Abstract
The cycle of life and death and Earth’s carbon cycle(s) are intimately linked, yet how bacterial cells, one of the largest pools of biomass on Earth, are recycled back into the carbon cycle remains enigmatic. In particular, no bacteria capable of scavenging dead cells in oxygen-depleted environments have been reported thus far. In this study, we discover the first anaerobes that scavenge dead cells and the two isolated strains use distinct strategies. Based on live-cell imaging, transmission electron microscopy, and hydrolytic enzyme assays, one strain (designated CYCD) relied on cell-to-cell contact and cell invagination for degrading dead food bacteria where as the other strain (MGCD) degraded dead food bacteria via excretion of lytic extracellular enzymes. Both strains could degrade dead cells of differing taxonomy (bacteria and archaea) and differing extents of cell damage, including those without artificially inflicted physical damage. In addition, both depended on symbiotic metabolic interactions for maximizing cell degradation, representing the first cultured syntrophic Bacteroidota. We collectively revealed multiple symbiotic bacterial decomposition routes of dead prokaryotic cells, providing novel insight into the last step of the carbon cycle.
Subject terms: Bacteriology, Microbial ecology
Introduction
Bacteria comprise one of the largest pools of biomass on Earth (>70 Gt), only second to plants [1], and, thus, the recycling of elements and energy stored in dead bacterial cells is a significant process in Earth’s biogeochemical cycles [2, 3]. Prokaryotic cells can die through multiple routes, including predation (by eukaryotes or predatory bacteria), viral infection, and starvation/stress [4]. Such death can yield lysed cells with released cell contents or dead unlysed cells, leaving prokaryotic “necromass” as both released and cellular fractions. Inactive microbial cells with damaged cell membranes (via the LIVE/DEAD Baclight method) comprising the cellular necromass are observed across various environments (marine and freshwater, biofilm, soil) [5–7]. For oxic environments, there are reports of organisms degrading both the released compounds [8] and intact dead cells, with the latter investigated through degradation of inactive cells with damaged membranes (prepared via autoclaving or heat treatment) (e.g., Flavobacteria, Myxobacteria, and protists) [9–11]. As for anoxic environments, the contribution of dead cell scavenging by protists [12] to prokaryotic biomass turnover is assumed to be low (less than 0.1%) due to low abundances [13, 14], indicating that prokaryotes are the primary drivers of anaerobic necromass degradation. However, though previous studies identify some prokaryotes potentially involved in necromass decomposition, whether the organisms degrade necromass derived compounds (e.g., macromolecules) or intact dead cells has not been investigated [15–18]. In other words, organisms capable of directly degrading/scavenging intact dead cells have yet to be identified in anoxic habitats. This is a major knowledge gap in the anaerobic carbon cycle given that a major fraction (~90%) of Earth’s biomass resides below the planet’s surface. Here, using dead bacterial cells as the sole energy source, we successfully isolated anaerobic Bacteroidota (formerly Bacteroidetes) bacteria that degrade dead cells and unravel metabolic strategies and symbioses that facilitate the unique niche.
Results and discussion
Cultivation of anaerobic cell scavengers
We looked to one natural and one artificial environment to culture scavengers of dead cells (i.e., the cellular necromass fraction), herein, referred to as “cell scavengers”, a subsurface aquifer and sewage-treating anaerobic reactor, and attempted cultivation of cell scavengers using dead bacteria cells as the sole energy source. To increase the activity and abundance of cell scavengers we first cultured environmental microbial community samples on liquid basal media containing dead Bacteroides graminisolvens cells prepared through freezing and thawing freshly grown cells [12]. Though the freeze-thawed cells may release cell contents and stimulate non-cell-degrading organisms, we could consistently detect decreases in turbidity (i.e., clearing of cells) across repeated subcultures, indicating activity of cell-scavenging organisms in the cultures. Control incubations only containing dead Bacteroides cells did not show significant decreases in turbidity over time, so the dead cells did not passively lyse extensively (data not shown).
We aimed to isolate cell scavengers from the above liquid subcultures through cultivation on agar containing dead Bacteroides cells. Freshly grown Bacteroides cells were autoclaved to ensure that there is no interference from contaminating live food bacteria or their enzymes. Despite the high temperature and pressure used during autoclaving, roughly 70% of the turbidity is retained and unlysed cells are clearly microscopically visible (Supplementary Fig 1). We observed cell degradation as “cleared” zones on the agar plates inoculated with dead cell-fed cultures from the bioreactor or aquifer. One difference between the samples was plaques from aquifer cultures had visible colonies at the center (Fig. 1a, b). Plaques were not formed on agar when live Bacteroides cells were used as food (data not shown). After subculturing from the plaques three times, we obtained cultures predominated by cells of a single morphotype (slender rods, 0.2–0.3 µm wide) from both bioreactor and aquifer samples.
Fig. 1. Plaque formation by anaerobic dead cell scavengers.
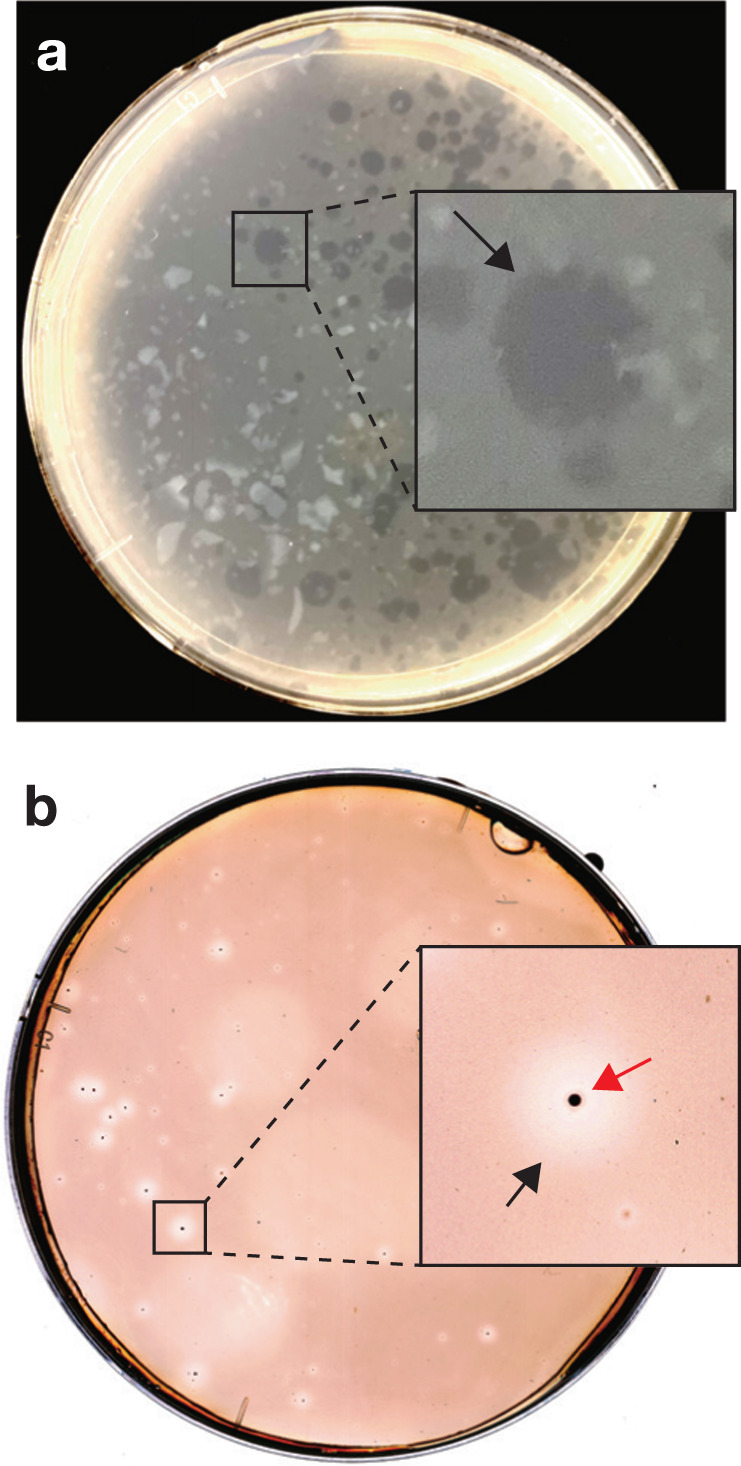
Cleared zones (plaques) formed on agar containing autoclaved B. graminisolvens cells from cultures from (a) bioreactor and (b) subsurface aquifer. In the inset, black arrows indicate plaques and red arrows indicate small colonies formed at the center of the plaques.
For the bioreactor-derived culture, we further purified the culture through filtration with 0.22 µm pore size filters (i.e., exclusion of larger/wider cells) and dilution to exclude other lower-abundance bacteria. We attempted cultivation with various soluble energy sources using the filtered and diluted cultures as inocula to obtain pure cultures without any contaminating live/dead bacteria. Using yeast extract as an energy source, we confirmed microscopically that we had obtained a pure culture of the target slender rod bacterium, designated as strain CYCD, albeit with poor growth. We also verified that strain CYCD could degrade dead bacterial cells, autoclaved Escherichia coli cells in this case (Fig. 2a).
