Abstract
Recent global events have drawn into focus the diversity of options for combatting disease across a spectrum of prophylactic and therapeutic approaches. The recent success of the mRNA-based COVID-19 vaccines has paved the way for RNA-based treatments to revolutionize the pharmaceutical industry. However, historical treatment options are continuously updated and reimagined in the context of novel technical developments, such as those facilitated through the application of synthetic biology. When it comes to the development of genetic forms of therapies and vaccines, synthetic biology offers diverse tools and approaches to influence the content, dosage, and breadth of treatment with the prospect of economic advantage provided in time and cost benefits. This can be achieved by utilizing the broad tools within this discipline to enhance the functionality and efficacy of pharmaceutical agent sequences. This review will describe how synthetic biology principles can augment RNA-based treatments through optimizing not only the vaccine antigen, therapeutic construct, therapeutic activity, and delivery vector. The enhancement of RNA vaccine technology through implementing synthetic biology has the potential to shape the next generation of vaccines and therapeutics.
Subject terms: Synthetic biology, Synthetic biology, Synthetic biology
Introduction
The pharmaceutical industry has long depended on synthetic chemistry approaches to develop novel therapeutics. These approaches, which have led to numerous breakthroughs in drug development, have typically relied on screening compound libraries populated by a range of molecules derived from a set of known and robust chemistry reactions in order to achieve improved efficacy, safety, and pharmacokinetic properties1. However, they often fall short when addressing complex diseases or diseases with genetic roots. Additionally, the cost and extended development timelines associated with these approaches have the potential to limit their application to emergent diseases such as SARS-CoV-2. Synthetic biology, a field that has already had a significant impact on the agricultural, environmental, and renewable biofuel industries2, has emerged as an attractive alternative drug development approach.
Synthetic biology is a scientific field that involves the rewiring of organisms or biomolecular parts for new and desired abilities. This encompasses diverse applications, from engineering new or improved activities in enzymes, heterologous production of commodity chemicals, assembly of genetic parts in a synthetic manner and developing cellular therapies. Although synthetic biology itself is a relatively new field that emerged in the early 2000’s, the tools required to engineer living systems were under development for decades before the field of synthetic biology was officially founded and its roots can be traced all the way back to the discovery and development of genetic engineering techniques in the 1970s3. These scientific breakthroughs allowed scientists to manipulate DNA in unprecedented ways and opened the door for a wave of innovation in genetic engineering, such as the invention of polymerase chain reaction in 19834, which allowed researchers to amplify DNA sequences for use in genetic manipulation. Not long after, in the 1990s genomics emerged as a field that involved sequencing entire genomes and provided researchers with complete maps of genomes5, punctuated by the completion of the human genome project in 20035,6. These technological advancements have since led to the creation of a vast and ever-growing catalog of cellular components and their interactions which, in-turn, has enabled a bottom-up approach to designing regulatory networks and circuits that can be used for the biosynthesis of various biochemical products (e.g., proteins and metabolites)7. Moreover, the modular components within this catalog can be assembled and tested to identify combinations that yield further advantages beyond each individually. One such modular component, a genetic toggle switch, was engineered into Escherichia coli in the early 2000’s8, which resulted in the creation of the first synthetic biological system8. This breakthrough paved the way for further advances such as bacteria that produce biofuels or cells programmed to target cancer cells.
In the past, synthetic biologists have primarily used DNA as the molecule of choice for designing synthetic systems. As a result, synthetic DNA had been instrumental in the development of artificial genes9, gene regulatory networks10, and even entire genomes11,12, enabling scientists to study complex biological processes and even create synthetic organisms2,13. However, with the rise of RNA therapeutics, there has been a growing interest in developing synthetic systems exploiting the unique attributes of RNA molecules. RNA is more than just a messenger—it has a direct role in regulating cellular behavior and, in the past few years, large libraries of RNA parts affecting almost every step of biological control have become available14. RNA-based systems constructed using these libraries are safer than those constructed from DNA as they do not integrate into the host genome, making them suitable for therapeutic applications with higher safety standards15. Unlike DNA-based systems, RNA-based systems, built from RNA devices and circuits, are fast-acting as they do not require transcription16. Using diverse libraries of RNA components, researchers have begun to generate RNA-based systems that couple environmental sensing with functional outputs for therapeutic synthetic biology applications17,18.
The recent success of mRNA COVID-19 vaccines has sparked a surge in interest in RNA therapeutic technology and how the field of synthetic biology can be employed. Particularly, there are some challenges associated with RNA technology which can be addressed using synthetic biology. For example, base modifications and mRNA circularization have been employed to reduce mRNA immunogenicity and improve mRNA’s lifespan respectively. Each of these represents the combination of various biomolecular features in novel and synthetic ways to enhance the native function of this malleable nature of RNA. As this technology advances, scientists are discovering new opportunities for combining an RNA platform with synthetic biology in creating novel therapeutics that could revolutionize the pharmaceutical and healthcare industries. This could even include combining various types of RNA to increase its specificity and enable it to become dynamically responsive. In this review, we will explore how synthetic biology can be employed in the context of RNA technology to create powerful therapies and treatments as researchers seek to combine the two disciplines in novel ways. Specific focus will be given to how synthetic biology principles can be used to advance RNA vaccines.
Biomedical applications of RNA in synthetic biology
RNA-based synthetic biological systems are composed of heterologous components capable of controlling gene expression in response to specific exogenous cues or endogenous metabolites. These components, referred to as RNA devices (e.g., RNA aptamers19, ribozymes20,21, and RNA switches22), can serve as sensors, regulators, or signal molecules. When undergoing the application of synthetic biology, these devices can either be improved through the incorporation of new parts or used in combination with other devices. When combined in a network-like structure, RNA devices can form synthetic circuits that can perform even more complex functions such as gene expression regulation23, signal amplification24, and logic operations25. Unsurprisingly, these devices and circuits, with their ability to control cell behavior, have already been extensively applied to the development of novel diagnostic strategies and therapeutics (Fig. 1 and Table 1)26–29. In addition to RNA devices, synthetic biology has also been applied to the development of RNA vaccines, including the COVID-19 mRNA vaccines developed by Moderna and Pfizer that introduced the general global population to RNA technology30.
Fig. 1. Overview of RNA-based diagnostics, therapeutics, and living therapeutics.
A Schematic representation of RNA-based diagnostics, highlighting toehold switches detecting a trigger RNA to activate fluorescent protein expression, and aptamers recognizing a specific target molecule to generate a fluorescent signal. B Illustration of the cellular mechanism of action of various RNA therapeutics and vaccines, including Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Cas9 with guide RNA, antisense oligonucleotides (ASOs), aptamers, small interfering RNAs (siRNAs), ribozymes, and mRNA vaccines. C Depiction of engineered RNA devices or circuits transforming cells into living factories producing therapeutic outputs, such as the expression of cell surface molecules like chimeric antigen receptors (CARs), the production of therapeutic proteins, or the initiation of cell migration. These engineered cells can also be designed to respond to small molecules as a switch to regulate therapeutic output. “Created with BioRender.com”.
Table 1.
