Abstract
Stuttering is a common speech disorder that interrupts speech fluency and tends to cluster in families. Typically, stuttering is characterized by speech sounds, words or syllables which may be repeated or prolonged and speech that may be further interrupted by hesitations or ‘blocks’. Rare variants in a small number of genes encoding lysosomal pathway proteins have been linked to stuttering. We studied a large four-generation family in which persistent stuttering was inherited in an autosomal dominant manner with disruption of the cortico-basal-ganglia-thalamo-cortical network found on imaging. Exome sequencing of three affected family members revealed the PPID c.808C>T (p.Pro270Ser) variant that segregated with stuttering in the family. We generated a Ppid p.Pro270Ser knock-in mouse model and performed ex vivo imaging to assess for brain changes. Diffusion-weighted MRI in the mouse revealed significant microstructural changes in the left corticospinal tract, as previously implicated in stuttering. Quantitative susceptibility mapping also detected changes in cortico-striatal-thalamo-cortical loop tissue composition, consistent with findings in affected family members. This is the first report to implicate a chaperone protein in the pathogenesis of stuttering. The humanized Ppid murine model recapitulates network findings observed in affected family members.
Keywords: chaperone, cyclophilin-40, brain MRI, PPID gene, stuttering
Morgan et al. identify a mutation in a gene encoding a chaperone protein that segregates with stuttering in a large four-generation family. A mouse model with the same mutation shows changes in the cortico-striatal-thalamo-cortical loop similar to those seen in affected family members.
See Szepetowski (https://doi.org/10.1093/brain/awad369) for a scientific commentary on this article.
Introduction
Stuttering affects around 1% of the population.1 Similar to other neurodevelopmental disorders, more males are affected.2–4 A clinical diagnosis of stuttering is based on the frequency of stuttering behaviours during conversational speech or reading.2 These difficulties typically begin in early childhood during the rapid development of speech and language skills.3 In over two-thirds of cases with childhood onset, stuttering eventually resolves; however, the remainder have persistent developmental stuttering.4
Evidence from twin and adoption studies (reviewed by Frigerio-Domingues and Drayna3 and Kang and Drayna5) points to genetic aetiologies for stuttering. Although this evidence is strong, including consistently higher concordance for stuttering in monozygotic compared to dizygotic twins and high (often >0.8) heritability estimates, the genetic architecture underlying stuttering remains poorly understood. Monogenic contributions are rare with the only molecular pathways implicated in stuttering consisting of rare variants in lysosomal targeting pathway genes (GNPTAB, GNPTG and NAGPA), which were initially identified in consanguineous Pakistani families with non-syndromic persistent stuttering, and subsequently in unrelated Pakistani and North American individuals.6 More recently, rare loss of function mutations in AP4E1 have been described in stuttering cases from Cameroon and Pakistan; this gene encodes a protein that is functionally related to the lysosomal targeting pathway.7 The relevance of these four genes to a broader population with stuttering is not known.
Here, we study a large family with developmental stuttering and implicate a new molecular pathway. Phenotypic, genetic and imaging analyses of the family, and a humanized mouse model, support a pathogenic variant in PPID as a novel cause for stuttering.
Materials and methods
Family and sporadic cases
We studied 27 members of a four-generation Australian Caucasian family with persistent developmental stuttering of whom 16 were genotyped for linkage mapping, as described previously (Fig. 1).8 We also studied 46 unrelated Australian Caucasian individuals with stuttering and 558 healthy controls. The Human Research Ethics Committee of the Royal Children’s Hospital, Melbourne, Australia (HREC 37353) and the Walter and Eliza Hall Institute of Medical Research (HREC G18/06) approved this study. Written informed consent was obtained from family members or their parents or legal guardians in the case of minors according to the Declaration of Helsinki. Resequencing was completed in 977 individuals with stuttering, and 809 gender and age-matched controls from four populations (Brazilian, Cameroonian, Caucasian and Pakistani) at the US National Institutes of Health enrolled with written informed consent under Institutional Review Board-approved protocol 97-DC-0057.
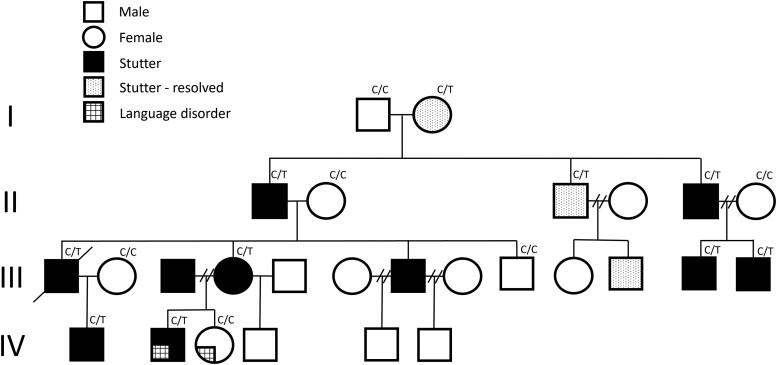
Figure 1.
Family with developmental stuttering and the PPID variant. Pedigree showing segregation of the c.808C>T (p.Pro270Ser) missense variant in PPID.
Biological samples
Venous blood or saliva was obtained from family members and sporadic patients. For blood, genomic DNA was extracted using the Qiagen QIAamp DNA Maxi Kit. For saliva obtained using the Oragene kit, genomic DNA was extracted using the prepIT·L2P kit (DNA Genotek Inc.).
Exome sequencing
Exome sequencing was performed using 3 μg of venous blood-derived genomic DNA from three family members (Individuals III-12, IV-1 and IV-2; Fig. 1A). Genomic DNA was sonicated to approximately 200 base pair fragments and adaptor-ligated to make a library for paired-end sequencing. Following amplification and barcoding, the libraries were hybridized to biotinylated complementary RNA oligonucleotide baits from the SureSelect Human All Exon 62Mb Kit (Agilent Technologies) and purified using streptavidin-bound magnetic beads as described previously.9 Sequencing was performed on the Illumina HiSeq 2000 system at 50-fold on-target depth. Exome sequencing reads were aligned with Novoalign version 3.02.00 (http://www.novocraft.com/) to the human assembly hg19. PCR duplicates were removed using MarkDuplicates from Picard (http://picard.sourceforge.net). Variant calling was performed with GATK (v.3.5) HaplotypeCaller and variant annotation run with ANNOVAR.10 Exome variants were filtered and selected according to the following criteria: location within a linkage region, genotype fitting the heterozygous dominant disease model, a minor allele frequency ≤ 0.01 according to the gnomAD (http://gnomad.broadinstitute.org) database, and variant type (missense, nonsense, coding indel or splice site variant). Twenty-eight variants from the exome analysis including PPID c.808C>T (p.Pro270Ser) were studied further (Supplementary Table 1).
PCR and Sanger sequencing
Primers were designed to PCR amplify and Sanger sequence the 28 variants in family members. We performed sequencing to detect the PPID c.808C>T (p.Pro270Ser) variant located in exon 7 in all family members and 558 healthy controls. Amplification reactions were cycled using a standard protocol on a Veriti Thermal Cycler (Applied Biosystems). Bidirectional sequencing of all exons and flanking regions (∼50 bp including splice site regions) was completed with a BigDyeTM v3.1 Terminator Cycle Sequencing Kit (Applied Biosystems), according to the manufacturer’s instructions. Sequencing products were resolved using a 3730xl DNA Analyzer (Applied Biosystems). All sequencing chromatograms were compared to published cDNA sequence; nucleotide changes were detected using Codon Code Aligner (CodonCode Corp.).