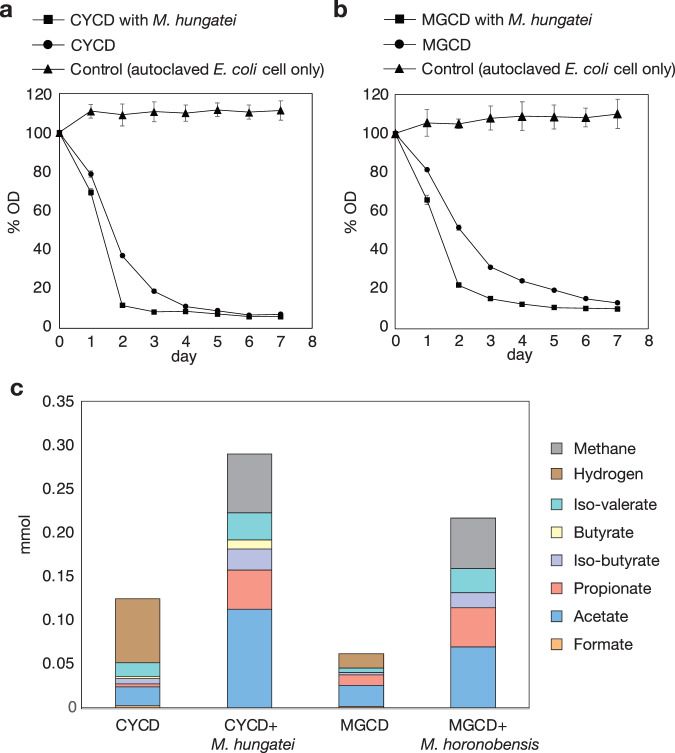
Fig. 2. Degradation of dead bacterial cells by strains CYCD and MGCD.
a Cultures of CYCD with and without Methanospirillum hungatei JF-1 and b cultures of MGCD with and without Methanoculleus horonobensis T10 were fed with autoclaved E. coli cells and changes in optical density (OD600) were monitored. For each culture, the optical densities are shown as percentages of the initial density. The data are means of three individual incubations; error bars represent standard deviation of these triplicates. c Metabolic byproducts formed during degradation of autoclaved E. coli cells.
Given the significant differences in growth rates between the pre-isolation mixed cultures and isolated CYCD, it is clear that axenic growth is not optimal for the strain. One major difference is that axenic cultures of CYCD accumulate H2 (Fig. 2c). Under anoxic conditions without favorable electron acceptors, organisms often couple oxidation of organic matter with reduction of H+ to H2, but accumulation of H2 is thermodynamically inhibitory [18]. In microbial communities, H2-generating organisms circumvent such inhibition through symbiotic interactions with H2-consuming partners (e.g., methane-generating archaea) [19]. To enhance growth of CYCD, we added a partner H2-consuming methane-generating organism (Methanospirillum hungatei JF-1). Strain CYCD could degrade both yeast extract and dead cells (autoclaved E. coli) to a much greater extent in the presence of M. hungatei (Fig. 2c and Supplementary Fig. 2). Comparing axenic and syntrophic cultures, CYCD in syntrophy could reduce turbidity faster (1.4x) and produce byproducts at higher concetrations (fatty acids and H2) when fed the same amount of cells, implying higher metabolic activity (Fig. 2a, c). This indicates that CYCD does not require but benefits from a H2-mediated metabolic symbiosis, a phenomenon known as semi-syntrophy [19, 20] (symbioses that involve an obligate dependency is referred to as “syntrophy” [18, 21]).
We examined whether CYCD co-cultures with M. hungatei can scavenge cells of differing levels of cell damage using dead Bacteroides cells prepared via (i) a non-phyiscal approach exposing the obligate anaerobe to air and UV for 30 min, (ii) freeze-thaw and (iii) autoclaving. One would expect that each approach would likely generate (i) minimally physically damaged cells, (ii) cells with damaged cell walls/membranes, and (iii) cells with damaged cell walls/membranes and denatured proteins, indicating that autoclaving would likely incur more damage to the cells than freeze-thaw and freeze-thaw more than air/UV treatment. The CYCD co-cultures could degrade all cell preparations (Supplementary Fig. 3). We did not detect any decreases of turbidity in incubations only containing the methanogen and dead cells. The extent of cell degradation by CYCD depended on the extent of damage associated with the cell-killing methods. We suspect that decreases in cell integrity (e.g., membrane damage by freeze-thaw or heat) increase the accessibility of energy sources (e.g., proteins/peptides) and thermal protein denaturation improves the hydrolyzability of proteins and peptides [22–24]. Thus, cell/protein integrity does not influence whether CYCD can degrade the cells but contributed to how easily the cells could be degraded (as observable by the extents of degradation and the degradation rates).
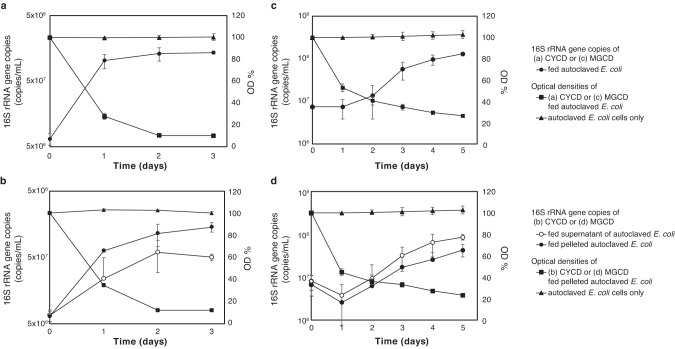
Strain CYCD showed clear growth in association with degradation of dead cells, as measured by quantitative PCR (qPCR) (Fig. 3a). As the feed (dead cells) presumably contained both released compounds and intact dead cells, we further examined which fraction(s) CYCD can utilize/grow on. Here, we left autoclaved E. coli cells at room temperature for one day to allow for leakage of cellular contents and then separated it into two fractions via centrifugation at 15,000 rpm. Given the increased turbidity of the pelleted fraction and the lack of turbidity in the supernatant fraction, we suspect the former primarily consists of cell debris and intact cells and the latter dissolved cell contents (e.g., peptides, amino acids, sugars, and nucleotides). We confirmed that CYCD could grow on both pelleted (cell) and supernatant (released compound) fractions, verifying that the strain can directly catabolize both dead cells and compounds released from necromass (Fig. 3c).
Fig. 3. Growth of strains CYCD and MGCD on autoclaved E. coli cells.
Growth curves of (a) CYCD and (b) MGCD fed with autoclaved E. coli cells as determined by qPCR. Changes in optical density (OD600) were monitored. Growth curves of (c) CYCD and (d) MGCD fed with autoclaved E. coli cells supernatant amd pelleted fraction as determined by qPCR. Changes of autoclaved E. coli pelleted fraction in optical density (OD600) were monitored. As partner methane-generating organism, M. hungatei JF-1 and M. horonobensis T10 were added to culture for CYCD and MGCD, respectively. The data are means of three individual incubations; error bars represent standard deviation of these triplicates.
For the aquifer-derived cultures, we were able to obtain a pure culture colony through cultivation on agar plates and basal medium supplemented with tryptone and yeast extract. The cultured organism was designated as strain MGCD. We confirmed that MGCD could grow on autoclaved E. coli cells as the sole energy source, as evidenced by the decreased turbidity, increased MGCD cell numbers (detected using qPCR), and accumulation of H2, a final metabolic byproduct of anaerobic organotrophy (Figs. 2b, c and 3b). Like CYCD, cell degradation was stimulated by a H2-scavenging partner (Methanoculleus horonobensis T10), as shown by faster degradation (1.5x) and the relative increase in fatty acids and H2 (as estimated from the CH4 production) (Fig. 2c). As observed for strain CYCD, MGCD could grow on both released and cellular necromass fractions (Fig. 3d) and could degrade cells regardless of the extent of cell wall/membrane/protein integrity (Supplementary Fig. S3).
We further evaluated the range of food cells CYCD can access and found that the strain degrades Gram-negative (B. graminisolvens and E. coli) and Gram-positive (Lactococcus lactis, Lactobacillus helveticus, and Micrococcus lutes) bacteria, archaea (M. hungatei and M. horonobensis), and yeast (Saccharomyces cerevisae), all only when inactivated (see Supplementary Table 1). No degradation of live cells was observed. Strain MGCD could degrade most of the above dead cells, except for yeast cells. Given that close relatives of CYCD and MGCD (>95% 16S rRNA sequence identity) are detected at relative abundances of 1–5% in diverse habitats (e.g., groundwater, oil reservoirs, marine sediments, anoxic soils, cave biofilms, and bioreactors) [25–30], these lineages may play important roles in scavenging dead cells in various anoxic environments.
Phylogenetic comparison with related Bacteroidota
Phylogenetic analysis (see Supplementary Notes) showed that the bioreactor- and aquifer-derived strains each affiliate with the genus Blvii28 wastewater-sludge group and species Perlabentimonas gracilis [31], which both belong to the Bacteroidota family Tenuifilaceae. We confirmed that P. gracilis M08_MB, isolated from a marine sediment environment, is capable of degrading dead (autoclaved) cells (Supplementary Fig. 4). The family Tenuifilaceae is divisible into two subgroups, one including CYCD and the other Perlabentimonas (Supplementary Figs. 5 and 6), and the capacity to scavenge dead cells is observed in both. The family is detectable in freshwater, marine, subsurface, host-associated, and artificial (i.e., bioreactors) habitats suggesting that the family may contribute to dead cell degradation across diverse ecosystems (Supplementary Table 2). For example, Tenuifilaceae members are often found as a highly dominant population in bioreactor systems that are fed dead cells as the primary energy source (i.e., anaerobic digesters) [18, 32]. However, further cultivation of other Tenuifilaceae members is necessary to determine the distribution of cell degradation in the family as one member of the family, Tenuifilum thalassicum, lacked the capacity (Supplementary Fig. 4). Bacteroidota members closely related to but outside of Tenuifilaceae did not show dead cell-degrading activity (Supplementary Fig. 4). In any case, this is the first report of cultured Bacteroidota bacteria capable of syntrophic interaction.