Technology readiness of RNA-based diagnostics, living therapeutics, RNA therapeutics and vaccines.
| Technology | Mechanism | Examples | Readiness Levela |
|---|---|---|---|
| RNA therapeutics |
mRNA degradation Genome editing Protein repression |
siRNAs miRNAs CRISPR Aptamers |
TLR9 |
| RNA vaccines | Expression of antigenic protein to generate an immune response | COVID-19 vaccines | TLR9 |
| RNA diagnostics | Structural change upon ligand binding that produces a detectable signal |
Aptamer Toehold switch |
TLR8 |
| Living Therapeutics | Engineered cells that direct the immune system towards a specific target | CAR-T Cells | TLR8 |
CRISPR clustered regularly interspaced short palindromic repeats, miRNA MicroRNA, siRNA small interfering RNA, TLR technology readiness level.
aThe TLR used is that defined by BARDA137.
Diagnostic tools
Early detection of disease is crucial for increasing patient survival and reducing the burden on the healthcare industry. Conventional diagnostic tools have relied on antibody-based strategies that, while highly sensitive, are costly to produce, and slow26. Synthetic biology, with access to a diverse catalog of RNA-based sensors and signal molecules and with its expedited development cycles, offers solutions that can overcome these limitations. In fact, a number RNA-based diagnostics have already been developed for a wide range of diseases with the potential to decrease the cost and time associated with conventional approaches with commentary on their technology readiness level listed in Table 131,32.
While different RNA devices can be employed in these tools, the general strategy remains the same: design an RNA molecule that can bind to a sequence of pathogenic nucleic acid sequence, or other non-nucleic acid targets, which then results in the generation of a detectable signal (e.g., fluorescence or color change)33. For example, toehold switches rely on a conformation change in the structure of this RNA device following its binding to the target sequence which triggers translation of a downstream reporter gene (e.g., gfp). These devices have been engineered to detect a wide range of viral pathogens, including SARS-CoV-2 and other coronavirueses34,35, West Nile Virus36, Zika virus37, and Ebola virus38. In recent years, aptamer-based sensors, or aptasensors, have also gained prominence for the detection of cancer biomarkers (e.g., human epidermal growth factor receptor-2 [HER2])39,40, cardiovascular disease (e.g., C-reactive protein [CRP])41,42, neurological disease (amyloid β)43,44, and infectious disease45,46.
Living therapeutics
The use of RNA devices allows for the engineering of living therapeutics, such as cell therapies, that have the ability to sense and respond to environmental factors that provide information about their location, relevant disease states, and the therapeutic window timing47. Such cell therapies include circulating cells, implantable cells, and tissue resident human cells47. For example, there has been growing interest in using RNA devices to control engineered T cells (i.e., CAR T cells) to improve the safety profile and therapeutic outcomes of CAR-T therapy17. CAR-T cells represent a form of immunotherapy where CARs have been engineered to recognize and eliminate cells expressing tumor antigens. This approach has demonstrated profound clinical efficacy against hematological cancers48–50. Despite promising clinical results, severe adverse events, such as cytokine release syndrome, have complicated treatment and have occasionally proven fatal51. RNA devices, such as RNA switches, can be engineered to respond to cues such as changes in temperature or the presence of a small molecule to trigger the deactivation of the engineered T cells52. Such devices can also be designed to be activated in the presence of a specific molecule or environmental cue, triggering the expression of the CAR and enabling the T cells to target cancer cells52. This approach has the potential to reduce the risk of adverse events associated with CAR-T cell therapy and improve patient outcomes.
RNA therapeutics and vaccines
RNA devices have also become increasingly popular as a means of modulating gene expression and have enabled researchers to develop gene therapies for a range of diseases. As such, significant efforts have been made to manipulate the expression or activity of therapeutic targets utilizing three main approaches (1) ASOs, (2) siRNAs, and (3) CRISPR. Each of these strategies utilize different mechanisms to repress gene expression.
ASOs are single-stranded RNA that are complementary to mRNA and that regulate gene expression by inhibiting translation and promoting degradation of RNA53. ASO therapeutics can be broken down into two main categories, (1) those that cleave target mRNA and (2) those that regulate the splicing of pre-mRNA54. The first category of drugs cleaves target mRNA by binding to the target mRNA sequence, cleaving the sequence between the DNA and RNA duplex via RNase H activity, and thus promoting the degradation of the target mRNA54. FDA-approved drugs using this mechanism of action (MoA) include mipomersen and inotersen, which are used to treat homozygous familial hypercholesterolemia55 and hereditary transthyretin amyloidosis56, respectively. The second category of ASO therapeutics uses a steric hindrance-based mechanism to regulate the splicing of pre-mRNAs54. This MoA has been of particular interest in the treatment of inheritable diseases and the drugs golodirsen and eteplirsen, both of which target Duchenne muscular dystrophy57, and nusinersen, which targets spinal muscular atrophy, have already been approved58,59.
siRNA make use of the RNA interference (RNAi) pathway, in which siRNA interacts with Argonaute (AGO) protein to form RNA-induced silencing complexes (RICS) that suppress target mRNA expression60. Unlike ASOs and most other RNA pharmaceuticals, siRNA molecules are double-stranded, which facilitates their activity without chemical modification. Currently, there are only three FDA-approved drugs that take advantage of siRNA; these drugs include patisiran, givosiran, and lumasiran, which target hereditary transthyretin-mediated (hATTR) amyloidosis61, acute hepatic porphyria62, and primary hypercholesterolemia or mixed dyslipidemia63, respectively.
RNA aptamers also provide an effective way to control gene expression by engineering them to bind only to specific proteins that are engaged in the process of gene expression, such as transcription factors. RNA aptamers offer several advantages due to their ability to target both intracellular and extracellular molecules. Unlike other RNA-based therapeutics that require entry into the cell to perform their functions, RNA aptamers can directly bind to extracellular targets and hinder or, in some cases, stimulate their functions32. This method has been extensively studied and successfully employed in the treatment of age-related macular degeneration (AMD) through the use of Vascular Endothelial Growth Factor-binding Pegaptanib64.
The development of the CRISPR-Cas system has been revolutionary within the field of gene therapy by empowering direct and specific genome editing. This is achieved through using guide RNA (gRNA) to direct introduction of a double-stranded break in DNA65. To date, synthetic biology has been applied to this system in several ways. For example, RNA devices which use small molecule- and light-responsive aptamers to regulate the spacer region of a gRNA can be devised66. In one case, an RNA aptamer was designed to bind to an small molecule, resulting in a conformation change that prevented Cas9 from binding to target DNA67. Upon the removal of stimuli, however, the RNA structure of the aptamer is changed into an active state that allows a catalytically inert ‘dead’ CRISPR-Cas9 (dCas9) to bind and regulate gene expression. Other systems use riboswiches in the 5′ region of the gRNA functioning cis-acting ribozyme that exposes the spacer sequence in the presence of the aptamer ligand68. Furthermore, combining a CRISPR effector with an anti-CRISPR gene and microRNA (miRNA) response elements (MREs) in the 3′ untranslated region has enabled researchers to restrict gene editing to certain tissues69. With this method, tissue-specific MREs direct repression of the anti-CRISPR proteins and enable tissue-specific inhibition or activation of the CRISPR effector.
