Abstract
Cell–cell adhesion is at the center of structure and dynamics of epithelial tissue. E-cadherin–catenin complexes mediate Ca2+-dependent trans-homodimerization and constitute the kernel of adherens junctions. Beyond the basic function of cell–cell adhesion, recent progress sheds light the dynamics and interwind interactions of individual E-cadherin–catenin complex with E-cadherin superclusters, contractile actomyosin and mechanics of the cortex and adhesion. The nanoscale architecture of E-cadherin complexes together with cis-interactions and interactions with cortical actomyosin adjust to junctional tension and mechano-transduction by reinforcement or weakening of specific features of the interactions. Although post-translational modifications such as phosphorylation and glycosylation have been implicated, their role for specific aspects of in E-cadherin function has remained unclear. Here, we provide an overview of the E-cadherin complex in epithelial cell and tissue morphogenesis focusing on nanoscale architectures by super-resolution approaches and post-translational modifications from recent, in particular in vivo, studies. Furthermore, we review the computational modelling in E-cadherin complexes and highlight how computational modelling has contributed to a deeper understanding of the E-cadherin complexes.
Keywords: Nanoscale architectures, Phosphorylation, Glycosylation, Computational modelling
Introduction
Adhesion molecules and complexes mediating specific cell–cell contacts are central to the evolution of metazoan species. Formation, maintenance, dynamics, and specificity of such adhesion complexes are key to tissue development, organization, morphogenesis, homeostasis, and function. In epithelial tissues conserved complexes of E-cadherin provide cell adhesion (Fig. 1A, B).
Fig. 1.

E-cadherin–catenin complex. A Illustration of intercellular junctions. Human intercellular junctions consist of tight junctions, adherens junctions, and desmosomes. Intercellular junctions in Drosophila include adherens junctions and septate junctions. They share a conserved structure and function, and both contain adherens junctions. In contrast to human tight junctions, Drosophila has some subapical complex function similar to those of tight junctions. Both the tight and septate junctions serve as paracellular diffusion barriers despite their differences. B Schematic illustration of the E-cadherin–catenin complex in humans and Drosophila. The E-cadherin contains five extracellular Cadherins (EC) repeats in the cytoplasmic domain, a transmembrane domain (TM), and a cytoplasmic domain (CP). DE-cadherin is made up of seven extracellular Cadherins (EC) repeats a nonchordate-specific classical Cadherin domain (NC), a cysteine-rich EGF-like domain (CE), a laminin globular domain (LG), a transmembrane domain (TM) and a cytoplasmic domain (CP). E-cadherin interacts with the acton–myosin network via β-catenin, α-catenin, and Vinculin. C Schematic illustration of the conformation changes of α-catenin under tension. α-Catenin is in closed conformation under low or without tension. α-Catenin is in open conformation under high tension, meanwhile, Vinculin and Afadin/Canoe are recruited. D E-cadherin cis- and trans-interaction (left). cis interactions typically take place in the EC1 and EC2, while trans interactions happen between EC1 in vertebrates. E-cadherin cis clusters are formed by cis interactions and are stabilized by actin filaments. A prediction of unpaired and paired cis clusters (right)
The single span transmembrane protein E-cadherin binds to a corresponding E-cadherin molecule in the plasma membrane of the adjacent cells, termed a trans complex. The homophilic complex relies on Ca2+-dependent homodimerization of one of its extracellular domains (Fig. 1). On the cytoplasmic side, the short C-terminal tail of E-cadherin binds to β-catenin and p120-catenin. β-Catenin further binds to α-catenin. Given the binding sites in α-catenin to components of the actomyosin cytoskeleton, the cadherin–catenin complexes bridge the force-generating actomyosin cortices of two adhering cells and are at the center of force transmission between cells. Cadherin–catenin complexes together with their associated components play multiple functions in cell adhesion, adhesion tension, cortical link and mechano-signaling. All these functions and their regulation mechanisms have been investigated in wide range of experimental systems and species, including mammalian cell cultures and model organisms (Bertocchi et al. 2017; Lecuit et al. 2011; Wang et al. 2012; Zaidel-Bar 2013). Evolutionary rate covariation analysis indicates that α-catenin, p120-catenin and E-cadherin evolved under similar selective pressures during evolution between Drosophila species (Raza et al. 2019). The key interactions of the cadherin complexes are conserved between bilaterians and cnidarians. It was demonstrated that the cadherin–catenin complex are conserved in the Sea Anemone Nematostella vectensis (Clarke et al. 2016). In N. vectensis, the heterodimer of α-catenin and β-catenin bind N. vectensis Cadherin-1 and -2 forming the cadherin complexes. The complexes bind F-actin via α-catenin. The architecture of cadherin–catenin complexes influences their functions, and these functions often regulate and influence each other. Furthermore, post-translational modifications contribute to and regulate the functions of E-cadherin as all other transmembrane proteins. Post-translational modifications may, therefore, alter the architecture, stabilization, adhesion tension, and mechano-signaling functions.
There has been a series of excellent and comprehensive reviews discussing the adherens junctions from various perspectives. Here, we review variations of the stereotypic cadherin–catenin complex and findings from in vivo studies focusing on nanoscale architectures by super-resolution approaches and post-translational modifications. We end this review by highlighting how computational modelling has contributed to a deeper understanding of the E-cadherin complexes.
E-cadherin complexes
E-cadherin was initially identified as an 84-kDa glycoprotein (gp80) protein mediating Ca2+-dependent cell adhesion and embryonic compaction in mouse embryos (Boller et al. 1985; Hyafil et al. 1980, 1981; Yoshida and Takeichi 1982). Evolutionary conservation was revealed by cloning and characterization of the shotgut gene, which encodes Drosophila E-cadherin (DE-cadherin) (Oda et al. 1994; Tepass et al. 1996; Uemura et al. 1996). Despite similarities in function and structure, E-cadherin in vertebrates and invertebrates displays distinct structural differences. Mature human E-cadherin protein contains an extracellular domain with five extracellular cadherins (EC) repeats (Fig. 1B) (Van Roy and Berx 2008). In contrast, the extracellular region of DE-cadherin contains seven EC repeats, followed by a cysteine-rich region, an EGF-like region and a laminin G domain. Within the cysteine-rich region, DE-cadherin is proteolytically cleaved into two polypeptides that remain associated via noncovalent interactions and functionally behave similar to the mouse E-cadherin molecule (Fig. 1B) (Oda and Tsukita 1999). A second variation concerns the trans complex (Fig. 1B). In vertebrates the distal most extracellular domain 1 (EC1) mediates Ca2+-dependent dimerization (Harrison et al. 2011), whereas medial EC3 and EC4 are responsible for trans-dimerization in Drosophila (Fig. 1B) (Nishiguchi et al. 2016). The function of the distal EC1 and EC2 domains of DE-cadherin has remained unclear.
The E-cadherin cytoplasmic tail contains a binding site for p120-catenin and β-catenin (Armadillo/Arm in Drosophila) (Pai et al. 1996; Yap et al. 1998). β-Catenin forms a stable complex with the vinculin homology domain 1 (VH1) of α-catenin and in this way assembles the stereotypic trimeric cadherin–catenin complex. One β-catenin molecule is associated with one α-catenin molecule (Aberle et al. 1994). The stoichiometry of α-catenin with E-cadherin appears to depend on the specific conditions and may be influenced by the F-actin association. α-Catenin can form a homodimer, which more strongly binds to F-actin than monomeric a-catenin or a trimeric cadherin–catenin complex (Drees et al. 2005).
Although not essential for viability, Drosophila p120-catenin and its binding site within E-cadherin support cell adhesion (Myster et al. 2003; Pettitt 2005). P120 influences E-cadherin turnover in the human cell line SW48 (Davis et al. 2003). Similarly, p120-catenin is involved in endocytosis of dynamic E-cadherin and Bazooka complexes in Drosophila embryos (Bulgakova and Brown 2016) and in E-cadherin turnover and epithelial viscoelasticity in Drosophila wing epithelium (Iyer et al. 2019).
Typical tight junctions are absent in invertebrates (Fig. 1A). The epithelial barrier function is provided by septate junctions. Despite this absence, the homolog of ZO-1/2 in Drosophila, Polychaetoid (Pyd), is expressed in the embryonic epidermis (Schmidt et al. 2023). Pyd colocalizes with adherens junctions to the sub-apical region, however, exhibited a distinct localization to strands that extend out from the region occupied by core junction proteins. Pyd reinforced cell junctions and facilitates cell rearrangements during germband extension.
The nanoscale architecture of E-cadherin complexes
Cadherin-based cell adhesions is mediated by complexes with changing composition and stoichiometry. In addition to the trans complex mentioned above, the trimeric of E-cadherin, β-catenin and a-catenin complex multimerizes to clusters of up to several hundred molecules, designated cis complexes. The cis cluster are assumed to come in two types: as paired clusters engaged in trans homophilic binding and as unpaired cluster with no juxta-positioned cluster in the other cell. Being in at the nano meter scale, analysis of the paired and unpaired cis clusters requires super-resolution microscopy (Schermelleh et al. 2019). Beside homomeric clusters, E-cadherin complexes engage in dynamic interactions with numerous of junction- and cortex-associated proteins, with diverse and often cell-type-specific functions for cell adhesion, cell and tissue tension, and signaling during cell and tissue morphogenesis (Fig. 2). Understanding the dynamics and stoichiometry of these multi-protein assemblies is essential for understanding the physiological role of E-cadherin during cell and tissue morphogenesis.
Fig. 2.

Examples of E-cadherin complexes’ (green) dynamic and functions for tissue morphogenesis. A Schematic representation of the cell sorting due to the different types of adhesion molecules. B Schematic representation of the E-cad complex generating cell adhesion and adhesion tension in epithelial cells. C–E Examples of E-cadherin complexes’ dynamic and functions in Drosophila epithelial tissues. C During Drosophila germband extension, E-cad localizes on tricellular junctions to stable cell vertex, while the asymmetrically distributed E-cad mediates the Myo II flow for junction collapse; E-cad generates cell adhesion to stabilize the newly formed junctions at junction elongation phase. D Decrease of E-cad concentration locally on the cell under cytokinesis mediates the neighbors’ response to cytokinesis forces, directing the actomyosin flows and contributing to MyoII accumulation in neighboring cells during Drosophila notum epithelium cytokinesis. Graph adapted from Pinheiro et al. 2017. E Schematic representation of viscoelasticity regulating by mechanosensitive E-cad turnover during Drosophila wing epithelium morphogenesis. p120-catenin is released from E-cad complexes due to mechanical stress; endocytic E-cad turnover is increased consequently. After junctional remodeling, p120-catenin relocalizes to E-cad complexes. Graph adapted from Iyer et al. 2019
The nanoscale architecture of adherens junction was analyzed in detail by transmission electron microscopy for several epithelial cell types (Buckley et al. 2014; Yonemura et al. 1995). Despite the availability of immunocolloidal gold technology, the composition and distribution of specific components within the junctions is difficult to image given the complex sample preparation process (Tepass and Hartenstein 1994). In contrast, fluorescence microscopy provides labelling specificity, double labelling, and sufficient sensitivity for detection of adherens junctions components (Sahl et al. 2017). The resolution of diffraction-based fluorescence microscopy is limited by Abbe's law to about 200–250 nm in lateral direction and 500–700 nm in axial direction (Shao et al. 2011). Super-resolution techniques allow imaging below the diffraction limit, in principle towards the single digit nanometer scale. Several studies have applied super-resolution microscopy techniques to the nanoscale architecture of E-cadherin complexes in vivo and in cultured cells during last decade (Fig. 3). The techniques, their advantages, challenges, and potential in vivo applications are discussed in Box 1.
