Abstract
Genetic variants conferring risks for Parkinson’s disease have been highlighted through genome-wide association studies, yet exploration of their specific disease mechanisms is lacking. Two Parkinson’s disease candidate genes, KAT8 and KANSL1, identified through genome-wide studies and a PINK1-mitophagy screen, encode part of the histone acetylating non-specific lethal complex. This complex localizes to the nucleus, where it plays a role in transcriptional activation, and to mitochondria, where it has been suggested to have a role in mitochondrial transcription. In this study, we sought to identify whether the non-specific lethal complex has potential regulatory relationships with other genes associated with Parkinson’s disease in human brain.
Correlation in the expression of non-specific lethal genes and Parkinson’s disease-associated genes was investigated in primary gene co-expression networks using publicly-available transcriptomic data from multiple brain regions (provided by the Genotype-Tissue Expression Consortium and UK Brain Expression Consortium), whilst secondary networks were used to examine cell type specificity. Reverse engineering of gene regulatory networks generated regulons of the complex, which were tested for heritability using stratified linkage disequilibrium score regression. Prioritized gene targets were then validated in vitro using a QuantiGene multiplex assay and publicly-available chromatin immunoprecipitation-sequencing data.
Significant clustering of non-specific lethal genes was revealed alongside Parkinson’s disease-associated genes in frontal cortex primary co-expression modules, amongst other brain regions. Both primary and secondary co-expression modules containing these genes were enriched for mainly neuronal cell types. Regulons of the complex contained Parkinson’s disease-associated genes and were enriched for biological pathways genetically linked to disease. When examined in a neuroblastoma cell line, 41% of prioritized gene targets showed significant changes in mRNA expression following KANSL1 or KAT8 perturbation. KANSL1 and H4K8 chromatin immunoprecipitation-sequencing data demonstrated non-specific lethal complex activity at many of these genes.
In conclusion, genes encoding the non-specific lethal complex are highly correlated with and regulate genes associated with Parkinson’s disease. Overall, these findings reveal a potentially wider role for this protein complex in regulating genes and pathways implicated in Parkinson’s disease.
Keywords: GWAS, NSL complex, Parkinson’s disease, gene co-expression networks
KANSL1 and KAT8 are Parkinson’s disease candidate genes that encode part of the histone acetylating non-specific lethal (NSL) complex. Hicks et al. show that these genes regulate the expression of many other genes and pathways implicated in Parkinson’s disease, suggesting that the NSL complex may be a useful drug target.
Introduction
An in-depth understanding of the genetic and pathophysiological mechanisms underlying neurodegenerative diseases is necessary to develop effective disease-modifying treatments. In the case of Parkinson’s disease, although 90–95% of cases are sporadic, historically much of the research into its genetic basis has focused on family-based linkage studies. Indeed, the identification of at least 23 genes with highly penetrant effects on Parkinson’s disease risk has succeeded in elucidating multiple biological pathways involved in its pathology. In particular, mitochondrial dysfunction and impaired protein degradation pathways are common themes. More recently, genome-wide association studies (GWASs) have identified 90 independent risk signals linked to Parkinson’s disease. Several of these had already appeared in familial studies, thereby highlighting important commonalities in the processes driving both types of the disease.1 However, a broader understanding of the molecular relationships between Parkinson’s disease loci is still lacking. Genes causally linked to the disease and involved in transcriptional regulation have the potential to provide such insights and shed light on key disease-relevant gene networks.
One such transcriptional regulator with strong links to Parkinson’s disease is KAT8. This gene was first linked to the disease through the identification of a risk signal on chromosome 16 (rs14235) with subsequent expression quantitative trait loci (eQTL) analysis suggesting the risk allele results in lower KAT8 mRNA levels.2,3 Further GWAS analyses have again highlighted KAT8 as a candidate gene, with recent co-localization and transcriptome-wide analyses strengthening the evidence for KAT8’s contribution to Parkinson’s disease.1,4 Importantly, KAT8 functions within two multiprotein complexes that regulate its activity and specificity, namely the male specific lethal (MSL) and non-specific lethal (NSL) complexes.5
Although the KAT8-encoded acetyl-transferase is thought to be the main catalytic driver in both complexes, differences in lysine specificity and in the genomic regions which are targeted can likely be attributed to subunits aside from KAT8 itself.6 This makes the other components of the MSL and NSL complexes of potential interest, with the latter particularly important in Parkinson’s disease, as it contains KAT8 Regulatory NSL Complex Subunit 1 (KANSL1), another protein encoded by a Parkinson’s disease candidate gene.3,7KANSL1 is contained within the 970 kb inversion polymorphism on chromosome 17q21, located within a linkage disequilibrium (LD) block of approximately 2 Mb, which gives rise to H1/H2 haplotype variation.8 The H1 haplotype has well established links to neurodegenerative disease, specifically progressive supranuclear palsy, Alzheimer’s disease and Parkinson’s disease.9–11 The precise mechanism underlying the link to Parkinson’s disease is disputed, with this risk frequently attributed to the adjacent tau-encoding MAPT, as well as, more recently, a putative enhancer RNA expressed from within KANSL1.12,13 Moreover, the first GWAS of short tandem repeats in Parkinson’s disease found the strongest signal within KANSL1.14
Furthermore, both KAT8 and KANSL1 have been linked to mitophagy, the process by which defective mitochondria are identified and degraded and a key pathway implicated in Parkinson’s disease. Accumulation of mitophagy marker phospho-ubiquitin (pUb, serine 65) has been detected in post-mortem diseased brains, whilst deficient mitophagy has been found in both sporadic and Mendelian patient-derived induced pluripotent stem cell (iPSC) models and even suggested to play a direct role in α-synuclein accumulation.15–17 Proteins involved in mitophagy, in particular PTEN-induced putative kinase 1 (PINK1) and parkin, are associated with early-onset autosomal recessive forms of the disease through mutations in their encoding genes, PINK1 and PRKN.18,19 A biological screening assay of Parkinson’s disease GWAS candidate genes which measured PINK1-mediated mitophagy in neuroblastoma cells demonstrated significantly reduced pUb accumulation, parkin recruitment and phosphorylation, as well as lysosomal localization of mitochondria following knockdown (KD) of both KAT8 and KANSL1, thus demonstrating an important role of the NSL complex in mitochondrial quality control and Parkinson’s disease.4,20 There is evidence that components of the NSL complex can localize to mitochondria, though the most established function of the complex is in the nucleus, where it is involved in chromatin regulation.21 Indeed, both KAT8 and KANSL1 KD reduced PINK1 mRNA levels in multiple cell types.4 Thus, KAT8 and KANSL1 could operate to regulate the risk of Parkinson’s disease in multiple sub-cellular compartments.
In this study, we focused on the role of the NSL complex within the nucleus and tested our hypothesis that this complex operates as a master regulator of Parkinson’s disease risk. This idea is supported by existing evidence implicating KAT8-dependent lysine acetylation of primarily histone 4 (H4Kac) in the regulation of a range of cellular processes, including DNA damage repair and autophagy.6,22–27 To pursue this idea, we performed a series of in silico analyses which successfully predicted gene regulatory relationships between the chromatin-modulating NSL complex and genes associated with Parkinson’s disease. These findings suggest a role for the NSL complex in modulating multiple pathological pathways and provide a useful framework for investigating potential gene regulatory mechanisms underlying disease risk associated with loci highlighted through GWASs.
Materials and methods
Gene selection
We collated three lists of genes, namely the NSL genes, genes causally associated with Mendelian forms of Parkinson’s disease and genes nominated through GWASs (Table 1). The nine genes encoding the NSL complex are widely published.5 An expert-curated list of genes linked to Mendelian forms of Parkinson’s disease and complex parkinsonism was obtained from PanelApp.28 Supplementary data published alongside a Parkinson’s disease GWAS was filtered for genes nominated by Mendelian randomization.1 Full gene lists are available in the Supplementary material and at https://github.com/amyrosehicks/NSL_PD_relationships (doi:10.5281/zenodo.7525823).29
Table 1.
