Abstract
Pathogenic variants impacting upon assembly of mitochondrial respiratory chain Complex IV (Cytochrome c Oxidase or COX) predominantly result in early onset mitochondrial disorders often leading to CNS, skeletal and cardiac muscle manifestations. The aim of this study is to describe a molecular defect in the COX assembly factor gene COX18 as the likely cause of a neonatal form of mitochondrial encephalo-cardio-myopathy and axonal sensory neuropathy. The proband is a 19-months old female displaying hypertrophic cardiomyopathy at birth and myopathy with axonal sensory neuropathy and failure to thrive developing in the first months of life. Serum lactate was consistently increased. Whole exome sequencing allowed the prioritization of the unreported homozygous substitution NM_001297732.2:c.667 G > C p.(Asp223His) in COX18. Patient’s muscle biopsy revealed severe and diffuse COX deficiency and striking mitochondrial abnormalities. Biochemical and enzymatic studies in patient’s myoblasts and in HEK293 cells after COX18 silencing showed a severe impairment of both COX activity and assembly. The biochemical defect was partially rescued by delivery of wild-type COX18 cDNA into patient’s myoblasts. Our study identifies a novel defect of COX assembly and expands the number of nuclear genes involved in a mitochondrial disorder due to isolated COX deficiency.
Subject terms: Neuromuscular disease, Rare variants, Genetics research, Disease genetics, Congenital heart defects
Introduction
Cytochrome c oxidase (COX or Complex IV) is the terminal enzyme in the mitochondrial respiratory chain. Mammalian COX is a multisubunit enzyme composed of 14 subunits: the three core subunits MT-CO1, MT-CO2 and MT-CO3 are encoded by mitochondrial DNA (mtDNA), while the remaining structural subunits are imported into mitochondria after the cytosolic translation of the respective nuclear genes [1].
COX deficiency (MIM#220110) is a primary mitochondrial presentation associated with severe isolated reduction of Complex IV activity leading to impaired OXPHOS metabolism in the affected tissues. Molecular defects are mainly identified in nuclear or mtDNA genes encoding for structural COX subunits or in nuclear genes encoding for proteins involved in the maturation and assembly of the COX holocomplex. Pathogenic variants in several COX assembly genes mainly result in neonatal or childhood onset disorders featuring severe COX deficiency in muscle, brain and, rarely, liver [2]. Hypertrophic cardiomyopathy has been also frequently observed in these disorders (Supplementary Table 1).
COX18 encodes for a mitochondrial protein proposed to play a role in the maturation of MT-CO2 (COX-II) subunit [3, 4]. Genetic inactivation of COX18 in eukaryotic models abolished COX assembly and activity [4], but no COX18 variants have been detected in patients with isolated COX deficiency so far [5].
Here we describe the first association between a molecular defect in COX18 and a mitochondrial disorder characterized by neonatal hypertrophic cardiomyopathy followed by signs of infantile myopathy and axonal polyneuropathy with predominant affection of sensory fibers.
Methods
The subject underwent several pediatric metabolic evaluations, neurological examinations, cardiological assessments, brain MRI, and neurophysiological studies. Blood samples and a muscle biopsy were collected. The Ethics Committee of the IRCCS Foundation Ca’ Granda Ospedale Maggiore Policlinico (Milan, Italy) approved the study. Written informed consent for publication of clinical details and images were obtained from patient’s parents.
Histological and immunohistochemical analysis
Tissue specimen was frozen in isopentane-cooled liquid nitrogen and processed according to standard techniques, as previously described [6]. For histological analysis, 8 µm-thick cryosections were picked and processed for routine staining with Haematoxylin and Eosin (H&E), Modified Gomori Trichrome (MGT), myosin ATPase (pH 9.4-4.6-4.3), cytochrome c oxidase (COX), succinate dehydrogenase (SDH), phosphatase acid, NADH, Oil Red O, Periodic Acid Schiff (PAS). Images fields were acquired at 40X using optical microscope Leica DM4000B equipped with DFC420C camera.
Electron microscopy
For ultrastructural examination a small part of muscle sample was fixed in 2.5% glutaraldehyde (pH 7.4), post fixed in 2% osmium tetroxide and then, after dehydration in a graded series of ethanol, embedded in Epon’s resin. Finally, ultrathin sections were stained with lead citrate and uranyl acetate and examined with Zeiss EM109 transmission electron microscope.
