Abstract
The production of the acidic exopolysaccharide succinoglycan (EPS I) by Rhizobium meliloti exoP* mutants expressing an ExoP protein lacking its C-terminal cytoplasmic domain and by mutants characterized by specific amino acid substitutions in the proline-rich motif (RX4PX2PX4SPKX9IXGXMXGXG) located from positions 443 to 476 of the ExoP protein was analyzed. The absence of the C-terminal cytoplasmic ExoP domain (positions 484 to 786) and the substitution of both arginine443 by isoleucine443 and proline457 by serine457 within the proline-rich motif resulted in enhanced production of low-molecular-weight (LMW) EPS I at the expense of high-molecular-weight (HMW) EPS I. The ratios of HMW to LMW EPS I of the wild type and mutant strains increased with osmolarity.
The gram-negative soil bacterium Rhizobium meliloti SU47 (Sinorhizobium meliloti SU47) and its derivative strains Rm2011 and Rm1021 produce the acidic exopolysaccharide succinoglycan (EPS I), which plays an important role in the invasion of Medicago sativa root nodules by R. meliloti (26). EPS I is a polymer of octasaccharide repeating units. Each repeating unit contains seven glucose molecules and one galactose molecule joined by β-1,4, β-1,3, and β-1,6 glycosidic linkages and can be decorated by acetyl, succinyl, and pyruvyl groups (1, 29). R. meliloti Rm1021 produces a high-molecular-weight (HMW) and a low-molecular-weight (LMW) form of EPS I (4). Breedveld et al. (10) reported that an increase of the osmotic pressure resulted in enhanced production of HMW EPS I at the expense of LMW EPS I.
Twenty-one exo and 2 exs genes involved in the biosynthesis of EPS I are located in a 27-kb gene cluster on megaplasmid 2 (7, 26). The combination of genetic and biochemical approaches allowed the assignment of functions to most of the exo gene products and resulted in a detailed model for the biosynthesis of the EPS I repeating unit (26, 31). The membrane-associated proteins ExoP, ExoQ, and ExoT were determined to be involved in polymerization and export of EPS I (6, 31). Recently, González et al. (21) reported evidence that the ExoQ protein is involved in the biosynthesis of HMW EPS I, whereas ExoT was suggested to be involved in the synthesis of EPS I octasaccharide trimers and tetramers. Moreover, ExoP was found to be essential for the synthesis of HMW and LMW EPS I octasaccharide multimers (21).
ExoP consists of 786 amino acids and can be divided into an N-terminal domain (positions 1 to 481), mainly located in the periplasm, and a C-terminal cytoplasmic domain (positions 482 to 786) (6). The C-terminal cytoplasmic domain contains a putative ATP binding motif (6). R. meliloti Rm2011 exoP* mutants characterized by a deletion of the 3′ portion of exoP encoding the C-terminal cytoplasmic domain produced a reduced amount of EPS I. In addition, the ratio of HMW to LMW EPS I secreted by these mutants was decreased (6).
Similarities of the N-terminal domain of ExoP to CLD (Rol and Wzz) proteins involved in the determination of O-antigen chain length (33) together with the phenotype of exoP* mutants suggested that ExoP might be involved in regulating the size distribution of EPS I (6). Although the amino acid sequences of the N-terminal ExoP domain and the similar proteins were only weakly conserved, these proteins displayed structural similarities, since they were characterized by two putative transmembrane helices and a conserved proline-rich amino acid motif located on the periplasmic side, very close to the second membrane helix (2, 6, 28, 32). Bastin et al. (2) proposed that the regulation of O-antigen chain length by CLD proteins might involve the interaction of these proteins with the O-antigen polymerase. They indicated that the proline-rich segment of CLD proteins together with conserved glycine residues of the putative second transmembrane helix may be involved in a protein-protein interaction of this region with an integral membrane protein. To date no experimental evidence for the functional significance of the conserved proline-rich segment has been reported.