Genome comparison showed no differences in lysozymes and putative adhesion proteins between cell-degading and non-cell-degrading Tenuifilaceae, suggesting these are not involved in manifestation of the cell degradation phenotype (Supplementary Table 3 and 4). On the other hand, strain CYCD and P. gracilis strains all possessed peptidoglycan endopeptidases (M23 family peptidases) with remote homology to other Tenuifilaceae isolates. M23 peptidases are typically involved in building/remodeling of peptidoglycan (an essential function for cell replication), so we suspect that those shared and closely related among cell-degrading and non-cell-degrading Tenuifilaceae (e.g., CYCD_09420 and A0A7D4CSX0 of Tenuifilum) likely serve in cell growth/maintenance while those distant from Tenuifilum (CYCD_23720 and Perlabentimonas WP_166360992) likely have a distinct function (Supplementary Table 4). Further investigation is necessary to verify the involvement of such enzymes in dead cell degradation.
Cell degradation strategies
We further explored what cell degradation strategies strains MGCD and CYCD employ to accomplish decomposition of dead cells and whether they differ. The supernatant of CYCD cultured on dead cells neither harbored cell-lysing nor peptide-hydrolyzing activity (Supplementary Fig. 7), suggesting their dead cell-scavenging strategy involves cell-to-cell contact and not excretion of cell-detached lytic enzymes. Live-cell imaging (bright-field) confirmed that strain CYCD attached to and degraded autoclaved E. coli cells (Supplementary Video 1). In the same culture, autoclaved E. coli cells without CYCD attachment were not degraded (Supplementary Video 2), reflecting the assays above. Though CYCD benefits from syntrophic interactions with methanogens, live-cell imaging showed that the strain did not form physical interactions with their partners, perhaps reflecting their semi-syntrophic nature. In transmission electron microscopy (TEM) micrographs, we found invaginations in many CYCD cells at the site of attachment with food bacteria (50% or 9 out of 18 food bacteria-associated cells; note that this method only shows a single cross-section so the other cells may also have similar structures; Fig. 4a–d). In contrast, CYCD cell invagination was absent in cells grown on substrates other than dead cells (e.g., yeast extract, Fig. 4i, j). Live-cell imaging and TEM both showed that cell degradation by CYCD leaves behind empty food bacteria, “ghost” cells, observable as cells retaining cell structure but with decreased electron density (lighter in TEM) or decreasing image intensity (darker in bright-field) of the intracellular area compared to control autoclaved E. coli cells (Fig. 4g, h and Supplemenatry Video 2). As for strain MGCD, unlike CYCD, cell degradation involves extracellular hydrolytic enzymes (Supplementary Fig. 7), food bacteria cell lysis (Fig. 4e, f), and neither physical attachment nor invagination. Though to a lesser extent, we also observed that MGCD cell degradation decreased food bacteria cell contents (i.e., electron density in TEM images).
Fig. 4. Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) images of degrader of dead cells.
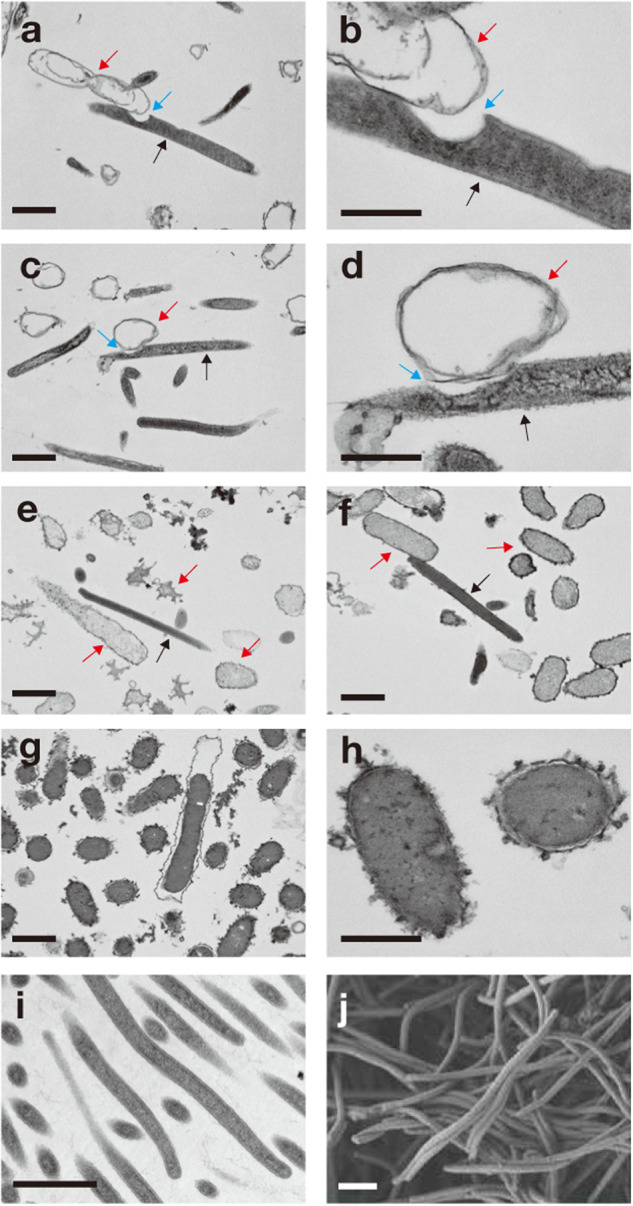
TEM images of strains (a–d) CYCD and (e, f) MGCD degrading autoclaved E. coli cells (imaged in g and h). In panels a through d, black, red, and blue arrows respectively indicate CYCD cells, empty food bacteria cells (i.e., “ghost” cells), and invaginations at the site of attachment between CYCD and food bacteria cells. In panels e and f, black and red arrows respectively indicate cells of strain MGCD and food bacteria. Cultures during cell degradation (1 day for incubation; see Fig. 2a, b) were sampling for TEM analysis. (i and j) TEM (i) and SEM (j) images of strain CYCD grown axenically on 0.1% w/v yeast extract. Scale bar, 1.0 µm.
Known cell degradation strategies involve (i) cell lysis via extracellular hydrolytic enzymes (e.g., Lysobacter) [33], (ii) cell lysis via cell-to-cell contact (e.g., Myxobacteria) [9], (iii) a phagocytosis-like behavior (Ca. Uab) [34], (iv) host-specific invasion into target cells (e.g., Bdellovibrio) [35, 36], or (v) host-specific fusion with target cells membranes (e.g., Vampirovibrio) [37]. Cell degradation by MGCD is similar to Lysobacter. Cell degradation by CYCD involves direct cell-to-cell contact with food cells like the other strategies, but has some distinct features. Although CYCD’s cell invagination during cell degradation somewhat physically resembles the phagocytosis-like behavior of Ca. Uab, CYCD likely does not engulf target cells as such behavior may be too energetically costly under anaerobic conditions, assuming it involves a system as high-cost as phagocytosis [38]. We suspect that the invagination simply increases cell surface area in contact with food cells, though there may be other benefits. In addition, though physical association during cell degradation also resembles the strategy Vampirovibrio and some Bdellovibrio takes, CYCD neither implements membrane fusion nor targets specific food species. Despite the clear differences, schematically, CYCD’s approach and the latter three aerobic predation strategies all involve physical association and direct consumption of intracellular material without release into the extracellular milieu. Reflecting this, CYCD, Vampirovibrio, and Bdellovibrio leave ghost cells behind (though some Bdellovibrio species lyse the ghost cell when leaving the food cell) [35–37]. These shared features may allow for efficient cell degradation through minimizing (i) cost of hydrolytic metabolism by avoiding release of extracellular enzymes into the environment and (ii) loss of energy sources through dispersion of cytosolic organic material from dead cells by retaining the cell’s “shell” during cell content degradation.
Substrate utilization
Bacterial cell consist of proteins, polynucleotides, polysaccharides, and lipids [39]. To clarify what cellular components CYCD can utilize, we further investigated the capacities of strain CYCD to catabolize the primary polymeric constituents of dead cells. In testing specific feed compounds, we found that strain CYCD strictly depended on protein or peptides as energy sources (casein, tryptone, and soy protein digest). In the presence of peptides, CYCD can utilize monosaccharides, polysaccharides, and polynucleotides (Supplementary Table 5). Growth on peptides was possible as long as H2 was scavenged either by a partner methanogen (M. hungatei) or headspace sparging with N2 periodically (Supplementary Fig. 8), indicating that CYCD peptide degradation requires syntrophic interaction. We further observed that (i) peptide-fed CYCD could grow axenically (albeit very poorly) when supplemented with glucose (Supplementary Fig. 8) and (ii) peptide-fed CYCD co-cultures could produce more methane when supplemented with glucose (1 or 10 mM) or DNA (0.01% or 0.1% w/v DNA) (Supplementary Fig. 9). The use of protein as the primary energy source and polynucleotides and polysaccharides as ancillary but supportive nutrients reflects the nutritional composition of cells (roughly at a ratio of 15:7:2 [39]).
CYCD could also interact with a major cell wall component, peptidoglycan. Peptidoglycan-fed CYCD produced a small amount of H2, suggesting the ability to degrade the polymer (Supplementary Fig. 10). Peptidoglycan-fed co-cultures of CYCD and M. hungateii produced H2 but no methane, indicating CYCD somehow inhibits its partner when peptidoglycan is present. Peptidoglycan did not inhibit methane production in H2-fed axenic cultures of M. hungatei (data not shown). In other words, though the exact response/mechanism remains unknown, CYCD reacts to the presence of peptidoglycan, which is consistent with its lifestyle as a dead cell scavenger. We suspect that CYCD is using the peptide fraction of peptidoglycan, given addition of peptidoglycan-associated monosaccharides, N-acetylmuramic acid and N-acetylglucosamine, to yeast extract-fed cultures did not increase growth (Supplementary Table 5). Other cell-derivable polymers, lipids (olive oil) and lipopolysaccharides (LPS), neither enhanced growth nor methane production. These results are in agreement with our observation that CYCD leaves the food cell membranes roughly intact.