Genetic studies
After written informed consent, genomic DNA was extracted from peripheral blood samples of proband and parents using standard procedures. The exonic regions and flanking splice junctions of the genome were captured using the Clinical Research Exome v.2 kit (Agilent Technologies, Santa Clara, CA). Sequencing was done on a NextSeq500 Illumina system with 150 bp paired end reads. Reads were aligned to human genome build GRCh37/UCSC hg19 and analyzed for sequence variants using a custom-developed analysis tool [7]. Additional sequencing technology and variant interpretation protocol have been previously described [7].
RNA studies
Total RNA was isolated from primary cells by using Reliaprep RNA cell miniprep system (Promega). Control tissues RNA were obtained from the Human Multiple Tissue Panel (Clontech). cDNA was obtained through reverse transcription polymerase chain reaction (RT-PCR) using the RT Maxima Reverse Transcription Master Mix (Thermo Fisher). Transcripts of interests were evaluated by Taqman-based quantitative RT-PCR on an Applied 7500 instrument. 18 S transcript was used for normalization. All the primers are available upon request.
Cell culture
The control muscle biopsies and cell cultures used in this study were derived from the Telethon Bank of DNA, nerve and muscle tissues (GTF01001). Primary myoblasts were cultured in skeletal muscle growth cell medium (PromoCell GmbH, Heidelberg, Germany) at 37 °C in 5% CO2. COX18 complementary DNA (cDNA) FLAG(DDK)-tagged was obtained from Origene and its transfection was carried out in patient and control primary myoblasts via Lipofectamine 3000 protocol (Thermo Fisher) and selection was performed through the addition of G418 to the culture medium (500 μg/ml). We also evaluated the role of COX18 by RNA interference (RNAi) in HEK293 cells, which were transfected with scrambled or RNA sequences against COX18 RNA (hs.Ri.COX18.13.1/2 from IDT) by using Lipofectamine RNAiMAX (Invitrogen) at a final concentration of 10 nM The transfections were repeated on days 5 and 10, and the cells were harvested and analyzed on day 15.
Biochemical and protein studies
Enzymatic activities of mitochondrial respiratory chain complexes I–IV and citrate synthase (CS) were measured by spectrophotometry in the mitochondrial fractions isolated from patient’s and controls’ myoblasts. The specific activity of each complex was normalized to that of citrate synthase. Each experiment was performed in triplicate.
A cocktail of antibodies was used to assess mitochondrial respiratory chain subunits (abcam ab110411, 1:1000). The mitochondrial PORIN (VDAC) was assayed by using a specific antibody (abcam ab15895, 1:1500). The amount of proteins was detected using florescent secondary antibodies (LI-COR IR-DYE 800 -680 CW). Actin (Sigma A2066) was used for normalization purpose.
Blue Native–PAGE was performed as previously described [8]. Briefly, isolated mitochondria were solubilized with 2% DDM (N-Dodecil–beta–D–Maltoside). Twenty micrograms of mitochondrial protein were separated on 4 to 16% NativePage gradient gels and transferred onto PVDF membranes. Membranes were incubated with primary antibodies targeting NDUFA9 (459100, 1:500, Thermo Fisher Scientific) for complex I, SDHA (ab14715, 1:10000, Abcam) for complex II, UQCRC1 ab14745, (1:2000, abcam) for complex III, and subunit IV (A21347, 1:1000, Life technologies) for complex IV. The amount of each fully assembled complexes I, III or IV was estimated by densitometry (LiCor Odissey FC) of two independent BN-PAGE immunoblots using assembled complex II as a control. In-gel activity of complex IV was performed with an incubation of the native gel in Diaminobenzidine and cytochrome C in 50 mM phosphate buffer pH 7.4 at 37 °C overnight.
Results
Clinical features
Our patient was the third-born child of consanguineous parents from Egypt (Fig. 1A). Her second cousin had died in neonatal age due to fatal hypertrophic cardiomyopathy. DNA sample from this subject was not available for molecular testing.
