Abstract
Soils harbor highly diverse microbial communities that are critical to soil health, but agriculture has caused extensive land use conversion resulting in negative effects on critical ecosystem processes. However, the responses and adaptations of microbial communities to land use conversion have not yet been understood. Here, we examined the effects of land conversion for long-term crop use on the network complexity and stability of soil microbial communities over 19 months. Despite reduced microbial biodiversity in comparison with native tallgrass prairie, conventionally tilled (CT) cropland significantly increased network complexity such as connectivity, connectance, average clustering coefficient, relative modularity, and the number of species acting at network hubs and connectors as well as resulted in greater temporal variation of complexity indices. Molecular ecological networks under CT cropland became significantly more robust and less vulnerable, overall increasing network stability. The relationship between network complexity and stability was also substantially strengthened due to land use conversion. Lastly, CT cropland decreased the number of relationships between network structure and environmental properties instead being strongly correlated to management disturbances. These results indicate that agricultural disturbance generally increases the complexity and stability of species “interactions”, possibly as a trade-off for biodiversity loss to support ecosystem function when faced with frequent agricultural disturbance.
Subject terms: Microbial ecology, Biodiversity, Soil microbiology
Introduction
Land use conversion, largely due to agricultural expansion, has considerably impacted ecosystem structure and function [1, 2]. Grasslands often have deep, rich soils that support increased soil carbon, making them targets for conversion for agricultural cultivation [3]. Temperate grasslands in the central U.S. have undergone one of the greatest anthropogenic transformations with habitat conversion greatly exceeding habitat protection [4]. From 2008 to 2012, roughly 77of new croplands in the U.S. were originally grasslands [5], and in the Southern Great Plains, these new croplands replaced approximately 11,000 km2 of grasslands with winter wheat (Triticum aestivum L.) alone [6], the dominant crop in this area. This extensive ecosystem conversion has resulted in significant declines in soil health, which also includes the effects on the soil biota and biotic processes [7].
Soil microorganisms are essential for providing many ecosystem services needed for agricultural production, but they are also very sensitive to land use changes and management disturbances [8]. Numerous studies examining the responses of microbial communities to agricultural land use and management consistently showed that increasing land use intensification significantly decreased microbial community diversity and shaped microbial community composition [9–11]. In addition, these studies also revealed that land use conversion substantially changed intrinsic soil properties such as soil moisture, pH, and nutrient status, all of which are known to further affect microbial community dynamics [12–14]. While many types of agricultural management exist, tillage is one of the most common practices that causes the largest disturbance and has led to the greatest degradation of soil ecosystems [15]. Tillage physically disturbs the soil, breaks down soil structure, causes nutrient loss [15, 16], and leaves the soil more vulnerable to climatic differences resulting in more perturbation to soil microbial communities. While previous studies have been valuable for describing the impact of agriculture on community composition, diversity, and the role of biotic and abiotic factors in shaping communities, few have investigated the associations among soil microorganisms which are likely more important to the functioning of complex ecosystems [17].
Individual populations of microbial species do not exist alone, but instead interact to form complex microbial communities [18, 19], and these interactions represent a crucial dimension of microbial community ecology. The widely used method of ecological network analysis has proven to be a powerful tool to examine the associations and organization of microbial communities [19–21]. It also provides a way to study community complexity and stability [22, 23], and serves as a basis to quantify the contribution of microbial interactions to ecosystem functions and services. The topological features of these networks have been shown to change with environmental conditions [24–26] and can be used to reflect the ability of the ecosystem to respond to such changes [27]. Recently, studies have investigated the associations of complex microbial systems in response to anthropogenic activities, including groundwater pollution [24], deforestation [26], nitrogen addition [28], and climate warming [22], but the effects on network associations due to converting native land for long-term cropland is still largely unknown. Yet, it is expected that the introduced disturbances will significantly affect the assembly and overall composition of the soil microbial community [29], emphasizing the importance of preserving biotic interactions that are equally at risk as individual species of extinction due to anthropogenic disturbances [30].
For these reasons, we set out to understand whether and how native land use conversion for long-term cropland affects the complexity and stability of soil microbial community networks by examining the temporal dynamics of soil microbial communities in native tallgrass prairie (TGP) and conventionally tilled (CT) winter wheat site in the U.S. Southern Plains in El Reno, Oklahoma. While previous studies from this area have shown that land use and sampling time impact bacterial abundance [31] and bacterial community diversity and composition [32] with increased management intensification having the greatest impact, it is not clear if the network dynamics of the microbial communities will be similarly affected. In this study, we aimed to address: (1) how does land use conversion from the native ecosystem to cropland impact the complexity and stability of the molecular ecological networks (MENs) over time? (2) does land use conversion from the native ecosystem to cropland change the relationship between the complexity and stability of the MENs?, and (3) are the relationships between complexity and stability of the MENs with environmental factors altered due to land use conversion and management practices? We hypothesized that increasing habitat disturbance under cropland would increase the complexity of species associations resulting in a more complex and stable network.
Results and discussion
Overall characteristics of the constructed molecular ecological networks
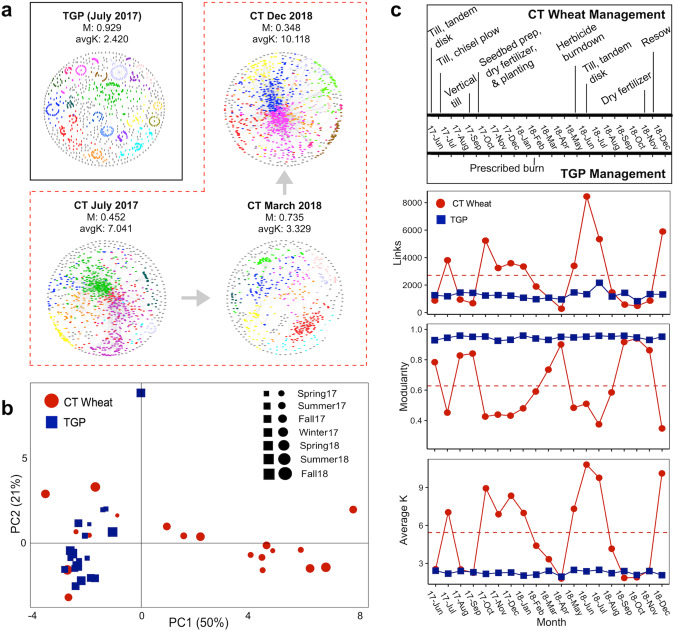
Molecular ecological networks were constructed for each sampling month resulting in 19 networks per land use (Fig. 1a, Fig. S1) [18]. In general, the empirical MENs were significantly different from the random MENs (Table S1). The overall topological properties (Table S1) revealed that the degrees of distribution (i.e., connectivity) fitted well with the power-law model with R2 values for CT wheat (0.75–0.95) and the native TGP (0.72–0.85), indicative of scale-free networks (Supplementary Text B). Networks also exhibited small-world properties with average path lengths (geodesic distance, GD) ranging from 3.3 to 8.3 for CT wheat and from 3.2 to 10.4 for the native TGP. The short path length between nodes enables efficient, rapid communication between network members and allows disturbances to spread quickly through the network for swift reactions [18], which is critical for responding to environmental changes. In addition, properties such as modularity can also be important for minimizing the impacts of disturbance by containing the disturbance and damage at a local level [33]. Modularity values for CT wheat and the native TGP were significantly greater (p = 0.04 and p < 0.001, respectively) than the corresponding modularity values for the randomized networks and the relative modularity was > 0, which is evidence of modular networks. Together, the architecture of these networks enables efficient communication between network members, which has important implications for microbial community dynamics in response to land use conversion.
Fig. 1. Temporal dynamics of soil microbial networks.
a Visualization of soil microbial networks. Difference of molecular ecological networks (MENs) due to land conversion represented by single tallgrass prairie (TGP) MEN since was relatively stable over sampling time. The other networks depict temporal differences of MENs for the conventional till (CT) wheat land use. Large modules with ≥10 node are shown in different colors, and smaller modules are shown in gray. Colors are not conserved between networks. Visual representations of all CT networks are found in Fig. S1. b Twenty-two network topological parameters were used for principal component analysis (PCA) to show differences in overall network properties over 19-month sampling period. Sampling times groups by season. c Temporal changes of select network topological parameters, including links, average K, and Modularity. Red circles represent networks under CT wheat land use and blue squares represent native TGP (control) land use. The dashed red line represents the mean of the properties for CT wheat land use.
