Abstract
INTRODUCTION:
Most Alzheimer disease (AD) loci were discovered in European ancestry individuals.
METHODS:
We applied principal component analysis using Gaussian mixture models and an Ashkenazi Jewish (AJ) reference genome-wide association study (GWAS) dataset to identify AJs ascertained in GWAS (n=42,682), whole genome sequencing (WGS, n=16,815) and whole exome sequencing (WES, n=20,504) datasets. Association of AD was tested genome-wide (GW) in the GWAS and WGS datasets and exome-wide (EW) in all three datasets (EW). Gene-based analyses were performed using aggregated rare variants.
RESULTS:
In addition to APOE, GW analyses (1,355 cases and 1,661 controls) revealed associations with TREM2 R47H (P=9.66x10−9), rs541586606 near RAB3B (P=5.01x10−8) and rs760573036 between SPOCK3 and ANXA10 (P=6.32x10−8). In EW analyses (1,504 cases and 2,047 controls), study-wide significant association was observed with rs1003710 near SMAP2 (P=1.91x10−7). A significant gene-based association was identified with GIPR (P=7.34x10−7).
DISCUSSION:
Our results highlight the efficacy of founder populations for AD genetic studies.
Keywords: Alzheimer disease, Ashkenazi Jews, genome-wide association study, founder population
1. Introduction
Alzheimer disease (AD), the most common neurodegenerative disorder in the world, affects individuals of all races and ethnicities; however, most genetic research for AD has been performed in individuals of European ancestry (EAs) [1, 2] with a limited number of large-scale genetic studies in other populations [1, 3-5]. Trans-ethnic studies have shown that population differences in genetic background can be leveraged to make novel discoveries that might require a sample size several orders of magnitude larger to achieve similar success studying a single population [5, 6]. Similarly, studies of small samples from founder populations (i.e., ethnic or religious groups whose origins can be traced to a limited number of ancestors and thus have a more homogeneous genetic background) have successfully detected robust and subsequently validated associations of AD with several genes [6-8].
For many centuries, Ashkenazi Jews (AJs) lived in communities in Eastern Europe and were genetically isolated from their non-Jewish neighbors. Because some rare autosomal recessive disorders manifesting in childhood (including Tay-Sachs disease, Gaucher disease, familial dysautonomia, Canavan disease, Bloom syndrome and spinal muscular atrophy), as well as particular gene mutations conferring high risk of common disorders such as early-onset breast cancer [9] and multiple gastrointestinal cancers [10-13], are found predominantly or at a much higher frequency in AJs compared to other populations, we hypothesized that some AD susceptibility variants are much more frequent, and thus more likely to show statistically significant associations, in an AJ sample compared to much larger and more genetically heterogeneous EA cohorts. We conducted a genome-wide association study (GWAS) of AD in a group of AJs who were discerned from large EA cohorts of AD cases and controls.
2. Methods
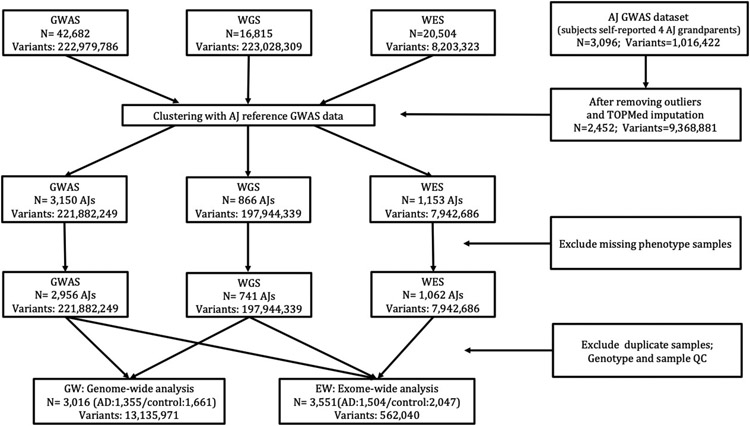
An overview of the study design and analysis workflow is shown in Figure 1.
Figure 1.
Overview of study design and analysis workflow. Number of subjects and variants are shown in each step. GWAS = genome-wide association study, WGS = whole genome sequencing, WES = whole exome sequencing, AJs = Ashkenazi Jews.
2.1. Study participants
Genome-wide variant and phenotypic data were obtained from the Azheimer’s Disease Genetic Consortium (ADGC) [14], Genomic Research at Fundació Ace (GR@ACE) [15] and Azheimer’s Disease Sequencing Project (ADSP) [16, 17]. The study sample includes GWAS data from 35 ADGC cohorts (35,273 non-Hispanic EAs [1, 18] and 7,409 Spanish participants of the GR@ACE GWAS study (https://ega-archive.org/studies/EGAS00001003424)). Genotyping in these datasets was performed as previously described [14, 15] and genotypes were imputed using the TOPMed reference panel [19] and retained if the imputation quality score (r2) was >0.3. Genotypes for subjects in the ADSP whole genome sequencing (WGS, n=16,815) and whole exome sequencing (WES, n=20,504) datasets (NG00067.v5) were joint called using its single nucleotide polymorphism (SNP) / Indel Variant Calling Pipeline and data management tool [20]. Ascertainment, diagnostic procedures, and previous studies using these data are described elsewhere [16, 21-23]. Characteristics of the GWAS, WGS and WES datasets are shown in Table S1. Studies of individual cohorts were approved by the appropriate institutional review boards, and all participants provided written informed permission or, for those with substantial cognitive impairment, consent was provided by a caregiver, legal guardian, or other proxy. A GWAS dataset including 3,096 AJs selected from the Hebrew University Genetic Resource (http://hugr.huji.ac.il) for a GWAS of schizophrenia [24] was obtained from dbGaP (phs000448.v1.p1) and used as a reference panel for clustering purposes. These subjects reported that all four grandparents were Ashkenazi.
2.2. Pre-processing AJ reference data
The AJ reference sample was genotyped with Illumina HumanOmni1-Quad arrays with A/B coding. We used GenGen [25] software to convert the coding to ATGC format and liftOver python scripts (https://github.com/knmkr/lift-over-vcf) to convert the genome positions to NCBI build 38. Genotypes were imputed using the TOPMed reference panel [19] and those with r2 >0.3 were retained. Reference sample outliers who likely have a genetic background with substantial admixture with non-AJs were identified with Aberrant [26] and excluded from subsequent analyses, after which 2,452 subjects remained for clustering analysis.
2.3. Clustering analysis
Population clustering was conducted sequentially for the AJ GWAS reference dataset combined separately with the GWAS, WGS and WES datasets by principal components (PC) analysis [27] using Gaussian Mixture Models after excluding related individuals in each dataset (GWAS:1,158; WGS:1,114; WES:1,514) identified by IBD () and variants with missingness >10%, divergence from Hardy-Weinberg Equilibrium (P<10−6), and minor allele frequency (MAF) <5%. The genetic profile of each individual was summarized as a PC coordinate and assigned to k Gaussian distributions, each representing a subpopulation (cluster) with a certain mean vector and variance matrix, that were determined based on the individuals assigned to the cluster using the Expectation-Maximization algorithm implemented in the Python sklearn.mixture package [28]. The optimal k was determined based on the minimum number of Gaussian distributions that maximized the number of reference AJ subjects in a single cluster. Individuals assigned to the subpopulation containing the AJ reference subjects were included in subsequent analyses, noting that none of the related individuals in the parent GWAS, WGS and WES datasets who were excluded from the clustering procedure were related to the sample identified as AJ. We also applied the ancestry estimation method implemented in the ADMIXTURE software [29] to assess the effect of particular clustering approaches on the group of individuals identified as AJ.
2.4. Association Analysis
To maximize power, genome-wide (GW) and exome-wide (EW) datasets were created by combining the AD datasets according to the type of variants they contained. The GW dataset included AJs in the GWAS and WGS datasets, and the EW dataset included AJs in all three datasets and variants called from exome capture. Duplicate individuals across datasets were identified by IBD analysis () and the genetic data retained for analysis were selected according to the following priority scheme: GW analyses: WGS > GWAS; EW analyses: WGS > WES > GWAS. Separate quality control (QC) criteria were applied to the GW and EW datasets to account for the distinct types of genotype data. PLINK [30] was used to filter out individuals with >10% missing genotypes and variants with >10% missing or not in Hardy-Weinberg equilibrium (HWE, p<1.0x10−6 in the GW dataset, and variants that were monomorphic or not in HWE in the EW dataset. After QC, the GW dataset included 13,135,971 variants and 3,016 individuals, and the EW dataset included 562,040 variants and 3,551 individuals. Among these individuals, 2,044 were common to both datasets.