Gene lists collated for frontal cortex gene co-expression network analysis
| List | Source | Number of genes | ||
|---|---|---|---|---|
| Total | Within GTEx GCN | Within UKBEC GCN | ||
| NSL complex | Sheikh et al.5 | 9 | 9 | 8 |
| Mendelian Parkinson’s disease | PanelApp: Parkinson disease and complex Parkinsonism version 1.68 | 43 | 42 | 42 |
| Sporadic Parkinson’s disease | Genes nominated by Mendelian randomization1 | 151 | 135 | 134 |
Genes missing from gene co-expression networks (GCNs) were not included in the search. GTEx = Genotype Tissue Expression; NSL = non-specific lethal; UKBEC = United Kingdom Brain Expression Consortium.
Expression weighted cell type enrichment analysis
Expression-weighted cell type enrichment (EWCE) analysis was used to test whether NSL complex encoding genes are more highly expressed in specific brain-related cell types than would be expected by chance (https://github.com/NathanSkene/EWCE.git).30 Specificity values, representing the proportion of the total expression of a gene in one cell type compared with others, were calculated from two independent single nucleus RNA-sequencing datasets derived from human substantia nigra and medial temporal gyrus tissue and were examined using the MarkerGenes github package.31–34 All data manipulation and visualization was performed in R (v 4.0.5; RRID:SCR_001905) using the packages described here: https://github.com/amyrosehicks/NSL_PD_relationships.29
Gene co-expression network analysis
We used two sources of public transcriptomic data for the generation of gene co-expression networks (GCNs), namely the Genotype Tissue Expression (GTEx, version 6) project (https://www.gtexportal.org/home/) and the United Kingdom Brain Expression Consortium (UKBEC, https://ukbec.wordpress.com/). GTEx data contain samples originating from 13 CNS regions as well as other non-diseased tissues provided by 544 donors in total, whilst UKBEC data contain samples from 12 CNS regions provided by 134 donors free of neurodegenerative disorders. Individual tissue sample numbers are summarized in Supplementary Table 1. GTEx samples were sequenced using Illumina sequencing, whilst UKBEC samples were assayed using Affymetrix arrays.35,36
Tissue-specific primary GCNs were built from these datasets using weighted gene co-expression network analysis (WGCNA), with k-means optimization followed by functional and cellular specificity annotation.34,37,38 The completed GCNs formed part of the CoExp R package suite.39 Four secondary GCNs were also examined, for which the gene multifunctionality in secondary co-expression network analysis (GMSCA) R package was used to remove the contribution of neuronal, microglial, astrocytic and oligodendrocytic cell types from the expression matrix before GCN reconstruction.40
Gene set enrichment analysis
For gene list enrichment analyses, we obtained P-values using Fisher’s exact tests comparing the overlap in input genes and genes contained within each module, with false discovery rate (FDR) corrections for multiple testing.39,40 GCNs were annotated with gene ontology (GO) terms, categorized into biological process (BP), molecular function (MF) or cellular component (CC), within the CoExp R packages. GO terms were reduced to parent terms using the Rutils github package to aid visualization (doi: 10.5281/zenodo.6127446).41 Further analyses of gene lists derived from a reverse engineering analysis were performed using the gProfiler2 R package (RRID:SCR_018190).42
Reverse engineering gene regulatory network analysis
Using the Algorithm for the Reconstruction of Accurate Cellular Networks with adaptive partitioning (ARACNe-AP, RRID:SCR_002180), we inferred regulatory relationships between NSL complex genes and genes within specific modules of the primary GCNs.43 The original Java package was adapted for use in R and executed with a P-value threshold of 1 × 10−8 and 10 000 bootstrap iterations. The strength of the association between regulators and regulons is denoted by mutual information values, which were compared using quantiles.44
Stratified linkage disequilibrium score regression
Heritability is defined as the proportion of variation in a trait that can be attributed to inherited genetic factors.45 More specifically to this project, single nucleotide polymorphism (SNP)-based heritability refers to the variance that can be explained by any set of SNPs, such as those derived from a GWAS.46 We used stratified linkage disequilibrium score regression (LDSC, v.1.0.1., https://github.com/bulik/ldsc/wiki) to evaluate the enrichment of common SNP-based heritability for Parkinson’s disease across individual regulons derived from a reverse engineering analysis and across regulon genes grouped according to cumulative frequency.1,47,48
The analysis was performed with parameters as described by Chen et al.49 Briefly, the baseline model of 97 annotations (v.2.2, GRCh37) to which annotations were added included only SNPs with minor allele frequencies over 5%. The major histocompatibility complex region was excluded due to its complicated LD patterns. Regression and LD reference panels utilized HapMap Project Phase 3 (https://www.sanger.ac.uk/data/hapmap-3/) and 1000 Genomes Project Phase 3 (https://www.internationalgenome.org/) European population SNPs, respectively.50,51 Gene coordinates were extended by 100 kb up- and downstream of their transcription start and end sites to capture potentially relevant regulatory elements.34 The resulting regression coefficients (contribution of annotation after controlling for all other categories in the model) were used to calculate two-tailed P-values.
Cell culture and siRNA treatment
We cultured wild-type (WT) SHSY5Y neuroblastoma cells (RRID:CVCL_0019) sourced from American Type Culture Collection (ATCC, RRID:SCR_00167) in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, 11995–065) containing fetal bovine serum (FBS, 10%, Gibco, 10500-064). For plating, cells were trypsinized, resuspended in culture media and counted using a Countess Automated Cell Counter.
Three types of siRNA were purchased as pre-designed siGENOME SMARTpools and transfected as per the manufacturer’s instructions: non-targeting/scrambled (SCR, D-001206-13), KAT8 (M-014800-00) and KANSL1 (M-031748-00). Each siRNA was diluted in FBS-free DMEM (final well concentration 50 nM) and mixed with DharmaFECT1 (Dharmacon, T-2001-03) before plating. After incubating at room temperature for 30 min, cell suspensions were prepared in culture media and added to respective wells on top of the siRNA mix. Following 72 h of incubation, media were removed and cells were lysed as per the manufacturer’s instructions for a QuantiGene multiplex assay.
QuantiGene multiplex assay
We used the QuantiGene multiplex system to simultaneously measure the expression of multiple genes in siRNA-treated SHSY5Y cell samples.52,53 Individual reagents and probe sets were purchased from ThermoFisher. Probes were directed against: (i) five housekeeping genes used for normalization; (ii) two NSL complex genes used to quantify the KD; (iii) all genes both causally-linked to Mendelian Parkinson’s disease and complex parkinsonism, and contained within NSL regulons in the GTEx dataset; and (iv) all genes nominated through GWASs which are expressed in brain and predicted to be regulated by at least three NSL complex genes (Table 2). CTSB was included as an exception, despite only appearing in two regulons, due to extensive literature implicating it in Parkinson’s disease.54–56 All GWAS-linked genes in this final list had mutual information values in the upper 50th regulon quantile (Table 2).
Table 2.