Fig. 3.
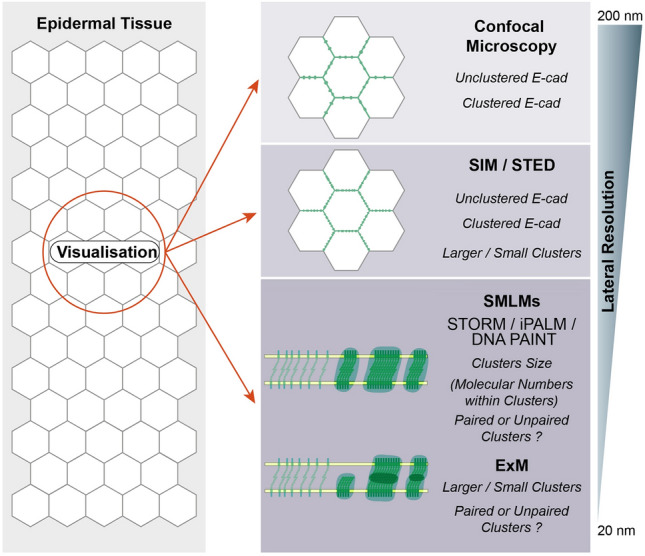
Visualization of E-cadherin complexes’ nanoscale architecture Schematic representation of the technologies for visualization of E-cadherin complexes’ (green) nanoscale architecture from epidermal tissues. The clustered and unclustered E-cad can be visualized by confocal microscopy. Large or small clusters can be distinguished via SIM or STED with improved resolution. The molecular numbers within clusters can be counted by SMLMs
With both SIM (Structured Illumination Microscopy) and STED (STimulated Emission Depletion) microscopy a lateral resolution below 100 nm can easily be achieved, and both benefit from the straightforward sample preparation and imaging acquisition (in Box 1). However, it is technically challenging to improve the resolution to 50 nm, the size of a trans complex, by STED and SIM. Cadherin and nectin, another adhesion molecule, were detected in separate clusters at adherens junctions in A431 cells (epidermoid carcinoma) and human keratinocytes (Indra et al. 2013). In cultured human epithelial cells, flotillin colocalized and formed complexes with cadherin at cell junctions in F-actin-rich regions (Guillaume et al. 2013). STED imaging was applied to dissect the molecular organization of focal adherens junctions and revealed that rather than part of the VE–cadherin complex, the F-BAR protein pacsin2 decorates around the VE–cadherin-based adhesion structure (Dorland et al. 2016). In MDCK cells, a SIM study revealed a direct structural connection of activated Vinculin with β-catenin (Bertocchi et al. 2019). Furthermore, a recent study combined the SIM with a calcium switch assay and visualized reassembly of the supramolecular actomyosin array at the zonula adherens. These findings support the model that myosin arrays bundles actin at mature junctions (Yu-Kemp et al. 2021). Clusters of E-cadherin at adherens junctions were visualized by STED microscopy at the lateral membrane of 3D MDCK cysts (Maraspini et al. 2019). In addition, E-cadherin was detected in nascent desmosomes via SIM, providing support for a role of E-cadherin in early desmosome assembly (Shafraz et al. 2018). A recent SIM study with Drosophila embryos during germband extension revealed that cadherin–catenin complex and Canoe/Afadin both localize to distinct puncta along the junctional membrane (Schmidt et al. 2023). Both methods allow rapid and multicolor imaging. Furthermore, by live-imaging with STED (Grimm et al. 2017), the nanoscale dynamics of adherens junctions proteins and actomyosin cytoskeleton can be visualized during tissue morphogenesis. STED can be combined with optogenetic and optochemical approaches (Kong and Großhans 2022; Krueger and De Renzis 2022), fluorescence correlation spectroscopy (FCS), and fluorescence lifetime imaging microscopy (FLIM) to investigate the in vivo function of adherens junctions components and associating proteins.
SMLMs (Single-Molecule Localization Microscopies) provide a lateral resolution of less than 40 nm and are, therefore, the method of choice for analysis of E-cadherin complexes with the distance between the two cytoplasmic tails of a trans E-cadherin complex of about 40 nm (Boggon et al. 2002). STORM (STochastic Optical Reconstruction Microscopy) imaging with cultured A431D cells revealed that loose clusters of approximately five E-cadherin molecules were delimited by a ‘‘fence’’ of F-actin (Wu et al. 2015). Employing a combination of iPALM (interferometric Photo Activated Localization Microscopy) and SIM in MDCK cells, the E-cadherin core complex could be distinctly imaged from the actin cortex and an interface zone containing Vinculin (Bertocchi et al. 2017). In this model, α-catenin determined the position of Vinculin. In the open conformation of α-catenin, Vinculin in a band of about 30 nm overlapped cadherin–catenin on the one side and F-actin on the other side. STORM data of human intestinal biopsies and cultured Caco2 cells indicate that nectins, rather than E-cadherin, are the major connectors of actin belts at the zonula adherens (Mangeol et al. 2019). Applying STORM to MDCK-II and PZ-HPV-7 cells, a recent study focused on the axial view of noncancerous junctions (Naser et al. 2022). In these samples, β-catenin and p120-catenin colocalized each other with cell junction proteins. Although through overexpression, the in vivo application of quantitative PLAM revealed E-cadherin cluster organization and molecular size in Drosophila embryos (Quang et al. 2013). The E-cadherin clusters were estimated to contain more than 50 molecules, and the distribution of cluster sizes followed power-law distributions with an exponential cutoff.
With these super-resolution studies, the uneven distribution and clustering of E-cadherin along the adherens junctions could be resolved. The two ends of the trans-complex are ~ 40 nm apart, yet so far they have not been imaged with an optical microscope (Boggon et al. 2002). As this distance is within the resolution of SMLMs methods, the two parallel sides of adherens junctions should be resolvable with a labeling the E-cadherin cytoplasmic tails or β-catenin. In this case, paired and unpaired E-cadherin clusters of adherens junctions could be distinguished, including their relationship to components of the actin cortex by multicolor imaging. In practice, however, we have to await such a visualization, yet. Another SMLMs technology, DNA–PAINT (DNA Points Accumulation for Imaging in Nanoscale Topography) has shown its strengths and multicolor imaging and promises to push forward the current limits in nanoscopic imaging of E-cadherin complexes (Cheng et al. 2021). In addition, ExM (Expansion Microscopy) and CLEM (Correlative Light and Electron Microscopy) provide alternative way to reveal the nanoscale architectures of adherens junction complexes.
Box 1 Super-resolution techniques.
1. In SIM (structured illumination microscopy): a spatially restricted fluorescence emission is produced by wide-field excitation of streak patterns (Schermelleh et al. 2008). By the Fourier transform in a process known as deconvolution, images with a lateral resolution of ~ 100 nm are processed from the raw data (Li et al. 2015). Such resolution is not an advantage for the visualization of the nanoscale structure of the AJ complexes. The advantages of SIM include using standard fluorophores and fluorescent proteins, fast imaging, and relatively low excitation power. Taking the same advantages, the Airyscan detector from ZEISS has made confocal imaging with improved signal-to-noise ratio and super-resolution to 150 nm lateral resolution with linear deconvolution (Huff 2015). The recent Airyscan 2 detector offers even higher imaging speed, combined with joined deconvolution processing, which is commoditized with user-friendly operation, making confocal images with lateral resolution below 100 nm in fixed samples (Lv et al. 2022).
2. STED (stimulated emission depletion) microscopy improved the lateral resolution to about 50 nm by optimizing imaging conditions, e.g., labelling density and photostable dyes (Vicidomini et al. 2011). A spatially restricted fluorescence emission is produced at the centre of a joint-focused excitation site surrounded by a ring-shaped depletion pattern in a STED microscopy (Hell and Wichmann 1994). The imaging detection is conducted in a confocal microscope by scanning the coaligned focused spots.
3. SMLMs are super-resolution methods based on single-molecule localization microscopy showing an improvement in terms of the lateral resolution to ~ 20 nm in dSTORM ((direct) stochastic optical reconstruction microscopy) and fPALM ((fluorescent) photo-activated localization microscopy) (Bates et al. 2007; Betzig et al. 2006; Rust et al. 2006). 3D resolution was reduced to 10 nm using two opposing objectives in a 4Pi geometry (Wang et al. 2021). There were few examples of successful demonstrations in live cells as well (Gustafsson et al. 2016; Jones et al. 2011; Wombacher et al. 2010). In SMLMs, small subsets of individual emitters are randomly activated or switched on/off in consecutive acquiring thousands of camera frames. With optimal imaging conditions, signals are sparse enough, and single molecule activating or switching events are identified in each frame. The signals are separated spatially in each frame by the location of a single molecule activating or switching events. They are separated temporally in thousands of camera frames during image acquisition. Determine centre positions with nanoscale precision of single molecules are detected computationally processed from each raw data frame. DNA–PAINT (DNA points accumulation for imaging in nanoscale topogra) is another SMLM. A DNA–PAINT system consists of a docking strand DNA and an imager strand DNA (Jungmann et al. 2016). The docking stand is conjugated with an antibody (or nanobody) targeting proteins of interest. The imager strand is conjugated to fluorescent dyes and diffuses freely in the imaging buffer, which is not camera detectable. A blinking event occurs when the imager strands transiently bind to docking strands. In this case, fluorescent dyes are located for an extended amount of time, emitting sufficient and camera-detectable photons. After accumulating this information from thousands of frames, a super-resolution image can be reconstructed. Because a signal super-resolution image requires thousands of times image acquisitions and computational processing, there are disadvantages of this technique are also obvious: long imaging time, stable and bright probe requirements, and biologist-unfriendly complex computationally processing. As the final image is reconstructed, it is difficult to judge immediately whether the final image is usable during several hours of image-processing.
4. ExM (expansion microscopy) is an approach to obtaining non-optical super-resolution by the physical expansion of the specimen (Chen et al. 2015).
5. CLEM (correlative light and electron microscopy) with chemically stable fluorescent proteins (Campbell et al. 2022) that combine the fluorescent labelling with electron microscopy enables to obtain a complete overview of a cell while at the same time analyzing biomolecules in that same cell on the nanometers scale.
Link to the cytoskeleton and cell cortex
Α-catenin is the linker of cadherin–catenin complexes to the cortical actomyosin cytoskeleton (Fig. 1C, D). Deletion of the α-catenin linker domain disrupts the cadherin–catenin complex and its interactions with F-actin (Watabe-Uchida et al. 1998). α-Catenin binds to F-actin either directly, or it forms complexes with actin-binding proteins, such as Formin, Arp2/3, Vinculin, and EPLIN (Desai et al. 2013; Kobielak and Fuchs 2004; Watabe-Uchida et al. 1998; Yonemura et al. 2010). Importantly, this link is dynamic and depends on the mechanics of adherens junctions leading to a reinforcement under mechanical tension. The cadherin–catenin complexes show stronger binding to actin filaments in an in vitro system when under tension (Buckley et al. 2014). A recent study revealed that α-catenin clustering together with intracellular tension engage a fluid-to-solid-phase transition at the membrane–cytoskeleton interface (Arbore et al. 2022).
