Abstract
Dravet syndrome is a severe epileptic encephalopathy, characterized by (febrile) seizures, behavioural problems and developmental delay. Eighty per cent of patients with Dravet syndrome have a mutation in SCN1A, encoding Nav1.1. Milder clinical phenotypes, such as GEFS+ (generalized epilepsy with febrile seizures plus), can also arise from SCN1A mutations. Predicting the clinical phenotypic outcome based on the type of mutation remains challenging, even when the same mutation is inherited within one family. This clinical and genetic heterogeneity adds to the difficulties of predicting disease progression and tailoring the prescription of anti-seizure medication. Understanding the neuropathology of different SCN1A mutations may help to predict the expected clinical phenotypes and inform the selection of best-fit treatments. Initially, the loss of Na+-current in inhibitory neurons was recognized specifically to result in disinhibition and consequently seizure generation. However, the extent to which excitatory neurons contribute to the pathophysiology is currently debated and might depend on the patient clinical phenotype or the specific SCN1A mutation.
To examine the genotype-phenotype correlations of SCN1A mutations in relation to excitatory neurons, we investigated a panel of patient-derived excitatory neuronal networks differentiated on multi-electrode arrays. We included patients with different clinical phenotypes, harbouring various SCN1A mutations, along with a family in which the same mutation led to febrile seizures, GEFS+ or Dravet syndrome.
We hitherto describe a previously unidentified functional excitatory neuronal network phenotype in the context of epilepsy, which corresponds to seizurogenic network prediction patterns elicited by proconvulsive compounds. We found that excitatory neuronal networks were affected differently, depending on the type of SCN1A mutation, but did not segregate according to clinical severity. Specifically, loss-of-function mutations could be distinguished from missense mutations, and mutations in the pore domain could be distinguished from mutations in the voltage sensing domain. Furthermore, all patients showed aggravated neuronal network responses at febrile temperatures compared with controls. Finally, retrospective drug screening revealed that anti-seizure medication affected GEFS+ patient- but not Dravet patient-derived neuronal networks in a patient-specific and clinically relevant manner.
In conclusion, our results indicate a mutation-specific excitatory neuronal network phenotype, which recapitulates the foremost clinically relevant features, providing future opportunities for precision therapies.
Keywords: epilepsy, MEA, hiPSC, Dravet syndrome
Van Hugte et al. reveal a functional excitatory neuronal network phenotype in the context of Dravet syndrome, which is dependent on mutation type (pore domain versus voltage sensing domain). The network phenotype recapitulates clinically relevant features of Dravet syndrome, providing opportunities for precision therapies.
Introduction
Dravet syndrome (DS) is a severe epileptic encephalopathy characterized by recurrent and prolonged seizures, manifesting before the first year of life.1 Patients have a poor prognosis, and the phenotype progresses into complex seizures and neurodevelopmental delays, including behavioural, cognitive and motor impairments.1-4 In 80% of cases, DS is caused by mutations in SCN1A, encoding the voltage-gated Na+-channel Nav1.1.5,6 A milder clinical phenotype, generalized epilepsy with febrile seizures plus (GEFS+), can also arise from SCN1A mutations. Most patients with GEFS+ show normal development and spontaneously remitting seizures later in life.2
To date, more than 1500 different SCN1A mutations have been identified, most of which occur de novo,7,8 including missense mutations, frame shifts, splice site variants or insertions/deletions that lead to loss-of-function (LoF).9,10 Although evidence suggests that the nature of the mutation affects the phenotype,9 with LoF mutations conferring a higher risk,11 predictions based on individual mutations remain challenging. The same mutation can give rise to variable phenotypes, even if it segregates within one family.12 This clinical and genetic heterogeneity adds to the difficulties of predicting disease progression and prescribing patient-tailored anti-seizure medication (ASM).
Scn1a knockout mouse models for DS show that a loss of excitability, caused by reduced Na+-current density, predominantly affects parvalbumin- and somatostatin-positive inhibitory neurons.13-18 Disinhibition is considered the major pathogenic mechanism of DS, in turn leading to hyperexcitability and epileptic activity.19 However, depending on the strain or age of the mice, excitatory neurons contribute equally to the epileptic activity.20 Studies using human induced pluripotent stem cell (hiPSC)-derived neurons have provided further conflicting results. While some have reported deficits in Na+-current density or action potential firing in inhibitory neurons alone,21-24 others have described deficits in both excitatory and inhibitory neurons.25-27
These conflicting observations might be attributed to intrinsic functional differences in the mutations investigated or cell types studied. Some mutations result in gain-of-function (GoF), while others in (partial) LoF of Nav1.1, suggesting that different SCN1A variants might affect inhibitory or excitatory neurons to a different extent.8 However, it remains difficult to attribute the neuronal phenotype to either the patient clinical phenotype or the type of variant studied. Moreover, the effects of individual mutations at the neuronal network level have not been investigated systematically. A series of patient hiPSC-derived neurons, which includes multiple SCN1A variants and clinical phenotypes, may help to determine whether the excitatory neuronal phenotype is dependent on the patient clinical phenotype or the specific mutation being studied.
To this end, we investigated CRISPR/Cas9-edited SCN1A lines and a panel of hiPSC-derived lines from five patients with different clinical phenotypes harbouring different SCN1A variants and mapped the effects on excitatory neuronal network behaviour. In particular, we studied a family in which the same SCN1A mutation resulted in different clinical phenotypes.
Our data provide evidence that excitatory neurons are affected to different extents by various SCN1A mutations. Specifically, missense versus nonsense mutations of SCN1A and the location of the mutation in Nav1.1, i.e. pore domain versus voltage sensing domain, influence the excitatory network phenotype. Although we did not observe differences based on clinical severity in basal network phenotypes, clinically relevant DS triggers, such as febrile temperatures and ASM, elicited different responses in DS and GEFS+ patient-derived neuronal networks. To conclude, we provide further evidence for the role of excitatory neurons in DS.
Materials and methods
Patient inclusion and generation of human induced pluripotent stem cells
All experiments involving peripheral mononuclear blood cells (PBMCs) and hiPSCs from subjects were carried out after informed consent was obtained and with approval from the medical ethical committee of Radboud University Medical Center, Nijmegen (2018–4525). PBMCs were isolated from all subjects via blood samples taken during routine diagnostic testing. The clinical history of all patients is described in the Supplementary material.
Patient PAT001_DRAV was a 4-year-old female at the time of sampling. She had a missense mutation (c.4168G>A p.Val1390Met) in the pore domain of SCN1A. Patient PAT001_GEFS was a female patient who was 10 years of age at the time of sampling, with a missense mutation (c.2576G>A p.Arg859His) in the voltage-sensing domain. Patients FAM001_DRAV, FAM001_GEFS and FAM001_FS were from the same family, and all carried a mutation in c.3926T>G (p. Leu1309Arg) in the voltage sensing domain. FAM001_DRAV was the male sibling (16 years old at the time of sampling) and FAM001_GEFS was the female sibling (14 years old at the time of sampling). FAM001_FS was the father (49 years of age at the time of sampling), while CTRL1 was the unaffected mother (43 years of age at the time of sampling).