The association of AD with each bi-allelic common variant (MAF ≥ 0.01) was assessed in PLINK using logistic regression for an additive model including covariates for sex, age, and the first four PCs that were recalculated in the AJ dataset. A genome-wide significant (GWS) threshold was set at P=5.0x10−8 for the GW analyses and a study-wide significance (SWS) threshold was calculated as P=2.74 x10−7 for the EW analyses based on a Bonferroni correction for the number of tested variants. Gene-based rare variant association was tested for genes with ≥ 2 variants after excluding variants with MAF ≥ 0.01 using the same models as described above and the SKAT-O program in RVTESTS [31]. The Bonferroni-corrected significance threshold was set at P=2.13x10−6 for GW analyses and P=1.77x10−6 for EW analyses based on the number of tested genes. Regional visualization of genome-wide and exome-wide association scan results were visualized using LocusZoom [32].
2.5. Differential gene expression
Differential expression of genes at top-ranked loci was evaluated in dorsolateral prefrontal cortex area tissue from 627 participants (380 autopsy-confirmed AD cases and 247 controls) of the Religious Orders Study and Rush Memory and Aging Project, temporal cortex area tissue from 162 participants (82 autopsy-confirmed AD cases and 80 controls) of the Mayo Clinic Study of Aging, and frontal cortex tissue from 208 participants (64 autopsy-confirmed AD cases and 129 controls) of the Framingham Heart Study and Boston University Alzheimer’s Disease Research Center. Details regarding RNA sequencing, quality control procedures, quantification of gene expression, and differential gene expression analysis are described elsewhere [33]. Results across datasets were combined by a sample size-weighted meta-analysis with log2 of fold change (logFC) as direction using the software METAL [34].
3. Results
3.1. Identification of Ashkenazi Jews in Multi-ethnic Cohorts
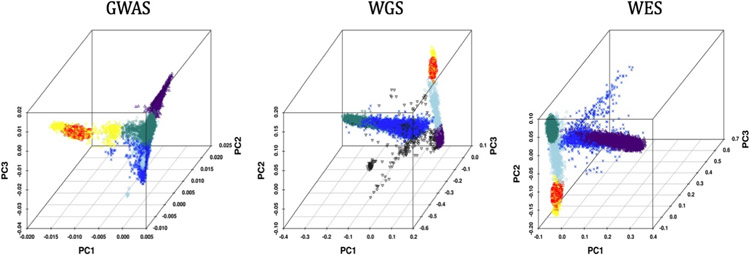
PC analysis conducted for each AD dataset combined with the AJ reference GWAS dataset revealed a subset of 5,169 individuals (GWAS −3,150; WGS - 866; WES - 1,153) who most closely aligned with the AJ reference group contained in one of five clusters (Figure 2), many of whom were identified in more than one dataset (Figure 3). AJs accounted for approximately 7.3% of individuals in the total GWAS dataset comprised of EAs only, but a slightly smaller proportion in the WGS (5.2%) and WES (5.6%) datasets which included cohorts of African Americans and Caribbean Hispanics who have substantial European and African ancestry. After excluding 574 and 1,170 duplicate individuals from the GW and EW datasets, respectively, and 426 and 448 individuals from the GW and EW datasets, respectively, who didn’t pass genotype and phenotype QC filtering, the final sample for the genome-wide analysis contained 1,355 AD cases and 1,661 controls, and for the exome-wide analysis included 1,504 AD cases and 2,047 controls (Table 1). Application of the ADMIXTURE method for identifying AJs resulted in a highly overlapping sample suggesting that the results from the clustering analysis using GMM are robust (Figure S1).
Figure 2.
3-dimensional principal component (PC) plots showing clustering of Ashkenazi Jews (AJs) in the GWAS, whole genome sequencing (WGS) and whole exome sequencing (WES) datasets. Distinct population clusters are color coded. Red dots indicate the AJ reference subjects. Yellow dots indicate the clustered AJ samples from each dataset.
Figure 3.
Venn diagram showing the overlap of Ashkenazi Jews among the GWAS, whole genome sequencing (WGS) and whole exome sequencing (WES) datasets. AJs = Ashkenazi Jews.
Table 1.
Distribution of Alzheimer disease (AD) cases and controls of detected Ashkenazi Jewish individuals in each dataset and analysis.
| Dataset | Genome-wide Analysis | Exome-wide Analysis | ||||
|---|---|---|---|---|---|---|
| Total | AD | Control | Total | AD | Control | |
| GWAS | 2,625 | 1,143 | 1,482 | 1,783 | 748 | 1,035 |
| WGS | 391 | 212 | 179 | 732 | 368 | 364 |
| WES | NA | NA | NA | 1,036 | 388 | 648 |
| Total | 3,016 | 1,355 | 1,661 | 3,551 | 1,504 | 2,047 |
GWAS: genome-wide association study; WGS: whole genome sequencing; WES: whole exome sequencing.
3.2. Genetic Associations with previously known and novel AD Loci
A genome-wide scan revealed several GWS and suggestive (P<1x10−6) associations with little evidence of genomic inflation (λ =1.044) (Figure S2). Among previously established AD loci, GWS associations were observed with many SNPs in the APOE region including the ε4 variant rs49358 (P=3.95x10−54) and the TREM2 R47H variant (rs75932628, P=9.66x10−9) (Table 2). Results were available for 57 of the top SNPs at 76 independent GWS loci in the GWAS conducted by Bellenguez et al. [2] (Table S2). Associations of AD with 11 of these SNPs (19.3%) were at least nominally significant (p<0.05) and seven were significant at the p<0.01 level including TREM2 R47H and BIN1 SNP rs6733839 (P=4.97x10−5). As one might expect, associations that were significant in the AJ sample had effect sizes larger than in the EA GWAS. Further comparisons between the two GWAS revealed that associations for 44 of the 57 loci (77.2%) with results in the AJ dataset, including all nominally significant ones, had the same effect direction and similar odds ratios. Multiple significant associations were also observed with novel loci. A borderline GWS association was observed with a SNP near RAB3B (rs541586606, P=5.01x10−8), as well as suggestive associations with 10 other novel loci (Table 2). This variant is not in LD with other SNPs in the region that may explain the modest corroborating evidence for this association (Figure S3).
Table 2.
Top-ranked associations in the genome-wide analysis.
| Gene | ID | Chr : position | Function | EA | RA | Minor Allele Frequency | OR | P-value * | ||
|---|---|---|---|---|---|---|---|---|---|---|
| AD | Control | EUR | ||||||||
| APOE | rs429358 | 19:44908684 | exonic | C | T | 0.30 | 0.12 | 0.16 | 3.07 | 3.95x10−54 |
| TREM2 | rs75932628 | 6:41161514 | exonic | T | C | 0.035 | 0.011 | 0.0023 | 3.17 | 9.66x10−9 |
| RAB3B | rs541586606 | 1:51996478 | intergenic | C | G | 0.011 | 0.035 | 0.0010 | 0.32 | 5.01x10−8 |
| SPOCK3/ANXA10 | rs760573036 | 4:167607429 | intergenic | CT | C | 0.0034 | 0.023 | 0.0014 | 0.15 | 6.32x10−8 |
| LRRC3B | rs6767457 | 3:26645626 | intronic | A | G | 0.17 | 0.12 | 0.20 | 1.49 | 8.09x10−8 |
| HFM1 | rs560945840 | 1:91344655 | intronic | G | A | 0.0037 | 0.023 | 0.0024 | 0.17 | 1.09x10−7 |
| JAKMIP2-AS1 | rs180825664 | 5:147562155 | intronic | G | A | 0.022 | 0.036 | 0.0008 | 0.35 | 1.33x10−7 |
| PPM1H/AVPR1A | rs559501118 | 12:63117195 | intergenic | T | A | 0.0055 | 0.028 | 0.0022 | 0.23 | 1.77x10−7 |
| CALCB/INSC | rs117925493 | 11:15092251 | intergenic | T | C | 0.0085 | 0.029 | 0.0067 | 0.29 | 2.39x10−7 |
| NCOA1 | rs572781170 | 2:24672133 | intronic | G | A | 0.010 | 0.033 | 0.0011 | 0.33 | 3.36x10−7 |
| NFX1/AQP7 | rs60257421 | 9:33378000 | intergenic | G | A | 0.12 | 0.079 | 0.088 | 1.56 | 4.46x10−7 |
| BZW2 | rs818581 | 7:16647544 | intronic | A | G | 0.44 | 0.50 | 0.62 | 0.76 | 4.55x10−7 |
| CNTN5/LOC100128386 | rs80135755 | 11:100624743 | intergenic | T | A | 0.059 | 0.033 | 0.095 | 1.92 | 4.90x10−7 |
| PIEZO2 | rs72865387 | 18:10955083 | intronic | G | A | 0.18 | 0.23 | 0.24 | 0.72 | 4.98x10−7 |
significance threshold P=5.0x10−8
Chr: chromosome; EA: effect allele; RA: reference allele; EUR: European reference population; OR: odds ratio.