Importance of “biotic interactions” in shaping molecular ecological networks
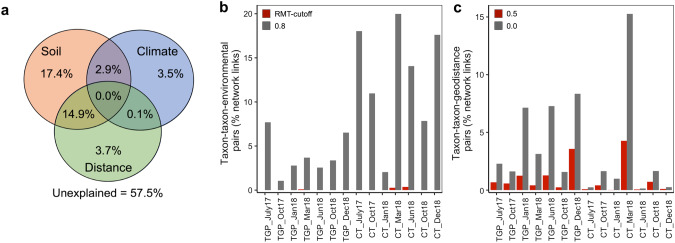
Although biotic interactions are a key part of regulating community assembly and disassembly [34], theoretically the observed species co-occurrence patterns in the MENs could also be largely due to environmental filtering and dispersal limitation [35]. Yet, it remains challenging to disentangle these mechanisms and determine the importance of biological interactions in ecological community assembly [36]. Therefore, we used multiple methods, such as CCA-based variation partitioning analysis (VPA) and the link test for environmental filtering and dispersal limitation (LTED) [22], to determine the relative contributions of these ecological processes to species co-occurrence in the MENs. While CCA results indicated that soil and climate variables had a significant (p ≤ 0.05) impact on the networked microbial communities (Supplementary Text C), VPA showed that over half of the variation (57.5%, Fig. 2a) could not be explained by the measured environmental variables (i.e., environmental filtering effect), and distance between samples only had a noticeable effect (14.9%) when considering interaction with soil properties. LTED suggested similar results of minor contributions (< 1% of links on average) from environmental filtering considered taxon-taxon-environmental covariates using the network correlation cutoff (Fig. 2b). When the correlation threshold (|r|) was lowered, links due to taxon-taxon-environment covariates increased in the CT wheat on average, but were still relatively minimal (2.1–20.0%). Additionally, dispersal limitations impacted less than 5% of links (Fig. 2c) in the networks on average (p ≤ 0.05, r > 0) based on LTED, and only 1.14 and 0.81% of the links on average were considered significant strong correlations (p ≤ 0.05, r ≥ 0.5) due to dispersal limitations. Collectively, these results indicated that biotic interactions could be the major driver shaping MENs in this study. Nevertheless, soils are highly heterogenous environments making it exceptionally difficult to determine the involvement of unmeasured environmental variations, especially in agriculturally managed systems which are rapidly fluctuating environments [8]. For this reason, the estimated “biotic interactions” via co-occurrence-based analyses should be at most considered putative biotic interactions [22] as well as interpreted with great caution [37].
Fig. 2. The relative contributions of different ecological processes to the observed network links.
a Variation partitioning analysis (VPA) based on significant (p ≤ 0.01) CCA model for networked microbial community. Soil category includes soil temperature, soil water content (SWC), soil pH, topsoil nitrate (NO3-), ammonia (NH4+), soil organic matter (OM), and available phosphorus (AP). Climate category includes average rainfall and average air temperature. Details of the CCA model can be found in Supplementary Table 8. b and c Analyses using the Link Test for Environmental filtering or Dispersal limitation (LTED). b The links in the molecular ecological networks (MENs) were tested with the 12 soil and climatic variables at the network correlation cutoff (St = 0.96) and a lower correlation threshold cutoff of |r | ≥ 0.8. In short, if a link between two taxa was caused by their covariation with environmental conditions, strong correlations between each taxon and the responsible environmental variable should be observed. c If dispersal limitation simultaneously affects the abundance distribution of two species across space, the abundances of both species are expected to covary with spatial distance. Therefore, assuming dispersal limitation was the only factor governing community assembly, the further away the sampling locations, the larger difference in the observed species abundances. For a pair of linked nodes in a network, it was tested whether significant (p ≤ 0.05, r > 0), and significant strong positive (p ≤ 0.05, r ≥ 0.5) correlations were observed simultaneously between the pairwise distance among sampling locations, and the difference in their relative abundance among samples based on Pearson correlation.
Networked community structure
The next critical question was how land use conversion impacted the composition and structure of the MENs. The number of ASVs used for network construction was on average 39% greater in the native TGP than CT wheat, and the resulting constructed networks were 24% larger. By contrast, 43% of ASVs made it into the constructed CT wheat networks compared to 34% of ASVs in the native TGP networks. Also, when considering ASVs in large modules (≥10 nodes), 72% of the nodes were in large modules in the CT wheat networks compared to 63% of nodes in the native TGP networks. Together, these results suggested that the CT wheat microbial taxa might associate more closely with each other than those of the native TGP.
The composition of the networked microbial communities significantly differed between the CT wheat cropland and the native TGP (Supplementary Text D). The two clusters representing CT wheat cropland and the native TGP communities were separate from each other as shown by principal coordinate analysis (Fig. S2a). Similarly, three non-parametric dissimilarity analyses (MRPP, ANOSIM, and Adonis; Table 1) confirmed that the networked microbial communities significantly (p = 0.001) differed by land use as well as sampling time. The diversity under CT wheat land use was lower for both the whole and networked communities. Meanwhile, compared with the whole community, the biodiversity of the networked microbial communities significantly (p < 0.001) decreased by more than half (0.27–0.47 for CT wheat and 0.27–0.45 for the native TGP) as measured by richness, phylogenetic diversity, and effective number of species from Shannon index. This suggests there might be a substantially reduced species pool from which the networked communities could draw (Fig. S2b). Management intensification and the resulting environmental changes likely acted as a deterministic filtering factor generating dynamic changes to the microbial communities and their network structure, which agreed with previous studies showing that land use strongly impacted microbial community structure [14, 38, 39].
Table 1.
Significance tests of the networked communities between conventional till (CT) wheat and native tallgrass prairie (TGP) land use.
| Dataset | Factor | MRPP | ANOSIM | Adonis | |||
|---|---|---|---|---|---|---|---|
| δ | p value | r | p value | F | p value | ||
| All | Field | 0.579 | 0.001 | 0.943 | 0.001 | 182.6 | 0.001 |
| Month | 0.693 | 0.001 | 0.144 | 0.001 | 2.156 | 0.001 | |
| TGP | Month | 0.457 | 0.001 | 0.726 | 0.001 | 4.043 | 0.001 |
| CT Wheat | Month | 0.533 | 0.001 | 0.443 | 0.001 | 3.778 | 0.001 |
Three different permutation tests were performed (MRPP, ANOSIM, and Adonis) on the basis of Bray–Curtis distance.
Bold values indicated significant p values.
Differences in complexity of MENs
To determine how land conversion for long-term cropland affected microbial network complexity, we closely examined several network topological properties. Based on 22 different network topological properties, the microbial MENs under CT wheat land use displayed noticeable variation in network structure compared to the native TGP over the 19-month sampling period (Fig. 1b; Figure S1). Network size (p = 0.001, W = 73) and modularity (M; p < 0.001, W = 5.5) significantly decreased under CT wheat, while average connectivity (avg K; p = 0.001, W = 219), connectance (con; p < 0.001, W = 345), and average clustering coefficient (avg CC; p = 0.077, W = 242) strongly increased under CT wheat (Fig. 1c, Table S1). The majority of the native TGP network topological properties remained stable over the 19-month sampling period compared to CT wheat cropland properties that had observable temporal variations. Furthermore, the relative modularity (how modular a network is as compared with the mean expected modularity, RM) of MENs was calculated as it is considered more meaningful for comparing modularity across networks (Supplementary Text E). The RM was significantly greater (p = 0.05) under CT wheat compared to the native TGP. These results indicated that MENs under CT wheat were on average more complex and experienced substantially more temporal variation, which coincided with and was likely due to the management (Fig. 1c) that occurred under CT wheat land use.
Variations in the structure of the microbial MENs could affect the network organization principles (i.e., modularity). Networks under CT wheat consisted of 189 large modules (modules with ≥10 nodes) accounting for 34.4–91.2% of the node in each MENs while the native TGP networks had 430 large modules totaling 52.4–71.1% of the networked nodes (Table S1). Between CT wheat and the native TGP, there were no preserved modular pairs (Table S3). In short, preserved module pairs are modules that contain a significantly large proportion of shared nodes when two modules in different networks are compared [20]. The native TGP also did not have any preserved module pairs over the sampling period in comparison with 67 preserved module pairs for CT wheat, suggesting that CT wheat land use resulted in greater similarities in module identity.