In this study, we analyzed the size distribution of EPS I produced by R. meliloti exoP* mutants lacking the C-terminal cytoplasmic ExoP domain and mutants characterized by specific amino acid substitutions in the proline-rich motif of ExoP.
Production of HMW EPS I by R. meliloti exoP* is induced by increasing the osmotic pressure.
Sodium chloride-induced variations of HMW and LMW EPS I production by the R. meliloti wild-type strain Rm2011 (14) and mutant RmΔP*1 (6) were analyzed (Table 1; Fig. 1). Mutant RmΔP*1 contained the exoP* gene encoding a truncated ExoP protein that comprises amino acids 1 to 484 instead of the complete ExoP protein (Fig. 2). In GMS medium (34) supplemented with sodium chloride to an osmolarity of 0.09 to 0.48 osmol/liter no significant difference between the growth rates of these two strains was observed. Culture supernatants of the exoP* mutant RmΔP*1 contained approximately 10 to 30% of the total amount of EPS I produced by the wild type (Table 1).
TABLE 1.
NaCl-induced osmotic effects on production of HMW EPS I and LMW EPS I by R. meliloti Rm2011 mutants characterized by alterations in the ExoP proteina
| Strain/plasmid | Mutation | Glucose equivalents (mg/liter), HMW EPS:LMW EPS (%) at osmolarity (osmol/liter):
|
||||
|---|---|---|---|---|---|---|
| 0.09 | 0.18 | 0.29 | 0.38 | 0.48 | ||
| Rm2011 | None (wild type) | 790, 31:69 | 802, 58:42 | 782, 72:28 | 785, 79:21 | 766, 77:23 |
| RmΔP*1 | exoP* | 90, 0:100 | 250, 11:89 | 265, 27:73 | 250, 49:51 | 243, 51:49 |
| RmΔexoP/pExoP | None (wild type) | 781, 30:70 | 792, 57:43 | 782, 73:27 | 786, 78:22 | 770, 77:23 |
| RmΔexoP/pExoP-R443L | exoP, R443L | 777, 3:97 | 788, 22:78 | 782, 50:50 | 784, 54:46 | 766, 59:41 |
| RmΔexoP/pExoP-P457S | exoP, P457S | 803, 2:98 | 807, 20:80 | 805, 52:48 | 789, 58:42 | 775, 60:40 |
The data represent averages of at least five independent experiments. Strains were grown for 10 days in GMS medium supplemented with sodium chloride to the osmolarity specified. Standard deviations were equal to or less than 8 and 4% for the determination of the total amount of extracellular carbohydrates in glucose equivalents and the ratio of HMW to LMW EPS, respectively.
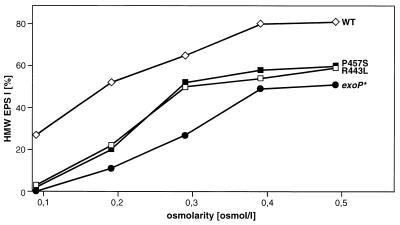
FIG. 1.
HMW EPS I production of different R. meliloti strains. The HMW EPS I production of the R. meliloti Rm2011 wild-type strain (WT), the exoP* mutant RmΔP*1 (exoP*), and strain RmΔexoP carrying the plasmids pExoP-R443L (R443L) and pExoP-P457S (P457S) in GMS medium supplemented with sodium chloride to osmolarities of 0.09 to 0.48 osmol/liter is shown. The percentage of the HMW EPS I fraction in relation to the total amount of EPS I is given. Values are averages of at least five independent experiments. Standard deviations were equal to or less than 4%.
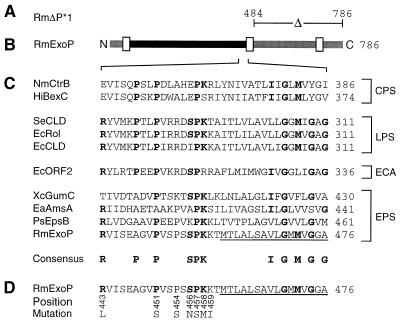
FIG. 2.
