Abstract
Hepatitis B protein x (HBx) has been reported to promote tumorigenesis in hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC), but the mechanism awaits further investigation. In this study, we found that cFAM210A (a circular RNA derived from the third exon of transcript NM_001098801 of the FAM210A gene; CircBase ID: hsa_circ_0003979) can be silenced by HBx. cFAM210A expression was downregulated and negatively correlated with tumorigenesis in patients with HBV-related HCC. Furthermore, cFAM210A reduced the proliferation, stemness, and tumorigenicity of HCC cells. Mechanistically, HBx increased the N6-methyladenosine (m6A) level of cFAM210A by promoting the expression of RBM15 (an m6A methyltransferase), thus inducing the degradation of cFAM210A via the YTHDF2-HRSP12-RNase P/MRP pathway. cFAM210A bound to YBX1 and inhibited its phosphorylation, suppressing its transactivation function toward MET. These findings suggest the important role of circular RNAs in HBx-induced hepatocarcinogenesis and identify cFAM210A a potential target in the prevention and treatment of HBV-related HCC.
Subject terms: Liver cancer, Oncogenes
Circular RNA cFAM210A: a new hope in HBV-related HCC treatment
A study has identified a circular RNA, cFAM210A, that inhibits the growth and stemness of hepatocellular carcinoma (HCC) cells. Researchers found that the hepatitis B virus (HBV) protein HBx promotes the degradation of cFAM210A through a process called N6-methyladenosine modification. This leads to increased tumour growth and stemness in HCC cells. By inhibiting the transactivation function of a protein called YBX1 on another protein called MET, cFAM210A suppresses tumour progression. These findings highlight the potential of cFAM210A as a target for the prevention and treatment of HBV-related HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and lethal cancers worldwide, and hepatitis B virus (HBV) infection is the main factor leading to HCC, especially in China1. Hepatitis B protein x (HBx) is essential in promoting tumorigenesis in HBV-related HCC2,3, but the underlying mechanisms have not been fully elucidated.
Circular RNAs (circRNAs), a specific series of noncoding RNAs with covalently closed, single-stranded structures and lacking a 5’ cap and 3’ poly(A) tail4, have long been considered “waste” produced during RNA splicing. In recent years, a large number of circRNAs in mammalian cells have been identified through high-throughput sequencing approaches5. Numerous studies have shed light on the important roles of circRNAs in the formation and development of almost all tumours, including those of gastric cancer6, breast cancer7, glioma8 and colorectal cancer9. In previous studies, we found that circRNA-cSMARCA5 can inhibit the growth and metastasis of HCC by sponging microRNAs10, and a plasma circRNA panel (containing three circRNAs) could be used to diagnose HBV-related HCC more accurately than the alpha-fetoprotein level11. However, whether HBx can promote tumorigenesis in HCC through circRNAs remains largely unknown.
In this study, we sequenced and characterized circRNAs in HBx-overexpressing (HBx-oe) and negative control (NC) HCC cells. Herein, we identified a set of differentially expressed circRNAs, among which cFAM210A (a circular RNA derived from the third exon of transcript NM_001098801 of the FAM210A gene; hg19, chr18:13681604-13682104; circBase ID: hsa_circ_0003979) exhibited the most notable decrease in expression. In addition, cFAM210A downregulation was confirmed to facilitate HBV-related hepatocarcinogenesis in clinical samples and promote stemness and tumorigenicity in HCC cells. Mechanistically, we revealed that HBx can increase N6-methyladenosine (m6A) modification of cFAM210A and facilitate its recognition and degradation. Using circRNA pull-down and RNA immunoprecipitation (RIP) assays, we discovered that cFAM210A can bind to the YBX1 protein, thus inhibiting its transactivation function.
Methods
Patients and samples
In total, 20 healthy liver tissues (distal healthy liver tissues from patients with hemangioma of the liver; Cohort 1) and 100 pairs of HCC and corresponding ANL tissues (divided into Cohorts 2 and 3) were obtained via surgical resection from patients with no preoperative treatment at Eastern Hepatobiliary Surgery Hospital (Shanghai, China). Human specimen collection was approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital. Written informed consent forms were signed by each patient according to the policies of the committee. The samples in Cohorts 1, 2 and 3 were obtained in 2020, 2020, and 2015–2016, respectively. The detailed clinicopathological features are described in Supplementary Table 5.
CircRNA pull-down assay
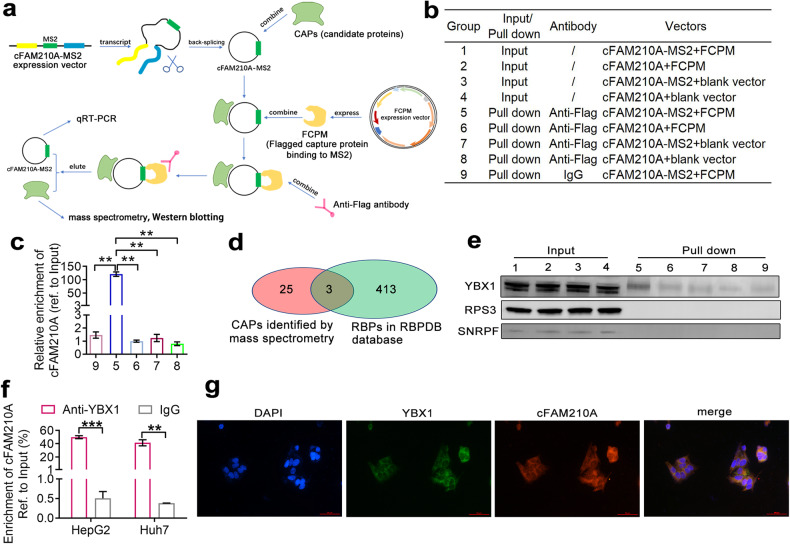
The circRNA pull-down assay was performed as previously described12. Briefly, the vector overexpressing MS2-fused cFAM210A (cFAM210A-MS2) and the vector overexpressing a Flag-tagged capture protein binding to MS2 (FCPM) were cotransfected into HepG2 cells. Then, the FCPM-cFAM210A-MS2-CAP (candidate protein) complexes were enriched by immunoprecipitation with an anti-Flag antibody (Fig. 7a, b, Group 5). The vectors without the MS2 or FCPM sequence were used as controls (Fig. 7b, Groups 6–8).
Fig. 7. cFAM210A can bind to YBX1.
a Schematic of the circRNA pull-down assay. b Grouping scheme used for circRNA pull-down assays. c qRT‒PCR following circRNA pull-down showed the enrichment of cFAM210A in Group 5. d Screening of proteins binding to cFAM210A. e Western blot results validating the candidate proteins binding to cFAM210A. f RIP-qPCR using an anti-YBX1 antibody in HCC cell lines. g Dual RNA fluorescence in situ hybridization (FISH) and immunofluorescence assay results showing the colocalization of cFAM210A and YBX1 in HepG2 cells. Scale bar, 100 μm. CAPs candidate proteins, FCPM flag-tagged capture protein binding to MS2. For (e), Student’s t test was used. ns not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Animal studies
The animal experiments in this study conformed to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines (http://www.nc3rs.org.uk/arrive-guidelines) and were approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital (Shanghai, China). The BALB/c nude mice used in this study were all male, 5 weeks old and purchased from Laboratory Animal Resources, Chinese Academy of Sciences (Beijing, China). All mice were housed in laminar flow cabinets under specific pathogen-free conditions at room temperature on a 12 h light/dark cycle, with food and water available ad libitum. The details of the in vivo extreme limiting dilution assay are described in Supplementary Data 2.