Fewer associations were found in the analysis of the exome-wide scan (Figure S4) and these included the same APOE and TREM2 variants (P=4.52x10−52 and P=2.64x10−10, respectively) identified in the genome-wide scan (Table 3). In addition, the association with SMAP2 SNP rs1003710 (P=1.91x10−7) was SWS and the association with ZNF890P SNP rs200698976 (P=3.49x10−7) was nearly SWS (Table 3, Figures S5 and S6). These associations were evident but less significant in the smaller GW dataset. There were also suggestive associations (P<1x10−5) with SNPs in five other loci. Further inspection of the top-ranked results showed that the minor alleles of most SNPs were greater than four times more frequent in AJ controls than in the general European ancestry population. The differences were at least 9-fold for the variants at the RAB3B, HFM1, NCO1, TRAPPC8, and OR7C2 loci, and may explain why associations with these variants, especially those showing a protective effect, were not detected in much larger GWAS samples.
Table 3.
Top-ranked associations in the exome-wide analysis
| Gene | ID | Chr : position | Function | EA | RA | Minor Allele Frequency | OR | P-value * | ||
|---|---|---|---|---|---|---|---|---|---|---|
| AD | Control | EUR | ||||||||
| APOE | rs429358 | 19:44908684 | exonic | C | T | 0.28 | 0.12 | 0.16 | 2.79 | 4.52x10−52 |
| TREM2 | rs75932628 | 6:41161514 | exonic | T | C | 0.036 | 0.011 | 0.0023 | 3.16 | 2.64x10−10 |
| SMAP2 | rs1003710 | 1:40374681 | 5’ UTR | A | G | 0.40 | 0.46 | 0.44 | 0.77 | 1.91x10−7 |
| ZNF890P | rs200698976 | 7:5132414 | intronic | TG | T | 0.28 | 0.34 | 0.31 | 0.76 | 3.49x10−7 |
| TRAPPC8 | rs149840013 | 18:31839471 | intronic | A | G | 0.0042 | 0.028 | 0.0022 | 0.16 | 1.53x10−6 |
| DMWD | rs138963867 | 19:45786302 | exonic | A | G | 0.021 | 0.0077 | 0.0013 | 2.98 | 1.78x10−6 |
| OR7C2 | rs113508813 | 19:14942161 | exonic | A | G | 0.033 | 0.054 | 0.000071 | 0.55 | 1.81x10−6 |
| SLC43A3 | rs117212150 | 11:57414602 | intronic | T | C | 0.099 | 0.068 | 0.015 | 1.52 | 2.19x10−6 |
| DOCK9 | rs2296984 | 13:98805177 | exonic | G | T | 0.21 | 0.17 | 0.20 | 1.34 | 3.26x10−6 |
significance threshold P=2.74x10−7
Chr: chromosome; EA: effect allele; RA: reference allele; EUR: European reference population; OR: odds ratio
Aggregated rare variant gene-based tests revealed a GWS association with GIPR (P=7.34x10−7, Table S3) and a nearly exome-wide significant association with MAT2B (P=7.26x10−6, Table S4).
3.3. Differential gene expression at AD-associated loci
Differential expression in brain between AD cases and controls was evaluated for 25 genes containing or closest to the 21 showing loci GWS or highly suggestive evidence for association with AD (Table 2, Table 3). Expression data were unavailable for JAKMIP2-AS1, LOC100128386, and OR7C2. Significant differences (Padj<0.0022) were found for genes at more than half of the loci (10/19). Higher expression in AD cases was observed for NCOA1 (P=1.50x10−5), TREM2 (P=2.00x10−4), SLC43A3 (P=1.20x10−11) and PIEZO2 (P=3.70x10−6), whereas expression of for SMAP2 (P=1.80x10−6), RAB3B (P=3.10x10−6), BZW2 (P=8.30x10−8), CALCB (P=2.70x10−5), CNTN5 (P=1.00x10−4), and PPM1H (P=6.70x10−5) was significantly reduced (Table 4).
Table 4.
Differential expression in brain of genes at top-ranked loci between AD cases and controls.
| Gene | Chr: pos | Z-score | P-value * |
|---|---|---|---|
| SMAP2 | 1:40344850 | −4.77 | 1.80x10−6 |
| RAB3B | 1:51907956 | −4.66 | 3.10x10−6 |
| HFM1 | 1:91260766 | −2.04 | 0.042 |
| NCOA1 | 2:24491914 | 4.34 | 1.50x10−5 |
| LRRC3B | 3:26622806 | −0.11 | 0.91 |
| SPOCK3 | 4:166733384 | 1.45 | 0.15 |
| ANXA10 | 4:168092515 | 1.62 | 0.11 |
| TREM2 | 6:41158506 | 3.71 | 2.00x10−4 |
| ZNF890P | 7:5121239 | −2.32 | 0.020 |
| BZW2 | 7:16646131 | −5.36 | 8.30x10−8 |
| NFX1 | 9:33290511 | −0.76 | 0.45 |
| AQP7 | 9:33383179 | 0.24 | 0.81 |
| CALCB | 11:14904997 | −4.20 | 2.70x10−5 |
| INSC | 11:15112424 | 0.14 | 0.89 |
| SLC43A3 | 11:57406954 | 6.79 | 1.20x10−11 |
| CNTN5 | 11:99020953 | −3.88 | 1.00x10−4 |
| PPM1H | 12:62643982 | −3.99 | 6.70x10−5 |
| AVPR1A | 12:63142759 | 1.88 | 0.060 |
| DOCK9 | 13:98793429 | 1.94 | 0.052 |
| PIEZO2 | 18:10666483 | 4.63 | 3.70x10−6 |
| TRAPPC8 | 18:31829173 | −0.66 | 0.51 |
| APOE | 19:44908684 | 2.06 | 0.039 |
| DMWD | 19:44905754 | −0.26 | 0.80 |
Chr: chromosome; Pos: start position.
significance threshold P=0.0022
4.0. Discussion
4.1. Novel AD loci identified in Ashkenazi Jews
We identified approximately 3,500 individuals whose ancestry is almost exclusively Ashkenazi Jewish among much larger European ancestry samples using a robust clustering approach that compared genetic signatures with members of an AJ reference GWAS dataset. Genome-wide and exome-wide scans for AD conducted in this AJ sample identified GWS associations with established AD loci including APOE and TREM2. Notably, the association with the TREM2 R47H variant has a similar p-value (P= 2.64 x10−10) and shows a much stronger effect in the AJ sample (OR=3.16, [2.14-4.70]) compared to a very large AD GWAS sample containing undifferentiated European ancestry individuals (n=82,771, OR=2.39, [2.09-2.73]) [18]. The strength of this finding in the AJ sample is likely due to the MAF of this variant in the AJ controls which is about five times higher than in other European ancestry populations. We also detected significant associations with several novel loci including RAB3B, SMAP2 and ZNF890P.
Confidence in these results is bolstered by evidence for association of AD with 10 other previously established AD loci and similar pattern of association for SNPs at 82% (47/57) of the GWS loci identified by GWAS in a sample of approximately 789,000 EAs [2]. Not surprisingly, few of the known loci were highly significant in the AJ sample that had little power for detecting associations with even the most common variants having odds ratios <1.2 (i.e., accounting for nearly all of the common variant locus associations). Our findings are also supported by evidence for significantly expression differences in brain between AD cases and controls for genes at more than one-half of the top-ranked loci including two novel ones that were GWS (RAB3B and SMAP2).