Characteristic features of different R. meliloti ExoP mutant proteins. (A) The part deleted in the ExoP protein encoded by the exoP* gene of mutant RmΔP*1 (6) is indicated. (B) Linear scheme of the R. meliloti wild-type ExoP protein. Putative transmembrane or membrane-associated segments (6) are marked by white boxes. Parts of ExoP probably located in the periplasm and the cytoplasm are indicated in black and grey, respectively. (C) Alignment of partial sequences of ExoP and similar proteins involved in the biosynthesis of capsular polysaccharides (CPS), lipopolysaccharides (LPS), entobacterial common antigen (ECA), and exopolysaccharides (EPS). Residues identical in at least four proteins of three different groups are printed in bold letters and are included in the consensus sequence. (D) Single substitutions of amino acid residues of the ExoP protein. Numbers below the partial ExoP sequence indicate the positions of the amino acid residues replaced by the residues indicated. Amino acid residues of the ExoP protein which are probably part of a membrane spanning helix are underlined. Numbers to the right indicate amino acid positions. Abbreviations: PsEpsB, Pseudomonas solanacearum EpsB (23); EaAmsA, Erwinia amylovora AmsA (11); EcCLD, E. coli O111 CLD (2); EcORF2, E. coli K-12 ORF2 protein (27); EcRol, E. coli O75 Rol (3); HiBexC, Haemophilus influenzae BexC (24); NmCtrB, Neisseria meningitidis CtrB (18); SeCLD, Salmonella enterica LT2 CLD; RmExoP, R. meliloti ExoP (5); XcGumC, Xanthomonas campestris GumC (6, 13).
After dialysis (molecular weight cutoff, 1,000 Da) of the culture supernatants and lyophilization, the HMW and LMW EPS fractions were separated by gel permeation chromatography on Nucleogel columns (2 × GFC 4000-8, 1 × GFC 300-8, 300 by 7.7 mm; Machery-Nagel, Germany; flow rate, 0.8 ml/min; 200 mM sodium chloride–200 mM sodium phosphate buffer [pH 7.0]). EPS fractions were detected by using a differential refraction index detector, and total carbohydrates were quantified by the HCl–l-cysteine method (15). In accordance with the observations of Breedveld et al. (10) an increase in the osmotic pressure of the culture medium supplemented with sodium chloride resulted in enhanced production of HMW EPS I at the expense of LMW EPS I in the wild-type strain, Rm2011 (Table 1; Fig. 1). Culture supernatants of the exoP* mutant contained significantly less HMW EPS I than wild-type cultures under all osmotic conditions tested (Table 1; Fig. 1). In medium of high osmolarity (0.48 osmol/liter) the exoP* mutant RmΔP*1 produced approximately 50% HMW EPS I in relation to the total amount of EPS I.
To verify that EPS I was exclusively secreted by the R. meliloti strains under the growth conditions used, the glucose and galactose contents of the EPS fractions were quantified by enzymatic assays (9, 25) after hydrochloric acid hydrolysis and neutralization. All EPS fractions tested contained glucose and galactose in ratios of 6.9:1 to 7.1:1, indicating that these fractions solely contained EPS I.
The osmotically induced production of HMW EPS I by exoP* mutants exclusively expressing the N-terminal ExoP domain indicates that the C-terminal cytoplasmic ExoP domain and therefore the putative binding and hydrolysis of ATP are not essential for HMW EPS I production. In addition, the C-terminal domain is not essential for the osmotically induced changes in the ratio of HMW to LMW EPS I. If ExoP plays a role in the osmotically induced alteration of this ratio, the presence of the N-terminal ExoP domain is sufficient to promote this change. High salt concentrations may influence the biosynthesis or the export of HMW or LMW EPS I. This influence may be exerted on the level of protein expression or stability.
Substitution of specific amino acid residues in the proline-rich motif of the ExoP protein.
