hovatta, Erkki Isometsä, Hanna Ollila, Jaana Suvisaari, Thomas Damm Als, Antti Mäkitie, Argyro Bizaki-Vallaskangas, Sanna Toppila-Salmi, Tytti Willberg, Elmo Saarentaus, Antti Aarnisalo, Elisa Rahikkala, Kristiina Aittomäki, Fredrik Åberg, Mitja Kurki, Aki Havulinna, Juha Mehtonen, Priit Palta, Shabbeer Hassan, Pietro Della Briotta Parolo, Wei Zhou, Mutaamba Maasha, Susanna Lemmelä, Aoxing Liu, Arto Lehisto, Andrea Ganna, Vincent Llorens, Henrike Heyne, Joel Rämö, Rodos Rodosthenous, Satu Strausz, Tuula Palotie, Kimmo Palin, Javier Gracia-Tabuenca, Harri Siirtola, Tuomo Kiiskinen, Jiwoo Lee, Kristin Tsuo, Kati Kristiansson, Kati Hyvärinen, Jarmo Ritari, Katri Pylkäs, Minna Karjalainen, Tuomo Mantere, Eeva Kangasniemi, Sami Heikkinen, Nina Pitkänen, Samuel Lessard, Clément Chatelain, Lila Kallio, Tiina Wahlfors, Eero Punkka, Sanna Siltanen, Teijo Kuopio, Anu Jalanko, Huei-Yi Shen, Risto Kajanne, Mervi Aavikko, Helen Cooper, Denise Öller, Rasko Leinonen, Henna Palin, Malla-Maria Linna, Masahiro Kanai, Zhili Zheng, L. Elisa Lahtela, Mari Kaunisto, Elina Kilpeläinen, Timo P. Sipilä, Oluwaseun Alexander Dada, Awaisa Ghazal, Anastasia Kytölä, Rigbe Weldatsadik, Kati Donner, Anu Loukola, Päivi Laiho, Tuuli Sistonen, Essi Kaiharju, Markku Laukkanen, Elina Järvensivu, Sini Lähteenmäki, Lotta Männikkö, Regis Wong, Auli Toivola, Minna Brunfeldt, Hannele Mattsson, Sami Koskelainen, Tero Hiekkalinna, Teemu Paajanen, Kalle Pärn, Mart Kals, Shuang Luo, Shanmukha Sampath Padmanabhuni, Marianna Niemi, Mika Helminen, Tiina Luukkaala, Iida Vähätalo, Jyrki Tammerluoto, Sarah Smith, Tom Southerington, and Petri Lehto
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-47441-w.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen FC, van-Overeem-Hansen T, Sorensen CS. Hereditary breast and ovarian cancer: New genes in confined pathways. Nat. Rev. Cancer. 2016;16:599–612. doi: 10.1038/nrc.2016.72. [DOI] [PubMed] [Google Scholar]
- 3.Breast Cancer Association, C et al. Breast cancer risk genes—association analysis in more than 113,000 Women. N Engl J Med. 2021;384:428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michailidou K, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoniou AC, et al. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br. J. Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weren RD, et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat. Genet. 2015;47:668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 7.Rivera B, Castellsague E, Bah I, van Kempen LC, Foulkes WD. Biallelic NTHL1 mutations in a woman with multiple primary tumors. N. Engl. J. Med. 2015;373:1985–1986. doi: 10.1056/NEJMc1506878. [DOI] [PubMed] [Google Scholar]
- 8.Belhadj S, et al. Delineating the phenotypic spectrum of the NTHL1-associated polyposis. Clin. Gastroenterol. Hepatol. 2017;15:461–462. doi: 10.1016/j.cgh.2016.09.153. [DOI] [PubMed] [Google Scholar]
- 9.Grolleman JE, et al. Mutational signature analysis reveals NTHL1 deficiency to cause a multi-tumor phenotype. Cancer Cell. 2019;35:256–266. doi: 10.1016/j.ccell.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Boulouard F, et al. Further delineation of the NTHL1 associated syndrome: A report from the French Oncogenetic Consortium. Clin. Genet. 2021;99:662–672. doi: 10.1111/cge.13925. [DOI] [PubMed] [Google Scholar]
- 11.Weatherill CB, et al. Six case reports of NTHL1-associated tumor syndrome further support it as a multi-tumor predisposition syndrome. Clin. Genet. 2023;103:231–235. doi: 10.1111/cge.14242. [DOI] [PubMed] [Google Scholar]
- 12.Salo-Mullen EE, et al. Prevalence and Characterization of biallelic and monoallelic NTHL1 and MSH3 variant carriers from a pan-cancer patient population. JCO Precis. Oncol. 2021;5:455. doi: 10.1200/PO.20.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck SH, et al. Intestinal and extraintestinal neoplasms in patients with NTHL1 tumor syndrome: A systematic review. Fam. Cancer. 2022;21:453–462. doi: 10.1007/s10689-022-00291-3. [DOI] [PubMed] [Google Scholar]
