Abstract
Hyperthermophile microorganisms have been discovered worldwide, and several studies regarding biodiversity and the potential biotechnological applications have been reported. In this work, we describe for the first time the diversity of hyperthermophile communities in the Calientes Geothermal Field (CGF) located 4400 m above sea level in Tacna Region, Perú. Three hot springs were monitored and showed a temperature around 84 to 88 °C, for the microbiome analyzed was taken by sampling of sediment and water (pH 7.3–7.6). The hyperthermophile diversity was determined by PCR, DGGE, and DNA sequencing. The sediments analyzed showed a greater diversity than water samples. Sediments showed a more abundant population of bacteria than archaea, with the presence of at least 9 and 5 phylotypes, respectively. Most interestingly, in some taxa of bacteria (Bacillus) and archaea (Haloarcula and Halalkalicoccus), any of operational taxonomic units (OTUs) have not been observed before in hyperthermophile environments. Our results provide insight in the hyperthermophile diversity and reveal the possibility to develop new biotechnological applications based on the kind of environments.
Keywords: Hyperthermophiles, Bacteria, Archaea, Geothermal field, DGGE
Introduction
The earth hosts various unique, unknown and extreme niches, among all of them, that have hyperthermophile environments, these hot spots could be found in the deep of the sea [1], and many terrestrial geothermal systems, including along tectonic boundaries, spreading centers, or “hot spots,” where magma bodies may reach within a few kilometers of the Earth’s surface [2–6]. The reports of these thermic habitats are terrestrial geothermal fields caused mainly by volcanism [7–11]. Some examples are known, such as geothermal fields reported from the Devonian Drummond Basin (Australia), the Devonian Rhynie cherts (Scotland), and the Yellowstone-style geothermal landscapes of Patagonia, Argentina ([12]).
These geothermal fields are also found in the Peruvian Andes. Here, geothermal activity is usually manifested on the surface as hot springs, geysers, and fumaroles. Water in these pools is often recycled through the geysers and back to the pool. Most of the hot springs in the Peruvian Andes discharge alkaline water; however, pH can vary between 1 and 9 [13]. The typical discharge of these springs is 5 L s, but discharges of up to 60 L s have been measured in the CGF [13].
The hot springs’s compositions have been monitored by geologic drivers (climate, tectonics, heat input) that a role as distal factors, such as water-rock interaction, hardness of water, conductivity, microbial metabolism, mixing, and other physicochemical characteristics could be important for the study of the environments [4]. The physicochemical characteristics in the hot springs occur in soils rich in iron and calcium salts, which generate yellow and whitish colorations. The chemical conditions in these sites facilitate the growth of organisms that can oxidize hydrogen, sulfur, hydrogen sulfide, and thiosulfate [14]; reduce sulfur [15], nitrates, or nitrites [16]; or reduce iron in a photosynthesis-dependent way [17]. Therefore, microorganisms dominate the biota associated with these sites, their associated high temperatures, and unique characteristics [14]. Hyperthermophile microorganisms can be found living in geothermal fields, which are fairly accessible and therefore constitute one of the most studied habitats in terms of hyperthermophile diversity [2, 8, 18].
For these environments, a variety of classification have been used to define the microorganisms that live in that kind of temperature habitat, the most common classification have been used on optimal growth temperature, psychrophiles growth temperature of 20 °C [19], mesophiles having a 20 to 45 °C [20], thermophiles having an optimal growth temperature ranging from 45 to 80 °C [21], and hyperthermophiles having an optimal growth temperature of 80 °C [21]. For the thermophiles and hyperthermophiles, the clades most commonly studied are the bacterial and archaeal phylum [17], based on experience focused on study on the microbial communities by culture-based approaches initially used to study microbial diversity [21]. Microbial exploration due to the development of molecular biological techniques has greatly improved in the recent decades. Advances in cultivation-independent methods for examining uncultured microbes, including single-cell genomics and deep sequencing of environmental samples, have begun yielding complete or near-complete genomes from many novel lineages [21].
Additional terms are commonly used to describe polyextremophiles, such as thermoacidophiles, capable of growth at high temperature and low pH, halophilic thermoalkaliphiles, capable of growth at high temperature, high salt, and high pH, and thermophilic piezophiles, capable of growth under high temperature and pressure [6, 22–24]. A technique culture-independent that has been described as denaturing gradient gel electrophoresis (DGGE) analysis of 16 S rRNA gene segments has been used to profile complex microbial communities and to infer the phylogenetic affiliation of the community members [25].
Since the application of the Taq polymerase from the hyperthermophile Thermus aquaticus [26], the biological application for the microorganisms on the hot springs environments has made a paradigm shift in the industry. The potential biotechnological of the thermophiles and hyperthermophiles or their enzyme applications are a variety of advantages as bioremediation [27], the production of biomolecules [28], production of biofuels [29], biomining [30], in agriculture, biosurfactants could substitute chemical surfactants as adjutants in herbicide and pesticide formulations [31], or a microorganism for a biomarker of the wasted o extreme environment [32]. This application of the thermophiles or their enzymes is not surprising. They have a remarkable capacity to work in environmental fluctuations such as pH, temperature, and other possibilities [33].
Here, we analyzed for the first time the microbial diversity in the Calientes Geothermal Field (CGF) of the Peruvian Andes, the highest-known geothermal field. We collected water and sediment samples from three hot springs in the CGF, and the abundance and diversity of bacteria and archaea were determined. These data allowed us to describe, for the first time, the hyperthermophile diversity present in the CGF and opened the possibility to develop novel biotechnological applications based on the bioresources present in the unique hyperthermophilic microorganisms inhabiting this environment.
Materials and methods
Sample collection
The CGF is located in the Cordillera Occidental in Candarave Province, Tacna Region, Peru (17 15’30” S, 70 9’ W) at 4400 m above sea level (Fig. 1 a, b), as they represent the most likely places to identify hyperthermophiles. A total of four water (W) samples (3 L) were collected, one from each hot spring at a depth of 1 m (samples G1 (W), G2 (W), and G3 (W)). Also, a water sample G2S (W) was collected from the surface of G2. Each sample was filtered on-site using sterile filter units (0.2 m, 25 mm diameter, Millipore Sterivex, Darmstadt, Germany). Sediments on the filters were retained, and filtered water was discarded. Pool temperature and pH were also measured. A total of five sediment (S) samples were collected using sterile spatulas, one from each pool at a depth of less than 10 cm (samples G1 (S), G2 (S), and G3 (S)). Two additional sediment samples were taken from G1 and G2 at a depth greater than 10 cm (G1T (S) and G2T (S)). The collected sediment was placed in sterilized 50-mL Falcon tubes. Filters and sediment samples were stored at room temperature until DNA extraction.
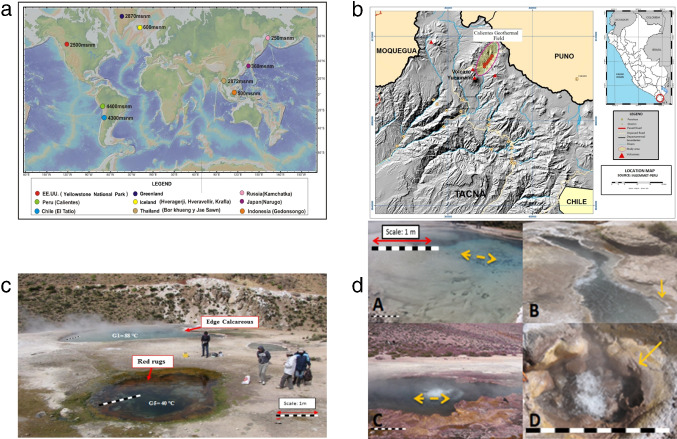
Fig. 1.