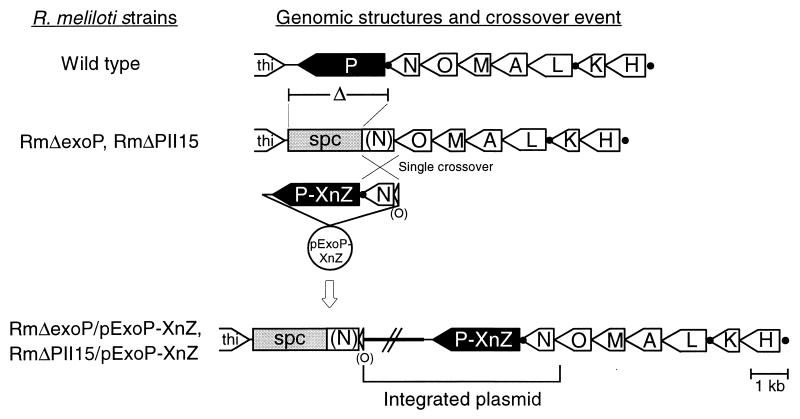
To analyze the relevance of the proline-rich segment of the R. meliloti Rm2011 ExoP protein, R. meliloti Rm2011 mutants expressing ExoP proteins characterized by specific amino acid substitutions in this segment were constructed. These substitutions were localized in the periplasmic portion of the ExoP protein (Fig. 2D). The exoP mutant genes were expressed in an R. meliloti exoP deletion mutant to exclude interference of ExoP proteins encoded by the endogenous gene and ExoP proteins encoded by the exoP mutant genes. exoP mutants are usually characterized by a slow growth rate, which is most likely due to an accumulation of lipid-linked intermediates of the EPS I biosynthetic pathway (30). Consistent with this observation, R. meliloti mutants lacking the exoP gene grew slowly and were not viable in media of low osmolarities (<0.1 osmol/liter) (data not shown). Therefore, mutant RmΔexoP, characterized by a deletion comprising 40 nucleotides of the 3′ terminus of the exoN coding region, the intergenic region between exoN and exoP, the complete exoP coding region, and 262 nucleotides downstream of the exoP coding region, was constructed (Fig. 3). The spectinomycin resistance cassette derived from pHP45Ω (16) was inserted into the NcoI site located 25 nucleotides downstream of the deletion site. Mutant RmΔexoP displayed a growth rate comparable to the growth rate of the wild type and grew in media of low osmolarities. Mutations in exoN cause a reduction of EPS I production, since this gene encodes a UDP-glucose pyrophosphorylase that participates in the synthesis of the nucleotide sugar precursors for EPS I biosynthesis (5, 20). In exoN mutants the activity of another pyrophosphorylase that can substitute for ExoN may account for the synthesis of enough UDP-glucose to produce the reduced amount of EPS I. Therefore, the additional mutation in the exoN gene probably reduced the deleterious accumulation of EPS I biosynthetic intermediates induced by the mutation of exoP.
FIG. 3.
Strategy for site-directed mutagenesis of the exoP gene from R. meliloti. At the top the structure of the operon comprising the genes exoH to exoP of the exo gene cluster from R. meliloti Rm2011 (5) is shown. Promoters directing the transcription of exoP (5) are indicated by black dots. Mutants RmΔexoP and RmΔPII15 were constructed by deletion of the complete exoP gene and a part of the exoN coding region of wild-type R. meliloti Rm2011 and the expA1 mutant RmAR1015 (8), respectively. The deleted fragment was replaced by a spectinomycin resistance cassette (spc). Due to the integration of pExoP-XnZ plasmids into the genomes of these mutants by homologous recombination, the native structure of the exoN-exoP region was restored. Incomplete genes are printed in parentheses. The pExoP-XnZ plasmids contained mutated exoP genes carrying single base pair substitutions causing the replacement of amino acid residue X in position n of the ExoP protein for residue Z (Fig. 2D).
Since R. meliloti Rm2011 has the cryptic ability to produce EPS II (19), the interposon insertion expA1#1015-lacZ-aacC1 (8) blocking the biosynthesis of EPS II was transferred to RmΔexoP by φM12-mediated transduction (17). The resulting mutant, RmΔPII15, and mutant RmΔexoP were used as recipient strains to analyze the effects of alterations in the proline-rich motif of ExoP on the production of EPS I.