- 14.Lim ET, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyne HO, et al. Mono- and biallelic variant effects on disease at biobank scale. Nature. 2023;613:519–525. doi: 10.1038/s41586-022-05420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nurmi AK, et al. Pathogenic Variant spectrum in breast cancer risk genes in Finnish patients. Cancers (Basel) 2022;14:6158. doi: 10.3390/cancers14246158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vehmanen P, et al. Low proportion of BRCA1 and BRCA2 mutations in Finnish breast cancer families: Evidence for additional susceptibility genes. Hum. Mol. Genet. 1997;6:2309–2315. doi: 10.1093/hmg/6.13.2309. [DOI] [PubMed] [Google Scholar]
- 18.Syrjakoski K, et al. Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J. Natl. Cancer Inst. 2000;92:1529–1531. doi: 10.1093/jnci/92.18.1529. [DOI] [PubMed] [Google Scholar]
- 19.Koivuluoma S, et al. Exome sequencing identifies a recurrent variant in SERPINA3 associating with hereditary susceptibility to breast cancer. Eur. J. Cancer. 2021;143:46–51. doi: 10.1016/j.ejca.2020.10.033. [DOI] [PubMed] [Google Scholar]
- 20.de Winter JP, et al. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat. Genet. 1998;20:281–283. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 21.Demuth I, et al. Spectrum of mutations in the Fanconi anaemia group G gene, FANCG/XRCC9. Eur. J. Hum. Genet. 2000;8:861–868. doi: 10.1038/sj.ejhg.5200552. [DOI] [PubMed] [Google Scholar]
- 22.Douglas SPM, et al. ERCC6L2 defines a novel entity within inherited acute myeloid leukemia. Blood. 2019;133:2724–2728. doi: 10.1182/blood-2019-01-896233. [DOI] [PubMed] [Google Scholar]
- 23.Hakkarainen M, et al. The clinical picture of ERCC6L2 disease: From bone marrow failure to acute leukemia. Blood. 2023;141:2853–2866. doi: 10.1182/blood.2022019425. [DOI] [PubMed] [Google Scholar]
- 24.Kurki MI, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–518. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li N, et al. Evaluation of the association of heterozygous germline variants in NTHL1 with breast cancer predisposition: An international multi-center study of 47,180 subjects. NPJ Breast Cancer. 2021;7:52. doi: 10.1038/s41523-021-00255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elsayed FA, et al. Monoallelic NTHL1 loss-of-function variants and risk of polyposis and colorectal cancer. Gastroenterology. 2020;159:2241–2243. doi: 10.1053/j.gastro.2020.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aspinwall R, et al. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl. Acad. Sci. U. S. A. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dizdaroglu M, Karahalil B, Senturker S, Buckley TJ, Roldan-Arjona T. Excision of products of oxidative DNA base damage by human NTH1 protein. Biochemistry. 1999;38:243–246. doi: 10.1021/bi9819071. [DOI] [PubMed] [Google Scholar]
- 30.Weren RD, et al. NTHL1 and MUTYH polyposis syndromes: Two sides of the same coin? J. Pathol. 2018;244:135–142. doi: 10.1002/path.5002. [DOI] [PubMed] [Google Scholar]
- 31.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drost J, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–238. doi: 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belhadj S, et al. NTHL1 biallelic mutations seldom cause colorectal cancer, serrated polyposis or a multi-tumor phenotype, in absence of colorectal adenomas. Sci. Rep. 2019;9:9020. doi: 10.1038/s41598-019-45281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantere T, et al. Finnish Fanconi anemia mutations and hereditary predisposition to breast and prostate cancer. Clin. Genet. 2015;88:68–73. doi: 10.1111/cge.12447. [DOI] [PubMed] [Google Scholar]