All lines were reprogrammed using episomal vectors with the Yamanaka transcription factors Oct4, c-Myc, Sox2 and Klf428 and had normal karyotypes (Supplementary Fig. 1A). All mutations were confirmed via Sanger sequencing. CTRL2 was an external control (a 30-year-old male, previously characterized29) and tested for genetic integrity using SNP assay.30
CRISPR/Cas9 gene editing of SCN1A
From CTRL2, two SCN1A CRISPR lines were generated (SCN1A+/− C3 and SCN1A+/− C7) with a heterozygous deletion in exon 21, leading to a premature stop codon at amino acid position 18. SCN1A+/− C3 had a heterozygous mutation at position c.500_501insAC (p.Thr18*) and SCN1A+/− C7 at position c.500_501delGTCAinsTACTC (p.Thr18*). A short guide RNA (sgRNA) was designed (GTCAAACCTGTCACCAGTTG) and cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene, #62988) according to a previously published protocol.31 In brief, 8 × 105 hiPSCs in single-cell suspension were nucleofected with 5 µg SpCas9-sgRNA plasmid using the P3 Primary Cell 4D-Nucleofector kit (Lonza, #V4XP-3024) in combination with the 4D Nucleofector Unit X (Lonza, #AAF-1002X). Cells were resuspended in E8 Flex supplemented with Revitacell (10 μg/ml, Thermo Fisher, #A2644501) and seeded on Biolaminin 521 LN (20 μg/ml, BioLamina, #LN521-05) pre-coated wells. Twenty-four hours after nucleofection, 0.5 µg/ml puromycin (Sigma-Aldrich, #P9620) was added for 24 h. Resistant colonies were picked manually and sent for Sanger sequencing to ensure heterozygous editing of exon 21.
Human induced pluripotent stem cell culturing and neuronal differentiation
All hiPSC lines were cultured on Matrigel (Corning, #356239) in Essential 8 (E8) flex medium (Gibco, #A2858401) supplemented with primocin (0.1 μg/ml, Invivogen, #ant-pm-2). For a quality check, hiPSCs were tested for pluripotency using qPCR or flow cytometry (Supplementary Fig. 1B and D). HiPSCs were infected, according to a previously published protocol, with lentiviral constructs encoding rtTA combined with Ngn2 to generate doxycycline-inducible excitatory neurons.32,33 Selection was performed by adjusting the concentrations of G418 (100–250 μg/ml, Sigma-Aldrich, #G8168), for Ngn2, and puromycin (1–2 μg/ml), for rtTA. Cells were passaged twice a week using ReLeSR passaging agent (Stemcell, #05872) and not kept for more than 10 passages. Every 2 weeks, cells were checked for mycoplasma contamination using MycoAlert PLUS (Lonza, #LT07-703) (Supplementary Fig. 1C).
HiPSCs were differentiated to glutamatergic neurons by overexpression of Ngn2 and cultured in a 1:1 ratio with E18 rodent astrocytes according to a previously published protocol and guidelines.32,33 Cell plating was adjusted to ensure equal cell densities (Supplementary Fig. 1E and F). Wells with unequal cell densities, unequally distributed cells or cell clumping were excluded. A detailed neuronal differentiation protocol can be found in the Supplementary material.
Immunocytochemistry
Coverslips were washed once with ice-cold PBS. Unless stated otherwise, subsequent steps were performed at room temperature and solutions were made in PBS. Cells were fixed with 4% paraformaldehyde and 4% sucrose for 15 min. Fixed coverslips were washed three times and stored in PBS at 4°C until further use. Cells were permeabilized with Triton X-100 (0.2%, Sigma-Aldrich, #T8787) for 10 min, followed by 1 h of blocking with normal goat serum (NGS) (5%, Invitrogen, #10000C). Primary antibodies raised against either peripherin (PRPH) (1:500, Millipore, #MAB1527), SATB2 (1:500, Abcam, #ab51502), synapsin (1:200, Millipore, #MAB1543P), homer (1:200, Synaptic Systems, #160011), Nav1.1 (1:500, Alomone Labs, #ASC-001), MAP2 (1:1000, Synaptic Systems, #188004), vesicular glutamate transporter 1 (vGlut1) (1:500, Synaptic Systems, #135311), ankyrin-G (AnkG, 1:1000, Invitrogen, #33-800) or cleaved caspase 3 (1:300, Cell Signaling, #9661S), diluted in 1% NGS, were incubated overnight at 4°C. After 10 PBS washes, coverslips were incubated in secondary antibodies diluted in 1% NGS. Coverslips were washed 10 times with PBS and stained with Hoechst (0.01%, ThermoFisher, #H3570). The coverslips were mounted in DAKO (Agilent, #S3023) on microscope slides and imaged on a Zeiss Axio Imager Z2 at ×63 magnification for quantification of Nav1.1 expression and synapse density, or at ×20 magnification for quantification of cell density or PRHP expression.
Western blot
For western blot, a total of 350 000 cells were seeded on a six-well plate. At 49 days in vitro (DIV), proteins were extracted in cold RIPA lysis buffer, and supernatant was used to determine protein concentration using the Pierce BCA protein assay kit (ThermoFisher). Fifty micrograms of protein were loaded on Mini-PROTEAN TGX stain-free gels before transfer to Nitrocellulose membranes (Bio-Rad). The membrane was blocked with blotto blocking buffer (Santa Cruz, #sc-2333) for 1 h at room temperature and incubated with primary antibody overnight at 4°C against Nav1.1 (1:500), using GAPDH (1:1000, Cell Signaling, #2118S) as a loading control. Membranes were washed three times in TBS-T followed by incubation for 1 h with goat anti-rabbit IgG (H+L) secondary antibody, horseradish peroxidase (1:50 000, ThermoFisher, #G-21234) at room temperature. Membranes were washed five times in TBS-T, before ECL detection with the Supersignal West Femto kit (ThermoFisher, #34095), then imaged using the Biorad Gel Imaging system (Chemidoc) and subsequently analysed with ImageJ.34
RNA sequencing
Cells were harvested at different DIV for RNA extraction using the Quick-RNA Microprep kit (Zymo Research, #ZY-R1051), following the manufacturer’s instructions. RNA concentration and integrity were measured using a Tapestation system (RNA High Sensitivity ScreenTape and Reagents, #5067–5579/80). All samples had an RNA integrity number (RIN)>8. cDNA libraries were generated with iScript (Bio-Rad, #1708890) and sequenced on an Illumina NovaSeq 6000 platform in paired-end at GenomeScan B.V. Leiden. The average number of reads was >50 million. Human and rat reads were mapped to both the rat (Rattus_norvegicus.mRatBN7.2) and human (Homo_sapiens.GRCh38) reference genomes, and split by organism with BBmap (v38.95) according to the best mapping. Then, human reads were aligned to the GRCh38.p13 reference genome with HISAT2 v2-2.2, sorted with samtools v1.10 and counted with HTSeq v2.0.2. Raw count matrices were loaded in R v4.2.1. For ensemble IDs that mapped to the same gene symbols, we only considered the IDs with the highest expression per sample. Count data were normalized using the size factors method with DESeq235 and lowly expressed genes (<15 normalized counts) were filtered.
