Abstract
Despite the significant role of the gut microbiota in infant health and development, little is known about the ecological processes determining gut microbial community assembly. According to ecology theory, the timing and order of arrival of microbial species into an ecosystem affect microbial community assembly, a phenomenon termed priority effects. Bifidobacterium species are recognized as highly abundant early colonizers of the infant’s gut, partly due to their ability to selectively utilize human milk oligosaccharides (HMOs) from breast milk. However, the role of priority effects in Bifidobacterium community assembly remains unclear. Here, we investigated the Bifidobacterium community assembly in the gut of 25 breastfed Danish infants longitudinally sampled throughout the first 6 months of life. Our results showed that the breastfed infants were often initially, but temporarily, dominated by suboptimal HMO-utilizing Bifidobacterium taxa, such as B. longum subsp. longum, before more efficient HMO-utilizers such as B. longum subsp. infantis, replaced the first colonizer as the dominant Bifidobacterium taxon. Subsequently, we validated this observation using gnotobiotic mice sequentially colonized with B. longum subsp. longum and B. longum subsp. infantis or vice versa, with or without supplementation of HMOs in the drinking water. The results showed that in the absence of HMOs, order of arrival determined dominance. Yet, when mice were supplemented with HMOs the strength of priority effects diminished, and B. longum subsp. infantis dominated regardless of colonization order. Our data demonstrate that the arrival order of Bifidobacterium taxa and the deterministic force of breast milk-derived HMOs, dictate Bifidobacterium community assembly in the infant’s gut.
Subject terms: Ecosystem ecology, Microbial communities, Next-generation sequencing, Bacteriology
Inadequate development of the infant gut microbiota has been linked with multiple adverse conditions such as asthma and allergies [1, 2], autoimmune diseases [3, 4], inflammatory bowel diseases [5], and poor neurological development and growth [6]. However, our understanding of the processes that govern microbial community assembly in the gut during infancy remains incomplete [7]. According to ecological theory, timing and order of arrival of species into a specific ecosystem influence the composition and function of that particular community [8]. This phenomenon is known as “priority effects”, and its potential significance in the gut microbiota assembly process during early life has reached considerable attention in recent years [7, 9, 10]. Priority effects occur in the gut when initial colonizing species either pre-empt or modify the ecological niche, resulting in either inhibition or facilitation of later arriving species [7]. Several conditions make strong priority effects more likely to occur [11]. Priority effects are favoured when the early-arriving species exhibit a large effect on the environment (e.g. early-arriving species deplete the niche of specific nutrients), when late-arriving species have high environmental requirements (e.g. late-arriving species have high demands for survival and growth in the niche), and when early-arriving and late-arriving species exhibit a high niche overlap (e.g. closely related taxa and/or taxa competing for the same resources in the niche). Priority effects have been demonstrated in gnotobiotic mouse models with selected Bacteroides species [12] or mouse faecal communities [9], and in hospital-associated preterm infants [13] with limited microbial exposure, confined to mainly skin and hospital-associated microbes. However, the previously studied species-interactions may not necessarily reflect interactions in the gut of the healthy term infant, which is often dominated by Bifidobacterium species [14]. The role of priority effects in Bifidobacterium species community assembly has to our knowledge only been explored in vitro using batch fermentations [15], and has not been validated in continuous culture systems or animal models. Nor has infant gut microbiota data with sufficient longitudinal sampling and taxonomical resolution been applied to understand priority effects in the context of the term infant´s gut. In theory, priority effects should be affected by community assembly principles such as selection [7]. Breastfeeding is one of the strongest deterministic factors for infant gut microbiota species selection [16–18], in part due to its high content of human milk oligosaccharides (HMOs), which are selectively consumed in the infant gut by key members of the Bifidobacterium genus [19]. Especially B. longum subsp. infantis is an efficient HMO-consuming species associated with health benefits for the infant [20–22]. However, the importance of priority effects in Bifidobacterium community assembly, as well as the potential deterministic role of HMOs in this process both remain elusive.