Details of QuantiGene 24-plex assay detecting changes in Parkinson’s disease-associated genes following NSL complex knockdown
| Symbol | Accession number | Type | Regulon frequency | Regulon MI quantile range | ||
|---|---|---|---|---|---|---|
| GTEx | UKBEC | GTEx | UKBEC | |||
| ATP5B | NM_001686 | Housekeeping | – | – | – | – |
| CANX | NM_001746 | Housekeeping | – | – | – | – |
| EIF4A2 | NM_001967 | Housekeeping | – | – | – | – |
| HPRT1 | NM_000194 | Housekeeping | – | – | – | – |
| PPIB | NM_000942 | Housekeeping | – | – | – | – |
| KANSL1 | NM_015443 | NSL | – | – | – | – |
| KAT8 | NM_032188 | NSL | – | – | – | – |
| PINK1 | NM_032409 | PD-Mendelian | 2 | – | 0.544–0.648 | |
| ATN1 | NM_001940 | PD-Mendelian | 1 | 1 | 0.942 | 0.966 |
| ATXN2 | NM_002973 | PD-Mendelian | 4 | 1 | 0.572–0.913 | 0.782 |
| PARK7 | NM_007262 | PD-Mendelian | 2 | – | 0.260 | – |
| PLA2G6 | NM_003560 | PD-Mendelian | 6 | – | 0.118–0.169 | – |
| WDR45 | NM_007075 | PD-Mendelian | 1 | – | 0.424 | – |
| BIN3 | NM_018688 | PD-sporadic | 4 | – | 0.457–0.754 | – |
| CCAR2 | NM_021174 | PD-sporadic | 3 | – | 0.729–0.839 | – |
| CTSB | NM_001908 | PD-sporadic | 2 | – | 0.825–0.899 | – |
| DGKQ | NM_001347 | PD-sporadic | 3 | – | 0.520–0.976 | – |
| GAK | NM_005255 | PD-sporadic | 3 | – | 0.864–0.977 | – |
| NCKIPSD | NM_016453 | PD-sporadic | 4 | – | 0.308–0.919 | – |
| PGS1 | NM_024419 | PD-sporadic | 3 | – | 0.651–0.805 | – |
| POLR2A | NM_000937 | PD-sporadic | 3 | 1 | 0.463–0.533 | 0.617 |
| QRICH1 | NM_017730 | PD-sporadic | 5 | – | 0.312–0.828 | – |
| SETD1A | NM_014712 | PD-sporadic | 4 | 2 | 0.294–0.869 | 0.0443–0.953 |
| SH2B1 | NM_015503 | PD-sporadic | 5 | – | 0.385–0.978 | – |
All Mendelian disease-linked genes appearing in NSL regulons in the GTEx dataset were included, as well as genome-wide association study genes expressed in brain and appearing in three or more regulons (CTSB was included as an exception due to extensive literature implicating it in Parkinson’s disease). GTEx = Genotype Tissue Expression; MI = mutual information; NSL = non-specific lethal; PD = Parkinson’s disease; UKBEC = United Kingdom Brain Expression Consortium.
Samples were prepared as per the manufacturer’s instructions: the working lysis mixture was prepared by diluting 1 µl proteinase K per 100 µl lysis mixture; cells were lysed by pipetting 150 µl warm working lysis mixture per well (1:2 working lysis mixture to culture media); then, plates were snap-frozen on dry ice and stored at −80°C. The remainder of the assay was performed as indicated in the manufacturer’s protocol, with the exception that the Streptavidin R-Phycoerythrin conjugate (SAPE) binding step was completed at 51°C.53 Plates were read using a Magpix (Luminex). This protocol is available on protocols.io (doi:dx.doi.org/10.17504/protocols.io.kqdg39ew7g25/v1).
Data analysis was performed by first subtracting the background, then normalizing the signals obtained for the genes of interest to the geometric mean of the five housekeeping gene signals. Technical duplicates were included for each experimental repeat, and outliers were identified and excluded using the ROUT method in GraphPad Prism (version 9, RRID:SCR_002798).57 Remaining duplicates were averaged and normalized to the SCR-treated sample mean.
Results
Components of the NSL complex are highly expressed across all CNS regions and cell types
Using transcriptomic data provided by GTEx, the expression of genes encoding the NSL complex across 13 CNS regions was analysed. This analysis demonstrated expression of all nine members of the NSL complex in all CNS regions, though we noted that KAT8 and OGT were most highly expressed in the cerebellum (median transcripts per million = 107.8 and 181.5) (Supplementary Fig. 1A). Differences in cell type-specific expression were also examined within two brain regions, namely the substantia nigra and medial temporal gyrus, using an EWCE analysis and based on single-nuclei transcriptomic data.31,32 Consistent with expectation, the NSL complex genes displayed no overt differences in cell type-specific gene expression when considered collectively (Supplementary Fig. 1B).
Components of the NSL complex cluster in gene co-expression modules derived from human brain data
Given that there was no clear specificity of expression of NSL genes in CNS tissues or, more importantly, cell types, gene co-expression analysis was used to investigate the possibility of regional differences in co-expression that could explain selective neuronal vulnerability in Parkinson’s disease. This approach was based on the fact that genes with highly correlated expression tend to share biological relationships, and so GCN analysis can reveal otherwise hidden patterns in expression that reflect molecular and cellular processes.39 With this in mind, we used transcriptomic data generated by GTEx and covering 13 CNS regions to generate tissue-specific GCNs. Each GCN consisted of 10–79 gene co-expression modules containing an average of 530 genes per module. Module membership (MM) values ranged between 0 and 1, with higher values indicating that gene expression was highly correlated with the module eigengene (ME).59
To identify the gene co-expression modules of most interest, we investigated the enrichment of NSL genes within all 489 modules across all 13 GTEx CNS-relevant GCNs. We identified significant enrichment of the NSL genes within the cerebellar hemisphere and cerebellum ‘yellow’ modules (FDR range = 3.19 × 10−2–4.64 × 10−2, MM range = 0.731–0.829), frontal cortex ‘red’ module (FDR = 5.86 × 10−3, MM range = 0.775–0.904), hippocampus ‘pink’ module (FDR = 9.15 × 10−5, MM range = 0.842–0.906), hypothalamus ‘red’ module (FDR = 3.94 × 10−2, MM range = 0.785–0.845) and spinal cord ‘darkgreen’ module (FDR = 4.04 × 10−3, MM range = 0.878–0.887) (Fig. 1A and Supplementary Fig. 2). Importantly, the enrichment of NSL genes within both frontal cortex and cerebellum GCNs was replicated in the corresponding UKBEC GCN within the ‘black’ module (FDR = 3.84 × 10−4, MM range = 0.495–0.857) and ‘grey60’ module, respectively (Supplementary Fig. 3). Of note was the significant enrichment of NSL genes in GCN modules derived from other disease-relevant tissues, including UKBEC substantia nigra (‘green’ and ‘black’ modules, FDR range = 1.82 × 10−2–3.49 × 10−2, MM range = 0.287–0.769) and putamen (‘purple’ module, FDR = 4.98 × 10−3, MM range = 0.587–0.815), as well as the nominal significance (FDR < 0.1) in one GTEx caudate module (‘salmon’ module, FDR = 5.28 × 10−2, MM range = 0.786–0.842) (Supplementary Figs 2 and 3).
Figure 1.
Exploration of GTEx frontal cortex gene co-expression network. (A) NSL gene enrichment analysis across GCN modules constructed using GTEx datasets. (B) GO BP, MF and CC term enrichments for GCN modules containing NSL genes. Each term has been uniformly reduced to a parent term in order to group together similar terms. Colour corresponds to the P-value of the most significantly enriched child term within the parent term. (C) Parkinson’s disease-associated gene set enrichment analysis, filtered for GCN modules containing NSL genes. Fisher’s exact test, displayed over a log scale (FDR-corrected P-values). ns = FDR > 0.1; +FDR < 0.1; *FDR < 0.05; **FDR < 0.01. BP = Biological process; CC = cellular compartment; FDR = false discovery rate; GCN = gene co-expression network; GO = gene ontology; GTEx = Genotype Tissue Expression; MF = molecular function; NSL = non-specific lethal; PD = Parkinson’s disease.