Among the novel genome-wide or study-wide significant findings, SMAP2 encodes a GTPase-activating protein (GAP) for Arf1 with a putative clathrin-binding domain that binds the clathrin adaptor protein complex 1 (AP-1) [35]. Clathrin adaptor AP-1-mediated Golgi export of amyloid precursor protein is crucial for the production of neurotoxic amyloid fragments [36]. Previous AD GWAS identified other genes in this pathway including PICALM and BIN1 [37]. RAB3B encodes one of four closely related small GTP-binding proteins (Rab3A, Rab3B, Rab3C and Rab3D) that are believed to involved in presynaptic vesicle trafficking and priming steps for regulating synaptic transmission and plasticity, an idea supported by patch-clamp electrophysiology experiments conducted in neuronal cultures from Rab3-deficient mouse hippocampus [38]. RAB3B overexpression improves dopamine handling and storage and confers protection to dopaminergic neurons [39]. The function of ZNF890P is unknown, however a GWS association was reported in a Japanese cohort between a SNP located approximately 11kb from ZNF890P and elevated high-sensitivity cardiac troponin level [40]. Troponin is a well-known plasma marker of myocardial injury and has been associated with cognitive decline in the elderly and incident AD or dementia [41]. We also obtained a GWS association of AD with aggregated rare variants in GIPR which encodes a G-protein coupled receptor for gastric inhibitory polypeptide. GIPR stimulates insulin resistance in the presence of elevated glucose. Activation of GIPR has been incorporated in the development of compounds to treat type 2 diabetes [42], a major risk factor for AD [43]. Multiple studies showed that GIPR knockout mice sustained impairment of synaptic plasticity and cognitive deficits [44, 45].
Several loci showing suggestive evidence for association also are relevant to AD. SPOCK3 encodes a member of a family of calcium-binding proteoglycan proteins that is strongly expressed in cerebral cortex and hippocampus and may have roles in regulation of action potential in neurons, neurotransmitter uptake, memory, and neuroimmunity [46]. Variants in SPOCK3 have been associated with verbal declarative memory in a GWAS [46] and variability in cytokine secretion in response to smallpox vaccine [47]. The rs117925493 SNP near INSC that was associated with AD in this study was recently reported as a GWS association with several cognitive traits and a significant expression quantitative trait locus for INSC [48]. NCOA1 has been associated with the Aβ42/Aβ40 ratio measured in plasma via a gene-based test including five rare variants in 370 middle aged African American participants of the Atherosclerosis Risk in Communities-Neurocognitive Study [49]. Contactin 5 (CNTN5) is a neuronal membrane protein that contributes to axonal targeting, synaptic formation and plasticity. Biffi et.al identified an FDR-corrected significant association of a CNTN5 SNP with white matter lesion volume in a sample of 746 ADNI participants [50].
4.2. Advantages of founder populations for AD gene discovery
Our study illustrates the greatly increased power for detection of GWS associations in founder communities in which disease-associated variants may be much more frequent compared to samples ascertained from large admixed populations (e.g., 4.6-771 fold difference for 12 of the highly significant findings shown in Tables 2 and 3). Moreover, some genetic association signals for complex diseases like AD are likely to be stronger in founder populations that are relatively genetically homogeneous. There are numerous examples of successful genome-wide scans for complex diseases and traits in religious and geographic isolates including the Amish [51-54], Hutterites [8, 55], and Icelanders [56-58]. Previously, we identified a GWS association (P=3.4x10−9) of AD with a two-SNP haplotype in ACE in 258 AD cases and controls from an Israeli-Arab community of >81,500 people who trace their ancestors to 14 founder families [7], many years before this locus emerged as GWS in a much larger dataset [37]. The advantage of founder populations for AD gene discovery was recently demonstrated in a study that identified GWS association with MGMT independently in a small portion of a Hutterite kindred and in the large ADGC GWAS sample [8]. The most pronounced finding in an ADSP study focused on rare functional variants was a missense mutation in NOTCH3 that was observed in 10 AD cases but no controls [59]. Previously, more than 125 NOTCH3 mutations had been linked to a rare dementia syndrome called Cerebral Autosomal Dominant Arteriopathy with Sub-cortical Infarcts and Leukoencephalopathy (CADASIL), but not to AD [60]. Subsequent analysis of the NOTCH3 mutation carriers showed that they all had a single haplotype and a high frequency of two mitochondrial DNA haplotypes found primarily in AJs [59]. This mutation is exceedingly rare in all populations surveyed except AJs.
4.3. Study limitations
This study has several caveats. Most notably, the sample size is small compared to other AD GWAS conducted in genetically heterogeneous EA samples containing several ancestral populations, but relatively few AJs. We applied relatively stringent limits on the PC boundaries of the AJ cluster that was derived from large EA datasets thereby excluding some individuals with substantial AJ ancestry and further reducing the sample. However, this strategy likely increased the genetic homogeneity of the sample which increased the sensitivity for detecting associations enhanced by a founder effect. In addition, because the ADGC GWAS dataset contained multiple independently ascertained and genotyped cohorts that necessitated genotype imputation to be performed for each cohort separately, relatively infrequent variants may not have been imputed well, particularly in the smaller cohorts and thus excluded by the variant QC procedure. The sample size was further reduced for genome-wide analyses because a portion of the data was derived from whole exome sequencing. Despite these limitations, we identified several GWS and SWS associations with common and infrequent variants in previously established and novel loci. Future studies of larger AJ samples will be required to detect associations with additional loci and variants that are rare in AJs.
4.4. Conclusions
Our results highlight the efficacy of conducting GWAS for AD in founder populations which may have a significantly higher frequencies for some variants that are rare in genetically heterogeneous populations, and can lead to effective discovery of genetic associations for specific ancestry populations.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: The authors reviewed the literature using traditional (eg., PubMed) as well as preprinted (e.g., medRxiv) sources on genetic association in Alzheimer disease (AD), genetics of AD and other diseases in founder populations, as well as the role of novel loci in AD.
Interpretation: We show that APOE, the TREM2 R47H mutation, and variants in several novel genes including RAB3B, SMAP2, ZNF890P and GIPR are associated with risk of AD in Ashkenazi Jews. These findings highlight the efficacy of conducting GWAS for AD in founder populations.
Future directions: Follow-up studies in independent cohorts of Ashkenazi Jews to confirm these findings and discovery additional loci are warranted. Future research should also focus on mechanisms that explain the association of AD with the novel genes and provide insight into potential novel therapeutic approaches.
Acknowledgements
The Alzheimer's Disease Sequencing Project (ADSP) is comprised of two Alzheimer's Disease (AD) genetics consortia and three National Human Genome Research Institute (NHGRI)-funded Large-Scale Sequencing and Analysis Centers (LSAC). The two AD genetics consortia are the Alzheimer's Disease Genetics Consortium (ADGC), funded by National Institute on Aging (NIA) (U01 AG032984), and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) funded by NIA (R01 AG033193), the National Heart, Lung, and Blood Institute (NHLBI), other National Institutes of Health (NIH) institutes, and other foreign governmental and nongovernmental organizations. The discovery phase analysis of sequence data is supported through UF1AG047133 (to L.A.F., J.L.H., E.R.M., M.A.P.-V., and G.D.S.); U01AG049505 to Dr. Seshadri; U01AG049506 to Dr. Boerwinkle; U01AG049507 to Dr. Wijsman; and U01AG049508 to Dr. Goate and the discovery extension phase analysis is supported through U01AG052411 to Dr. Goate, U01AG052410 to M.A.P.-V.; and U01 AG052409 to Drs. Seshadri and Fornage.
Sequencing for the Follow Up Study (FUS) is supported through U01AG057659 (to M.A.P.-V., R.M., and B.V.) and U01AG062943 (to M.A.P.-V. and R.M.). Data generation and harmonization in the Follow-up Phase is supported by U54AG052427 (to G.D.S. and L.-S.W.). The FUS Phase analysis of sequence data is supported through U01AG058589 (to Drs. Destefano, Boerwinkle, De Jager, Fornage, Seshadri, and Wijsman), U01AG058654 (to Drs. J.L.H., W.S.B., L.A.F., E.R.M., and M.A.P.-V.), U01AG058635 (to Dr. Goate), RF1AG058066 (to J.L.H., M.A.P.-V., and Dr. Scott), RF1AG057519 (to L.A.F. and Dr. Jun), R01AG048927 (to L.A.F.), and RF1AG054074 (to M.A.P.-V. and Dr. Beecham).