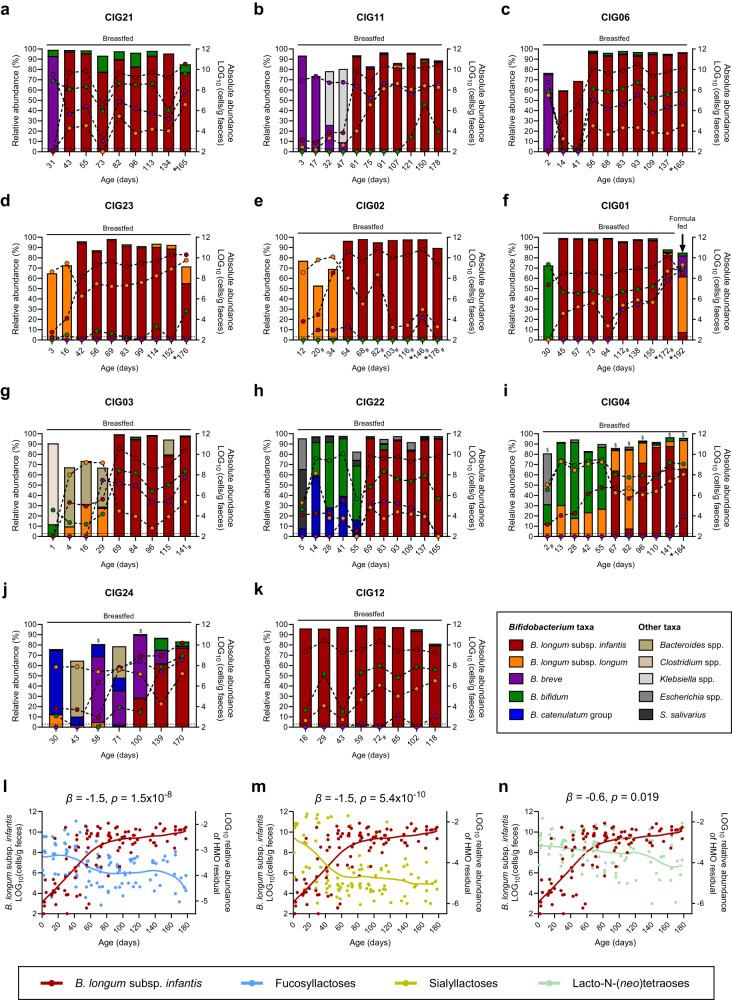
To study Bifidobacterium community assembly, we took advantage of our previously established infant cohort [23], Copenhagen Infant Gut (CIG) comprising 25 healthy breast- and mixed fed infants longitudinally sampled from birth until 6 months of age (9–11 samplings per infant). We previously analysed the gut microbiota by 16 S rRNA gene amplicon sequencing and found, in accordance with others [24, 25], that Bifidobacterium was by far the most abundant genus (64.3% of all reads), representing mainly B. longum (38.5%), B. breve (9.1%), and B. bifidum (8.0%) as the three most abundant taxa in the dataset [23]. By use of species/subspecies specific qPCR, we further quantified the absolute abundance of B. breve, B. bifidum, B. longum subsp. longum, and B. longum subsp. infantis [23]. All infants were breastfed for at least four months, except CIG15, who shifted to formula milk around one month of age. Three infants received a single course of oral antibiotics (CIG05, CIG10 and CIG16), two infants were born pre-term by C-section (CIG08 and CIG09), and three infants displayed poor colonization (CIG07, CIG18 and CIG19) with these Bifidobacterium species (Supplementary Table 1) [23]. Given that antibiotics [26], pre-term birth [27], and high degree of formula exposure [28] can disrupt infant gut microbiota assembly, we did not consider these further in our analyses. We focused on the remaining 16 infants that were all breastfed, term and vaginally born and studied the longitudinal dynamics of Bifidobacterium species with respect to priority effects within each infant. We noticed that the aforementioned dominant Bifidobacterium species exhibited different abundance patterns across individuals (Fig. 1 and Supplementary Fig. 1). We found that B. longum subsp. infantis ended up vastly dominating the gut microbiota over time in 11 of the 16 infants (44% of the whole cohort), reaching a relative abundance of 84.9 ± 12.8% (mean ± s.d.) after efficiently colonizing these infants somewhere between age 14 to 100 days depending on the individual (Fig. 1). Apart from one infant that was solely dominated by B. longum subsp. infantis throughout the entire sampling period, almost all of the infants were initially dominated by other Bifidobacterium species or combinations thereof. Some infants were initially dominated by B. breve (Fig. 1a–c), others were dominated by B. longum subsp. longum (Fig. 1d, e), B. bifidum (Fig. 1f), or combinations of B. longum subsp. longum, B. bifidum, B. breve, B. catenulatum group, and Bacteroides species (Fig. 1g–j). Evidently, in the very first sample from these infants (age range 1–31 days), B. longum subsp. infantis was either not detected (below limit of detection; 2 × 102 cells/g faeces), as for CIG03, CIG06 and CIG21, or present in counts below 104 cells/g faeces as for CIG02, CIG04, CIG11, CIG22, CIG24 and CIG23. One exception was CIG01, where