However, given the replication across datasets and the importance of the region in Parkinson’s disease progression and dementia, we focused on frontal cortex GCNs in particular and analysed the correlations between modules containing NSL genes.60–63 We found that several frontal cortex GCN modules containing NSL genes, including the aforementioned GTEx ‘red’ module along with the ‘darkred’ module (FDR = 0.205, MM range = 0.719–0.849) had highly correlated expression, as visualized in a ME dendrogram plot (Supplementary Fig. 4A and C). Again, this was replicated in the frontal cortex UKBEC GCN, where we noted high correlations in the expression of aforementioned ‘black’ module and the ‘darkgreen’ module (FDR = 0.270, MM = 0.506) (Supplementary Fig. 4B and D). Taken together, these findings suggest that genes encoding the NSL complex are significantly co-expressed in the frontal cortex, a finding which is consistent with the importance of this brain region in Parkinson’s disease.
A role for the NSL complex in regulating chromatin and mitochondrial function
Next, we focused on co-expression modules containing NSL complex genes to better understand their function in human brain. This was achieved by examining GO term enrichment within five modules of interest in the GTEx frontal cortex GCN, with the primary focus being the ‘red’ module. In total, these five modules were significantly enriched for 2899 GO terms (FDR < 0.05). Following term reduction, we noted enrichments of terms representing both the well-characterized nuclear-based and lesser-known mitochondrial-based role of the NSL complex. Consistent with expectation, nucleus-related terms, such as histone modification (GO:0016570, FDR range = 1.48 × 10−4–1.65 × 10−2), were enriched within the ‘red’ module, whilst the ‘darkred’ module was enriched for a range of mitochondria-related terms, including cytochrome c oxidase activity (GO:0004129, FDR range = 1.10 × 10−8–7.05 × 10−5) and mitochondrial inner membrane (GO:0005743, FDR range = 5.10 × 10−51–2.89 × 10−3) (Fig. 1B). Also of note was the enrichment of nucleus-related terms such as histone acetylation (GO:0016573, FDR range = 3.17 × 10−8–3.18 × 10−2) in the GO term enrichment analysis of caudate GCN ‘salmon’ module, contrasting to terms like mitochondrial ATP synthesis coupled proton transport (GO:0042776, FDR range = 1.54 × 10−29–3.46 × 10−6) in the ‘darkred’ module (Supplementary Fig. 6A). The UKBEC frontal cortex GCN was also significantly enriched for several nuclear terms in the ‘black’ module but lacked any mitochondrial terms (Supplementary Fig. 5A). This was also the case for the putamen GCN, with many nucleus-related terms enriched within the ‘purple’ module; however, the substantia nigra GCN contained several mitochondria-related terms particularly enriched within the ‘brown’ module (Supplementary Fig. 5B and C). Together these results indicated that both the chromatin- and mitochondria-related functions of the NSL complex were captured in these GCNs, indicating that these functions are likely to be active within human brain regions relevant to Parkinson’s disease.
Components of the NSL complex cluster together with Parkinson’s disease-associated genes
Next, we explored the possibility that NSL genes are functionally related to genes associated with Parkinson’s disease. Parkinson’s-related genes were divided into those causally linked to Mendelian forms of the disease and those associated with sporadic Parkinson’s disease through the identification of GWAS risk loci in close proximity. The GTEx frontal cortex GCN was then tested for enrichment of the two disease lists. Gene set enrichment analysis revealed a significant enrichment of GWAS genes in the ‘red’ GTEx GCN module (FDR = 2.69 × 10−2, MM range = 0.553–0.907) (Fig. 1C). This gene list was also significantly enriched in the ‘black’ module derived from UKBEC substantia nigra GCN (FDR = 4.82 × 10−2, MM range = 0.280–0.673), as well as reaching nominal significance in both the ‘salmon’ module within the GTEx caudate GCN (FDR = 8.51 × 10−2, MM range = 0.484–0.938) and the ‘purple’ UKBEC putamen GCN module (FDR = 7.51 × 10−2, MM range = 0.294–0.656) (Supplementary Figs 5D and E and 6B). These results point to the possibility that important gene regulatory links exist between the NSL complex and genes associated with sporadic Parkinson’s disease.
NSL complex activity in Parkinson’s disease may be most important in neuronal cell types
Although the genes encoding the NSL complex are expressed ubiquitously across different cell types, their gene regulatory relationships may nonetheless be cell type-specific, and only in specific cell types might there be a relationship with Parkinson’s disease genes. We used the GMSCA tool to test this possibility and generated secondary frontal cortex GCNs from GTEx data, from which the contribution of four major cell types (neurons, astrocytes, microglia and oligodendrocytes) was removed in turn prior to network construction.40 As might be expected, this correction altered the clustering of NSL genes and their co-expression patterns with Parkinson’s disease-associated genes across secondary GCN modules (Fig. 2A and C). The sporadic Parkinson’s disease gene list was significantly enriched alongside KANSL1—as in the primary frontal cortex GCN—within the oligodendrocyte-corrected ‘green’ module (FDR = 4.81 × 10−3 and 5.87 × 10−3, respectively) alone, whilst each of the neuron-, microglia- and astrocyte-corrected networks disrupted the relationship between NSL and disease genes (Fig. 2A and C). This suggested that the relationship between these genes is likely to be active in the neurons, microglia and/or astrocytes.
Figure 2.
Exploration of GTEx frontal cortex secondary gene co-expression networks. (A) NSL gene enrichment analysis of secondary GCN modules constructed using GTEx datasets. (B) Cell type enrichment analysis of primary (labelled ‘none’) and secondary (labelled ‘-corrected’) GCN modules containing NSL genes. NSL genes found within modules devoid of any cell type enrichment were not included. (C) Mendelian and sporadic Parkinson’s disease-associated gene enrichment analysis of secondary GCN modules, filtered for those containing KAT8 and KANSL1 (gene in brackets, no box indicates no genes present). Fisher’s exact test, displayed over a log scale (FDR-corrected P-values). ns = FDR > 0.05; *FDR < 0.05; **FDR < 0.01; ***FDR < 0.001; ****FDR < 0.0001. FDR = false discovery rate; GCN = gene co-expression network; GTEx = Genotype Tissue Expression; NSL = non-specific lethal; PD = Parkinson’s disease.
To identify which of the three cell types was most important for NSL-Parkinson's disease co-expression, we examined the enrichment of cell type markers across all primary and secondary GCN modules. The ‘darkred’ module containing KAT8 and MCRS1 was enriched for markers of dopaminergic neuronal signalling (P = 3.23 × 10−5) in the GTEx primary GCN (Fig. 2B). MCRS1 remained predominantly in modules enriched for different neuronal markers following the correction of microglial, astrocytic and oligodendrocytic signatures (P-value range = 3.92 × 10−16–7.42 × 10−3, MM range = 0.8452–0.8529) (Fig. 2B). The primary GCN ‘grey60’ module containing PHF20 lacked any cell type enrichment, contrasting to the secondary GCN ‘salmon’ and ‘greenyellow’ modules, which were both enriched for multiple neuronal cell types following the correction of microglial and oligodendrocytic signatures respectively (P-value range = 3.99 × 10−11–5.11 × 10−3, MM range = 0.8539–0.8544) (Fig. 2B). Although KANSL1-containing modules had no cell type enrichments, these results suggest the gene regulatory links between the NSL complex and genes associated with Parkinson’s disease may be most important in neuronal cell types.