RNA vaccines represent an exciting new opportunity for the application of synthetic biology as they are designed to introduce synthetic RNA encoding for pathogen-specific antigens, such as the SARS-CoV-2 spike protein, into cells to trigger an immune response against target pathogens70. Compared to conventional vaccines, such as virus-, viral-vector-, and protein-based vaccines, the synthetic nature of RNA vaccines offers advantages in terms of speed of development, scalability, safety, and immunogenicity71. For example, their synthetic, cell-free production method enable mRNA vaccines can be produced quickly and at a large scale making them particularly well-suited for addressing emerging infectious diseases, such as COVID-1972. In fact, mRNA vaccines from Pfizer/BioNTech and Moderna were conceived of, designed, clinically tested, and granted Emergency Use Authorization in under a year73.
The uses of mRNA vaccines are not limited to COVID-19. Recent studies have revealed the potential of mRNA vaccines to be a powerful means of protective immunization against a variety of infectious diseases, such as influenza74, dengue virus75, herpes simplex virus type 2 (HSV-2)76, rabies77, and Zika virus78. There have also been efforts to develop mRNA vaccines to protect against bacterial diseases such as Mycobacterium tuberculosis (M.tb), one of the world’s leading infectious killers79. However, using host synthetic machinery to produce bacterial proteins can lead to issues such as difficulties in folding, transport, and post-translational modifications80. There is also currently no way to produce more complex biomolecule antigens, such as the polysaccharides found in the Streptococcus pneumoniae vaccines (e.g., Prevnar and Pneumovax) in human cells using mRNA vaccines, limiting their ability to compete with well-established bacterial vaccines. Additionally, mRNA vaccines are being investigated for potential use in cancer therapy81. Initial studies have demonstrated the ability of mRNA vaccines to induce an immune response against cancer cells, enabling the body’s own defense system to recognize and attack them, with promising results for the treatment of breast cancer82 and prostate cancer83.
Using synthetic biology to advance RNA vaccines
By taking advantage of the existing protein synthesis machinery in a transfected cell, an mRNA-based vaccine approach can turn human cells into factories for theoretically any protein antigen or therapeutic84. However, factors such as gene size or number, organism-dependent codons, protein confirmation, and posttranslational modifications can, in practice, limit which antigens can be effectively delivered using mRNA vaccines. However, the synthetic nature of mRNA vaccines offers many opportunities to apply synthetic biology principles to overcome such potential impediments (Fig. 2).
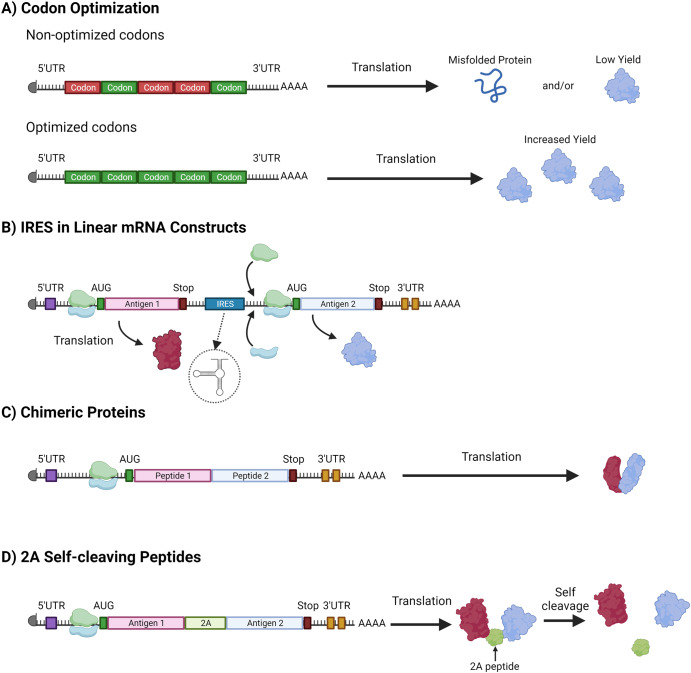
Fig. 2. Synthetic biology tools for engineering RNA.
A Codon optimization tackles issues with non-optimized codons that can lead to misfolded antigens or low yield when expressing antigens from non-human organisms in human cells. By substituting non-optimized codons (red) with synonymous codons optimized for human expression (green), this strategy significantly enhances the yield of correctly folded, functional antigens. B internal ribosome entry sites (IRES) elements function as a novel entry point for ribosomes, allowing them to bind at locations beyond the traditional 5’ Untranslated Region (UTR) on the mRNA. This feature facilitates the expression of multiple distinct antigens from a single mRNA construct. C The strategy for the creation of chimeric proteins fuses two distinct peptides to form a single, functional protein, enhancing the diversity of potential antigenic constructs. D Inclusion of 2A self-cleaving peptides serve as a molecular ‘comma’ during protein synthesis. They induce a ‘ribosome skipping’ event during translation, leading to the cleavage of the polypeptide at the 2A site. This allows for the independent expression of multiple antigens from a single open reading frame, thereby enabling multi-antigen expression from a single construct. “Created with BioRender.com”.
One such tool is codon optimization, the process by which the codons within a synthetic construct are optimized for the environment in which protein expression will occur. Each cellular environment (e.g., bacteria, viral, and eukaryotic cells) has a unique codon distribution linked with cognate tRNA molecules85,86. Thus, a codon bias from one cell type may result in expression challenges within another cellular environment (i.e., expressing a prokaryotic protein in a eukaryotic cell). Codon optimization can increase and improve the translation of RNA, therefore increasing antigen production. This process has been greatly advanced by the use of computational tools, such as deep learning87. However, there are some risks associated with codon optimization that should be considered when applying this process to vaccine design. For example, too rapid translation can result in misfolded proteins88. Moreover, there is evidence indicating that codon optimization is context dependent, impacted by both adjacent codons and the cellular metabolic state89. This suggests that further information can be gathered to enhance the application of codon optimization within pharmaceutical applications.
Due to size constraints associated with mRNA vaccines, design of vaccines for bacterial pathogens can be particularly challenging due to the need for multiple antigens to account for the complexity of the pathogen physiology or pathology. Synthetic biology principles developed for recombinant protein expression can also be exploited to overcome such impediments which will result in increased cost and complexity and may result in reduced delivery efficacy. For example, size restrictions of genetic vaccine constructs can be sidestepped by encoding for chimeric proteins, or proteins made up of immunogenic epitopes of multiple proteins90,91. In this way, epitopes from proteins associated with different strains, disease phases, or even different pathogens can be combined to provide more comprehensive protection. This principle is already being applied to some mRNA vaccines, including SARS-CoV-2 mRNA vaccines that encode for a chimeric spike protein92. Furthermore, a chimeric mRNA vaccine is under development that combines the SARS-CoV-2 spike with the influenza hemagglutinin matrix protein 1 proteins93.