The ADGC cohorts include: Adult Changes in Thought (ACT) (UO1 AG006781, UO1 HG004610, UO1 HG006375, U01 HG008657), the Alzheimer's Disease Centers (ADC) (P30 AG019610, P30 AG013846, P50 AG008702, P50 AG025688, P50 AG047266, P30 AG010133, P50 AG005146, P50 AG005134, P50 AG016574, P50 AG005138, P30 AG008051, P30 AG013854, P30 AG008017, P30 AG010161, P50 AG047366, P30 AG010129, P50 AG016573, P50 AG016570, P50 AG005131, P50 AG023501, P30 AG035982, P30 AG028383, P30 AG010124, P50 AG005133, P50 AG005142, P30 AG012300, P50 AG005136, P50 AG033514, P50 AG005681, and P50 AG047270), the Chicago Health and Aging Project (CHAP) (R01 AG11101, RC4 AG039085, K23 AG030944), Indianapolis Ibadan (R01 AG009956, P30 AG010133), the Memory and Aging Project (MAP) (R01 AG17917), Mayo Clinic (MAYO) (R01 AG032990, U01 AG046139, R01 NS080820, RF1 AG051504, P50 AG016574), Mayo Parkinson's Disease controls (NS039764, NS071674, 5RC2HG005605), University of Miami (R01 AG027944, R01 AG028786, R01 AG019085, IIRG09133827, A2011048), the Multi-Institutional Research in Alzheimer's Genetic Epidemiology Study (MIRAGE) (R01 AG09029, R01 AG025259), the National Cell Repository for Alzheimer's Disease (NCRAD) (U24 AG21886), the National Institute on Aging Late Onset Alzheimer's Disease Family Study (NIA-LOAD) (R01 AG041797), the Religious Orders Study (ROS) (P30 AG10161, R01 AG15819), the Texas Alzheimer's Research and Care Consortium (TARCC) (funded by the Darrell K Royal Texas Alzheimer's Initiative), Vanderbilt University/Case Western Reserve University (VAN/CWRU) (R01 AG019757, R01 AG021547, R01 AG027944, R01 AG028786, P01 NS026630, and Alzheimer's Association), the Washington Heights–Inwood Columbia Aging Project (WHICAP) (RF1 AG054023), the University of Washington Families (VA Research Merit Grant, NIA: P50AG005136, R01AG041797, NINDS: R01NS069719), the Columbia University HispanicEstudio Familiar de Influencia Genetica de Alzheimer (EFIGA) (RF1 AG015473), the University of Toronto (UT) (funded by Wellcome Trust, Medical Research Council, Canadian Institutes of Health Research), and Genetic Differences (GD) (R01 AG007584). The CHARGE cohorts are supported in part by National Heart, Lung, and Blood Institute (NHLBI) infrastructure grant HL105756 (Dr. Psaty), RC2HL102419 (Dr. Boerwinkle), and the neurology working group is supported by the National Institute on Aging (NIA) R01 grant AG033193.
The CHARGE cohorts participating in the ADSP include the following: Austrian Stroke Prevention Study (ASPS), ASPS-Family study, and the Prospective Dementia Registry-Austria (ASPS/PRODEM-Aus), the Atherosclerosis Risk in Communities (ARIC) Study, the Cardiovascular Health Study (CHS), the Erasmus Rucphen Family Study (ERF), the Framingham Heart Study (FHS), and the Rotterdam Study (RS). ASPS is funded by the Austrian Science Fond (FWF) grant number P20545-P05 and P13180 and the Medical University of Graz. The ASPS-Fam is funded by the Austrian Science Fund (FWF) project I904), the EU Joint Programme—Neurodegenerative Disease Research (JPND) in frame of the BRIDGET project (Austria, Ministry of Science) and the Medical University of Graz and the Steiermärkische Krankenanstalten Gesellschaft. PRODEM-Austria is supported by the Austrian Research Promotion agency (FFG) (Project No. 827462) and by the Austrian National Bank (Anniversary Fund, project 15435). ARIC research is carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). Neurocognitive data in ARIC is collected by U01 2U01HL096812, 2U01HL096814, 2U01HL096899, 2U01HL096902, 2U01HL096917 from the NIH (NHLBI, NINDS, NIA, and NIDCD), and with previous brain MRI examinations funded by R01-HL70825 from the NHLBI. CHS research was supported by contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grants U01HL080295 and U01HL130114 from the NHLBI with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by R01AG023629, R01AG15928, and R01AG20098 from the NIA. FHS research is supported by NHLBI contracts N01-HC-25195 and HHSN268201500001I. This study was also supported by additional grants from the NIA (R01s AG054076, AG049607, and AG033040) and NINDS (R01NS017950). The ERF study as a part of EUROSPAN (European Special Populations Research Network) was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community's Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the programme “Quality of Life and Management of the Living Resources” of 5th Framework Programme (no. QLG2-CT-2002-01254). High-throughput analysis of the ERF data was supported by a joint grant from the Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); and the municipality of Rotterdam. Genetic data sets are also supported by the Netherlands Organization of Scientific Research NWO Investments (175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), and the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project 050-060-810. All studies thank their participants, faculty, and staff. The content of these manuscripts is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the U.S. Department of Health and Human Services.
The FUS cohorts include: the Alzheimer's Disease Centers (ADC) (P30 AG019610, P30 AG013846, P50 AG008702, P50 AG025688, P50 AG047266, P30 AG010133, P50 AG005146, P50 AG005134, P50 AG016574, P50 AG005138, P30 AG008051, P30 AG013854, P30 AG008017, P30 AG010161, P50 AG047366, P30 AG010129, P50 AG016573, P50 AG016570, P50 AG005131, P50 AG023501, P30 AG035982, P30 AG028383, P30 AG010124, P50 AG005133, P50 AG005142, P30 AG012300, P50 AG005136, P50 AG033514, P50 AG005681, and P50 AG047270), Alzheimer's Disease Neuroimaging Initiative (ADNI) (U19AG024904), Amish Protective Variant Study (RF1AG058066), Cache County Study (R01AG11380, R01AG031272, R01AG21136, RF1AG054052), Case Western Reserve University Brain Bank (CWRUBB) (P50AG008012), Case Western Reserve University Rapid Decline (CWRURD) (RF1AG058267, NU38CK000480), CubanAmerican Alzheimer's Disease Initiative (CuAADI) (3U01AG052410), Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA) (5R37AG015473, RF1AG015473, R56AG051876), Genetic and Environmental Risk Factors for Alzheimer Disease Among African Americans Study (GenerAAtions) (2R01AG09029, R01AG025259, 2R01AG048927), Gwangju Alzheimer and Related Dementias Study (GARD) (U01AG062602), Hussman Institute for Human Genomics Brain Bank (HIHGBB) (R01AG027944, Alzheimer's Association "Identification of Rare Variants in Alzheimer Disease"), Ibadan Study of Aging (IBADAN) (5R01AG009956), Mexican Health and Aging Study (MHAS) (R01AG018016), Multi-Institutional Research in Alzheimer's Genetic Epidemiology (MIRAGE) (2R01AG09029, R01AG025259, 2R01AG048927), Northern Manhattan Study (NOMAS) (R01NS29993), Peru Alzheimer's Disease Initiative (PeADI) (RF1AG054074), Puerto Rican 1066 (PR1066) (Wellcome Trust [GR066133/GR080002], European Research Council [340755]), Puerto Rican Alzheimer Disease Initiative (PRADI) (RF1AG054074), Reasons for Geographic and Racial Differences in Stroke (REGARDS) (U01NS041588), Research in African American Alzheimer Disease Initiative (REAAADI) (U01AG052410), Rush Alzheimer's Disease Center (ROSMAP) (P30AG10161, R01AG15819, R01AG17919), University of Miami Brain Endowment Bank (MBB), and University of Miami/Case Western/North Carolina A&T African American (UM/CASE/NCAT) (U01AG052410, R01AG028786).
The four LSACs are: the Human Genome Sequencing Center at the Baylor College of Medicine (U54 HG003273), the Broad Institute Genome Center (U54HG003067), The American Genome Center at the Uniformed Services University of the Health Sciences (U01AG057659), and the Washington University Genome Institute (U54HG003079).