Peptide uptake and amino acids degradation catabolism
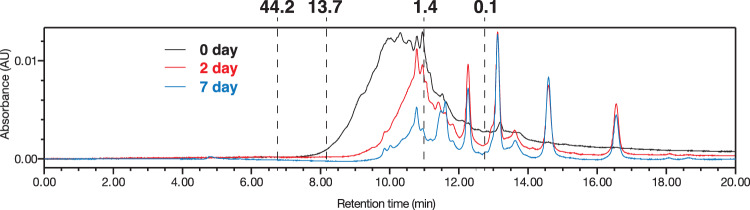
When grown on the soy protein digest, which contains peptides across a wide range of molecular weights (on the order of 1–10 kDa), we observed that strain CYCD could utilize peptides across the entire range (Fig. 5 and Supplementary Fig. 11). CYCD could also uptake peptides as short as di- and tri-peptides (Supplementary Fig. 12). We further found that strain CYCD could not catabolize free amino acids (AAs) (i.e., casamino acids or mixture of free AAs), showing clear specialization towards peptides. Given that CYCD can catabolize peptide-derived AAs, the inability to grow on free AAs is very likely due to the lack of the capacity to uptake individual AAs. To verify this, we took advantage of strain CYCD’s inability to synthesize tryptophan and tested whether CYCD could utilize (i.e., uptake) free tryptophan when grown in the presence of gelatin (a tryptophan-deficient protein substrate). Indeed, strain CYCD could neither grow in the presence of gelatin nor gelatin and supplementary free tryptophan, but could grow well when provided gelatin and small amounts of soy protein digest, which presumably includes tryptophan-containing peptides (Supplementary Fig. 13).
Fig. 5. Degradation of protein by strains CYCD with M. hungatei.
Protein molecular weight distribution (measured by UPLC) of 0.01% w/v soy protein digest-fed cultures of strain CYCD with M. hungatei on 0, 2, and 7 days. Ovalbumin (44 kDa), ribonuclease A (13.7 kDa), vancomycin (1.4 kDa), and uracil (0.1 kDa) was used as standard for molecular weight.
Genomic and transcriptomic analyses showed that strain CYCD possesses transporters for a wide range of peptides (RagAB) [40] (Supplementary Fig. 14 and Supplementary Table 6) and lacks transporters specific to short oligopeptides (≤5 AAs; e.g., OppABCDF [P06202]) and single AAs. Transporting peptides rather than free AAs may minimize the energetic cost of absorbing AAs – active transport of peptides via the TonB-like system of RagAB (one proton per peptide transported) [41] is much more cost-efficient that that of single AAs (at least one proton per AA) [42, 43]. This may be especially helpful under anaerobic conditions, in which anaerobic peptide degradation has ATP yields only amounting to 5.2% of that of aerobic degradation (0.6–2.3 vs 15-43 ATP per AA, Supplementary Table 7). Reflecting specialization towards utilization of a wide range of peptides, the strain possesses diverse extracellular peptidases (Supplementary Table 8). Examination of the strain’s gene expression when grown on soy protein digest (supplemented with either glucose or DNA) or autoclaved E. coli cells showed that the strain consistently expresses genes encoding peptide transporters and a variety of extracellular peptidases (34 total, 16 families).
In the transcriptomes, we also found that CYCD expressed pathways for degrading 15 types of AAs simultaneously (Supplementary Fig. 15 and Supplementary Table 9). Accordingly, we detected a variety of fatty acid byproducts of AA degradation in soy protein digest- and E. coli-fed cultures: acetate, propionate, butyrate, iso-butyrate, and iso-valerate (Supplementary Fig. 16). Simultaneous degradation of many AAs is theoretically more logical than diauxie (step-wise degradation in order of preference [44–46]) for peptide degraders as it takes advantage of the diverse AAs that are indiscriminately taken up in the form of peptides. Mixed substrate utilization has also been proposed to beneficial/favored under low substrate concentrations [47, 48]. As for the individual AA degradation pathways, CYCD utilizes non-fermentative pathways (i.e., non-reductive and H2-generating) rather than fermentative non-H2-generating pathways that sacrifice ATP recovery at the expense of electron disposal. For example, serine degradation recovers 1.33 and 0.83 ATP respectively from H2-generating and fermentative pathways [49] and, similarly, glutamate recovers 1.67 and 1.5 ATP respectively [50]. Thus, the symbiotic H2-generating lifestyle likely also helps CYCD maximize energy recovery from peptides.
Comparing CYCD with other anaerobes that reportedly specialize towards peptides over free amino acids (Coprothermobacter proteolyticus, Porphyromonas gingivalis, and Salinivirga cyanobacteriivorans) [40, 51, 52], we find that the other organisms all encode AA transporters, indicating that CYCD takes a unique lifestyle. In addition, among the above anaerobic peptide degraders, only CYCD and C. proteolyticus benefit from syntrophic interactions with methanogens. Notably, CYCD likely specializes to the cell- and polymer-degrading growth mode given that its gene expression in co-cultures did not change significantly when grown with a different carbohydrate (glucose or DNA) or peptide source (soy protein digest or dead cells) (Supplementary Table 8).
Summary
We discovered anaerobic scavenging of dead cells and obtained the first isolates with this capacity from a subsurface aquifer and bioreactor, each using different strategies for degrading dead bacterial cells – lysis and cell-to-cell contact. While lysis is a strategy often employed by aerobic degradation of cells (both dead and live), degradation of dead cells via cell-to-cell contact has only been reported for aerobic Myxobacteria. However, the cell-to-cell-contact dependent strategy we find is anaerobic, non-host-specific, involves cell invagination, and leaves ghost cells behind. The strain performing such unique cell degradation, CYCD, was also characterized with high degradation efficiency/rates, possibly supported by physical attachment to food cells (including cell invagination at the site of contact), syntrophic symbiosis, and a peptide degradation strategy that maximizes ATP yield. Given the physiological and phylogenetic novelty of the strain, we propose strain CYCD as a new species, Necrotrophus endoclepta gen. nov., sp. nov (see Supplementary Note). Degradation of dead cells is the last step of the global carbon cycle and, here, we provide the first insight into what strategies may be involved in accomplishing this in the anoxic microbial world.
Methods
Sample collection
Samples for cultivation were collected from (i) a sewage-treating upflow anaerobic sludge blanket (UASB) reactor containing methanogenic granular sludge [53] and (ii) a settling pond placed downstream of a commercial well recovering gas- and water from a gas-bearing aquifer in Mobara, Chiba containing biogenic methane [20] and consisting of alternating beds of sandstone and mudstone deposited in deep marine environments during the Plio-Pleistocene periods [54].
Cultivation and isolation
The samples above were inoculated in 25-ml glass tubes with basal medium and frozen Bacteroides graminisolvens JCM 15093 cells (thawed prior to addition). For the reactor and aquifer samples, a freshwater and saline mineral medium and incubation temperatures of 25 and 45 °C were used respectively [13, 55]. The culture tubes were flushed with N2/CO2 gas (80:20 v/v) and sealed with butyl rubber stoppers and aluminum seals. Cell degradation was monitored by measuring optimal density at a wavelength of 600 nm (OD600) with Ultrospec 500 Pro visible spectrophotometer (GE Healthcare Life Sciences, Buckinghamshire, UK). The food bacteria cells, B. graminisolvens, were cultivated anoxically at 37°C in anaerobic basal medium containing 10 mM glycose and 0.03% w/v yeast extract as described previously [56], harvested by centrifugation at 8000 rpm, washed three times with 0.1 M phosphate-buffered saline (PBS), and resuspended in 0.1 M PBS (1% volume of the original culture) prior to storage at –20 °C.
Dead cell scavengers were further selected for from the above cultures using the double-layer plating method. The bottom layer was made with 1.5% w/v agar, and the top layer with 0.5% w/v agar, in basal mineral medium. The agar for the top layer was prepared by mixing 8–10 mL of basal mineral medium containing 0.5% w/v agar heated to 55 °C, the inoculum (1 mL of liquid media culture sample), and food B. graminisolvens (1 mL), prepared by culturing/collecting B. graminisolvens as described above and autoclaved at 121 °C for 20 min. The agar solution for the bottom layer was dispensed first and, after this layer solidified, the above mixture was poured over this. The double-layer plates were incubated at 37 °C for 4 weeks. The plaques were randomly selected and purified by three consecutive subculturing of individual plaques. For the reactor-derived cultures, the final plaques were resuspended in 1 mL of basal medium and filtered by 0.22 µm pore size filter to further select for thin cells observed to predominate in the plaques. The filtrate was inoculated into liquid basal medium containing 0.1% w/v yeast extract and these cultures were incubated at 37 °C. Serial dilutions of the cultures yielded a pure culture, designated as strain CYCD. For the aquifer-derived cultures, the final plaques were resuspended in basal medium and spread onto agar plates (basal medium with 0.1 % w/v yeast extract, 0.1 % w/v tryptone, and 1.5 % w/v agar) for colony isolation. After incubation at 37 °C for 20 days, the harvested colonies were subcultured into a liquid basal medium with 0.1% w/v yeast extract and 0.1% w/v tryptone. Serial dilution yielded a pure culture of a bacterium designated as strain MGCD. Agar plates were incubated under anaerobic conditions using Anaero Pack systems (Mitsubishi Gas Chemical, Tokyo, Japan) and, to minimize exposure to oxygen during subculture inoculation, handled in anaerobic chambers under an atmosphere of N2:CO2:H2 (92:5:3 v/v) (Bactron, Sheldon Manufacturing, Cornelius, OR, USA). Purity of each culture was verified by microscopy and the lack of detection of any sequences derived from other organisms in the 16S ribosomal RNA (rRNA) gene amplicon sequencing data.