Additional strategies to load multiple antigens onto a genetic vaccine construct include the use internal ribosome entry sites (IRESs) and self-cleaving 2A peptides. IRESs are RNA sequences used to initiate translation at an internal location within an mRNA construct, thereby facilitating the coding of polycistronic RNA. This is particularly important when designing circular RNA (circRNA) vaccines which do not have a 5’ cap to support classical cap-dependent translation94,95. However, there are some disadvantages to using IRESs, including size (>500 bps) and inefficient downstream gene expression that is dependent on the sequence of the gene upstream96–99. Self-cleaving 2A peptides represent an attractive alternative to IRESs due to their smaller size (<100 bps) and high gene expression efficiency96,97. These peptides, when encoded directly upstream of glycine, inhibit ribosomes from forming a peptide bond between the glycine and subsequent amino acid, thereby facilitating the expression of two separate protein antigens100. However, the cleavage is not 100%, which can have implications on therapeutic protein stability and activity101.
RNA vaccine constructs
Synthetic biology can also be applied to the RNA construct to overcome some of the existing limitations of mRNA vaccines, such as the poor stability and lifespan of mRNA within a physiological environment, which has limited success of native mRNA in pharmaceutical applications (Fig. 3). For example, modified RNA (modRNA), in which the mRNA sequence has been subjected several modifications (e.g., codon optimization, nucleic acid modification, and polyadenylation) to improve stability and expression following administration has been successfully implemented102. Further modification of mRNA can be done through encoding a replicase within the nucleotide sequence. This enables the mRNA to self-amplify (i.e., self-amplifying RNA (saRNA)), therefore reducing the required dose103. One of the more promising developments in mRNA technology has been the development of circRNA. This technology utilizes self-splicing introns to circularize mRNA into a ribonucleic acid “plasmid”. This can also be achieved through enzymatic means104. This provides similar improvements in stability as compared to the chemical modifications in modRNA102. The application of this approach has demonstrated promising results in various systems. For example, a circRNA vaccine encoding for the SARS-CoV-2 receptor-binding domain was found to induce a more potent and prolonged immune response than an equivalent dose of modRNA105. Importantly, the circRNA construct was also found to be highly heat stable and could be stored at room temperature for up to 2 weeks, which could facilitate easier distribution to geographical regions where a cold chain is difficult to maintain105. In addition to these mechanisms, origami-based approaches can be employed to generate nanoscale molecular RNA structures. Such structures have shown the ability to improve delivery and stability of such constructs106,107.
Fig. 3. Comparative overview of linear mRNA, saRNA, and circRNA constructs.
A For traditional mRNA vaccines, the antigenic or immunomodulatory sequence is present between 5’ and 3’ UTRs and translated directly from linear, non-replicating mRNA transcripts. B saRNA encodes additional replicase sequences within their construct that facilitate in vivo replication of entire mRNA construct. C circRNA constructs display a covalently closed loop structure created via back-splicing events that connect the 3’ end of an upstream exon to the 5’ end of a downstream exon. D CircRNA constructs can also be constructed enzymatically using enzymes that catalyze the formation of covalent bonds between moieties located at the 5’ and 3’ ends of the sequence. These constructs are notably stable due to their resistance to exonucleases, leading to potential prolonged antigenic protein expression within the cell. “Created with BioRender.com”.
These advances in RNA technology potentially offer exciting opportunities to express additional factors that influence vaccine efficacy. Signal proteins, for example, can redirect the immune response of an intracellularly produced antigen, which would typically be loaded on to a major histocompatibility (MHC) class I molecule108. This is because signal proteins, when fused with the antigen, can lead to the excretion and subsequent loading of the antigen onto an MHC class II molecule. Additional tags, such as secretory or organelle-targeting tags can be implemented to impart delivery to specific locations as desired. Protein adjuvants can also be encoded in such a way to fuse the adjuvant with the antigen, thus leading to a more effective adaptive and innate immune response109–112.
The ability of saRNA and circRNA constructs to reduce the required vaccine dose while still expressing multiple antigens on the same construct may one day lead to the expression of complex processes within the host cell, such as those required to facilitate pathogen-specific posttranslational modifications or produce pathogen polysaccharides. Moreover, further application of synthetic biology to these constructs has the potential to further enhance or otherwise modulate antigen expression to address diverse applications.
RNA vaccine delivery
Synthetic biology principles can be applied not only to the genetic content of an RNA vaccine, but also to the vehicle in which it is delivered. The choice of delivery system for mRNA vaccines is particularly important, as its size (~1.7 MDa) can make intracellular delivery difficult113,114. Additionally, the negatively charged mRNA is repulsed by the negatively charged cell membrane, and is susceptible to degradation by extracellular ribonucleases114,115. As such, a delivery vehicle for mRNA vaccines should facilitate cellular uptake and protect mRNA from degradation115,116.
Characterizing biological delivery vectors, such as viruses and bacteria, allows for the use of synthetic biology tools to make targeted modifications, therefore enhancing desired delivery characteristics. One of the most notable examples of a biologic delivery vector for genetic vaccines is the adeno-associated virus (AAV), which has been used to deliver RNA encoding for the SARS-CoV-2 spike protein117,118. Tools have been developed to control AAV functions and have been described in detail previously119. In brief, these tools include synthetic biological switches (e.g., chemical, protease, or optogenetic switches) that can control cellular processes such as receptor activation, transgene expression, and protein trafficking. Gene expression following AAV delivery can also be controlled by tailoring the regulatory parts of the inverted terminal repeat-flanked expression cassette within the AAV capsid119. Interestingly, AAVs can also be hybridized with other viral delivery vectors, such as parvoviruses and bacteriophages, and with extracellular vesicles (i.e., exosomes) from eukaryotic cells.
Bacterial vectors, such as Escherichia coli, represent another potential category of delivery vehicles for genetic vaccines. A key driver of this approach is the compatibility between the molecular biology underpinnings of synthetic biology and the microbiology advantages and knowledge collected for associated microbes. E. coli, for example, has perhaps the most expansive knowledge base and associated molecular biology tools of any microorganism120–122. As a result, advanced efforts in vaccine design can be accomplished using this carrier system while taking advantage of its ability to serve as a natural adjuvant. Attenuated, nonpathogenic strains of E. coli represent attractive options for use in vaccine design, as many tools are available to influence antigen number, expression level, and type using this host. Moreover, other recombinant features enabled by synthetic biology tools could serve in ways to assist with antigen delivery. For example, certain endosomal lysis proteins such as listeriolysin, natively associated with the intracellular pathogenic bacterium Listeria monocytogenes, facilitate the endosomal escape and cytosolic trafficking required to better deliver antigenic content to antigen presentation cells during administration of an engineered bacterial-based vaccine carrier123–127.
One drawback to antigen delivery within the confines of a microbial delivery vehicle is its biological complexity. Though bacterial or viral particles offers potential advantages with regards to adjuvant patterning and synthetic biology tools, there are other biological features that do not enhance vaccine efficacy. These same features may have a negative impact on the overall vaccine design, as extraneous elements of the microbial vaccine carrier system may spur unwanted immune reactions or cause undesirable toxicity. Furthermore, bacteria are prone to the degradation of mRNA and are thus not suited for its delivery. However, by using synthetic biology tools (e.g., DNA synthesis), it is possible to construct a synthetic, minimalized bacterial genome within a hollowed-out cellular framework11,128,129. Such a synthetic cell would be designed for a specific biological purpose with minimal extraneous cellular features.