Differences in network complexity could also impact the role of individual members within the network with the identity of the keystone nodes differing between land uses (Supplementary Text D). The roles of each node were classified based on the within-module connectivity (Zi) and among-module connectivity (Pi) [18]. A total of 433 and 637 module hubs were identified for the CT wheat and the native TGP networks (Table S4–S7), respectively. The CT wheat networks also consisted of 38 network hubs and 456 connectors. However, the native TGP networks had no network hubs and only one connector for all networks. Together, module hubs, network hubs, and connectors are considered keystone nodes or nodes that play critical roles in shaping network structure [40] and drive community composition regardless of their abundance. Of the 1226 unique ASVs that acted as keystone nodes among all MENs, only 18 (1.5%) were found to be shared between both land uses. Additionally, of the keystone nodes within each land use, 17.7% acted as keystones in two or more of the CT wheat networks compared to only 3.8% in the native TGP networks. Taken together, CT wheat land use altered the roles of members within the networks and resulted in a greater number of temporally preserved keystone nodes.
Although the microbial community was more diverse and the networks were larger under the native TGP land use, the resulting networks were less complex, suggesting that greater diversity does not necessarily mean greater complexity [41]. This observation could arise for various reasons. For example, tallgrass prairies harbor greater aboveground plant species diversity (i.e., combination of several C3 and C4 species), providing more diverse nutrient and energy sources for the belowground microbial communities compared to croplands where nitrogen fertilizers provide the majority of the nutrients [42]. Therefore, the more diverse environmental nutrient and energy may support the microbial community instead of supplies through complex species interactions [41]. Another potential explanation could be greater functional redundancy due to higher microbial biodiversity in tallgrass prairies. Microbes often interact through function/metabolite preference [43], and higher diversity and functional redundancy of the microbial community reduces reliance on a few taxa and provides more opportunity for microbes to generate relationships within neighborhoods (i.e. modules). This could lead to greater modularity, reduced complexity, and the lack of persevered module pairs and keystone taxa as observed under native land use. In addition, the greater modularity in native land use is likely linked to stronger niche differentiation [44, 45] as the soils in native tallgrass prairies are generally a more heterogeneous and disconnected habitat compared to soils that are mixed by tilling creating a more homogeneous soil structure.
Impacts on the stability of MENs
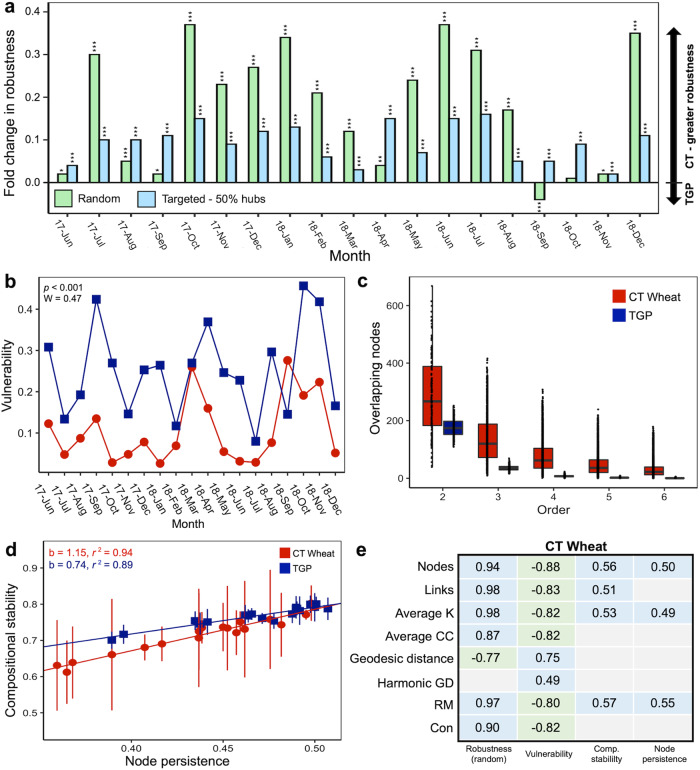
To determine whether and how land use conversion affected MENs stability, multiple stability indices were calculated based on simulations and empirical data. First, robustness or the resistance to node loss [46] of the MENS was calculated by simulating species extinction. Under random species loss (Fig. 3a), the MENs had significantly higher robustness (p < 0.001, W = 317) under CT wheat land use than the native TGP. When five module hubs were targeted for removal (Fig. S5a), there was no significant difference (p = 0.246, W = 221) in robustness. Yet, when 50% of module hubs were removed (Fig. 3a), robustness of the MENs was significantly greater (p < 0.001, W = 361) under CT wheat land use than the native TGP. Second, the vulnerability or the maximum decrease in efficiency when a single node was deleted from the network [47] was significantly lower (p < 0.001, W = 47) under CT wheat land use (Fig. 3b). Third, while the temporal invariability of the community composition [48] was greater (p < 0.001, W = 6825) under the native TGP based on consecutive monthly comparisons (Fig. S5b), more of the same nodes were present under CT wheat than the native TGP when any two pairs of networks were compared (p = 0.02, W = 53.5; Fig. 3c). This held true for comparisons up to any six networks (p < 0.005). The compositional stability and node persistence strongly correlated under both CT cropland (p < 0.001, rho = 0.94) and the native TGP (p < 0.001, rho = 0.89; Fig. 3d), but the slope was significantly greater (p < 0.001) under CT wheat. The constancy (inverse of temporal variations) of nodes (Figure S5e) was greater (p = 0.001) under the native TGP land use, while the constancy of links (Fig. S5f) was greater (p < 0.001) under CT wheat land use. Overall, the networked CT wheat microbial community was more stable and consistent over time with significantly more shared nodes between networks, conserved modules, and conserved keystone nodes compared to the native TGP land use (Supplementary Text E). Nevertheless, for the whole community, CT wheat had a much lower number of overlapping ASVs than the native TGP when comparing any two sampling times (Fig. S5c), indicating low retention of species over time in the species pool for the networked CT wheat to draw from. Similar to macroorganisms [49, 50], land use conversion can cause biotic homogenization of microbial communities [26, 51], which leads to greater similarity of communities over time and/or space [52]. This could be a cause for concern if biotic homogenization is a result of the loss of endemic taxa as these taxa tend to have unique traits, and homogenization of these traits likely alters ecosystem function and reduces ecosystem resilience [49, 53].
Fig. 3. Temporal dynamics of network stability.
a Fold change in robustness measured by randomly removing 50% of taxa from each of the empirical molecular ecological networks (MENs) and by removing 50% of module hubs from each of the empirical MENs. Robustness for each timepoint was compared between conventional till (CT) wheat and native tallgrass prairie (TGP, control) land use using a two-sided t-test. Significances are expressed as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. b The network vulnerability of the empirical MENS measured by maximum node vulnerability in each network. c The number of overlapping nodes under CT wheat and native TGP land use among different numbers of networks (that is, orders). For example, for order = 2, the overlapping nodes were between any two pairs of networks; for order = 3, they were among any three networks. d Relationship between compositional stability and node persistence for CT wheat and native TGP land use based on linear regression (p < 0.001). Slopes (b) and adjusted r2 values shown. e Spearman correlations between network stability and network complexity indices under CT wheat land use. Significant correlations (p ≤ 0.05) are shown in blue for positive correlations and green for negative correlations. Inside the cells are the corresponding correlation coefficients. Non-significant correlations are shown in gray.
Significant correlations were detected between network stability and network complexity that differed with land use. Overall, robustness, compositional stability, and node persistence significantly (p ≤ 0.05) positively correlated with several network complexity indices under CT wheat (Fig. 3e), while only robustness had significant positive correlations with network complexity for the native TGP (Fig. S5d). Consistently, network stability indices under CT wheat significantly positively correlated with nodes, average connectivity, and relative modularity. Network vulnerability had a significant negative relationship with the majority of network complexity indices for CT wheat compared to no significant correlations for the native TGP. This was further supported by SEM analysis which showed a significant negative relationship between complexity and vulnerability (Fig. 4c). In general, native TGP land use conversion for CT wheat cropland enhanced the relationship between network stability and complexity. For an ecological system, relationships between complexity and stability often have important functional implications [54, 55]. Greater complexity could produce differential effects on stability creating a more resistant [56, 57] but less resilient system [58]. Hence, while the CT cropland developed stable ecological networks after many years of cultivation, the networks also heavily rely on interactions to maintain stability, potentially leaving the networks vulnerable to cascade effects [59], which could disrupt these interactions (i.e. complexity) and the network stability.