Statistical analysis
All statistical analyses were performed using SPSS version 27.0 software (SPSS, Inc., Chicago, IL). For comparisons, the chi-square test, Student’s t test, the Wilcoxon signed-rank test or the Mann‒Whitney U test was used, as appropriate. Correlations were evaluated by Pearson correlation analysis. Survival curves were generated using the Kaplan‒Meier method, and differences in survival were assessed by the log-rank test. The estimated stem cell frequency in the extreme limiting dilution assays was calculated with ELDA (http://bioinf.wehi.edu.au/software/elda/). Statistical significance was indicated by P values of less than 0.05.
The remaining methods are described in Supplementary Data 2.
Results
cFAM210A is the main target of HBx
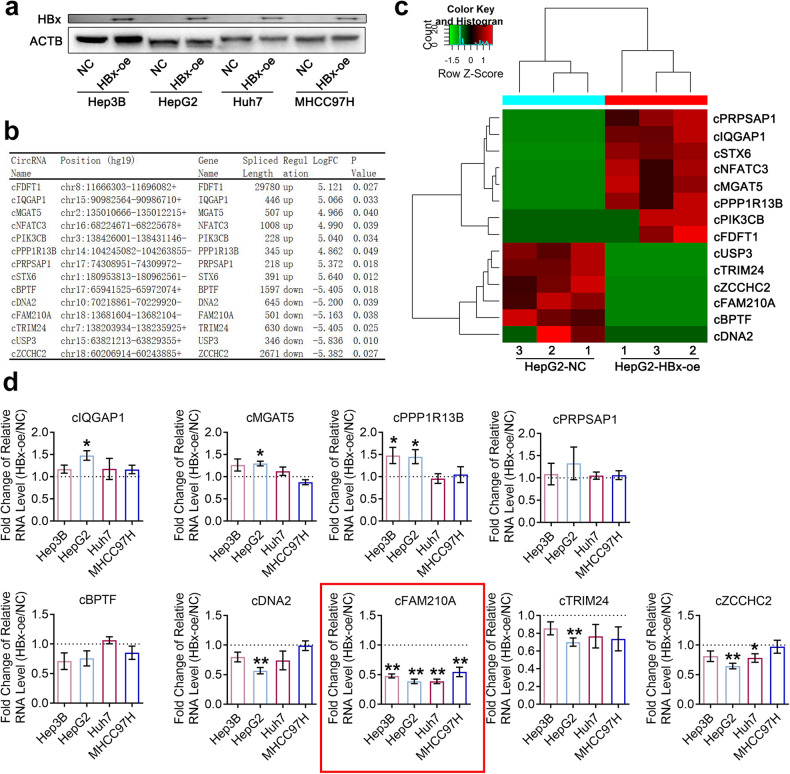
First, the HBx protein was overexpressed in four human HCC cell lines (Hep3B, HepG2, Huh7 and MHCC97H) using lentivirus expressing HBx (Fig. 1a), which has been confirmed to promote HCC13. We then performed whole-transcriptome sequencing and identified differentially expressed circRNAs in HBx-overexpressing (HBx-oe) and NC (negative control) HepG2 cells. Among the 14 differentially expressed circRNAs after induction of HBx expression, 8 were upregulated, while 6 were downregulated (Supplementary Table 1 and Fig. 1b, c). By quantitative reverse transcription PCR (qRT‒PCR) using circRNA-specific divergent primers, agarose gel electrophoresis, Sanger sequencing and RNase R treatment assays10, we successfully validated the existence of 4 of the upregulated circRNAs (cIQGAP1, cMGAT5, cPPP1R13B and cPRPSAP1) and 5 of the downregulated circRNAs (cBPTF, cDNA2, cFAM210A, cTRIM24 and cZCCHC2) in HepG2 cells (Supplementary Fig. 1). Next, we measured the expression of these 9 circRNAs in HBx-oe and NC HCC cell lines. The expression of cFAM210A was the most obviously changed in all four HCC cell lines (Fig. 1d).
Fig. 1. Screening and identification of HBx-regulated circRNAs.
a Western blot analysis to measure the expression of HBx protein in HCC cell lines. ACTB was used as an endogenous control. b CircRNA-seq results showing the differentially expressed circRNAs between HBx-oe and NC HepG2 cells. c Cluster heatmap of differentially expressed molecules in the HBx-oe and NC groups. The horizontal axis shows the names of the circRNAs, and the vertical axis shows the NC and HBx-oe cell samples (3 replicates). d qRT‒PCR results validating the expression differences in nine circRNAs in four HCC cell lines. ACTB was used as an endogenous control. Student’s t test was used. NC negative control, HBx-oe HBx-overexpressing. *P < 0.05; **P < 0.01; ***P < 0.001.
Therefore, we assumed that cFAM210A was the main target of HBx.
cFAM210A expression is downregulated and negatively correlated with tumorigenesis in patients with HBV-related HCC
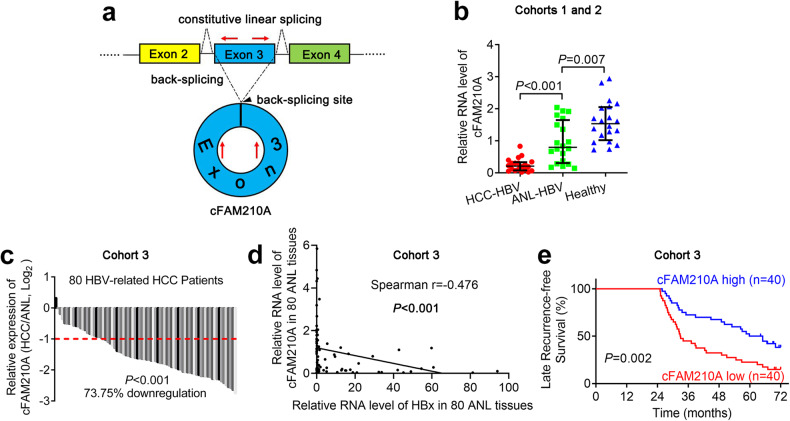
cFAM210A is derived from the third exon of the NM_001098801 transcript of the FAM210A gene by back-splicing (Fig. 2a), and its length is 501 nucleotides. We then measured the expression of cFAM210A in 20 healthy liver tissues (distal healthy liver tissues from patients with cavernous hemangioma of the liver; Cohort 1) and 20 paired HBV-related HCC and adjacent noncancerous liver (ANL) tissues (Cohort 2). The expression of cFAM210A in HCC tissues was lower than that in ANL tissues, indicating an antitumor role of cFAM210A (Fig. 2b). Importantly, the expression of cFAM210A in HBV-related ANL tissues was lower than that in healthy tissues, indicating that cFAM210A may be downregulated by HBV infection (Fig. 2b).
Fig. 2. Clinical significance of cFAM210A expression.
a Diagram of the pattern of cFAM210A biogenesis, with red arrows representing specific primers. b qRT‒PCR results showing the expression of cFAM210A in 20 healthy liver tissues (Cohort 1) and 20 paired HBV-related HCC and ANL tissues (Cohort 2). For comparisons between HCC-HBV and ANL-HBV samples, the Wilcoxon signed-rank test was used. For comparisons between ANL-HBV and healthy control samples, the Mann‒Whitney U test was used. c qRT‒PCR results comparing cFAM210A expression in HBV-associated HCC samples and non-HBV-related HCC samples. The Wilcoxon signed-rank test was used. d The correlation between the expression of HBx and that of cFAM210A in 80 ANL tissues was analyzed. e Kaplan‒Meier curve indicating the correlation between cFAM210A expression and cumulative late recurrence-free survival. The log-rank test was used. For (b), (c) and (d), ACTB was used as an endogenous control.