Genetic payloads can also be delivered in fully chemical carrier systems, such as liposomes. Liposomal delivery systems have often accompanied studies in gene and drug delivery130,131, but the approach gained wider recognition due to their use in the COVID-19 mRNA vaccines132,133. Such liposomes represent an intersection of antigen delivery with synthetic biology as they can be further modified to impart delivery specificity. For example, there have been multiple strategies developed for the attachment of proteins to liposomal surfaces134–136. These mechanisms can be adapted to surface localization of an antibody or similar targeting ligand to directly deliver RNA to a specific tissue. This would enable directed RNA therapy in an approach like radioimmunotherapy.
Outlook
The union of synthetic biology and RNA technology has led to remarkable advances in medical treatments, allowing us to create novel therapeutics and vaccines that hold the potential to revolutionize global health care. However, the application of this technology is still in its early stages and has not come close to reaching its full potential.
We are currently in the midst of rapidly advancing artificial intelligence (AI) and machine learning that will further expand the horizons of RNA therapeutics and vaccines. AI and advanced computer-based models can be used to suggest modifications to RNA sequences for enhanced efficacy, identify novel disease targets, and computationally design specialized RNA sequences, such as aptamers and encoded antibodies, with precise specificity to unique targets. For example, we can imagine the use of AI to analyze large datasets of genetic information and proposing RNA construct designs for the expression of complex, multidomain proteins such as monoclonal antibodies. Antibody sequence and binding information can be utilized to train AI to propose antibody sequences with desired target or neutralizing abilities. We can also envision the use of AI to identify novel regulatory and ribozyme functions that could be incorporated into RNA medical products in order to detect specific disease states such as an increase in blood glucose levels in diabetes patients. Such regulatory and ribozyme functions could be used to develop an “on/off switch” that would then stimulate the production of insulin, thereby providing precise blood sugar level control for patients.
The application of AI and machine learning to synthetic biology can also potentially be used to overcome current limitations of mRNA vaccines, such as an inability to express multidomain antigens or polysaccharide antigens. Without the capability to produce these antigens, RNA vaccines will remain limited in the diseases it can target and will not be able to fully replace traditional vaccines. AI could potentially help design complex circuits that could carry out the multistage processes required for production of such antigens and that could be administered as an RNA vaccine, thereby opening new possibilities for vaccine development.
While there are still many challenges to be tackled when it comes to using synthetic biology for developing vaccines and RNA therapeutics, the potential is undeniable. With continued research and innovation, we may soon be able to see groundbreaking new technologies that can improve the lives of millions. Through careful consideration and creative design, synthetic biology can provide an effective and safe platform to develop novel treatments for a range of diseases. In the years ahead, we are sure to see amazing advances in this field that could change the face of healthcare forever.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was funded by Pfizer.
Author contributions
B.A.P. developed the outline. B.A.P., M.B. and J.B. researched sources, and drafted and edited the manuscript. A.H. provided strategic input, as well as drafting and editing support. C.H.J. conceptualized the publication and provided strategic oversight and editing. A.H. and M.B. are employees of Bulmore Consulting, which received funding from Pfizer in connection with the development of this manuscript. This work was funded by Pfizer.
Competing interests
C.H.J. reports that they are an employee of Pfizer Inc. and may hold stock or stock options in the company. C.H.J. reports that they are an employee of Pfizer Inc. and may hold stock or stock options in the company. M.B. and A.B.H. reports that they received financial compensation for consultancy and technical writing services.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41540-023-00323-3.
References
- 1.Campos KR, et al. The importance of synthetic chemistry in the pharmaceutical industry. Science. 2019;363:eaat0805. doi: 10.1126/science.aat0805. [DOI] [PubMed] [Google Scholar]
- 2.Meng F, Ellis T. The second decade of synthetic biology: 2010–2020. Nat. Commun. 2020;11:1–4. doi: 10.1038/s41467-020-19092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen SN, Chang AC, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro. PNAS. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erlich, H. A. PCR Technology - Principles and Applications for DNA Amplification. Vol. 246 (Palgrave Macmillan London, 1989).
- 5.Giani AM, Gallo GR, Gianfranceschi L, Formenti G. Long walk to genomics: history and current approaches to genome sequencing and assembly. Comput. Struct. Biotechnol. J. 2020;18:9–19. doi: 10.1016/j.csbj.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins FS, Morgan M, Patrinos A. The human genome project: lessons from large-scale biology. Science. 2003;300:286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 7.Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 8.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 9.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinform. 2006;7:1–8. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaman F, Bhatia S, Adler A, Densmore D, Beal J. Automated selection of synthetic biology parts for genetic regulatory networks. ACS Synth. Biol. 2012;1:332–344. doi: 10.1021/sb300032y. [DOI] [PubMed] [Google Scholar]
- 11.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 12.Fredens J, et al. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569:514–518. doi: 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang TC, et al. Materials design by synthetic biology. Nat. Rev. Mater. 2021;6:332–350. doi: 10.1038/s41578-020-00265-w. [DOI] [Google Scholar]
- 14.Kushwaha M, Rostain W, Prakash S, Duncan JN, Jaramillo A. Using RNA as molecular code for programming cellular function. ACS Synth. Biol. 2016;5:795–809. doi: 10.1021/acssynbio.5b00297. [DOI] [PubMed] [Google Scholar]
- 15.Kawasaki S, Ono H, Hirosawa M, Saito H. RNA and protein-based nanodevices for mammalian post-transcriptional circuits. Curr. Opin. Biotechnol. 2020;63:99–110. doi: 10.1016/j.copbio.2019.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell. Biol. 2014;15:95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dykstra PB, Kaplan M, Smolke CD. Engineering synthetic RNA devices for cell control. Nat. Rev. Genet. 2022;23:215–228. doi: 10.1038/s41576-021-00436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim CM, Smolke CD. Biomedical applications of RNA-based devices. Curr. Opin. Biomed. Eng. 2017;4:106–115. doi: 10.1016/j.cobme.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adachi T, Nakamura Y. Aptamers: a review of their chemical properties and modifications for therapeutic application. Molecules. 2019;24:4229. doi: 10.3390/molecules24234229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SV, et al. Catalytic RNA, ribozyme, and its applications in synthetic biology. Biotechnol. Adv. 2019;37:107452. doi: 10.1016/j.biotechadv.2019.107452. [DOI] [PubMed] [Google Scholar]