The Calientes Geothermal Field (CGF) geographical characteristics. a Geographical world location of CGF and comparison with other GF. b Location of the CGF in Peru, South America. c The arrows indicate the location where water and sediment samples were taken (Source: INGEMMET-Peru). Panoramic view of hot springs in the CGF. Biofilms are visible in pool G5; calciferous borders are visible in pool G1. The bar approximates 1 m. d Sampling locations in the CGF. (A) Hot spring 1 (G1), arrows indicate where water sample (G1 (W)) was collected; (B) G1, arrow indicates where sediment sample (G1 (S)) was collected; (C) G2, arrows indicate where water samples (G2 (W) and G2S (W)) were obtained; (D) G3, arrow indicates the calciferous sediment. Scale bars represents 1 m of length
Physicochemical analysis of the CGF water
The composition and content of chemical elements in the water of three geysers G1, G2, and G3 were evaluated. For this, 1 L of water was collected from the surface of each geyser in a glass flask, and the pH was brought to 2 with nitric acid (1 N) and hermetically covered. The three samples were sent to a private laboratory for physicochemical analysis of total solids, total hardness, conductivity, and the composition of the trace elements (EPA 200), and anions (EPA 300).
Genomic DNA extraction and amplification of 16 S rRNA
Genomic DNA was extracted from sediment (600 mg) or approximately 1 cm of the filter from each sample using the MOBIO’s Power Soil DNA isolation kit (QIAGEN, California, USA.) according to the manufacturer’s instructions. Amplification of the 16 S rRNA gene from Bacteria and Archaea was performed by nested PCR. For archaea analysis, the first PCR included the primers Ar4F and Un1492 and produced fragments of 1500 bp (Table 1). These were used as templates for the nested PCR with the primers Ar3F (positions 7–26) and Ar9R (positions 906–927) [34], which generated an 880 bp fragment. For bacterial analysis, primers Eub9-27F and Eub1542R were used for the first PCR [35] (Table 1). PCR (25 L final volume) conditions were as follows: 2 mM MgCl (Roche, Switzerland), 200 M dNTPs (Promega, Wisconsin, USA), 1 pmol of each oligonucleotide, 2.5 U Gotaq ((Promega, Wisconsin, USA ), and 10–100 ng template DNA. DNA was denatured for 5 min at 94 C, followed by 35 cycles of 94 C for 30 s, 55 C for 45 s, and 72 C for 78 s. Fragments were further used for denaturing gradient gel eletrophoresis (DGGE).
Table 1.
List of primers used in this study
| Name | Domain | Position | Sequence (5’-3’) |
|---|---|---|---|
| Ar4Fa | Archaea | 8–25 | TCY GGT TGA TCC TGC CRG |
| Un1492Rb | Universal | 1492–1510 | GGT TAC CTT GTT ACG ACT T |
| Ar3Fc | Archaea | 7–26 | TTC CGG TTG ATC CTG CCG GA |
| Ar9Rd | Archaea | 906–927 | CCC GCC AAT TCC TTT AAG TTT C |
| Ar344 Fe | Archaea | 344–363 | TCG CGC CTG CTG CTC CCC GT |
| Ar915Rf | Archaea | 915–934 | GTG CTC CCC CGC CAA TTC CT |
| 27Fg | Bacteria | 8–27 | AGA GTT TGA TCC TGG CTC AG |
| 1542Rh | Bacteria | 1542–1525 | AGA AAG GAG GTG ATC CAG CC |
| 341 F-GC(P3)i | Bacteria | 111–130 | GGA ATC TTC CAC AAT GGG CG |
| 534 R(P2)j | Bacteria | 361–380 | TTC CCC ACG CGT TAC TCA CC |
| 344F-GCk | Archaea | 111–130 | ACG GGG CGC AGC AGG CGC GA |
| 915Rl | Archaea | 361–380 | GTG CTC CCC CGC CAA TTC CT |
| G+C Clamp | CGC CCG CCG CGC CCC GCG CCC | ||
| GTC CCG CCG CCC CCG CCC C |
Denaturing gradient gel electrophoresis (DGGE)
DGGE analysis was performed according to [41]. PCR products from bacterial 16 S rRNA were generated with primers 341F-GC and 534R (Table 1) [39]. For archaea, primers 344F-GC [40] and 915R [40] (Table 1) were used. Primer 344F-GC contains a 5’ 40-nucleotide GC clamp that provides stability during DGGE [39]. PCR products were loaded on 7.5% polyacrylamide gels (MERCK, USA) containing a denaturing linear gradient from 30 to 60% for bacteria and 20 to 70% for Archaea according to Green et al. (2017) [25]). Urea (7 M) and 40% formamide were defined as 100%. Separations were achieved at 60 C and 200 V for 6 h (BioRad D Gene, California, Estados Unidos). Gels were developed with SYBR Gold (Invitrogen; final concentration 2.5×). Bands were observed by UV translumination (Vilber Lourmat, Germany).
Re-PCR of DGGE bands
The brightest DGGE bands (Fig. 2 a, b) were cut out of the gel with a sterile scalpel and placed in Eppendorf tubes containing 100 L of distilled sterile water (MERCK, USA). Samples were incubated at 37 C for 1 h and 4 C for 24 h. Recovered DNA (5 L) from each band was amplified by PCR in 25 L. The primers used for re-PCR had the same sequence as the original amplification but lacked the GC clamp.
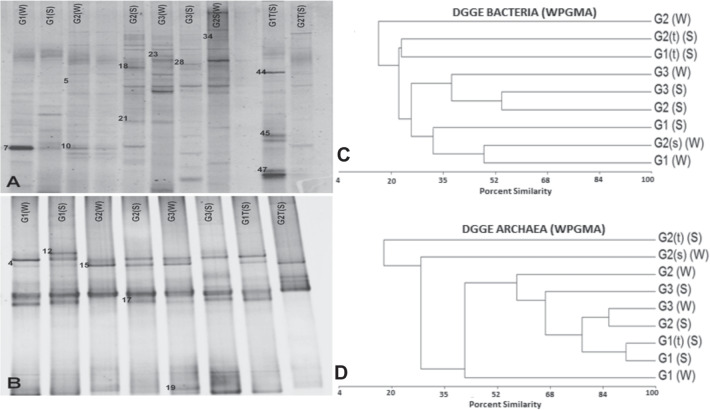
Fig. 2.