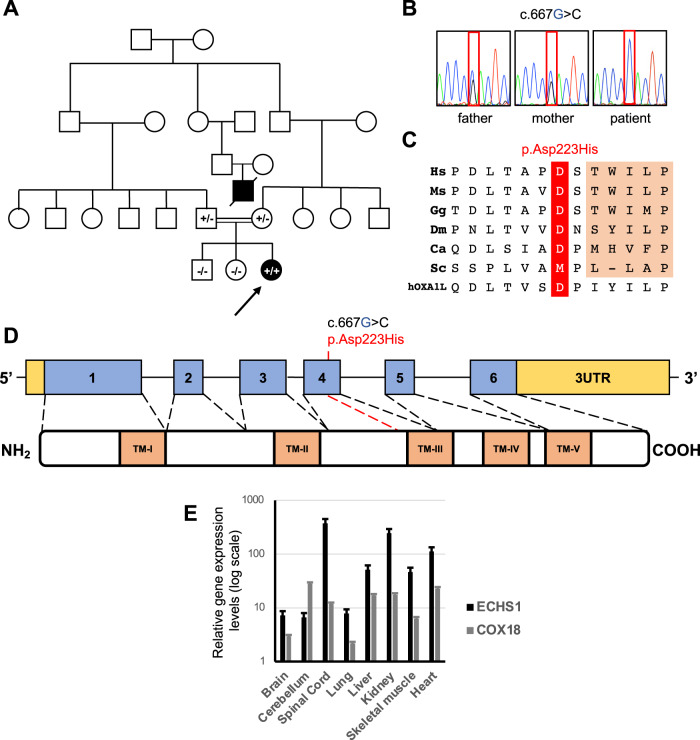
Fig. 1. Instrumental findings and sequence analysis in the Patient.
A Pedigree of the investigated family. Affected patients are indicated with black symbols. Arrow indicates the proband described in the report. The genotype for the COX18 variant c.667 G > C is indicated in the symbol of genotyped subjects (+/+ homozygous, +/– heterozygous carrier, –/– absence of the variant). B Sequence electropherograms showing the COX18 c.667 G > C variant in the patient and her unaffected parents. C Conservation of the human Asp223 residue across species and in the human OXA1L insertase protein. D Scheme of the COX18 gene and its encoded gene product displaying transmembrane (TM) domains. E Relative gene expression levels in human control tissues of COX18 compared to ECHS1 (encoding a mitochondrial matrix protein).
The patient was born at term with normal delivery (APGAR 9, weight at birth 2660 g). During pregnancy at the 35th gestational week a hypertrophic cardiomyopathy was detected. This finding was confirmed at an echocardiography which was performed at few days of life. Electrocardiography documented conduction abnormalities suggestive for Wolff-Parkinson-White syndrome. Moreover, thorax CT scan revealed the presence of aortic arc vascular anulus malformation.
Neurological examination showed hypertonia and poor sucking with normal deep tendon reflexes. Brain MRI was normal as far as ophthalmological examinations and audiological screening. EEG appeared mildly reduced in amplitude with poor organization during sleep. Due to insufficient sucking, a nasogastric tube was used for supplementary nutrition. Increased blood lactate levels were detected (4.1 mmol/L, cut off <1.8) with an increase of alanine (497 µmol/L, range 239–345 µmol/L) to lysine (90 µmol/L, range 107–163 µmol/L) ratio at plasmatic aminoacidic profile (5.5, cut off <3) and an increase excretion of fumaric acid (85 mmol/mol creatinine, range 4–45 mmol/mol creatinine) and 2 ketoglutaric acid (736 mmol/mol creatinine, range 66–552 mmol/mol creatinine) in the urinary organic acids. In the following months the plasma lactate was always increased (range 2.3–8 mmol/L) with the same constant alteration in the plasma aminoacidic profile and the presence of lactate (97 mmol/mol creatinine, range 21–38 mmol/mol creatinine) and pyruvate (82 mmol/mol creatinine, range 3–19 mmol/mol creatinine) in the urine organic acid in addition to the other organic acids related to the Krebs cycle. Because of the presence of acidosis with bicarbonate deficiency at the venous blood gas analysis, a correction with bicarbonate 2 meq/kg/day was started at 3 months of age.
The growth after birth dropped at 3rd percentile for weight and below the 3rd percentile for height.
At last assessment (19 months of life) the cardiological involvement was stable and confirmed a moderate left ventricular hypertrophy with normal systolic function, while a moderate-severe psychomotor delay with prominent motor impairment was observed. Neurological examination showed slight facial weakness, dysphagia, generalized hypotonia, muscle weakness with proximal-distal gradient, and mildly reduced deep tendon reflexes. She was not able to sit or control the head but could raise the four limbs against gravity. Nerve conductions studies showed a marked reduction of compound sensory action potential amplitudes and needle electromyography showed mild chronic neurogenic changes of motor unit action potentials suggesting an axonal polyneuropathy with predominant involvement of sensory fibers. She constantly needed the nasogastric tube to complete the meals. The study of swallowing demonstrated an insufficiency oral phase with incomplete tongue movements. The baby is waiting for placement of percutaneous gastrostomy tube.