There are two types of HCC recurrence after hepatectomy: early recurrence (within 2 years after surgery), which arises from residual HCC cells disseminated in the remnant liver; and late recurrence (more than two years after surgery), constituting de novo tumorigenesis independent of the completely resected primary tumor14. Gene expression analysis of ANL tissue can predict the risk of late recurrence of HCC following hepatectomy14. To determine whether cFAM210A is associated with de novo tumorigenesis of HCC, we collected HCC and ANL tissues as well as prognostic data from 80 patients with HBV-related HCC with late recurrence following surgery (Cohort 3). qRT‒PCR analysis of the tissues in Cohort 3 confirmed the downregulation of cFAM210A in HCC (Fig. 2c) and showed that the expression of cFAM210A was negatively correlated with the level of HBx in ANL tissues (Fig. 2d, Spearman r = −0.476, P < 0.001). Based on the median cFAM210A expression level in ANL tissues, we divided Cohort 3 into the cFAM210A-high group (n = 40) and the cFAM210A-low group (n = 40). Kaplan‒Meier survival analysis showed that the cFAM210A-low group had poorer late recurrence-free survival (P = 0.002) (Fig. 2e).
Taken together, these results showed that cFAM210A expression was negatively correlated with tumorigenesis in patients with HBV-related HCC.
HBx promotes the degradation of cFAM210A by increasing its m6A level
The abundance of a circRNA is determined by its biogenesis and degradation. CircRNAs are produced from precursor mRNAs (pre-mRNAs) by back-splicing, which can be affected by cis-acting elements (long flanking introns and intronic complementary sequences) and trans-acting factors (ADAR1, DHX9, FUS, NF90/NF110 and QKI)5,15. On the other hand, circRNAs can be degraded in four ways15: (i) circRNAs can be degraded by RNase L upon cellular infection with a double-stranded RNA (dsRNA) virus16; (ii) highly structured circRNAs can be undergo decay mediated by UPF1 and G3BP117; (iii) circRNAs can be degraded after being bound by microRNAs in an AGO2-dependent manner in unique situations18; and (iv) m6A-modified circRNAs can be degraded through an endoribonucleolytic cleavage pathway, that is, the YTHDF2-HRSP12-RNase P/MRP pathway19. In this pathway, YTHDF2 (an m6A reader), HRSP12 (an adaptor that bridges YTHDF2 and RNase P/MRP) and POP1 (a component of the endoribonuclease RNase P/MRP) are key proteins19.
First, we explored whether HBx can affect the biogenesis of cFAM210A. qRT‒PCR and Western blot analyses showed that the level of the FAM210A pre-mRNA (pFAM210A) and the mRNA and protein levels of FAM210A and the tested trans-acting factors did not show significant changes after HBx overexpression (Supplementary Fig. 2), indicating that HBx may not affect the transcription of either the FAM210A gene or the trans-acting factors of cFAM210A. In addition, the HBx protein could not directly affect the cis-acting elements of cFAM210A due to its inability to bind to nucleic acids independently20. Therefore, we assumed that HBx could not affect the biogenesis of cFAM210A.
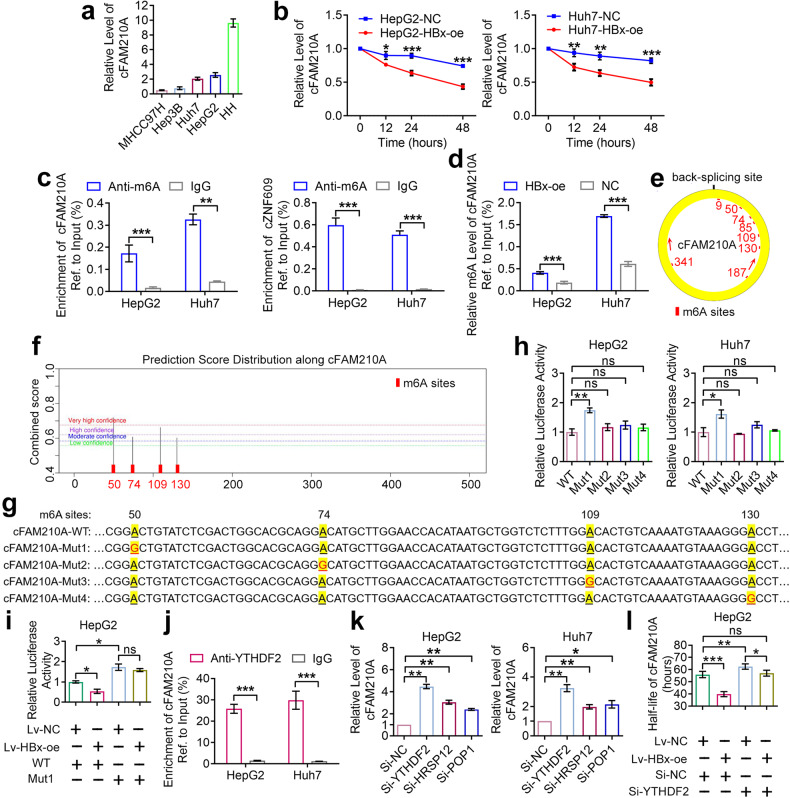
Then, we explored whether HBx can affect the degradation of cFAM210A. qRT‒PCR analysis showed that the expression of cFAM210A in primary human hepatocytes (HHs) was higher than that in HCC cells (HepG2, Huh7, Hep3B and MHCC97H), while its expression in HepG2 and Huh7 cells was higher than that in Hep3B and MHCC97H cells (Fig. 3a). After treatment with actinomycin D to block the biogenesis of cFAM210A, HBx overexpression resulted in significant downregulation of cFAM210A expression (Fig. 3b), indicating that HBx could promote the degradation of cFAM210A. As HBV is not a dsRNA virus, cFAM210A is not a highly structured circRNA (defined as a circRNA with a −ΔG/nt value ≥ 0.300; the −ΔG/nt value of cFAM210A was 0.254)17, and cFAM210A could not bind to AGO2 (Supplementary Fig. 3a); thus, the first three mechanisms of degradation are not applicable to cFAM210A.
Fig. 3. HBx promotes the degradation of cFAM210A by increasing its m6A level.
a qRT‒PCR results showing the expression of cFAM210A in human hepatocytes (HHs) and four HCC cell lines. b qRT‒PCR results showing the expression of cFAM210A after treatment with actinomycin D at the indicated time points. c MeRIP-qPCR analysis of m6A-modified cFAM210A in HCC cell lines. cZNF609 was used as a positive control. d MeRIP-qPCR analysis of m6A-modified cFAM210A in HBx-oe and NC cell lines. e, f The m6A sites of cFAM210A were predicted by CircPrimer 2.0 software (e) and SRAMP (f). The red arrows represent the primers used for MeRIP-qPCR. g Schematic representation of mutated m6A sites in cFAM210A. h Dual-luciferase assay result showing the relative luciferase activity of the wild-type (WT) and mutant (Mut)1/2/3/4 reporters. i Rescue assay results demonstrating that the relative luciferase activity of the WT reporter but not the Mut1 reporter was decreased by overexpression of HBx. j RIP-qPCR using an anti-YTHDF2 antibody was performed in HCC cell lines. k qRT‒PCR results showing the expression of cFAM210A after knockdown of YTHDF2, HRSP12 or POP1. l The actinomycin D treatment assay demonstrated that the half-life of cFAM210A was rescued by HBx overexpression in YTHDF2-depleted HepG2 cells. Student’s t test was used. NC negative control, HBx-oe HBx-overexpressing, ns not significant. *P < 0.05; **P < 0.01; ***P < 0.001. For (a) and (k), ACTB was used as an endogenous control.