DGGE diversity profiles. A PCR-DGGE analysis of the predominant bacterial/archaeal communities of water and sediment samples obtained from the CGF. Total DNA was extracted from samples, and the 16 S rRNA gene was amplified by nested PCR using primers specific for bacteria (A) or archaea (B). Each labeled well represents a separate sample with a denaturing linear gradient from 30 to 60% for bacteria and 20 to 70% for archaea. The bands had been sequenced and compared in the NCBI database, the best results are shown in Table 4. B Numbered bands were submitted for sequencing. UPGMA (unweighted pair-group method using arithmetic averages) dendrogram generated from bacterial denaturing gradient gel electrophoresis (DGGE) profiles. Samples are from water (W), sediment (S), C bacteria, and D archea. G1, G2, and G3 are the hot springs used for sample collection. (W), water sample collected at 1 m of depth; (s) (W), water sample collected at the surface; (S), sediment sample collected at less than 10 cm of depth; (t) (S), sediment sample collected at more than 10 cm of depth
Sequencing of DGGE bands
DNA samples were sent to Macrogen Inc. (Seoul, South Korea) for sequencing in an ABI PRISM 3700 (Applied Biosystems). The forward and reverse primers were 341F and 907R for bacteria and 344F and 915R for archaea.
Denaturing gradient gel electrophoresis (DGGE)
To analyze the diversity of microbes in the samples, a matrix of the distribution of DNA bands was prepared in a way to clarify the presence or absence of different microbial species. This relationship was represented graphically by cluster analysis (cluster-WPGMA), based on the similarity percentage between samples (Multi-Variate Statistical Package, version 3.12d; Kovach Computing Services, Wales, UK).
Sequence analysis
A total of 16 S rRNA sequences were compared with available sequences in the database of the National Center for Biotechnology Information (NCBI), using BlastN. The cladograms were made in MEGA X [42].
Results
Ambiental and physical chemistry characteristics of the CGF
There were more than 87 geothermal features in the CGF, whose water temperature ranged around 37 and 88 C and pH almost neutral (7). Vents with temperatures close to 40 C contained layers of red, green, and gray biofilms, some of which were exposed to the air (Fig. 1c). The three hot springs chosen for this study had the highest water temperatures (G1 = 88 C; G2 = 88 C; G3 = 84 C), and pH was close to neutral (G1 = 7.3; G2 = 7.14 and G3 = 7.6). Coherent to most thermal water found in the CGF, these pools had a distinct white-yellow coloration and a hard bottom, which is characteristic of carbonate and other mineral precipitates (Fig. 1 c and d; [13]).
The composition chemical of water showed certain similarities among the total solids, sulfates, and bicarbonates concentration in G1 and G2, with a similarity above 99% in all chemical characteristics (Tables 2 and 3). While trace element composition showed a more difference between G1 and G2, aluminum, calcium, magnesium, and manganese with similarities around 23.4%, 79.3%, 76.82%, and 35.4% within G2 have detected more concentration than G1 in these trace elements. However, G3 revealed the most variations in its chemical parameters about G1 and G2, within more bicarbonate over 188%, but a total solid lesser of the other two points (<50%). The trace element has been different in all of the components only the calcium reported moderate similarity with G2 (95.9) (Table 3).
Table 2.
General characteristics of the three hot springs in the CGF selected for biodiversity analysis
| Hot springs number | Activity | Parameters | Visual description | ||
|---|---|---|---|---|---|
| Temperature (C) | pH | Diameter (m) | |||
| G1 | +++ | 88 | 7.3 | 10 | Whitish calcareous sediment |
| G2 | ++ | 88 | 7.4 | 4.5 | Stony and gray |
| G3 | ++ | 84 | 7.6 | 0.89 | Chalky whitish beige |
The air temperature was 11 C when the samples were collected
++, active
+++, very active
Table 3.
Physico-chemical analysis of the CGF water of the three hot springs in the CGF selected for biodiversity analysis
| Elements | Geysers (mg L) | ||
|---|---|---|---|
| G1 | G2 | G3 | |
| Aluminum | 0.128 | 0.030 | <0.001 |
| Arsenic | 12.1338 | 12.0930 | 7.3290 |
| Cadmium | <0.0002 | <0.0002 | <0.0002 |
| Calcium | 37.260 | 46.998 | 49.421 |
| Cobalt | <0.00007 | <0.00007 | <0.00007 |
| Copper | <0.001 | <0.001 | <0.001 |
| Chromium | <0.002 | <0.002 | <0.002 |
| Phosphorus | <0.2 | <0.2 | <0.2 |
| Iron | 0.023 | <0.001 | 0.046 |
| Lithium | 8.2425 | 8.9223 | 5.5001 |
| Magnesium | 0.285 | 0.371 | 4.275 |
| Manganese | 0.0540 | 0.1414 | 0.6112 |
| Mercury | 0.00270 | 0.00164 | 0.00081 |
| Molybdenum | 0.06133 | 0.06397 | 0.04687 |
| Potassium | 85.6 | 86.4 | 50.1 |
| Selenium | <0.002 | <0.002 | <0.002 |
| Sodium | 1426.07 | 1510.39 | 856.95 |
| Zinc | 0.0055 | 0.0030 | 0.0016 |
| Sulfate | 85.80 | 85.12 | 91.41 |
| Total Solids | 3920 | 3950 | 1930 |
| Bicarbonatesa | 88.6 | 99.8 | 188.2 |
| Total hardnessa | 96.2 | 124.7 | 145.5 |
| Conductivityb | 6970.00 | 7020.00 | 4440.00 |
a, unity (mg HCO3/L); b, unity (S/cm)
Composition and diversity of microbial communities in CGF
To analyze the bacterial and archaeal diversity of the hot springs, DGGE was used to visualize several PCR-amplified 16 S rRNA gene fragments. A 30–60% urea-formamide denaturing gradient gel was used to separate the bacterial PCR products obtained from primers 341F-GC and 534R (Table 1; Fig. 2A), and a 20–70% gel was used to separate the archaeal PCR products obtained from primers 344F-GC and 915R (Fig. 2B). Next, cluster analysis (WPGMA) of DGGE bands was used to look for similarities in bacterial and archaeal composition between samples and hot springs. Bacterial samples (Fig. 2C) showed high diversity. Indeed, the most similar clusters, samples G2 (S) and G3 (S), had less than 60% similarity. Analysis of archaeal samples (Fig. 2D) revealed both diversity and similarity. The highest percentage of similarity was between G1 (S) and G1 (t) (S) (90%), followed by G2 (S) and G3 (W) (85%), and by the G3 group, which showed an equal similarity among them (65%). All other samples exhibited high diversity, with less than 60% similarity.