Activity of cell degradation
Both strains’ degradative behavior of autoclaved E. coli DH5α competent cells (Invitrogen, Waltham, MA, USA) (cultured on LB medium and collected, resuspended, and autoclaved as described for B. graminisolvens) were evaluated in axenic cultures and co-cultures with partner methanogens – M. hungatei JF-1 (JCM 10133) with strain CYCD and Methanoculleus horonobensis T10 (JCM 15517) with strain MGCD. Downstream experiments were performed in triplicate. Strains CYCD and MGCD cultured with 0.1% w/v yeast extract (and mineral medium) and grown to a density of OD600 0.05 were used as inocula. Autoclaved E. coli cells were provided at a concentration equivalent to 10x that of cell suspensions with an OD600 of 1.0. Cell degradation was monitored by measuring OD600 of samples diluted by a factor of 5. M. hungatei and M. horonobensis were cultivated at 37 °C using the same medium for CYCD and MGCD, respectivery, except that H2/CO2 (80:20, headspace) and 10 mM acetate were added to used for the energy and carbon sources.
Cell scavenging capacity of cocultures CYCD and MGCD with methanogen were investigate using dead Bacteroides cells prepared with exposure to UV and air, freeze-thaw and autoclaving. For deactivation by UV and air, cell suspentions of B. graminosolvens (obligate anaerobe) were put into petri dish and exposured air under UV light (GL15; Toshiba, Tokyo, Japan) for 30 min. Preparation of freeze-thaw and autoclaved B. graminosolvens cells was described above.
For further evaluation of the range of cells each strain can degrade, co-cultures of CYCD and axenic cultures of MGCD (inoculated at 5% v/v from cultures grown with 0.1% w/v yeast extract) were incubated in triplicate with autoclaved cells of E. coli DH5α, B. graminisolvens (JCM 15093), Lactococcus lactis (JCM 5805), Lactobacillus helveticus (JCM 1004), Microccous lutes (dried cells purchased from Wako, Tokyo, Japan), M. hungatei JF-1 (JCM 10133), M. horonobensis T10 (JCM 15517), and Saccharomyces cerevisiae strain VL6-48 (ATCC MYA-3666). During incubation at 37°C for one week, cell degradation was monitored as decrease in OD600. B. graminisolvens, E. coli, M. hungatei, and M. horonobensis were cultivated as described above. L. lactis and L. helveticus were pre-cultivated in Gifu anaerobic medium (GAM broth; Nissui, Tokyo, Japan) at 37 °C. S. cerevisiae was cultivated in YPD medium containing 2.5% glucose, 2.0% peptone, and 1.0% yeast extract at 30°C with shaking. Food cells were collected, centrifuged, washed, resuspended, and autoclaved as described above.
To evaluate cell-degrading activity of other members of the family Tenuifilaceae and closely related clades, Perlabentimonas gracilis (DSM 110720), Tenuifilum thalassicum (DSM 100343), and Acetobacteroides hydrogenigenes (JCM 17603) pre-cultured on corresponding DSMZ culture media [31, 57, 58] were inoculated in triplicates into modified DSMZ culture media prepared without organic carbon/energy sources and instead supplemented with autoclaved E. coli cells. The cell degradation was confirmed using the same method as described above. For each strain, culture conditions were adjusted according to the optimum temperature, salinity, and pH defined in the DSMZ media.
Growth monitoring using qPCR
Genomic DNAs was extracted by using the DNEasy Ultra Clean Microbial kit (Qiagen, Hilden, Germany). For the quantitative analysis, a QuantStadio 3 Real-Time PCR System (Thermo Fisher Scientific) with a PowerUp SYBR Green Master Mix (ThermoFisher Scientific, Waltham, MA, USA) was used. Specific primers pair Bact934F/Bact1060R and qPCR program were used for amplification of 16S rRNA genes of Bacteroidota as previously described [59]. To construct a template standard for the primer set, we used a dilution series of the 16 S rRNA gene amplicon of strain CYCD and MGCD, which were obtained using a bacterial primer pair EUB338F/1496R [59].
Assays for the activity of extracellular hydrolytic enzymes
Activity of exoenzymes capable of cell lysis or peptide hydrolysis were measured in CYCD co-cultures and MGCD axenic cultures grown on autoclaved E. coli cells for two to three days. Extracellular peptidase activity was also measured for cultures grown with 0.1% w/v yeast extract. Culture samples were centrifuged for 10 minutes at 8000 rpm and the supernatant was filtered using 0.1 µm pore size filters (Membrane Solutions LLC, Plano, TX, USA).Assays were performed in triplicate. For cell lysis activity, the filtrate (2 mL) was mixed with 20 mL basal medium containing autoclaved E. coli cells and incubated at 37 °C for one week. For peptidase activity, the Amplite Universal Fluorimetric Protease Activity Assay Kit, Green Fluorescence (AAT Bioquest, Sunnyvale, CA, USA) was used according to the standard protocol. The above filtrate (50 μL) and a solution of fluorescent casein conjugate (50 μL) were mixed and incubated at 37°C for one hour in a 96-well solid microplate. The signal was read by a microplate reader (Spark 10 M, Tecan Ltd., Kanagawa, Japan) with excitation and emission at 490 nm and 525 nm.
Microscopic analyses
Cell morphology and structure of strain CYCD was observed via phase-contrast microscopy (Axio Observer. Z1, Carl Zeiss, Germany), scanning electron microscopy (SEM) (S-4500; Hitachi, Tokyo, Japan), and transmission electron microscopy (TEM) (H-7600; Hitachi, Tokyo, Japan). Co-cultures of strain CYCD and M. hungatei JF-1 grown on 0.1% w/v yeast extract in exponential phase were used for microscopic observation. For SEM and TEM observation, cells were prepared as described previously [20].
For live-cell imaging, to follow that strain CYCD attached and degrade autoclaved E. coli cells, we first inoculated coculture CYCD with M.hungatei and autoclaved E. coli into chamber filled with basal medium containg 0.5% agar made using 1 cm × 1 cm Gene Frames (Thermo Fisher Scientific) between a slide and a coverglass. The slide and edge of coverglass were fixed by epoxy resin for minimizing exposure to oxygen. This operations for live cell imaging was handled in anaerobic chambers under an atmosphere of N2:CO2:H2 (92:5:3 v/v) (Bactron, Sheldon Manufacturing, Cornelius, OR, USA). Images were taken on an Axio Observer. Z1 (Carl Zeiss, Oberkochen, Germany) at 10 min intervals for 22 h.
For TEM observation of cell degradation by strains CYCD and MGCD, cells were fixed between copper plates and frozen with liquid propane at –175 °C. The cells were freeze-substituted with 2% glutaraldehyde, 1% tannic acid in ethanol and 2% distilled water at –80 °C for 48 h. The samples were gradually adjusted to a higher temperature by incubation at −20 °C for 3 h and subsequently 4 °C for 3 h. Next, samples were dehydrated in anhydrous ethanol three times for 30 min, and then once more overnight. Dehydrated samples were incubated with propylene oxide (PO) two times for 30 min at room temperature and subsequently placed in a 70:30 mixture of PO and resin (Quetol-812; Nisshin EM Co., Tokyo, Japan) for one hour. To allow the propylene oxide to vaporize, sample tubes were left open overnight. The samples were embedded in 100% Quetol-812 resin at 60 °C for 48 h. Ultrathin sections (70 nm) were cut with an ultramicrotome (Ultracut UCT; Leica Vienna, Austria), mounted on copper grids, and stained with 2% uranyl acetate for 15 min and lead stain solution (Sigma-Aldrich, MO, USA) for three minutes at room temperature. The grids were observed through TEM (JEM-1400 Plus; JEOL Ltd., Tokyo, Japan) using a CCD camera (EM-14820RUBY2; JEOL Ltd., Tokyo, Japan).
Phenotypic characterization of strain CYCD
All phenotypic experiments were performed in triplicate and the growth was monitored by measuring OD600 and gas production. The strain’s temperature, pH, and salinity (i.e., NaCl concentration) growth ranges and electron acceptor utilization were evaluated under the same medium used for isolation. To evaluate the optimum temperature for growth, the strain CYCD was cultivated at 4, 10, 15, 20, 30, 35, 37, 40, 45, 50, and 60 °C. The pH range was tested between pH of 4.0 and 9.0 (increment of 0.5) by adjusting the pH of the culture medium with sterile N2-purged solutions of 5 N HCl or 5 N NaOH prior to inoculation. Suitable buffers were implemented for each pH: Acetate buffer (for pH 4.0, 4.5, and 5.0), MES (5.5, 6.0, and 6.5), carbonate buffer (7.0 and 7.5) and TAPS (8.0, 8.5, and 9.0). The NaCl range for growth was determined by incubation at 0 to 3.0% w/v (0.5% increment). Substrate utilization was tested in co-culture with M. hungatei at optimum condition (37 °C, pH 7.0, and 0% w/v NaCl).
To investigate peptide utilization by strain CYCD, we prepared an oligopeptide mixture solution through digesting soy protein (Wako, Tokyo, Japan) with papain and then with trypsin as described previously [60]. No free amino acids were detected in the soy protein digest through measurement by ultra-performance liquid chromatography (UPLC; see later description for precise methodology).
Chemical analysis
Methane and hydrogen gas production were detected using gas chromatography (GC-8A and GC-2014, Shimadzu, Kyoto, Japan) fitted with a ShinCarbon ST packed column (50/80 mesh; length, 2 m; diameter, 3.0 mm, Shinwa Chemical Industries LTD, Kyoto, Japan). Argon was used as a carrier gas. The temperatures of the injection port/detector and the column were 150 and 130 °C, respectively.