Multi-electrode array recording and analysis
To record spontaneous network activity, multi-well multi-electrode arrays (MEAs) were used, which consisted of 24 individual wells (Multichannel systems, MCS) according to a previous published protocol.36 In brief, each MEA well was embedded with 12 gold electrodes spaced 300 µm apart with a diameter of 30 µm. Baseline neuronal network activity was recorded for 10 min at DIV49, after a 10-min acclimatization period, in a recording chamber maintained at 37°C/95%O2/5%CO2. For temperature experiments, the amplifier temperature was set to 40°C. When this temperature was reached, the cultures were allowed to acclimate for 10 min before a 10 min recording at 40°C. The same method was applied for the recovery recording at 37°C. For all recordings, the raw signal was sampled at 10 kHz and filtered with a high-pass 2nd order Butterworth filter with a 100 Hz cut-off and a low-pass 4th order Butterworth filter with a 3500 Hz cut-off. The noise threshold for individual spike detection was set to ±4.5 standard deviations (SD).
Pharmacology
All reagents were prepared into stocks and stored at −20°C. A set of five proconvulsive compounds were chosen to induce seizurogenic responses of the neuronal network at DIV49: NMDA (100 µM working concentration, Tocris, #6384-92-5), linopirdine (1.5 µM, Sigma, #105431-72-9), 4-aminopyridine (4-AP,10 µM, Tocris, #504-24-5), phenytoin (25 µM, Sigma, #57-41-0), kainic acid (KA, 10 µM, Sigma #58002-62-3) and picrotoxin (100 µM, Tocris, #1128). The working dilution was added directly to the wells during MEA recordings. A set of four ASMs was used to rescue the neuronal network phenotypes: valproic acid (Tocris, #1069-66-5), levetiracetam (Tocris, #102767-28-2), topiramate (Tocris, #97240-79-4) and carbamazepine (Tocris, #298-46-4). Cultures were treated from DIV13 onwards, and the ASM concentration was kept constant at 10 µM. The working dilution was first diluted into the medium before the regular medium change. For all pharmacological dilutions, the amount of DMSO in the cell culture medium was ≤0.5% v/v in each experiment.
Data analysis using Multiwell-Analyzer
To guarantee sufficient experimental quality, we adhered to a set of published guidelines.36 We included experiments with a minimum of nine wells per hiPSC line, across at least two independent batches. Networks that did not show network bursts (NBs) at DIV27, or a mean firing rate (MFR) <0.1 Hz and burst rate <0.4 bursts/min, were excluded. Wells that displayed low cell densities or cell clumping were discarded. All experiments were carried out at DIV49. Off-line data analysis was performed using Multiwell-Analyzer software (Multichannel systems) to extract spike-trains and a custom-made in-house code developed in MATLAB (The Mathworks, Natick, MA, USA). Spikes were detected using a threshold of ±4.5 SD from the baseline noise. The MFR was calculated for each well individually by averaging the firing rate of each separate channel by the total number of active channels of the well. The Multiwell-Analyzer build-in burst detection algorithm was used to detect bursts and defined bursts as four spikes in close proximity, with a maximum 50 ms inter-spike interval (ISI) to start a burst, a maximum 50 ms ISI to end a burst and a minimum 100 ms inter-burst interval (IBI). The mean burst rate (MBR) was calculated by averaging the burst rate across all electrodes. NBs were defined making use of the previously mentioned single burst detection as an intermediate step according to Mendis et al.37 An NB was defined when these two qualifications applied: the number of distinct bursting channels was at least 50%, and at some point during the sequence, at least 50% of the channels were bursting simultaneously. Simultaneous bursting was defined as any burst that had a temporal overlap >0 ms with another burst. Therefore, to classify as an NB, at least 50% of all channels needed to have bursts with a temporal overlap of >0 ms with each other burst in the sequence. The start of the NB was detected at the first point where any of the participating channels were bursting simultaneously. It ended with the last bursting channel in the temporally overlapping sequence of bursts. The NB rate (NBR) was derived by dividing all NBs in the recording by the length of the recording in minutes. The percentage of random spikes (PRS) was defined by calculating the percentage of isolated spikes that did not belong to a burst. The IBI and NB IBI (NIBI) were calculated by subtraction of the time stamp at the beginning from the time stamp at the end of each burst or NB. The mean IBI per channel was calculated and averaged across all channels. The NIBI was averaged for each well. A detailed description of the calculation of these network parameters can be found in Mossink et al.36 High frequency bursts (HFBs) within an NB period were detected by decreasing the burst detection ISI to 5 ms, with a maximum IBI of 10 ms, to individually detect each HFB. An in-house MATLAB script quantified the number of HFBs inside one NB period using a maximum inter-HFB-interval of 1500 ms.
Single cell electrophysiology
Experiments were performed in a recording chamber at DIV49 and visualized using an Olympus BX51WI upright microscope (Olympus Life Science) and a DAGE-MTI IR−1000E (DAGE- MTI) camera. Data were acquired using a Digidata 1440-A digitizer and a Multiclamp 700B amplifier (Molecular Devices), sampled at 20 000 Hz and filtered using a low-pass 1 kHz filter. Filamented patch pipettes were pulled from borosilicate glass (Science Products GmbH) with a PC-10 micropipette puller (Narishige), with an open tip resistance between 5 and 7 MΩ. The recording chamber was continuously perfused with artificial CSF (ACSF) containing (in mM): 124 NaCl, 1.25 NaH2PO4, 3 KCl, 26 NaHCO3, 11 glucose, 2 CaCl2, 1 MgCl2 (adjusted to pH 7.4) and maintained constant at 37°C/5% CO2. Recordings were excluded if the series resistance was above 20 MΩ or if the series resistance to membrane resistance was below a 1:10 ratio.
Intrinsic properties
For intrinsic properties, pipettes contained a potassium-based internal solution containing (in mM): 130 K-gluconate, 5 KCl, 10 HEPES, 2.5 MgCl2, 2 Na2-ATP, 0.4 Na3-ATP, 10 Na-phosphocreatine, 0.6 EGTA (adjusted to pH 7.25 and osmolarity 290 mOsmol). Resting membrane potential (Vrmp) was determined immediately after reaching whole cell configuration in the current clamp. Both active and passive membrane properties were determined at a holding potential of −60 mV. Action potentials were elicited by a stepwise current injection protocol, ranging from −30 pA to +50 pA, and determined by analysing the first action potential that was elicited. All intrinsic properties were analysed using Clampfit 10.7 (Molecular Devices).