Regulation of Parkinson’s disease-associated genes by members of the NSL complex
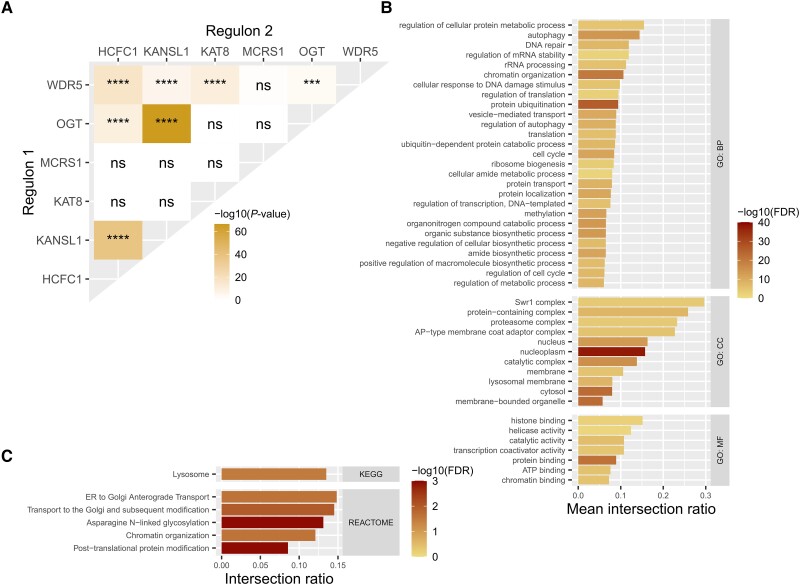
The genetic interactions modelled in GCNs are typically undirected in that causality is unassigned.38 However, it is already known that the NSL complex is highly important in the regulation of gene expression, suggesting that at least a proportion of the genes co-expressed with the NSL complex are regulated by it.5 We formally tested this possibility with the ARACNe-AP tool, which uses expression data to reverse engineer gene regulatory networks.43,64 By applying ARACNe-AP, we predicted the genes most likely to be regulated by the NSL complex amongst those contained within the ‘red’ and ‘darkred’ GTEx frontal cortex GCN modules (Supplementary Table 2).43 The resulting target gene lists, termed regulons, produced by this analysis ranged from 491 genes predicted to be regulated by KANSL1 to 1788 predicted to be regulated by WDR5 in the GTEx dataset. As expected for genes encoding a protein complex that regulates gene expression, regulons showed significant overlaps with each other. The regulons of the four NSL genes contained within the ‘red’ GTEx GCN module all significantly overlapped (FDR range = 3.53 × 10−67–3.76 × 10−4), whilst the regulon of KAT8 significantly overlapped only with that of WDR5 (FDR = 2.98 × 10−11) (Fig. 3A).
Figure 3.
Characterization of NSL complex regulons, derived from ARACNe-AP analysis of ‘red’ and ‘darkred’ GTEx primary gene co-expression network modules. (A) Overlaps of regulons of each NSL gene. (B) GO BP, MF and CC term enrichments for genes appearing in three or more NSL regulons. Each term has been uniformly reduced to a parent term in order to group together similar terms. Colour corresponds to the P-value of the most significantly enriched child term within the parent term and the x-axis denotes the mean ratio of genes intersecting with the term to total genes within each term. (C) REACTOME and KEGG term enrichments for genes appearing in three or more NSL regulons, filtered for those genetically linked to Parkinson’s disease. The x-axis denotes the ratio of genes intersecting with the term to total genes within each term. Fisher’s exact tests, displayed over a log scale (gene set enrichment P-values are FDR corrected). ns = P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. BP = Biological process; CC = cellular compartment; FDR = false discovery rate; GCN = gene co-expression network; GO = gene ontology; GTEx = Genotype Tissue Expression; MF = molecular function; NSL = non-specific lethal; PD = Parkinson’s disease.
To reduce the impact of noise and focus on genes most representative of NSL complex activity, genes appearing in three or more regulons were collated and termed the NSL regulon (n = 1101). We then assessed this gene set for its role in Parkinson’s disease risk. Firstly, we noted that two Mendelian disease-associated genes (ATXN2 and PLA2G6) and 12 sporadic disease-associated genes (BIN3, CCAR2, DGKQ, GAK, GBAP1, IGSF9B, NCKIPSD, PGS1, POLR2A, QRICH1, SETD1A, SH2B1) were contained within this dataset, although no significant enrichment was observed (P = 0.843 and 0.619, respectively). Furthermore, we used stratified LDSC to assess the enrichment of Parkinson’s disease SNP-based heritability amongst all genes in the NSL regulon.1 Despite the relatively small gene set, we found that the regression coefficient—a stringent measure of the contribution to SNP-based heritability which accounts for underlying contributions of genetic architecture captured within the baseline model—reached nominal significance (P = 5.19 × 10−2; Supplementary Table 3). This hinted at a link between Parkinson’s disease heritability and NSL complex activity.
Next, we assessed the NSL regulon for the enrichment of pathways of potential relevance to Parkinson’s disease. Using a gene set enrichment analysis using parent terms to reduce redundancy, we found that genes contained within the NSL regulon were enriched for a range of terms including: autophagy (GO:0006914, FDR range = 3.11 × 10−12–3.54 × 10−2), regulation of autophagy (GO:0010506, FDR range = 2.90 × 10−4–1.23 × 10−2) and chromatin organization (GO:0006325, FDR range = 7.50 × 10−9–4.83 × 10−2) (Fig. 3B). Gene set enrichment analysis was also completed on individual regulons, revealing many of the same parent terms were enriched across different regulons (Supplementary Fig. 7). To assess the association of Parkinson’s disease-relevant pathways with the regulons more directly, we specifically asked whether our enriched gene sets overlapped with a list of 46 gene sets genetically implicated in the disease through common genetic variation.65 This highlighted the enrichment of five disease-linked terms, including KEGG:04142 Lysosome (FDR = 3.21 × 10−2) and REAC:R-HSA-4839726 chromatin organization (FDR = 2.42 × 10−2), suggesting that the NSL regulon in frontal cortex is enriched for genes causally implicated in Parkinson’s disease (Fig. 3C).