Seven independent base pair substitutions affecting the amino acid sequence of the proline-rich segment of ExoP were introduced into plasmid pSDM-P. This plasmid resulted from the insertion of the internal 876-bp XhoI-EcoRI fragment of exoP into the vector pHIP2 (8). A mutagenic primer (Table 2) carrying the mutation to be inserted into the exoP gene and a selective primer (Table 2) characterized by a substitution that eliminates the internal EcoRV site of the aacC1 gene of pSDM-P were simultaneously annealed to one strand of the denaturated plasmid. After completion of the second strand, using T7 DNA polymerase and T4 DNA ligase, nonmutated plasmids were excluded from transformation due to linearization by EcoRV restriction. Plasmids were introduced into Escherichia coli XLmutS (Stratagene) by transformation, and nonmutated plasmids were again eliminated from plasmid DNA isolated from the pool of transformants by EcoRV restriction. After transformation of E. coli XL1-Blue (12) with this plasmid pool, single clones were tested for the desired mutation in exoP by DNA sequencing. Subsequently, the 371-bp BglII-EcoRI wild-type fragment of the exoP gene of pExoP was replaced by the corresponding fragment of pSDM-P carrying the base pair substitution. Mutations in exoP carried by pExoP derivative plasmids were verified by DNA sequencing. Plasmid pExoP carried the fragment deleted from the exoP-exoN region of strains RmΔexoP and RmΔPII15. Additionally, it contained the first part of the exoN gene and a part of the 3′ terminus of the exoO coding region (Fig. 3). Therefore, integration of pExoP containing the wild-type exoP gene and its derivative plasmids carrying the exoP mutant genes into the genomes of RmΔexoP and RmΔPII15 by homologous recombination occurred upstream of the deletion site, thereby restoring the native structure of the exoN-exoP region (Fig. 3). The genomic structures of the exoO-exoN-exoP regions of the mutants were verified by Southern hybridization. Since ExoP may interact with other proteins involved in EPS I biosynthesis, the copy number of exoP and the regulation of exoP gene expression is probably critical for EPS I production. The effect of specific amino acid substitutions in ExoP was therefore analyzed using the R. meliloti strains which carried a single exoP mutant gene at the native site in the genome.
TABLE 2.
Selective and mutagenic primers
| Primer | Sequencea |
|---|---|
| Selective | CACTTTGACATCGACCCAAGTACC |
| Mutagenic primers | |
| R443L | GCCAAGGCTCTCGTCATCTCC |
| P451S | GCCGGCGTGTCCGTGTCGCCG |
| P454S | CCCGTGTCGTCGTCGAGCCCC |
| S456N | TCGCCGTCGAACCCCAAGAAA |
| P457S | CCGTCGAGCTCCAAGAAAACC |
| K458M | TCGAGCCCCATGAAAACCATG |
| K459I | AGCCCCAAGATAACCATGACT |
Substitutions in the selective and the mutagenic primers are underlined.
In Fig. 2C the amino acid sequences of the proline-rich segment of ExoP and a selection of proteins similar to the N-terminal domain of ExoP are compared. These proteins are involved in the biosynthesis of either capsular polysaccharides, lipopolysaccharides, enterobacterial common antigens, or exopolysaccharides. The substituted seven amino acid residues localized in the periplasmic portion of the ExoP protein (Fig. 2D) are either invariant or at least strongly conserved in the proline-rich segment of ExoP and similar proteins.
Size distribution of succinoglycan produced by strains characterized by specific mutations in the proline-rich motif of the ExoP protein.
Since the osmotic pressure of the culture medium influences the ratio of HMW to LMW EPS I produced by R. meliloti (10), the size distributions of EPS I produced by the wild-type Rm2011, the expA1 mutant RmAR1015, and the strains characterized by specific mutations in the proline-rich motif of ExoP in media of various osmolarities were compared. After cultivation of the merodiploid R. meliloti strains the presence of the plasmid carrying the various exoP genes was verified by comparison of the total cell titer to the titer of neomycin-resistant cells. In all cases the total cell number corresponded to the number of neomycin-resistant cells. This indicates that during cultivation the plasmid was not lost in a significant number of cells by homologous recombination.