Subsequently, we tried to investigate whether cFAM210A can be degraded by HBx in an m6A-dependent manner. The results of methylated RNA immunoprecipitation (MeRIP)-qPCR showed that cFAM210A was m6A modified (Fig. 3c; cZNF609, a well-known21,22 circular RNA, was used as a positive control) and that its relative m6A level was increased after HBx overexpression (Fig. 3d). Furthermore, four m6A sites (50, 74, 109 and 130) were predicted both by CircPrimer 2.023 (Fig. 3e) and SRAMP24 (Fig. 3f). As a dual-luciferase assay can be used to determine the key m6A sites25, we generated dual-luciferase reporter vectors by inserting the sequence of wild-type (WT) cFAM210A or mutant (Mut)1/2/3/4 (A to G; Mut1: site 50, Mut2: site 74; Mut3: site 109; Mut4: site 130) cFAM210A (Fig. 3g). The relative luciferase activity of the Mut1 (but not the Mut2, Mut3 or Mut4) reporter was higher than that of the WT reporter in both HepG2 and Huh7 cells (Fig. 3h), indicating that m6A site 50 is the key m6A site in cFAM210A. Moreover, the relative luciferase activity of the WT reporter but not the Mut1 reporter was decreased by overexpression of HBx (Fig. 3i). Therefore, we concluded that HBx could promote cFAM210A degradation by increasing its m6A level at m6A site 50.
Importantly, cFAM210A could be enriched by YTHDF2 (Fig. 3j) and was significantly upregulated upon knockdown of YTHDF2, HRSP12 or POP1 (Fig. 3k, Supplementary Fig. 3b). Furthermore, the half-life of cFAM210A was significantly increased after knockdown of YTHDF2 in HepG2 cells, and this increase was reversed by HBx overexpression (Fig. 3l, Supplementary Fig. 3c). Finally, HBx did not affect the expression of YTHDF2, HRSP12 or POP1 (Supplementary Fig. 3d, e).
In addition, the results of m6A dot blot assays showed that HBx increased the global m6A level of RNA extracted from HCC cells (Supplementary Fig. 4).
In summary, HBx can promote the m6A modification of cFAM210A at m6A site 50, and m6A-modified cFAM210A can be degraded via the YTHDF2-HRSP12-RNase P/MRP pathway.
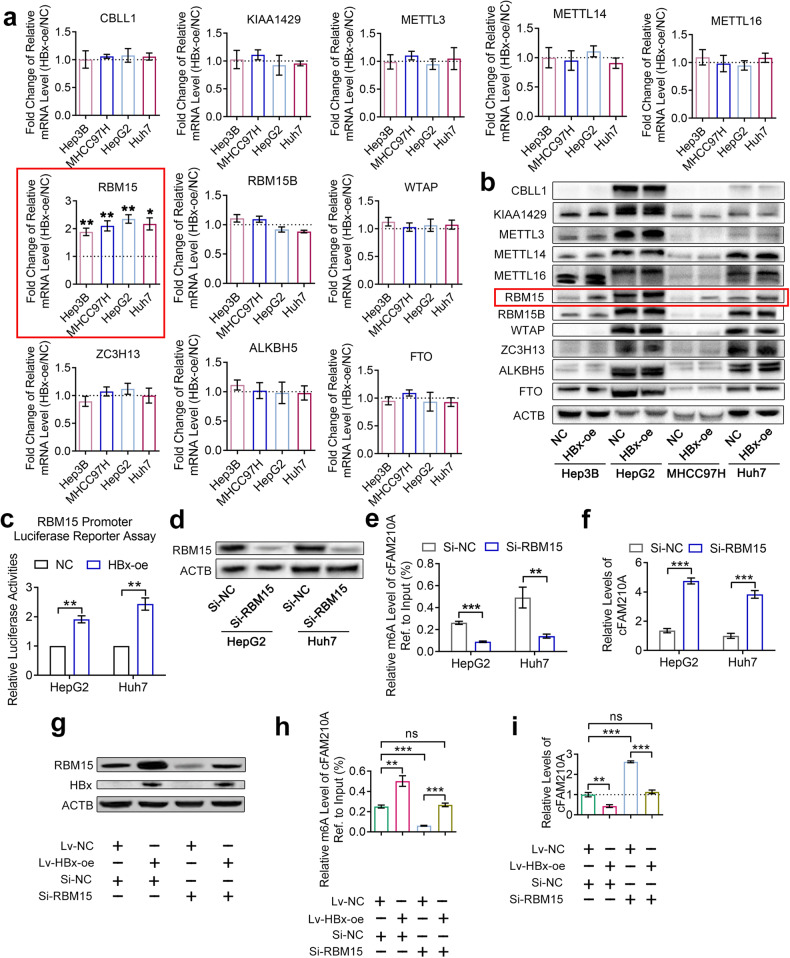
HBx increases the m6A level of cFAM210A by promoting the expression of RBM15
It has been reported that m6A methyltransferases (CBLL1 (HAKAI), KIAA1429 (VIRMA), METTL3, METTL14, METTL16 (METT10D), RBM15, RBM15B, WTAP and ZC3H13) can increase the m6A level of RNA, while m6A demethylases (ALKBH5 and FTO) can decrease the m6A level of RNA26. qRT‒PCR (Fig. 4a) and Western blot (Fig. 4b) analyses demonstrated that RBM15 but not the other methyltransferases or demethylases was significantly upregulated after HBx overexpression in HCC cell lines. In the dual-luciferase assay with a reporter containing the RBM15 promoter, overexpression of HBx promoted RBM15 transcription (Fig. 4c). Therefore, we concluded that HBx promoted the expression of RBM15 by inducing its transcription.
Fig. 4. HBx increases the m6A level of cFAM210A by promoting the expression of RBM15.
a, b qRT‒PCR (a) and Western blot (b) results showing the mRNA and protein levels of m6A regulators in HBx-oe and NC cells. c Dual-luciferase assay results indicating the activity of the RBM15 gene promoter in HBx-oe and NC cells. d Western blot results showing the knockdown efficiency of si-RBM15 in HCC cells. e, f MeRIP-qPCR (e) and qRT‒PCR (f) results showing the m6A level and expression of cFAM210A in si-RBM15 and si-NC cells. g Western blot results showing the knockdown efficiency of si-RBM15 and overexpression efficiency of the HBx-oe vector in HepG2 cells. h, i MeRIP-qPCR (h) and qRT‒PCR (i) results in HepG2 cells showing that the effects of si-RBM15 were abolished by overexpressing HBx. For (a), (f) and (i), ACTB was used as an endogenous control. For (a), (c), (e), (f), (h) and (i), Student’s t test was used. NC negative control, HBx-oe HBx-overexpressing, ns not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
After knocking down RBM15 (Fig. 4d), both the m6A level (Fig. 4e) and the expression (Fig. 4f) of cFAM210A were significantly decreased. Importantly, this effect was abolished by overexpressing HBx simultaneously (Fig. 4g–i).
In summary, HBx can increase the m6A level of cFAM210A by promoting the expression of RBM15 through transcriptional induction.