Phylogenetic diversity of bacteria and archaea in CGF
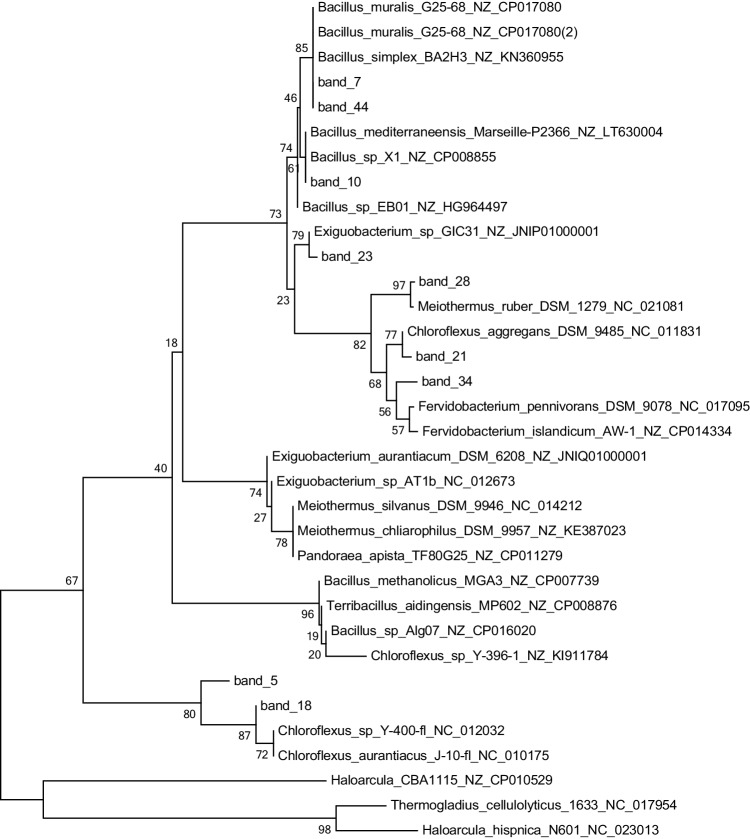
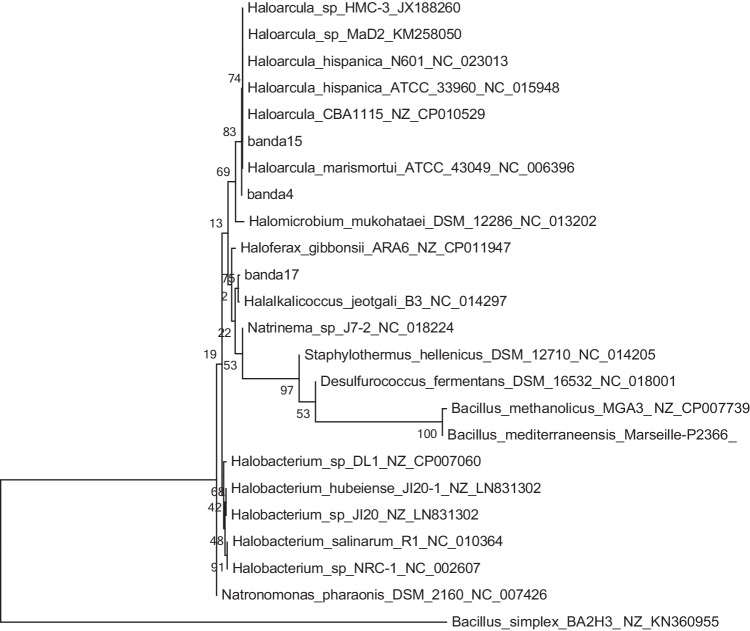
To identify the phylotypes present in the CGF, 16 S rRNA sequences obtained from the brightest DGGE bands were compared with the NCBI database using BLASTN. The threshold for similarity was set at greater than or equal to 97%, which excluded 80.4% and 79.2% of bacterial and archaeal sequence results, respectively. Sequence analysis revealed 124 operational taxonomic units (OTUs) in sediment for bacteria and 54 OTUs for archaea. The analysis of bacterial sequences revealed the presence of the phylotypes Firmicutes, Deinococci, Gammaproteobacteria, and Chloroflexi (Fig. 3, Table 4). The blast hits obtained (except for Enterobacteriales) were associated with high salinity environments; however, the bands 21, 28, and 34 are high affinity with thermophilic microorganisms like Meiothermus spp. and Fervidobacterium spp. associated with geothermal environment [43, 44], and the bands 7, 10, 23, and 44 within a relationship with the clade of Bacillus spp. included Bacillus simplex and Bacillus muralis [45, 46]. However, bands 5, 18, 21, and 23 are associated with Exiguobacterium species; this clade is not reported to live above 80 °C, although relationship in a hot spring or thermophilic application [47, 48], and Chloroflexus spp. are able to live until 70 °C [49, 50] This suggests the potential formation of microenvironments in the niches and/or the potential presence of more thermophilic variants of these species. For the archaea domain, the sequences belonged to the phylotype Halobacteria (Fig. 4, Table 4). Sequences from Band4 and Band15 were related to Haloarcula spp., a species that has not been reported to grow at high temperatures [51]. Meanwhile, Band17 is closed Halalkalicoccus spp., a genus commonly grown in mesophilic environments; these have been more associated with subterranean water environments with high salinity (>6%) [52–54]. Band19 was related to Haloarcula spp., another halophilic archaeon, also not reported to grow at high temperatures, and more “alkaliphilic” than Haloarcula [23]; however, Band12 and Band19 are associated with a high similarity (>97%) to uncultured archaeon.
Fig. 3.
Phylogenetic tree of bacterial diversity in the CGF based on partial 16 S rRNA gene sequences. The brightest bands obtained from the DGGE were cut, re-amplified, and sequenced. Sequences were compared with BLASTN to the NCBI database. Sequences from this study are in red, indicating the corresponding DGGE band number. Reference sequences were chosen to represent the greatest diversity of bacteria. Scale bar at the bottom left represents 0.2 (20%) nucleotide sequence difference
Table 4.
BLASTN phylogenetic analysis of bacterial and archaeal 16 S rRNA gene sequences obtained from DGGE
| Sequence | Hot springs number | Accession | Best match | % Identity |
|---|---|---|---|---|
| Bacteria | ||||
| Band05 | G2 (W) | NZ_CP007739 | Bacilllus methanolicus strain MGA3 | 100.0 |
| Band07 | G1 (W) | KY615350 | Bacillus simplex strain E204 | 100.0 |
| Band10 | G2 (W) | NR_144741 | Bacillus mediterraneensis strain Marseille-P2366 | 100.0 |
| Band18 | G2 (S) | NR_074263 | Chloroflexus aurantiacus strain J-10-fl | 97.7 |
| Band21 | G2 (S) | NR_074226 | Chloroflexus aggregans strain DSM 9485 | 97.8 |
| Band23 | G3 (W) | NZ_JNIP01000001 | Exiguobacterium sp. E11_27 | 98.3 |
| Band28 | G3(S) | NR_145943 | Meiothermus roseus strain YIM 71031 | 100.0 |
| Band34 | G2S (W) | MK299259 | Uncultured Chthonomonadales bacterium | 99.0 |
| Band44 | G1T (S) | AY795693 | Bacterium Schreyahn_K9.Sali | 98.4 |
| Band45 | G1T (S) | EU876657 | Uncultured proteobacterium clone DB2 | 99.3 |
| Band47 | G1T (S) | EU919218 | Uncultured Raoultella sp. clone QRSYY3 | 99.4 |
| Archeae | ||||
| Band04 | G1 (W) | JX188260 | Haloarcula sp. HMC-3 | 99.6 |
| Band12 | G1 (S) | HM234400 | Uncultured archaeon clone A3-14 | 97.2 |
| Band15 | G2 (W) | AB074561 | Halobacterium sp. NCIMB 714 | 99.8 |
| Band17 | G2 (S) | NC_014297 | Uncultured Halalkalicoccus sp | 99.4 |
| Band19 | G3 (W) | HQ425124 | archaeon BC32 | 97.7 |
Fig. 4.