Resequencing studies
PPID (OMIM ID: 601753) was amplified and Sanger sequenced as described above using specific primers (Supplementary Table 2) designed to each of the 10 exons in the reference human gene transcript (NM_005038, NCBI Gene; http://www.ncbi.nlm.nih.gov/) in 46 Australian Caucasian individuals with stuttering. Sequence data from 977 individuals with stuttering and 809 matched controls from four populations (Brazilian, Cameroonian, Caucasian, Pakistani) was generated by exome capture with Illumina TruSeq Rapid Exome (45 Mb) Capture kit at an average of 70-fold coverage, as described previously (Supplementary Table 3).6
Generation of Ppid knock-in mice
The humanized knock-in Ppid c.808C>T (p.Pro270Ser) mice were generated by the MAGEC laboratory (WEHI) as previously described11 on a C57BL/6J background. To generate the mice, 20 ng/μl of Cas9 mRNA, 10 ng/μl of sgRNAs (GAGCTGTACATAATTTGTAT and GGGTCTCTCTCGGCAAAGGA) and 5 ng/μl of plasmid targeting vector (see Supplementary Fig. 1 for sequence) were injected into the pronucleus of fertilized one-cell stage embryos. Twenty-four hours later, two-cell stage embryos were transferred into the oviducts of pseudo-pregnant female mice. Viable offspring were genotyped by PCR at weaning. Mice were housed and studied according to the approval of the Florey Institute of Neuroscience and Mental Health Animal Ethics Committee (Project No. 19-054-FINMH).
MRI acquisition
Excised brains from n = 8 wild-type and n = 8 heterozygous Ppid c.808C>T, p.Pro270Ser (Ppid Het) mice were scanned using a Bruker 9.4 T MRI and actively decoupled volume transmit and four-channel receive surface cryocoil. Multi-gradient echo images were acquired in the axial plane with minimum echo time (TE) = 5 ms and echo spacing = 5 ms. Ten echoes were acquired with the following imaging parameters: repetition time (TR) = 62 ms; field of view (FOV) = 16.32 × 12.24 × 7.14 mm3; matrix size = 192 × 144 × 84; and spatial resolution = 85 × 85 × 85 μm3. Diffusion weighted imaging (DWI) was performed with imaging parameters: TR/TE = 10 000/38 ms; δ = 4 ms; Δ = 14 ms; b = 2000 and 4000 s/mm2; 126 diffusion directions, two minimum diffusion images (b0), FOV = 19.2 × 12.8 mm2; matrix size = 96 × 64; and 34 slices with slice thickness = 200 μm.
MRI analysis
Diffusion weighted images were preprocessed for fixel-based analysis (FBA) using the multi-shell multi-tissue constrained spherical deconvolution pipeline using MRTrix3 v3.0.3, supplemented by FMRIB Software Library v6.0 and Advanced Normalisation Tools v2.3.5 toolboxes. In short, the images were denoised, de-ringed, motion and eddy current corrected, and bias field corrected. White matter, grey matter and CSF response functions were estimated and averaged. Whole-brain tractography was performed using a population-specific white matter fibre orientation distribution template generated from all images from both groups. Twenty million streamlines were seeded and subsequently reduced to 2 million using spherical deconvolution informed filtering. Three fixel-based metrics were computed: fibre cross-section (logFC), fibre density and the combined fibre density and cross-section.
The corpus callosum and bilateral corticospinal tracts were extracted for further a priori analyses. Both fixel-based and diffusion tensor imaging (DTI)-metrics were sampled at 20 equidistant points along the corpus callosum and 30 equidistant points along the left and right corticospinal tracts. Metrics derived from subject fixels, DTI, quantitative susceptibility mapping (QSM) and rate of darkening (R2*) images were sampled at each point and outliers identified and removed using Gibbs’ tests (α = 0.05). For each brain, the area under the curve (AUC; or more accurately in some cases, area above the curve) was calculated for comparison.
Fractional anisotropy (FA), axial diffusivity (AD), radial diffusivity (RD) and apparent diffusion coefficient (ADC) images were estimated using MRTrix3. In addition to corpus callosum and pyramidal tract-of-interest based analyses, DTI metrics were also assessed using tract-based spatial statistics with a skeleton threshold = 0.25.
QSM images were generated from transverse relaxation time (T2*) data using MATLAB (r2017, MathWorks, Natick, Massachusetts) with V-SHARP for background removal and non-linear dipole inversion12 for dipole inversion. To reduce the effects of streaking on susceptibility maps, white matter and grey matter maps of stable susceptibility regions were derived from the wild-type group’s mean QSM image in a similar approach to that previously described.13,14 The accepted thresholds were: coefficient of variation < 0.8, χWM < −0.01 ppm for white matter regions, and 0 < χGM < 0.02 ppm for grey matter regions. All CSF regions were excluded, and the mean bulk susceptibilities calculated for group-wise comparison.
As the cortico-striatal-thalamo-cortical (CSTC) loop has been implicated in stuttering,15 we undertook a hypothesis driven analysis of six CSTC specific regions of interest: thalamus, substantia nigra, globus pallidus, caudoputamen, primary motor cortex (M1) and primary somatosensory cortex (S1). Regions of interest were delineated using the Waxholm Space mouse brain atlas.16 As M1 and S1 are undefined in the mouse atlas, we utilized M1 and S1 segmentations from the Sprague-Dawley version of the atlas to define these regions. Affine and 3D symmetric diffeomorphic registrations between individual and atlas space were determined using ANTs and MRTrix3 with cross-correlation as the similarity metric.
Vocalization recordings
Mice ultrasonic vocalizations (USVs) were elicited, recorded and analysed to evaluate communication. USVs were elicited using the pup separation assay, as previously described.17 Briefly, pups were removed from the dam prior to testing at postnatal Day 4. The dam was removed from the cage and placed in a new clean cage away from the home cage. The home cage containing pups was placed in an incubator at 34°C. Before testing, the auxiliary temperature of the pups was taken with a flexible thermistor on the back of the animal. Ten minutes after the dam’s removal from the home cage, each pup was placed into the test chamber alone. Pup USVs were recorded for 3 min. Pups were weighed and marked on the back with an odourless ink to allow for subsequent identification and returned to the dam post recording. An Ultrasound Microphone (Avisoft UltraSoundGate condenser microphone capsule CM16, Avisoft Bioacoustics) recorded USVs at a sampling rate of 250 kHz. The microphone was connected to a PC via an UltraSoundGate 116 H recording interface and audio was captured using Avisoft-RECORDER USGH software (Version 4.2.29).
USVs were analysed using Avisoft SASLab Pro (Version 5.2.14). USVs were first identified visually.18 Call type was allocated as previously described19 into one of nine predefined call categories including: (i) complex; (ii) two components; (iii) upward; (iv) downward; (v) chevron; (vi) short; (vii) composite; (viii) frequency step; and (ix) flat (see Vogel et al.18 for indicative spectrum constellation of the call types). Calls were examined for count, inter-utterance duration and categorized into complex and simple types (Supplementary Tables 5–7 and Supplementary Fig. 2).
Call counts, call lengths, and inter-call durations were compared between heterozygous Ppid p.Pro270Ser mice and wild-type littermates. Call counts were defined per mouse, as the number of recorded calls. Call lengths were calculated as the end minus the start time. Calls with length = 0 were excluded from analyses of call length. Inter-call durations were calculated as the length of time between the end time of a call, and the start time of the following call. Inter-call durations were classified based on the categories of the calls either side of the gap: ‘Simple, Simple’; ‘Simple, Complex’, ‘Complex, Simple’, ‘Complex, Complex’ (simple calls include flat, upward, downward, short; complex calls were characterized by harmonic or fluctuating frequencies including chevron, two-components, composite, frequency steps, complex).18 For Inter-call durations with calculated length < 0, the duration was set to 0. The duration between the start of recording and the first call, and the duration between the final call and the end of recording were excluded.
For each of the three outcome measures, simple univariable associations with genotype, sex, litter, weight and temperature were first undertaken. Generalized linear mixed models were then fitted, testing associations between genotype and each outcome, with adjustment for relevant covariates (P < 0.05 in the univariable associations). For call counts, negative binomial models were fitted with a random intercept for litter. For call lengths and inter-call durations, linear mixed models were used, with each outcome log-transformed, and litter and mouse as nested random effects. For call lengths, sex was included as an additional fixed effect covariate. All models were fitted separately for each call (or inter-call) category. An additional model was fitted for all calls, with category included as an additional fixed effect covariate.