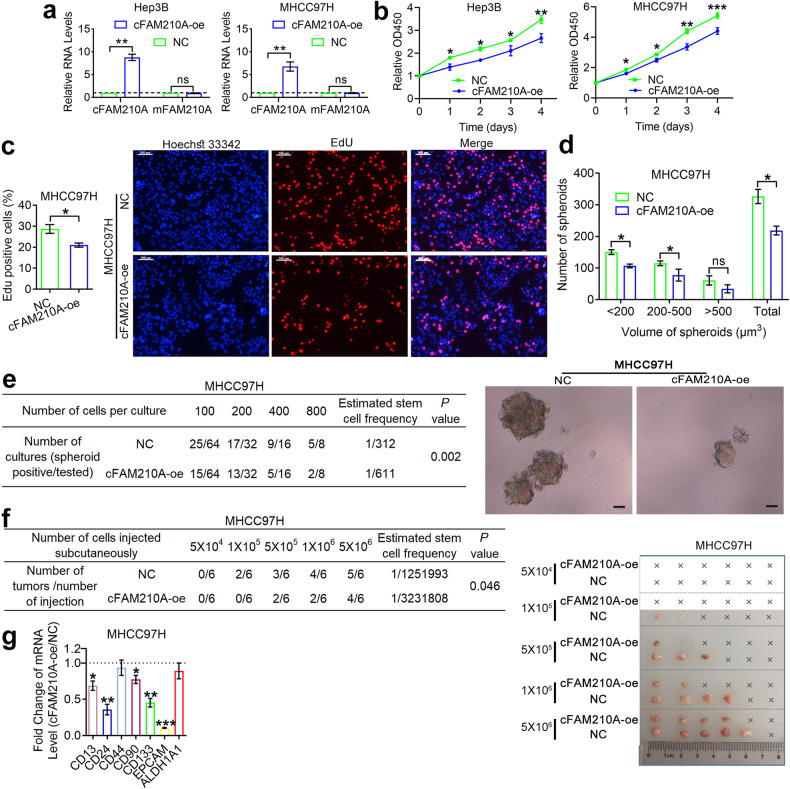
cFAM210A inhibits the proliferation, stemness, and tumorigenicity of HCC cells
To explore the biological functions of cFAM210A in HCC, we overexpressed cFAM210A in Hep3B and MHCC97H cells using a lentiviral vector (Fig. 5a). Notably, the mRNA level of FAM210A (mFAM210A) did not show significant changes after cFAM210A overexpression (Fig. 5a). The CCK-8 (Fig. 5b) and EdU incorporation (Fig. 5c) assay results showed that the proliferation ability of HCC cells was significantly reduced after cFAM210A overexpression. The results of spheroid formation assays (Fig. 5d) and in vitro (Fig. 5e) and in vivo (Fig. 5f) limiting dilution assays demonstrated that forced expression of cFAM210A suppressed the stemness and tumorigenicity of HCC cells. Furthermore, among the seven markers (CD13, CD24, CD44, CD90, CD133, EPCAM and ALDH1A1)27 of cancer stem cells (CSCs) in HCC, five were downregulated after cFAM210A overexpression (Fig. 5g).
Fig. 5. Overexpressing cFAM210A inhibits the proliferation, stemness, and tumorigenicity of HCC cells.
a qPCR results showing the expression of cFAM210A and mFAM210A after infection with cFAM210A-oe lentivirus and its negative control. b CCK-8 assays were performed in cFAM210A-oe HCC cells and their negative controls. c EdU incorporation assays were performed in MHCC97H cells (left). Representative images; scale bar, 100 μm (right). d Spheroid formation assays were performed in MHCC97H cells (top). Representative images (bottom). e, f In vitro (e) and in vivo (f) limiting dilution assays. The estimated stem cell frequency of each group was calculated by ELDA (http://bioinf.wehi.edu.au/software/elda/). g qRT‒PCR showing the expression of seven markers of cancer stem cells in HCC. For (a) and (g), ACTB was used as an endogenous control. For (a–d) and (g), Student’s t test was used. NC negative control, cFAM210A-oe cFAM210A-overexpressing lentivirus, ns not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
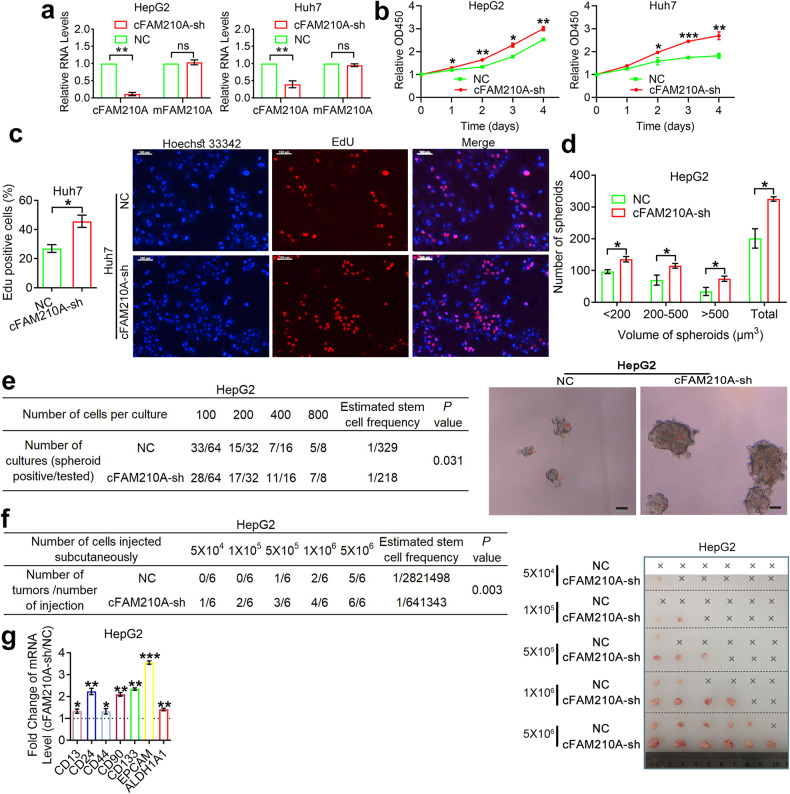
Next, we successfully knocked down cFAM210A in HepG2 and Huh7 cells (Fig. 6a). Consistent with the above findings, the CCK-8 (Fig. 6b) and EdU incorporation (Fig. 6c) assay results indicated the proliferation-inhibiting ability of cFAM210A. The results of spheroid formation assays (Fig. 6d) and in vitro (Fig. 6e) and in vivo (Fig. 6f) limiting dilution assays showed that the stemness and tumorigenicity of HCC cells were enhanced by knockdown of cFAM210A. Moreover, all seven detected markers of CSCs in HCC were upregulated after silencing of cFAM210A (Fig. 6g).
Fig. 6. Knocking down cFAM210A promotes the proliferation, stemness, and tumorigenicity of HCC cells.
a qPCR results showing the expression of cFAM210A and mFAM210A after using cFAM210A-sh lentivirus and its negative control. b CCK-8 assays were performed in cFAM210A-sh HCC cells and their negative controls. c EdU incorporation assays were performed in Huh7 cells (left). Representative images; scale bar, 100 μm (right). d Spheroid formation assays were performed in HepG2 cells (top). Representative images (bottom). e, f In vitro (e) and in vivo (f) limiting dilution assays. The estimated stem cell frequency of each group was calculated by ELDA (http://bioinf.wehi.edu.au/software/elda/); scale bar, 50 μm. g qRT‒PCR showing the expression of seven markers of cancer stem cells in HCC. For (a) and (g), ACTB was used as an endogenous control. For (a–d) and (g), Student’s t test was used. NC negative control, cFAM210A-sh, lentivirus-mediated short hairpin RNA against cFAM210A. ns not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Collectively, these findings indicated that cFAM210A inhibited the proliferation, stemness, and tumorigenicity of HCC cells in vitro and in vivo.
cFAM210A can bind to YBX1
It has been reported that circRNAs can function in the following three ways15: (i) binding to microRNAs in an AGO2-dependent manner5; (ii) being translated into proteins or peptides; and (iii) binding to proteins.