Na+ currents
A CsCl-based internal pipette solution was used, containing (in mM): 115 CsMeSO3, 20 CsCl, 10 HEPES, 2.5 MgCl2, 4 Na2ATP, 0,4 Na3GTP, 10 Na-phosphocreatine and 0.6 EGTA (pH adjusted to 7.24, osmolarity 294 mOsmol). CNQX (40 μM, Tocris #0190) was used to block synaptic activity during recording. TTX (1 μM, Tocris, #1069) was used as a negative control. Cells were measured in whole cell voltage clamp configuration including P/8 leak subtraction. Using a stepwise protocol, cell membrane potential was increased from a holding potential of −90 mV in 10 mV increments to 60 mV for 100 ms. Peak currents were determined using Clampfit 10.7 and divided by the cell capacitance. Conductance was calculated using Equation 1, where G is the conductance, I the peak current amplitude, V the holding potential of each pulse and Vrev the Na+ reversal potential. Vrev was extrapolated from the experimental data using an in-house MATLAB script, and data were fitted to Equation 2, where I is the normalized current, Imax the maximum current, V the holding potential of each pulse, V1/2 the half maximum of activation and k the slope.
| (1) |
| (2) |
Statistics
Statistical analysis was performed using Graphpad PRISM 9.0.0 (GraphPad Software, Inc., CA, USA). All values are reported as mean ± standard error of the mean (SEM), unless stated otherwise. In all figures, P-values are indicated as follows: *P<0.05, ** P <0.005, ***P <0.0005, **** P <0.0001, two-tailed. In each figure legend, n refers to the number of wells/independent differentiations. Wells from the same independent differentiation were differentiated on the same day (same differentiation round). Gaussian distribution and equal variances were ensured using Kolmogorov–Smirnov or Levene’s test. When comparing two groups with non-Gaussian distributed data, a Mann–Whitney test was used (Fig. 2 and Supplementary Fig. 3). When comparing three or more groups without repeated measures and non-Gaussian distributed data, a one-way ANOVA with Kruskal–Wallis test was used (Figs 3–5 and Supplementary Figs 4 and 5) and Dunn’s correction for multiple comparisons was used when necessary. A repeated measures one-way ANOVA was used with Friedman correction when comparing repeated measures with one factor (Fig. 4) from non-Gaussian distributed data or Dunnett’s correction when comparing Gaussian distributed data. For comparison of two or more factors with repeated measures, we used a repeated measures two-way ANOVA with Dunn–Šidák correction for multiple testing (Fig. 1 and Supplementary Fig. 1). Outliers were only excluded if they met the exclusion criteria described above (extreme cell clustering or clumping, unequal or low cell density). Principal component analysis (PCA) was performed on data from eight parameters [MFR, PRS, MBR, burst duration (BD), burst spike rate (BSR), NBR, network burst duration (NBD) and HFB] using Graphpad PRISM 9.0.0. Values were standardized and principal components were selected based on parallel analysis using Monte Carlo simulation on random data of equal dimension to the input data, generated with a random seed.
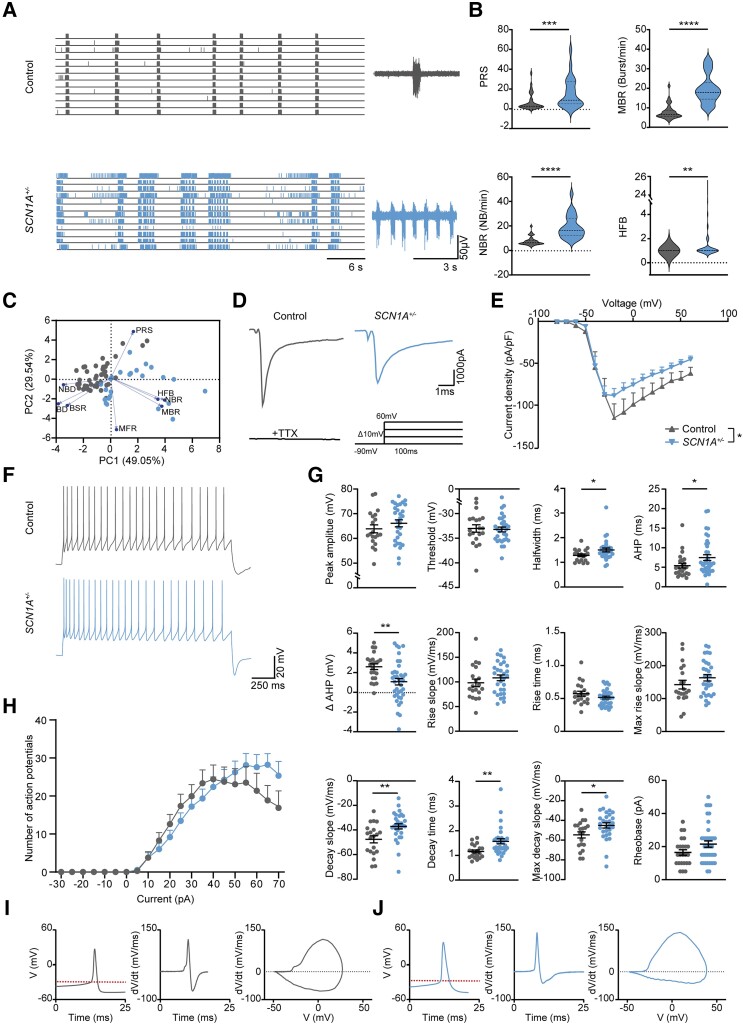
Figure 2.
SCN1A +/−-deficient neurons depict hyperactive, desynchronized and irregular firing patterns. (A) Representative raster plots of MEA recordings at DIV49 from control and SCN1A+/−-deficient neuronal networks, with a 6 s closer view (right) of the activity of one electrode. (B) Quantification of network parameters, including percentage of random spikes (PRS), mean burst rate (MBR), network burst rate (NBR) and number of high frequency bursts (HFB), quantified as the number of HFBs inside an NB period. Dashed line represents median, dotted line represents quartiles. Mann–Whitney test. (C) PCA plot of eight MEA parameters, including mean firing rate (MFR), MBR, NBR, PRS, burst duration (BD), NBD, HFB, burst spike rate (BSR), showing parameters that explain the differences in network behaviour between control and SCN1A+/−-deficient neuronal networks. Blue arrows indicate loadings. For all MEA data, n = number of wells/independent differentiations: control n = 33/5 (CTRL2) and SCN1A+/−n = 24/2 (SCN1A+/− C3: 8/2, SCN1A+/− C7:16/2). (D) Representative Na+-current traces of control and SCN1A+/−-deficient neurons at DIV49. Stimulation paradigm (inset): stepwise protocol from a −90 mV holding potential to a maximum test-pulse of 60 mV in increments of 10 mV, Na+-currents were tetrodotoxin (TTX)-sensitive. (E) Current-density plot of Na+-current recordings from n = cells/batches Control n = 18/3 (CTRL2) and SCN1A+/−n = 22/3 (SCN1A+/− C3: 11/3, SCN1A+/− C7:11/3). Repeated measures ANOVA with Dunnett’s multiple comparisons test. (F) Representative firing patterns of control and SCN1A+/− neurons, measured by a step-wise current injection protocol. (G) Analysis of active and passive membrane properties, including peak amplitude, threshold, half-width, after hyper polarization time (AHP), ΔAHP, defined as the difference between the AHP of the first action potential and the second action potential in the same sweep, rise slope, rise time, maximum rise slope, decay slope, decay time, max decay slope, rheobase. (H) Quantification of the number of action potentials per current injection in a 2-s time window. (I) Representative plots depicting control action potential amplitude and time course, dV/dt versus time and a phase plot of dV/dt versus V. (J) Representative plots depicting SCN1A+/−-deficient action potential amplitude and time course, dV/dt versus time and a phase plot of dV/dt versus V. Red dotted line represents the action potential threshold. For all action potential properties, n = cells/independent differentiations: control n = 24/3 (CTRL2) and SCN1A+/−n = 39/3, (SCN1A+/− C3: 21/3, SCN1A+/− C7:18/3). Mann–Whitney test. Data represented as mean ± SEM. *P = 0.05, **P = 0.01, ***P = 0.001, ****P < 0.0001. All means, SEM and P-values of significant differences are listed in Supplementary Table 1. Data from separate SCN1A+/−-deficient lines are listed in Supplementary Table 2.