Experimental data confirm the regulation of Parkinson’s disease-associated genes by the NSL complex
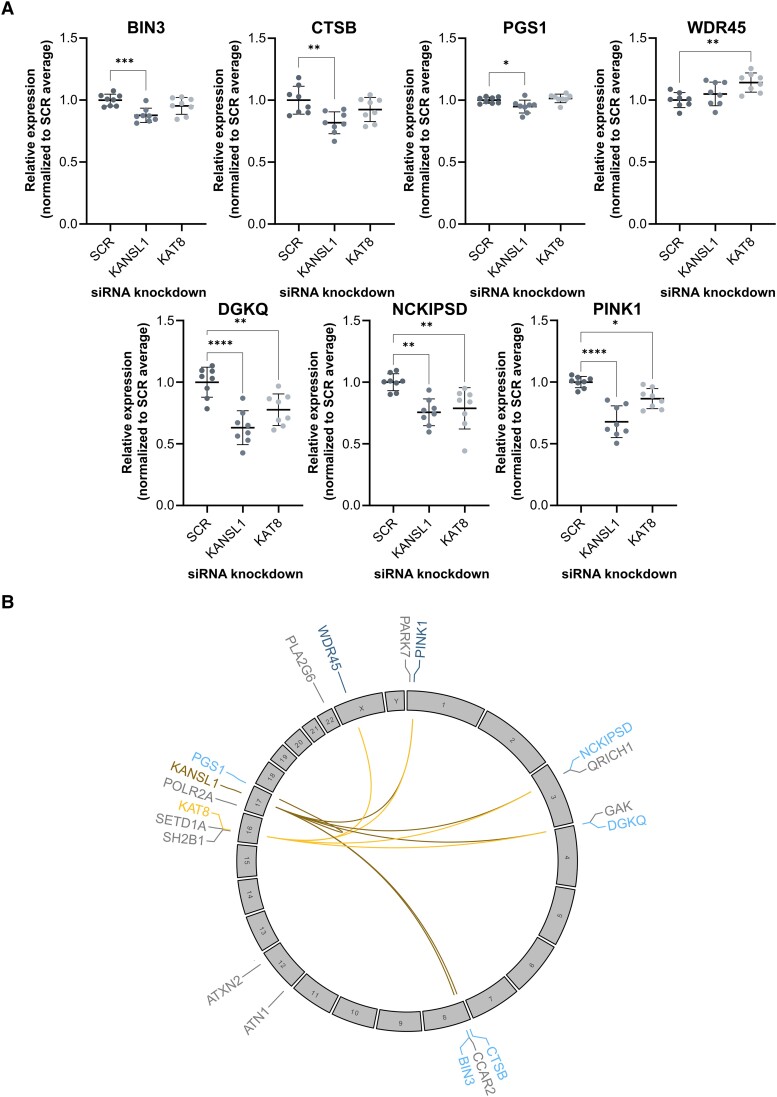
Given the success of our in silico analyses, we wanted to validate some of the regulatory relationships identified in vitro, in particular focusing on genes causally associated with Parkinson’s disease and contained within the NSL regulon (Table 2). With this in mind, we measured the expression of 17 genes of interest in response to KANSL1 and KAT8 siRNA KD in an SHSY5Y cell line, a cell model regularly used for Parkinson’s disease research (Fig. 4A and Supplementary Fig. 8).4 After confirming that KANSL1 and KAT8 KDs did not affect housekeeping gene expression levels, we found that both KDs significantly reduced PINK1 expression (P < 1 × 10−4 and = 1.51 × 10−2, respectively), consistent with previous reports (Supplementary Fig. 8A and B).4BIN3, CTSB, DGKQ, NCKIPSD and PGS1 were also significantly reduced following KANSL1 KD alone (P-value range < 1 × 10−4–2.95 × 10−2), with reductions in DGKQ and NCKIPSD following KAT8 KD also reaching significance (P = 4.55 × 10−3 and 4.25 × 10−3, respectively). Interestingly, WDR45 expression followed an inverse pattern, with KAT8 KD resulting in a significant increase in expression (P = 3.31 × 10−3) (Fig. 4A). Thus, 7 of the 17 genes (41.2%) predicted to be regulated by the NSL complex using in silico analyses were indeed found to show significant changes in expression when KAT8 or KANSL1 expression was suppressed (Fig. 4B).
Figure 4.
Gene regulatory relationships detected between NSL complex and Parkinson’s disease-associated genes. (A) Changes in mRNA levels of Parkinson’s disease-associated genes following suppression of NSL complex genes, measured using QuantiGene multiplex assay, normalized to average SCR control. (B) Representation of cross-genome regulatory relationships detected. Genes highlighted were tested using QuantiGene multiplex assay. Colours correspond to classification of gene (blue, genes linked to Parkinson’s disease with NSL regulation detected; grey, genes linked to Parkinson’s disease without NSL regulation detected; lightest yellow, KAT8 regulation; darkest yellow, KANSL1 regulation). One-way ANOVA with Dunnett’s correction for multiple comparisons, n = 8. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. NSL = non-specific lethal; SCR = scrambled.
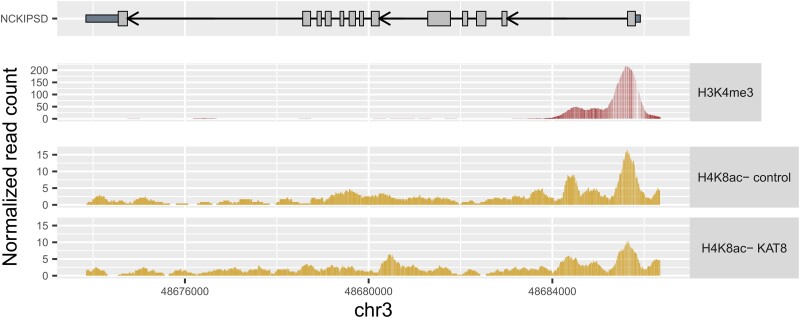
To explore the possibility that the NSL complex regulates these targets directly through its chromatin-modulating activity, we used two public chromatin immuno-precipitation (ChIP)-sequencing datasets. The first, containing KANSL1 ChIP-sequencing conducted in HeLa cells, highlighted 3637 genes bound by KANSL1 at transcription start sites across three replicate experiments.66 We found three of the seven genes identified through the QuantiGene multiplex assay, namely WDR45, PGS1 and BIN3, were bound directly by KANSL1. Interestingly, four genes which did not validate in the QuantiGene multiplex assay, namely CCAR2, SH2B1, POLR2A and SETD1A, also had evidence of KANSL1 binding at their transcription start sites. Secondly, we examined the effects of CRISPRi-mediated KD of KAT8 in THP-1 cells, as measured by H4K8 acetylation peak identification.6 We noted that H4K8 acetylation peaks at three of the seven hit genes, namely DGKQ, NCKIPSD and CTSB, were reduced following KAT8 KD (Fig. 5 and Supplementary Fig. 9). Taken together, these findings provide direct experimental support for NSL complex activity at six Parkinson’s disease-associated genes, thus offering a mechanistic understanding of their co-regulation.
Figure 5.
Changes in epigenetic modifications of NCKIPSD following KAT8 knockdown. H4K8ac peaks indicating NSL complex activity and H3K4me3 peaks indicating a promotor region at the NCKIPSD gene, produced using ChIP-sequencing data published by Radzisheuskaya et al.6 H3K8me3 was measured in wild-type THP-1 cells and H4K8ac in THP-1 cells treated with control or KAT8-targeting CRISPRi guides. ChIP = chromatin immuno-precipitation; chr = chromosome; H4K8ac = histone 4 lysine 8 acetylation; H3K4me3 = histone 3 lysine 4 trimethylation; NSL = non-specific lethal; WT = wild-type.
Discussion
This project used publicly available transcriptomic data from human brain tissue to characterize the expression patterns of genes encoding the NSL complex and their relationships to genes genetically-linked to Parkinson’s disease. First, NSL genes were found to cluster together with Parkinson’s-associated genes in GCN modules annotated for both chromatin- and mitochondria-related functions. Second, these co-expression relationships appeared to be most associated with neuronal cell types. Third, a number of Parkinson’s-associated genes predicted to be regulated directly by multiple components of the NSL complex were validated in a relevant cell model and using public sequencing data.
Despite no clear specificity of expression being detected across tissues, the NSL genes showed specific co-expression patterns across GCNs produced from different brain regions. These results are consistent with the theory of the NSL complex having context-specific gene regulatory functions. Within multiple GCNs derived from disease-relevant tissues, NSL genes clustered within modules significantly enriched for nuclear GO terms as expected for a chromatin-regulating protein complex. The enrichment of mitochondrial GO terms within modules containing NSL genes adds supporting evidence for the less well-characterized role of the NSL complex based at mitochondria.4,21 Furthermore, the enrichment of genes associated with sporadic Parkinson’s disease alongside NSL genes highlights a high level of correlation in gene expression. These results in particular support a functional interaction with the chromatin-regulating NSL complex and point to a biological link between multiple genes highlighted through GWASs. It would be difficult to investigate such interactions in any way other than using post-mortem tissue, given that this method captures the full background of the disease. However, this also means the results depend heavily on the numbers of samples available as this may limit the power of analyses. Particularly in the context of Parkinson’s disease, it may have been most interesting to focus on gene regulatory interactions within the substantia nigra, where much of the pathophysiological features of the disease are observed.67 However, within the GTEx dataset studied here, there are only 63 substantia nigra samples compared with 108 frontal cortex samples, thus complicating this analysis due to lack of power.36
Secondary frontal cortex GCNs allowed us to examine the activity of the genes encoding the NSL complex within different cell type contexts.40 Cell type enrichments annotated to modules containing NSL genes when present were almost always neuronal. These findings support in vitro experimental results demonstrating changes in multiple markers of mitochondrial dysfunction following NSL KD in neuronal models.4 Differences in activity in non-neuronal cell types needs to be validated further, perhaps by examining single-cell RNA-sequencing datasets.