Phylogenetic tree of archaeal diversity in the CGF based on partial 16 S rRNA gene sequences. Sequences were compared with BLASTN to the NCBI database. Sequences from this study are in red, indicating the corresponding DGGE band number. Reference sequences were chosen to represent the greatest diversity of archaea. Scale bar at the bottom left represents 0.2 (20%) nucleotide sequence difference
Discussion
Geothermal fields are excellent sites for the study of hyperthermophile microorganisms, which are of great interest to biotechnology. Here, we characterized for the first time the bacterial and archaeal diversity of the CGF, the highest-elevation geothermal field in the world (Fig. 1). Although other CGF has been reported around the world, as Lardello, Italy (160 to 860 m a.s.l.) [55]; Rotokawa, New Zealand ( 500 m a.s.l) [56]; Hengill, Iceland ( 800 m a.s.l.) [57]; Yellowstone park, USA ( 2.500 m a.s.l) [58]; and GCF in Latin America as Copahue, Argentina ( 1600 m a.s.l) [24], Tatio, Chile ( 4.200) [22], in this study showed an environment diversity assay with an elevation around 4.400 m a.s.l, this could give an advantage and restriction in this environment that will be presented in discussion.
About the physicochemical, the pH in this CGF is moderately alkaline in the three hot springs around 7.3 to 7.6, against other CGF with evidence of chemotroph metabolism (Fig. 1b), like Copahue or Yellowstone with a pH around 2 to 5 [24, 58], but similar pH to hot spring with the presence of high concentration of microorganism photosynthetic [59]. This could be explaining to the dessert that it is more heavy metal-free and that these alkalize the water [60], this could be supported due to the fact that Tatio hot spring in Chile has been reported similar pH (Table 3) [22]. The conductivity is similar to that reported for other authors with present chemotrophic microorganisms, and the temperature consistency among the three hot springs was studied (Tabla 2), which is a similar temperature for other CGF [24].
Microbial diversity in the CGF was abundant and varied among samples and collection sites. The WPGMA cluster analysis of Bacteria in sediment and water samples did not show a clear cluster (Fig. 2), indicating high diversity in this environment. These results confirm that each testing site and type of sample presents different physical and chemical conditions that resulted in adaptation of the microbes present in those sites (Tables 2 and 3) [14–17, 61]. Abiotic factors such as the ion cocktail (nutrients) of each site, as well as the temperature that determines the microbial communities, were observed in a particular location [62, 63]. Although, samples such as G2 (S) and G3 (S) showed reasonably high similarity. Differences in the environment surrounding geothermal fields may also influence microbial diversity. For example, the Yellowstone National Park has more abundant flora and fauna than the CGF or any hot spring in a desertic zone, and animals such as birds and mammals may facilitate the spread of bacteria among thermal features [8, 14, 64, 65].
PCR-DGGE finger printer in bacterial and archaea showed different taxas in the environment samples in both phylum (Fig. 2) and have no representative difference in the sediments in bacteria (Fig. 2b); however, the archaea revealed a more significative distribution according to the CGFs location (Fig. 1 c and d). Furthermore, the abundance in the CGF samples was not considered because this intensity in the bands may have been obtained by an amplification artifact [66].
DGGE and sequence analysis revealed a higher diversity of bacteria than archaea. Additionally, 80.4% of bacteria and 79.2% of archaea sequences had a BLASTN similarity of less than 97%, constituting a variety of unknown microorganisms that could serve as a platform for new studies and hyperthermophile research. This match was similar to other experiences from other hot springs reported in the literature [18, 67, 68].
Here, we demonstrated for the first time the presence of different taxa of hyperthermophiles (Table 3) in the GCF. Also, we found a new clade of hyperthermophile bacteria represented by bands 5, 18, 45, 47, and 48 (Fig. 3) and observed new branches (bands 28 and 21), which are not related to known sequences in GenBank; in this case, the bands associated with uncultured microorganism and proteobacteria are commonly associated with mesophilic and pathogenic but was reported to geothermal environments, which suggest a new clade with environment enzyme like as metal resistance associate with antibiotic resistance capacity [46, 51, 69]. It is possible that these new hyperthermophile bacterial species may be endemic to the CGF, indicating a high level of endemism for bacteria in this site [70–72]. Likewise, we identified a new clade of Archaea in bands 12 and 19 (Fig. 4) Several taxa reported in this study, such as Bacillus spp., Gammaproteobacteria, Chloroflexus aggregans, Meiothermus, and Archaeas have been previously reported in this type of environment [73]. However, we also determined the presence of some taxa that have not been described in thermal springs before. These include uncultured Exiguobacterium spp., Halobacterium spp., and Halalkalicoccus spp. These taxa may have adapted physiologically and metabolically to take advantage of the thermal spring environment [17, 74–76].
The biodiversity analysis of the three evaluated geysers shows interesting results. It was determined that there is a similarity between the prokaryotic members identified in the geysers (G1 and G2), between bacterial members (Bacillus muralis, B. simplex, Bacillus mediterraniensis, and Chloroflexus) and archaea (Halobacterium sp, Halalkalicoccus sp., and band 12, which would be related by its proximity to halophilic members); on the other hand, the diversity analysis of the geyser (G3) shows different members of the bacteria domain such as Exiguobacterium and Meiothermus ruber, as well as in archaea the appearance of a band (19) distant from the halophilic group. These results added to the fact that there are similarities in the temperature and pH data (Table 2), and concentrations of certain metals (Table 3) would lead us to suppose that geysers (G1 and G2) tend to have a common origin and G3 a different origin.
In summary, the three sites tested had similar pH and water temperature, but each one of them had unique microbial diversity. Entire new clades of archaea and bacteria were identified, while other taxa of archaea and bacteria have uniquely diversified to live in these springs, which showed a high degree of microbial endemism. These results, the first of their kind in the CGF, will set the stage for continued research into the metabolic strategies, use, and identification of hyperthermophiles. This study is also important on biotechnological applications from the singular metabolism at a high temperature, which would have industrial benefits and improve the circular economy.
Acknowledgements
We would like to thank Dr. Jose Perez (Andrés Bello University and Center for Bioinformatics and Genome Biology, Fundación Ciencia & Vida, Chile) for his contribution to experimental development and manuscript preparation, respectively; Vicentina Cruz for map preparation (INGEMMET, Peru); and Dr. Albert Saavedra for his help with sample collection.
Funding
This work has been co-funded by CONCYTEC-PERU, Project No 357/2012 (Roberto Castellanos).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Silvia Valdez and Fabián Veliz de la Vega contributed equally to this work.