As a control, the EPS I production of the wild-type Rm2011 and the expA1 mutant RmAR1015 was compared to that of the exoP deletion mutant RmΔexoP and the exoP deletion-expA1 mutant RmΔPII15, both carrying plasmid pExoP with the wild-type exoP gene. No significant differences between the growth rates, the total amount of EPS I in the culture supernatant, and the ratio of HMW to LMW EPS I secreted to the medium were observed for the wild-type Rm2011 and RmΔexoP carrying pExoP (Table 1) or for RmAR1015 and RmΔPII15 containing pExoP (data not shown). In addition, all EPS fractions obtained from culture supernatants of these strains contained glucose and galactose in the ratios of 6.9:1 to 7.1:1, indicating that the sole exopolysaccharide in these fractions was EPS I. These results show that the mutants RmΔexoP and RmΔPII15 in combination with derivative plasmids of pExoP containing exoP mutant genes can be used to analyze the effects of the different mutations in exoP on the production of EPS I.
Compared to strains RmΔexoP and RmΔPII15, containing an integrated wild-type copy of exoP, these strains displayed no significant differences in growth rate and the total amount of EPS I isolated from the culture supernatants when carrying the exoP mutant genes. The ratios of glucose to galactose of 6.9:1 to 7.1:1 determined for the HMW and LMW EPS I fractions of these strains indicated that EPS I was the sole exopolysaccharide in these fractions.
Virtually the same ratio of HMW to LMW EPS I was determined for RmΔexoP carrying pExoP plasmids with exoP mutant genes as for RmΔPII15 carrying the identical plasmids. With respect to the ratio of HMW to LMW EPS I, these strains could be grouped into two classes. Substitution of arginine443 by leucine443 (R443L) and proline457 by serine457 (P457S) resulted in enhanced production of LMW EPS I at the expense of HMW EPS I (Table 1, Fig. 1). On the other hand, strains carrying the mutation R443L or P457S produced more HMW EPS I than did mutant RmΔP*1 (Fig. 1) and as much EPS I as the wild type (Table 1). The phenotype of these mutants therefore supports the hypothesis that the reduced EPS I production by exoP* mutants results from alterations in the stability or the amount of either exoP* transcripts or the N-terminal ExoP domain. These alterations might be explained either by the absence of the part of the exoP gene encoding the C-terminal domain or by the absence of this domain.
In contrast to the substitutions R443L and P457S, substitutions of proline451 by serine451 (P451S), proline454 by serine454 (P454S), serine456 by asparagine456 (S456N), lysine458 by methionine458 (K458M), and lysine459 by isoleucine459 (K459I) resulted in a ratio of HMW EPS I to LMW EPS I similar to that determined for the wild type (data not shown).
Two of seven amino acid substitutions in the proline-rich motif of ExoP affected EPS I production, indicating that most of the conserved residues of this motif can be exchanged without influencing the function of ExoP. This is in accordance with the observation that in the periplasmic portion of ExoP and the homologous proteins identified, only one residue of the proline-rich motif is invariant. Interestingly, the exchange of this invariant residue, proline457, with serine457 (P457S), which is located within the highly conserved central SPK motif (Fig. 2), resulted in enhanced production of LMW EPS I at the expense of HMW EPS I. Essentially the same effect resulted from the exchange of arginine443 with leucine443 (R443L). The positively charged arginine443 is highly conserved in the proteins involved in lipopolysaccharide, enterobacterial common antigen, and exopolysaccharide biosynthesis but not in proteins involved in the production of capsular polysaccharides.
The cytoplasmic localization of the C-terminal ExoP domain is not affected by the mutations R443L and P457S.