α-Catenin reversibly switches to an open conformation when mechanical force is applied. In the context of adherens junctions the pulling force between two adhering cells is sensed by the number of open conformations of α-catenin (Fig. 1C) (Yao et al. 2014). The force-dependent conformational change exposes binding sites for Vinculin, at least, and thus reinforces the cortical link or triggers mechanotransduction pathways (Bertocchi et al. 2017; Kong et al. 2019). Vinculin activation and positioning depends on tension or tyrosine phosphorylation, for which α-catenin plays a vital role (Bertocchi et al. 2017). A series of in vivo studies using Drosophila embryos supports this view. As Drosophila Vinculin is recruited to α-catenin in adhesion complexes, the ratio between Vinculin and E-cadherin provides a quantitative readout for mechanical forces at adherens junctions during tissue morphogenesis (Cavey et al. 2008; Engl et al. 2014). The Vinculin D1 domain, which binds to the open conformation of α-catenin, may serve as an in vivo tension reporter. Such a Vinculin D1-GFP reporter is rapidly recruited to cell junctions in the contracting cells (Bertocchi et al. 2017; Lv et al. 2022; Kong and Großhans 2022; Kong et al. 2019; Krueger and De Renzis 2022). A recent study reports that the M region of α-catenin is required for cell adhesion during morphogenesis, in particular the M2 domain at contacts that experience higher tension in Drosophila embryos. It has been revealed that Vinculin, Jub, and Canoe/Afadin are recruited differentially to enhance adhesion by three distinct tension states reading of α-catenin mechano-sensing during tissue morphogenesis (Sheppard et al. 2022). Cadherin–catenin complex associated protein srGAP was revealed to bind to the M domain of α-catenin, where it strengthens cadherin-dependent adhesion during C. elegans morphogenesis (Serre et al. 2022).
In addition, α-catenin interacts with several proteins and their regulators of cell–cell adhesion by less defined mechanisms. Cadherin–catenin complex associated proteins (CAPs) interact with Cadherin–catenin complex also outside of adherens junctions in A431 cells (Troyanovsky et al. 2021), where they are organized in separate cluster, such as Scribble and Erbin that are two scaffolding proteins belonging to the LAP protein family. The authors suggest that CAP clustering may organize cytoplasmic proteins into distinct domains and in this way may synchronize signaling networks between adhering cells. Similarly, Afadin was found to regulate actomyosin organization through α-catenin at adherens junctions in EpH4 cells (Sakakibara et al. 2020). Canoe/Afadin localizes to puncta along the junctional membrane differently than cadherin–catenin complex in Drosophila embryonic epithermal during tissue elongation (Schmidt et al. 2023).
The cortical interactions represent an essential part in the dynamics and functions of E-cadherin–catenin complexes (Fig. 1C, D). In vitro studies with cultured cells have allowed us to assess the time course of a variety of processes and events leading to E-cadherin-based initial cell–cell contacts and mature adherens junctions. Adjacent cells make initial contacts with Ca2+-dependent and E-cadherin-based trans-interactions between protrusions, which involve branched, Arp2/3-dependent actin networks (Kovacs et al. 2002; Le Clainche and Carlier 2008; Verma et al. 2004). The protrusions depend on actin polymerization at apical junctions to maintain and stabilize cell–cell adhesion (Li et al. 2020, 2021). Maturation into stable adherens junctions requires mechanical tension and actomyosin contractility (Heuzé et al. 2019; Mège and Ishiyama 2017; Said et al. 2022). Consistent with this view from cultured cells are studies in vivo. Studies in Zebrafish embryonic primary cells indicate a role of cadherins in contact expansion and cell sorting. During zebrafish gastrulation cells of different germ layers segregate due to differences in cortical tension at the cell–cell contacts; this study challenged the conventional view that differential cell adhesion is at the center-stage of cell sorting (Maître et al. 2012). A recent study from zebrafish supports the role of cortical tension by revealing a nonlinear relationship between cortex tension and cell–cell contact size (Slováková et al. 2022); the authors revealed a linear relation at low tension, while small contact areas were promoted at high cortical tensions. In Drosophila embryos, both knockdown of α-catenin and decreased junctional contractility via RhoGEF2 RNAi reduced E-cadherin levels at cell junctions (Cavey et al. 2008; Kale et al. 2018). More in vivo studies are required for clarification of mechanisms leading to maturation of adherens junctions.
E-cadherin–catenin complexes form spot-like structures with an uneven distribution along the cell–cell contact both in vertebrate and invertebrate epidermis (Fig. 1D) (Harris and Peifer 2004; Hong et al. 2010; Müller and Wieschaus 1996; Patel et al. 2006). The E-cadherin spots represent nano-clusters with diameters in the range of 50 nm and micro-clusters with diameters of 1–2 μm (micrometers) (Yap et al. 2015). The E-cadherin clusters require cis-interactions, binding of molecules within the plasma membrane of the same cells (Quang et al. 2013; Takeichi 1991; Zhu et al. 2003). Cis-interaction in mammalian cells require the extracellular EC1 and EC2 domains (Brasch et al. 2011). Cis and trans interactions are interdependent as they mutually promote the respective interactions (Thompson et al. 2021). Although cis-complexes were observed and characterized in Drosophila embryos and primary hemocytes (Chandran et al. 2021; Quang et al. 2013), the structural requirements with respect to extracellular domains have not yet been defined. In addition, the intracellular p120–catenin protein appears to contribute to cis interactions (Vu et al. 2021). Studies both in mammalian cells in culture and Drosophila primary hemocytes suggest that extracellular cis-interactions initiate E-cadherin oligomeric complexes, whereas cortical actomyosin promotes higher order nanoclusters, which are necessary for stable cell adhesions (Chandran et al. 2021; Shewan et al. 2005; Smutny et al. 2010; Wu et al. 2015). The degree of clustering can be assayed by fluorescence recovery after photobleaching (FRAP) in vivo. In FRAP experiments, fluorescence of E-cadherin within clusters does not recover within many minutes, both in Drosophila embryos and suspension cultured cell doublets (Cavey et al. 2008; Engl et al. 2014), indicating a slow exchange of molecules within clusters. The clustering and immobilization of E-cadherin requires α-catenin or the cortical contractility in zebrafish embryos (Slováková et al. 2022).
Link of E-cadherin complexes with mechano-gated ion channels
Beside interaction with cortical actomyosin, E-cadherin may interact with mechanogated ion channels as an alternative mechanism of mechano-transduction (Roy Choudhury et al. 2021). The precedence for such a mechanism comes from hair cells in auditory sensory systems, where Cadherin-15 is tightly associated with the putative mechanogated channel Tmc (Choudhary et al. 2020; Jeong et al. 2022). Study in keratinocytes HEK293 cells revealed that Transient Receptor Potential (TRP) channel TRPV4 channel expression is required for the normal cell–cell junctions of skin epithelium (Sokabe et al. 2010). Recently, an interaction was found between E-cadherin–β-catenin complexes and the mechano-gated channel Piezo in the context of F-actin-dependent gating (Wang et al. 2022). Ca2+ entry via the mechanosensitive channel Piezo assembles E-cad at invasive protrusions during cell dissemination in Drosophila (Cabrera et al. 2021). In addition to the mechano-gated ion channels, TMEM16A, a calcium-activated chloride channel, represses E-cadherin expression in gastric cancer cells (Liu et al. 2015). However, E-cadherin staining was reduced in TMEM16A knock-out mouse inner medullary collecting duct cells (He et al. 2017). It will promote a better understanding of the morphodynamics to investigate how E-cadherin complexes coordinate mechano-gated ion channels for mechanotransduction during tissue morphogenesis.
Post-translational modifications of E-cadherin complexes
As most, if not all, proteins passing through the secretory pathway of ER and Golgi, E-cadherin is glycosylated on its extracellular part. In addition, further post-translational modifications have been detected, such as phosphorylation and ubiquitination. Several studies from the last decades have investigated functional implication of these modifications on cell–cell adhesion, protein trafficking, carcinoma progression, and tissue homeostasis. However, a comprehensive reviews of the corresponding literature is lacking. In the following paragraphs, we focus on the phosphorylation and glycosylation to summaries the recent studies about Post-translational modifications of E-cadherin complexes.
Phosphorylation
Multiple phosphorylation sites have been identified in the intracellular portion of E-cadherin (Fig. 4A) (McEwen et al. 2014) going back to initial studies about E-cadherin phosphorylation during compaction in the mouse embryo (Sefton et al. 1992). Eight phosphorylation sites have been identified in human E-cadherin, with five serine residues (Ser 770, 793, 838, 840, and 846, Uniprot P12830) and three tyrosine residues (Tyr 753, 754, and 755, Uniprot P12830) (Bian et al. 2014; Mukherjee et al. 2012). Five sites (Ser 2839, 2909, 2915, 2918, threonine 2912, Uniprot Q967F4) were reported in C. elegans (Choi et al. 2015). There are five potential phosphorylation sites of serine residues (Ser 1454, 1457, 1459, 1460 and 1463, Uniport 12,830) in Drosophila E-cadherin (Chen et al. 2017), among which one site (Ser 1493, Uniprot Q24298) has been conformed (Fig. 4A) (Zhai et al. 2008).
Fig. 4.
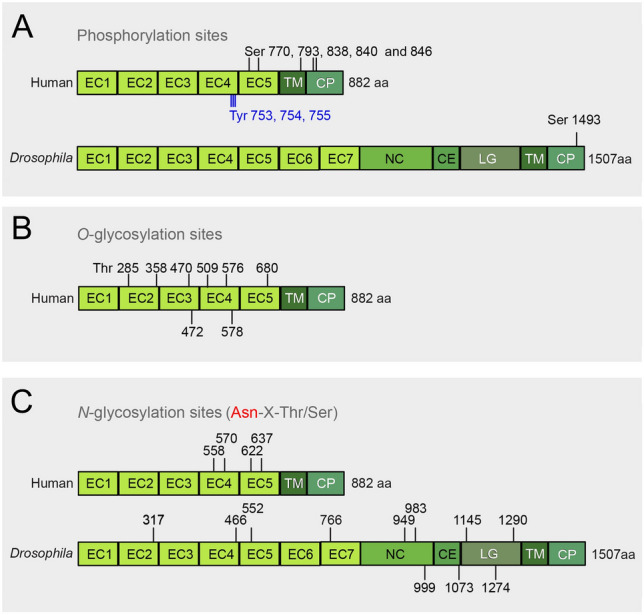
E-cadherin post-translational modifications. A Schematic illustration of E-cadherin phosphorylation sites in humans (Ser 770, 793, 838, 840, and 846, Tyr 753, 754, 755) and Drosophila (Ser 1493). B Schematic representation of E-cadherin O-glycosylation sites in humans (Thr 285, 358, 470, 472, 509, 576, 578, and 680). C Schematic illustration of E-cadherin N-glycosylation sites in humans (Asn 558, 570, 622, and 637) and Drosophila (Asn 317, 466, 552, 766, 949, 983, 999, 1073, 1145, 1274, and 1290)
Phosphorylation of Ser appears to be involved in binding of β-catenin, as mutation to alanine reduced complex formation (McEwen et al. 2014; Stappert and Kemler 1994). Casein kinase II (CK2) and glycogen synthase kinase 3 (GSK-3β) are potential protein kinases for these sites, as an impact has been observed on enhanced β-catenin binding, reduced endocytosis, and increased junction formation (Choi et al. 2006; Lickert et al. 2000; Serres et al. 2000). Mutation of serine 1212 in C. elegans impairs cell adhesion and embryonic development (Choi et al. 2015). Studies in Drosophila suggested that phosphorylation of E-cadherin is not required for adherens junctions formation but does enhance the recruitment of β-catenin by E-cadherin in vivo (Chen et al. 2017).
Src is a candidate protein kinase for the tyrosine sites. Phosphorylation of tyrosine 754 by Src kinase as well as over-activation of c-Met and EGFR lead to an interaction with Hakai, a ubiquitin-ligase (E3) and subsequent protein internalization and ubiquitin-dependent degradation in mammalian cell lines (Figueroa et al. 2009; Fujita et al. 2002; Gong et al. 2010; Kaido et al. 2009; Mukherjee et al. 2012; Shen et al. 2008). A recent study reveals that Src42A controls E-cadherin residence time in Drosophila embryos during axis elongation (Chandran et al. 2023). Similarly, phosphorylation of β-catenin suppresses the cell–cell contacts for N-cadherin. In contrast, phosphorylation of p120-catenin promotes cell–cell contacts (Piedra et al. 2003; Pinho et al. 2011; Rhee et al. 2007).