References
- 1.English JM, English KL, Dunphy RB, Blake S, Walsh J, Raine R, Vafeas NA, Salgado PR. An overview of deep geothermal energy and its potential on the Island of Ireland. First Break. 2023;41(2):33–43. doi: 10.3997/1365-2397.fb2023009. [DOI] [Google Scholar]
- 2.Song Z-Q, Wang F-P, Zhi X-Y, Chen J-Q, Zhou E-M, Liang F, Xiao X, Tang S-K, Jiang H-C, Zhang CL, et al. Bacterial and archaeal diversities in Yunnan and Tibetan Hot Springs, China. Environ Microbiol. 2013;15:1160–1175. doi: 10.1111/1462-2920.12025. [DOI] [PubMed] [Google Scholar]
- 3.Fu Q, Fukushima N, Maeda H, Sato K, Kobayashi H. Bioelectrochemical analysis of a hyperthermophilic microbial fuel cell generating electricity at temperatures above 80 C. Biosci Biotechnol Biochem. 2015;79:1200–1206. doi: 10.1080/09168451.2015.1015952. [DOI] [PubMed] [Google Scholar]
- 4.Hedlund BP, Thomas SC, Dodsworth JA, Zhang CL (2016) Life in high-temperature environments. Manual of Environmental Microbiology 4–3
- 5.Antranikian G, Suleiman M, Schäfers C, Adams MW, Bartolucci S, Blamey JM, Birkeland N-K, Bonch-Osmolovskaya E, da Costa MS, Cowan D, et al. Diversity of bacteria and archaea from two shallow marine hydrothermal vents from Vulcano Island. Extremophiles. 2017;21:733–742. doi: 10.1007/s00792-017-0938-y. [DOI] [PubMed] [Google Scholar]
- 6.Orellana R, Macaya C, Bravo G, Dorochesi F, Cumsille A, Valencia R, Rojas C, Seeger M. Living at the frontiers of life: extremophiles in Chile and their potential for bioremediation. Front Microbiol. 2018;9:2309. doi: 10.3389/fmicb.2018.02309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/JB.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozubal MA, Romine M, Jennings RD, Jay ZJ, Tringe SG, Rusch DB, Beam JP, McCue LA, Inskeep WP. Geoarchaeota: a new candidate phylum in the archaea from high-temperature acidic iron mats in Yellowstone National Park. ISME J. 2013;7:622–634. doi: 10.1038/ismej.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hynek BM, Rogers KL, Antunovich M, Avard G, Alvarado GE. Lack of microbial diversity in an extreme Mars analog setting: Poás volcano, Costa Rica. Astrobiology. 2018;18:923–933. doi: 10.1089/ast.2017.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavalazzi B, Barbieri R, Gómez F, Capaccioni B, Olsson-Francis K, Pondrelli M, Rossi A, Hickman-Lewis K, Agangi A, Gasparotto G, et al. The dallol geothermal area, northern Afar (Ethiopia)–an exceptional planetary field analog on earth. Astrobiology. 2019;19:553–578. doi: 10.1089/ast.2018.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colman DR, Lindsay MR, Harnish A, Bilbrey EM, Amenabar MJ, Selensky MJ, Fecteau KM, Debes RV, Stott MB, Shock EL, et al. Seasonal hydrologic and geologic forcing drive hot spring geochemistry and microbial biodiversity. Environ Microbiol. 2021;23:4034–4053. doi: 10.1111/1462-2920.15617. [DOI] [PubMed] [Google Scholar]
- 12.Campbell KA, Guido DM, Gautret P, Foucher F, Ramboz C, Westall F. Geyserite in hot-spring siliceous sinter: window on Earth’s hottest terrestrial (Paleo) environment and its extreme life. Earth Sci Rev. 2015;148:44–64. doi: 10.1016/j.earscirev.2015.05.009. [DOI] [Google Scholar]
- 13.Cruz Pauccara V, Vargas Rodríguez V, Matsuda K. Geochemical characterization of thermal waters in the Calientes Geothermal Field. South of Peru: Tacna; 2010. [Google Scholar]
- 14.Colman DR, Feyhl-Buska J, Robinson KJ, Fecteau KM, Xu H, Shock EL, Boyd ES (2016) Ecological differentiation in planktonic and sediment-associated chemotrophic microbial populations in Yellowstone hot springs. FEMS Microbiol Ecol 92 [DOI] [PubMed]
- 15.Kublanov I, Prokofeva M, Kostrikina N, Kolganova T, Tourova T, Wiegel J, Bonch-Osmolovskaya E. Thermoanaerobacterium aciditolerans sp. nov., a moderate thermoacidophile from a Kamchatka hot spring. Int J Syst Evol Microbiol. 2007;57:260–264. doi: 10.1099/ijs.0.64633-0. [DOI] [PubMed] [Google Scholar]
- 16.Khelifi N, Ben Romdhane E, Hedi A, Postec A, Fardeau M-L, Hamdi M, Tholozan J-L, Ollivier B, Hirschler-Réa A. Characterization of Microaerobacter geothermalis gen. nov., sp. nov., a novel microaerophilic, nitrate-and nitrite-reducing thermophilic bacterium isolated from a terrestrial hot spring in Tunisia. Extremophiles. 2010;14:297–304. doi: 10.1007/s00792-010-0308-5. [DOI] [PubMed] [Google Scholar]
- 17.Chaban B, Ng SY, Jarrell KF. Archaeal habitats-from the extreme to the ordinary. Can J Microbiol. 2006;52:73–116. doi: 10.1139/w05-147. [DOI] [PubMed] [Google Scholar]
- 18.Strazzulli A, Iacono R, Giglio R, Moracci M, Cobucci-Ponzano B (2017) Metagenomics of hyperthermophilic environments: biodiversity and biotechnology. In: Microbial Ecology of Extreme Environments. pp 103–135
- 19.Collins T, Margesin R. Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Appl Microbiol Biotechnol. 2019;103:2857–2871. doi: 10.1007/s00253-019-09659-5. [DOI] [PubMed] [Google Scholar]
- 20.Eskandari F, Shahnavaz B, Mashreghi M. Optimization of complete RB-5 Azo dye decolorization using novel cold-adapted and mesophilic bacterial consortia. J Environ Manage. 2019;241:91–98. doi: 10.1016/j.jenvman.2019.03.125. [DOI] [PubMed] [Google Scholar]
- 21.Lattanzi P, Benesperi R, Morelli G, Rimondi V, Ruggieri G. Biomonitoring studies in geothermal areas: a review. Front Environ Sci. 2020;8:579343. doi: 10.3389/fenvs.2020.579343. [DOI] [Google Scholar]
- 22.Nicolau C, Reich M, Lynne B. Physico-chemical and environmental controls on siliceous sinter formation at the high-altitude El Tatio geothermal field, Chile. J Volcanol Geoth Res. 2014;282:60–76. doi: 10.1016/j.jvolgeores.2014.06.012. [DOI] [Google Scholar]
- 23.Mallik S, Kundu S. Molecular interactions within the halophilic, thermophilic, and mesophilic prokaryotic ribosomal complexes: clues to environmental adaptation. J Biomol Struct Dyn. 2015;33:639–656. doi: 10.1080/07391102.2014.900457. [DOI] [PubMed] [Google Scholar]
- 24.Urbieta MS, Willis Porati G, Segretín AB, González-Toril E, Giaveno MA, Donati ER. Copahue geothermal system: a volcanic environment with rich extreme prokaryotic biodiversity. Microorganisms. 2015;3(3):344–363. doi: 10.3390/microorganisms3030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green SJ, Leigh MB, Neufeld JD. Denaturing gradient gel electrophoresis (DGGE) for microbial community analysis. Community Profiling and Array Approaches, Hydrocarbon and Lipid Microbiology Protocols: Microbial Quantitation; 2017. pp. 77–99. [Google Scholar]