- 21.Zhan Y, Cao C, Li A, Mei H, Liu Y. Enhanced RNA knockdown efficiency with engineered fusion guide RNAs that function with both CRISPR-CasRx and hammerhead ribozyme. Genome Biol. 2023;24:9. doi: 10.1186/s13059-023-02852-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt CM, Smolke CD. RNA switches for synthetic biology. Cold Spring Harb. Perspect. 2019;11:a032532. doi: 10.1101/cshperspect.a032532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent R, Dixon N. Contemporary tools for regulating gene expression in bacteria. Trends Biotechnol. 2020;38:316–333. doi: 10.1016/j.tibtech.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Chen P, Wang L, Qin P, Yin BC, Ye BC. An RNA-based catalytic hairpin assembly circuit coupled with CRISPR-Cas12a for one-step detection of microRNAs. Biosens. Bioelectron. 2022;207:114152. doi: 10.1016/j.bios.2022.114152. [DOI] [PubMed] [Google Scholar]
- 25.Matsuura S, et al. Synthetic RNA-based logic computation in mammalian cells. Nat. Commun. 2018;9:4847. doi: 10.1038/s41467-018-07181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slomovic S, Pardee K, Collins JJ. Synthetic biology devices for in vitro and in vivo diagnostics. PNAS. 2015;112:14429–14435. doi: 10.1073/pnas.1508521112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao N, et al. Synthetic biology-inspired cell engineering in diagnosis, treatment, and drug development. Signal Transduct. Target Ther. 2023;8:112. doi: 10.1038/s41392-023-01375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan X, Letendre JH, Collins JJ, Wong WW. Synthetic biology in the clinic: engineering vaccines, diagnostics, and therapeutics. Cell. 2021;184:881–898. doi: 10.1016/j.cell.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andries O, Kitada T, Bodner K, Sanders NN, Weiss R. Synthetic biology devices and circuits for RNA-based ‘smart vaccines’: a propositional review. Expert Rev. Vaccines. 2015;14:313–331. doi: 10.1586/14760584.2015.997714. [DOI] [PubMed] [Google Scholar]
- 30.Rappuoli R, et al. Vaccinology in the post-COVID-19 era. PNAS. 2021;118:e2020368118. doi: 10.1073/pnas.2020368118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pieve CD, Perkins AC, Missailidis S. Anti-MUC1 aptamers: radiolabelling with (99m)Tc and biodistribution in MCF-7 tumour-bearing mice. Nucl. Med. Biol. 2009;36:703–710. doi: 10.1016/j.nucmedbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Germer K, Leonard M, Zhang X. RNA aptamers and their therapeutic and diagnostic applications. Int. J. Biochem. Mol. Biol. 2013;4:27–40. [PMC free article] [PubMed] [Google Scholar]
- 33.Thavarajah W, Hertz LM, Bushhouse DZ, Archuleta CM, Lucks JB. RNA engineering for public health: innovations in RNA-based diagnostics and therapeutics. Annu. Rev. Chem. Biomol. Eng. 2021;12:263–286. doi: 10.1146/annurev-chembioeng-101420-014055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt JP, et al. Towards detection of SARS-CoV-2 RNA in human saliva: a paper-based cell-free toehold switch biosensor with a visual bioluminescent output. N. Biotechnol. 2022;66:53–60. doi: 10.1016/j.nbt.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park S, Lee JW. Detection of coronaviruses using RNA toehold switch sensors. Int. J. Mol. Sci. 2021;22:1772. doi: 10.3390/ijms22041772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giakountis A, et al. Development of Toehold Switches as a Novel Ribodiagnostic Method for West Nile Virus. Genes. 2023;14:237. doi: 10.3390/genes14010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pardee K, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 38.Kaushik A, Tiwari S, Jayant RD, Marty A, Nair M. Towards detection and diagnosis of Ebola virus disease at point-of-care. Biosens. Bioelectron. 2016;75:254–272. doi: 10.1016/j.bios.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong HY, et al. Development of HER2-specific aptamer-drug conjugate for breast cancer therapy. Int. J. Mol. Sci. 2020;21:9764. doi: 10.3390/ijms21249764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poturnayová A, Dzubinová Ľ, Buríková M, Bízik J, Hianik T. Detection of breast cancer cells using acoustics aptasensor specific to HER2 receptors. Biosensors. 2019;9:72. doi: 10.3390/bios9020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amouzadeh Tabrizi M, Acedo P. Highly sensitive RNA-based electrochemical aptasensor for the determination of C-reactive protein using carbon nanofiber-chitosan modified screen-printed electrode. Nanomaterials. 2022;12:415. doi: 10.3390/nano12030415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ali GK, Omer KM. Ultrasensitive aptamer-functionalized Cu-MOF fluorescent nanozyme as an optical biosensor for detection of C-reactive protein. Anal. Biochem. 2022;658:114928. doi: 10.1016/j.ab.2022.114928. [DOI] [PubMed] [Google Scholar]
- 43.Obata Y, et al. Detection of Amyloid β Oligomers with RNA Aptamers in AppNL-GF/NL-GF Mice: A Model of Arctic Alzheimer’s Disease. ACS Omega. 2020;5:21531–21537. doi: 10.1021/acsomega.0c02134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, et al. Advances in aptamers against Aβ and applications in Aβ detection and regulation for Alzheimer’s disease. Theranostics. 2022;12:2095. doi: 10.7150/thno.69465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu LS, Wang F, Ge Y, Lo PK. Recent developments in aptasensors for diagnostic applications. ACS Appl. Mater. Interfaces. 2021;13:9329–9358. doi: 10.1021/acsami.0c14788. [DOI] [PubMed] [Google Scholar]
- 46.Wan Q, Liu X, Zu Y. Oligonucleotide aptamers for pathogen detection and infectious disease control. Theranostics. 2021;11:9133–9161. doi: 10.7150/thno.61804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cubillos-Ruiz A, et al. Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov. 2021;20:941–960. doi: 10.1038/s41573-021-00285-3. [DOI] [PubMed] [Google Scholar]
- 48.Frigault MJ, Maus MV. State of the art in CAR T cell therapy for CD19+ B cell malignancies. J. Clin. Investig. 2020;130:1586–1594. doi: 10.1172/JCI129208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park JH, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. NEJM. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chavez JC, Bachmeier C, Kharfan-Dabaja MA. CAR T-cell therapy for B-cell lymphomas: clinical trial results of available products. Ther. Adv. Hematol. 2019;10:2040620719841581. doi: 10.1177/2040620719841581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schubert ML, et al. Side-effect management of chimeric antigen receptor (CAR) T-cell therapy. Ann. Oncol. 2021;32:34–48. doi: 10.1016/j.annonc.2020.10.478. [DOI] [PubMed] [Google Scholar]
- 52.Wong RS, Chen YY, Smolke CD. Regulation of T cell proliferation with drug-responsive microRNA switches. Nucleic Acids Res. 2018;46:1541–1552. doi: 10.1093/nar/gkx1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zogg H, Singh R, Ro S. Current Advances in RNA therapeutics for human diseases. Int. J. Mol. Sci. 2022;23:2736. doi: 10.3390/ijms23052736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YK. RNA therapy: rich history, various applications and unlimited future prospects. Exp. Mol. Med. 2022;54:455–465. doi: 10.1038/s12276-022-00757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parham JS, Goldberg AC. Mipomersen and its use in familial hypercholesterolemia. Expert Opin. Pharmacother. 2019;20:127–131. doi: 10.1080/14656566.2018.1550071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benson MD, et al. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Eng. J. Med. 2018;379:22–31. doi: 10.1056/NEJMoa1716793. [DOI] [PubMed] [Google Scholar]