Molecular studies
Trio-WES analysis did not detect variants in known disease genes but identified the novel homozygous nucleotide substitution chr4:73064834 C > G (GRCh38), corresponding to the c.667 G > C transversion in the COX18 gene (NM_001297732.2) in the patient. Unaffected parents were heterozygous carriers for the variant which was absent in the healthy siblings (Fig. 1B). Mitochondrial DNA sequence analysis (haplogroup L3e) revealed no pathogenic variant.
The c.667 G > C variant results in the substitution of the conserved aspartate at codon 223 (Fig. 1C) with histidine p.(Asp223His), in the proximity of the third transmembrane domain involved in the insertion of COX18 protein into the inner mitochondrial membrane (Fig. 1D). This position is also maintained in the distant homolog OXA1L, an inner membrane mitochondrial enzyme belonging to the highly conserved family of proteins which favor the insertion of substrates into membranes (insertases) [9].
The variant was absent in publicly available population databases, and several predictors supported its pathogenic burden (Supplementary Table 2). The variant was submitted to Clinvar (accession number: 2506394).
The analysis of COX18 cDNA in patient’s myoblasts did not display any evident effect of the variant on COX18 splicing or transcript stability (not shown). COX18 is highly expressed in liver, skeletal muscle and heart (Fig. 1E), as previously published [3].
Histological and ultrastructural studies in skeletal muscle
Muscle sample was taken from the quadriceps femoris of the Patient at 2 months of age. On Modified Gomori Trichrome staining, neither ragged-red fibers nor nemaline bodies and nuclear centralizations were seen (Fig. 2A). However, a severe reduction in COX activity, reflecting the almost total absence of Complex IV, was uniformly seen in patient’s muscle (Fig. 2B) compared to control age-matched muscle (Fig. 2C). EM studies showed the presence of some enlarged mitochondria with disorganization of the cristae, arranged to form concentric layers of membranes (Fig. 2D–F), and the presence of osmiophilic inclusions (Fig. 2F).
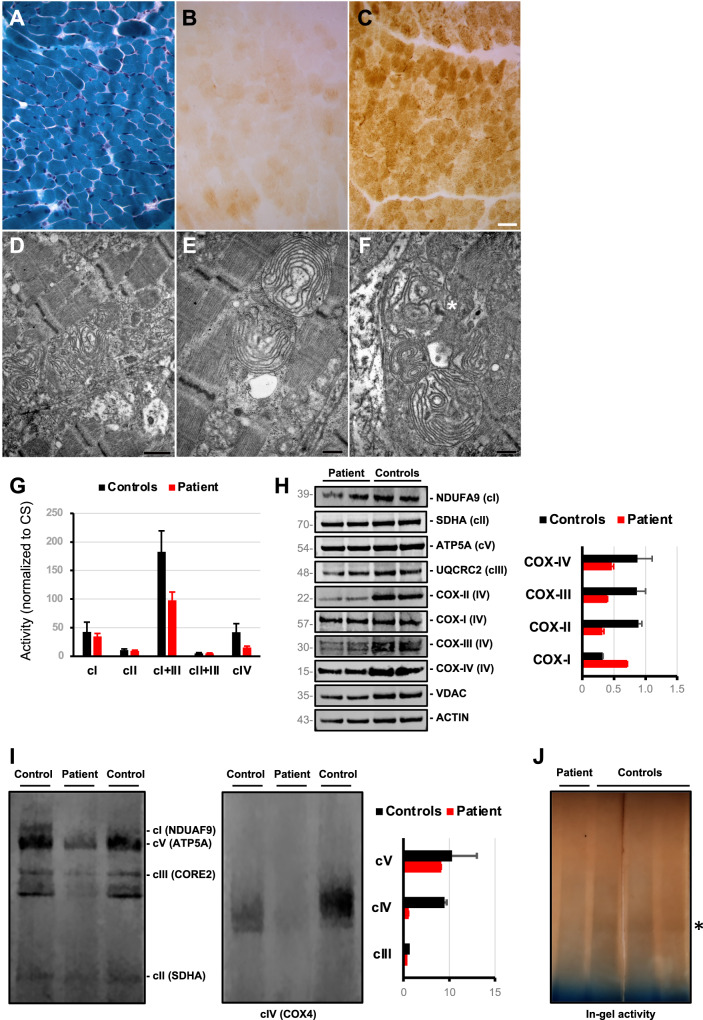
Fig. 2. Muscle histology and biochemical studies in myoblasts of the COX18-mutated patient.