Calls were then analysed objectively via acoustic analysis protocols.18 One hundred acoustic features included measures of quality, duration, amplitude and entropy across each call. Dimensionality reduction of features was undertaken using principal component analysis, with components selected based on examination of a scree plot. Associations between principal components and genotype were examined using linear mixed models, with litter and mouse as nested random effects.
Behavioural studies
Prior to each behavioural experiment mice were allowed to acclimatize in their home cages in the behavioural room for at least 1 h.
Open-field exploratory locomotion assay
Mice were placed in a square 27.3 × 27.3 × 20.3 cm3 open-field arena and were allowed to move freely for 1 h, during which infrared rays tracked their movement. Data indicating distance travelled, time travelled, and other parameters were recorded and compiled using MED Associates Activity Monitor software (Med Associates Inc.).
Rotarod assay
Mice were trained to walk on the rotarod over three sessions: two 2 min sessions at a fixed speed of 4 rpm, and one 2 min session accelerating from 4 to 20 rpm. Mice were then tested on the rotarod for three 10-min trials (with at least 30 min between trials). Each trial consisted of 5 min where the rotarod accelerated from 4 to 40 rpm, followed by 5 min at a fixed speed of 40 rpm. Latency to fall was recorded. Average time on the rotarod across the three trials was analysed.
Light-dark transition test
A square mouse open field arena was divided into two zones of equal size (27.3 × 13.7 cm2). One half of each arena was illuminated at 750 ± 25 lx using a small lamp (the ‘light’ zone), while the other half was enclosed with a black acrylic box insert with a lid and a small opening on one side (the ‘dark’ zone). Mice were individually placed in the dark zone and allowed to freely explore both zones for 10 min. Data indicating the duration of time spent in each zone was recorded and compiled using MED Associates Activity Monitor software (Med Associates Inc.).
Elevated plus maze
The elevated plus maze consists of a plus-shaped platform raised 40 cm from the floor. Two of the arms, on opposite sides, are enclosed with 55 cm high walls extending from the centre, with the other two arms remaining open with no walls. Mice were individually placed in the centre of the maze facing one of the open arms and were allowed to freely roam the maze for 10 min, and their movements were tracked by a ceiling-mounted video camera. Entries into each arm, along with their respective durations, were measured using tracking software (CleverSys TopScan Lite). Percentage of time spent in the open arm was calculated as:
| (1) |
Y maze
The Y maze consists of a Y-shaped Perspex enclosure with three 7.5 × 30 × 14 cm3 arms each containing a distinct visual cue at the end. The test was performed in two phases: a training session and a test session, with a 1-h interval in between. During the training session, access to one of the three arms (the ‘novel’ arm) was blocked by an opaque Perspex barrier. Mice were allowed to explore the two accessible arms (the ‘home’ arm—where they are first placed—and the ‘familiar’ arm) of the maze for 10 min. During the test session, the barrier blocking the novel arm was removed and mice were placed into the home arm and allowed to explore all three arms for 5 min. The time the mouse spent in each of the three arms was measured using tracking software (CleverSys TopScan Lite). Mice with intact spatial working memory would be expected to spend more time in the novel arm than the home or familiar arms during the test session.
Three-chamber social interaction assay
This test was conducted in a 43 × 39 × 11 cm3 enclosure made of transparent plastic and divided into three chambers linked by small rectangular openings that mice could freely move through. Metal mesh cages (16 × 10 × 11 cm3) were placed at one end of each of the two lateral chambers. The assay consisted of two trials, habituation and test, which occurred successively. During the habituation phase, mice were individually placed into the central chamber and allowed to freely explore all three chambers for 10 min. Their movements were tracked using a ceiling-mounted video camera and measured using tracking software (CleverSys TopScan Lite). The side where each mouse preferentially spent more time was noted. During the test phase, an unfamiliar age- and sex-matched ‘stranger’ mouse was placed into the mesh cage on the opposite side to the test mouse’s preference. The test mouse was then returned to the enclosure and allowed to explore all three chambers for 10 min. Time spent interacting with the stranger mouse was quantified using tracking software (CleverSys TopScan Lite).
RNA sequencing analysis
Brain tissue was harvested from 10 mice, five Ppid p.Pro270Ser knock-in and five wild-type mice. RNA was extracted and libraries were prepared for RNA sequencing using the Illumina TruSeq strand-specific kit (Illumina). RNA sequencing was performed on the NovaSeq 6000 generating 151 bp, paired-end reads, with a mean of 39.4 million paired-end reads per sample (range 33.8 to 46.6 million).
RNA-seq reads were aligned to the mm10 reference genome using STAR (v2.7.9a)20 and the number of reads mapping to each gene was quantified using salmon (v1.5.2)21 as implemented in the nf-core/rnaseq pipeline (v3.6).22 Statistical analysis was undertaken to search for genes that were differentially expressed between knock-in and wild-type mice. We compared the expression of Ppid and 12 other genes (Ahsa1, Esr1, Fkbp4, Fkbp5, Hsp90aa1, Hsp90ab1, Mapk1, Ppp5c, Ptges3, S100a1, Stip1, Vdac1) that cluster with Ppid in the STRING database of protein-protein interactions (Supplementary Fig. 3).23 Genome-wide differential expression analysis was performed for 14 367 genes remaining after genes with low expression (counts per million < 0.5 in more than one sample) and genes on the sex chromosomes were excluded. We used the edgeR Bioconductor package24 to fit a generalized linear model including covariates for mouse sex, which was inferred based on expression of all Y chromosome genes. We accounted for multiple testing by controlling the false discovery rate (FDR) using the Benjamini–Hochberg procedure, with a threshold of FDR < 0.05 required for a gene to be considered statistically significant.
Molecular dynamics simulations and docking calculations
Structures of murine Cyp40 and the [Pro270Ser]-Cyp40 variant were modelled using AlphaFold2.25,26 MD simulations of both proteins were performed with GROMACS 2022.5 using the CHARMM36m force field.27 Each protein was solvated using 47472 TIP3P28 water molecules in a dodecahedral box, with a minimum distance of 1.5 nm between any protein atom to the box edge. Energy minimization was performed using the steepest-descent gradient method. For subsequent MD simulations, time steps of 2 fs were used. Bond lengths were constrained using the LINCS algorithm29 and long-range electrostatic forces were calculated using the particle-mesh Ewald scheme (PME).30 A cut-off radius of 1.2 nm for Coulomb and van der Waals potentials were used for the calculation of short-range nonbonded interactions. Equilibration simulations were performed in which all non-hydrogen atoms were positionally restrained, firstly using a canonical (NVT) ensemble followed by an isothermal-isobaric ensemble (NPT) for 0.1 ns each. For the equilibration simulations, the temperature was maintained at 310 K with a modified Berendsen thermostat,31 and pressure kept at 1.0 bar with the Parrinello–Rahman barostat.32 Subsequently, unrestrained production simulations were carried out for 340 ns for each system, with the temperature maintained at 310 K using the Bussi velocity-rescaling thermostat,33 while the pressure was maintained at 1.0 bar with the Parrinello–Rahman barostat. The stabilities of the MD simulation trajectories were analysed using root mean square deviation (RMSD). Visual Molecular Dynamics (1.9.3)34 was used to examine the trajectories and produce graphical molecular representations.
Autodock Vina (1.2.0)35,36 was subsequently used to perform molecular docking calculations to predict the binding pose of the Hsp90 C-terminal peptide, Ace-MEEVD, on the final simulation frame of both wild-type Cyp40 and the [Pro270Ser]-Cyp40 variant. The Hsp90 peptide was extracted from the RCSB structure with PDBID 3KD7.37 AutodockTools38 was used to convert the Hsp90 peptide, Cyp40 and [P270S]-Cyp40 to pdbqt format. All torsions angles, with the exception of those associated with backbone peptide bonds, were set to be freely rotatable. PyRx (version 0.8)39 was used to prepare the docking input files. Boxes centred on the Cɑ atom of position 270 with dimensions of 32.56 × 22.12 × 38.86 Å3 were used for both docking calculations. An exhaustiveness value of 48 was used for both docking calculations.