- 35.Dennis J, et al. Rare germline copy number variants (CNVs) and breast cancer risk. Commun. Biol. 2022;5:65. doi: 10.1038/s42003-021-02990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukasaki K, et al. Mutations in the mitotic check point gene, MAD1L1, in human cancers. Oncogene. 2001;20:3301–3305. doi: 10.1038/sj.onc.1204421. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Oliva AB, et al. USP45 deubiquitylase controls ERCC1-XPF endonuclease-mediated DNA damage responses. EMBO J. 2015;34:326–343. doi: 10.15252/embj.201489184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waller RG, et al. Novel pedigree analysis implicates DNA repair and chromatin remodeling in multiple myeloma risk. PLoS Genet. 2018;14:e1007111. doi: 10.1371/journal.pgen.1007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danecek P, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:2. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashburner M, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carbon S, et al. AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.UniProt Consortium UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–D531. doi: 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown GR, et al. Gene: A gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43:D36–42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunningham F, et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lefter M, et al. Mutalyzer 2: Next generation HGVS nomenclature checker. Bioinformatics. 2021;37:2811–2817. doi: 10.1093/bioinformatics/btab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Higgins J, et al. Verifying nomenclature of DNA variants in submitted manuscripts: Guidance for journals. Hum. Mutat. 2021;42:3–7. doi: 10.1002/humu.24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiiski JI, et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15172–15177. doi: 10.1073/pnas.1407909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mantere T, et al. Case-control analysis of truncating mutations in DNA damage response genes connects TEX15 and FANCD2 with hereditary breast cancer susceptibility. Sci. Rep. 2017;7:681. doi: 10.1038/s41598-017-00766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinsella RJ, et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database (Oxf.) 2011;2011:030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kilpivaara O, et al. Correlation of CHEK2 protein expression and c.1100delC mutation status with tumor characteristics among unselected breast cancer patients. Int. J. Cancer. 2005;113:575–580. doi: 10.1002/ijc.20638. [DOI] [PubMed] [Google Scholar]
- 58.Fagerholm R, et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat. Genet. 2008;40:844–853. doi: 10.1038/ng.155. [DOI] [PubMed] [Google Scholar]
- 59.Vahteristo P, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am. J. Hum. Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eerola H, Blomqvist C, Pukkala E, Pyrhonen S, Nevanlinna H. Familial breast cancer in southern Finland: How prevalent are breast cancer families and can we trust the family history reported by patients? Eur. J. Cancer. 2000;36:1143–1148. doi: 10.1016/s0959-8049(00)00093-9. [DOI] [PubMed] [Google Scholar]
- 61.Kaunisto MA, et al. Pain in 1,000 women treated for breast cancer: A prospective study of pain sensitivity and postoperative pain. Anesthesiology. 2013;119:1410–1421. doi: 10.1097/ALN.0000000000000012. [DOI] [PubMed] [Google Scholar]
- 62.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/ (2022).
- 63.Mbatchou J, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 2021;53:1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For the Helsinki and Tampere BC series, the data that support the findings of our study are available on reasonable request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions. Instructions on accessing the FinnGen data are available at https://www.finngen.fi/en/access_results.