A Modified Gomori Trichrome B Severe reduction of muscle fibers staining positive for cytochrome c oxidase (COX) is seen in the COX18 subject, whereas normal COX staining is shown in a healthy control (C) for comparison (Scale bar 25 µm). D–F Representative images of the alterations observed in mitochondria. Inner cristae were arranged to form concentric layers. (Asterisk in F indicates an osmiophilic inclusion. Scale bars D = 926 nm, E–F = 463 nm). G Spectrophotometric analysis of respiratory chain complexes activities normalized to citrate synthase levels in patient’s and control myoblasts. H Immunoblot analysis of myoblasts protein with antibodies directed against respiratory chain subunits. The mitochondrial porin (VDAC) and the cytosolic Actin proteins were used as a loading control. Densitometry (right) shows the reduction of steady state levels of COX subunits COX-II, COX-III and COX-IV in patient’s compared to controls’ myoblasts. I Blue-native polyacrylamide gel electrophoresis (BN-PAGE) analysis of myoblasts protein shows absence of fully assembled COX in the COX18 subject. Each of the five OXPHOS complexes (I–V) was visualised with a subunit-specific antibody that recognizes the native complex as follows: Complex I (NDUFA9), Complex II (SDHA), Complex III (UQCRC1), Complex IV (COX4), Complex V (ATP5A1). Densitometry after SDHA normalization (right) shows the prevalent reduction of fully assembled Complex IV. J In-gel activity assay showing the severe reduction of Complex IV activity in patient’s cells compared to controls.
Biochemical studies in patient’s myoblasts
Mitochondrial respiratory chain activities of complex I + III and IV were reduced in patient’s myoblasts with residual levels of 53% and 36%, respectively, compared to controls (Fig. 2G).
Western blotting of representative subunits of respiratory chain complexes showed the selective reduction of steady-state levels of the COX subunits COX-II, COX-III, and COX-IV (36%, 46% and 54% compared to controls, respectively) (Fig. 2H).
Blue native-PAGE analysis revealed a pronounced reduction in the amount of fully assembled COX in patient’s myoblasts while the other respiratory chain complexes, normalized to Complex II, were spared (Fig. 2I). We found no evidence for the accumulation of a subassembly in the subject investigated. In-gel analysis of COX activity also showed a severe impairment in patient’s but not in controls’ cells (Fig. 2J).
These findings suggest that the very low levels of holocomplex IV in patient’s myoblast result in destabilization of individual complex IV subunits, in particular COX-II.
Effects of COX18 silencing and upregulation in cellular models
To evaluate the role of COX18 in the maturation of Complex IV holocomplex, we examined the effects of COX18 downregulation in HEK293 cells and the consequences of COX18 overexpression in patient’s myoblasts. Suppression of COX18 in HEK293 cells by RNA interference (Fig. 3A) resulted in the instability of COX subunits, in particular COX-II (Fig. 3B), leading to an overall reduction of 60% of COX activity in silenced HEK293 cells compared to control cells (Fig. 3C). In addition, COX18 silencing resulted in both a COX-assembly defect (Fig. 3D) and reduced in-gel COX activity (Fig. 3E). These findings parallel those obtained in patient’s cells as well as in previously generated knock-out COX18 cells [4], confirming the importance of COX18 for the assembly of a fully active Complex IV.
Fig. 3. Biochemical effects of COX18 downregulation and overexpression.
A Quantitative gene expression levels of COX18 in HEK293 treated with scrambled or RNAi sequences directed against COX18. B Immunoblot analysis of selected respiratory chain subunits in COX18-silenced HEK293 cells, showing the reduction of COX subunits COX-II and COX-IV (densitometry showed on the right). C Activities of respiratory chain Complex II and IV in control or COX18-silenced HEK293 cells. D BN-PAGE analysis and (E) in-gel activity assay of respiratory chain complexes in control or COX18-silenced HEK293 cells, documenting a severe Complex IV reduction. F Immunoblot analysis showing the rescue of COX-II protein levels and Complex IV activity (G) in patient’s myoblasts after COX18 overexpression (visualized by fusion tag FLAG-DDK band, PatientCOX18wt) compared to mock-transfected patient’s myoblasts and control cells. H Profound decrease of COX histochemical reaction, visualized in patient’s myoblasts, compared to control cells, was partially rescued after COX18 overexpression (PatientCOX18wt). Scale bar 20 µm.