B. longum subsp. infantis reached above 107 cells/g in the first sample (day 30), yet it was 100 fold lower in abundance compared to the dominating species B. bifidum. Thus, these data show that other Bifidobacterium species often colonize the breastfed infant’s gut before B. longum subsp. infantis, enabling them to initially dominate the community. However, this dominance is only transient as B. longum subsp. infantis eventually takes over. We also found that while CIG01 stopped being breastfed before the last sampling, B. longum subsp. infantis suddenly no longer dominated the community and other Bifidobacterium species reached higher absolute abundances (Fig. 1f), suggesting that B. longum subsp. infantis only dominates the community as long as the infant is primarily breastfed. In the case of CIG12, B. longum subsp. infantis consistently dominated throughout the entire sampling period (Fig. 1k), suggesting that this subspecies was the first Bifidobacterium colonizer in this particular infant. However, we cannot exclude the possibility that other Bifidobacterium species actually dominated before the first sample was collected (day 16), as seen with B. breve in CIG06 (Fig. 1c). Finally, we never detected B. longum subsp. infantis in two infants (CIG17 and CIG25), which were instead colonized mainly by B. longum subsp. longum alone or in combination with B. breve and B. bifidum (Supplementary Fig. 1a, b).
Fig. 1. B. longum subsp. infantis eventually dominates the breastfed infant’s gut due to efficient utilization of human milk oligosaccharides.
a–k Longitudinal relative abundance (bars) and absolute abundance (dots connected by dashed lines) of the major bacterial taxa detected in faeces of eleven full term, vaginally delivered, antibiotics naïve, breastfed infants from the Copenhagen Infant Gut cohort, as measured by 16 S rRNA gene amplicon sequencing (bars) and qPCR (dots), respectively. Only the abundant Bifidobacterium species, B. longum subsp. longum, B. longum subsp. infantis, B. breve and B. bifidum were quantified by qPCR. Dashed line illustrate the limit of detection (LOD) of the qPCR assay. *time points where solid foods have been consumed. #time points where breastfeeding was supplemented with formula milk. $Inconsistency between qPCR and 16 S rRNA gene amplicon sequence data regarding the dominant Bifidobacterium species. l, m, n Longitudinal absolute abundance of B. longum subsp. infantis and residual HMOs in faeces across all eleven infants displayed in panel (a–k) (the last sample from CIG01 was excluded due to cessation of breastfeeding). Statistical significance of the associations between B. longum subsp. infantis and (l) Fucosyllactoses, (m) Sialyllactoses and (n) Lacto-N-(neo)tetraoses were evaluated by linear mixed models, with β denoting the subject adjusted association coefficient. Locally weighted scatterplot smoothing (LOWESS) curves were fitted to the data points.