First, the results of RIP-qPCR using an anti-AGO2 antibody showed that cFAM210A could not bind to AGO2 (Supplementary Fig. 3a), meaning that cFAM210A may not bind to microRNAs. Furthermore, although circRNADb (http://reprod.njmu.edu.cn/cgi-bin/circrnadb/circRNADb.php) predicted that cFAM210A could be translated into a 159 AA protein, this protein could not be detected by Western blotting (Supplementary Fig. 5). In addition, SProtP Human (http://reprod.njmu.edu.cn/cgi-bin/sprotp/api.py) indicated that the estimated half-life of the predicted cFAM210A protein, even if it did exist, was very short (<30 min). Therefore, we concluded that cFAM210A may not function through translation into a protein or peptide.
To explore whether cFAM210A can bind to proteins (candidate proteins, CAPs), we performed a circRNA pull-down assay as previously described12. Briefly, a vector overexpressing MS2-fused cFAM210A (cFAM210A-MS2) and a vector overexpressing a Flag-tagged capture protein binding to MS2 (FCPM) were cotransfected into HepG2 cells. Then, the FCPM-cFAM210A-MS2-CAP complexes were enriched by immunoprecipitation with an anti-Flag antibody (Fig. 7a, b, Group 5). The vectors without the MS2 or FCPM sequence were used as controls (Fig. 7b, Groups 6–8). Subsequent Western blotting using an anti-Flag antibody showed that FCPM was successfully pulled down in Groups 5 and 6 (Supplementary Fig. 6a, b), while qRT‒PCR analysis indicated that cFAM210A was successfully pulled down only in Group 5 (Fig. 7c). The enriched complexes in Groups 5 and 6 were then analyzed by mass spectrometry. We detected 77 proteins in Group 5 (Supplementary Table 2) and 138 proteins in Group 6 (Supplementary Table 3). Among the detected proteins, 28 (Supplementary Table 4) were detected only in Group 5 (but not in Group 6) and were thought to be candidate proteins binding to cFAM210A.
We then overlapped these 28 candidate proteins with 416 established RNA binding proteins (RBPs) (http://rbpdb.ccbr.utoronto.ca)28,29 and obtained three candidate proteins: YBX1, RPS3 and SNRPF (Fig. 7d). CircRNA pull-down followed by western blotting (Fig. 7e), RIP-qPCR using antibodies against these three proteins (Fig. 7f, Supplementary Fig. 6c) and dual RNA fluorescence in situ hybridization (FISH) and immunofluorescence staining (Fig. 7g) demonstrated that cFAM210A bound to YBX1 but not PRS3 or SNRPF.
Moreover, RIP-qPCR using an anti-YBX1 antibody showed that mFAM210A cannot bind to YBX1 (Supplementary Fig. 7a). Compared to the sequence of mFAM210A, the sequence of cFAM210A differs only in the region spanning its back-splice site (BSS). Therefore, we assumed that the sequence spanning the BSS of cFAM210A was the key sequence for its binding to YBX1. To test this hypothesis, the vector overexpressing MS2-fused cFAM210A and lacking the 50 nucleotides spanning the BSS (cFAM210A-MS2-△BSS50) (Supplementary Fig. 7b) and the FCPM vector were cotransfected into HepG2 cells. Subsequent circRNA pull-down followed by Western blotting showed that YBX1 could not be pulled down (Supplementary Fig. 7c). These experiments demonstrated that the key YBX1-binding sequence in cFAM210A was located in the 50 nucleotides spanning its BSS.
In summary, cFAM210A can function by binding to the YBX1 protein but not by other mechanisms.
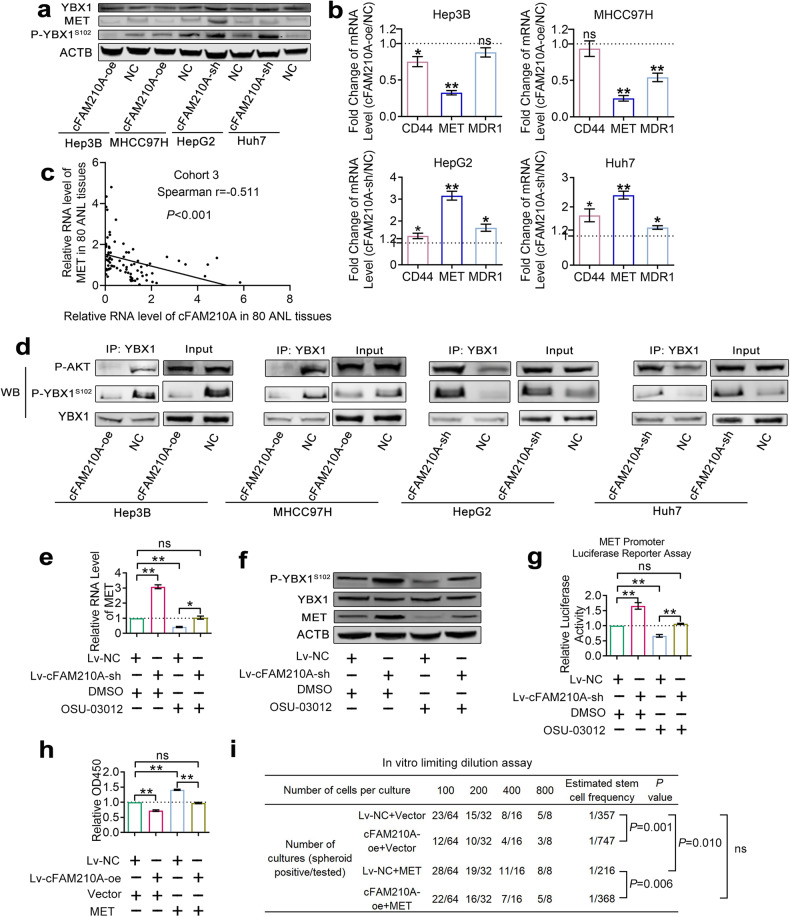
cFAM210A inhibits the phosphorylation of YBX1, suppressing its transactivation function toward MET
CircRNAs can bind to proteins, resulting in their translocation (from the nucleus to the cytoplasm, or vice versa)30 or affecting their functions30 or stability31,32. YBX1 can interact directly with promoters and induce the transcription of various genes33–36. Among these genes, CD4437, MET37 and MDR138 have been reported to play important roles in HCC stemness.
Western blotting showed that the expression (Fig. 8a) and localization (Supplementary Fig. 8) of YBX1 did not change significantly after overexpression or knockdown of cFAM210A. qRT‒PCR showed that cFAM210A inhibited the mRNA expression of YBX1-transactivated genes (CD44, MET and MDR1) (Fig. 8b). Among these genes, MET exhibited the most obvious change (Fig. 8b). Furthermore, the protein level of MET was decreased by cFAM210A (Fig. 8a). Importantly, the RNA levels of cFAM210A and MET were positively correlated in ANL tissues of patients with HBV-related HCC (Fig. 8c). Therefore, we concluded that cFAM210A could inhibit the ability of YBX1 to transactivate MET.