Finally, we attempted the rescue of COX activity introducing wildtype COX18 cDNA into patient’s myoblasts.
After 2 weeks, robust overexpression of COX18 was associated with the partial recovery of normal amount of MT-CO2 subunit (Fig. 3F) and of COX activity (Fig. 3G) in mutant cells. A moderate increase of Complex IV activity was also observed by COX histochemical staining in treated versus untreated patient’s myoblasts (Fig. 3H–J).
Discussion
Our study links a molecular defect in COX18 with a neonatal form of mitochondrial hypertrophic cardiomyopathy, lactic acidosis, failure to thrive and neurological involvement associated with severe skeletal muscle COX deficiency.
COX18 encodes for a mitochondrial inner membrane protein involved in the synthesis of mitochondrial respiratory chain. The role of COX18 and its homologs in COX assembly has been investigated in human cells [4] and yeast [10]. COX18 partially shares structure and topology of the human OXA1L insertase and acts as an Oxa1-independent enzyme for the translocation of the C-term domain of the MT-CO2 subunit across the inner mitochondrial membrane, allowing its exposure to intermembrane mitochondrial space [4]. This event is functional to the copper uptake at CuA metal center and the stabilization of MT-CO2 subunit. Despite their conserved functions, human COX18 and S.cerevisiae orthologue ScCOX18 cannot functionally complement each other [11]. This fact and the poor homology of human and yeast COX18 orthologues (25% of identity and 45% of similarity) hampers the modeling of COX18 variants in yeast.
The following data are consistent with COX18 c.667 G > C variant being the cause of the COX activity defect in our patient: (i) the variant is unreported in population database; (ii) the predicted change affects an highly conserved aminoacidic position; (iii) BN-PAGE and SDS-PAGE analyses showed that the decreased enzymatic activity of complex IV is associated with lower abundance of holo-complex IV and destabilization of COX subunits in patient’s myoblasts; (iv) downregulation of COX18 in a human cellular line produces the same biochemical defect observed in patient’s cells and in cellular models established after genetic inactivation of COX18 [4]; (v) the delivery of COX18 cDNA into patient’s myoblasts rescues the biochemical COX defect. By applying ACMG guidelines [12], the c.667 G > C variant should be classified as of uncertain significance (PM2 + PP3). However, considering the strong pathogenic score (0.973) of the meta-predictor MetaRNN [13] and the functional studies described in this study (PS3) we suggest to re-classify the c.667 G > C variant as pathogenic.
The homozygous COX18 variant NM_001300729.1:c.914 G > A, corresponding to the NM_001297732.2:c.908 G > C p.(Arg302His) change, has been recently reported in two Kurd twins presenting non-syndromic hearing loss [14]. Three other affected family members were found mutated in TMIE, whose biallelic variants are an established cause of deafness. Since hearing loss is characterized by huge genetic heterogeneity, the presence of distinct molecular defects in the same family is not rare. However, audiological examination in our patient was normal and the lack of functional studies does not support a definitive pathogenic role for the reported c.914 G > A variant.
COX18 has been previously considered a candidate gene for COX assembly defects, but no pathogenic variants have been so far detected in patients with isolated COX deficiency [5]. It is likely that severe COX18 defects might result in embryonic lethality, escaping the chance for detection. The residual COX activity (about 35% of controls) detected in patient’s cells supports the hypothesis of a permissive behavior for the p.(Arg223His) change. This has been previously observed for other COX assembly genes (e.g. COX10 and COX15) which are exclusively affected by hypomorphic alleles that, not abolishing the function of the gene product, are sufficient to produce a clinical phenotype [2].
The assembly of COX holocomplex is a complex pathway in which modular components merge into fully assembled COX through sequential steps involving more than 30 proteins. The three mtDNA-encoded subunits MT-CO1, MT-CO2, and MT-CO3 form the highly conserved structural core of the enzyme, in which the redox reaction takes place. The remaining 11 nuclear-encoded subunits are believed to confer structural stability and modulate enzyme activity. Additional nuclear encoded factors are required for the maturation and assembly of individual subunits into the functional holocomplex [2].