We then sought to couple these observations with in vivo HMO-utilization. We compared absolute abundance of B. longum subsp. infantis with the residual faecal levels of the major HMO structures (i.e. fucosyllactoses (2’FL and 3FL), sialyllactoses (3’SL and 6’SL), and lacto-N-(neo)tetraoses (LNT and LNnT)) in the eleven infants with dominant B. longum subsp. infantis colonization (Fig. 1a–k). The obvious bloom of B. longum subsp. infantis occurring within the first months of life in these infants coincided with a rapid decline in the faecal residuals of fucosyllactoses and sialyllactoses, whereas lacto-N-(neo)tetraoses were less prominently affected (Fig. 1l–n). These associations withstood adjustment of infant age and were, except for weaker associations with B. bifidum, not observed for the other Bifidobacterium species (Supplementary Table 2). This is largely consistent with the broad and efficient HMO-utilization capability of B. longum subsp. infantis strains enabling them to consume a range of fucosylated and sialylated HMOs [29–31], a more variable and less efficient HMO-utilization among B. bifidum strains [31, 32], and a limited capacity in B. breve and B. longum subsp. longum strains often confined to LNT and LNnT [33, 34]. However, in three infants (CIG13, CIG14 and CIG20), we did not observe a bloom in B. longum subsp. infantis over time, even though this subspecies was detected in low quantities throughout the sampling period. Instead, these infants became dominantly colonized with a combination of B. breve and B. bifidum (Supplementary Fig. 2c–e), which are known to be capable of cross-feeding HMOs efficiently [35, 36]. We speculate that this combination hindered B. longum subsp. infantis from taking over in these infants and/or that the B. longum subsp. infantis strains in these infants lack key genes to metabolize HMOs [37]. Indeed, in these three infants, faecal residuals of HMOs correlated negatively with the abundance of B. breve/B. bifidum and not B. longum subsp. infantis (Supplementary Table 2). Together, these data suggest that the initial Bifidobacterium colonizers are able to temporarily benefit from priority effects, but the strength of these priority effects are over time modified by breastfeeding selecting for efficient HMO-utilizers. Our data suggest that this is most commonly B. longum subsp. infantis, but it might also be other taxa or combinations thereof, as indicated in the three infants eventually dominated by B. breve/B. bifidum.
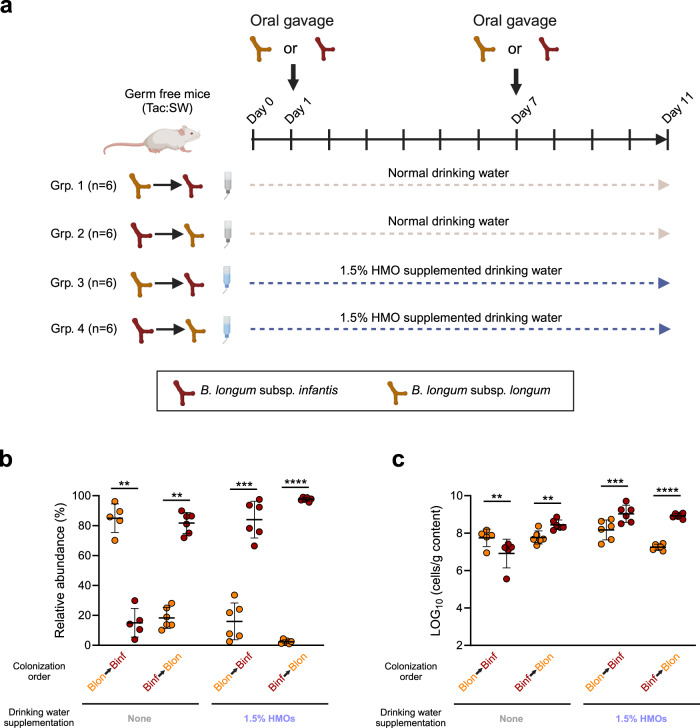
To demonstrate priority effects and validate the observations in our cohort with respect to key Bifidobacterium members, we designed an animal experiment, using sequential inoculation of the type strains of B. longum subsp. infantis DSM 20088 and B. longum subsp. longum DSM 20219 into germ free (GF) mice with or without supplying a 1.5% (w/v) mixture of HMOs (an equal mixture of 2’FL, 3FL, LNT, LNnT, 3’SL and 6’SL) into the drinking water as model of breastfeeding (Fig. 2a). We chose these strains given our observations of shifts in dominance between exactly these two subspecies within multiple infants (Fig. 1d, e, g, i), and since it is common that infants are initially colonized with maternal (vertically acquired) B. longum subsp. longum strains [38], but later colonized preferentially with (horizontally acquired) B. longum subsp. infantis strains [39]. Furthermore, a higher niche overlap would be expected between closely related taxa, such as strains of same species, facilitating priority effects. However, since B. longum subsp. longum versus B. longum subsp. infantis strains generally differ substantially in their HMO-degradation profiles [30], we would expect to be able to modify the strength of priority effects in the presence of HMOs. We first confirmed the differences in HMO-utilization between the type strains of these two subspecies by culturing them in vitro individually on 2’FL, 3FL, LNT, LNnT, 3’SL and 6’SL (Supplementary Fig. 2). B. longum subsp. longum DSM 20219 grew mainly on LNT and LNnT, whereas B. longum subsp. infantis DSM 20088 grew well on all six HMOs, which is highly consistent with the general picture of HMO-utilization reported on infant isolates of the two subspecies [31, 34]. Nonetheless, since it has been reported that a fraction of B. longum subsp. longum strains isolated from breastfed infants are capable of efficiently utilizing fucosyllactoses [31, 40], we cannot rule out that B. longum subsp. longum strains with HMO-utilization capability superior to the type strain may exist in our cohort. However, we did not find any significant associations between abundance of B. longum subsp. longum and faecal residuals of fucosyllactoses in the 11 B. longum subsp. infantis dominated infants (Fig. 1 and Supplementary Table 2). Together, this suggest that the HMO-utilization profiles of the type strains are well representing HMO-degradation capabilities of the strains found in the CIG infants.