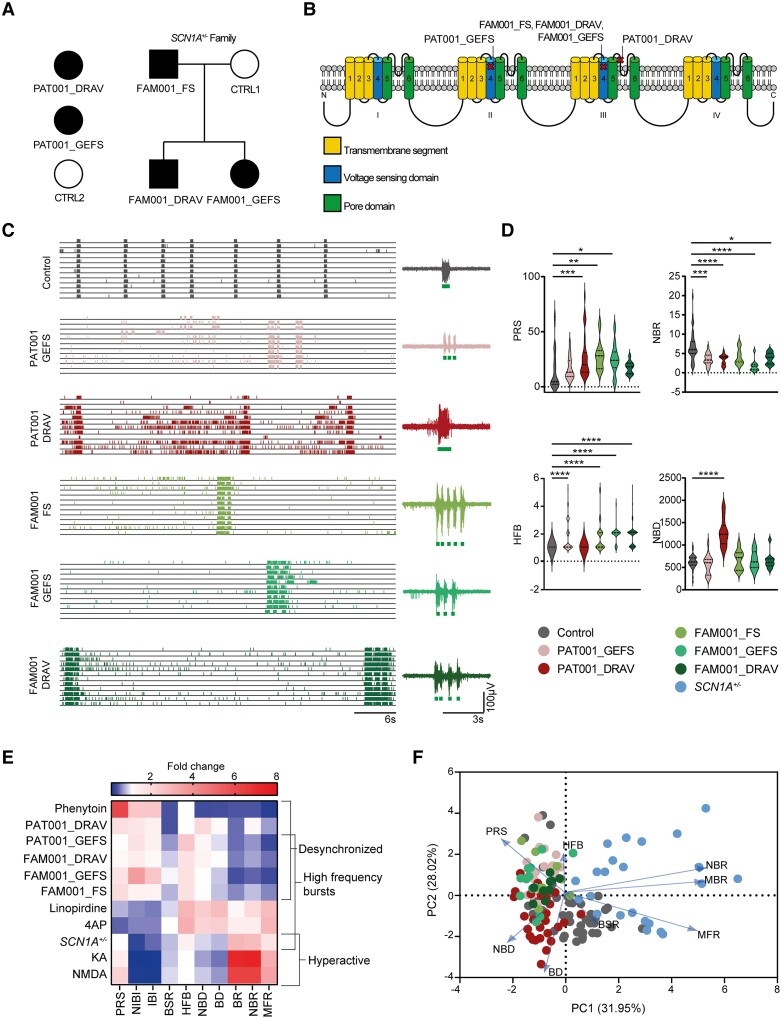
Figure 3.
Dravet syndrome patient-derived neuronal networks show mutation-specific network fingerprints. (A) Schematic overview of patient inclusion, including pedigree of one family, where the same SCN1A mutation is inherited via the paternal line. Filled black symbol represents Dravet syndrome (DS), three quarters black symbol represents GEFS+ syndrome, one quarter black symbol represents febrile seizures (FS). (B) Overview of affected locations in the Nav1.1 protein. (C) Representative raster plots of multi-electrode array (MEA) recordings from control and patient-derived networks, with a 6 s closer view (right) of the activity of one electrode. (D) Quantification of network parameters, including percentage of random spikes (PRS), network burst rate (NBR), number of high frequency bursts (HFB) quantified as the number of HFBs inside a network burst period and network burst duration (NBD). Dashed line represents median, dotted line represents quartiles. (E) Heat map of all MEA parameters for all compound-treated recordings, normalized to pretreatment baseline, and all SCN1A -deficient lines normalized to control. Parameters include mean firing rate (MFR), mean burst rate (MBR), NBR, PRS, burst duration (BD), NBD, HFB, burst spike rate (BSR), inter-burst interval (IBI) and network inter-burst interval (NIBI). (F) PCA plot of eight MEA parameters, including MFR, MBR, NBR, PRS, BD, NBD, BSR and HFB, showing parameters that explain the differences in network behaviour between control and patient lines. Blue arrows indicate loadings. For all MEA data, n = number of wells/independent differentiations: CTRL1 n = 9/2, CTRL2 n = 36/5, PAT001_GEFS n = 22/4, PAT001_DRAV n = 33/6, FAM001_FS n = 9/2, FAM001_GEFS n = 11/3, FAM001_DRAV n = 11/3, SCN1A+/−n = 23/3; one way ANOVA with Kruskal–Wallis test with Dunn’s correction for multiple comparisons. *P = 0.05, **P = 0.01, ***P = 0.001, ****P < 0.0001. All means, SEM and P-values of significant differences are listed in Supplementary Table 1.
Figure 5.