O-glycosylation
E-cadherin is O-glycosylated within the secretory pathway in Golgi that targets the extracellular part and by cytoplasmic enzymes that target the intracellular tail. Different from phosphorylation, O-glycosylation at the cytoplasmic tail weakens p120 binding and cell–cell adhesion (Chihara and Nance 2012; Pinho et al. 2011; Zhu et al. 2001). O-glycosylation reduces its cell surface trafficking during apoptosis (Zhu et al. 2001). Human E-cadherin contains eight potential O-glycosylation sites for N-acetylglucosamine: threonine 285, 358, 470, 472, 509, 576, 578, 680, Uniprot P12830) (Fig. 4B). For mouse E-cadherin O-glycosylation for both serine and threonine residues has been reported (Thr 360, 472, 474, 511, 578, 580, 582, Ser 282 and 287, Uniprot P09803) (Larsen et al. 2017). The situation in Drosophila is unclear, but cell culture studies indicate that O-glycosylation serves a role in cell adhesion, even though no E-cadherin O-glycosylation sites have been reported (Schwientek et al. 2007; Zhang et al. 2008).
N-glycosylation
N-glycosylation is restricted to the ER and involves a stereotypic and conserved N-glycan that is transferred co-translationally to nascent proteins by an oligosaccharide transferase (OST) (Helenius and Aebi 2004). The stereotypic N-glycans may be modified in the Golgi. The consensus motif for N-glycosylation is Asn-X-Ser/Thr, where X can be any amino acid other than proline (Kornfeld and Kornfeld 1985; Pinho et al. 2011). Within the ER N-glycans are involved in folding and quality control, while they increase the hydrophilicity of the mature proteins. The majority of the E-cadherin N-glycosylation sites are conserved within closely related species (Pinho et al. 2011). For instance, three of the four potential N-glycosylation sites are conserved in both humans and canines (Pinho et al. 2011). Human E-cadherin has two sites in EC4 (Asn 558 and 570, Uniprot P12830) and two in EC5 (Asn 622 and 637, Uniprot P12830) (Fig. 4C) (Pinho et al. 2011). Canine E-cadherin shares the two sites in EC5 but has only one side in EC4. An additional site in EC5 was found in a carcinoma cell line (Pinho et al. 2011).
More sites have been found for Drosophila E-cadherin (Fig. 4C). Out of the predicted eleven sites (Asn 317, 466, 552, 766, 949, 983, 999, 1073, 1145, 1274, and 1290, Uniprot Q24298), nine (Asn 317, 466, 949, 983, 999, 1073, 1145, 1274, and 1290, Uniprot Q24298) have been verified by proteome-wide mass spectrometry (Zielinska et al. 2012). E-cadherin of C. elegans contains 18 potential sites (Asn 72, 243, 253, 339, 508, 658, 685, 715, 826, 1177, 1417, 1646, 1935, 2224, 2232, 2307, 2332, and 2623, Uniprot Q967F4) (Fan et al. 2005; Kaji et al. 2003).
In the Golgi, N-glycans are processed mainly by three glycosyltransferases, namely, N-acetylglucosaminyltransferase III (GnT-III), N-acetylglucosaminyltransferase V (GnT-V), and fructosyltransferase (FUT8). The GnT-III and GnT-V add bisecting GlcNAc (β1,4 GlcNAc) and β1,6 GlcNAc branched GlcNAc to the N-glycan stem. FUT8 is essential for α1,6 fucose N-glycan biosynthesis (Brockhausen et al. 1988; Narasimhan 1982; Pinho et al. 2011; Uozumi et al. 1996). The role of these enzymes and N-glycan processing for E-cadherin function is unclear. Whereas, GnT-III has been implicated in tumor suppression, GnT-V appears to facilitate tumor proliferation (Kimura et al. 2012; Yoshimura et al. 1995).
The literature is unclear concerning the functions of specific N-glycan residues. N-glycan may increase the general hydrophilicity of the protein or be involved in unspecific interactions with the extracellular matrix. Evidence for specific functions is sparse. Mutation of Asn 554 shows a protective role in human gastric cancer cell lines by affecting cellular localization, cis-dimer formation, and molecular stability (Carvalho et al. 2016). N-glycan removal at Asn 633 has been implicated in E-cadherin trafficking, folding, and quality control in the human MDA-MB-435 cell line; The Asn 633 mutant E-cadherin folded incorrectly and arrested in endoplasmic reticulum and then sequentially degraded by endoplasmic reticulum associated degradation (Zhao et al. 2008; Zhou et al. 2008).
Beside site-directed mutagenesis of specific sites, N-glycosylation can be affected by mutants of ER enzymes involved in N-glycan precursor synthesis. The Drosophila mutants wollknäuel (wol), garnystan (gny), xiantuan (xit) encode enzymes for addition of the last three glucosyl to the dolichol precursor (Jamal et al. 2009; Liwosz et al. 2006; Palovuori and Eskelinen 2000; Zhang et al. 2014). In all three mutants, E-cadherin is hypoglycosylated, as detected by an incomplete bandshift in comparison to glycosidase F treated extracts. These are lethal mutations for embryogenesis, in which germband extension and cell intercalation were delayed. The phenotypic consequences are morphological defects and undulated cell junctions during gastrulation in the xiantuan mutant, to which other N-glycosylated proteins may also contribute besides E-cadherin (Zhang et al. 2009; Zhao et al. 2008). In cultured CHO and carcinoma cells, hypo-N-glycosylated E-cadherin enhanced the cell adhesion and showed increased association with γ-catenin-p120 and Vinculin (Liwosz et al. 2006; Palovuori and Eskelinen 2000).
There is no understanding as to why O-glycosylation of E-cadherin reduces binding to p120-catenin (Chen et al. 2016a), whereas N-glycosylation promotes cell–cell contact (Jamal et al. 2009; Liwosz et al. 2006; Palovuori and Eskelinen 2000; Zhang et al. 2014).
Computational modelling of E-cadherin complexes
The complex, context-dependent and dynamic architecture, dynamics, and functions of the adhesion complexes presents a challenge when experimentally investigating E-cadherin–catenin complexes, especially during animal development and tissue morphogenesis. Computational modelling is a powerful and rapid approach for validation and prediction. In this section, we will highlight the impact of computational models on the field. Mathematical theories yield quantitative predictions and generate new questions that can be tested experimentally (Fig. 5) (Hale et al. 2015; Katsamba et al. 2009; Smyrek et al. 2019; Vanderleest et al. 2018). Modelling also allows us to look for general principles (National Academies of Sciences and Medicine 2022), so below we first consider general Cadherin dynamics before diving into E-cadherin specifics.
Fig. 5.
Physics–biology nexus: insights from the Ising model in cell adhesion. A Universality of mathematics enables the application of well-understood principles from physics to biological phenomena, including E-cadherin dynamics and cell adhesion. Simultaneously, exploring biological problems using math can yield novel insights into physics. Notably, recent research (Blom and Godec 2021) highlights the interconnection between physics and biology as follows. B Ising model, a fundamental and extensively studied concept in modern physics, elucidates the emergence of collective behavior through a phase transition at a critical temperature Tc. Analogous to fruit flies being a widely studied model system in biology, the Ising model serves as the quintessential system for phase transitions—the "Drosophila" of phase transitions. C When applied to cell adhesion, the Ising model explains the formation of collective behavior among cell adhesion molecules at a critical membrane rigidity. The interaction strength J between neighboring adhesion sites, inversely related to the cell membrane's rigidity, acts as the order parameter. D Ising model is further used to investigate the kinetics of adhesion molecule association and dissociation. Surprisingly, while more adhesion molecules intuitively translate to stronger adhesion and smaller dissociation rates, there exists an optimum depending on membrane rigidity. A recently identified "dynamical critical point" at critical coupling Jd minimizes the mean formation and dissolution times between fully associated and dissociated adhesion clusters. This novel discovery, arising from cell adhesion modeling, suggests its existence in magnetization reversal times as well and, therefore, demonstrates how interdisciplinary approaches can be mutually beneficial for both fields. Graph adapted from Blom and Godec 2021
Modeling of cadherin dynamics aims to understand how, and under which conditions, macroscopic cell and tissue properties, such as adhesion, emerge from basic mechanics, Cadherin dynamics, and interactions, such as with the actin cortex. One approach is to use continuum mechanics, including Navier–Stokes or other fluid mechanics theory, such as the immersed boundary method (Dallon et al. 2009). Applications of these techniques have, for example, investigated the active role of the actin cortex for adhesive cells and cell adhesion in the context of cell sorting (Armstrong et al. 2006; Dallon et al. 2009; Murakawa and Togashi 2015). Hamiltonian dynamics was used to model catch bonds in the cadherin–catenin–actin complex, where forces induce conformational changes that lead to regulation of interaction strength between catenin and F-actin (Adhikari et al. 2018). A thermodynamic Markov model approach explains how generic self-stabilization through adhesion growth works under mechanical loads which is the type of stabilization that occurs in adhesive system with adaptor proteins, such as Vinculin and Talin (Braeutigam et al. 2022). Another common approach is to use kinetic reaction-rate models, for example, used to investigate interactions of E-cadherin, β-catenin and N-glycosylation (Vargas et al. 2016).
One specific property of E-cadherin, and cadherins in general, is their nature to form clusters. Theoretical considerations can predict power law distributions that match experimental measurements (Quang et al. 2013). In this model, E-cadherin clusters and actin filaments control the fission, while dynamin-dependent endocytosis is assumed to target only large clusters. A kinetic model showed that cadherin mediated adhesion and clustering can both inhibit and promote wnt signaling (Chen et al. 2014). More general models of Cadherin clustering use a similar kinetic approach (Chen et al. 2016b).Using stochastic Brownian dynamics, a more recent study suggests that E-cadherin binding probability increases with forces, and therefore, actomyosin generated tension determines cluster size (Chen et al. 2021). Despite these studies, no consensus exists on a quantitative, mechanistic understanding of Cadherin clustering, and consequently more combined experimental and theoretical studies are needed (Thompson et al. 2020; Yu et al. 2022).
Cadherins also play a role for mechanotransduction and signaling, including establishing planar cell polarity (PCP) (Leckband and De Rooij 2014; Levayer and Lecuit 2013). Computational models, using chemical reactions and diffusion, have shown that asymmetric intercellular complexes can lead to PCP (Le Garrec et al. 2006). In addition, other Cadherins, for example, interactions of the atypical Cadherins Fat and Dachsous have been modeled computationally and shown to lead to asymmetric localization (Hale et al. 2015; Jolly et al. 2014) as well as having an impact on cell morphology (Kumar et al. 2020). In the Drosophila eye, the dynamics of N-cadherin together with E-cadherin leads to asymmetric distribution of myosin and the control of cell shape as well as rearrangements, which has been modeled using energy functionals (Chan et al. 2017; Gemp et al. 2011). E-cadherin mediated mechanotransduction has also been directly linked to population growth in tissues using agent-based simulations (Walker et al. 2010).
In addition, computational models have been utilized for investigations regarding general Cadherin dynamics and properties, for example, solving a measurement problem for binding affinities (Wu et al. 2011), computational design of peptidomimetics of E-cadherin using molecular dynamics simulations (Civera et al. 2019; Doro et al. 2015), or sequencing (Hill et al. 2001). Another field where modeling of E-cadherin dynamics plays a role is Cancer research and models from there could also have an impact on tissue dynamics (Jolly et al. 2019; Ramis-Conde et al. 2008).