Fig. 2. The strength of priority effects in the mouse gut are modified by human milk oligosaccharide supplementation.
a Experimental design of the study. Germ free mice, consuming either normal drinking water or drinking water supplemented with HMOs, were sequentially colonized with B. longum subsp. infantis DSM 20088 and B. longum subsp. longum DSM 20219 or vice versa at day 1 and day 7 and caecal contents were sampled after euthanization at day 11. b, c Relative and absolute abundances of B. longum subsp. infantis DSM 20088 and B. longum subsp. longum DSM 20219 in caecal contents of the mice at day 11, quantified by subspecies-specific qPCR. Relative abundances were calculated by dividing the qPCR estimated counts of the given subspecies by the sum of the counts of the two and multiplying with hundred. Data represent mean ± s.d. and statistical significance was evaluated by two tailed paired T-tests. Data from one mouse in group 1 was excluded due to very poor B. longum subsp. longum colonization (counts < LLOQ of 20 copies per reaction in the qPCR assay).
We then divided twenty-four GF mice into four groups and kept them on drinking water with or without HMOs, starting 24 h before oral gavage with approximately 5 × 108 CFU/ml of either B. longum subsp. longum DSM 20219 or B. longum subsp. infantis DSM 20088 on day 1. The mice were subsequently orally gavaged with approximately 5 × 108 CFU/ml of the other strain on day 7 and euthanized on day 11 (Fig. 2a). Clear priority effects were observed when the mice consumed normal drinking water, with the order of arrival determining the relative and absolute abundance of the two species in caecal contents on day 11 (Fig. 2b, c). In contrast, when the mice consumed the HMO-supplemented drinking water, B. longum subsp. infantis dominated regardless of the colonization order. Notably, the dominance of B. longum subsp. infantis was most pronounced when it colonized first (Fig. 2b, c). These data demonstrate priority effects in vivo using closely related Bifidobacterium subspecies highly prevalent in the infant gut microbiota and show that the consumption of HMOs can strongly modify the strength of priority effects in an experimental setting, explaining the observations from our infant cohort.
Our results has important conceptual consequences for the way we view infant gut microbiota assembly as it underlines the importance of both order of colonization, and breastfeeding (HMOs) as a selective force. Considering that efficient HMO-utilizers such as B. longum subsp. infantis have been associated to lower prevalence of atopic disease [20], less gut inflammation [41], improved immune system development [22], and improved growth among malnourished infants [21], our insights emphasise the role of breastfeeding to modify the strength of priority effects in early life. Although B. longum subsp. infantis was highly abundant in 11 out of 25 of the studied Danish infants, this subspecies has been reported to be present at a very low prevalence, even in breastfed infants, in many Western populations, raising concerns about its possible extinction [39, 42]. However, clinical trials with oral supplementation of B. longum subsp. infantis EVC001 in breastfed neonates have demonstrated abundant and persistent colonization of this strain [43, 44]. Thus, in light of our observations, promoting and supporting breastfeeding remains a key priority, which may be complemented with early probiotic administration to ensure prominent gut colonization with key Bifidobacterium taxa during early infancy.