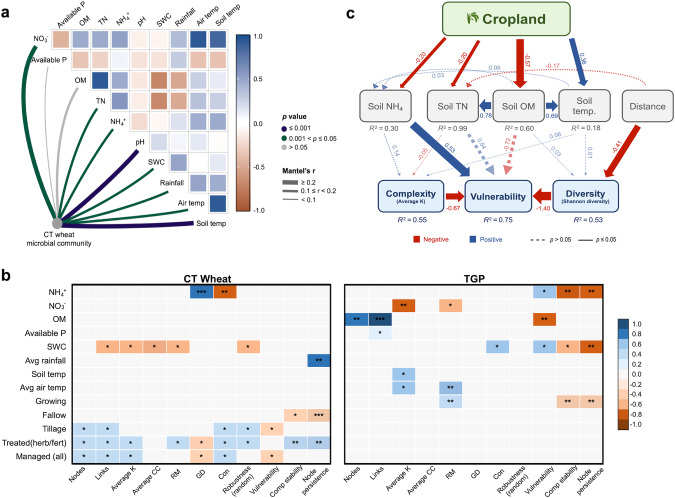
Fig. 4. Associations between network indices, environmental properties, and management.
a Correlations of the networked community structures (Bray-Curtis distance) and soil and climate properties for the conventional till (CT) wheat land use. Edge width corresponds to Mantel’s r value and the edge color represents the statistical significance. Pairwise spearman correlations of the variables are shown with a color gradient representing the correlation coefficients. b Spearman correlations between network stability and network complexity indices under CT wheat land use and native tallgrass prairie (TGP, control) land use. Spearman’s rho for significant correlations is depicted in a color gradient. The p-values of correlations are shown in the color squares expressed as *0.5 < p < 0.1, **p ≤ 0.01, ***p ≤ 0.001. Non-significant correlations are shown in gray. c Structural equation model (SEM) showing the relationships among treatment, soil variables, sampling distance, microbial diversity of MENs, and MENs complexity and stability. Blue and red arrows indicate positive and negative relationships, respectively. Solid or dashed lines indicate significant (p ≤ 0.05) or nonsignificant relationships, respectively. Numbers near the pathway arrow indicate the standard path coefficients. R2 represents the proportion of variance explained for every dependent variable. The model parameters include p = 0.78 (large p-value indicates that the predicted model and observed data are equal, that is, good model fitting), CFI = 0.99, SRMR = 0.008, and RMSE < 0.01. Native tallgrass prairie used as control and CT wheat cropland represents the treatment. Network complexity represented by the complexity index average K, network stability represented by vulnerability, and network microbial diversity represented by Shannon diversity index.
Interactions between complexity, stability, and the environment
An important following question is whether the relationships between the complexity and stability of the networks with the environment are altered due to land use conversion. Soil microbial communities are the most sensitive indicators of land use conversion and disturbance often being altered by soil properties [60], climate [61], land use intensity [62], and plant communities [63]. Land conversion for CT wheat land use resulted in less importance of various environmental factors in shaping the networked community structure than for the native TGP (Fig. 4a, Fig. S6). The TGP networked community structure was strongly correlated to most environmental factors. In comparison, the networked community under CT wheat was only strongly correlated with soil temperature, soil pH, and nitrate. CT wheat land use and feasibly its associated management also resulted in more negative correlations between pairwise comparisons of environmental factors.
Similar to network community structure, environmental factors played a less important role in influencing network complexity and stability of CT wheat land use than of the native TGP. The TGP network complexity and stability were influenced by multiple environmental factors (Fig. 4b). Overall, organic matter (OM), air temperature, and soil temperature had positive correlations with network complexity, while nitrate negatively correlated with complexity. Increases in soil water content (SWC) and ammonium (NH4+) decreased the TGP network stability by reducing node persistence, composition stability, and increasing vulnerability. The TGP received minimal management, including cattle grazing and a prescribed burn, but no significant correlations were detected. Likely, plant activity had a substantial influence on the complexity and stability of the native TGP MENs as plants are important to microbial community dynamics in natural ecosystems due to the co-evolution of plant-microorganism interactions [42]. Plant productivity interacts with all the factors important to shaping the native TGP networks which in turn could, directly and indirectly, affect microbial interactions. For instance, grasslands are often nitrogen limited with the productivity of many plant communities relying on nitrogen availability [64], and aboveground net primary production in grasslands generally has a large response to increased water availability [65]. This was further supported by the decrease in stability during periods of active vegetation growth under native land use.
While nutrient content and water availability can act as robust environmental filters to strongly select for microbial communities [66, 67], management disturbance could be equally if not more important to shaping microbial communities. Overall, the complexity and stability of the MENs in the CT cropland were more strongly correlated to management input (Fig. 4b). Summer-fallow decreased network stability, while the frequent disturbance of tillage, herbicide, and fertilizer input generally increased complexity and stability. Meanwhile, frequent fertilizer use in CT wheat cropland affects nitrogen (NH4+) content which was important for influencing CT wheat cropland network complexity [44, 68]. Similar results were observed for SEM with CT land use significantly impacting soil properties, but only NH4+ had a significant direct impact on stability (Fig. 4c). Additionally, water availability is frequently limited in areas where wheat is grown, and summer-fallow wheat ecosystems generally have reduced water use efficiency [65]. Thus, increased precipitation in the CT cropland community might disrupt existing microbial associations. Although land use and the changes in soil properties may have changed the microbial community structure and diversity, repeated management disturbance in the CT cropland greatly influenced the ecological networks, generating more complex and stable MENs presumably because greater interactions are needed for the microbial community to quickly respond to management disturbances.
Concluding remarks
Determining the extent of microbial associations and how they are mediated by land use conversion and management disturbance is a difficult issue to address and remains understudied. Our study provides insights into the impacts of land conversion on microbial ecological network dynamics. First, network features of the MENs differed due to land conversion and these changes increased the complexity of the MENs of the CT wheat cropland compared to the native ecosystem. The increased complexity of CT wheat MENs may have resulted from decreased microbial diversity, increased biotic homogenization, and/or greater niche sharing related to the more homogenous soil habitat of croplands than native land use; while the increased temporal variability coincided with management activity which was strongly temporally dependent. Second, the stability of the MENs was also greater under CT wheat and had an enhanced relationship with complexity. Similar results have been observed under other disturbance scenarios with greater network complexity contributing to greater stability [22]. Yet, these types of communities tend to have greater susceptibility to cascading biodiversity loss [69]. Third, our study showed that while the MENs complexity and stability under native land use were strongly influenced by various environmental factors, disturbances in the form of different management inputs were the driving force shaping complexity and stability of the MENs under CT wheat land use.
Together, our results have several important implications on the impacts of land conversion and intensive management on soil microbial communities. On the one hand, comparable to biodiversity, microbial ecological networks are also shaped by land use and are temporally dynamic. Changes in network structure could have important ecological consequences. For example, frequent management disturbances stimulated dynamic responses that led to greater complexity and stability of microbial ecological networks, making the ecosystem potentially less vulnerable to further disturbances. However, it remains unclear how resilient the community and the links between microorganisms would be to non-management related disturbances. On the other hand, the negative impacts of biodiversity loss due to land conversion could far exceed the positive effects of greater complexity and stability of microbial networks, resulting in more vulnerable ecosystems to both management and non-management related disturbances. Considering the increasing intensity of anthropogenic disturbances and environmental changes, preserving both microbial biodiversity and “interactions” could be vital to maintaining critical ecosystem functions.
Materials and methods
Study site and sampling strategy
This study included a native tallgrass prairie (64 ha) as the control site and a conventionally tilled winter wheat field (20.5 ha) as the treatment site located at the United States Department of Agriculture, Agricultural Research Service (USDA-ARS), Grazinglands Research Laboratory (GRL) in El Reno, Oklahoma, USA (35° 34.1’ N, 98° 03.6’ W). Both sites are included in the Southern Plains site of the Long-Term Agroecosystem Research (LATR) network [70, 71]. El Reno, Oklahoma, has a temperate continental climate with summer months generally hot and dry and most rainfall occurring during May-June and September-October. The average daily maximum and minimum air temperature of the study sites were 23 ± 8.7 °C and 8.9 ± 6.4 °C, respectively, with an average total annual rainfall of 855 mm ± 44.7 mm over a 30-year period (1980–2010) [72]. Detailed site descriptions can be found in Supplemental Text A.