- 26.Raghunathan G, Marx A. Identification of Thermus aquaticus DNA polymerase variants with increased mismatch discrimination and reverse transcriptase activity from a smart enzyme mutant library. Sci Rep. 2019;9(1):590. doi: 10.1038/s41598-018-37233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sar P, Kazy SK, Paul D, Sarkar A (2013) Metal bioremediation by thermophilic microorganisms. Thermophilic Microbes in Environmental and Industrial Biotechnology: Biotechnology of Thermophiles 171–201
- 28.Anwar UB, Zwar IP, de Souza AO (2020) Biomolecules produced by extremophiles microorganisms and recent discoveries. In: New and Future Developments in Microbial Biotechnology and Bioengineering. Elsevier, pp 247–270
- 29.Mukhtar S, Aslam M (2020) Biofuel synthesis by extremophilic microorganisms. Biofuels Production–Sustainability and Advances in Microbial Bioresources 115–138
- 30.Orell A, Remonsellez F, Arancibia R, Jerez CA (2013) Molecular characterization of copper and cadmium resistance determinants in the biomining thermoacidophilic archaeon Sulfolobus metallicus. Archaea 2013 [DOI] [PMC free article] [PubMed]
- 31.Schultz J, Rosado AS. Extreme environments: a source of biosurfactants for biotechnological applications. Extremophiles. 2020;24:189–206. doi: 10.1007/s00792-019-01151-2. [DOI] [PubMed] [Google Scholar]
- 32.Seckbach J, Chela-Flores J. Extremophiles and chemotrophs as contributors to astrobiological signatures on Europa: a review of biomarkers of sulfate-reducers and other microorganisms. Instruments, Methods, and Missions for Astrobiology X. 2007;6694:294–304. [Google Scholar]
- 33.Kohli I, Joshi NC, Mohapatra S, Varma A. Extremophile-an adaptive strategy for extreme conditions and applications. Curr Genomics. 2020;21(2):96–110. doi: 10.2174/1389202921666200401105908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jurgens G, Glöckner F-O, Amann R, Saano A, Montonen L, Likolammi M, Münster U. Identification of novel archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol Ecol. 2000;34:45–56. doi: 10.1111/j.1574-6941.2000.tb00753.x. [DOI] [PubMed] [Google Scholar]
- 35.Palkova L, Tomova A, Repiska G, Babinska K, Bokor B, Mikula I, Minarik G, Ostatnikova D, Soltys K. Evaluation of 16s rRNA primer sets for characterisation of microbiota in paediatric patients with autism spectrum disorder. Sci Rep. 2021;11:1–13. doi: 10.1038/s41598-021-86378-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo H, Yu Z, Thompson IP, Zhang H. A contribution of hydrogenotrophic methanogenesis to the biogenic coal bed methane reserves of southern Qinshui Basin, China. Appl Microbiol Biotechnol. 2014;98:9083–9093. doi: 10.1007/s00253-014-5908-z. [DOI] [PubMed] [Google Scholar]
- 37.Xing P, Li H, Liu Q, Zheng J. Composition of the archaeal community involved in methane production during the decomposition of microcystis blooms in the laboratory. Can J Microbiol. 2012;58:1153–1158. doi: 10.1139/w2012-097. [DOI] [PubMed] [Google Scholar]
- 38.Stackebrandt E, Liesack W, Goebel B. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16s rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Ma T, Zhao L, Lv J, Li G, Liang F, Liu R. PCR-DGGE method for analyzing the bacterial community in a high temperature petroleum reservoir. World J Microbiol Biotechnol. 2008;24:1981–1987. doi: 10.1007/s11274-008-9694-6. [DOI] [Google Scholar]
- 40.Adrados B, Sánchez O, Arias C, Becares E, Garrido L, Mas J, Brix H, Morató J. Microbial communities from different types of natural wastewater treatment systems: vertical and horizontal flow constructed wetlands and biofilters. Water Res. 2014;55:304–312. doi: 10.1016/j.watres.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Casamayor EO, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–508. doi: 10.1128/AEM.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Habib N, Khan IU, Hussain F, Zhou E-M, Xiao M, Dong L, Zhi X-Y, Li W-J. Meiothermus luteus sp. nov., a slightly thermophilic bacterium isolated from a hot spring. Int J Syst Evol Microbiol. 2017;67:2910–2914. doi: 10.1099/ijsem.0.002040. [DOI] [PubMed] [Google Scholar]
- 44.Kang E, Jin H-S, La JW, Sung J-Y, Park S-Y, Kim W-C, Lee D-W. Identification of keratinases from Fervidobacterium Islandicum AW-1 using dynamic gene expression profiling. Microb Biotechnol. 2020;13:442–457. doi: 10.1111/1751-7915.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee KC-Y, Dunfield PF, Morgan XC, Crowe MA, Houghton KM, Vyssotski M, Ryan JL, Lagutin K, McDonald IR, Stott MB. Chthonomonas calidirosea gen. nov., sp. nov., an aerobic, pigmented, thermophilic micro-organism of a novel bacterial class, Chthonomonadetes classis nov., of the newly described phylum Armatimonadetes originally designated candidate division OP10. Int J Syst Evol Microbiol. 2011;61:2482–2490. doi: 10.1099/ijs.0.027235-0. [DOI] [PubMed] [Google Scholar]
- 46.Jardine JL, Abia ALK, Mavumengwana V, Ubomba-Jaswa E. Phylogenetic analysis and antimicrobial profiles of cultured emerging opportunistic pathogens (phyla Actinobacteria and Proteobacteria) identified in hot springs. Int J Environ Res Public Health 14:1070 [DOI] [PMC free article] [PubMed]
- 47.Kasana RC, Pandey C. Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit Rev Biotechnol. 2018;38:141–156. doi: 10.1080/07388551.2017.1312273. [DOI] [PubMed] [Google Scholar]
- 48.White RA, III, Soles SA, Gavelis G, Gosselin E, Slater GF, Lim DS, Leander B, Suttle CA. The complete genome and physiological analysis of the eurythermal firmicute Exiguobacterium chiriqhucha strain RW2 isolated from a freshwater microbialite, widely adaptable to broad thermal, pH, and salinity ranges. Front Microbiol. 2019;9:3189. doi: 10.3389/fmicb.2018.03189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zadvornyy OA, Boyd ES, Posewitz MC, Zorin NA, Peters JW. Biochemical and structural characterization of enolase from Chloroflexus aurantiacus: evidence for a thermophilic origin. Front Bioeng Biotechnol. 2015;3:74. doi: 10.3389/fbioe.2015.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawai S, Martinez JN, Lichtenberg M, Trampe E, Kühl M, Tank M, Haruta S, Nishihara A, Hanada S, Thiel V. In-situ metatranscriptomic analyses reveal the metabolic flexibility of the thermophilic anoxygenic photosynthetic bacterium Chloroflexus aggregans in a hot spring cyanobacteria-dominated microbial mat. Microorganisms. 2021;9:652. doi: 10.3390/microorganisms9030652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yadav AN, Verma P, Kaushik R, Dhaliwal H, Saxena A. Archaea endowed with plant growth promoting attributes. EC Microbiol. 2017;8:294–298. [Google Scholar]
- 52.Xue Y, Fan H, Ventosa A, Grant WD, Jones BE, Cowan DA, Ma Y. Halalkalicoccus tibetensis gen. nov., sp. nov., representing a novel genus of haloalkaliphilic archaea. Int J Syst Evol Microbiol. 2005;55:2501–2505. doi: 10.1099/ijs.0.63916-0. [DOI] [PubMed] [Google Scholar]