Results
Family report
A four-generation Australian family of Caucasian origin with stuttering was studied (Fig. 1), as described previously.8 Thirteen of 27 family members had a history of stuttering, which was persistent in 10/13 members, and self-reported as resolved in 3/13 (Individuals I-2, II-3, III-11). We previously established8 the neural phenotype using quantitative MRI, revealing that affected family members had failed to follow the typical trajectory of age related thinning of Broca’s area revealed in controls. Anomalies were also present bilaterally in the middle frontal gyrus and globus pallidus. A larger right globus pallidus was correlated with more severe stuttering. The lack of typical development of these structures reflects the anatomical basis of the abnormal inhibitory control network between Broca’s area and the striatum underpinning stuttering.8 Parametric linkage analysis was used to map genetic loci with peak logarithm of the odds (LOD) scores of 3.0088 on chromosomes 1 and 4.8
Genotyping of family with exome sequencing analysis
We performed exome sequencing on three affected family members with persistent stuttering (Individuals III-12, IV-1, IV-2; Fig. 1) and interrogated variation in the previously identified broad linkage regions on chromosomes 1 and 4.8 We conducted segregation analysis of 28 variants detected on exome sequencing (Supplementary Table 1). The peptidyl-prolyl isomerase D (PPID) gene variant (c.808C>T, p.Pro270Ser) segregated with stuttering in the family (Fig. 1) and was absent from 558 Caucasian healthy controls and the gnomAD database of 141 456 individuals at time of analysis in 2017. It is now present in gnomAD (v3.1.2, 2023) and has been observed once in 152 162 alleles (minor allele frequency of ∼0.000007; rs1203927418). This allele frequency is very low, unsurprising in the context of stuttering which affects ∼1% of the population.3 The only other variant identified to segregate with stuttering in the family was in RPTN on chromosome 1. We considered digenic inheritance, and while we cannot exclude this possibility, the RPTN variant (chr1:152129405; A>T; p.Arg57Asn; rs200282084) is present in 287 of 152 212 alleles with a much higher minor allele frequency of 0.001886 in gnomAD (v3.1.2, 2023) compared to the PPID p.Pro270Ser variant.
We next analysed exome, genome or Sanger sequencing data from 46 Australian Caucasian individuals with stuttering, and 977 individuals with stuttering and 809 population-matched controls from four populations (Brazilian, Cameroonian, Caucasian, Pakistani), as outlined in Supplementary Table 3. Interrogation of the 10 coding exons and splice sites of the PPID gene revealed 48 variants in stuttering cases and 38 variants in controls without stuttering (Supplementary Table 3). However, comparison of PPID variation between stuttering cases and controls divided into the four population sub-groups did not reveal any statistically significant differences in the frequency of variants in PPID (Supplementary Table 3). Although 16 different coding variants in PPID were discovered, the c.808C>T (p.Pro270Ser) variant was the only pathogenic one.
MRI analysis of knock-in humanized Ppid mice
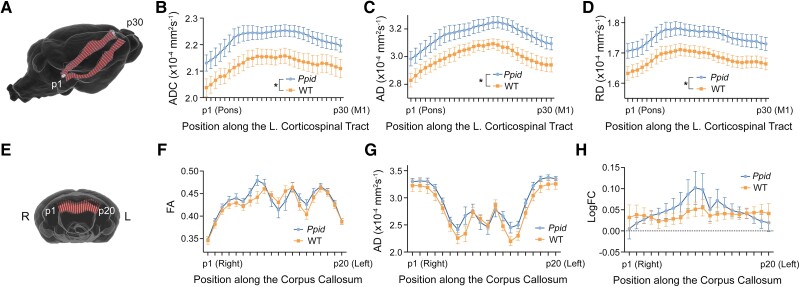
Excised brains from wild-type and Ppid Het (p.Pro270Ser) knock-in mice (n = 8 per group) were scanned using a Bruker 9.4 T MRI to investigate for structural changes due to the humanized Ppid missense substitution. We performed exploratory whole-brain and tract-of-interest analyses of the white matter using DWI, an MRI technique sensitive to changes in the tissue microstructure. No differences were observed in any diffusion metric using connectivity-based fixel enhancement or tract-based spatial statistics; however, analysis of the corticospinal tracts (Fig. 2A) revealed significant group-wise differences in DTI metrics. Ppid Het mice had significantly increased ADC [F(1,13) = 6.350, P = 0.0256; Fig. 2B], AD [F(1,13) = 5.976, P = 0.0295; Fig. 2C] and RD [F(1,13) = 6.673, P = 0.0227; Fig. 2D] in the left corticospinal tract when compared to wild-type (n = 7 following outlier testing). There were no significant differences in fractional anisotropy (FA) or any metric in the right corticospinal tract. In the corpus callosum (Fig. 2E), there were interactions between genotype and position along the tract for mean FA [F(19,266) = 2.470, P = 0.0008; Fig. 2F], mean AD [F(19,266) = 1.727, P = 0.0319; Fig. 2G] and fibre cross-section [logFC; F(19,249) = 2.551, P = 0.0005; Fig. 2H].
Figure 2.
Diffusion-weighted imaging (DWI) reveals significant microstructural white matter changes in Ppid mice when compared to wild-type (WT). (A) The corticospinal tracts were segmented by selecting streamlines that connected the primary motor cortex (M1) and pons. Diffusion metrics were sampled at 30 points along the left and right corticospinal tracts from the pons (p1) to M1 (p30). Significant differences were observed in the left corticospinal tract for (B) apparent diffusion coefficient (ADC), (C) axial diffusivity (AD) and (D) radial diffusivity (RD). (E) A similar segmentation and analysis of the corpus callosum was performed from the right (p1) to left (p20) hemispheres, with significant interactions observed between genotype and position for (F) fractional anisotropy (FA), (G) AD and (H) fibre cross-section (FC). Graphs show mean ± SEM with Ppid shown in blue and WT in orange. *Significant difference between Ppid and WT, P < 0.05.
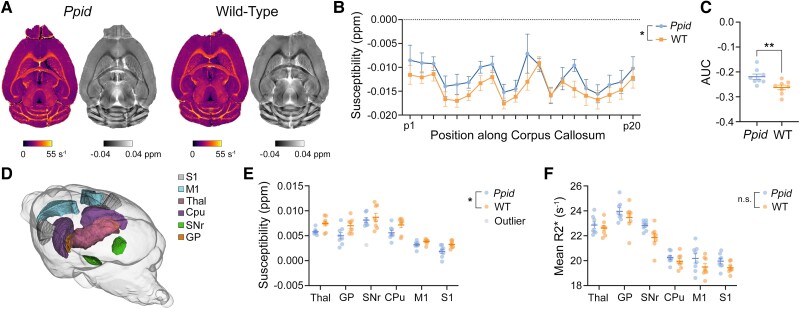
Using high-resolution transverse relaxation time (T2*)-weighted imaging we assessed QSM images and rate of darkening (R2*) values along the DWI-defined corticospinal tracts and corpus callosum. Representative QSM and R2* images are shown in Fig. 3A. While no statistically significant changes in bulk susceptibility were observed in either of the corticospinal tracts, we found a significant increase in Ppid Het QSM values of the corpus callosum when compared to wild-type mice [F(1,14) = 8.166, P = 0.0127; Fig. 3B]. Calculation of the area below the curve (AUC) also yielded significant susceptibility differences between the two groups [t(14) = 3.003, P = 0.0095; Fig. 3C]. No significant differences were reported for R2* values in any of the fibre bundles.
Figure 3.
T2*-weighted imaging reveals significant changes in Ppid mouse brain tissue susceptibility. (A) Representative R2* (left, pseudocolour) and quantitative susceptibility map (QSM, right, greyscale) images for Ppid and wild-type (WT) mice. (B) Analysis of mean susceptibility along the corpus callosum revealed significantly increased values in Ppid mice (blue line) compared to WT (orange line) and (C) a significant difference in the area under the curves (AUC). (D) We assessed six brain structures of the cortico-striatal-thalamo-cortical (CSTC) loop, shown here in a glass brain: S1 = primary somatosensory cortex; M1 = primary motor cortex; Thal = thalamus; Cpu = caudoputamen; SNr = substantia nigra; and GP = globus pallidus. (E) Mean susceptibility values were significantly decreased in Ppid mice when compared to WT. (F) Mean R2* values in the CSTC loop tended to be elevated in Ppid mice; however, this did not reach significance. Graphs show mean ± SEM. Significant difference between Ppid and WT, *P < 0.05, **P < 0.01.