Supplementary information
Acknowledgements
We thank the children and families participating in the CIG cohort. We thank Marlene Danner Dalgaard at the Technical University of Denmark in-house facility (DTU Multi-Assay Core, DMAC) for performing the 16 S rRNA gene sequencing, and Glycom A/S for kindly donating the HMOs. This work was supported by Augustinus Fonden (grant no. 17-2003 to HMR), Hørslev Fonden (grant no. 203866 to HMR), Beckett Fonden (grant no. 17-2-0551 to HMR) Ejnar og Aase Danielsens Fond (grant no. 10-002019 to HMR), the Independent Research Fund Denmark (MOTILITY; grant no. 0171-00006B to HMR), and the Novo Nordisk Foundation (NNF19OC0056246; PRIMA—toward Personalized dietary Recommendations based on the Interaction between diet, Microbiome and Abiotic conditions in the gut).
Author contributions
HMR and MFL conceived and designed the CIG cohort. MFL prepared the samples for sequencing/qPCR and analyzed the sequencing/qPCR data. HMR prepared the samples for faecal metabolome analyses and performed the LC-MS to measure HMO residuals. MFL cultured Bifidobacterium species and assessed growth on HMOs. MFL designed and performed the animal experiment and analysed the data. Both authors contributed to the writing of the manuscript.
Funding
Open access funding provided by Technical University of Denmark.
Data availability
16 S rRNA gene amplicon sequencing data has been deposited in the Sequence Read Archive (SRA) under BioProject PRJNA554596.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Martin F. Laursen, Email: mfrla@food.dtu.dk
Henrik M. Roager, Email: hero@nexs.ku.dk
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01525-7.
References
- 1.Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. 2018;9:1–10. doi: 10.1038/s41467-017-02573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–91. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vatanen T, Kostic AD, D’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–53. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562:589–94. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres J, Hu J, Seki A, Eisele C, Nair N, Huang R, et al. Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut. 2020;69:42–51. doi: 10.1136/gutjnl-2018-317855. [DOI] [PubMed] [Google Scholar]
- 6.Gehrig JL, Venkatesh S, Chang HW, Hibberd MC, Kung VL, Cheng J, et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019;365:1–12. doi: 10.1126/science.aau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018;15:197–205. doi: 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukami T. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst. 2015;46:1–23. doi: 10.1146/annurev-ecolsys-110411-160340. [DOI] [Google Scholar]
- 9.Martínez I, Maldonado-Gomez MX, Gomes-Neto JC, Kittana H, Ding H, Schmaltz R, et al. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. Elife. 2018;7:1–26. doi: 10.7554/eLife.36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debray R, Herbert RA, Jaffe AL, Crits-Christoph A, Power ME, Koskella B. Priority effects in microbiome assembly. Nat Rev Microbiol. 2021;20:109–21. doi: 10.1038/s41579-021-00604-w. [DOI] [PubMed] [Google Scholar]
- 11.Vannette RL, Fukami T. Historical contingency in species interactions: towards niche-based predictions. Ecol Lett. 2014;17:115–24. doi: 10.1111/ele.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–9. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR, Rakoff-Nahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021;591:633–8. doi: 10.1038/s41586-021-03241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laursen MF, Bahl MI, Michaelsen KF, Licht TR. First foods and gut microbes. Front Microbiol. 2017;8:1–8. doi: 10.3389/fmicb.2017.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ojima MN, Jiang L, Arzamasov AA, Yoshida K, Odamaki T, Xiao J, et al. Priority effects shape the structure of infant-type Bifidobacterium communities on human milk oligosaccharides. ISME J. 2022;16:2265–79. doi: 10.1038/s41396-022-01270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–8. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galazzo G, van Best N, Bervoets L, Dapaah IO, Savelkoul PH, Hornef MW, et al. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology. 2020;158:1584–96. doi: 10.1053/j.gastro.2020.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Laursen MF, Pekmez CT, Larsson MW, Lind MV, Yonemitsu C, Larnkjær A, et al. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME Commun. 2021;1:1–13. doi: 10.1038/s43705-021-00021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao JZ, et al. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: Prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. 2020;12:1–21. doi: 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seppo AE, Bu K, Jumabaeva M, Thakar J, Choudhury RA, Yonemitsu C, et al. Infant gut microbiome is enriched with Bifidobacterium longum ssp. infantis in Old Order Mennonites with traditional farming lifestyle. Allergy. 