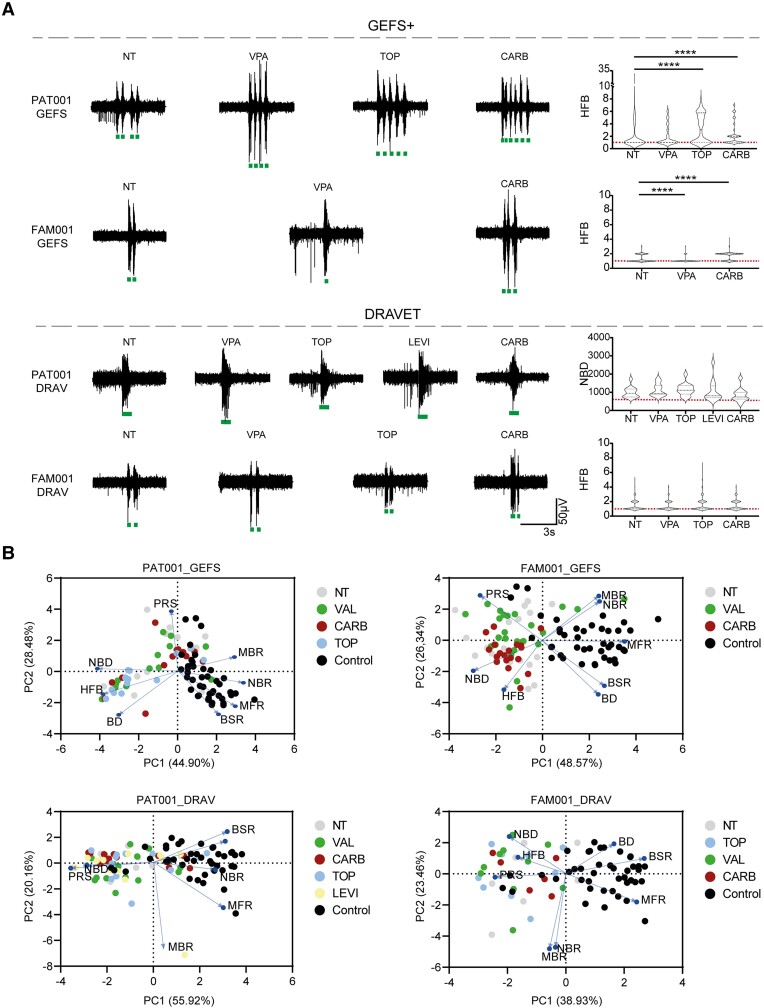
GEFS+, but not Dravet syndrome-derived neuronal networks, respond to anti-seizure medication. (A) Representative burst traces of GEFS+ patients (top) and Dravet syndrome (DS) patients (bottom), including quantification of number of high frequency bursts (HFB) for PAT001_GEFS, FAM001_GEFS and FAM001_DRAV lines, and network burst duration (NBD) for PAT001_DRAV in non-treated (NT) patients or those treated with 10 µM topiramate (TOP), valproic acid (VPA), levetiracetam (LEVI) or carbamazepine (CARB). HFB quantification for PAT001_DRAV was not included, since this line does not show HFB. Dashed line represents median, dotted line represents quartiles. Red dashed line represents control median. (B) Principal component analysis (PCA) plot of eight multi-electrode array (MEA) parameters, including mean firing rate (MFR), mean burst rate (MBR), network burst rate (NBR), percentage of random spikes (PRS), burst duration (BD), NBD, burst spike rate (BSR) and HFB, showing parameters that explain the differences in network behaviour between control and patient lines, in either non-treated patients or those treated with anti-seizure medication. Blue arrows indicate loadings. For all MEA data, n = number of wells/independent differentiations: PAT001_GEFSNTn = 13/5, PAT001_GEFSVALn = 13/5, PAT001_GEFSTOPn = 13/5, PAT001_GEFSCARBn = 13/5, PAT001_DRAVNTn = 13/5, PAT001_DRAVVALn = 15/5, PAT001_DRAVTOPn = 14/6, PAT001_DRAVLEVIn = 12/5, PAT001_DRAVCARBn = 13/4, FAM001_GEFSNTn = 18/6, FAM001_GEFSVALn = 24/6, FAM001_GEFSCARBn = 19/4, FAM001_DRAVNTn = 7/3, FAM001_DRAVTOPn = 9/3, FAM001_DRAVVALn = 10/3 FAM001_DRAVCARBn = 6/1. Significance calculated using ANOVA with Kruskal–Wallis test and Dunn’s correction for multiple comparisons. *P = 0.05, **P = 0.01, ***P = 0.001, ****P < 0.0001. All means, SEM and P-values of significant differences are listed in Supplementary Table 1.
Figure 4.
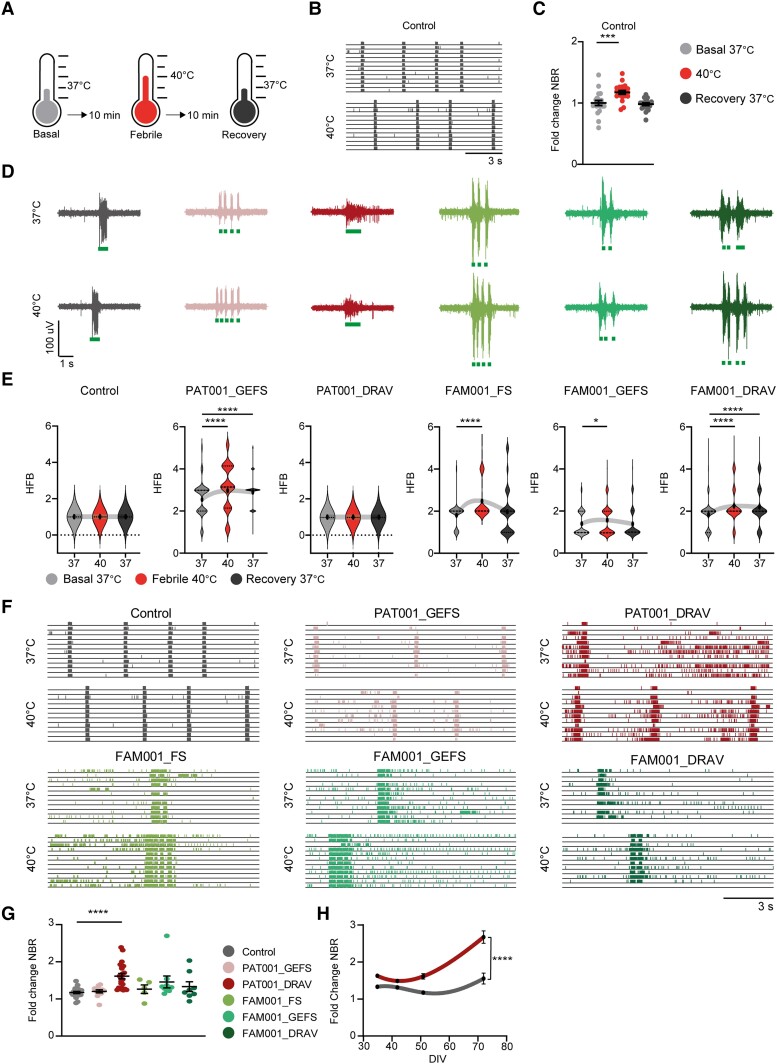
Febrile temperatures lead to altered neuronal network organization. (A) Schematic stimulation paradigm for febrile temperature recordings. (B) Representative raster plots from multi-electrode array (MEA) recordings of control neuronal networks, recorded at 37°C and 40°C. (C) Network burst rate (NBR) from control neuronal networks, normalized to the basal recording at 37°C. Data is shown as mean ± SEM. Repeated measures ANOVA with Friedmans correction for multiple testing. (D) Representative burst shapes of control and patient lines green lines indicate high frequency burst (HFB) detection. (E) Number of HFB, quantified as the number of HFBs inside a network burst period, during basal (37°C), febrile (40°C) and recovery (37°C) recordings. Dashed line represents median, dotted line represents quartiles, Kruskall–Wallis test. (F) Representative raster plots from MEA recordings of control and patient neuronal networks, recorded at 37°C and 40°C. (G) NBR from control and patient neuronal networks, normalized to the basal recording at 37°C. Data are shown as mean ± SEM, Kruskall–Wallis test. (H) NBR from control and PAT001_DRAV neuronal networks over developmental days in vitro (DIV), normalized to the basal recording at 37°C, two-way ANOVA. For all MEA data, n = number of wells/independent differentiations: control n = 20/3 (CTRL2), PAT001_GEFS n = 12/2, PAT001_DRAV n = 22/3 FAM001_FS n = 5/1, FAM001_GEFS n = 9/2, FAM001_DRAV n = 8/2, all data were recorded at DIV49. *P = 0.05, **P = 0.01, ***P = 0.001, ****P < 0.0001. All means, SEM and P-values of significant differences are listed in Supplementary Table 1.