Soil samples were collected monthly from the native TGP and CT winter wheat site from June 2017 to December 2018. To collect soil samples representative of each field, eight soil samples were taken 20 meters apart along a diagonal transect in each field. Each replicate soil sample consisted of four pooled soil cores. In total, 304 soil samples were collected consisting of eight replicates for individual sampling times for each field. Soil samples were taken using a 2.5 cm-diameter soil probe at a depth of 0–15 cm. Soils were passed through a 2 mm sieve to remove debris and stored at −80 °C until analysis.
Soil properties and climate data
Weather data were collected from the Oklahoma Mesonet station (http://www.mesonet.org/index.php/weather/local/elre) in El Reno (ELRE), Oklahoma. The Mesonet tower is located on the native tallgrass prairie used in this study (35° 32.9’ N, 98° 02.2’ W). Mesonet data included rainfall, maximum air temperature, average air temperature, and minimum air temperature. Soil chemical analyses were performed at the Oklahoma State University Soil, Water and Forage Analytical Laboratory (https://agriculture.okstate.edu/departments-programs/plant-soil/soil-testing/publications.html). Tests included topsoil nitrate (NO3-), soil organic matter (OM), soil total nitrogen (TN), ammonium (NH4+), and available phosphorus (AP). Gravimetric soil water content (SWC) and soil pH were measured in the lab. The SWC was determined by oven drying for ≥24 h at 65 °C or until the weight no longer changed [73]. Soil pH was measured with a pH meter using soil:water (w/v) = 1:5 [74]. Soil properties were measured for seven of the 19 sampling times being representative of different seasons over the study period. Soil properties were measured for all eight replicates.
Soil DNA extraction, amplicon sequencing, and analysis
DNA was extracted from 0.5 g of individual soil samples using an established protocol involving bead mill and SDS lysis [75] combined with the MoBio PowerSoil DNA isolation kit (MoBio Laboratories, a QIAGEN company, Carlsbad, CA, USA). The quality of DNA was assessed based on 260/280 nm and 260/230 nm absorbance ratios using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). DNA concentrations were quantified based on PicoGreen using a FLUOstar Optima fluorescence plate reader (BMG Labtech, Jena, Germany). For microbial community profiling, the V4 hypervariable regions of 16 S rRNA genes were amplified using the common primer pair 515 F (5’- GTGCCAGCMGCCGCGGTAA-3’) and 806 R (5’- GGACTACHVGGGTWTCTAAT-3’). A two-step PCR protocol was used and carried out in triplicate to minimize amplification bias as previously described [76]. PCR products from triplicate reactions were then pooled and quantified using PicoGreen. An equal amount of DNA for each sample was further pooled and purified with Qiagen QIAqick gel extraction kit. Sequencing was carried out on a MiSeq platform (Illumina Inc., San Diego, CA, USA) using a 2 × 250 pair-end format.
Raw amplicon sequencing data were processed through a pipeline (http://zhoulab5.rccc.ou.edu:8080) by the Institute for Environmental Genomics at the University of Oklahoma [25] to check read quality, demultiplex reads, and remove primers. Reads were then processed using USEARCH-UNOISE3 [77, 78] which has been shown to have a good balance between resolution and specificity for amplicon sequencing process [79]. Reads were merged as suggested by USEARCH documentation for 2 × 250 pairs with longer overlaps. Reads were quality filtered using 1 as the maximum expected error threshold and unique reads identified. UNOISE3 was then used for ASV-level denoising based on the default level minimum abundance. An ASV table was generated and resampled to the same sequencing depth across all samples (27,000 sequences per sample). Taxonomy was assigned using the USEARCH suggested RDP v18 training set.
Network construction and characterization
Correlation networks using a Random Matrix Theory (RMT)-based approach [17, 18, 22] were constructed for all individual sampling times resulting in a total of 19 networks for each site using the Molecular Ecological Network Analysis Pipeline (MENAP) available at the Institute for Environmental Genomics, University of Oklahoma (http://ieg4.rccc.ou.edu/MENA/). RMT distinguishes system-specific, nonrandom associations from random associations and thus yields association networks that are robust to random noise. Each of the networks was constructed independently, with nodes representing ASVs and edges representing tentative association relationships based on correlation between the abundance profiles of connected nodes. To increase the reliability of the predicted association relationships, only ASVs in at least six of the eight replicates were used for the network construction. In short, ASV abundance data was centered-log-ratio transformed to mitigate the effects of compositional bias [80, 81], and Pearson correlations were used to calculate the correlation matrix followed by an RMT-based approach [17, 18, 22]. In order to compare network topologies under the same condition, a uniform cutoff value (St) was used to generate microbial networks. The best cutoff value for all networks was determined by a scheme based on the generalized Brody distribution [82]. Then, an adjacency matrix was generated, containing only the correlations whose absolute values of coefficient (correlation strengths) were larger or equal to St. Nodes in isolation after the cut (no correlation strength to other nodes ≥St) were removed from the network. iDIRECT was further applied to these networks to reduce the influence of indirect relationships [83].
The potential contributions of environmental filtering or dispersal limitation in shaping network topology were tested. First, we determined the importance of soil factors, climate variables, and spatial distance between samples on the networked community structure using a CCA model followed by VPA. Next, we used a publicly available pipeline using R and Python 3 script [22] to detect taxon–taxon–environment co-variation links [84] and links possibly caused by dispersal limitation. While such analyses could provide insights on the relative importance of biotic interactions in shaping MENs, it is still not possible to prove that the links are truly due to biotic interactions. For this reason, MENs are best for making relative comparisons between conditions or treatments [20, 85]. Therefore, this study focused on comparing network differences between native land (TGP) converted for long-term cropland (CT wheat).
A total of twenty-two network topological indices were calculated using the MENAP to characterize network topological structure [17]. We focused on several indices, including nodes, the total number of links, average connectivity (average links per node, avgK), average clustering coefficient (the extent to which nodes are clustered, avgCC), average path distance (geodesic distance, GD), connectance (the proportion of realized links in all possible ones, Con), and modularity (M). All network properties were calculated individually for each random network. To test the significance of the constructed empirical MENs, 100 random networks corresponding to each network were generated. The numbers of nodes and links in random networks were constant, but link positions were rewired randomly so that the rewired network was comparable to the empirical network [86]. The same suite of network topological properties was calculated with each randomization. The means and standard deviations of these properties from the 100 randomizations were calculated and compared with those from the corresponding empirical MENs. Networks were visualized using Cytoscape 3.8.2 [87].
Network size and connectivity considerably varied among the MENs especially under CT wheat land use, and therefore relative modularity (RM) was calculated. RM is considered to be more meaningful for comparing modular structures across different networks by measuring how modular a network is compared to the mean expected modularity [22, 88]. RM was calculated as the ratio of the difference between the modularity of an empirical network and the mean of modularity from the random networks over the mean of modularity from the random network [88].
Each node was grouped into a topological role in the network based on its within-module connectivity (Zi) and among-module connectivity (Pi) [89]. As used in previous studies [20, 22, 90], nodes were classified as network hubs (highly connected nodes within the entire network, Zi > 2.5 and Pi > 0.62), module hubs (highly connected nodes within the modules, Zi > 2.5 and Pi <≤ 0.62), connectors (nodes that connect the modules, Zi ≤ 2.5 and Pi > 0.62), and peripherals (nodes connected in the modules with few links, Zi ≤ 2.5 and Pi ≤ 0.62). Module hubs, connectors, and network hubs are referred to as keystone nodes [40, 91]
Statistical analyses for network complexity comparisons
To evaluate the differences of MENs over time in both land uses, the 22 topological indices calculated for each empirical MEN were used for principal component analysis using the ‘prcomp’ function in the stats package in R [92]. The overall differences of network topological properties between land uses were compared using a Mann–Whitney U test in the stats package in R [92]. To examine differences in module composition, Fischer’s exact test was performed to identify preserved module pairs in networks [22] (1) under CT wheat land use or native TGP land use over time and (2) between CT wheat and native TGP land use. P-values from the exact tests were adjusted using the Bonferroni procedure within each network. In short, if two modules in different networks consisted of large proportions of shared nodes (adjusted p ≤ 0.05), they were considered preserved module pairs [20]. The exact tests were performed in R with the ‘fisher.test’ function in the stats package, and p-value adjustment was done with the ‘p.adjust’ function in the stats package in R [92].
In order to assess the differences of the networked communities under CT wheat and native TGP land use, three non-parametric multivariate analyses of dissimilarity were performed including MRPP, ANOSIM, and Adonis based on Bray-Curtis distance using the R package ‘vegan’ [93] and visualized using principal coordinate analysis (PCoA). Mantel tests were also performed between networked community structures and soil and climate variables using the R package ‘vegan’ [93]. The taxonomic composition of the networked communities under CT wheat and native TGP land use was analyzed at the phylum and class levels. Mann–Whitney U tests were used to evaluate the changes in the average relative abundance of each taxon due to land conversion.