Samples were filtered through 0.2 μm pore-size membranes prior to preservation and analysis of fermentation products, amino acids, and peptides. Fermentation products were measured with high-performance liquid chromatography (HPLC) (LC-40AD, SIL-40C, CTO-40C, CBM40, DGU403, and CDD-10AVP; Shimadzu, Kyoto, Japan) equipped with two tandem ion-exclusion chromatography columns (Shim-pack Fast-OA; 7.8 mm × 100 mm; 5 μm; Shimadzu, Kyoto, Japan). Two solutions, 5 mM p-toluenesulfonic acid and 5 mM p-toluenesulfonic acid with 20 mM Bis-Tris and 0.1 mM EDTA (Shimadzu, Kyoto, Japan), were used as mobile phases. Both mobile phases were pumped at 0.8 ml/min. The column temperature was maintained at 40 °C.
Molecular weight range of peptide and protein was measured by ACQUITY UPLC H-Class PLUS system (Waters, Milford, USA) with photodiode array (PDA) detector equipped with ACQUITY UPLC Protein BEH SEC column (200 Å, 1.7 µm, 4.6 mm × 300 mm, Waters, Milford, USA). Mobile phase was 100 mM sodium phosphate buffer. The column temperature was set at 25 °C, and the mobile phase flow rate was maintained at 0.3 mL/min. The detection wavelength of the UV detector was set at 220 nm.
For detection of amino acids, free amino acids were derivatized using a AccQ-Tag Ultra Derivatization Kit (Waters, Milford, USA) according to the manufacturer’s instructions. Briefly, 10 µL standard amino acids mixtures or the samples was mixed with 20 μL derivatization reagent and 70 μL AccQ-Tag Ultra Borate Buffer and incubated at 55 °C for 10 min. Single amino acid concentrations of derivatized samples were measured by ACQUITY UPLC H-Class PLUS system (Waters, Milford, USA) with PDA detector equipped with ACCQ-TAG ULTRA C18 (1.7 μm, 2.1 × 100 mm column, Waters, Milford, USA). The mobile phase was prepared by mixing the four following solutions at different ratios throughout the program: AccQ-Tag Ultra Eluent A (Waters, Milford, USA), AccQ-Tag Ultra Eluent B (Waters, Milford, USA), highly pure liquid chromatograph (LC)-grade water (Sigma-Aldrich, MO, USA), and AccQ-Tag Ultra Eluent B 10-times diluted with LC-grade water (see Supplementary Table 10 for details). The column temperature was set at 43 °C, and the combined mobile phase flow rate was maintained at 0.7 mL/min.
For cellular fatty acids and quinones analyses, cells of strain CYCD were grown in basal medium with 0.1 % w/v yeast extract, 0.1 % w/v tryptone and harvested in late exponential growth phase. Cellular fatty acid compositions were identified and quantified using fatty acid methyl ester analysis (Sherlock Microbial Identification System version 6.0; Microbial ID; MIDI, USA). Respiratory quinones were determined using the ACQUITY UPLC H-Class system (Waters, Milford, USA) with PDA detector equipped with BEH C18 (130 Å, 1.7 µm, 2.1 mm × 150 mm, Waters, Milford, USA). The mobile phase was methanol/isopropanol (70:30 v/v). The column temperature was set at 35 °C, and the combined mobile phase flow rate was maintained at 0.3 mL/min.
Genomic and transcriptomic analyses
Genomic DNA of isolate strain CYCD was extracted using enzyme-based DNA extraction methods as described previously [61]. In brief, lysozyme, achromopeptidase, and proteinase K were used to lyse cells, followed by genomic DNA purification using the phenol-chloroform method. Genomic DNA concentrations were determined using a Qubit dsDNA HS assay kit with a Qubit fluorometer (Thermo Fisher Scientific, MA, USA). Genomic DNA was sequenced using DNBseq (MGI Tech, Shenzhen, China) and GridION X5 (Oxford Nanopore Technologies, Oxford, UK) at Bioengineering Lab. Co., Ltd., Sagamihara, Japan. A hybrid assembly using short- and long-read sequencing was carried out using Unicycler v0.4.7 [62] with default parameters, and the assembly was polished using Pilon v.1.22 [63]. Gene identification and annotations were performed using annotated by Prokka v1.14.6 [64] and eggnog-mapper (v2.0.1; eggNOG database v5.0) [65]. The conserved domains of each protein were searched using CD-Search (CDD v.3.20) [66]. Carbohydrate-active enzymes and peptidases were identified using dbCAN2 [67] and MEROPS [68], respectively. Transmembrane proteins and proteins containing signal peptides were predicted by TMHMM v2.0 [69] and SignalP 6.0 [70], respectively. In addition, blast searches were performed against the genes/proteins with verified function UniProtKB/SwissProt (2022_04) [71, 72].
RNA was extracted from cultures under exponential growth by cell lysis using bead beating method and purified using the phenol-chloroform method. In co-cultures with M. hungatei, 0.01% w/v soy protein digest with 10 mM glucose or 0.1% w/v DNA, or autoclaved E. coli cell were used as substrates. Cells were collected through centrifugation (14,000 rpm) for 5 min, resuspended in 450 μl lysis buffer (60 mM EDTA, 400 mM tris HCl, 40 mM NaCl, 10% SDS) and 50 µL 2-mercaptoethanol, transferred to lysing matrix E tube and lysed using a FastPrep-24 instrument for 40 s at 5.5 m/s. After addition of 500 µL acid phenol-chloroform (pH 4.5, Thermo Fisher Scientific, Waltham, MA, USA), samples were vortexed. The samples were then incubated for 5 min at room temperature, centrifuged for 5 min at 4 °C for 14,000 rpm, and the aqueous layer was transferred to a new sterile microtube. RNA was precipitated with equal volumes of cold isopropanol for 1 h at –20 °C. Samples were centrifuged for 15 min at 4 °C for 14,000 rpm. The RNA pellet was washed, precipitated with cold 70% ethanol, and air-dried. Air-dried RNA pellets were resuspended in RNase-free water (Ambion, Austin, TX). Any remaining DNA was digested using Turbo DNA-free DNase kit (Ambion, Austin, TX, USA). The extracted RNA was applied to a NucleoSpin RNA XS Kit (Takara, Shiga, Japan) for purification and concentration. The sequencing libraries were prepared from extracted RNA using Universal Prokaryotic RNA-Seq, Prokaryotic AnyDeplete (NuGEN, San Carlos, CA, USA). Sequencing was performed on a NovaSeq 6000 (Illumina, San Diego, CA, USA) to obtain 2 ×150 bp (paired end) reads. Low quality RNA sequences were trimmed using Fastp v0.20.1 [73] (-W 6 -M 30) and mapped to the assembled genome using BBmap v38.96 (https://sourceforge.net/projects/bbmap/; minid = 0.99) to calculate the gene expression levels, as reads per kilobase transcript per million reads (RPKM). For comparison across different treatments, gene expression levels (RPKM) were normalized to the average expression levels of ribosomal proteins of each sample.
Phylogenetic analysis
NCBI blastn analysis (http://www.ncbi.nlm.nih.gov/BLAST/) of the 16S rRNA gene sequences obtained from genome of strain CYCD was used to identify closely related species. A maximum-likelihood tree based on 16S rRNA gene sequences was constructed using sequences aligned by MAFFT v7.49 [74] with default settings and trimmed with trimAl v.1.4 [75] (--automated1) and tree calculation through RaxmlHPC-PTHREADS-AVX2 version 8.2.12 [76] with the GTRGAMMA model and 100 bootstrap replicates. Bootstrap values were recalculated using BOOSTER [77].
For analyzing the phylogenetic relationship of strain CYCD with other Bacteroidota members, we performed maximum-likelihood estimation of a Bacteroidota phylogenetic tree using a concatenated alignment of DNA-/RNA-binding proteins conserved among most bacteria (i.e., a subset of the GTDB r207 marker proteins; Supplementary Table 11). Sequences were collected from GTDB r207, aligned using MAFFT, concatenated, and trimmed using BMGE v1.12 [78] with the BLOSUM30 matrix and maximum gap rate of each position of 0.67 and minimum length of selected regions of 3. Maximum-likelihood estimation was performed using IQ-TREE v2.2.0.3 [79] with the universal distribution mixture (UDM) model with 64 components and LCLR transformation (Schrempf D, Lartillot N, Szöllősi G. Scalable empirical mixture models that account for across-site compositional heterogeneity [80] and 1000 ultrafast bootstrap replicates (-B 1000). Bootstrap values were recalculated using BOOSTER [77].
For analysis of SusC/RagA, homologs were collected through blastp [81] analysis of sequences obtained from genome of CYCD against the UniProt database [82] (2022_04). Of homologs with sequence similarity ≥35% and coverage ≥80%, representative sequences were selected using CD-HIT v.4.8.1 with a clustering cut-off of 40% similarity (default settings otherwise). Known and predicted SusC/RagA proteins involved in uptake of polypeptides, α-glucan, alginate, pectin, and laminarin described previously [83, 84] were also obtained from the UniProt database (2022_04). The protein sequences were aligned using MAFFT v7.49 [74] with default settings and trimmed using trimAl v.1.4 [75] (--automated1). The phylogenetic tree was generated using a maximum likelihood-based approach using RaxmlHPC-PTHREADS-AVX2 version 8.2.12 [76] with PROTGAMMAAUTO for proteins and 100 bootstrap replicates. Bootstrap values were recalculated using BOOSTER [77].
Supplementary information
Acknowledgements
This research was supported financially by Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science 18H03367 to MKN and 22K18046 to YH. We thank Dr. Bernhard Schink and Dr. Aharon Oren for their help with the nomenclature of the species.