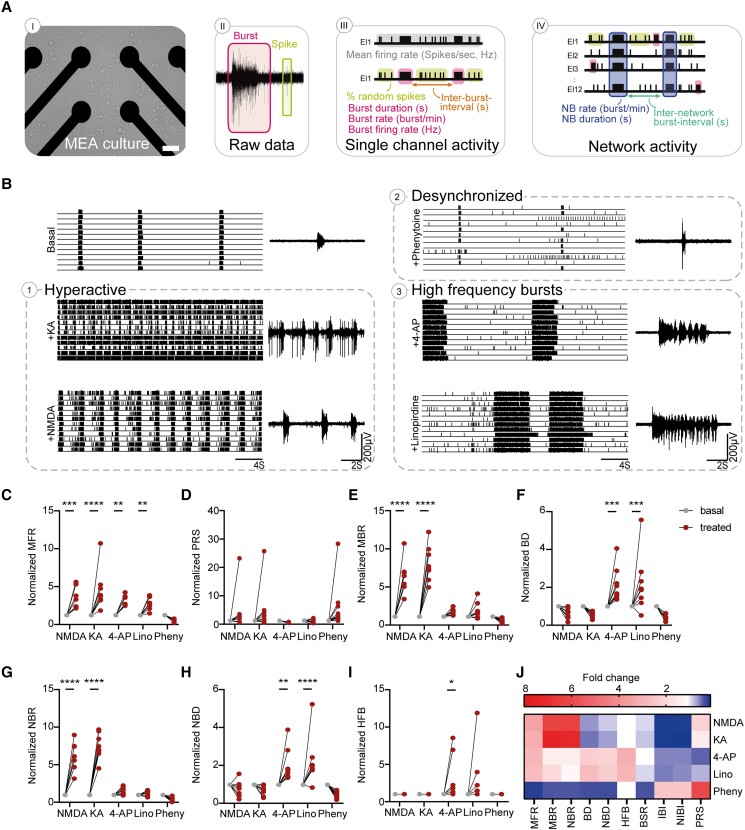
Figure 1.
Seizure liability screening reveals two distinctive seizure prediction patterns. (A) Schematic overview of a multi-electrode array (MEA) culture, raw recording from a representative electrode and raster plot, including the measured MEA parameters on both single channel and network levels. Scale bar = 100 µm (B) Representative raster plots of MEA recordings from control networks and control networks acutely treated with different compounds: NMDA (100 µM), 4-aminopyridine (4-AP,10 µM), kainic acid (KA, 10 µM), linopirdine (Lino, 1.5 µM) and phenytoin (Pheny, 25 µM) with an 8 s closer view (right) of the activity of one electrode. Quantification of network parameters normalized to pretreatment baseline including: (C) mean firing rate (MFR), (D) percentage of random spikes (PRS), (E) mean burst rate (MBR), (F) burst duration (BD), (G) network burst rate (NBR), (H) network burst duration (NBD) and (I) number of high frequency bursts (HFB). (J) Heat map of all MEA parameters for all compound-treated recordings, normalized to pretreatment baseline, including MFR, MBR, NBR, PRS, BD, NBD, HFB, burst spike rate (BSR), inter-burst interval (IBI) and network inter-burst interval (NIBI). For all MEA data, NMDA n = 7, KA n = 8, 4-AP n = 8, Lino n = 8 and Pheny n = 10 (number of pre-and post-treatment wells). All wells were derived from CTRL2. *P = 0.05, **P = 0.01, ***P = 0.001, ****P < 0.0001; repeated measures ANOVA with Dunn–Šidák multiple comparisons correction. All means, SEM and test statistics are listed in Table 1.
Results
Proconvulsive compounds induce three distinct phenotypes in control neuronal networks
Previously, we functionally benchmarked 10 control neuronal networks on MEAs by differentiating hiPSCs to electrophysiologically mature excitatory neurons through forced expression of Ngn2.32,33,36 Network activity consisted of spikes (single action potentials) and bursts (clustered action potentials at high frequency), which self-organized into synchronous NBs (bursts recorded from all electrodes) (Fig. 1A).36 Traditionally, proconvulsive compounds are used to elicit neuronal network responses as a predictor for seizurogenic network activity.38,39 To understand the applicability of excitatory neurons as a model for DS-related epilepsy, and to predict excitatory seizurogenic phenotypes, we selected five different seizurogenic compounds that affect excitatory neuronal mechanisms and functionally characterized the network properties (Fig. 1B–J and Supplementary Fig. 1G). NMDA and KA both led to a hyperactive neuronal network phenotype, reflected by a significant increase in MFR, MBR and NBR (Fig. 1B–I). To mimic mutations in SCN1A, we used the Na+-channel blocker phenytoin, traditionally used as an ASM, which can also represent seizure prediction patterns in healthy neuronal networks.38 Phenytoin led to a distinct seizure prediction pattern, with a general decrease in activity and synchronicity, reflected by a decreased MFR, MBR and NBR but increased PRS (isolated, a-synchronous spikes) (Fig. 1C–E and G). K+-channel blockers 4-AP and linopirdine showed a third distinct seizure prediction pattern (Fig. 1B and J). While the neuronal networks responded with a similar increase in MFR (Fig. 1C), the organization of the network differed and showed an irregular neuronal network pattern (Fig. 1B). Whereas the NBR was not significantly increased, the NBD increased, and the NBs consisted of several HFBs (Fig. 1G–I). Importantly, networks did not respond to picrotoxin, confirming their excitatory identity (Supplementary Fig. 1H). A heat map clearly showed three different seizure prediction patterns upon exposure to proconvulsive compounds (Fig. 1J) in analogy to neuronal networks in rodent cultures.39 Prediction Pattern 1, elicited by KA and NMDA, led to a hyperactive neuronal network phenotype. Prediction Pattern 2, elicited by phenytoin, led to a hypoactive, desynchronized network activity. Finally, Prediction Pattern 3, elicited by 4-AP and linopirdine, led to disorganization including NBs with HFBs.
Heterozygous loss-of-function mutations in SCN1A lead to a desynchronized and hyperactive network phenotype
Heterozygous LoF mutations in SCN1A predominantly result in a severe DS clinical phenotype and not in an intermediate GEFS+- or mild febrile seizure (FS)-like phenotype.11 To examine the effect of heterozygous LoF of SCN1A in mature excitatory neuronal networks, we generated two SCN1A+/−-deficient lines (SCN1A+/− C3 and SCN1A+/− C7). We used CRISPR/Cas9 genome editing in a curated control hiPSC-line to induce a frameshift mutation, leading to a heterozygous deletion in exon 21 of SCN1A, resulting in a premature stop codon at amino acid position 18 in both clones (Supplementary Fig. 2A–C). Western blot confirmed reduced Nav1.1 expression in SCN1A+/−-deficient lines (Supplementary Fig. 2D).