- 57.Frank DE, et al. Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology. 2020;94:e2270–e2282. doi: 10.1212/WNL.0000000000009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gidaro T, Servais L. Nusinersen treatment of spinal muscular atrophy: current knowledge and existing gaps. Dev. Med. Child Neurol. 2019;61:19–24. doi: 10.1111/dmcn.14027. [DOI] [PubMed] [Google Scholar]
- 59.Mendell JR, et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 60.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 61.Adams D, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Eng. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 62.Balwani M, et al. Phase 3 trial of RNAi therapeutic givosiran for acute intermittent porphyria. N. Eng. J. Med. 2020;382:2289–2301. doi: 10.1056/NEJMoa1913147. [DOI] [PubMed] [Google Scholar]
- 63.Scott LJ, Keam SJ. Lumasiran: first approval. Drugs. 2021;81:277–282. doi: 10.1007/s40265-020-01463-0. [DOI] [PubMed] [Google Scholar]
- 64.Ng EW, et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 65.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gangopadhyay SA, et al. Precision control of CRISPR-Cas9 using small molecules and light. Biochemistry. 2019;58:234–244. doi: 10.1021/acs.biochem.8b01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang W, Hu JH, Liu DR. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat. Commun. 2017;8:15939. doi: 10.1038/ncomms15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galizi R, Jaramillo A. Engineering CRISPR guide RNA riboswitches for in vivo applications. Curr. Opin. Biotechnol. 2019;55:103–113. doi: 10.1016/j.copbio.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 69.Lee J, et al. Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins. RNA. 2019;25:1421–1431. doi: 10.1261/rna.071704.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwon S, Kwon M, Im S, Lee K, Lee H. mRNA vaccines: the most recent clinical applications of synthetic mRNA. Arch. Pharm. Res. 2022;45:245–262. doi: 10.1007/s12272-022-01381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dolgin E. How COVID unlocked the power of RNA vaccines. Nature. 2021;589:189–192. doi: 10.1038/d41586-021-00019-w. [DOI] [PubMed] [Google Scholar]
- 72.Chaudhary N, Weissman D, Whitehead KA. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021;20:817–838. doi: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kashte S, Gulbake A, El-Amin Iii SF, Gupta A. COVID-19 vaccines: rapid development, implications, challenges and future prospects. Hum. Cell. 2021;34:711–733. doi: 10.1007/s13577-021-00512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chivukula S, et al. Development of multivalent mRNA vaccine candidates for seasonal or pandemic influenza. npj Vaccines. 2021;6:153. doi: 10.1038/s41541-021-00420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wollner CJ, et al. A dengue virus serotype 1 mRNA-LNP vaccine elicits protective immune responses. Virol. J. 2021;95:e02482–02420. doi: 10.1128/JVI.02482-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.LaTourette II, et al. Protection against herpes simplex virus type 2 infection in a neonatal murine model using a trivalent nucleoside-modified mRNA in lipid nanoparticle vaccine. Vaccine. 2020;38:7409–7413. doi: 10.1016/j.vaccine.2020.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J, et al. An mRNA-based rabies vaccine induces strong protective immune responses in mice and dogs. Virol. J. 2022;19:184. doi: 10.1186/s12985-022-01919-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Essink B, et al. The safety and immunogenicity of two Zika virus mRNA vaccine candidates in healthy flavivirus baseline seropositive and seronegative adults: the results of two randomised, placebo-controlled, dose-ranging, Phase 1 clinical trials. Lancet Infect. Dis. 2023;23:621–633. doi: 10.1016/S1473-3099(22)00764-2. [DOI] [PubMed] [Google Scholar]
- 79.Larsen SE, Baldwin SL, Coler RN. Tuberculosis vaccines update: Is an RNA-based vaccine feasible for tuberculosis? Int. J. Iinfect. Dis. 2023;130:S47–S51. doi: 10.1016/j.ijid.2023.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ozdilek A, Paschall AV, Dookwah M, Tiemeyer M, Avci FY. Host protein glycosylation in nucleic acid vaccines as a potential hurdle in vaccine design for nonviral pathogens. PNAS. 2020;117:1280–1282. doi: 10.1073/pnas.1916131117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miao L, Zhang Y, Huang L. mRNA vaccine for cancer immunotherapy. Mol. Cancer. 2021;20:1–23. doi: 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang X-t, Liu Q. mRNA vaccination in breast cancer: current progress and future direction. J. Cancer Res. Clin. Oncol. 2023;149:1–6. doi: 10.1007/s00432-023-04805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kübler H, et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J. Immunother. Cancer. 2015;3:1–14. doi: 10.1186/s40425-015-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qin S, et al. mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduct. Target. Ther. 2022;7:166. doi: 10.1038/s41392-022-01007-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iriarte A, Lamolle G, Musto H. Codon usage bias: an endless tale. J. Mol. Evol. 2021;89:589–593. doi: 10.1007/s00239-021-10027-z. [DOI] [PubMed] [Google Scholar]
- 86.Parvathy ST, Udayasuriyan V, Bhadana V. Codon usage bias. Mol. Biol. Rep. 2022;49:539–565. doi: 10.1007/s11033-021-06749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fu H, et al. Codon optimization with deep learning to enhance protein expression. Sci. Rep. 2020;10:17617. doi: 10.1038/s41598-020-74091-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pascolo S. Synthetic messenger RNA-based vaccines: from Scorn to Hype. Viruses. 2021;13:270. doi: 10.3390/v13020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lanza AM, Curran KA, Rey LG, Alper HS. A condition-specific codon optimization approach for improved heterologous gene expression in Saccharomyces cerevisiae. BMC Syst. Biol. 2014;8:33. doi: 10.1186/1752-0509-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Castro JC, et al. Vaccination with chimeric protein induces protection in murine model against ascariasis. Vaccine. 2021;39:394–401. doi: 10.1016/j.vaccine.2020.11.046. [DOI] [PubMed] [Google Scholar]
- 91.Harryvan TJ, de Lange S, Hawinkels LJAC, Verdegaal EME. The ABCs of antigen presentation by stromal non-professional antigen-presenting cells. Int. J. Mol. Sci. 2022;23:137. doi: 10.3390/ijms23010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martinez DR, et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Biswas A, Mandal RS, Chakraborty S, Maiti G. Tapping the immunological imprints to design chimeric SARS-CoV-2 vaccine for elderly population. Immunol. Rev. 2022;41:448–463. doi: 10.1080/08830185.2021.1925267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao J, et al. IRESbase: a comprehensive database of experimentally validated internal ribosome entry sites. Genom. Proteom. Bioinform. 2020;18:129–139. doi: 10.1016/j.gpb.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marques R, Lacerda R, Romão L. Internal Ribosome Entry Site (IRES)-mediated translation and its potential for novel mRNA-based therapy development. Biomedicines. 2022;10:1865. doi: 10.3390/biomedicines10081865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu, X., Ricci-Tam, C. & Sgro, A. E. Self-cleaving 2A peptides allow for expression of multiple genes in Dictyostelium discoideum. Preprint at https://www.biorxiv.org/content/10.1101/2022.03.08.482734v1.abstract (2022). [DOI] [PMC free article] [PubMed]