Author contributions
Conceptualization: YH, MKN; Methodology: YH, RM, MKN; Investigation: YH, KM, XM; Resources: YH, MH, TW, TY, TK, MKN; Visualization: YH, MKN; Writing – original draft: YH, MKN; Writing – review & editing: YH, RM, KM, TK, HT, XM, TW, TY, MH, MKN. All authors have read and approved the manuscript submission.
Data availability
The genome sequences and annotation data of strain CYCD are available in National Center for Biotechnology Information (NCBI) BioProject under accession number PRJDB15250.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuga Hirakata, Email: y-hirakata@aist.go.jp.
Masaru K. Nobu, Email: mnobu@jamstec.go.jp
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01538-2.
References
- 1.Bar-On YM, Phillips R, Milo R. The biomass distribution on Earth. Proc Natl Acad Sci USA. 2018;115:6506–11. doi: 10.1073/pnas.1711842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogawa H, Amagai Y, Koike I, Kaiser K, Benner R. Production of refractory dissolved organic matter by bacteria. Science. 2001;292:917–20. doi: 10.1126/science.1057627. [DOI] [PubMed] [Google Scholar]
- 3.Liang C, Amelung W, Lehmann J, Kästner M. Quantitative assessment of microbial necromass contribution to soil organic matter. Glob Chang Biol. 2019;25:3578–90. doi: 10.1111/gcb.14781. [DOI] [PubMed] [Google Scholar]
- 4.Camenzind T, Mason-Jones K, Mansour I, Rillig MC, Lehmann J. Formation of necromass-derived soil organic carbon determined by microbial death pathways. Nat Geosci. 2023;16:115–22. doi: 10.1038/s41561-022-01100-3. [DOI] [Google Scholar]
- 5.Tam L, Kevan PG, Trevors JT. Viable bacterial biomass and functional diversity in fresh and marine waters in the Canadian Arctic. Polar Biol. 2003;26:287–94. doi: 10.1007/s00300-002-0474-8. [DOI] [Google Scholar]
- 6.Miwa T, Takimoto Y, Hatamoto M, Kuratate D, Watari T, Yamaguchi T. Role of live cell colonization in the biofilm formation process in membrane bioreactors treating actual sewage under low organic loading rate conditions. Appl Microbiol Biotechnol. 2021;105:1721–1729. doi: 10.1007/s00253-021-11119-y. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Kim HS, Jo HY, Kwon MJ. Revisiting soil bacterial counting methods: optimal soil storage and pretreatment methods and comparison of culture-dependent and-independent methods. PloS One. 2021;16:e0246142. doi: 10.1371/journal.pone.0246142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apostel C, Herschbach J, Bore EK, Spielvogel S, Kuzyakov Y, Dippold MA. Food for microorganisms: position-specific 13C labeling and 13C-PLFA analysis reveals preferences for sorbed or necromass C. Geoderma. 2018;312:86–94. doi: 10.1016/j.geoderma.2017.09.042. [DOI] [Google Scholar]
- 9.Arend KI, Schmidt JJ, Bentler T, Lüchtefeld C, Eggerichs D, Hexamer HM, et al. Myxococcus xanthus predation of Gram-positive or Gram-negative bacteria is mediated by different bacteriolytic mechanisms. Appl Environ Microbiol. 2021;87:e02382–20.. doi: 10.1128/AEM.02382-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamaki H, Hanada S, Kamagata Y, Nakamura K, Nomura N, Nakano K, et al. Flavobacterium limicola sp. nov., a psychrophilic, organic-polymer-degrading bacterium isolated from freshwater sediments. Int J Syst Evol Microbiol. 2003;53:519–26. doi: 10.1099/ijs.0.02369-0. [DOI] [PubMed] [Google Scholar]
- 11.Ferrier-Pages C, Rassoulzadegan F. N remineralization in planktonic protozoa. Limnol Oceanogr. 1994;39:411–419. doi: 10.4319/lo.1994.39.2.0411. [DOI] [Google Scholar]
- 12.Shinzato N, Watanabe I, Meng XY, Sekiguchi Y, Tamaki H, Matsui T, et al. Phylogenetic analysis and fluorescence in situ hybridization detection of archaeal and bacterial endosymbionts in the anaerobic ciliate Trimyema compressum. Microb Ecol. 2007;54:627–36. doi: 10.1007/s00248-007-9218-1. [DOI] [PubMed] [Google Scholar]
- 13.Hirakata Y, Hatamoto M, Oshiki M, Watari T, Araki N, Yamaguchi T. Food selectivity of anaerobic protists and direct evidence for methane production using carbon from prey bacteria by endosymbiotic methanogen. ISME J. 2020;14:1873–85. doi: 10.1038/s41396-020-0660-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massana R, Pedrós-Alió C. Role of anaerobic ciliates in planktonic food webs: abundance, feeding, and impact on bacteria in the field. Appl Environ Microbiol. 1994;60:1325–34. doi: 10.1128/aem.60.4.1325-1334.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller AL, Pelikan C, De Rezende JR, Wasmund K, Putz M, Glombitza C, et al. Bacterial interactions during sequential degradation of cyanobacterial necromass in a sulfidic arctic marine sediment. Environ Microbiol. 2018;20:2927–40. doi: 10.1111/1462-2920.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong X, Greening C, Brüls T, Conrad R, Guo K, Blaskowski S, et al. Fermentative Spirochaetes mediate necromass recycling in anoxic hydrocarbon-contaminated habitats. ISME J. 2018;12:2039–50. doi: 10.1038/s41396-018-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geesink P, Taubert M, Jehmlich N, von Bergen M, Küsel K. Bacterial necromass is rapidly metabolized by heterotrophic bacteria and supports multiple trophic levels of the groundwater microbiome. Microbiol Spectr. 2022;10:e00437–22.. doi: 10.1128/spectrum.00437-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nobu MK, Narihiro T, Mei R, Kamagata Y, Lee PK, Lee PH, et al. Catabolism and interactions of uncultured organisms shaped by eco-thermodynamics in methanogenic bioprocesses. Microbiome. 2020;8:111. doi: 10.1186/s40168-020-00885-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun L, Toyonaga M, Ohashi A, Matsuura N, Tourlousse DM, Meng XY, et al. Isolation and characterization of Flexilinea flocculi gen. nov., sp. nov., a filamentous, anaerobic bacterium belonging to the class Anaerolineae in the phylum Chloroflexi. Int J Syst Evol Microbiol. 2016;66:988–96. doi: 10.1099/ijsem.0.000822. [DOI] [PubMed] [Google Scholar]
- 20.Katayama T, Nobu MK, Kusada H, Meng XY, Hosogi N, Uematsu K, et al. Isolation of a member of the candidate phylum ‘Atribacteria’reveals a unique cell membrane structure. Nat Commun. 2020;11:6381. doi: 10.1038/s41467-020-20149-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stams AJ, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol. 2009;7:568–77. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 22.Marquez UL, Lajolo FM. Composition and digestibility of albumin, globulins, and glutelins from Phaseolus vulgaris. J Agric Food Chem. 1981;29:1068–74. doi: 10.1021/jf00107a043. [DOI] [PubMed] [Google Scholar]
- 23.Kato Y, Watanabe K, Nakamura R, Sato Y. Effect of preheat treatment on tryptic hydrolysis of Maillard-reacted ovalbumin. J Agric Food Chem. 1983;31:437–41. doi: 10.1021/jf00116a063. [DOI] [PubMed] [Google Scholar]
- 24.Semino GA, Cerletti P. Effect of preliminary thermal treatment on the digestion by trypsin of lupin seed protein. J Agric Food Chem. 1987;35:656–60. doi: 10.1021/jf00077a006. [DOI] [Google Scholar]
- 25.Abram F, Enright AM, O’reilly J, Botting CH, Collins G, O’flaherty V. A metaproteomic approach gives functional insights into anaerobic digestion. J Appl Microbiol. 2011;110:1550–60. doi: 10.1111/j.1365-2672.2011.05011.x. [DOI] [PubMed] [Google Scholar]
- 26.Yamane K, Hattori Y, Ohtagaki H, Fujiwara K. Microbial diversity with dominance of 16S rRNA gene sequences with high GC contents at 74 and 98 ˚C subsurface crude oil deposits in Japan. FEMS Microbiol Ecol. 2011;76:220–35. doi: 10.1111/j.1574-6941.2011.01044.x. [DOI] [PubMed] [Google Scholar]
- 27.Cheng TW, Chang YH, Tang SL, Tseng CH, Chiang PW, Chang KT, et al. Metabolic stratification driven by surface and subsurface interactions in a terrestrial mud volcano. ISME J. 2012;6:2280–90. doi: 10.1038/ismej.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco Y, Rivas LA, Garcia-Moyano A, Aguirre J, Cruz-Gil P, Palacin A, et al. Deciphering the prokaryotic community and metabolisms in South African deep-mine biofilms through antibody microarrays and graph theory. PLoS One. 2014;9:e114180. doi: 10.1371/journal.pone.0114180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirakata Y, Oshiki M, Kuroda K, Hatamoto M, Kubota K, Yamaguchi T, et al. Effects of predation by protists on prokaryotic community function, structure, and diversity in anaerobic granular sludge. Microbes Environ. 2016;31:279–87. doi: 10.1264/jsme2.ME16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Key KC, Sublette KL, Duncan K, Mackay DM, Scow KM, Ogles D. Using DNA-stable isotope probing to identify MTBE- and TBA- degrading microorganisms in contaminated groundwater. Ground Water Monit Remediat. 2013;33:57–68. doi: 10.1111/gwmr.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khomyakova MA, Merkel AY, Slobodkin AI. Perlabentimonas gracilis gen. nov., sp. nov., a gliding aerotolerant anaerobe of the order Bacteroidales, isolated from a terrestrial mud volcano. Syst Appl Microbiol. 2021;44:126245. doi: 10.1016/j.syapm.2021.126245. [DOI] [PubMed] [Google Scholar]
- 32.Mei R, Nobu MK, Narihiro T, Kuroda K, Sierra JM, Wu Z, et al. Operation-driven heterogeneity and overlooked feed-associated populations in global anaerobic digester microbiome. Water Res. 2017;124:77–84. doi: 10.1016/j.watres.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 33.Stepnaya OA, Tsfasman IM, Logvina IA, Ryazanova LP, Muranova TA, Kulaev IS. Isolation and characterization of a new extracellular bacteriolytic endopeptidase of Lysobacter sp. XL1. Biochemistry. 2005;70:1031–1037. doi: 10.1007/s10541-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 34.Shiratori T, Suzuki S, Kakizawa Y, Ishida KI. Phagocytosis-like cell engulfment by a planctomycete bacterium. Nat Commun. 2019;10:5529. doi: 10.1038/s41467-019-13499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dwidar M, Monnappa AK, Mitchell RJ. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 2012;45:71–78. doi: 10.5483/BMBRep.2012.45.2.71. [DOI] [PubMed] [Google Scholar]
- 36.Herencias C, Salgado-Briegas S, Prieto MA, Nogales J. Providing new insights on the biphasic lifestyle of the predatory bacterium Bdellovibrio bacteriovorus through genome-scale metabolic modeling. PLoS Comput Biol. 2020;16:e1007646. doi: 10.1371/journal.pcbi.1007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganuza E, Sellers CE, Bennett BW, Lyons EM, Carney LT. A novel treatment protects Chlorella at commercial scale from the predatory bacterium Vampirovibrio chlorellavorus. Front Microbiol. 2016;7:848. doi: 10.3389/fmicb.2016.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin WF, Tielens AG, Mentel M, Garg SG, Gould SB. The physiology of phagocytosis in the context of mitochondrial origin. Microbiol Mol Biol Rev. 2017;81:10–1128.. doi: 10.1128/MMBR.00008-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connor CM, Adams JU, Fairman J. Essentials of cell biology. Cambridge, MA: NPG Education; 2010.