We also observed decreased bulk susceptibility in the CSTC (Fig. 3D) loop of Ppid mice when compared to wild-type [mixed-effects analysis, F(1,14) = 6.676, P = 0.0216, n = 8 Ppid Het, n = 8 wild-type except substantia nigra, where one out-lying Ppid Het value was excluded; Fig. 3E]. When corrected for multiple comparisons, the thalamus and primary somatosensory cortex showed trends of decreased susceptibility (P = 0.0694 and P = 0.0833, respectively). We also observed a trend toward increased R2* in the CSTC [F(1,14) = 3.897, P = 0.0684, n = 8 Ppid Het, n = 8 wild-type; Fig. 3F].
Complex vocalizations measured in Ppid pups
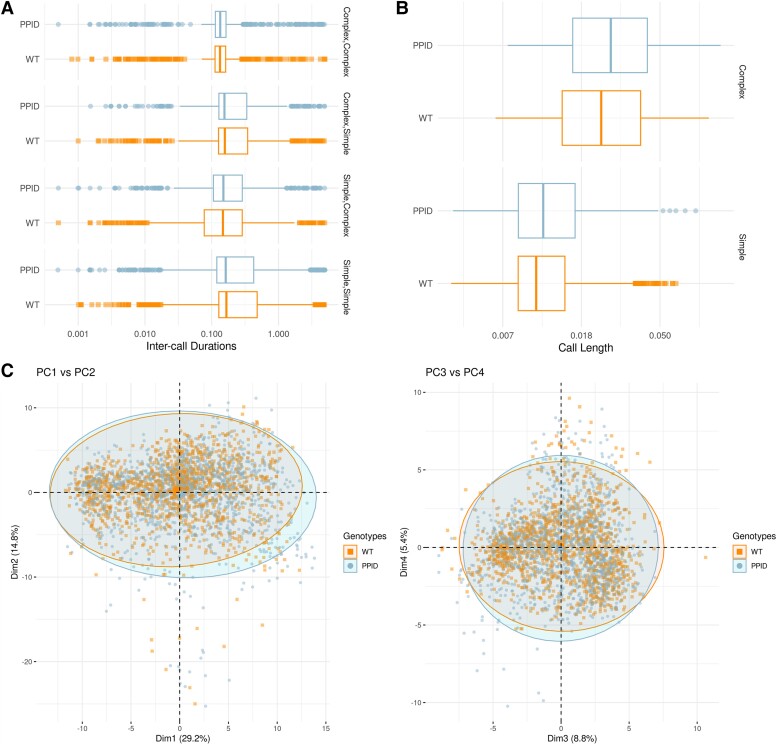
Heterozygous Ppid p.Pro270Ser mice and wild-type littermates pups were separated from dams at postnatal Day 4 to elicit USVs. Call types were identified by visual inspection of sonograms (Supplementary Fig. 2). Splitting acoustic metrics by call type (i.e. simple, complex; Supplementary Table 4) and examining inter-call durations did not reveal significant differences on multiple testing (Fig. 4A and Supplementary Table 5). Although there was a difference in inter-call durations when calls were categorized into simple and complex types (Supplementary Table 5), these data were not corrected for multiple comparisons. No significant differences in call lengths or call counts were observed (Fig. 4B and Supplementary Table 6). Principal component analysis of acoustic outcomes derived within a training set designed to classify call types18 suggested there was no evidence of association with genotype in the current cohort (Fig. 4C).
Figure 4.
Complex vocalizations measured in Ppid pups . ( A) Distribution of inter-call durations, by genotype and transition category. (B) Distribution of call lengths, by genotype and call category. (C) Principal component (PC) analysis of acoustic features of calls. PC1 versus PC2 and PC3 versus PC4 plotted, coloured by genotype. The first four principal components cumulatively explain 58% of the variance. WT = wild-type.
Behavioural studies of adult Ppid mice
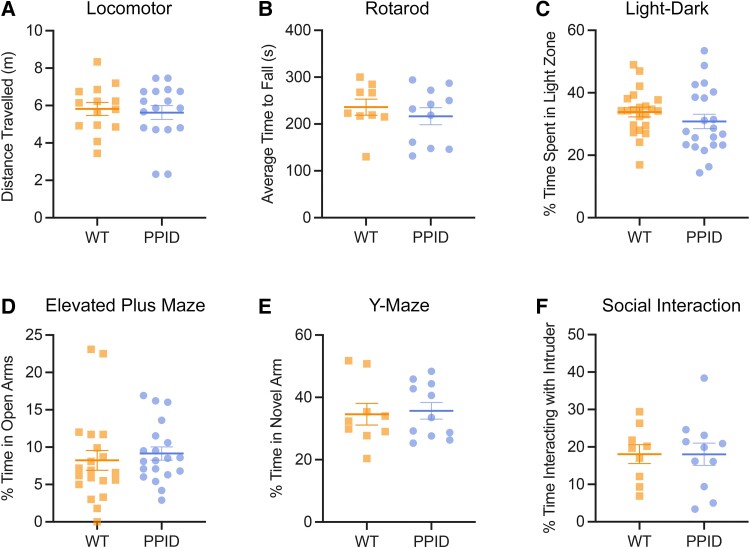
Heterozygous Ppid p.Pro270Ser mice and wild-type littermates were subjected to a battery of six standard behavioural tests including open-field exploratory locomotion assay, rotarod assay, light-dark transition test, elevated plus maze, Y maze and three-chamber social interaction assay. No significant differences in behaviours were observed on any of these tests (Fig. 5).
Figure 5.
Behavioural testing of adult Ppid mice. (A) There were no significant differences in distance travelled between wild-type (WT) littermates and Ppid mice in the open field locomotor assay over a 60-min period (n = 14 WT, n = 17 PPID), P = 0.2266. (B) The average time to fall (s) across three trials on the rotarod was not significantly different between WT and PPID mice (n = 9 WT, n = 11 PPID), P = 0.4527. (C) There were no significant differences between genotypes in the time spent in the light zone of the light-dark box (n = 22 WT, n = 21 PPID), P = 0.2763. (D) Time spent in the open arm of the elevated plus maze was not significantly different between WT and PPID mice (n = 20 WT, n = 20 PPID), P = 0.5596. (E) There were no significant differences in time spent in the novel of the Y maze (n = 9 wild-type, n = 11 PPID), P = 0.8006. (F) Time spent interacting with an age- and sex-matched stranger mouse in the three-chamber social interaction was not significantly different between genotypes (n = 9 WT, n = 11 PPID), P = 0.9919.
Brain transcriptomic analyses of Ppid pups
RNA sequencing was performed on brain tissue harvested from five heterozygous Ppid p.Pro270Ser and five wild-type mice. We compared the expression for Ppid and 12 other genes that cluster with Ppid in the STRING protein-protein interaction database.23 No genes had a significant difference in expression between Ppid Het knock-in and wild-type mice. Similarly, genome-wide differential expression analysis did not identify any genes that were significantly different (Supplementary Fig. 3).
P270S-induced changes in secondary structure and Hsp90 peptide binding mechanism
Molecular dynamics simulation of Cyp40 revealed a stable hinge bending motion within the TPR2 helix, involving the relative flexing of the short helical segment comprising residues Ile260 to Pro270 (Supplementary Video 1). This motion is also evidenced by the abrupt increase in RMSD at 100 ns, followed by recovery to a baseline RMSD level by ∼150 ns, as shown in Supplementary Fig. 4 (blue line). Such helical bending is typical of those observed for other Pro-containing helices. Pro270 imparts a degree of helix flexibility which enables it to retain its helical structure across the hinge region. In contrast, simulation of [P270S]-Cyp40 indicates a substantial unravelling of the Ile260-Ser270 segment (Supplementary Video 2), which remains disordered for the rest of the trajectory. This unfolding is captured in the RMSD time series plot in Supplementary Fig. 4 (red line), with a discrete jump in RMSD occurring at ∼225 ns until the end of the simulation at 340 ns. These trajectories therefore suggest a localized, but marked, difference in helical stability due to Pro270Ser mutation.