2021;76:3489–503. doi: 10.1111/all.14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barratt MJ, Nuzhat S, Ahsan K, Frese SA, Arzamasov AA, Sarker SA, et al. Bifidobacterium infantis treatment promotes weight gain in Bangladeshi infants with severe acute malnutrition. Sci Transl Med. 2022;14:1–17. doi: 10.1126/scitranslmed.abk1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884–98. doi: 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 23.Laursen MF, Sakanaka M, von Burg N, Mörbe U, Andersen D, Moll JM, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol. 2021;6:1367–82. doi: 10.1038/s41564-021-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukuda N, Yahagi K, Hara T, Watanabe Y, Matsumoto H, Mori H, et al. Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J. 2021;15:2574–90. doi: 10.1038/s41396-021-00937-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012;56:5811–20. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forsgren M, Isolauri E, Salminen S, Rautava S. Late preterm birth has direct and indirect effects on infant gut microbiota development during the first six months of life. Acta Paediatr. 2017;106:1103–9. doi: 10.1111/apa.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 29.LoCascio RG, Niñonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, et al. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Micro Biotechnol. 2009;2:333–42. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76:7373–81. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB, Mills DA. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep. 2015;5:1–18. doi: 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotoh A, Katoh T, Sakanaka M, Ling Y, Yamada C, Asakuma S, et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep. 2018;8:1–14. doi: 10.1038/s41598-018-32080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz-Moyano S, Totten SM, Garrido DA, Smilowitz JT, German JB, Lebrilla CB, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol. 2013;79:6040–9. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, et al. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep. 2016;6:1–18. doi: 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egan M, O’Connell Motherway M, Kilcoyne M, Kane M, Joshi L, Ventura M, et al. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014;14:1–14. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Centanni M, Ferguson SA, Sims IM, Biswas A, Tannocka GW. Bifidobacterium bifidum ATCC 15696 and Bifidobacterium breve 24b metabolic interaction based on 2’-O-Fucosyl-lactose studied in steady-state cultures in a freter-style chemostat. Appl Environ Microbiol. 2019;85:1–17. doi: 10.1128/AEM.02783-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duar RM, Casaburi G, Mitchell RD, Scofield LNC, Ortega Ramirez CA, Barile D, et al. Comparative genome analysis of Bifidobacterium longum subsp. infantis strains reveals variation in human milk oligosaccharide utilization genes among commercial probiotics. Nutrients. 2020;12:1–22. doi: 10.3390/nu12113247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makino H, Kushiro A, Ishikawa E, Muylaert D, Kubota H, Sakai T, et al. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl Environ Microbiol. 2011;77:6788–93. doi: 10.1128/AEM.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taft DH, Lewis ZT, Nguyen N, Ho S, Masarweh C, Dunne-Castagna V, et al. Bifidobacterium species colonization in infancy: a global cross-sectional comparison by population history of breastfeeding. Nutrients. 2022;14:1–20. doi: 10.3390/nu14071423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawson MAE, O’Neill IJ, Kujawska M, Gowrinadh Javvadi S, Wijeyesekera A, Flegg Z, et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020;14:635–48. doi: 10.1038/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henrick BM, Chew S, Casaburi G, Brown HK, Frese SA, Zhou Y, et al. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr Res. 2019;86:749–57. doi: 10.1038/s41390-019-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casaburi G, Duar RM, Brown H, Mitchell RD, Kazi S, Chew S, et al. Metagenomic insights of the infant microbiome community structure and function across multiple sites in the United States. Sci Rep. 2021;11:1472. doi: 10.1038/s41598-020-80583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. 2017;2:1–15. doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen M, Holdbrooks H, Mishra P, Abrantes MA, Eskew S, Garma M, et al. Impact of probiotic B. infantis EVC001 feeding in premature infants on the gut microbiome, nosocomially acquired antibiotic resistance, and enteric inflammation. Front Pediatr. 2021;9:1–19. doi: 10.3389/fped.2021.618009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16 S rRNA gene amplicon sequencing data has been deposited in the Sequence Read Archive (SRA) under BioProject PRJNA554596.