Several computational studies have been conducted that address E-cadherin dynamics only implicitly, for example, as a source for adhesion and, therefore, coarse grain the microscopic dynamics. Such models might describe E-cadherin explicitly as Hookean springs (Nestor-Bergmann et al. 2022), in terms of friction between cell interfaces (Metzcar et al. 2019) or address higher level descriptions, such as in vertex models, where the impact of microscopic molecular dynamics at the cell–cell interface is described by a line-tension parameter (Fletcher et al. 2014). In addition, active continuum models that describe the rheology of tissues include parameters that ultimately emerge from the microscopic dynamics (Jülicher et al. 2018). Further studies are still needed to better understand how macroscopic tissue properties emerge from the subcellular microscopic dynamics of E-cadherin and other molecules, as well as the collective behavior of cells.
Conclusion and perspectives
After decades of research, a good understanding emerges of the structure of the adherens junction in relation to its function in morphogenesis, but simultaneously also a growing awareness of the complexity of its structure and dynamics. Understanding the nanoscale architectures of the adherens junctions complexes, their interplay with cadherin–catenin-associated proteins, and their cytoskeleton, which are still poorly documented, are essential for a real understanding of these issues. For instance, studies are needed on: (1) the interplay and relation of trans and cis complexes of E-cadherin especially the function of unpaired cis complexes for adhesion; (2) the role of α-catenin as a dynamic, versatile and mechanoresponsive linker to the cortex; and (3) the interplay of structure with mechanical forces from the cortex for assembly and disassembly of adherens junctions, both in resilient and dynamic tissues during animal development and tissues morphogenesis.
Visualizing and comparing nanoscale architectures of the adherens junctions proteins (e.g., in different development stages during epithelial tissue morphogenesis or between species) will show possible variations of a central theme. Visualizing nanoscale architectures in mutants in comparison to phenotypes will reveal the mechanism underlying the signaling pathways and morphogenesis. The nanoscale architecture of cadherin-based cell adhesions from cultured mammalian cells has begun to be uncovered by super-resolution methods. However, applying super-resolution methods in the developmental tissues is a further goal. The rapid and effective gene editing techniques, such as CRISPR/Cas9 (Nyberg and Carthew 2022) and prime editing (Bosch and Perrimon 2022), allow labeling the AJ components with photoactivable and photo-switchable tags in the gene locus, which can help to visualize the endogenous proteins. The combination of super-resolution methods and gene editing techniques is the way to understand the nanoscale architectures of adherens junctions complexes in vivo.
Box 2 Notes.
Contact expansion is a process which two epidermal cells establish and expand cellular junctions between cells. Cortical tension is mainly generated by the non-muscle myosin II motor, which produces contractile stress by pulling actin filaments against each other. Cell sorting is a process that the different cell types are clustered separately from each other by homophilic cell junctions. Tissue elongation is a process that the tissue become longer in one direction during development and morphogenesis. Germband extension is an embryonic morphological process in Drosophila melanogaster in which the germ-band approximately doubles in length along the anterior–posterior axis while subsequently narrowing along the dorsal–ventral axis. Mechano-gated ion channels are proteins found in eukaryotic and prokaryotic cell membranes that open in response to mechanical forces. Transient Receptor Potential (TRP) channels are a family of ion channels that are expressed in various organisms and tissues involving in sensing and regulating different stimuli. Arp2/3, Actin-related protein 2/3, the Arp2/3 complex is a central actin nucleator promoting the growth of new filaments into branches that form a complex network of cortical actin. EPLIN, Epithelial Protein Lost In Neoplasm is an actin-binding protein. Flotillins are membrane proteins that form microdomains in the plasma membrane. Formins are conserved as actin polymerization machines. srGAP, slit-robo GTPase-activating protein. RhoGEF2, Rho protein guanine exchange factor. Nectin, a family of cell-adhesion molecules.
Acknowledgements
This work was supported by the grants of the Deutsche Forschungsgemeinschaft (GR1945/10-1and GR1945/10-2 to JG)
Author contributions
NZ, MH and DK: wrote the manuscript and drew the figures; FW: helped improve the writing; JG and DK improved and revised the manuscript.
Data availability
This article does not contain any original research data.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Animal and human rights statement
No animal or human rights are involved in this article.
Footnotes
Special Topic: EvoDevo.
Na Zhang and Matthias Häring share co-first authorship.
References
- Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- Adhikari S, Moran J, Weddle C, Hinczewski M. Unraveling the mechanism of the cadherin-catenin-actin catch bond. PLoS Comput Biol. 2018;14:e1006399. doi: 10.1371/journal.pcbi.1006399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbore C, Sergides M, Gardini L, Bianchi G, Kashchuk A, Pertici I, Bianco P, Pavone F, Capitanio M. α-catenin switches between a slip and an asymmetric catch bond with F-actin to cooperatively regulate cell junction fluidity. Nat Commun. 2022;13:1146. doi: 10.1038/s41467-022-28779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong NJ, Painter KJ, Sherratt JA. A continuum approach to modelling cell–cell adhesion. J Theor Biol. 2006;243:98–113. doi: 10.1016/j.jtbi.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertocchi C, Wang Y, Ravasio A, Hara Y, Wu Y, Sailov T, Baird MA, Davidson MW, Zaidel-Bar R, Toyama Y. Nanoscale architecture of cadherin-based cell adhesions. Nat Cell Biol. 2017;19:28–37. doi: 10.1038/ncb3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertocchi C, Ravasio A, Ong HT, Toyama Y, Kanchanawong P. Mechanical roles of vinculin/β-catenin interaction in adherens junction. bioRxiv. 2019;7:770735. [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bian Y, Song C, Cheng K, Dong M, Wang F, Huang J, Sun D, Wang L, Ye M, Zou H. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteomics. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- Blom K, Godec A. Criticality in cell adhesion. Phys Rev X. 2021;11:031067. [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Boller K, Vestweber D, Kemler R. Cell-adhesion molecule uvomorulin is localized in the intermediate junctions of adult intestinal epithelial cells. J Cell Biol. 1985;100:327–332. doi: 10.1083/jcb.100.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch JA, Perrimon N. Prime editing for precise genome engineering in Drosophila. In: Dahmann C, editor. Drosophila: methods and protocols. US, New York: Springer; 2022. pp. 113–134. [DOI] [PubMed] [Google Scholar]
- Braeutigam A, Simsek AN, Gompper G, Sabass B. Generic self-stabilization mechanism for biomolecular adhesions under load. Nat Commun. 2022;13:1–9. doi: 10.1038/s41467-022-29823-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch J, Harrison OJ, Ahlsen G, Carnally SM, Henderson RM, Honig B, Shapiro L. Structure and binding mechanism of vascular endothelial cadherin: a divergent classical cadherin. J Mol Biol. 2011;408:57–73. doi: 10.1016/j.jmb.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhausen I, Narasimhan S, Schachter H. The biosynthesis of highly branched N-glycans: studies on the sequential pathway and functional role of N-actylglucosaminyltransferases I, II, III, IV, V and VI. Biochimie. 1988;70:1521–1533. doi: 10.1016/0300-9084(88)90289-1. [DOI] [PubMed] [Google Scholar]
- Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346:1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakova NA, Brown NH. Drosophila p120-catenin is crucial for endocytosis of the dynamic E-cadherin–Bazooka complex. J Cell Sci. 2016;129:477–482. doi: 10.1242/jcs.177527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera AJ, Gumbiner BM, Kwon YV (2021) The roles of distinct Ca2+ signaling mediated by Piezo and inositol triphosphate receptor (IP3R) in the remodeling of E-cadherin during cell dissemination. bioRxiv: 2021.2011. 2010.467957
- Campbell BC, Paez-Segala MG, Looger LL, Petsko GA, Liu CF. Chemically stable fluorescent proteins for advanced microscopy. Nat Methods. 2022;19:1–10. doi: 10.1038/s41592-022-01660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Catarino TA, Dias AM, Kato M, Almeida A, Hessling B, Figueiredo J, Gärtner F, Sanches JM, Ruppert T, Miyoshi E, Pierce M, Carneiro F, Kolarich D, Seruca R, Yamaguchi Y, Taniguchi N, Reis CA, Pinho SS. Preventing E-cadherin aberrant N-glycosylation at Asn-554 improves its critical function in gastric cancer. Oncogene. 2016;35:1619–1631. doi: 10.1038/onc.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M, Rauzi M, Lenne P-F, Lecuit T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008;453:751–756. doi: 10.1038/nature06953. [DOI] [PubMed] [Google Scholar]
- Chan EH, Shivakumar PC, Clément R, Laugier E, Lenne P-F. Patterned cortical tension mediated by N-cadherin controls cell geometric order in the Drosophila eye. elife. 2017;6:e22796. doi: 10.7554/eLife.22796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran R, Kale G, Philippe J-M, Lecuit T, Mayor S. Distinct actin-dependent nanoscale assemblies underlie the dynamic and hierarchical organization of E-cadherin. Curr Biol. 2021;31:1736–1736. doi: 10.1016/j.cub.2021.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran L, Backer W, Schleutker R, Kong D, Beati SA, Luschnig S, Müller H-AJ. Src42A is required for E-cadherin dynamics at cell junctions during Drosophila axis elongation. Development. 2023;150:dev201119. doi: 10.1242/dev.201119. [DOI] [PubMed] [Google Scholar]