Biological samples and associated phenotypic data used in primary data analyses were stored at study investigator institutions and at the National Cell Repository for Alzheimer's Disease (NCRAD, U24AG021886) at Indiana University funded by NIA. Associated phenotypic data used in primary and secondary data analyses were provided by the study investigators, the NIA funded Alzheimer's Disease Centers (ADCs), the National Alzheimer's Coordinating Center (NACC, U01AG016976); and the National Institute on Aging Genetics of Alzheimer's Disease Data Storage Site (NIAGADS, U24AG041689) at the University of Pennsylvania, funded by NIA. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Library of Medicine. Contributors to the Genetic Analysis Data included the study investigators on projects that were individually funded by NIA and other NIH institutes, and by private US organizations, foreign governmental organizations, or nongovernmental organizations.
The Genome Research @ Fundació ACE project (GR@ACE) is supported by Grifols SA, Fundación bancaria ‘La Caixa,’ Fundació ACE, and CIBERNED. I.d.R. is supported by a national grant from the Instituto de Salud Carlos III (ISCIII) FI20/00215. A. Ruiz and M.B. receive support from the European Union / EFPIA Innovative Medicines Initiative joint undertaking ADAPTED and MOPEAD projects (grant numbers 115975 and 115985, respectively). M.B. and A. Ruiz are also supported by national grants PI13/02434, PI16/01861, PI17/01474, PI19/01240, PI19/01301 and PI22/01403. Acción Estratégica en Salud is integrated into the Spanish National R + D + I Plan and funded by ISCIII –Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER–‘Una manera de hacer Europa’). A. Ruiz is also funded by JPco-fuND-2 “Multinational research projects on Personalized Medicine for Neurodegenerative Diseases,” PREADAPT project (ISCIII grant: AC19/00097), and EURONANOMED III Joint Transnational call for proposals (2017) for European Innovative Research & Technological Development Projects in Nanomedicine (ISCIII grant: AC17/00100). Fundacio ACE researchers research receives support from Roche, Janssen, Life Molecular Imaging, Araclon Biotech, Alkahest, Laboratorio de Análisis Echevarne, and IrsiCaixa.
We also acknowledge the following investigators who assembled and characterized participants of cohorts included in this study.
Adult Changes in Thought: James D. Bowen, Paul K. Crane, Gail P. Jarvik, C. Dirk Keene, Eric B. Larson, W. William Lee, Wayne C. McCormick, Susan M. McCurry, Shubhabrata Mukherjee.
Katie Rose Richmire Atherosclerosis Risk in Communities Study: Rebecca Gottesman, David Knopman, Thomas H. Mosley, B. Gwen Windham.
Austrian Stroke Prevention Study: Thomas Benke, Peter Dal-Bianco, Edith Hofer, Gerhard Ransmayr, Yasaman Saba.
Cardiovascular Health Study: James T. Becker, Joshua C. Bis, Annette L. Fitzpatrick, M. Ilyas Kamboh, Lewis H. Kuller, W.T. Longstreth, Jr, Oscar L. Lopez, Bruce M. Psaty, Jerome I. Rotter.
Chicago Health and Aging Project: Philip L. De Jager, Denis A. Evans.
Erasmus Rucphen Family Study: Hieab H. Adams, Hata Comic, Albert Hofman, Peter J. Koudstaal, Fernando Rivadeneira, Andre G. Uitterlinden, Dina Voijnovic.
Estudio Familiar de la Influencia Genetica en Alzheimer: Sandra Barral, Rafael Lantigua, Richard Mayeux, Martin Medrano, Dolly Reyes-Dumeyer, Badri Vardarajan.
Framingham Heart Study: Alexa S. Beiser, Vincent Chouraki, Jayanadra J. Himali, Charles C. White.
Genetic Differences: Duane Beekly, James Bowen, Walter A. Kukull, Eric B. Larson, Wayne McCormick, Gerard D. Schellenberg, Linda Teri.
Mayo Clinic: Minerva M. Carrasquillo, Dennis W. Dickson, Nilufer Ertekin-Taner, Neill R. Graff-Radford, Joseph E. Parisi, Ronald C. Petersen, Steven G. Younkin.
Mayo PD: Gary W. Beecham, Dennis W. Dickson, Ranjan Duara, Nilufer Ertekin-Taner, Tatiana M. Foroud, Neill R. Graff-Radford, Richard B. Lipton, Joseph E. Parisi, Ronald C. Petersen, Bill Scott, Jeffery M. Vance.
Memory and Aging Project: David A. Bennett, Philip L. De Jager.
Multi-Institutional Research in Alzheimer's Genetic Epidemiology Study: Sanford Auerbach, Helan Chui, Jaeyoon Chung, L. Adrienne Cupples, Charles DeCarli, Ranjan Duara, Martin Farlow, Lindsay A. Farrer, Robert Friedland, Rodney C.P. Go, Robert C. Green, Patrick Griffith, John Growdon, Gyungah R. Jun, Walter Kukull, Alexander Kurz, Mark Logue, Kathryn L. Lunetta, Thomas Obisesan, Helen Petrovitch, Marwan Sabbagh, A. Dessa Sadovnick, Magda Tsolaki.
National Cell Repository for Alzheimer's Disease: Kelley M. Faber, Tatiana M. Foroud.
National Institute on Aging (NIA) Late Onset Alzheimer's Disease Family Study: David A. Bennett, Sarah Bertelsen, Thomas D. Bird, Bradley F. Boeve, Carlos Cruchaga, Kelley Faber, Martin Farlow, Tatiana M. Foroud, Alison M. Goate, Neill R. Graff-Radford, Richard Mayeux, Ruth Ottman, Dolly Reyes-Dumeyer, Roger Rosenberg, Daniel Schaid, Robert A. Sweet, Giuseppe Tosto, Debby Tsuang, Badri Vardarajan.
NIA Alzheimer Disease Centers: Erin Abner, Marilyn S. Albert, Roger L. Albin, Liana G. Apostolova, Sanjay Asthana, Craig S. Atwood, Lisa L. Barnes, Thomas G. Beach, David A. Bennett, Eileen H. Bigio, Thomas D. Bird, Deborah Blacker, Adam Boxer, James B. Brewer, James R. Burke, Jeffrey M. Burns, Joseph D. Buxbaum, Nigel J. Cairns, Chuanhai Cao, Cynthia M. Carlsson, Richard J. Caselli, Helena C. Chui, Carlos Cruchaga, Mony de Leon, Charles DeCarli, Malcolm Dick, Dennis W. Dickson, Nilufer Ertekin-Taner, David W. Fardo, Martin R. Farlow, Lindsay A. Farrer, Steven Ferris, Tatiana M. Foroud, Matthew P. Frosch, Douglas R. Galasko, Marla Gearing, David S. Geldmacher, Daniel H. Geschwind, Bernardino Ghetti, Carey Gleason, Alison M. Goate, Teresa Gomez-Isla, Thomas Grabowski, Neill R. Graff-Radford, John H. Growdon, Lawrence S. Honig, Ryan M. Huebinger, Matthew J. Huentelman, Christine M. Hulette, Bradley T. Hyman, Suman Jayadev, Lee-Way Jin, Sterling Johnson, M. Ilyas Kamboh, Anna Karydas, Jeffrey A. Kaye, C. Dirk Keene, Ronald Kim, Neil W. Kowall, Joel H. Kramer, Frank M. LaFerla, James J. Lah, Allan I. Levey, Ge Li, Andrew P. Lieberman, Oscar L. Lopez, Constantine G. Lyketsos, Daniel C. Marson, Ann C. McKee, Marsel Mesulam, Jesse Mez, Bruce L. Miller, Carol A. Miller, Abhay Moghekar, John C. Morris, John M. Olichney, Joseph E. Parisi, Henry L. Paulson, Elaine Peskind, Ronald C. Petersen, Aimee Pierce, Wayne W. Poon, Luigi Puglielli, Joseph F. Quinn, Ashok Raj, Murray Raskind, Eric M. Reiman, Barry Reisberg, Robert A. Rissman, Erik D. Roberson, Howard J. Rosen, Roger N. Rosenberg, Martin Sadowski, Mark A. Sager, David P. Salmon, Mary Sano, Andrew J. Saykin, Julie A. Schneider, Lon S. Schneider, William W. Seeley, Scott Small, Amanda G. Smith, Robert A. Stern, Russell H. Swerdlow, Rudolph E. Tanzi, Sarah E. Tomaszewski Farias, John Q. Trojanowski, Juan C. Troncoso, Debby W. Tsuang, Vivianna M. Van Deerlin, Linda J. Van Eldik, Harry V. Vinters, Jean Paul Vonsattel, Jen Chyong Wang, Sandra Weintraub, Kathleen A. Welsh-Bohmer, Shawn Westaway, Thomas S. Wingo, Thomas Wisniewski, David A. Wolk, Randall L. Woltjer, Steven G. Younkin, Lei Yu, Chang-En Yu.