GCN analysis was augmented by the use of ARACNe, a method for reverse engineering gene regulatory networks.64 By applying ARACNe-AP to members of the NSL complex, we identified significant overlaps between the predicted regulons, increasing our confidence in their representativeness of NSL complex function. Importantly, the NSL regulon, comprising genes predicted to be regulated by three or more NSL genes, was enriched for multiple pathways genetically-linked to Parkinson’s disease through common genetic variation and nominally enriched for heritability through stratified LDSC.65 This strengthens the notion that the NSL complex acts across the genome to regulate the expression of multiple different genes linked to Parkinson’s disease risk. eQTL analysis may be a way of testing this result more directly, by asking whether SNPs regulating the expression of NSL genes in cis also regulates the expression of genes relating to Parkinson’s disease in trans. However, very large sample numbers are required to power such an analysis.
Individual genes predicted to be regulated by the NSL complex served as prioritized targets for in vitro validation. Significant changes were detected in the expression of seven Parkinson’s disease-associated genes following KANSL1 and KAT8 KD, suggesting the NSL complex may act as a master regulator of multiple pathways implicated in disease pathology. Furthermore, existing publicly-available experimental data supported direct NSL complex activity at six of these genes, with additional disease-associated genes also found. We note that these regulatory relationships have been captured elsewhere in existing literature: RNA-sequencing of embryonic fibroblasts from KANSL1 knockout mice identified a decrease in PINK1, BIN3 and DGKQ, but found contrasting increases in CSTB and NCKIPSD.66 These results support the notion that the gene regulatory activity of the NSL complex has potential organism and cell type specificity.
In this study, we demonstrated the potential of in silico analyses to identify regulatory relationships between genes highlighted in GWASs and successfully validate such interactions in vitro. The NSL complex thus provides a potentially useful therapeutic target which could be used to simultaneously modulate a range of molecular pathways implicated in Parkinson’s disease, a strategy that might combat problematic compensatory responses that can occur when individual pathway components are targeted. Chromatin regulation is already an emerging target for the treatment of rare disorders, including specific cancers and developmental diseases, and we believe a similar approach may prove effective in neurodegenerative disorders.68–70
Supplementary Material
Acknowledgements
The authors would like to thank M.M. and C.G.P. from the lab of G.B. for their help setting up and running the QuantiGene Multiplex Assay.
Contributor Information
Amy R Hicks, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA.
Regina H Reynolds, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA; Department of Genetics and Genomic Medicine, Great Ormond Street Institute of Child Health, Bloomsbury, London WC1N 1EH, UK.
Benjamin O’Callaghan, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA.
Sonia García-Ruiz, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA; Department of Genetics and Genomic Medicine, Great Ormond Street Institute of Child Health, Bloomsbury, London WC1N 1EH, UK.
Ana Luisa Gil-Martínez, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA; Department of Genetics and Genomic Medicine, Great Ormond Street Institute of Child Health, Bloomsbury, London WC1N 1EH, UK; Department of Information and Communication Engineering, University of Murcia, Murcia 30100, Spain.
Juan Botía, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Department of Information and Communication Engineering, University of Murcia, Murcia 30100, Spain.
Hélène Plun-Favreau, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA.
Mina Ryten, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA; Department of Genetics and Genomic Medicine, Great Ormond Street Institute of Child Health, Bloomsbury, London WC1N 1EH, UK; NIHR GOSH Biomedical Research Centre, Great Ormond Street Institute of Child Health, Bloomsbury, London WC1N 1EH, UK.
Data availability
Raw data used to generate specificity matrices from substantia nigra and medial temporal gyrus are available at https://github.com/RHReynolds/MarkerGenes. Primary GCNs are available in the CoExpNets package (https://github.com/juanbot/CoExpNets) or on the CoExp website (www.rytenlab.com/coexp/Run/Catalog/). Details of the construction of secondary GCNs using the GMSCA package are available at https://github.com/drlaguna/GMSCA. Stratified LDSC analysis used the LDSCforRyten package (https://github.com/RHReynolds/LDSCforRyten). Instructions for the use of ARACNe-AP were utilized from https://github.com/califano-lab/ARACNe-AP and the Java package adapted for R is available alongside all code used for this paper at https://github.com/amyrosehicks/NSL_PD_relationships.29 Previously published, publicly available ChIP-sequencing datasets for H3K4me3 (GEO accession number: GSM4809272) and H4K8ac (GEO accession numbers: GSM4809295, GSM4809296, GSM4809297, GSM4809298) were downloaded from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE158736 and data were visualized using the ggtranscript R package (https://github.com/dzhang32/ggtranscript.git).6,58
Funding
A.H. was supported by the award of an Eisai-Leonard Wolfson Doctoral Training programme in Neurodegeneration. This research was funded in part by Aligning Science Across Parkinson’s (grant number ASAP 000478) through the Michael J. Fox Foundation for Parkinson’s Research (MJFF).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nalls MA, Pankratz N, Lill CM, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang D, Nalls MA, Hallgrímsdóttir IB, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soutar MPM, Melandri D, O’Callaghan B, et al. Regulation of mitophagy by the NSL complex underlies genetic risk for Parkinson’s disease at 16q11.2 and MAPT H1 loci. Brain. 2022;145:4349–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sheikh BN, Guhathakurta S, Akhtar A. The non-specific lethal (NSL) complex at the crossroads of transcriptional control and cellular homeostasis. EMBO Rep. 2019;20:e47630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Radzisheuskaya A, Shliaha PV, Grinev VV, et al. Complex-dependent histone acetyltransferase activity of KAT8 determines its role in transcription and cellular homeostasis. Mol Cell. 2021;81:1749–1765.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. International Parkinson Disease Genomics Consortium; Nalls MA, Plagnol V, et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soto-Beasley AI, Walton RL, Valentino RR, et al. Screening non-MAPT genes of the Chr17q21 H1 haplotype in Parkinson’s disease. Parkinsonism Relat Disord. 2020;78:138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skipper L, Wilkes K, Toft M, et al. Linkage disequilibrium and association of MAPT H1 in Parkinson disease. Am J Hum Genet. 2004;75:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Höglinger GU, Melhem NM, Dickson DW, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Allen M, Kachadoorian M, Quicksall Z, et al. Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alzheimers Res Ther. 2014;6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wray S, Lewis PA. A tangled web-tau and sporadic Parkinson’s disease. Front Psychiatry. 2010;1:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dong XJ, Liao ZX, Gritsch D, et al. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat Neurosci. 2018;21:1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bustos BI, Billingsley K, Blauwendraat C, et al. Genome-wide contribution of common short-tandem repeats to Parkinson’s disease genetic risk. Brain. 2022;146:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shiba-Fukushima K, Ishikawa KI, Inoshita T, et al. Evidence that phosphorylated ubiquitin signaling is involved in the etiology of Parkinson’s disease. Hum Mol Genet. 2017;26:3172–3185. [DOI] [PubMed] [Google Scholar]
- 16. Chung SY, Kishinevsky S, Mazzulli JR, et al. Parkin and PINK1 patient iPSC-derived midbrain dopamine neurons exhibit mitochondrial dysfunction and α-synuclein accumulation. Stem Cell Reports. 2016;7:664–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hsieh CH, Shaltouki A, Gonzalez AE, et al. Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell. 2016;19:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. [DOI] [PubMed] [Google Scholar]
- 19. Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. [DOI] [PubMed] [Google Scholar]