The influence of P270S mutation on the interaction between Cyp40 and Hsp90 was predicted using molecular docking calculations focusing on the known binding locale for the Hsp90 C-terminal binding motif, MEEVD.37 Docking calculations show a single preferred binding location in the vicinity of position 270 for wild-type Cyp40 within the TPR2 domain (Supplementary Fig. 5A), with the Glu residues of MEEVD supported by several salt bridge interactions below the P270 hinge, including K227, K235 and R312. In contrast, for [P270S]-Cyp40, two locations were found to be equally favourable (Supplementary Fig. 5B). Pose #1 (yellow) is similar to that of wild-type Cyp40, albeit closer to the N-terminal end of TPR2 (‘above’ Pro270 with respect to the orientation shown in Supplementary Fig. 5B), enabling the peptide to form direct contact with S270. A second position (Pose #2, green) is located at the N-terminal tip of TPR3, supported by a salt bridge interaction with K308. Thus, docking calculations suggest that P270S mutation of Cyp40 may result in a greater diversity in binding locations for the MEEVD peptide, with the secondary site (Pose #2) potentially competing with the primary site, weakening the latter’s interaction. This change in binding mechanism may have consequences for subsequent Cyp40 interactions with Hsp90 and other chaperones.
Discussion
We studied a large family with developmental stuttering across four generations and identified an ultra-rare missense variant (c.808C>T, p.Pro270Ser) in PPID mapped to chromosome 4 (LOD = 3.0088). Sanger sequencing revealed complete co-segregation with the stuttering phenotype. Resequencing of a large cohort of individuals with stuttering and age-matched controls from four populations did not reveal pathogenic variants in PPID suggesting mutation of this gene is not a frequent cause of developmental stuttering. This scenario is likely typical for the genetic architecture of stuttering as only a small fraction of cases have been explained by rare genetic variants to date.5,40
Several lines of evidence suggest a pathogenic role for this variant in stuttering. First, the Pro270 residue resides in the key tetratricopeptide repeat 2 (TPR2) functional domain of the cyclophilin-40 (CYP40) protein encoded by PPID. Second, the variant introduces a serine residue which are often sites of phosphorylation for regulation of protein function. Third, our 3D structural modelling predicts the Pro270Ser mutation may disrupt binding of Cyp40 with Hsp90 and other chaperones. Fourth, the PPID gene is expressed in the striatum and globus pallidus (Allen Brain Atlas, https://www.brain-map.org/), brain regions that are critical for speech and have been reported to be affected in individuals with persistent stuttering including members of our family.8,41,42 Other studies of individuals with stuttering have implicated changes on imaging in the cortico-striatal-thalamic network41 or reduced activation of the globus pallidus in adults.42
DWI revealed significantly increased diffusivity in the left corticospinal tract of Ppid knock-in mice. These findings are consistent with reduced white matter integrity which may involve oedema, demyelination and Wallerian degeneration.43–46 The left corticospinal tract has also been implicated in previous DWI studies of adults who stutter41,47 with reduced FA observed in a study of developmental stuttering.48 While these findings are consistent with a left hemisphere bias for white matter alterations in people who stutter, bilateral structural abnormalities might be expected, particularly for developmental disorders48 and reduced FA has been observed in the right corticospinal tract of people who stutter previously.49 Another recent study reported increased rate of darkening (R2*) values, thought to reflect increased non-heme iron, in the cortico-basal ganglia-thalamocortical loop of adults who stutter.15 Although we observed a trend toward increased R2* in Ppid mouse brain structures of the CSTC loop, we found that these regions exhibited an unexpected but significant decrease in tissue susceptibility, inconsistent with an increase in paramagnetic non-heme iron. Instead, our results suggest a possible increase in diamagnetic zinc and or calcium,50 both of which have been implicated in neurodegeneration. Ppid Het knock-in mice also had increased susceptibility along the corpus callosum when compared to wild-type mice, consistent with demyelination of this fibre bundle.51,52 Microstructural changes to the corpus callosum have been observed in developmental stuttering,48,52 and also in other persistent developmental speech disorders.53
Altered number and timing of vocalizations have previously been reported in rodent models of stuttering.47 Our preliminary data suggests there may be differences in inter-call durations when calls are categorized into simple and complex types, but further work is required. We did not see differences in call count between genotypes as previously reported.17 Likewise, the acoustic feature profile of each group was similar when principal components were explored.54
Our Ppid knock-in mouse brain transcriptomics analysis did not identify any differentially expressed genes. The lack of differentially expressed genes from our brain RNA-seq analysis could be due to the limitations of this technology. RNA sequencing can identify changes in transcription, however, the functional effects of this missense variant in Cyp40 may only manifest at a post-translational level as suggested by our 3D protein modeling studies.
Despite being a relatively frequent speech disorder a paucity of genes have been discovered for stuttering; here we implicate PPID in this debilitating disorder, albeit as a likely rare cause.
Supplementary Material
Acknowledgements
We thank the families for their participation in this study. We thank the National Computational Infrastructure (NCI) and The Pawsey Supercomputing Centre in Australia, which are both supported by the Australian Government’s National Collaborative Research Infrastructure Strategy (NCRIS). A.T.M., T.S.S., A.P.V., C.A.R., I.E.S., D.K.W., Me.B. and M.S.H., initiated and directed the project. M.S.H., C.F-D., Ed.S., K.A.R., M.A.C., J.G., R.J.M. and D.D. performed molecular genetics experiments. T.S.S., C.F.-D., Ed.S., V.E.J., M.F.B. and M.B. performed bioinformatics analyses. A.H. performed molecular dynamic simulations and docking calculations. S.J.T., S.R., S.B., E.K., O.V., D.J.A., I.H., D.G.Y.L., F.J.L., I.E.S., D.D. and A.T.M. conducted clinical phenotyping or imaging. A.P.V., M.Q., V.E.J., E.L.B., C.A.R., C.M., T.J.F., K.L.R., A.K., M.J.H. and D.K.W. generated or performed functional studies on the mouse model. A.T.M. and M.S.H. wrote the paper. All authors reviewed the manuscript.
Contributor Information
Angela T Morgan, Murdoch Children’s Research Institute, Parkville 3052, Australia; Department of Audiology and Speech Pathology, University of Melbourne, Parkville 3052, Australia.
Thomas S Scerri, Murdoch Children’s Research Institute, Parkville 3052, Australia; The Walter and Eliza Hall Institute of Medical Research, Parkville 3052, Australia; Department of Medical Biology, University of Melbourne, Parkville 3052, Australia.
Adam P Vogel, Department of Audiology and Speech Pathology, University of Melbourne, Parkville 3052, Australia; Centre for Neuroscience of Speech, The University of Melbourne, Parkville 3053, Australia; Clinical Trials, Redenlab Inc., Melbourne 3000, Australia.
Christopher A Reid, Florey Institute of Neuroscience and Mental Health, University of Melbourne, 3052, Parkville 3052, Australia; Department of Medicine, Epilepsy Research Centre, University of Melbourne, Heidelberg 3084, Australia.
Mara Quach, Department of Neuroscience, Central Clinical School, Monash University, Melbourne 3004, Australia.
Victoria E Jackson, The Walter and Eliza Hall Institute of Medical Research, Parkville 3052, Australia; Department of Medical Biology, University of Melbourne, Parkville 3052, Australia.
Chaseley McKenzie, Florey Institute of Neuroscience and Mental Health, University of Melbourne, 3052, Parkville 3052, Australia.
Emma L Burrows, Florey Institute of Neuroscience and Mental Health, University of Melbourne, 3052, Parkville 3052, Australia.