Network stability analyses
To determine whether and how land use conversion affects the stability of the constructed MENs, several indices were used to characterize network stability. Detailed descriptions of the calculations can be found in Supplementary Table 8.
Network stability based on simulation includes robustness and vulnerability. The robustness of a MEN is defined as the proportion of the remaining species in the network after random or targeted node removal [46, 94]. For simulations of random removal, robustness was measured when 50% of random nodes were removed from each MEN. For simulations of targeted removal, robustness was compared when five module hubs were removed and when half of the module hubs were removed since the number of module hubs differed greatly between networks. Vulnerability of each node measures the relative contribution of the node to the global efficiency. The vulnerability of a network is indicated by the maximal vulnerability of nodes in the network [17] and the global efficiency of a graph was calculated as the average of the efficiencies over all pairs of nodes. In ecological networks, efficiency explains the ability to spread information within a network and is important to determine how quickly the effect of biological/ecological events spread to parts or the entire network [22].
Network stability based on empirical data includes node constancy, link constancy, node overlap, node persistence, and compositional stability. Constancy measures the temporal stability of species. It is defined as μ/σ, where μ is the mean of abundance over time and σ is the standard deviation [95]. The constancy of node i was calculated as μi/σi. The abundance of species i at a certain time point was positive only if species i was in the MEN at that time point. Otherwise, the abundance of species i was considered zero for that time point and removed from subsequent analyses. The average of all the node constancy values was reported. Similar procedure was used to calculate link constancy. We let lij+ = 1 if nodes i and j were positively linked in a network, lij- = 1 if nodes i and j were negatively linked in a network, and lij+ = lij- = 0 if there was no link between i and j [22]. Again, nonfinite values were removed from subsequent analyses. The average of all the link constancy values was reported. The number of overlapping nodes among multiple networks was calculated following previous methods by Hui et al. [96] where the higher numbers of overlapping nodes among networks indicated slower turnover of species composition in the networks with time points being referred to as “orders” [22]. The node persistence is defined as the proportion of coexisting species (over the total number of species) at an ecological regime [57]. Node persistence was calculated as the percentage of nodes present in the network in consecutive monthly comparisons. The compositional stability evaluates the change in community structure over time [48]. The compositional stability for the networked microbial communities was calculated using the sample × ASV matrix. If community structure does not change, the stability index is equal to 1; while if community structure is completely different among time points if stability index is 0. Stability was addressed as consecutive monthly comparisons as done with node persistence.
The overall differences in stability indices between CT wheat and native TGP land use were determined using Mann–Whitney U test. The relationship between node persistence and compositional stability for each land use was tested based on Spearman correlation. Spearman correlations were also used to associate soil properties, climate variables, and management input with network stability and complexity indices. The correlations with management data were calculated using Spearman’s generalized equation due to repetitive values in the coded management data. Structural equation modeling (SEM) was also performed to further discern the environmental drivers on network stability and complexity using the ‘lavaan’ R package. All reasonable pathways were included then non-significant pathways were sequentially eliminated unless the pathways were biologically informative. Pathways were added based on the residual correlations. The procedure was repeated until the model showed sufficient fitting with the p values of χ2 test larger than 0.05 and the root mean square error of approximation (RMSE) < 0.08 [97].
Supplementary information
Acknowledgements
This study was funded by the USDA National Institute of Food and Agriculture (NIFA) award 2016-68002-24967. It was also supported in part by the USDA-ARS Office of National Programs (Project number: 3070-21610-003-00D) and USDA-LTAR (Long-Term Agroecosystem Research) Network. USDA is an equal opportunity provider and employer. Computing for this project was performed at the OU Supercomputing Center for Education & Research (OSCER) at the University of Oklahoma (OU).
Author contributions
JZ and XX designed research; CRC collected soil samples; PW provided site data; CRC and YZ performed research; CRC, YZ, DN, and NX analyzed data; CRC and YZ wrote paper; DN, NX, PW, JZ, and XX reviewed and edited paper.
Code availability
16S rRNA gene sequences were deposited to the Sequence Read Archive (SRA) under the project accession number PRJNA954023. The R scripts and Python 3 scripts were adapted from the publicly available code on GitHub at https://github.com/Mengting-Maggie-Yuan/warming-network-complexity-stability with the identifier 10.5281/zenodo.4383469.
Competing interests
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Carolyn R. Cornell, Ya Zhang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01521-x.
References
- 1.Verchot LV. Impacts of forest conversion to agriculture on microbial communities and microbial function. In: Dion P, editor. Soil Biology and Agriculture in the Tropics. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. pp. 45–63. [Google Scholar]
- 2.Shaoqiang W, Jiyuan L, Guirui Y, Yuanyuan P, Qingmei C, Kerang L, et al. Effects of land use change On the storage of soil organic carbon: A case study of the Qianyanzhou forest experimental station in China. Clim Change. 2004;67:247–55. doi: 10.1007/s10584-004-2847-1. [DOI] [Google Scholar]
- 3.Lark TJ, Larson B, Schelly I, Batish S, Gibbs HK. Accelerated conversion of native prairie to cropland in Minnesota. Environ Conserv. 2019;46:155–62. doi: 10.1017/S0376892918000437. [DOI] [Google Scholar]
- 4.Hoekstra JM, Boucher TM, Ricketts TH, Roberts C. Confronting a biome crisis: global disparities of habitat loss and protection. Ecol Lett. 2005;8:23–9. doi: 10.1111/j.1461-0248.2004.00686.x. [DOI] [Google Scholar]
- 5.Lark TJ, Salmon JM, Gibbs HK. Cropland expansion outpaces agricultural and biofuel policies in the United States. Environ Res Lett. 2015;10:044003. doi: 10.1088/1748-9326/10/4/044003. [DOI] [Google Scholar]
- 6.Bajgain R, Xiao X, Basara J, Wagle P, Zhou Y, Mahan H, et al. Carbon dioxide and water vapor fluxes in winter wheat and tallgrass prairie in central Oklahoma. Sci Total Environ. 2018;644:1511–24. doi: 10.1016/j.scitotenv.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Doran JW, Zeiss MR. Soil health and sustainability: managing the biotic component of soil quality. Appl Soil Ecol. 2000;15:3–11. doi: 10.1016/S0929-1393(00)00067-6. [DOI] [Google Scholar]
- 8.Trivedi P, Delgado-Baquerizo M, Anderson IC, Singh BK. Response of soil properties and microbial communities to agriculture: Implications for primary productivity and soil health indicators. Front Plant Sci. 2016;7:990. doi: 10.3389/fpls.2016.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauber CL, Strickland MS, Bradford MA, Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem. 2008;40:2407–15. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- 10.Ding GC, Piceno YM, Heuer H, Weinert N, Dohrmann AB, Carrillo A, et al. Changes of soil bacterial diversity as a consequence of agricultural land use in a semi-arid ecosystem. PLoS ONE. 2013;8:e59497. doi: 10.1371/journal.pone.0059497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishaq SL, Seipel T, Yeoman CJ, Menalled FD. Soil bacterial communities of wheat vary across the growing season and among dryland farming systems. Geoderma. 2020;358:113989. doi: 10.1016/j.geoderma.2019.113989. [DOI] [Google Scholar]
- 12.Bainard L, Hamel C, Gan Y. Edaphic properties override the influence of crops on the composition of the soil bacterial community in a semiarid agroecosystem. Appl Soil Ecol. 2016;105:160–8. doi: 10.1016/j.apsoil.2016.03.013. [DOI] [Google Scholar]