Fig. 8. cFAM210A inhibits the phosphorylation of YBX1, suppressing its transactivation function toward MET.
a Western blot results showing the protein levels of MET, YBX1 and P-YBX1S102 upon overexpression or silencing of cFAM210A. b qRT‒PCR results showing the expression of CD44, MET and MDR1 upon overexpression or silencing of cFAM210A. c The correlation between the RNA level of MET and that of cFAM210A in 80 ANL tissues was analyzed. d Western blot analysis following immunoprecipitation (IP) after overexpression or silencing of cFAM210A. e–g qRT‒PCR (e), Western blot (f) and MET promoter luciferase reporter assay (g) results showing that OSU-03012 suppressed the phosphorylation of YBX1S102 and downregulated the expression of MET, while these effects were abolished by silencing cFAM210A. h–i The CCK-8 assay (OD450 was measured on the third day) (h) and in vitro limiting dilution assay (i) results demonstrated that MET promoted the proliferation and stemness of HCC cells, while these effects were abolished by overexpressing cFAM210A. For (b), (e), (g) and (h), Student’s t test was used. For (i), the estimated stem cell frequency of each group was calculated by ELDA (http://bioinf.wehi.edu.au/software/elda/). NC negative control; cFAM210A-oe cFAM210A-overexpressing lentivirus, cFAM210A-sh lentivirus-mediated short hairpin RNA against cFAM210A, ns not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Then, we tried to explore the related mechanism. It has been reported that phosphoinositide dependent kinase-1 (PDK-1) can activate AKT by phosphorylating it and that phosphorylated AKT (P-AKT) can bind to and phosphorylate YBX1 at Ser102 (YBX1S102)39,40. YBX1 can bind to the promoter of MET, and phosphorylation of YBX1S102 could strengthen its transactivation function toward MET35. Furthermore, treatment with OSU-03012 (a PDK1 inhibitor) was found to decrease the phosphorylation of YBX1S102,35,40, thus inhibiting the transcription of MET, in normal and malignant human mammary cells35.
Interestingly, we found that cFAM210A inhibited the phosphorylation of YBX1S102 (Fig. 8a). Furthermore, immunoprecipitation (IP) experiments using an anti-YBX1 antibody showed that the enrichment of P-AKT decreased after overexpressing cFAM210A but increased after knockdown of cFAM210A (Fig. 8d). This result indicated that cFAM210A may prevent P-AKT from binding to YBX1. Moreover, qRT‒PCR analysis (Fig. 8e), Western blot analysis (Fig. 8f) and the MET promoter luciferase reporter assay (Fig. 8g) showed that OSU-03012 treatment suppressed the phosphorylation of YBX1S102 and downregulated the transcription and expression of MET, while these effects were abolished by silencing cFAM210A. Therefore, we concluded that cFAM210A suppressed the transcription of MET by inhibiting the phosphorylation of YBX1S102.
In addition, the results of CCK-8 (Fig. 8h) and in vitro limiting dilution (Fig. 8i) assays demonstrated that MET promoted the proliferation and stemness of HCC cells, while these effects were attenuated by overexpressing cFAM210A.
Finally, in HBV-infected HepG2.2.1541,42 and HepAD3843,44 cells, knockdown of HBx led to downregulation of RBM15 and MET, suppression of YBX1S102 phosphorylation and upregulation of cFAM210A (Supplementary Fig. 9a, b), while overexpression of HBx had the opposite effects (Supplementary Fig. 9b, c). These results further confirmed our findings.
In conclusion, cFAM210A, which is degraded in response to HBx-mediated m6A modification, inhibited tumorigenesis in HCC by suppressing the transactivation function of YBX1 toward MET. The graphical abstract of this study is shown in Supplementary Fig. 10.
Discussion
FAM210A has been reported to increase bone and muscle strength45 and to influence cardiac remodelling during heart failure46, but the role of FAM210A in tumours has not been mentioned before. In this study, we found that a circRNA derived from the FAM210A gene inhibited tumorigenesis in HBV-related HCC, demonstrating the function of the FAM210A gene in another aspect.
The expression levels of circRNAs are determined by multiple factors, and the modes of action of circRNAs vary15. In the present research, by a process of elimination, we determined that the downregulation of cFAM210A after HBx overexpression was attributed to m6A-dependent degradation via the YTHDF2-HRSP12-RNase P/MRP pathway and that cFAM210A inhibited tumorigenesis in HCC by suppressing the transactivation function of YBX1 toward MET but not by other mechanisms. To the best of our knowledge, this is the first report on the function of cFAM210A.
Rao et al.47. found that HBx could upregulate the expression of METTL3. However, they investigated only one m6A methyltransferase (METTL3) in a single cell line (HepG2), and the mechanism was not explored. In the present study, we investigated all known m6A methyltransferases and m6A demethylases in four HCC cell lines and found that RBM15 but not METTL3 was upregulated in all four cell lines. Mechanistically, the dual-luciferase assay results showed that HBx promoted the transcription of RBM15. Consistent with this finding, transcriptome sequencing data from a public database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64875)48 also suggested that the expression of RBM15 in HBx-oe HepG2 cells was higher than that in negative control cells (adjusted P = 0.021, fold change = 1.547).
A few studies47,49 have reported that m6A modification can affect the back-splicing of a subset of circRNAs, resulting in higher expression of these circRNAs and lower expression of the corresponding pre-mRNAs and mRNAs. However, in this research, after HBx overexpression, the m6A level of cFAM210A was increased, facilitating its degradation via the YTHDF2-HRSP12-RNase P/MRP pathway, while the expression of pFAM210A and mFAM210A did not show significant changes. These findings suggest the multifaceted roles of m6A modification of circRNAs50.
YBX1 is a well-known RBP with multiple functions33. YBX1 has been shown to be upregulated51, to be associated with poorer prognosis52, to promote tumorigenesis52 and to mediate sorafenib resistance53 in HCC. Several studies have reported that circRNAs can bind to YBX1 to affect its stability, location or function30. For example, circRNA-SORE can sequester YBX1 in the cytoplasm and prevent it from being degraded by nuclear PRP1931, while circNfix can mediate the degradation of Ybx1 by inducing its ubiquitination in mice32. CircAnks1a can recruit Ybx1 to the promoter of Vegfb and activate its transcription in rodents54. CircACTN4 can recruit YBX1 to the promoter of FZD7 and activate its transcription in human intrahepatic cholangiocarcinoma cells55. In this research, we first reported that circRNA-cFAM210A can inhibit the phosphorylation of YBX1 to suppress its transactivation function, uncovering another mode of action linking circRNAs and YBX1. Notably, YBX1 can function in various processes, including transactivation, DNA repair, DNA replication, pre-mRNA splicing, and disassembly of nucleoli33. However, whether cFAM210A can affect the functions of YBX1, except for transactivation, in other ways needs to be further explored.
In addition, the mechanism by which cFAM201A is involved in the phosphorylation of YBX1 awaits further investigation.
In summary, this study suggests the important role of circular RNAs in HBx-induced hepatocarcinogenesis, making cFAM210A a potential target in the prevention and treatment of HBV-related HCC.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81972657, 81521091, 81802983, 81830085, 81972575, 82002458, 82073031 and 82273342), Clinical Research Plan of SHDC (Nos. SHDC2020CR5007 and SHDC22020213), San Hang Program of Naval Medical University and Meng Chao’s talent training program for young doctors.
Data availability
The Gene Expression Omnibus accession number for the whole-transcriptome sequencing data in HBx-overexpressing (HBx-oe) and NC (negative control) HepG2 cells is GSE215232.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jian Yu, Wen Li, Guo-jun Hou, Da-peng Sun.
Contributor Information
Gang Huang, Email: squaror@163.com.
Wei-ping Zhou, Email: ehphwp3@126.com.
Fu Yang, Email: yangfusq1997@smmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s12276-023-01108-8.