- 20. Biedler JL, Helson L, Spengler BA. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 1973;33:2643–2652. [PubMed] [Google Scholar]
- 21. Chatterjee A, Seyfferth J, Lucci J, et al. MOF Acetyl transferase regulates transcription and respiration in mitochondria. Cell. 2016;167:722–738 e23. [DOI] [PubMed] [Google Scholar]
- 22. Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dias J, Van Nguyen N, Georgiev P, et al. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev. 2014;28:929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. [DOI] [PubMed] [Google Scholar]
- 25. Taipale M, Rea S, Richter K, et al. HMOF Histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharma GG, So S, Gupta A, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Füllgrabe J, Lynch-Day MA, Heldring N, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin AR, Williams E, Foulger RE, et al. Panelapp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet. 2019;51:1560–1565. [DOI] [PubMed] [Google Scholar]
- 29. Hicks AR. amyrosehicks/NSL_PD_relationships: v1. Published 11 January 2023. doi:10.5281/zenodo.7525823
- 30. Skene NG, Grant SG. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front Neurosci. 2016;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hodge RD, Bakken TE, Miller JA, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agarwal D, Sandor C, Volpato V, et al. A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat Commun. 2020;11:4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chia R, Sabir MS, Bandres-Ciga S, et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. 2021;53:294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mencacci NE, Reynolds R, Ruiz SG, et al. Dystonia genes functionally converge in specific neurons and share neurobiology with psychiatric disorders. Brain. 2020;143:2771–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferrari R, Forabosco P, Vandrovcova J, et al. Frontotemporal dementia: Insights into the biological underpinnings of disease through gene co-expression network analysis. Mol Neurodegener. 2016;11:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group , et al. Genetic effects on gene expression across human tissues. Nature. 2017; 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Botía JA, Vandrovcova J, Forabosco P, et al. An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks. BMC Syst Biol. 2017;11:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. García-Ruiz S, Gil-Martínez AL, Cisterna A, et al. Coexp: a web tool for the exploitation of co-expression networks. Front Genet. 2021;12:630187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanchez JA, Gil-Martinez AL, Cisterna A, et al. Modeling multifunctionality of genes with secondary gene co-expression networks in human brain provides novel disease insights. Bioinformatics. 2021;37:2905–2911 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reynolds RH. Common utility functions. https://github.com/RHReynolds/rutils
- 42. Kolberg L, Raudvere U, Kuzmin I, Vilo J, Peterson H. Gprofiler2 – an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res. 2020;9:709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lachmann A, Giorgi FM, Lopez G, Califano A. ARACNe-AP: gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics. 2016;32:2233–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics. 2002;18(Suppl 2):S231–S240. [DOI] [PubMed] [Google Scholar]
- 45. Visscher PM, Hill WG, Wray NR. Heritability in the genomics era–concepts and misconceptions. Nat Rev Genet. 2008;9:255–266. [DOI] [PubMed] [Google Scholar]
- 46. Yang J, Bakshi A, Zhu Z, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Finucane HK, Bulik-Sullivan B, Gusev A, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen Z, Reynolds RH, Pardiñas AF, et al. The contribution of Neanderthal introgression and natural selection to neurodegenerative diseases. bioRxiv. [Preprint] doi: 10.1101/2022.04.29.490053 [DOI] [PubMed]
- 50. International HapMap 3 Consortium, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.1000 Genomes Project Consortium, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Flagella M, Bui S, Zheng Z, et al. A multiplex branched DNA assay for parallel quantitative gene expression profiling. Anal Biochem. 2006;352:50–60. [DOI] [PubMed] [Google Scholar]
- 53. Papadopoulou AS, Gomez-Paredes C, Mason MA, Taxy BA, Howland D, Bates GP. Extensive expression analysis of Htt transcripts in brain regions from the zQ175 HD mouse model using a QuantiGene multiplex assay. Sci Rep. 2019;9:16137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grenn FP, Kim JJ, Makarious MB, et al. The Parkinson’s disease genome-wide association study locus browser. Mov Disord. 2020;35:2056–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blauwendraat C, Reed X, Krohn L, et al. Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia. Brain. 2020;143:234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kia DA, Zhang D, Guelfi S, et al. Identification of candidate Parkinson disease genes by integrating genome-wide association study, expression, and epigenetic data sets. JAMA Neurol. 2021;78:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gustavsson EK, Zhang D, Reynolds RH, Garcia-Ruiz S, Ryten M.. ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics. 2022;38:3844–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reynolds RH, Botia J, Nalls MA, et al. Moving beyond neurons: the role of cell type-specific gene regulation in Parkinson's disease heritability. NPJ Parkinsons Dis. 2019;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hawkes CH, Del Tredici K, Braak H. Parkinson’s disease: a dual-hit hypothesis. Neuropathol Appl Neurobiol. 2007;33:599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Parker WD Jr., Parks JK, Swerdlow RH. Complex I deficiency in Parkinson’s disease frontal cortex. Brain Res. 2008;1189:215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thomas RR, Keeney PM, Bennett JP. Impaired complex-I mitochondrial biogenesis in Parkinson disease frontal cortex. J Parkinsons Dis. 2012;2:67–76. [DOI] [PubMed] [Google Scholar]
- 63. Butler PM, Chiong W. Neurodegenerative disorders of the human frontal lobes. Handb Clin Neurol. 2019;163:391–410. [DOI] [PubMed] [Google Scholar]
- 64. Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bandres-Ciga S, Saez-Atienzar S, Kim JJ, et al. Large-scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease. Acta Neuropathol. 2020;140:341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li T, Lu D, Yao C, et al. Kansl1 haploinsufficiency impairs autophagosome-lysosome fusion and links autophagic dysfunction with Koolen-de Vries syndrome in mice. Nat Commun. 2022;13:931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ye H, Robak LA, Yu M, Cykowski M, Shulman JM. Genetics and pathogenesis of Parkinson’s syndrome. Annu Rev Pathol. 2022;18:95–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pallotta MM, Di Nardo M, Sarogni P, Krantz ID, Musio A. Disease-associated c-MYC downregulation in human disorders of transcriptional regulation. Hum Mol Genet. 2022;31:1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mastoraki A, Schizas D, Vlachou P, et al. Assessment of synergistic contribution of histone deacetylases in prognosis and therapeutic management of sarcoma. Mol Diagn Ther. 2020;24:557–569. [DOI] [PubMed] [Google Scholar]
- 70. McCabe MT, Creasy CL. EZH2 As a potential target in cancer therapy. Epigenomics. 2014;6:341–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data used to generate specificity matrices from substantia nigra and medial temporal gyrus are available at https://github.com/RHReynolds/MarkerGenes. Primary GCNs are available in the CoExpNets package (https://github.com/juanbot/CoExpNets) or on the CoExp website (www.rytenlab.com/coexp/Run/Catalog/). Details of the construction of secondary GCNs using the GMSCA package are available at https://github.com/drlaguna/GMSCA. Stratified LDSC analysis used the LDSCforRyten package (https://github.com/RHReynolds/LDSCforRyten). Instructions for the use of ARACNe-AP were utilized from https://github.com/califano-lab/ARACNe-AP and the Java package adapted for R is available alongside all code used for this paper at https://github.com/amyrosehicks/NSL_PD_relationships.29 Previously published, publicly available ChIP-sequencing datasets for H3K4me3 (GEO accession number: GSM4809272) and H4K8ac (GEO accession numbers: GSM4809295, GSM4809296, GSM4809297, GSM4809298) were downloaded from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE158736 and data were visualized using the ggtranscript R package (https://github.com/dzhang32/ggtranscript.git).6,58