- 13.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–90. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 14.Lacerda-Júnior GV, Noronha MF, Cabral L, Delforno TP, de Sousa STP, Fernandes-Júnior PI, et al. Land use and seasonal effects on the soil microbiome of a Brazilian dry forest. Front Microbiol. 2019;10:648. doi: 10.3389/fmicb.2019.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu XB, Zhang XY, Wang YX, Sui YY, Zhang S, Herbert S, et al. Soil degradation: A problem threatening the sustainable development of agriculture in Northeast China. Plant Soil Environ. 2010;56:87–97. doi: 10.17221/155/2009-PSE. [DOI] [Google Scholar]
- 16.Six J, Elliott ET, Paustian K. Aggregate and soil organic matter dynamics under conventional and no-tillage systems. Soil Sci Soc Am J. 1999;63:1350–8. doi: 10.2136/sssaj1999.6351350x. [DOI] [Google Scholar]
- 17.Deng Y, Jiang YH, Yang Y, He Z, Luo F, Zhou J. Molecular ecological network analyses. BMC Bioinform. 2012;13:113. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. Functional molecular ecological networks. mBio. 2010;1:e00169–10. doi: 10.1128/mBio.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barberan A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–51. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Deng Y, Luo F, He Z, Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio. 2011;2:e00122–11. doi: 10.1128/mBio.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–50. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 22.Yuan MM, Guo X, Wu L, Zhang Y, Xiao N, Ning D, et al. Climate warming enhances microbial network complexity and stability. Nat Clim Change. 2021;11:343–8. doi: 10.1038/s41558-021-00989-9. [DOI] [Google Scholar]
- 23.Montoya JM, Pimm SL, Solé RV. Ecological networks and their fragility. Nature. 2006;442:259–64. doi: 10.1038/nature04927. [DOI] [PubMed] [Google Scholar]
- 24.Deng Y, Zhang P, Qin Y, Tu Q, Yang Y, He Z, et al. Network succession reveals the importance of competition in response to emulsified vegetable oil amendment for uranium bioremediation. Environ Microbiol. 2016;18:205–18. doi: 10.1111/1462-2920.12981. [DOI] [PubMed] [Google Scholar]
- 25.Wu L, Yang Y, Chen S, Zhao M, Zhu Z, Yang S, et al. Long-term successional dynamics of microbial association networks in anaerobic digestion processes. Water Res. 2016;104:1–10. doi: 10.1016/j.watres.2016.07.072. [DOI] [PubMed] [Google Scholar]
- 26.Tian J, He N, Kong W, Deng Y, Feng K, Green SM, et al. Deforestation decreases spatial turnover and alters the network interactions in soil bacterial communities. Soil Biol Biochem. 2018;123:80–6. doi: 10.1016/j.soilbio.2018.05.007. [DOI] [Google Scholar]
- 27.Jia M, Gao Z, Gu H, Zhao C, Liu M, Liu F, et al. Effects of precipitation change and nitrogen addition on the composition, diversity, and molecular ecological network of soil bacterial communities in a desert steppe. PLoS ONE. 2021;16:e0248194. doi: 10.1371/journal.pone.0248194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B-B, Roley SS, Duncan DS, Guo J, Quensen JF, Yu H-Q, et al. Long-term excess nitrogen fertilizer increases sensitivity of soil microbial community to seasonal change revealed by ecological network and metagenome analyses. Soil Biol Biochem. 2021;160:108349. doi: 10.1016/j.soilbio.2021.108349. [DOI] [Google Scholar]
- 29.Yan Y, Kuramae EE, de Hollander M, Klinkhamer PG, van Veen JA. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 2017;11:56–66. doi: 10.1038/ismej.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pocock MJO, Evans DM, Memmott J. The robustness and restoration of a network of ecological networks. Science. 2012;335:973–7. doi: 10.1126/science.1214915. [DOI] [PubMed] [Google Scholar]
- 31.Cornell CR, Zhang Y, Nostrand JDV, Wagle P, Xiao X, Zhou J. Temporal changes of virus-like particle abundance and metagenomic comparison of viral communities in cropland and prairie soils. mSphere. 2021;6:e01160–20. doi: 10.1128/mSphere.01160-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornell CR, Zhang Y, Ning D, Wu L, Wagle P, Steiner Jean L, et al. Temporal dynamics of bacterial communities along a gradient of disturbance in a U.S. southern plains agroecosystem. mBio. 2022;13:e03829–21. doi: 10.1128/mbio.03829-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–37. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 34.Galiana N, Lurgi M, Claramunt-Lopez B, Fortin MJ, Leroux S, Cazelles K, et al. The spatial scaling of species interaction networks. Nat Ecol Evol. 2018;2:782–90. doi: 10.1038/s41559-018-0517-3. [DOI] [PubMed] [Google Scholar]
- 35.Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–51. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J, Ning D. Stochastic community assembly: Does it matter in microbial ecology? Microbiol Mol Biol Rev. 2017;81:e00002–17. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanchet FG, Cazelles K, Gravel D. Co-occurrence is not evidence of ecological interactions. Ecol Lett. 2020;23:1050–63. doi: 10.1111/ele.13525. [DOI] [PubMed] [Google Scholar]
- 38.Montecchia MS, Tosi M, Soria MA, Vogrig JA, Sydorenko O, Correa OS. Pyrosequencing reveals changes in soil bacterial communities after conversion of Yungas forests to agriculture. PLoS ONE. 2015;10:e0119426. doi: 10.1371/journal.pone.0119426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauber CL, Ramirez KS, Aanderud Z, Lennon J, Fierer N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013;7:1641–50. doi: 10.1038/ismej.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee S, Schlaeppi K, van der Heijden MGA. Reply to ‘Can we predict microbial keystones?’. Nat Rev Microbiol. 2019;17:194. doi: 10.1038/s41579-018-0133-x. [DOI] [PubMed] [Google Scholar]
- 41.Tu Q, Yan Q, Deng Y, Michaletz ST, Buzzard V, Weiser MD, et al. Biogeographic patterns of microbial co-occurrence ecological networks in six American forests. Soil Biol Biochem. 2020;148:107897. doi: 10.1016/j.soilbio.2020.107897. [DOI] [Google Scholar]
- 42.Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol. 2013;11:789–99. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 43.Tu Q, Zhou X, He Z, Xue K, Wu L, Reich P, et al. The diversity and co-occurrence patterns of N2-fixing communities in a CO2-enriched grassland ecosystem. Micro Ecol. 2016;71:604–15. doi: 10.1007/s00248-015-0659-7. [DOI] [PubMed] [Google Scholar]
- 44.Zhang B, Zhang J, Liu Y, Shi P, Wei G. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol Biochem. 2018;118:178–86. doi: 10.1016/j.soilbio.2017.12.011. [DOI] [Google Scholar]
- 45.Shi S, Nuccio EE, Shi ZJ, He Z, Zhou J, Firestone MK. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol Lett. 2016;19:926–36. doi: 10.1111/ele.12630. [DOI] [PubMed] [Google Scholar]
- 46.Montesinos-Navarro A, Hiraldo F, Tella JL, Blanco G. Network structure embracing mutualism–antagonism continuums increases community robustness. Nat Ecol Evol. 2017;1:1661–9. doi: 10.1038/s41559-017-0320-6. [DOI] [PubMed] [Google Scholar]
- 47.Banerjee S, Schlaeppi K, van der Heijden MGA. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16:567–76. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 48.Zelikova TJ, Blumenthal DM, Williams DG, Souza L, LeCain DR, Morgan J, et al. Long-term exposure to elevated CO2 enhances plant community stability by suppressing dominant plant species in a mixed-grass prairie. Proc Natl Acad Sci USA. 2014;111:15456–61. doi: 10.1073/pnas.1414659111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKinney ML, Lockwood JL. Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends Ecol Evol. 1999;14:450–3. doi: 10.1016/S0169-5347(99)01679-1. [DOI] [PubMed] [Google Scholar]
- 50.Smart SM, Thompson K, Marrs RH, Le Duc MG, Maskell LC, Firbank LG. Biotic homogenization and changes in species diversity across human-modified ecosystems. Proc R Soc B: Biol Sci. 2006;273:2659–65. doi: 10.1098/rspb.2006.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodrigues JLM, Pellizari VH, Mueller R, Baek K, Jesus EDC, Paula FS, et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc Natl Acad Sci USA. 2013;110:988. doi: 10.1073/pnas.1220608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olden JD, Poff NL. Toward a mechanistic understanding and prediction of biotic homogenization. Am Nat. 2003;162:442–60. doi: 10.1086/378212. [DOI] [PubMed] [Google Scholar]
- 53.Olden JD, LeRoy Poff N, Douglas MR, Douglas ME, Fausch KD. Ecological and evolutionary consequences of biotic homogenization. Trends Ecol Evol. 2004;19:18–24. doi: 10.1016/j.tree.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 54.MacArthur R. Fluctuations of animal populations and a measure of community stability. Ecology. 1955;36:533–6. doi: 10.2307/1929601. [DOI] [Google Scholar]
- 55.May RM. Stability and complexity in model ecosystems. Princeton NJ, USA: Princeton University Press; 2001.