- 97.Wang X, Marchisio MA. Synthetic polycistronic sequences in eukaryotes. Synth. Syst. Biotechnol. 2021;6:254–261. doi: 10.1016/j.synbio.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hennecke M, et al. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29:3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bochkov YA, Palmenberg AC. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques. 2006;41:283–292. doi: 10.2144/000112243. [DOI] [PubMed] [Google Scholar]
- 100.Donnelly ML, et al. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J. Gen. Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 101.Liu Z, et al. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017;7:2193. doi: 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bloom K, van den Berg F, Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2021;28:117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Müller S, Appel B. In vitro circularization of RNA. RNA Biol. 2017;14:1018–1027. doi: 10.1080/15476286.2016.1239009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qu L, et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185:1728–1744.e1716. doi: 10.1016/j.cell.2022.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Parsons MF, et al. 3D RNA-scaffolded wireframe origami. Nat. Commun. 2023;14:382. doi: 10.1038/s41467-023-36156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu M, et al. Lantern-shaped flexible RNA origami for Smad4 mRNA delivery and growth suppression of colorectal cancer. Nat. Commun. 2023;14:1307. doi: 10.1038/s41467-023-37020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim J, Eygeris Y, Gupta M, Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/j.addr.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kumar S, Sunagar R, Gosselin E. Bacterial protein toll-like-receptor agonists: a novel perspective on vaccine adjuvants. Front. Immunol. 2019;10:1144. doi: 10.3389/fimmu.2019.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Díaz-Dinamarca DA, et al. Protein-based adjuvants for vaccines as immunomodulators of the innate and adaptive immune response: current knowledge, challenges, and future opportunities. Pharmaceutics. 2022;14:1671. doi: 10.3390/pharmaceutics14081671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bouazzaoui A, et al. Strategies for vaccination: conventional vaccine approaches versus new-generation strategies in combination with adjuvants. Pharmaceutics. 2021;13:140. doi: 10.3390/pharmaceutics13020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mishra RPN, et al. Structural and immunological characterization of E. coli derived recombinant CRM(197) protein used as carrier in conjugate vaccines. Biosci. Rep. 2018;38:BSR20180238. doi: 10.1042/BSR20180238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Damase TR, et al. The limitless future of RNA therapeutics. Front. Bioeng. Biotechnol. 2021;9:628137. doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wadhwa A, Aljabbari A, Lokras A, Foged C, Thakur A. Opportunities and Challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wei J, Hui A-M. The delivery of mRNA vaccines for therapeutics. Life. 2022;12:1254. doi: 10.3390/life12081254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van den Berg AIS, Yun CO, Schiffelers RM, Hennink WE. Polymeric delivery systems for nucleic acid therapeutics: Approaching the clinic. J. Controll. Release. 2021;331:121–141. doi: 10.1016/j.jconrel.2021.01.014. [DOI] [PubMed] [Google Scholar]
- 117.Madhi SA, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 Variant. N. Eng. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sadoff J, et al. Final analysis of efficacy and safety of single-dose Ad26.COV2.S. N. Eng. J. Med. 2022;386:847–860. doi: 10.1056/NEJMoa2117608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wagner HJ, Weber W, Fussenegger M. Synthetic biology: emerging concepts to design and advance adeno-associated viral vectors for gene therapy. Adv. Sci. 2021;8:2004018. doi: 10.1002/advs.202004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baneyx F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 1999;10:411–421. doi: 10.1016/S0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 121.Choi, K. R., Shin, J. H., Cho, J. S., Yang, D. & Lee, S. Y. Systems metabolic engineering of Escherichia coli. EcoSal Plus7 (2016). [DOI] [PMC free article] [PubMed]
- 122.Li L, Liu X, Wei K, Lu Y, Jiang W. Synthetic biology approaches for chromosomal integration of genes and pathways in industrial microbial systems. Biotechnol. Adv. 2019;37:730–745. doi: 10.1016/j.biotechadv.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 123.Higgins DE, Shastri N, Portnoy DA. Delivery of protein to the cytosol of macrophages using Escherichia coli K-12. Mol. Microbiol. 1999;31:1631–1641. doi: 10.1046/j.1365-2958.1999.01272.x. [DOI] [PubMed] [Google Scholar]
- 124.Hu PQ, et al. Escherichia coli expressing recombinant antigen and Listeriolysin O stimulate class I-Restricted CD8+ T Cells following uptake by human APC. J. Immunol. 2004;172:1595–1601. doi: 10.4049/jimmunol.172.3.1595. [DOI] [PubMed] [Google Scholar]
- 125.Radford KJ, et al. A recombinant E. coli vaccine to promote MHC class I-dependent antigen presentation: application to cancer immunotherapy. Gene Ther. 2002;9:1455–1463. doi: 10.1038/sj.gt.3301812. [DOI] [PubMed] [Google Scholar]
- 126.Parsa S, Wang Y, Rines K, Pfeifer BA. A high-throughput comparison of recombinant gene expression parameters for E. coli-mediated gene transfer to P388D1 macrophage cells. J. Biotechnol. 2008;137:59–64. doi: 10.1016/j.jbiotec.2008.07.1815. [DOI] [PubMed] [Google Scholar]
- 127.Kayal S, Charbit A, Listeriolysin O. a key protein of Listeria monocytogenes with multiple functions. FEMS Microbiol. Rev. 2006;30:514–529. doi: 10.1111/j.1574-6976.2006.00021.x. [DOI] [PubMed] [Google Scholar]
- 128.Lartigue C, et al. Genome transplantation in bacteria: changing one species to another. Science. 2007;317:632–638. doi: 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- 129.Hutchison CA, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 130.Olusanya TOB, et al. Drug delivery systems and anticancer drugs. Molecules. 2018;23:907. doi: 10.3390/molecules23040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Abu Lila AS, Ishida T. Liposomal delivery systems: design optimization and current applications. Biol. Pharm. Bull. 2017;40:1–10. doi: 10.1248/bpb.b16-00624. [DOI] [PubMed] [Google Scholar]
- 132.Polack FP, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Eng. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Baden LR, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Eng. J. Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shao S, et al. Functionalization of cobalt porphyrin–phospholipid bilayers with his-tagged ligands and antigens. Nat. Chem. 2015;7:438–446. doi: 10.1038/nchem.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jones CH, et al. Comprehensive vaccine design for commensal disease progression. Sci. Adv. 2017;3:e1701797. doi: 10.1126/sciadv.1701797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hill AB, Beitelshees M, Nayerhoda R, Pfeifer BA, Jones CH. Engineering a next-generation glycoconjugate-like streptococcus pneumoniae vaccine. ACS Infect. Dis. 2018;4:1553–1563. doi: 10.1021/acsinfecdis.8b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Countermeasures, M. Integrated Technology Readiness Levels (TRLs) for Medical Countermeasure Products (Drugs and Biologics), https://medicalcountermeasures.gov/trl/integrated-trls/ (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.