Religious Orders Study: David A. Bennett, Philip L. De Jager.
Rotterdam Study: Kamran Ikram, Frank J. Wolters.
Texas Alzheimer's Research and Care Consortium: Perrie Adams, Alyssa Aguirre, Lisa Alvarez, Gayle Ayres, Robert C. Barber, John Bertelson, Sarah Brisebois, Scott Chasse, Munro Culum, Eveleen Darby, John C. DeToledo, Thomas J. Fairchild, James R. Hall, John Hart, Michelle Hernandez, Ryan Huebinger, Leigh Johnson, Kim Johnson, Aisha Khaleeq, Janice Knebl, Laura J. Lacritz, Douglas Mains, Paul Massman, Trung Nguyen, Sid O'Bryant, Marcia Ory, Raymond Palmer, Valory Pavlik, David Paydarfar, Victoria Perez, Marsha Polk, Mary Quiceno, Joan S. Reisch, Monica Rodriguear, Roger Rosenberg, Donald R. Royall, Janet Smith, Alan Stevens, Jeffrey L. Tilson, April Wiechmann, Kirk C. Wilhelmsen, Benjamin Williams, Henrick Wilms, Martin Woon.
University of Miami: Larry D. Adams, Gary W. Beecham, Regina M. Carney, Katrina Celis, Michael L. Cuccaro, Kara L. Hamilton-Nelson, James Jaworski, Brian W. Kunkle, Eden R. Martin, Margaret A. Pericak-Vance, Farid Rajabli, Michael Schmidt, Jeffery M Vance.
University of Toronto: Ekaterina Rogaeva, Peter St. George-Hyslop.
University of Washington Families: Thomas D. Bird, Olena Korvatska, Wendy Raskind, Chang-En Yu.
Vanderbilt University: John H. Dougherty, Harry E. Gwirtsman, Jonathan L. Haines.
Washington Heights-Inwood Columbia Aging Project: Adam Brickman, Rafael Lantigua, Jennifer Manly, Richard Mayeux, Christiane Reitz, Nicole Schupf, Yaakov Stern, Giuseppe Tosto, Badri Vardarajan.
GR@ACE: Itziar de Rojas, Sonia Moreno-Grau, Pablo Garcia-Gonzalez, Emilio Alarcón-Martín, Montserrat Alegret, Ana Espinosa, Pablo García-González, Marta Marquié, Urszula Bojaryn, Ester Esteban de Antonio, Diana Ariton, Mario Ricciardi, Laura Montrreal, Adelina Orellana, Gemma Ortega, Alba Pérez-Cordón, Raquel Puerta, Natalia Roberto, Maitée Rosende-Roca, Ángela Sanabria, Oscar Sotolongo-Grau, Juan Pablo Tartari, Lluís Tárraga, Sergi Valero, Ana Mauleón, Ana Pancho, Anna Gailhajenet, Asunción Lafuente, Elvira Martín, Esther Pelejà, Liliana Vargas, Mar Buendia, Marina Guitart, Mariona Moreno, Marta Ibarria, Nuria Aguilera, Pilar Cañabate, Silvia Preckler, Susana Diego, Nuria Aguilera, Amanda Cano, Pilar Cañabate, Raúl Nuñez-Llaves, Clàudia Olivé, Ester Pelejá, Anaïs Corma-Gómez, Marta Fernández-Fuertes, Juan Macías, Juan A. Pineda, Luis M. Real & Juan Macias, Olalla Maroñas Ángel Carracedo, Inés Quintela, Vanesa Pytel, Mercè Boada & Agustín Ruiz
Funding
This work was supported in part by NIH grants R01-AG048927, U54-AG052427, U01-AG058654, U01-AG032984, RF1-AG057519, U01-AG062602, U19-AG068753, and P30-AG072878.
Abbreviations
- AD
Alzheimer disease
- ADGC
Alzheimer’s Disease Genetics Consortium
- ADSP
Alzheimer’s Disease Sequencing Project
- AJ
Ashkenazi Jewish
- EA
European ancestry
- EW
exome-wide
- GW
genome-wide
- GWAS
genome-wide association study
- GWS
genome-wide significant
- HWE
Hardy-Weinberg equilibrium
- MAF
minor allele frequency
- QC
quality control
- SNP
single nucleotide polymorphism
- SWS
study-wide significant
- WES
whole exome sequencing
- WGS
whole genome sequencing
Footnotes
Conflicts
AR and IDR acknowledge research support from Grifols SA (Spain), Fundacion Bacaria LaCaixa (Spain), ISCIII Ministry of Health (Spain), Roche, and Janssen. AR received consulting fees and honoraria from Landsteiner Genmed SL, Grifols SA, and Janssen; support for attending meetings from Grifols SA; and stock options from Landsteiner genmed SL. MB is an employee of ACE Alzheimer Center and a member of the Advisory Boards for Grifols SA, Roche, Lilly, Araclon Biotech , Merck, Zambon, Biogen, Novo-Nordisk, Bioiberica, Eisai, Servier, and Schwabe Pharma. GDS received meeting travel support from LASI-DAD. The rest of the authors do not have anything to disclose
Consent Statement
All participants provided written informed permission or, for those with substantial cognitive impairment, consent was provided by a caregiver, legal guardian, or other proxy
Data Availability
ADGC GWAS data, ADSP WGS and WES data, and summarized results are available from the National Institutue on Aging Genetics of Alzheimer Disease Storage site (NIAGADS; https://www.niagads.org).