References
- 1.Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 2016;64:S84–s101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi VK, et al. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb. Pathog. 2019;128:184–194. doi: 10.1016/j.micpath.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Adelman K, Egan E. Non-coding RNA: more uses for genomic junk. Nature. 2017;543:183–185. doi: 10.1038/543183a. [DOI] [PubMed] [Google Scholar]
- 5.Kristensen LS, et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 6.Sun HD, et al. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci. Rep. 2018;8:10111. doi: 10.1038/s41598-018-27837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sang Y, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol. Ther. 2019;27:1638–1652. doi: 10.1016/j.ymthe.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J. Exp. Clin. Cancer Res. 2019;38:398. doi: 10.1186/s13046-019-1376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachmayr-Heyda A, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, et al. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: a large-scale, multicenter study. Int. J. Cancer. 2020;146:1754–1763. doi: 10.1002/ijc.32647. [DOI] [PubMed] [Google Scholar]
- 12.Zheng R, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol. Cancer. 2022;21:49. doi: 10.1186/s12943-021-01471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu QG, et al. A novel HBx genotype serves as a preoperative predictor and fails to activate the JAK1/STATs pathway in hepatocellular carcinoma. J. Hepatol. 2019;70:904–917. doi: 10.1016/j.jhep.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Utsunomiya T, et al. Molecular signatures of noncancerous liver tissue can predict the risk for late recurrence of hepatocellular carcinoma. J. Gastroenterol. 2010;45:146–152. doi: 10.1007/s00535-009-0164-1. [DOI] [PubMed] [Google Scholar]
- 15.Yang L, Wilusz JE, Chen LL. Biogenesis and Regulatory Roles of Circular RNAs. Annu. Rev. Cell Dev. Biol. 2022;38:263–289. doi: 10.1146/annurev-cellbio-120420-125117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu CX, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880.e821. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 17.Fischer JW, Busa VF, Shao Y, Leung AKL. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell. 2020;78:70–84.e76. doi: 10.1016/j.molcel.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen TB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park OH, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell. 2019;74:494–507.e498. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 20.Maguire HF, Hoeffler JP, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, et al. Emerging roles of Circ-ZNF609 in multiple human diseases. Front. Genet. 2022;13:837343. doi: 10.3389/fgene.2022.837343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian Y, et al. circ-ZNF609: a potent circRNA in human cancers. J. Cell Mol. Med. 2021;25:10349–10361. doi: 10.1111/jcmm.16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong S, Feng J. CircPrimer 2.0: a software for annotating circRNAs and predicting translation potential of circRNAs. BMC Bioinform. 2022;23:215. doi: 10.1186/s12859-022-04705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, et al. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11:2578. doi: 10.1038/s41467-020-16306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji J, Wang XW. Clinical implications of cancer stem cell biology in hepatocellular carcinoma. Semin Oncol. 2012;39:461–472. doi: 10.1053/j.seminoncol.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011;39:D301–D308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol. Med. 2019;11:e10835. doi: 10.15252/emmm.201910835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou WY, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol. Cancer. 2020;19:172. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Sig. Trans. Target Ther. 2020;5:298. doi: 10.1038/s41392-020-00375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc) 2011;76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 34.To K, et al. Y-box binding protein-1 induces the expression of CD44 and CD49f leading to enhanced self-renewal, mammosphere growth, and drug resistance. Cancer Res. 2010;70:2840–2851. doi: 10.1158/0008-5472.CAN-09-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finkbeiner MR, et al. Profiling YB-1 target genes uncovers a new mechanism for MET receptor regulation in normal and malignant human mammary cells. Oncogene. 2009;28:1421–1431. doi: 10.1038/onc.2008.485. [DOI] [PubMed] [Google Scholar]
- 36.Sengupta S, Mantha AK, Mitra S, Bhakat KK. Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene. 2011;30:482–493. doi: 10.1038/onc.2010.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rani B, et al. Galunisertib suppresses the staminal phenotype in hepatocellular carcinoma by modulating CD44 expression. Cell Death Dis. 2018;9:373. doi: 10.1038/s41419-018-0384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. CXCL11 promotes self-renewal and tumorigenicity of α2δ1(+) liver tumor-initiating cells through CXCR3/ERK1/2 signaling. Cancer Lett. 2019;449:163–171. doi: 10.1016/j.canlet.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Sutherland BW, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 40.To, K. et al. The phosphoinositide-dependent kinase-1 inhibitor 2-amino-N-[4-[5-(2-phenanthrenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]phenyl]-acetamide (OSU-03012) prevents Y-box binding protein-1 from inducing epidermal growth factor receptor. Mol. Pharmacol. 72, 641–652 (2007). [DOI] [PubMed]
- 41.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl Acad. Sci. USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, et al. Chronic ethanol consumption and HBV induce abnormal lipid metabolism through HBx/SWELL1/arachidonic acid signaling and activate Tregs in HBV-Tg mice. Theranostics. 2020;10:9249–9267. doi: 10.7150/thno.46005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King RW, Ladner SK. Hep AD38 assay: a high-throughput, cell-based screen for the evaluation of compounds against Hepatitis B virus. Methods Mol. Med. 2000;24:43–50. doi: 10.1385/1-59259-245-7:43. [DOI] [PubMed] [Google Scholar]
- 44.Stadelmayer B, et al. Full-length 5’RACE identifies all major HBV transcripts in HBV-infected hepatocytes and patient serum. J. Hepatol. 2020;73:40–51. doi: 10.1016/j.jhep.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka KI, et al. FAM210A is a novel determinant of bone and muscle structure and strength. Proc. Natl. Acad. Sci. USA. 2018;115:E3759–e3768. doi: 10.1073/pnas.1719089115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J, et al. MicroRNA-574 regulates FAM210A expression and influences pathological cardiac remodeling. EMBO Mol. Med. 2021;13:e12710. doi: 10.15252/emmm.202012710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao X, et al. N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life. 2021;73:408–417. doi: 10.1002/iub.2438. [DOI] [PubMed] [Google Scholar]
- 48.Chan C, et al. Global re-wiring of p53 transcription regulation by the hepatitis B virus X protein. Mol. Oncol. 2016;10:1183–1195. doi: 10.1016/j.molonc.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Timoteo G, et al. Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 2020;31:107641. doi: 10.1016/j.celrep.2020.107641. [DOI] [PubMed] [Google Scholar]
- 50.Lin H, Wang Y, Wang P, Long F, Wang T. Mutual regulation between N6-methyladenosine (m6A) modification and circular RNAs in cancer: impacts on therapeutic resistance. Mol. Cancer. 2022;21:148. doi: 10.1186/s12943-022-01620-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gunasekaran VP, Ganeshan M. Inverse correlation of ribosomal protein S27A and multifunctional protein YB-1 in hepatocellular carcinoma. Clin. Biochem. 2014;47:1262–1264. doi: 10.1016/j.clinbiochem.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Chao HM, et al. Y-box binding protein-1 promotes hepatocellular carcinoma-initiating cell progression and tumorigenesis via Wnt/β-catenin pathway. Oncotarget. 2017;8:2604–2616. doi: 10.18632/oncotarget.13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao LZ, et al. Y-box binding protein-1 promotes epithelial-mesenchymal transition in sorafenib-resistant hepatocellular carcinoma cells. Int. J. Mol. Sci. 2020;22:224. doi: 10.3390/ijms22010224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang SB, et al. CircAnks1a in the spinal cord regulates hypersensitivity in a rodent model of neuropathic pain. Nat. Commun. 2019;10:4119. doi: 10.1038/s41467-019-12049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Q, et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J. Hepatol. 2022;76:135–147. doi: 10.1016/j.jhep.2021.08.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Gene Expression Omnibus accession number for the whole-transcriptome sequencing data in HBx-overexpressing (HBx-oe) and NC (negative control) HepG2 cells is GSE215232.