By analyzing translational fusions to reporter genes, we demonstrated that the C-terminal domain of ExoP is located in the cytoplasm (6). To test whether the mutations R443L and P457S, which caused enhanced production of LMW EPS I, affected the cytoplasmic localization of the C-terminal ExoP domain, a lacZ reporter gene was inserted into plasmids pExoP, pExoP-R443L, and pExoP-P457S, resulting in translational fusions to the exoP genes. The fusion site of the ExoP proteins and the LacZ monitor protein was located at amino acid position 773 of the ExoP proteins in their cytoplasmic domains. R. meliloti RmΔexoP carrying plasmid pExoP, pExoP-R443L, or pExoP-P457S with the exoP-lacZ translational fusions displayed β-galactosidase activities of 80 to 86 U. These activities correspond to the activity previously determined for a strain carrying the LacZ monitor protein fused to amino acid 773 of the wild-type ExoP protein (6). Since the LacZ monitor protein displays β-galactosidase activity only when fused to cytoplasmic portions of membrane proteins (22), these results indicate that the amino acid substitutions R443L and P457S did not affect the cytoplasmic localization of the C-terminal ExoP domain.
The alteration of the ratio of HMW to LMW EPS I produced by the exoP* mutant might result from a structural alteration of the N-terminal ExoP domain or from an alteration in the amount or the stability of the N-terminal ExoP domain due to the deletion. Mutations R443L and P457S might cause comparable alterations. The virtually identical β-galactosidase activities mediated by ExoP-R443L-LacZ, ExoP-P457S-LacZ, and wild-type ExoP-LacZ fusion proteins argue against an alteration of the amount or the stability of the ExoP protein and in favor of structural alterations of the protein, although it cannot be excluded that the lacZ fusion stabilized either the exoP mRNA or the ExoP-LacZ fusion protein. The cytoplasmic localization of the C-terminal domain of the ExoP-R443L and ExoP-P457S mutant proteins indicate that a structural alteration of the ExoP protein would not involve the destruction of its second transmembrane helix.
Conclusions.
We were able to show that specific mutations within a proline-rich segment of ExoP which is conserved in a number of proteins involved in polysaccharide biosynthesis can drastically alter the ratio of HMW to LMW EPS I. This supports our previous proposition that ExoP influences the ratio of HMW to LMW EPS I in R. meliloti. This ratio may be altered by influencing the biosynthesis or the export of HMW or LMW EPS I. This may include the release of EPS I polymerization products from the polymerization apparatus.
Acknowledgments
This work was supported by grant Pu 28/19-1 from Deutsche Forschungsgemeinschaft.
We thank J. E. González for sharing unpublished results and H. Küster for critical reading of the manuscript.
REFERENCES
- 1.Aman P, McNeil M, Franzen L, Darvill A G, Albersheim P. Structural elucidation using HPLC-MS and GLC-MS of the acidic polysaccharide secreted by Rhizobium meliloti strain 1021. Carbohydr Res. 1981;95:263–282. [Google Scholar]
- 2.Bastin D A, Stevenson G, Brown P K, Haase A, Reeves P R. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosome and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 3.Batchelor R A, Alifano P, Biffali E, Hull S I, Hull R A. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol. 1992;174:5228–5236. doi: 10.1128/jb.174.16.5228-5236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battisti L, Lara J C, Leigh J A. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc Natl Acad Sci USA. 1992;89:5625–5629. doi: 10.1073/pnas.89.12.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker A, Kleickmann A, Arnold W, Keller M, Pühler A. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 6.Becker A, Niehaus K, Pühler A. Low molecular weight succinoglycan is predominantly produced by Rhizobium meliloti strains carrying a mutated ExoP protein characterized by a periplasmic N-terminal and a missing C-terminal domain. Mol Microbiol. 1995;16:191–203. doi: 10.1111/j.1365-2958.1995.tb02292.x. [DOI] [PubMed] [Google Scholar]
- 7.Becker A, Küster H, Niehaus K, Pühler A. Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: identification of the exsA gene encoding an ABC transporter protein, and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol Gen Genet. 1995;249:487–497. doi: 10.1007/BF00290574. [DOI] [PubMed] [Google Scholar]
- 8.Becker A, Rüberg S, Küster H, Roxlau A A, Keller M, Ivashina T, Cheng H-P, Walker G C, Pühler A. The 32-kb exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J Bacteriol. 1997;179:1375–1384. doi: 10.1128/jb.179.4.1375-1384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beutler H O. Lactose and d-galactose: UV-method. In: Bergmeyer H U, editor. Methods of enzymatic analysis. 3rd ed. Vol. 6. Weinheim, Germany: Verlag Chemie; 1984. pp. 104–112. [Google Scholar]
- 10.Breedveld M W, Zevenhuizen L P T M, Zehnder A J B. Osmotically induced oligo- and polysaccharide synthesis by Rhizobium meliloti SU47. J Gen Microbiol. 1990;136:2511–2519. [Google Scholar]
- 11.Bugert P, Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 12.Bullock W C, Fernandez J M, Short J M. XL1-Blue: a high efficient plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 13.Capage, M. A., D. H. Doherty, M. R. Betlach, and R. van der Slice. October 1987. Recombinant-DNA mediated production of xanthan gum. Patent International publication no. WO87/05938.