References
- [1].Kunkle BW, Schmidt M, Klein HU, Naj AC, Hamilton-Nelson KL, Larson EB, et al. Novel Alzheimer Disease Risk Loci and Pathways in African American Individuals Using the African Genome Resources Panel: A Meta-analysis. JAMA Neurol. 2021;78:102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, et al. New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat Genet. 2022;54:412–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, et al. SORL1 is genetically associated with late-onset Alzheimer's disease in Japanese, Koreans and Caucasians. PLoS One. 2013;8:e58618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang LS, et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ϵ4,and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jun GR, Chung J, Mez J, Barber R, Beecham GW, Bennett DA, et al. Transethnic genome-wide scan identifies novel Alzheimer's disease loci. Alzheimers Dement. 2017;13:727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Meng Y, Baldwin CT, Bowirrat A, Waraska K, Inzelberg R, Friedland RP, et al. Association of polymorphisms in the Angiotensin-converting enzyme gene with Alzheimer disease in an Israeli Arab community. Am J Hum Genet. 2006;78:871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chung J, Das A, Sun X, Sobreira DR, Leung YY, Igartua C, et al. Genome-wide association and multi-omics studies identify MGMT as a novel risk gene for Alzheimer's disease among women. Alzheimers Dement. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Levy-Lahad E, Catane R, Eisenberg S, Kaufman B, Hornreich G, Lishinsky E, et al. Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am J Hum Genet. 1997;60:1059–67. [PMC free article] [PubMed] [Google Scholar]
- [10].Laken SJ, Petersen GM, Gruber SB, Oddoux C, Ostrer H, Giardiello FM, et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet. 1997;17:79–83. [DOI] [PubMed] [Google Scholar]
- [11].Foulkes WD, Thiffault I, Gruber SB, Horwitz M, Hamel N, Lee C, et al. The founder mutation MSH2*1906G-->C is an important cause of hereditary nonpolyposis colorectal cancer in the Ashkenazi Jewish population. Am J Hum Genet. 2002;71:1395–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Niell BL, Long JC, Rennert G, Gruber SB. Genetic anthropology of the colorectal cancer-susceptibility allele APC I1307K: evidence of genetic drift within the Ashkenazim. Am J Hum Genet. 2003;73:1250–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zauber NP, Sabbath-Solitare M, Marotta S, Zauber AG, Foulkes W, Chan M, et al. Clinical and genetic findings in an Ashkenazi Jewish population with colorectal neoplasms. Cancer. 2005;104:719–29. [DOI] [PubMed] [Google Scholar]
- [14].Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Moreno-Grau S, de Rojas I, Hernandez I, Quintela I, Montrreal L, Alegret M, et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer's disease and three causality networks: The GR@ACE project. Alzheimers Dement. 2019;15:1333–47. [DOI] [PubMed] [Google Scholar]
- [16].Beecham GW, Bis JC, Martin ER, Choi SH, DeStefano AL, van Duijn CM, et al. The Alzheimer's Disease Sequencing Project: Study design and sample selection. Neurol Genet. 2017;3:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Crane PK, Foroud T, Montine TJ, Larson EB. Alzheimer's Disease Sequencing Project discovery and replication criteria for cases and controls: Data from a community-based prospective cohort study with autopsy follow-up. Alzheimers Dement. 2017;13:1410–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Leung YY, Valladares O, Chou YF, Lin HJ, Kuzma AB, Cantwell L, et al. VCPA: genomic variant calling pipeline and data management tool for Alzheimer's Disease Sequencing Project. Bioinformatics. 2019;35:1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bis JC, Jian X, Kunkle BW, Chen Y, Hamilton-Nelson KL, Bush WS, et al. Whole exome sequencing study identifies novel rare and common Alzheimer's-Associated variants involved in immune response and transcriptional regulation. Mol Psychiatry. 2020;25:1859–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang X, Zhu C, Beecham G, Vardarajan BN, Ma Y, Lancour D, et al. A rare missense variant of CASP7 is associated with familial late-onset Alzheimer's disease. Alzheimers Dement. 2019;15:441–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Belloy ME, Le Guen Y, Eger SJ, Napolioni V, Greicius MD, He Z. A Fast and Robust Strategy to Remove Variant-Level Artifacts in Alzheimer Disease Sequencing Project Data. Neurol Genet. 2022;8:e200012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shifman S, Johannesson M, Bronstein M, Chen SX, Collier DA, Craddock NJ, et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bellenguez C, Strange A, Freeman C, Donnelly P, Spencer CC, Consortium WTCC. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics. 2012;28:134–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. [DOI] [PubMed] [Google Scholar]
- [28].Pedregosa FaVGaGAaMVaTBaGOaBMaPPaWRa. Scikit-learn: Machine Learning in P ython. Journal of Machine Learning Research. 2011;12:2825–30. [Google Scholar]
- [29].Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Panitch R, Hu J, Chung J, Zhu C, Meng G, Xia W, et al. Integrative brain transcriptome analysis links complement component 4 and HSPA2 to the APOE ε2 protective effect in Alzheimer disease. Mol Psychiatry. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Matsudaira T, Niki T, Taguchi T, Arai H. Transport of the cholera toxin B-subunit from recycling endosomes to the Golgi requires clathrin and AP-1. J Cell Sci. 2015;128:3131–42. [DOI] [PubMed] [Google Scholar]
- [36].Januario YC, Eden J, de Oliveira LS, De Pace R, Tavares LA, da Silva-Januario ME, et al. Clathrin adaptor AP-1-mediated Golgi export of amyloid precursor protein is crucial for the production of neurotoxic amyloid fragments. J Biol Chem. 2022;298:102172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schlüter OM, Basu J, Südhof TC, Rosenmund C. Rab3 superprimes synaptic vesicles for release: implications for short-term synaptic plasticity. J Neurosci. 2006;26:1239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chung CY, Koprich JB, Hallett PJ, Isacson O. Functional enhancement and protection of dopaminergic terminals by RAB3B overexpression. Proc Natl Acad Sci U S A. 2009;106:22474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nasu T, Satoh M, Hachiya T, Sutoh Y, Ohmomo H, Hitomi S, et al. A genome-wide association study for highly sensitive cardiac troponin T levels identified a novel genetic variation near a RBAK-ZNF890P locus in the Japanese general population. Int J Cardiol. 2021;329:186–91. [DOI] [PubMed] [Google Scholar]
- [41].Schneider AL, Rawlings AM, Sharrett AR, Alonso A, Mosley TH, Hoogeveen RC, et al. High-sensitivity cardiac troponin T and cognitive function and dementia risk: the atherosclerosis risk in communities study. Eur Heart J. 2014;35:1817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Min T, Bain SC. The Role of Tirzepatide, Dual GIP and GLP-1 Receptor Agonist, in the Management of Type 2 Diabetes: The SURPASS Clinical Trials. Diabetes Ther. 2021;12:143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].James BD, Bennett DA. Causes and Patterns of Dementia: An Update in the Era of Redefining Alzheimer's Disease. Annu Rev Public Health. 2019;40:65–84. [DOI] [PubMed] [Google Scholar]
- [44].Faivre E, Gault VA, Thorens B, Holscher C. Glucose-dependent insulinotropic polypeptide receptor knockout mice are impaired in learning, synaptic plasticity, and neurogenesis. J Neurophysiol. 2011;105:1574–80. [DOI] [PubMed] [Google Scholar]
- [45].Lennox R, Moffett RC, Porter DW, Irwin N, Gault VA, Flatt PR. Effects of glucose-dependent insulinotropic polypeptide receptor knockout and a high-fat diet on cognitive function and hippocampal gene expression in mice. Mol Med Rep. 2015;12:1544–8. [DOI] [PubMed] [Google Scholar]
- [46].Debette S, Ibrahim Verbaas CA, Bressler J, Schuur M, Smith A, Bis JC, et al. Genome-wide studies of verbal declarative memory in nondemented older people: the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Biol Psychiatry. 2015;77:749–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kennedy RB, Ovsyannikova IG, Pankratz VS, Haralambieva IH, Vierkant RA, Poland GA. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Hum Genet. 2012;131:1403–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang K, Xu C, Smith A, Xiao D, Navia RO, Lu Y, et al. Genome-wide association study identified INSC gene associated with Trail Making Test Part A and Alzheimer's disease related cognitive phenotypes. Prog Neuropsychopharmacol Biol Psychiatry. 2021;111:110393. [DOI] [PubMed] [Google Scholar]
- [49].Simino J, Wang Z, Bressler J, Chouraki V, Yang Q, Younkin SG, et al. Whole exome sequence-based association analyses of plasma amyloid-beta in African and European Americans; the Atherosclerosis Risk in Communities-Neurocognitive Study. PLoS One. 2017;12:e0180046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, et al. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Shen H, Damcott CM, Rampersaud E, Pollin TI, Horenstein RB, McArdle PF, et al. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Arch Intern Med. 2010;170:1850–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lu W, Cheng YC, Chen K, Wang H, Gerhard GS, Still CD, et al. Evidence for several independent genetic variants affecting lipoprotein (a) cholesterol levels. Hum Mol Genet. 2015;24:2390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Montasser ME, Aslibekyan S, Srinivasasainagendra V, Tiwari HK, Patki A, Bagheri M, et al. An Amish founder population reveals rare-population genetic determinants of the human lipidome. Commun Biol. 2022;5:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Osterman MD, Song YE, Nittala M, Sadda SR, Scott WK, Stambolian D, et al. Genomewide Association Study of Retinal Traits in the Amish Reveals Loci Influencing Drusen Development and Link to Age-Related Macular Degeneration. Invest Ophthalmol Vis Sci. 2022;63:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yao TC, Du G, Han L, Sun Y, Hu D, Yang JJ, et al. Genome-wide association study of lung function phenotypes in a founder population. J Allergy Clin Immunol. 2014;133:248–55.e1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–5. [DOI] [PubMed] [Google Scholar]
- [57].Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009;41:734–8. [DOI] [PubMed] [Google Scholar]
- [58].Oskarsson GR, Magnusson MK, Oddsson A, Jensson BO, Fridriksdottir R, Arnadottir GA, et al. Genetic architecture of band neutrophil fraction in Iceland. Commun Biol. 2022;5:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Patel D, Mez J, Vardarajan BN, Staley L, Chung J, Zhang X, et al. Association of Rare Coding Mutations With Alzheimer Disease and Other Dementias Among Adults of European Ancestry. JAMA Netw Open. 2019;2:e191350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssière C, et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 1997;350:1511–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ADGC GWAS data, ADSP WGS and WES data, and summarized results are available from the National Institutue on Aging Genetics of Alzheimer Disease Storage site (NIAGADS; https://www.niagads.org).