COX deficiency is a frequent isolated respiratory chain defect, occurring in 19–27% of pediatric mitochondrial patients [15, 16]. Associated clinical presentations are heterogenous and include Leigh or Leigh-like syndrome, lactic acidosis, leukodystrophy, myopathy, SMA (Spinal muscular Atrophy)-like disease. Hypertrophic cardiomyopathy is the most frequent cardiac manifestation in mitochondrial disorders, and it has been reported in several COX assembly defects alone or accompanied by further symptoms, especially encephalopathy and muscular hypotonia, as in our patient [17]. Although hypertrophic cardiomyopathy can be the consequence of inter-myofibrillary proliferation of mitochondria, resulting in mechanical damage and deficit of sarcomere function, in respiratory chain assembly defects the severe impairment of OXPHOS activity in the high-energy demanding cardiac tissue is likely the main driver of disease.
Interestingly, hypertrophic cardiomyopathy has been repeatedly associated with molecular defects in genes taking part in the maturation and stabilization of MT-CO1 (COX10, COX15) and MT-CO2 (SCO1, SCO2, COA6) [17]. COX18, catalyzing the translocation of the C-term domain of MT-CO2 across the inner mitochondrial membrane, participates in the maturation of MT-CO2 after its insertion by the COX20 chaperone [18] and precedes the metalation of the binuclear copper center (CuA) bound to MT-CO2 which is performed by a complex in which SCO2 and COA6 physically interact, as previously demonstrated [19].
The hypertrophic cardiomyopathy resulted fatal in the second cousin of our patient. Although DNA specimen was not available for molecular studies, the reported consanguinity increases the likelihood of a common genetic defect underlying these presentations. The mortality rate in patients with neonatal mitochondrial disorders is high [16] and usually related to the acute or subacute multiorgan failure secondary to mitochondrial respiratory chain dysfunction exacerbated by fever, illness, stress, medications, or heat. These “mitochondrial crises” can be associated with striking elevations in lactate levels. The presence of cardiac disease may result in a worse prognosis [15]. Therefore, the apparent stable course of cardiomyopathy in our patient must be carefully monitored since rapid deterioration may occur.
Bourens and Barrientos hypothesized that molecular defects in COX18 and other factors taking part in the biogenesis of MT-CO2 module might result in clinical phenotypes featuring encephalomyopathy and cardiomyopathy [4]. Our findings confirm this hypothesis and prompt the inclusion of COX18 in the molecular screening of infantile forms of cardiomyopathies and other infantile neuromuscular disorders featuring isolated COX deficiency.
Supplementary information
Acknowledgements
We thank the patient and her family for their kind cooperation. This work was promoted within the European Reference Network (ERN) for Rare Neuromuscular Diseases. We thank the Associazione Centro Dino Ferrari for its support.
Author contributions
DR and GPC contributed to the conception and design of the study. DR, MG, FMa, FMe, MM, MFB, RD, VC, IP, FFu, VP, SS, LP, FFo, MB, DP, MR, SZ, LN, PC, MS, MI, GM, FMo contributed to the acquisition and analysis of data. DR, RD, SC, MI and GPC contributed to drafting the text and preparing the figures.
Funding
This study was (partially) funded by Italian Ministry of Health—Current research IRCCS Ca’ Granda Ospedale Maggiore Policlinico and by SEQMD project (IRCCS Cà Granda Ospedale Maggiore Policlinico, PI: Giacomo Comi). The case was part of the RARE (Rapid Analysis for Rapid carE) project using the exome analysis in an urgent setting in neonatal intensive care.
Data availability
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. Supplementary Table 1 (genes associated with clinical presentations featuring isolated Complex IV deficiency) and Supplementary Table 2 (predictors and in silico tools used in this study) are included as Supplementary Information.
Competing interests
The authors declare no competing interests.
Ethics approval
The studies involving human participants were reviewed and approved by the Comitato Etico Milano Area 2 Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico (Milan, Italy).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-023-01433-6.