- 56.Okuyama T, Holland JN. Network structural properties mediate the stability of mutualistic communities. Ecol Lett. 2008;11:208–16. doi: 10.1111/j.1461-0248.2007.01137.x. [DOI] [PubMed] [Google Scholar]
- 57.Landi P, Minoarivelo HO, Brännström Å, Hui C, Dieckmann U. Complexity and stability of ecological networks: a review of the theory. Popul Ecol. 2018;60:319–45. doi: 10.1007/s10144-018-0628-3. [DOI] [Google Scholar]
- 58.Pimm SL. The complexity and stability of ecosystems. Nature. 1984;307:321–6. doi: 10.1038/307321a0. [DOI] [Google Scholar]
- 59.Helbing D. Globally networked risks and how to respond. Nature. 2013;497:51–9. doi: 10.1038/nature12047. [DOI] [PubMed] [Google Scholar]
- 60.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–31. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao M, Rui J, Niu H, Heděnec P, Li J, He Z, et al. The differentiation of soil bacterial communities along a precipitation and temperature gradient in the eastern Inner Mongolia steppe. Catena. 2017;152:47–56. doi: 10.1016/j.catena.2017.01.007. [DOI] [Google Scholar]
- 62.Thomson BC, Tisserant E, Plassart P, Uroz S, Griffiths RI, Hannula SE, et al. Soil conditions and land use intensification effects on soil microbial communities across a range of European field sites. Soil Biol Biochem. 2015;88:403–13. doi: 10.1016/j.soilbio.2015.06.012. [DOI] [Google Scholar]
- 63.Guo X, Chen HYH, Meng M, Biswas SR, Ye L, Zhang J. Effects of land use change on the composition of soil microbial communities in a managed subtropical forest. Ecol Manag. 2016;373:93–9. doi: 10.1016/j.foreco.2016.03.048. [DOI] [Google Scholar]
- 64.Vitousek PM, Howarth RW. Nitrogen limitation on land and in the sea: How can it occur? Biogeochemistry. 1991;13:87–115. doi: 10.1007/BF00002772. [DOI] [Google Scholar]
- 65.Lauenroth W, Burke I, Paruelo J. Patterns of production and precipitation-use efficiency of winter wheat and native grasslands in the central great plains of the United States. Ecosystems. 2000;3:344–51. doi: 10.1007/s100210000031. [DOI] [Google Scholar]
- 66.Chase JM. Drought mediates the importance of stochastic community assembly. Proc Natl Acad Sci USA. 2007;104:17430. doi: 10.1073/pnas.0704350104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang M, Chai L, Jiang D, Zhang M, Jia W, Huang Y, et al. Dissolved organic matter (DOM) quality drives biogeographic patterns of soil bacterial communities and their association networks in semi-arid regions. FEMS Microbiol Ecol. 2021;97:fiab083. doi: 10.1093/femsec/fiab083. [DOI] [PubMed] [Google Scholar]
- 68.Lu L, Yin S, Liu X, Zhang W, Gu T, Shen Q, et al. Fungal networks in yield-invigorating and -debilitating soils induced by prolonged potato monoculture. Soil Biol Biochem. 2013;65:186–94. doi: 10.1016/j.soilbio.2013.05.025. [DOI] [Google Scholar]
- 69.Mougi A, Kondoh M. Diversity of interaction types and ecological community stability. Science. 2012;337:349–51. doi: 10.1126/science.1220529. [DOI] [PubMed] [Google Scholar]
- 70.Kleinman PJA, Spiegal S, Rigby JR, Goslee SC, Baker JM, Bestelmeyer BT, et al. Advancing the sustainability of US agriculture through long-term research. J Environ Qual. 2018;47:1412–25. doi: 10.2134/jeq2018.05.0171. [DOI] [PubMed] [Google Scholar]
- 71.Spiegal S, Bestelmeyer B, Archer D, Augustine D, Boughton E, Boughton R, et al. Evaluating strategies for sustainable intensification of US agriculture through the Long-Term Agroecosystem Research network. Environ Res Lett. 2018;13:034031. doi: 10.1088/1748-9326/aaa779. [DOI] [Google Scholar]
- 72.Bajgain R, Xiao X, Basara J, Doughty R, Wu X, Wagle P, et al. Differential responses of native and managed prairie pastures to environmental variability and management practices. Agric Meteorol. 2020;294:108137. doi: 10.1016/j.agrformet.2020.108137. [DOI] [Google Scholar]
- 73.Peterson BL, Hanna L, Steiner JL. Reduced soil disturbance: Positive effects on greenhouse gas efflux and soil N losses in winter wheat systems of the southern plains. Soil Tillage Res. 2019;191:317–26. doi: 10.1016/j.still.2019.03.020. [DOI] [Google Scholar]
- 74.Hendershot W, Lalande H, Duquette M. Soil reaction and exchangeable acidity. In: Carter MR, Gregorich EG, editors. Soil sampling and methods of analysis. 2nd ed. Boca Raton, FL, USA: CRC Press; 2007. pp. 173–8. [Google Scholar]
- 75.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–22. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ding J, Zhang Y, Deng Y, Cong J, Lu H, Sun X, et al. Integrated metagenomics and network analysis of soil microbial community of the forest timberline. Sci Rep. 2015;5:7994. doi: 10.1038/srep07994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 78.Edgar RC. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv. 2016:081257.
- 79.Prodan A, Tremaroli V, Brolin H, Zwinderman AH, Nieuwdorp M, Levin E. Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS ONE. 2020;15:e0227434. doi: 10.1371/journal.pone.0227434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carr A, Diener C, Baliga NS, Gibbons SM. Use and abuse of correlation analyses in microbial ecology. ISME J. 2019;13:2647–55. doi: 10.1038/s41396-019-0459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aitchison J. Principles of Compositional Data Analysis. Lecture Notes-Monograph Series. 24: Institute of Mathematical Statistics; 1994.24:73–81.
- 82.Sabri H, Hashemi SS, Maleki BR, Jafarizadeh MA. Generalization of Brody distribution for statistical investigation. Random Matricies: Theory Appl. 2014;03:1450017. doi: 10.1142/S2010326314500178. [DOI] [Google Scholar]
- 83.Xiao N, Zhou A, Kempher M, Zhou B, Shi Z, Yuan M, et al. Disentangling direct from indirect relationships in association networks. Proc Natl Acad Sci USA. 2022;119:e2109995119. doi: 10.1073/pnas.2109995119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Ocean plankton. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 85.Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. mBio. 2015;6:e02288–14. doi: 10.1128/mBio.02288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–3. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 87.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thebault E, Fontaine C. Stability of ecological communities and the architecture of mutualistic and trophic networks. Science. 2010;329:853–6. doi: 10.1126/science.1188321. [DOI] [PubMed] [Google Scholar]
- 89.Guimera R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olesen JM, Bascompte J, Dupont YL, Jordano P. The modularity of pollination networks. Proc Natl Acad Sci USA. 2007;104:19891. doi: 10.1073/pnas.0706375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rottjers L, Faust K. Can we predict keystones? Nat Rev Microbiol. 2019;17:193. doi: 10.1038/s41579-018-0132-y. [DOI] [PubMed] [Google Scholar]
- 92.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 93.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’Hara RB, et al. Vegan: Community ecology package. R Package Version. 2019;2:5–6. [Google Scholar]
- 94.Dunne JA, Williams RJ, Martinez ND. Food-web structure and network theory: The role of connectance and size. Proc Natl Acad Sci USA. 2002;99:12917–22. doi: 10.1073/pnas.192407699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hautier Y, Seabloom EW, Borer ET, Adler PB, Harpole WS, Hillebrand H, et al. Eutrophication weakens stabilizing effects of diversity in natural grasslands. Nature. 2014;508:521–5. doi: 10.1038/nature13014. [DOI] [PubMed] [Google Scholar]
- 96.Hui C, McGeoch MA, Susan H, Judith LB. Zeta diversity as a concept and metric that unifies incidence-based biodiversity patterns. Am Nat. 2014;184:684–94. doi: 10.1086/678125. [DOI] [PubMed] [Google Scholar]
- 97.Rosseel Y. lavaan: an R package for structural equation modeling. J Stat Softw. 2012;48:1–36. doi: 10.18637/jss.v048.i02. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene sequences were deposited to the Sequence Read Archive (SRA) under the project accession number PRJNA954023. The R scripts and Python 3 scripts were adapted from the publicly available code on GitHub at https://github.com/Mengting-Maggie-Yuan/warming-network-complexity-stability with the identifier 10.5281/zenodo.4383469.