- 53.Roh SW, Nam Y-D, Chang H-W, Sung Y, Kim K-H, Oh H-M, Bae J-W. Halalkalicoccus jeotgali sp. nov., a halophilic archaeon from shrimp jeotgal, a traditional Korean fermented seafood. Int J Syst Evol Microbiol. 2007;57:2296–2298. doi: 10.1099/ijs.0.65121-0. [DOI] [PubMed] [Google Scholar]
- 54.Liu B-B, Tang S-K, Zhang Y-G, Lu X-H, Li L, Cheng J, Zhang Y-M, Zhang L-L, Li W-J. Halalkalicoccus paucihalophilus sp. nov., a halophilic archaeon from Lop Nur region in Xinjiang, northwest of China. Antonie Van Leeuwenhoek. 2013;103:1007–1014. doi: 10.1007/s10482-013-9880-x. [DOI] [PubMed] [Google Scholar]
- 55.Loppi S, Frati L, Benedettini G, Pirintsos SA, Leonzio C. Biodiversity of epiphytic lichens as indicator of air pollution in the geothermal area of Lardello (Tuscany, Central Italy) Isr J Plant Sci. 2002;50(2):119–126. doi: 10.1560/WW0J-5LF2-NJA6-YG5U. [DOI] [Google Scholar]
- 56.McNamara DD, Sewell S, Buscarlet E, Wallis IC. A review of the Rotokawa Geothermal Field, New Zealand. Geothermics. 2016;59:281–293. doi: 10.1016/j.geothermics.2015.07.007. [DOI] [Google Scholar]
- 57.Grigoli F, Clinton JF, Diehl T, Kaestli P, Scarabello L, Agustsdottir T, Kristjansdottir S, Magnusson R, Bean CJ, Broccardo M, et al. Monitoring microseismicity of the Hengill Geothermal Field in Iceland. Sci Data. 2022;9(1):220. doi: 10.1038/s41597-022-01339-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker JJ, Spear JR, Pace NR. Geobiology of a microbial endolithic community in the Yellowstone Geothermal environment. Nature. 2005;434(7036):1011–1014. doi: 10.1038/nature03447. [DOI] [PubMed] [Google Scholar]
- 59.Goswami S, Pati P. Physico-chemical characteristics of thermal water and soil of Tarabalo and Attri geothermal province, Orissa, India. J Ecophysiol Occup Health. 2008;8:83–88. [Google Scholar]
- 60.Zhai Y, Liu X, Zhu Y, Peng C, Wang T, Zhu L, Li C, Zeng G. Hydrothermal carbonization of sewage sludge: the effect of feed-water pH on fate and risk of heavy metals in hydrochars. Biores Technol. 2016;218:183–188. doi: 10.1016/j.biortech.2016.06.085. [DOI] [PubMed] [Google Scholar]
- 61.Satrio S, Prasetio R, Syah BYCSS, Iskandarsyah TYWM, Muhammadsyah F, Hadian MSD, Hendarmawan H, et al. Isotope and geochemistry characterization of hot springs and cold springs of Sembalun-Rinjani area, East Lombok, West Nusa Tenggara-Indonesia. Indian J Chem. 2020;20:1347–1359. doi: 10.22146/ijc.50790. [DOI] [Google Scholar]
- 62.Mathur J, Bizzoco RW, Ellis DG, Lipson DA, Poole AW, Levine R, Kelley ST. Effects of abiotic factors on the phylogenetic diversity of bacterial communities in acidic thermal springs. Appl Environ Microbiol. 2007;73:2612–2623. doi: 10.1128/AEM.02567-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Massello FL, Chan CS, Chan K-G, Goh KM, Donati E, Urbieta MS. Meta-analysis of microbial communities in hot springs: recurrent taxa and complex shaping factors beyond PH and temperature. Microorganisms. 2020;8:906. doi: 10.3390/microorganisms8060906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bull AT, Asenjo de Leuze J, Goodfellow M, Gómez Silva B. The Atacama desert: technical resources and the growing importance of novel microbial diversity. Annu Rev Microbiol. 2016;70:215–234. doi: 10.1146/annurev-micro-102215-095236. [DOI] [PubMed] [Google Scholar]
- 65.Struneckỳ O, Kopejtka K, Goecke F, Tomasch J, Lukavskỳ J, Neori A, Kahl S, Pieper DH, Pilarski P, Kaftan D, et al. High diversity of thermophilic cyanobacteria in Rupite hot spring identified by microscopy, cultivation, single-cell PCR and amplicon sequencing. Extremophiles. 2019;23:35–48. doi: 10.1007/s00792-018-1058-z. [DOI] [PubMed] [Google Scholar]
- 66.Neilson JW, Jordan FL, Maier RM. Analysis of artifacts suggests DGGE should not be used for quantitative diversity analysis. J Microbiol Methods. 2013;92:256–263. doi: 10.1016/j.mimet.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mashzhan A, Javier-López R, Kistaubayeva A, Savitskaya I, Birkeland N-K. Metagenomics and culture-based diversity analysis of the bacterial community in the Zharkent geothermal spring in Kazakhstan. Curr Microbiol. 2021;78(8):2926–2934. doi: 10.1007/s00284-021-02545-2. [DOI] [PubMed] [Google Scholar]
- 68.Yin Z, Ye L, Jing C. Genome-resolved metagenomics and metatranscriptomics reveal that aquificae dominates arsenate reduction in Tengchong geothermal springs. Environ Sci Technol. 2022;56(22):16473–16482. doi: 10.1021/acs.est.2c05764. [DOI] [PubMed] [Google Scholar]
- 69.Pal C, Asiani K, Arya S, Rensing C, Stekel DJ, Larsson DJ, Hobman JL. Metal resistance and its association with antibiotic resistance. Adv Microb Physiol. 2017;70:261–313. doi: 10.1016/bs.ampbs.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Barbieri R, Cavalazzi B. How do modern extreme hydrothermal environments inform the identification of Martian habitability? The case of the El Tatio Geyser field. Challenges. 2014;5:430–443. doi: 10.3390/challe5020430. [DOI] [Google Scholar]
- 71.Gugliandolo C, Maugeri TL. Phylogenetic diversity of archaea in shallow hydrothermal vents of Eolian Islands. Italy. Diversity. 2019;11:156. doi: 10.3390/d11090156. [DOI] [Google Scholar]
- 72.Podar PT, Yang Z, Björnsdóttir SH, Podar M. Comparative analysis of microbial diversity across temperature gradients in hot springs from Yellowstone and Iceland. Front Microbiol. 2020;11:1625. doi: 10.3389/fmicb.2020.01625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takacs-Vesbach C, Mitchell K, Jackson-Weaver O, Reysenbach A-L. Volcanic calderas delineate biogeographic provinces among Yellowstone thermophiles. Environ Microbiol. 2008;10:1681–1689. doi: 10.1111/j.1462-2920.2008.01584.x. [DOI] [PubMed] [Google Scholar]
- 74.Keller MW, Lipscomb GL, Loder AJ, Schut GJ, Kelly RM, Adams MW. A hybrid synthetic pathway for butanol production by a hyperthermophilic microbe. Metab Eng. 2015;27:101–106. doi: 10.1016/j.ymben.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 75.Yilmazel YD, Zhu X, Kim K-Y, Holmes DE, Logan BE. Electrical current generation in microbial electrolysis cells by hyperthermophilic archaea Ferroglobus placidus and Geoglobus ahangari. Bioelectrochemistry. 2018;119:142–149. doi: 10.1016/j.bioelechem.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 76.Ebaid R, Wang H, Sha C, Abomohra AE-F, Shao W. Recent trends in hyperthermophilic enzymes production and future perspectives for biofuel industry: a critical review. J Clean Prod. 2019;238:117925. doi: 10.1016/j.jclepro.2019.117925. [DOI] [Google Scholar]