- Chen J, Xie Z-R, Wu Y. Computational modeling of the interplay between cadherin-mediated cell adhesion and Wnt signaling pathway. PLoS ONE. 2014;9:e100702. doi: 10.1371/journal.pone.0100702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-L, Wang S-H, Chan P-C, Shen M-R, Chen H-C. Phosphorylation of E-cadherin at threonine 790 by protein kinase Cδ reduces β-catenin binding and suppresses the function of E-cadherin. Oncotarget. 2016;7:37260. doi: 10.18632/oncotarget.9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Newhall J, Xie Z-R, Leckband D, Wu Y. A computational model for kinetic studies of cadherin binding and clustering. Biophys J. 2016;111:1507–1518. doi: 10.1016/j.bpj.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-J, Huang J, Huang L, Austin E, Hong Y. Phosphorylation potential of Drosophila E-cadherin intracellular domain is essential for development and adherens junction biosynthetic dynamics regulation. Development. 2017;144:1242–1248. doi: 10.1242/dev.141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Brasch J, Harrison OJ, Bidone TC. Computational model of E-cadherin clustering under force. Biophys J. 2021;120:4944–4954. doi: 10.1016/j.bpj.2021.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Allgeyer ES, Richens JH, Dzafic E, Palandri A, Lewków B, Sirinakis G, St Johnston D. A single-molecule localization microscopy method for tissues reveals nonrandom nuclear pore distribution in Drosophila. J Cell Sci. 2021;134:jcs259570. doi: 10.1242/jcs.259570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara D, Nance J. An E-cadherin-mediated hitchhiking mechanism for C. elegans germ cell internalization during gastrulation. Development. 2012;139:2547–2556. doi: 10.1242/dev.079863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-J, Huber AH, Weis WI. Thermodynamics of β-catenin-ligand interactions: the roles of the N-and C-terminal tails in modulating binding affinity. J Biol Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- Choi H-J, Loveless T, Lynch AM, Bang I, Hardin J, Weis WI. A conserved phosphorylation switch controls the interaction between cadherin and β-catenin in vitro and in vivo. Dev Cell. 2015;33:82–93. doi: 10.1016/j.devcel.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary D, Narui Y, Neel BL, Wimalasena LN, Klanseck CF, De-la-Torre P, Chen C, Araya-Secchi R, Tamilselvan E, Sotomayor M. Structural determinants of protocadherin-15 mechanics and function in hearing and balance perception. Proc Natl Acad Sci USA. 2020;117:24837–24848. doi: 10.1073/pnas.1920444117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civera M, Vasile F, Potenza D, Colombo C, Parente S, Vettraino C, Prosdocimi T, Parisini E, Belvisi L. Exploring E-cadherin-peptidomimetics interaction using NMR and computational studies. PLoS Comput Biol. 2019;15:e1007041. doi: 10.1371/journal.pcbi.1007041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DN, Miller PW, Lowe CJ, Weis WI, Nelson WJ. Characterization of the cadherin–catenin complex of the sea anemone Nematostella vectensis and implications for the evolution of metazoan cell–cell adhesion. Mol Biol Evol. 2016;33:2016–2029. doi: 10.1093/molbev/msw084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallon J, Newren E, Hansen MD. Using a mathematical model of cadherin-based adhesion to understand the function of the actin cytoskeleton. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79:031918. doi: 10.1103/PhysRevE.79.031918. [DOI] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, Tepass U. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat Cell Biol. 2013;15:261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- Dorland YL, Malinova TS, Van Stalborch A-MD, Grieve AG, Van Geemen D, Jansen NS, de Kreuk B-J, Nawaz K, Kole J, Geerts D. The F-BAR protein pacsin2 inhibits asymmetric VE-cadherin internalization from tensile adherens junctions. Nat Commun. 2016;7:1–18. doi: 10.1038/ncomms12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doro F, Colombo C, Alberti C, Arosio D, Belvisi L, Casagrande C, Fanelli R, Manzoni L, Parisini E, Piarulli U. Computational design of novel peptidomimetic inhibitors of cadherin homophilic interactions. Org Biomol Chem. 2015;13:2570–2573. doi: 10.1039/C4OB02538E. [DOI] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. α-catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engl W, Arasi B, Yap L, Thiery J, Viasnoff V. Actin dynamics modulate mechanosensitive immobilization of E-cadherin at adherens junctions. Nat Cell Biol. 2014;16:584–591. doi: 10.1038/ncb2973. [DOI] [PubMed] [Google Scholar]
- Fan X, She Y-M, Bagshaw RD, Callahan JW, Schachter H, Mahuran DJ. Identification of the hydrophobic glycoproteins of Caenorhabditis elegans. Glycobiology. 2005;15:952–964. doi: 10.1093/glycob/cwi075. [DOI] [PubMed] [Google Scholar]
- Figueroa A, Kotani H, Toda Y, Mazan-Mamczarz K, Mueller EC, Otto A, Disch L, Norman M, Ramdasi RM, Keshtgar M, Gorospe M, Fujita Y. Novel roles of hakai in cell proliferation and oncogenesis. Mol Biol Cell. 2009;20:3533–3542. doi: 10.1091/mbc.e08-08-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher AG, Osterfield M, Baker RE, Shvartsman SY. Vertex models of epithelial morphogenesis. Biophys J. 2014;106:2291–2304. doi: 10.1016/j.bpj.2013.11.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HEM, Behrens J, Sommer T, Birchmeier W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol. 2002;4:222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- Gemp IM, Carthew RW, Hilgenfeldt S. Cadherin-dependent cell morphology in an epithelium: constructing a quantitative dynamical model. PLoS Comput Biol. 2011;7:e1002115. doi: 10.1371/journal.pcbi.1002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong EY, Park E, Lee K. Hakai acts as a coregulator of estrogen receptor alpha in breast cancer cells. Cancer Sci. 2010;101:2019–2025. doi: 10.1111/j.1349-7006.2010.01636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods. 2017;14:987–994. doi: 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume E, Comunale F, Do Khoa N, Planchon D, Bodin S, Gauthier-Rouvière C. Flotillin microdomains stabilize cadherins at cell–cell junctions. J Cell Sci. 2013;126:5293–5304. doi: 10.1242/jcs.133975. [DOI] [PubMed] [Google Scholar]
- Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM, Henriques R. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat Commun. 2016;7:1–9. doi: 10.1038/ncomms12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale R, Brittle AL, Fisher KH, Monk NA, Strutt D. Cellular interpretation of the long-range gradient of Four-jointed activity in the Drosophila wing. elife. 2015;4:e05789. doi: 10.7554/eLife.05789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. Adherens junction-dependent and-independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol. 2004;167:135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Ye W, Wang W-J, Sison ES, Jan YN, Jan LY. Cytoplasmic Cl− couples membrane remodeling to epithelial morphogenesis. Proc Natl Acad Sci USA. 2017;114:E11161–E11169. doi: 10.1073/pnas.1714448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/OL.19.000780. [DOI] [PubMed] [Google Scholar]
- Heuzé ML, Narayana GHNS, d'Alessandro J, Cellerin V, Dang T, Williams DS, Van Hest JC, Marcq P, Mège R-M, Ladoux B. Myosin II isoforms play distinct roles in adherens junction biogenesis. elife. 2019;8:e46599. doi: 10.7554/eLife.46599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E, Broadbent ID, Chothia C, Pettitt J. Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J Mol Biol. 2001;305:1011–1024. doi: 10.1006/jmbi.2000.4361. [DOI] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc Natl Acad Sci USA. 2010;107:3528–3533. doi: 10.1073/pnas.0911027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff J. The Airyscan detector from ZEISS: confocal imaging with improved signal-to-noise ratio and super-resolution. Nat Methods. 2015;12:i–ii. doi: 10.1038/nmeth.f.388. [DOI] [Google Scholar]
- Hyafil F, Morello D, Babinet C, Jacob F. A cell surface glycoprotein involved in the compaction of embryonal carcinoma cells and cleavage stage embryos. Cell. 1980;21:927–934. doi: 10.1016/0092-8674(80)90456-0. [DOI] [PubMed] [Google Scholar]
- Hyafil F, Babinet C, Jacob F. Cell-cell interactions in early embryogenesis: a molecular approach to the role of calcium. Cell. 1981;26:447–454. doi: 10.1016/0092-8674(81)90214-2. [DOI] [PubMed] [Google Scholar]
- Indra I, Hong S, Troyanovsky R, Kormos B, Troyanovsky S. The adherens junction: a mosaic of cadherin and nectin clusters bundled by actin filaments. J Invest Dermatol. 2013;133:2546–2554. doi: 10.1038/jid.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer KV, Piscitello-Gómez R, Paijmans J, Jülicher F, Eaton S. Epithelial viscoelasticity is regulated by mechanosensitive E-cadherin turnover. Curr Biol. 2019;29:578–591. doi: 10.1016/j.cub.2019.01.021. [DOI] [PubMed] [Google Scholar]
- Jamal BT, Nita-Lazar M, Gao Z, Amin B, Walker J, Kukuruzinska MA. N-glycosylation status of E-cadherin controls cytoskeletal dynamics through the organization of distinct β-catenin-and γ-catenin-containing AJs. Cell Health Cytoskelet. 2009;2009:67. doi: 10.2147/chc.s5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Clark S, Goehring A, Dehghani-Ghahnaviyeh S, Rasouli A, Tajkhorshid E, Gouaux E. Structures of the TMC-1 complex illuminate mechanosensory transduction. Nature. 2022;610:796–803. doi: 10.1038/s41586-022-05314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly MK, Rizvi MS, Kumar A, Sinha P. Mathematical modeling of sub-cellular asymmetry of fat-dachsous heterodimer for generation of planar cell polarity. PLoS ONE. 2014;9:e97641. doi: 10.1371/journal.pone.0097641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly MK, Ware KE, Xu S, Gilja S, Shetler S, Yang Y, Wang X, Austin RG, Runyambo D, Hish AJ, DeWitt SB, George JT, Kreulen RT, Boss MK, Lazarides AL, Kerr DL, Gerber DG, Sivaraj D, Armstrong AJ, Dewhirst MW, et al. E-cadherin represses anchorage-independent growth in sarcomas through both signaling and mechanical mechanismse-cadherin represses anchorage-independent growth. Mol Cancer Res. 2019;17:1391–1402. doi: 10.1158/1541-7786.MCR-18-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Shim S-H, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–505. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jülicher F, Grill SW, Salbreux G. Hydrodynamic theory of active matter. Rep Prog Phys. 2018;81:076601. doi: 10.1088/1361-6633/aab6bb. [DOI] [PubMed] [Google Scholar]
- Jungmann R, Avendaño MS, Dai M, Woehrstein JB, Agasti SS, Feiger Z, Rodal A, Yin P. Quantitative super-resolution imaging with qPAINT. Nat Methods. 2016;13:439–442. doi: 10.1038/nmeth.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaido M, Wada H, Shindo M, Hayashi S. Essential requirement for RING finger E3 ubiquitin ligase Hakai in early embryonic development of Drosophila. Genes Cells. 2009;14:1067–1077. doi: 10.1111/j.1365-2443.2009.01335.x. [DOI] [PubMed] [Google Scholar]
- Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K-i, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- Kale GR, Yang X, Philippe J-M, Mani M, Lenne P-F, Lecuit T. Distinct contributions of tensile and shear stress on E-cadherin levels during morphogenesis. Nat Commun. 2018;9:1–16. doi: 10.1038/s41467-018-07448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell T, Ben-Shaul A. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci USA. 2009;106:11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Terao M, Kato A, Hanafusa T, Murota H, Katayama I, Miyoshi E. Upregulation of N-acetylglucosaminyltransferase-V by heparin-binding EGF-like growth factor induces keratinocyte proliferation and epidermal hyperplasia. Exp Dermatol. 2012;21:515–519. doi: 10.1111/j.1600-0625.2012.01515.x. [DOI] [PubMed] [Google Scholar]
- Kobielak A, Fuchs E. α-catenin: at the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Großhans J. Optochemical control of cell contractility in embryos Drosophila. In: Dahmann C, editor. Drosophila: methods and protocols. US, New York: Springer; 2022. pp. 285–299. [Google Scholar]
- Kong D, Lv Z, Häring M, Lin B, Wolf F, Großhans J. In vivo optochemical control of cell contractility at single-cell resolution. EMBO Rep. 2019;20:e47755. doi: 10.15252/embr.201947755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kovacs EM, Goodwin M, Ali RG, Paterson AD, Yap AS. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr Biol. 2002;12:379–382. doi: 10.1016/S0960-9822(02)00661-9. [DOI] [PubMed] [Google Scholar]
- Krueger D, De Renzis S. Optogenetic methods to control tissue mechanics in Drosophila. In: Dahmann C, editor. Drosophila: methods and protocols. US, New York: Springer; 2022. pp. 269–283. [DOI] [PubMed] [Google Scholar]