Mark F Bennett, The Walter and Eliza Hall Institute of Medical Research, Parkville 3052, Australia; Department of Medical Biology, University of Melbourne, Parkville 3052, Australia; Department of Medicine, Epilepsy Research Centre, University of Melbourne, Heidelberg 3084, Australia.
Samantha J Turner, Murdoch Children’s Research Institute, Parkville 3052, Australia.
Sheena Reilly, Murdoch Children’s Research Institute, Parkville 3052, Australia; Menzies Health Institute Queensland, Griffith University, 4215 Southport, Australia.
Sarah E Horton, Murdoch Children’s Research Institute, Parkville 3052, Australia; Department of Audiology and Speech Pathology, University of Melbourne, Parkville 3052, Australia.
Susan Block, Discipline of Speech Pathology, School of Allied Health, La Trobe University, Bundoora 3086, Australia.
Elaina Kefalianos, Murdoch Children’s Research Institute, Parkville 3052, Australia; Department of Audiology and Speech Pathology, University of Melbourne, Parkville 3052, Australia.
Carlos Frigerio-Domingues, Laboratory of Communication Disorders, National Institute on Deafness and Other Communication Disorders, National Institutes of Health, Bethesda, MD 20892-2320, USA.
Eduardo Sainz, Laboratory of Communication Disorders, National Institute on Deafness and Other Communication Disorders, National Institutes of Health, Bethesda, MD 20892-2320, USA.
Kristin A Rigbye, Department of Medicine, Epilepsy Research Centre, University of Melbourne, Heidelberg 3084, Australia.
Travis J Featherby, Florey Institute of Neuroscience and Mental Health, University of Melbourne, 3052, Parkville 3052, Australia.
Kay L Richards, Florey Institute of Neuroscience and Mental Health, University of Melbourne, 3052, Parkville 3052, Australia.
Andrew Kueh, The Walter and Eliza Hall Institute of Medical Research, Parkville 3052, Australia; Department of Medical Biology, University of Melbourne, Parkville 3052, Australia.
Marco J Herold, The Walter and Eliza Hall Institute of Medical Research, Parkville 3052, Australia; Department of Medical Biology, University of Melbourne, Parkville 3052, Australia.
Mark A Corbett, Adelaide Medical School, The University of Adelaide, Adelaide 5000, Australia; Neurogenetics Research Program, South Australian Health and Medical Research Institute, Adelaide 5000, Australia.
Jozef Gecz, Adelaide Medical School, The University of Adelaide, Adelaide 5000, Australia; Neurogenetics Research Program, South Australian Health and Medical Research Institute, Adelaide 5000, Australia.
Ingo Helbig, Department of Neurology, Children’s Hospital, Philadelphia, PA 19104, USA.
Daisy G Y Thompson-Lake, Developmental Neurosciences Department, UCL Great Ormond Street Institute of Child Health, London, UK.
Frédérique J Liégeois, Developmental Neurosciences Department, UCL Great Ormond Street Institute of Child Health, London, UK.
Robert J Morell, Laboratory of Molecular Genetics, National Institute on Deafness and Other Communication Disorders, National Institutes of Health, Bethesda, MD 20892, USA; Genomics and Computational Biology Core, National Institute on Deafness and Other Communication Disorders, National Institutes of Health, Bethesda, MD 20892, USA.
Andrew Hung, School of Science, STEM College, RMIT University, Melbourne 3001, Australia.
Dennis Drayna, Laboratory of Communication Disorders, National Institute on Deafness and Other Communication Disorders, National Institutes of Health, Bethesda, MD 20892-2320, USA.
Ingrid E Scheffer, Murdoch Children’s Research Institute, Parkville 3052, Australia; Florey Institute of Neuroscience and Mental Health, University of Melbourne, 3052, Parkville 3052, Australia; Department of Medicine, Epilepsy Research Centre, University of Melbourne, Heidelberg 3084, Australia; Department of Paediatrics, University of Melbourne, Royal Children's Hospital, Parkville 3052, Australia.
David K Wright, Department of Neuroscience, Central Clinical School, Monash University, Melbourne 3004, Australia.
Melanie Bahlo, The Walter and Eliza Hall Institute of Medical Research, Parkville 3052, Australia; Department of Medical Biology, University of Melbourne, Parkville 3052, Australia; School of Mathematics and Statistics, University of Melbourne, 3010 Parkville, Australia.
Michael S Hildebrand, Murdoch Children’s Research Institute, Parkville 3052, Australia; Department of Medicine, Epilepsy Research Centre, University of Melbourne, Heidelberg 3084, Australia.
Data availability
Data are available upon reasonable request. The PPID c.808C>T (p.Pro270Ser) variant has been submitted to ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
Funding
This study was supported by a National Health and Medical Research Council Centre of Research Excellence (1116976) and Project Grant (1127144) to A.T.M., S.E.F., I.E.S., M.B. and M.S.H., a Practitioner Fellowship to I.E.S. (1006110) and Investigator Grants, to A.T.M., I.E.S., M.B. and D.K.W (1105008; 1172897; 1195236; 1174040), a Senior Research Fellowship to M.B. (1102971), a R.D Wright Career Development Fellowship (1063799) to M.S.H, NHMRC Dementia (1135683) and ARC Future Fellowships (220100253) to A.P.V. D.D. was supported by NIH/NIDCD intramural research grant Z1A-000046-18. R.M and the Genomic & Computational Biology Core were supported by NIH/NIDCD intramural research grant ZIC-DC000086. This work was supported by the Victorian Government’s Operational Infrastructure Support Program and Australian Government National Health and Medical Research Council Independent Research Institute Infrastructure Support Scheme (NHMRC IRIISS). The generation of knock-in Ppid p.Pro270Ser mice used in this study was supported by Phenomics Australia and the Australian Government through the National Collaborative Research Infrastructure Strategy (NCRIS) program. The authors acknowledge the facilities and scientific and technical assistance of the National Imaging Facility (NIF), a National Collaborative Research Infrastructure Strategy (NCRIS) capability at Monash Biomedical Imaging, a Technology Research Platform at Monash University.