- 40.Madej M, White JB, Nowakowska Z, Rawson S, Scavenius C, Enghild JJ, et al. Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis. Nat Microbiol. 2020;5:1016–25. doi: 10.1038/s41564-020-0716-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratliff AC, Buchanan SK, Celia H. Ton motor complexes. Curr Opin Struct Biol. 2021;67:95–100. doi: 10.1016/j.sbi.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosgriff AJ, Brasier G, Pi J, Dogovski C, Sarsero JP, Pittard AJ. A study of AroP-PheP chimeric proteins and identification of a residue involved in tryptophan transport. J Bacteriol. 2000;182:2207–17. doi: 10.1128/JB.182.8.2207-2217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider F, Krämer R, Burkovski A. Identification and characterization of the main β-alanine uptake system in Escherichia coli. Appl Microbiol Biotechnol. 2004;65:576–82. doi: 10.1007/s00253-004-1636-0. [DOI] [PubMed] [Google Scholar]
- 44.Monod J. The growth of bacterial cultures. Annu Rev Microbiol. 1949;3:371–94. doi: 10.1146/annurev.mi.03.100149.002103. [DOI] [Google Scholar]
- 45.Perrin E, Ghini V, Giovannini M, Di Patti F, Cardazzo B, Carraro L, et al. Diauxie and co-utilization of carbon sources can coexist during bacterial growth in nutritionally complex environments. Nat Commun. 2020;11:3135. doi: 10.1038/s41467-020-16872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prüss BM, Nelms JM, Park C, Wolfe AJ. Mutations in NADH: ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol. 1994;176:2143–50. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harder W, Dijkhuizen L. Strategies of mixed substrate utilization in microorganisms. Philos. Trans R Soc Lond Ser B. 1982;297:459–80. doi: 10.1098/rstb.1982.0055. [DOI] [PubMed] [Google Scholar]
- 48.Lendenmann URS, Snozzi M, Egli T. Kinetics of the simultaneous utilization of sugar mixtures by Escherichia coli in continuous culture. Appl Environ Microbiol. 1996;62:1493–1499. doi: 10.1128/aem.62.5.1493-1499.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivakanesan R, Dawes EA. Anaerobic glucose and serine metabolism in Staphylococcus epidermidis. Microbiology. 1980;118:143–57. doi: 10.1099/00221287-118-1-143. [DOI] [PubMed] [Google Scholar]
- 50.Buckel W, Barker H. Two pathways of glutamate fermentation by anaerobic bacteria. J Bacteriol. 1974;117:1248–60. doi: 10.1128/jb.117.3.1248-1260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki K, Morita M, Sasaki D, Nagaoka J, Matsumoto N, Ohmura N, et al. Syntrophic degradation of proteinaceous materials by the thermophilic strains Coprothermobacter proteolyticus and Methanothermobacter thermautotrophicus. J Biosci Bioeng. 2011;112:469–72. doi: 10.1016/j.jbiosc.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Ben Hania W, Joseph M, Bunk B, Spröer C, Klenk HP, Fardeau ML, et al. Characterization of the first cultured representative of a Bacteroidetes clade specialized on the scavenging of cyanobacteria. Environ Microbiol. 2017;19:1134–48. doi: 10.1111/1462-2920.13639. [DOI] [PubMed] [Google Scholar]
- 53.Kubota K, Hayashi M, Matsunaga K, Iguchi A, Ohashi A, Li YY, et al. Microbial community composition of a down-flow hanging sponge (DHS) reactor combined with an up-flow anaerobic sludge blanket (UASB) reactor for the treatment of municipal sewage. Bioresour Technol. 2014;151:144–50. doi: 10.1016/j.biortech.2013.10.058. [DOI] [PubMed] [Google Scholar]
- 54.Kunisue S, Mita I, Waki F. Relationship between subsurface geology and productivity of natural gas and iodine in the Mobara gas field, Boso Peninsula, central Japan. J Jpn Assoc Pet Technol. 2002;67:83–96. doi: 10.3720/japt.67.1_83. [DOI] [Google Scholar]
- 55.Katayama T, Kamagata Y. Cultivation of Methanogens. In: McGenity TJ, Timmis KN, Nogales B (eds). Hydrocarbon and lipid microbiology protocols, isolation and cultivation. Berlin: Springer; 2016. pp 177–95.
- 56.Hatamoto M, Kaneshige M, Nakamura A, Yamaguchi T. Bacteroides luti sp. nov., an anaerobic, cellulolytic and xylanolytic bacterium isolated from methanogenic sludge. Int J Syst Evol Microbiol. 2014;64:1770–1774. doi: 10.1099/ijs.0.056630-0. [DOI] [PubMed] [Google Scholar]
- 57.Su XL, Tian Q, Zhang J, Yuan XZ, Shi XS, Guo RB, et al. Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol. 2014;64:2986–91. doi: 10.1099/ijs.0.063917-0. [DOI] [PubMed] [Google Scholar]
- 58.Podosokorskaya OA, Kochetkova TV, Novikov AA, Toshchakov SV, Elcheninov AG. Kublanov IV. Tenuifilum thalassicum gen. nov., sp. nov., a novel moderate thermophilic anaerobic bacterium from a Kunashir Island shallow hot spring representing a new family Tenuifilaceae fam. nov. in the class Bacteroidia. Syst Appl Microbiol. 2020;43:126126. doi: 10.1016/j.syapm.2020.126126. [DOI] [PubMed] [Google Scholar]
- 59.Guo X, Xia X, Tang R, Wang K. Real-time PCR quantification of the predominant bacterial divisions in the distal gut of Meishan and Landrace pigs. Anaerobe. 2008;14:224–228. doi: 10.1016/j.anaerobe.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Kimura H, Arai S. Oligopeptide mixtures produced from soy protein by enzymatic modification and their nutritional qualities evaluated by feeding tests with normal and malnourished rats. J Nutr Sci Vitaminol. 1988;34:375–86. doi: 10.3177/jnsv.34.375. [DOI] [PubMed] [Google Scholar]
- 61.Morinaga K, Kusada H, Sakamoto S, Murakami T, Toyoda A, Mori H, et al. Granulimonas faecalis gen. nov., sp. nov., and Leptogranulimonas caecicola gen. nov., sp. nov., novel lactate-producing Atopobiaceae bacteria isolated from mouse intestines, and an emended description of the family Atopobiaceae. Int J Syst Evol Microbiol. 2022;72:005596. doi: 10.1099/ijsem.0.005596. [DOI] [PubMed] [Google Scholar]
- 62.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 65.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rawlings ND, Barrett AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44(D1):D343–D350. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 70.Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD, et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40:1023–1025. doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. In: Edwards D (ed). Plant bioinformatics: methods and protocols. Totowa: Humana Press; 2007. pp 89–112.
- 72.Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lemoine F, Domelevo Entfellner JB, Wilkinson E, Correia D, Dávila Felipe M, De, et al. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature. 2018;556:452–456. doi: 10.1038/s41586-018-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–20. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 80.Nobu MK, Nakai R, Tamazawa S, Mori H, Toyoda A, Ijiri A, et al. Unique H2-utilizing lithotrophy in serpentinite-hosted systems. ISME J. 2023;17:95–104. doi: 10.1038/s41396-022-01197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. [DOI] [PMC free article] [PubMed]
- 83.Nagano K, Murakami Y, Nishikawa K, Sakakibara J, Shimozato K, Yoshimura F. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J Med Microbiol. 2007;56:1536–48. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- 84.Kappelmann L, Krüger K, Hehemann JH, Harder J, Markert S, Unfried F, et al. Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J. 2019;13:76–91. doi: 10.1038/s41396-018-0242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences and annotation data of strain CYCD are available in National Center for Biotechnology Information (NCBI) BioProject under accession number PRJDB15250.