- Kumar A, Rizvi MS, Athilingam T, Parihar SS, Sinha P. Heterophilic cell–cell adhesion of atypical cadherins fat and dachsous regulate epithelial cell size dynamics during Drosophila thorax morphogenesis. Mol Biol Cell. 2020;31:546–560. doi: 10.1091/mbc.E19-08-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen ISB, Narimatsu Y, Joshi HJ, Siukstaite L, Harrison OJ, Brasch J, Goodman KM, Hansen L, Shapiro L, Honig B. Discovery of an O-mannosylation pathway selectively serving cadherins and protocadherins. Proc Natl Acad Sci USA. 2017;114:11163–11168. doi: 10.1073/pnas.1708319114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Carlier M-F. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- Le Garrec JF, Lopez P, Kerszberg M. Establishment and maintenance of planar epithelial cell polarity by asymmetric cadherin bridges: a computer model. Dev Dyn. 2006;235:235–246. doi: 10.1002/dvdy.20617. [DOI] [PubMed] [Google Scholar]
- Leckband D, De Rooij J. Cadherin adhesion and mechanotransduction. Annu Rev Cell Dev Biol. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Lenne P-F, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- Levayer R, Lecuit T. Oscillation and polarity of E-cadherin asymmetries control actomyosin flow patterns during morphogenesis. Dev Cell. 2013;26:162–175. doi: 10.1016/j.devcel.2013.06.020. [DOI] [PubMed] [Google Scholar]
- Li D, Shao L, Chen B-C, Zhang X, Zhang M, Moses B, Milkie DE, Beach JR, Hammer JA, III, Pasham M. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:aab3500. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JXH, Tang VW, Brieher WM. Actin protrusions push at apical junctions to maintain E-cadherin adhesion. Proc Natl Acad Sci USA. 2020;117:432–438. doi: 10.1073/pnas.1908654117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JXH, Tang VW, Boateng KA, Brieher WM. Cadherin puncta are interdigitated dynamic actin protrusions necessary for stable cadherin adhesion. Proc Natl Acad Sci USA. 2021;118:e2023510118. doi: 10.1073/pnas.2023510118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Bauer A, Kemler R, Stappert J. Casein kinase II phosphorylation of E-cadherin increases E-cadherin/β-catenin interaction and strengthens cell-cell adhesion. J Biol Chem. 2000;275:5090–5095. doi: 10.1074/jbc.275.7.5090. [DOI] [PubMed] [Google Scholar]
- Liu F, Cao Q-H, Lu D-J, Luo B, Lu X-F, Luo R-C, Wang X-G. TMEM16A overexpression contributes to tumor invasion and poor prognosis of human gastric cancer through TGF-β signaling. Oncotarget. 2015;6:11585. doi: 10.18632/oncotarget.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liwosz A, Lei T, Kukuruzinska MA. N-glycosylation affects the molecular organization and stability of E-cadherin junctions. J Biol Chem. 2006;281:23138–23149. doi: 10.1074/jbc.M512621200. [DOI] [PubMed] [Google Scholar]
- Lv Z, Zhang N, Zhang X, Großhans J, Kong D. The lateral epidermis actively counteracts pulling by the amnioserosa during dorsal closure. Front Cell Dev Biol. 2022;10:865397. doi: 10.3389/fcell.2022.865397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maître J-L, Berthoumieux H, Krens SFG, Salbreux G, Jülicher F, Paluch E, Heisenberg C-P. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science. 2012;338:253–256. doi: 10.1126/science.1225399. [DOI] [PubMed] [Google Scholar]
- Mangeol P, Massey-Harroche D, Le Bivic A, Lenne P-F. Nectins rather than E-cadherin anchor the actin belts at cell-cell junctions of epithelia. bioRxiv. 2019;107:809343. [Google Scholar]
- Maraspini R, Wang C-H, Honigmann A. Optimization of 2D and 3D cell culture to study membrane organization with STED microscopy. J Phys D Appl Phys. 2019;53:014001. doi: 10.1088/1361-6463/ab45df. [DOI] [Google Scholar]
- McEwen AE, Maher MT, Mo R, Gottardi CJ. E-cadherin phosphorylation occurs during its biosynthesis to promote its cell surface stability and adhesion. Mol Biol Cell. 2014;25:2365–2374. doi: 10.1091/mbc.e14-01-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mège RM, Ishiyama N. Integration of cadherin adhesion and cytoskeleton at adherens junctions. Cold Spring Harb Perspect Biol. 2017;9:a028738. doi: 10.1101/cshperspect.a028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzcar J, Wang Y, Heiland R, Macklin P. A review of cell-based computational modeling in cancer biology. JCO Clin Cancer Inform. 2019;2:1–13. doi: 10.1200/CCI.18.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Chow SY, Yusoff P, Seetharaman J, Ng C, Sinniah S, Koh XW, Asgar NFM, Li D, Yim D. Structure of a novel phosphotyrosine-binding domain in Hakai that targets E-cadherin. EMBO J. 2012;31:1308–1319. doi: 10.1038/emboj.2011.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H, Wieschaus E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J Cell Biol. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa H, Togashi H. Continuous models for cell–cell adhesion. J Theor Biol. 2015;374:1–12. doi: 10.1016/j.jtbi.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Myster SH, Cavallo R, Anderson CT, Fox DT, Peifer M. Drosophila p120 catenin plays a supporting role in cell adhesion but is not an essential adherens junction component. J Cell Biol. 2003;160:433–449. doi: 10.1083/jcb.200211083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S. Control of glycoprotein synthesis. UDP-GlcNAc: glycopeptide beta 4-N-acetylglucosaminyltransferase III, an enzyme in hen oviduct which adds GlcNAc in beta 1–4 linkage to the beta-linked mannose of the trimannosyl core of N-glycosyl oligosaccharides. J Biol Chem. 1982;257:10235–10242. doi: 10.1016/S0021-9258(18)34010-9. [DOI] [PubMed] [Google Scholar]
- Naser AN, Guiler W, Lu Q, Chen YH. Nanoarchitecture and molecular interactions of epithelial cell junction proteins revealed by super-resolution microscopy. Ann NY Acad Sci. 2022;1516:175–187. doi: 10.1111/nyas.14855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine . Physics of Life. The National Academies Press; 2022. [Google Scholar]
- Nestor-Bergmann A, Blanchard GB, Hervieux N, Fletcher AG, Étienne J, Sanson B. Adhesion-regulated junction slippage controls cell intercalation dynamics in an apposed-cortex adhesion model. PLoS Comput Biol. 2022;18:e1009812. doi: 10.1371/journal.pcbi.1009812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi S, Yagi A, Sakai N, Oda H. Divergence of structural strategies for homophilic E-cadherin binding among bilaterians. J Cell Sci. 2016;129:3309–3319. doi: 10.1242/jcs.189258. [DOI] [PubMed] [Google Scholar]
- Nyberg KG, Carthew RW. CRISPR-/Cas9-mediated precise and efficient genome editing in Drosophila. In: Dahmann C, editor. Drosophila. Methods in molecular biology. New York: Humana; 2022. pp. 135–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Tsukita S. Nonchordate classic cadherins have a structurally and functionally unique domain that is absent from chordate classic cadherins. Dev Biol. 1999;216:406–422. doi: 10.1006/dbio.1999.9494. [DOI] [PubMed] [Google Scholar]
- Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- Pai LM, Kirkpatrick C, Blanton J, Oda H, Takeichi M, Peifer M. Drosophila alpha-catenin and E-cadherin bind to distinct regions of Drosophila Armadillo. J Biol Chem. 1996;271:32411–32420. doi: 10.1074/jbc.271.50.32411. [DOI] [PubMed] [Google Scholar]
- Palovuori R, Eskelinen S. Role of vinculin in the maintenance of cell-cell contacts in kidney epithelial MDBK cells. Eur J Cell Biol. 2000;79:961–974. doi: 10.1078/0171-9335-00120. [DOI] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Pettitt J (2005) The cadherin superfamily. In: The C. elegans Research Community (eds) WormBook. doi/10.1895/wormbook.1.50.1, http://www.wormbook.org/ [DOI] [PMC free article] [PubMed]
- Piedra J, Miravet S, Castaño J, Pálmer HG, Heisterkamp N, García de Herreros A, Duñach M. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate β-catenin Tyr-142 phosphorylation and β-catenin-α-catenin Interaction. Mol Cell Biol. 2003;23:2287–2297. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro D, Hannezo E, Herszterg S, Bosveld F, Gaugue I, Balakireva M, Wang Z, Cristo I, Rigaud SU, Markova O, Bellaïche Y. Transmission of cytokinesis forces via E-cadherin dilution and actomyosin flows. Nature. 2017;545:103–107. doi: 10.1038/nature22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho SS, Seruca R, Gärtner F, Yamaguchi Y, Gu J, Taniguchi N, Reis CA. Modulation of E-cadherin function and dysfunction by N-glycosylation. Cell Mol Life Sci. 2011;68:1011–1020. doi: 10.1007/s00018-010-0595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang B-AT, Mani M, Markova O, Lecuit T, Lenne P-F. Principles of E-cadherin supramolecular organization in vivo. Curr Biol. 2013;23:2197–2207. doi: 10.1016/j.cub.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Ramis-Conde I, Drasdo D, Anderson AR, Chaplain MA. Modeling the influence of the E-cadherin-β-catenin pathway in cancer cell invasion: a multiscale approach. Biophys J. 2008;95:155–165. doi: 10.1529/biophysj.107.114678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza Q, Choi JY, Li Y, O’Dowd RM, Watkins SC, Chikina M, Hong Y, Clark NL, Kwiatkowski AV. Evolutionary rate covariation analysis of E-cadherin identifies Raskol as a regulator of cell adhesion and actin dynamics in Drosophila. PLoS Genet. 2019;15:e1007720. doi: 10.1371/journal.pgen.1007720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J, Buchan T, Zukerberg L, Lilien J, Balsamo J. Cables links Robo-bound Abl kinase to N-cadherin-bound β-catenin to mediate Slit-induced modulation of adhesion and transcription. Nat Cell Biol. 2007;9:883–892. doi: 10.1038/ncb1614. [DOI] [PubMed] [Google Scholar]
- Roy Choudhury A, Großhans J, Kong D. Ion channels in epithelial dynamics and morphogenesis. Cells. 2021;10:2280. doi: 10.3390/cells10092280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl SJ, Hell SW, Jakobs S. Fluorescence nanoscopy in cell biology. Nat Rev Mol Cell Biol. 2017;18:685–701. doi: 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- Said R, Wang H, Pernier J, Narassimprakash H, Roméro S, Gautreau AM, Mège R-M, Le Clainche C. The adherens junction proteins α-catenin, vinculin and VASP cooperate to promote actin assembly. bioRxiv. 2022;261:10915. [Google Scholar]
- Sakakibara S, Mizutani K, Sugiura A, Sakane A, Sasaki T, Yonemura S, Takai Y. Afadin regulates actomyosin organization through αE-catenin at adherens junctions. J Cell Biol. 2020;219:e201907079. doi: 10.1083/jcb.201907079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Ferrand A, Huser T, Eggeling C, Sauer M, Biehlmaier O, Drummen GPC. Super-resolution microscopy demystified. Nat Cell Biol. 2019;21:72–84. doi: 10.1038/s41556-018-0251-8. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Finegan T, Häring M, Kong D, Fletcher AG, Alam Z, Grosshans J, Wolf F, Peifer M. Polychaetoid/ZO-1 strengthens cell junctions under tension while localizing differently than core adherens junction proteins. Mol Biol Cell. 2023;34:8. doi: 10.1091/mbc.E23-03-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwientek T, Mandel U, Roth U, Müller S, Hanisch FG. A serial lectin approach to the mucin-type O-glycoproteome of Drosophila melanogaster S2 cells. Proteomics. 2007;7:3264–3277. doi: 10.1002/pmic.200600793. [DOI] [PubMed] [Google Scholar]
- Sefton M, Johnson MH, Clayton L. Synthesis and phosphorylation of uvomorulin during mouse early development. Development. 1992;115:313–318. doi: 10.1242/dev.115.1.313. [DOI] [PubMed] [Google Scholar]
- Serre JM, Lucas B, Martin SC, Heier JA, Shao X, Hardin J. C elegans srGAP is an α-catenin M domain-binding protein that strengthens cadherin-dependent adhesion during morphogenesis. Development. 2022;149:dev200775. doi: 10.1242/dev.200775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serres M, Filhol O, Lickert H, Grangeasse C, Chambaz EM, Stappert J, Vincent C, Schmitt D. The disruption of adherens junctions is associated with a decrease of E-cadherin phosphorylation by protein kinase CK2. Exp Cell Res. 2000;257:255–264. doi: 10.1006/excr.2000.4895. [DOI] [PubMed] [Google Scholar]