- 14.Casse F, Boucher C, Julliot J S, Michel M, Dénarié J. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J Bacteriol. 1979;113:229–242. [Google Scholar]
- 15.Chaplin M F, Kennedy S F. Carbohydrate analysis. A practical approach. Washington, D.C: IRL Press; 1986. [Google Scholar]
- 16.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 17.Finan T M, Hartwieg E, LeMieux K, Bergman K, Walker G C, Signer E R. General transduction in Rhizobium meliloti. J Bacteriol. 1984;159:120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frosch M, Edwards U, Bousset K, Kraube B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 19.Glazebrook J, Walker G C. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 20.Glucksmann M A, Reuber T L, Walker G C. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol. 1993;175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González J E, York G M, Walker G C. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene. 1996;179:141–146. doi: 10.1016/s0378-1119(96)00322-8. [DOI] [PubMed] [Google Scholar]
- 22.Hennessey E S, Broome-Smith J K. Gene-fusion techniques for determining membrane-protein topology. Curr Opin Struct Biol. 1993;3:524–531. [Google Scholar]
- 23.Huang J, Schell M. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 24.Kroll J S, Loynds B, Brophy L M, Moxon E R. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol. 1990;4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 25.Kunst A, Draeger B, Ziegenhorn J. UV-methods with hexokinase and glucose-6-phosphate dehydrogenase. In: Bergmeyer H U, editor. Methods of enzymatic analysis. 3rd ed. Vol. 6. Weinheim, Germany: Verlag Chemie; 1984. pp. 163–172. [Google Scholar]
- 26.Leigh J A, Walker G C. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 27.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of entobacterial common antigen. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 28.Morona R, Van Den Bosch L, Manning P A. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J Bacteriol. 1995;177:1059–1068. doi: 10.1128/jb.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhold B B, Chan S Y, Reuber T L, Marra A, Walker G C, Reinhold V N. Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J Bacteriol. 1994;176:1997–2002. doi: 10.1128/jb.176.7.1997-2002.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuber T L, Long S, Walker G C. Regulation of Rhizobium meliloti exo genes in free-living cells and in planta by using TnphoA fusions. J Bacteriol. 1991;173:426–434. doi: 10.1128/jb.173.2.426-434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuber T L, Walker G C. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 32.Stevenson G, Kessler A, Reeves P R. A plasmid-borne O-antigen chain length determinant and its relationship to other chain length determinants. FEMS Microbiol Lett. 1995;125:25–30. doi: 10.1111/j.1574-6968.1995.tb07330.x. [DOI] [PubMed] [Google Scholar]
- 33.Whitfield C, Amor P A, Köplin R. Modulation of the surface architecture of Gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol Microbiol. 1997;23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 34.Zevenhuizen L P T M. Selective synthesis of polysaccharides by Rhizobium trifolii strain TA-1. FEMS Microbiol Lett. 1986;35:43–47. [Google Scholar]