References
- 1.Kadenbach B, Hüttemann M. The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion. 2015;24:64–76. doi: 10.1016/j.mito.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Brischigliaro M, Zeviani M. Cytochrome c oxidase deficiency. Biochim Biophys Acta Bioenerg. 2021;1862:148335. doi: 10.1016/j.bbabio.2020.148335. [DOI] [PubMed] [Google Scholar]
- 3.Sacconi S, Trevisson E, Pistollato F, Baldoin MC, Rezzonico R, Bourget I, et al. hCOX18 and hCOX19: two human genes involved in cytochrome c oxidase assembly. Biochem Biophys Res Commun. 2005;337:832–9. doi: 10.1016/j.bbrc.2005.09.127. [DOI] [PubMed] [Google Scholar]
- 4.Bourens M, Barrientos A. Human mitochondrial cytochrome c oxidase assembly factor COX18 acts transiently as a membrane insertase within the subunit 2 maturation module. J Biol Chem. 2017;292:7774–83. doi: 10.1074/jbc.M117.778514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacconi S, Salviati L, Trevisson E. Mutation analysis of COX18 in 29 patients with isolated cytochrome c oxidase deficiency. J Hum Genet. 2009;54:419–21. doi: 10.1038/jhg.2009.36. [DOI] [PubMed] [Google Scholar]
- 6.Napoli L, Crugnola V, Lamperti C, Silani V, Di Mauro S, Bresolin N, et al. Ultrastructural mitochondrial abnormalities in patients with sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1612–3. doi: 10.1001/archneur.68.12.1612. [DOI] [PubMed] [Google Scholar]
- 7.Pezzoli L, Pezzani L, Bonanomi E, Marrone C, Scatigno A, Cereda A, et al. Not Only Diagnostic Yield: Whole-Exome Sequencing in Infantile Cardiomyopathies Impacts on Clinical and Family Management. J Cardiovasc Dev Dis. 2021;9:2. doi: 10.3390/jcdd9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–31. doi: 10.1016/0003-2697(91)90094-A. [DOI] [PubMed] [Google Scholar]
- 9.Thompson K, Mai N, Oláhová M, Scialó F, Formosa LE, Stroud DA, et al. OXA1L mutations cause mitochondrial encephalopathy and a combined oxidative phosphorylation defect. EMBO Mol Med. 2018;10:e9060. doi: 10.15252/emmm.201809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saracco SA, Fox TD. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol Biol Cell. 2002;13:1122–31. doi: 10.1091/mbc.01-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaisne M, Bonnefoy N. The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast. FEMS Yeast Res. 2006;6:869–82. doi: 10.1111/j.1567-1364.2006.00083.x. [DOI] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pejaver V, Byrne AB, Feng BJ, Pagel KA, Mooney SD, Karchin R, et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am J Hum Genet. 2022;109:2163–77. doi: 10.1016/j.ajhg.2022.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohseni M, Babanejad M, Booth KT, Jamali P, Jalalvand K, Davarnia B, et al. Exome sequencing utility in defining the genetic landscape of hearing loss and novel-gene discovery in Iran. Clin Genet. 2021;100:59–78. doi: 10.1111/cge.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scaglia F, Towbin JA, Craigen WJ, Belmont JW, Smith EO, Neish SR, et al. Clinical spectrum, morbidity, and mortality in 113 pediatric patients with mitochondrial disease. Pediatrics. 2004;114:925–31. doi: 10.1542/peds.2004-0718. [DOI] [PubMed] [Google Scholar]
- 16.Debray FG, Lambert M, Chevalier I, Robitaille Y, Decarie JC, Shoubridge EA, et al. Long-term outcome and clinical spectrum of 73 pediatric patients with mitochondrial diseases. Pediatrics. 2007;119:722–33. doi: 10.1542/peds.2006-1866. [DOI] [PubMed] [Google Scholar]
- 17.Mazzaccara C, Mirra B, Barretta F, Caiazza M, Lombardo B, Scudiero O, et al. Molecular epidemiology of mitochondrial cardiomyopathy: a search among mitochondrial and nuclear genes. Int J Mol Sci. 2021;22:5742. doi: 10.3390/ijms22115742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott LE, Saracco SA, Fox TD. Multiple roles of the Cox20 chaperone in assembly of Saccharomyces cerevisiae cytochrome c oxidase. Genetics. 2012;190:559–67. doi: 10.1534/genetics.111.135665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pacheu-Grau D, Bareth B, Dudek J, Juris L, Vögtle FN, Wissel M, et al. Cooperation between COA6 and SCO2 in COX2 maturation during cytochrome c oxidase assembly links two mitochondrial cardiomyopathies. Cell Metab. 2015;21:823–33. doi: 10.1016/j.cmet.2015.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. Supplementary Table 1 (genes associated with clinical presentations featuring isolated Complex IV deficiency) and Supplementary Table 2 (predictors and in silico tools used in this study) are included as Supplementary Information.