- Shafraz O, Rübsam M, Stahley SN, Caldara AL, Kowalczyk AP, Niessen CM, Sivasankar S. E-cadherin binds to desmoglein to facilitate desmosome assembly. elife. 2018;7:e37629. doi: 10.7554/eLife.37629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Kner P, Rego EH, Gustafsson MG. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods. 2011;8:1044–1046. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- Shen Y, Hirsch DS, Sasiela CA, Wu WJ. Cdc42 regulates E-cadherin ubiquitination and degradation through an epidermal growth factor receptor to Src-mediated pathway. J Biol Chem. 2008;283:5127–5137. doi: 10.1074/jbc.M703300200. [DOI] [PubMed] [Google Scholar]
- Sheppard L, Green DG, Lerchbaumer G, Rothenberg KE, Fernandez-Gonzalez R, Tepass U. The α-catenin mechanosensing M region is required for cell adhesion during tissue morphogenesis. J Cell Biol. 2022;222:e202108091. doi: 10.1083/jcb.202108091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewan AM, Maddugoda M, Kraemer A, Stehbens SJ, Verma S, Kovacs EM, Yap AS. Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell–cell contacts. Mol Biol Cell. 2005;16:4531–4542. doi: 10.1091/mbc.e05-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slováková J, Sikora M, Arslan FN, Caballero-Mancebo S, Krens SG, Kaufmann WA, Merrin J, Heisenberg C-P. Tension-dependent stabilization of E-cadherin limits cell–cell contact expansion in zebrafish germ-layer progenitor cells. Proc Natl Acad Sci USA. 2022;119:e2122030119. doi: 10.1073/pnas.2122030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smutny M, Cox HL, Leerberg JM, Kovacs EM, Conti MA, Ferguson C, Hamilton NA, Parton RG, Adelstein RS, Yap AS. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat Cell Biol. 2010;12:696–702. doi: 10.1038/ncb2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrek I, Mathew B, Fischer S, Lissek S, Becker S, Stelzer E. E-cadherin, actin, microtubules and FAK dominate different spheroid formation phases and important elements of tissue integrity. Biol Open. 2019;8:bio037051. doi: 10.1242/bio.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokabe T, Fukumi-Tominaga T, Yonemura S, Mizuno A, Tominaga M. The TRPV4 channel contributes to intercellular junction formation in keratinocytes. J Biol Chem. 2010;285:18749–18758. doi: 10.1074/jbc.M110.103606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappert J, Kemler R. A short core region of E-cadherin is essential for catenin binding and is highly phosphorylated. Cell Commun Adhes. 1994;2:319–327. doi: 10.3109/15419069409014207. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Tepass U, Hartenstein V. The development of cellular junctions in the Drosophila embryo. Dev Biol. 1994;161:563–596. doi: 10.1006/dbio.1994.1054. [DOI] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Török T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- Thompson CJ, Su Z, Vu VH, Wu Y, Leckband DE, Schwartz DK. Cadherin clusters stabilized by a combination of specific and nonspecific cis-interactions. elife. 2020;9:e59035. doi: 10.7554/eLife.59035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CJ, Vu VH, Leckband DE, Schwartz DK. Cadherin cis and trans interactions are mutually cooperative. Proc Natl Acad Sci USA. 2021;118:e2019845118. doi: 10.1073/pnas.2019845118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky RB, Sergeeva AP, Indra I, Chen C-S, Kato R, Shapiro L, Honig B, Troyanovsky SM. Sorting of cadherin–catenin-associated proteins into individual clusters. Proc Natl Acad Sci USA. 2021;118:e2105550118. doi: 10.1073/pnas.2105550118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Oda H, Kraut R, Hayashi S, Kotaoka Y, Takeichi M. Zygotic Drosophila E-cadherin expression is required for processes of dynamic epithelial cell rearrangement in the Drosophila embryo. Genes Dev. 1996;10:659–671. doi: 10.1101/gad.10.6.659. [DOI] [PubMed] [Google Scholar]
- Uozumi N, Yanagidani S, Miyoshi E, Ihara Y, Sakuma T, Gao C-X, Teshima T, Fujii S, Shiba T, Taniguchi N. Purification and cDNA cloning of porcine brain GDP-L-Fuc: N-acetyl-β-D-glucosaminide α1→ 6fucosyltransferase. J Biol Chem. 1996;271:27810–27817. doi: 10.1074/jbc.271.44.27810. [DOI] [PubMed] [Google Scholar]
- Van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderleest TE, Smits CM, Xie Y, Jewett CE, Blankenship JT, Loerke D. Vertex sliding drives intercalation by radial coupling of adhesion and actomyosin networks during Drosophila germband extension. elife. 2018;7:e34586. doi: 10.7554/eLife.34586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DA, Sun M, Sadykov K, Kukuruzinska MA, Zaman MH. The integrated role of Wnt/β-catenin, N-glycosylation, and E-cadherin-mediated adhesion in network dynamics. PLoS Comput Biol. 2016;12:e1005007. doi: 10.1371/journal.pcbi.1005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Shewan AM, Scott JA, Helwani FM, den Elzen NR, Miki H, Takenawa T. Arp2/3 activity is necessary for efficient formation of E-cadherin adhesive contacts. J Biol Chem. 2004;279:34062–34070. doi: 10.1074/jbc.M404814200. [DOI] [PubMed] [Google Scholar]
- Vicidomini G, Moneron G, Han KY, Westphal V, Ta H, Reuss M, Engelhardt J, Eggeling C, Hell SW. Sharper low-power STED nanoscopy by time gating. Nat Methods. 2011;8:571–573. doi: 10.1038/nmeth.1624. [DOI] [PubMed] [Google Scholar]
- Vu V, Light T, Sullivan B, Greiner D, Hristova K, Leckband D. P120 catenin potentiates constitutive E-cadherin dimerization at the plasma membrane and regulates trans binding. Curr Biol. 2021;31:3017–3027.e7. doi: 10.1016/j.cub.2021.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D, Georgopoulos NT, Southgate J. Anti-social cells: predicting the influence of E-cadherin loss on the growth of epithelial cell populations. J Theor Biol. 2010;262:425–440. doi: 10.1016/j.jtbi.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Wang Y-C, Khan Z, Kaschube M, Wieschaus EF. Differential positioning of adherens junctions is associated with initiation of epithelial folding. Nature. 2012;484:390–393. doi: 10.1038/nature10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Allgeyer ES, Sirinakis G, Zhang Y, Hu K, Lessard MD, Li Y, Diekmann R, Phillips MA, Dobbie IM. Implementation of a 4Pi-SMS super-resolution microscope. Nat Protoc. 2021;16:677–727. doi: 10.1038/s41596-020-00428-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jiang J, Yang X, Zhou G, Wang L, Xiao B. Tethering Piezo channels to the actin cytoskeleton for mechanogating via the cadherin-β-catenin mechanotransduction complex. Cell Rep. 2022;38:110342. doi: 10.1016/j.celrep.2022.110342. [DOI] [PubMed] [Google Scholar]
- Watabe-Uchida M, Uchida N, Imamura Y, Nagafuchi A, Fujimoto K, Uemura T, Vermeulen S, Van Roy F, Adamson ED, Takeichi M. α-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J Cell Biol. 1998;142:847–857. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wombacher R, Heidbreder M, Van De Linde S, Sheetz MP, Heilemann M, Cornish VW, Sauer M. Live-cell super-resolution imaging with trimethoprim conjugates. Nat Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- Wu Y, Vendome J, Shapiro L, Ben-Shaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475:510–513. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Kanchanawong P, Zaidel-Bar R. Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev Cell. 2015;32:139–154. doi: 10.1016/j.devcel.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Yao M, Qiu W, Liu R, Efremov AK, Cong P, Seddiki R, Payre M, Lim CT, Ladoux B, Mege R-M. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat Commun. 2014;5:4525. doi: 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Gomez GA, Parton RG. Adherens junctions revisualized: organizing cadherins as nanoassemblies. Dev Cell. 2015;35:12–20. doi: 10.1016/j.devcel.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Itoh M, Nagafuchi A, Tsukita S. Cell-to-cell adherens junction formation and actin filament organization: similarities and differences between non-polarized fibroblasts and polarized epithelial cells. J Cell Sci. 1995;108:127–142. doi: 10.1242/jcs.108.1.127. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. α-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Yoshida C, Takeichi M. Teratocarcinoma cell adhesion: identification of a cell-surface protein involved in calcium-dependent cell aggregation. Cell. 1982;28:217–224. doi: 10.1016/0092-8674(82)90339-7. [DOI] [PubMed] [Google Scholar]
- Yoshimura M, Nishikawa A, Ihara Y, Si T, Taniguchi N. Suppression of lung metastasis of B16 mouse melanoma by N-acetylglucosaminyltransferase III gene transfection. Proc Natl Acad Sci USA. 1995;92:8754–8758. doi: 10.1073/pnas.92.19.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Kim T, Rajagopal V. Role of actin filaments and cis binding in cadherin clustering and patterning. PLoS Comput Biol. 2022;18:e1010257. doi: 10.1371/journal.pcbi.1010257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Kemp H-C, Szymanski RA, Cortes DB, Gadda NC, Lillich ML, Maddox AS, Peifer M. Micron-scale supramolecular myosin arrays help mediate cytoskeletal assembly at mature adherens junctions. J Cell Biol. 2021;221:e202103074. doi: 10.1083/jcb.202103074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R. Cadherin adhesome at a glance. J Cell Sci. 2013;126:373–378. doi: 10.1242/jcs.111559. [DOI] [PubMed] [Google Scholar]
- Zhai B, Villén J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Y, Ten Hagen KG. A mucin-type O-glycosyltransferase modulates cell adhesion during Drosophila development. J Biol Chem. 2008;283:34076–34086. doi: 10.1074/jbc.M804267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci USA. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kong D, Reichl L, Vogt N, Wolf F, Großhans J. The glucosyltransferase Xiantuan of the endoplasmic reticulum specifically affects E-Cadherin expression and is required for gastrulation movements in Drosophila. Dev Biol. 2014;390:208–220. doi: 10.1016/j.ydbio.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Zhao H, Liang Y, Xu Z, Wang L, Zhou F, Li Z, Jin J, Yang Y, Fang Z, Hu Y. N-Glycosylation affects the adhesive function of E-Cadherin through modifying the composition of adherens junctions (AJs) in human breast carcinoma cell line MDA-MB-435. J Cell Biochem. 2008;104:162–175. doi: 10.1002/jcb.21608. [DOI] [PubMed] [Google Scholar]
- Zhou F, Su J, Fu L, Yang Y, Zhang L, Wang L, Zhao H, Zhang D, Li Z, Zha X. Unglycosylation at Asn-633 made extracellular domain of E-cadherin folded incorrectly and arrested in endoplasmic reticulum, then sequentially degraded by ERAD. Glycoconj J. 2008;25:727–740. doi: 10.1007/s10719-008-9133-9. [DOI] [PubMed] [Google Scholar]
- Zhu W, Leber B, Andrews DW. Cytoplasmic O-glycosylation prevents cell surface transport of E-cadherin during apoptosis. EMBO J. 2001;20:5999–6007. doi: 10.1093/emboj/20.21.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Chappuis-Flament S, Wong E, Jensen IE, Gumbiner BM, Leckband D. Functional analysis of the structural basis of homophilic cadherin adhesion. Biophys J. 2003;84:4033–4042. doi: 10.1016/S0006-3495(03)75129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Schropp K, Wiśniewski JR, Mann M. Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol Cell. 2012;46:542–548. doi: 10.1016/j.molcel.2012.04.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article does not contain any original research data.