Competing interests
I.E.S. has served on scientific advisory boards for UCB, Eisai, GlaxoSmithKline, BioMarin, Nutricia, Rogcon, Chiesi, Encoded Therapeutics, Knopp Biosciences and Xenon Pharmaceuticals; has received speaker honoraria from GlaxoSmithKline, UCB, BioMarin, Biocodex, Chiesi, Liva Nova and Eisai; has received funding for travel from UCB, Biocodex, GlaxoSmithKline, Biomarin and Eisai; has served as an investigator for Zogenix, Zynerba, Ultragenyx, GW Pharma, UCB, Eisai, Xenon Pharmaceuticals, Anavex Life Sciences, Ovid Therapeutics, Epigenyx, Encoded Therapeutics and Marinus; and has consulted for Zynerba Pharmaceuticals, Atheneum Partners, Ovid Therapeutics, Care Beyond Diagnosis, Epilepsy Consortium and UCB; and is a Non-Executive Director of Bellberry Ltd. She may accrue future revenue on pending patent WO61/010176 (filed: 2008): Therapeutic Compound; has a patent for SCN1A testing held by Bionomics Inc. and licensed to various diagnostic companies; has a patent molecular diagnostic/theranostic target for benign familial infantile epilepsy (BFIE) [PRRT2] 2011904493 & 2012900190 and PCT/AU2012/001321 (TECH ID:2012-009). A.P.V. works for and owns stock in Redenlab Inc., an audio analysis company. The remaining authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Craig A, Hancock K, Tran Y, Craig M, Peters K. Epidemiology of stuttering in the community across the entire life span. J Speech Lang Hear Res. 2002;45:1097–1105. [DOI] [PubMed] [Google Scholar]
- 2. Riley GD. A stuttering severity instrument for children and adults. J Speech Hear Disord. 1972;37:314–322. [DOI] [PubMed] [Google Scholar]
- 3. Frigerio-Domingues C, Drayna D. Genetic contributions to stuttering: The current evidence. Mol Genet Genomic Med. 2017;5:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyce JO, Jackson VE, van Reyk O, et al. . Self-reported impact of developmental stuttering across the lifespan. Dev Med Child Neurol. 2022;64:1297–1306. [DOI] [PubMed] [Google Scholar]
- 5. Kang C, Drayna D. Genetics of speech and language disorders. Annu Rev Genomics Hum Genet. 2011;12:145–164. [DOI] [PubMed] [Google Scholar]
- 6. Kang C, Riazuddin S, Mundorff J, et al. . Mutations in the lysosomal enzyme-targeting pathway and persistent stuttering. N Engl J Med. 2010;362:677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raza MH, Mattera R, Morell R, et al. . Association between rare variants in AP4E1, a component of intracellular trafficking, and persistent stuttering. Am J Hum Genet. 2015;97:715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thompson-Lake DGY, Scerri TS, Block S, et al. . Atypical development of Broca's area in a large family with inherited stuttering. Brain. 2022;145:1177–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng J, Miller KK, Yang T, et al. . Carcinoembryonic antigen-related cell adhesion molecule 16 interacts with alpha-tectorin and is mutated in autosomal dominant hearing loss (DFNA4). Proc Natl Acad Sci U S A. 2011;108:4218–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kueh AJ, Pal M, Tai L, et al. . An update on using CRISPR/Cas9 in the one-cell stage mouse embryo for generating complex mutant alleles. Cell Death Differ. 2017;24:1821–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polak D, Chatnuntawech I, Yoon J, et al. . Nonlinear dipole inversion (NDI) enables robust quantitative susceptibility mapping (QSM). NMR Biomed. 2020;33:e4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koch KM, Meier TB, Karr R, Nencka AS, Muftuler LT, McCrea M. Quantitative susceptibility mapping after sports-related concussion. Am J Neuroradiol. 2018;39:1215–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wright DK, O'Brien TJ, Shultz SR. Sub-acute changes on MRI measures of cerebral blood flow and venous oxygen saturation in concussed Australian rules footballers. Sports Med Open. 2022;8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cler GJ, Krishnan S, Papp D, Wiltshire CEE, Chesters J, Watkins KE. Elevated iron concentration in putamen and cortical speech motor network in developmental stuttering. Brain. 2021;144:2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson GA, Badea A, Brandenburg J, et al. . Waxholm space: An image-based reference for coordinating mouse brain research. Neuroimage. 2010;53:365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barnes TD, Wozniak DF, Gutierrez J, Han TU, Drayna D, Holy TE. A mutation associated with stuttering alters mouse pup ultrasonic vocalizations. Curr Biol. 2016:S0960- 9822(16)30179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vogel AP, Tsanas A, Scattoni ML. Quantifying ultrasonic mouse vocalizations using acoustic analysis in a supervised statistical machine learning framework. Sci Rep. 2019;9:8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scattoni ML, Gandhy SU, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in the BTBR T+tf/J mouse model of autism. PLoS One. 2008;3:e3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dobin A, Davis CA, Schlesinger F, et al. . STAR: Ultrafast universal RNA-Seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias- aware quantification of transcript expression. Nat Methods. 2017;14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ewels PA, Peltzer A, Fillinger S, et al. . The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38:276–278. [DOI] [PubMed] [Google Scholar]
- 23. Szklarczyk D, Gable AL, Lyon D, et al. . STRING V11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson MD, McCarthy DJ, Smyth GK. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. Colabfold: Making protein folding accessible to all. Nat Methods. 2022;19:679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jumper J, Evans R, Pritzel A, et al. . Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang J, Rauscher S, Nawrocki G, et al. . CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 29. Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 30. Darden T, York D, Pedersen L. Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 31. Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 32. Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 33. Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. [DOI] [PubMed] [Google Scholar]
- 34. Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. [DOI] [PubMed] [Google Scholar]
- 35. Eberhardt J, Santos-Martins D, Tillack AF, Forli S. Autodock vina 1.2.0: New docking methods, expanded force field, and python bindings. J Chem Inf Model. 2021;61:3891–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trott O, Olson AJ. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cortajarena AL, Wang J, Regan L. Crystal structure of a designed tetratricopeptide repeat module in complex with its peptide ligand. FEBS J. 2010;277:1058–1066. [DOI] [PubMed] [Google Scholar]
- 38. Morris GM, Huey R, Lindstrom W, et al. . Autodock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. [DOI] [PubMed] [Google Scholar]
- 40. Kang C, Drayna D. A role for inherited metabolic deficits in persistent developmental stuttering. Mol Genet Metab. 2012;107:276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Etchell AC, Civier O, Ballard KJ, Sowman PF. A systematic literature review of neuroimaging research on developmental stuttering between 1995 and 2016. J Fluency Disord. 2018;55:6–45. [DOI] [PubMed] [Google Scholar]
- 42. Toyomura A, Fujii T, Kuriki S. Effect of an 8-week practice of externally triggered speech on basal ganglia activity of stuttering and fluent speakers. Neuroimage. 2015;109:458–468. [DOI] [PubMed] [Google Scholar]
- 43. Klawiter EC, Schmidt RE, Trinkaus K, et al. . Radial diffusivity predicts demyelination in ex vivo multiple sclerosis spinal cords. Neuroimage. 2011;55:1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pierpaoli C, Barnett A, Pajevic S, et al. . Water diffusion changes in Wallerian degeneration and their dependence on white matter architecture. Neuroimage. 2001;13:1174–1185. [DOI] [PubMed] [Google Scholar]
- 45. Wright DK, Gardner AJ, Wojtowicz M, et al. . White matter abnormalities in retired professional rugby league players with a history of concussion. J Neurotrauma. 2021;38:983–988. [DOI] [PubMed] [Google Scholar]
- 46. Wright DK, Johnston LA, Kershaw J, Ordidge R, O'Brien TJ, Shultz SR. Changes in apparent fiber density and track-weighted imaging metrics in white matter following experimental traumatic brain injury. J Neurotrauma. 2017;34:2109–2118. [DOI] [PubMed] [Google Scholar]
- 47. Kronfeld-Duenias V, Amir O, Ezrati-Vinacour R, Civier O, Ben-Shachar M. The frontal aslant tract underlies speech fluency in persistent developmental stuttering. Brain Struct Funct. 2016;221:365–381. [DOI] [PubMed] [Google Scholar]
- 48. Connally EL, Ward D, Howell P, Watkins KE. Disrupted white matter in language and motor tracts in developmental stuttering. Brain Lang. 2014;131:25–35. [DOI] [PubMed] [Google Scholar]
- 49. Cai S, Tourville JA, Beal DS, Perkell JS, Guenther FH, Ghosh SS. Diffusion imaging of cerebral white matter in persons who stutter: Evidence for network-level anomalies. Front Hum Neurosci. 2014;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lorio S, Sedlacik J, So PW, et al. . Quantitative MRI susceptibility mapping reveals cortical signatures of changes in iron, calcium and zinc in malformations of cortical development in children with drug-resistant epilepsy. Neuroimage. 2021;238:118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hametner S, Endmayr V, Deistung A, et al. . The influence of brain iron and myelin on magnetic susceptibility and effective transverse relaxation—A biochemical and histological validation study. Neuroimage. 2018;179:117–133. [DOI] [PubMed] [Google Scholar]
- 52. Liu C, Li W, Johnson GA, Wu B. High-field (9.4T) MRI of brain dysmyelination by quantitative mapping of magnetic susceptibility. Neuroimage. 2011;56:930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Civier O, Kronfeld-Duenias V, Amir O, Ezrati-Vinacour R, Ben-Shachar M. Reduced fractional anisotropy in the anterior corpus callosum is associated with reduced speech fluency in persistent developmental stuttering. Brain Lang. 2015;143:20–31. [DOI] [PubMed] [Google Scholar]
- 54. Luders E, Kurth F, Pigdon L, Conti-Ramsden G, Reilly S, Morgan AT. Atypical callosal morphology in children with speech sound disorder. Neuroscience. 2017;367:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request. The PPID c.808C>T (p.Pro270Ser) variant has been submitted to ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).