Immunocytochemistry confirmed the expression of Nav1.1 in excitatory neurons (Supplementary Fig. 2E). In line with the localization of Nav1.1 in inhibitory neurons, we found Nav1.1 expression in the soma and axon initial segment (AIS), with specific dense puncta [Supplementary Fig. 2F(ii)].16 In addition, Nav1.1 was expressed in the dendrites [Supplementary Fig. 2F(ii)]. The expression of SCN1A increased over time and was highest at DIV49 (Supplementary Fig. 2G); we therefore conducted all experiments at this time-point. We further confirmed that the identity of the generated neurons were mostly of cortical excitatory origin, in line with previous reports (Supplementary Fig. 2H–J).33
Control networks showed synchronous, rhythmic NBs at DIV49 (Fig. 2A), a hallmark of physiological network activity.36 In contrast, SCN1A+/−-deficient lines showed a hyperactive, desynchronous network state (Fig. 2A), where NB regularity was completely disrupted. The network activity in both SCN1A+/−-deficient lines showed blocks of intense high-frequency periods, consisting of multiple HFBs, followed by relatively silent periods, and a significant increase in PRS, NBR and MBR but a significantly decreased BD (Fig. 2B and Supplementary Fig. 3A). The PCA showed clear separation of SCN1A+/−-deficient and control networks (Fig. 2C). In conclusion, SCN1A+/−-deficient neuronal networks were differently organized than control neuronal networks, showing a hyperactive-desynchronized neuronal network phenotype, resembling the previously identified seizure Prediction Pattern 1 (Fig. 1J).
To determine the contribution of single cell properties to the phenotype observed in SCN1A+/−-deficient excitatory networks, we examined Na+-current and action potential properties (Fig. 2D–J and Supplementary Fig. 2L–O). Current/voltage plots showed a slight reduction in Na+-current density in SCN1A+/−- deficient neurons (Fig. 2D and E), without changes in the channel kinetics (Boltzmann-fitting) (Supplementary Fig. 2L). Action potentials in SCN1A+/−-deficient neurons had a significantly increased half-width (Fig. 2G), which is directly related to fast neuronal Na+-channel activation. Interestingly, this was not reflected by altered rise dynamics (Fig. 2G). Rather, action potentials in SCN1A+/−-deficient neurons showed a significant decline in decay dynamics (Fig. 2G, I and J), indicative of altered dynamics in voltage dependent potassium (Kv)-channels. Indeed, the after hyperpolarization (AHP) time was increased and inter-spike AHPs (ΔAHP), reflecting AHP adaptation, were more hyperpolarized (Fig. 2G). Because excitability could also be affected by changes in AIS properties,40 we quantified AIS length but found no significant differences (Supplementary Fig. 3F and G). Finally, SCN1A+/−-deficient neurons displayed remarkable irregular firing patterns, with a slightly increased frequency (Fig. 2H and Supplementary Fig. 2N and O). To conclude, excitatory neurons are affected by SCN1A+/−-deficiency, both at the neuronal network and single cell levels.
Patient-derived excitatory neuronal networks display mutation-specific phenotypes
Previous evidence in both hiPSC-derived neurons and mouse models has shown conflicting data regarding the contribution of excitatory neurons to the DS phenotype.14,16,20,21,26,27 We wondered whether the excitatory neuronal network phenotypes might segregate according to the type of mutation and/or the patient clinical phenotype (GEFS+ versus DS). To this end, we investigated the role of SCN1A missense mutations in a patient cohort that consisted of patients with severe (DS), intermediate (GEFS+) and mild (FS) phenotypes (Fig. 3A and B and Supplementary material). In particular, we studied a family where the same missense mutation in SCN1A resulted in different clinical phenotypes (Fig. 3A and B and Table 1). Our final cohort consisted of: two controls, CTRL1 and CTRL2; one independent GEFS patient: PAT001_GEFS; one independent DS patient: PAT001_DRAV; and the SCN1A family, consisting of control CTRL1, FS patient FAM001_FS, GEFS+ patient FAM001_GEFS and DS patient FAM001_DRAV (Fig. 3A and B).
Table 1.
Patient information and SCN1A variant position and amino acid changes
| ID | Sex | Age (years) | Mutation |
|---|---|---|---|
| PAT001_DRAV | Female | 4 | c.4168G>A (p.Val1390Met) |
| PAT001_GEFS | Female | 10 | c.2576G>A (p.Arg859His) |
| FAM001_FS | Male | 49 | c.3926T>G (p.Leu1309Arg) |
| FAM001_GEFS | Female | 14 | c.3926T>G (p.Leu1309Arg) |
| FAM001_DRAV | Male | 16 | c.3926T>G (p.Leu1309Arg) |
All patient-derived lines showed significantly different network phenotypes than control (Fig. 3C). PAT001_DRAV, PAT001_GEFS, FAM001_FS, FAM001_DRAV and FAM001_GEFS neuronal networks consisted of significantly less synchronous NBs and more random spikes (Fig. 3D) than control networks, consistent with seizure Prediction Pattern 2 (Fig. 1J). Moreover, we observed in PAT001_GEFS, FAM001_FS, FAM001_GEFS and FAM001_DRAV a burst phenotype that included NBs with HFBs, followed by relatively silent periods, reminiscent of the burst phenotype of SCN1A+/−-deficient lines and seizure Prediction Pattern 3 (Fig. 3E). In contrast, PAT001_DRAV neuronal networks did not show this particular HFB phenotype but presented with a significant increase in NBD (Fig. 3D). Overall, two different network phenotypes were distinguished. We found either a desynchronized neuronal network phenotype, specifically in patient-derived neuronal networks with missense mutations in SCN1A, or a desynchronized but hyperactive neuronal network phenotype, in neuronal networks with a heterozygous LoF in SCN1A. The segregation of these different network phenotypes is in line with previously reported seizure-prediction patterns,39 and the response of the control neuronal network to proconvulsive compounds in Fig. 1 (Fig. 3E). The PCA further revealed four clearly identifiable clusters based on control, SCN1A+/−-deficient and patient lines (Fig. 3F). More importantly, the patient lines subclustered into patients with a missense mutation in the pore domain of Nav1.1 (PAT001_DRAV) and patients with a missense mutation in the voltage sensing domain of Nav1.1 (PAT001_GEFS, FAM001_FS, FAM001_GEFS and FAM001_DRAV) (Fig. 3F). We conclude that the excitatory neuronal network phenotype is dependent on the type of SCN1A mutation but that different clinical phenotypes could not be distinguished.
In addition, we investigated if we could uncover mutation-specific phenotypes based on single cell properties (Supplementary Fig. 4A–E). In line with the biophysical properties of a mutation in the pore domain, PAT001_DRAV neurons displayed a significant reduction in Na+-current density (Supplementary Fig. 4A and B). On the other hand, PAT001_GEFS, FAM001_FS, FAM001_GEFS and FAM001_DRAV excitatory neurons did not show a significant different Na+-current dynamics, V1/2 or k (Supplementary Fig. 4B and C). Interestingly, the ΔAHP was significantly more hyperpolarized in all patient-derived neurons with a missense mutation in the voltage sensing domain (Supplementary Fig. 4E). We confirmed that the four different network phenotype clusters we observed were not a result of differences in synapse density (Supplementary Fig. 5A and B). Moreover, the PCA of the controls used here, combined with a larger control cohort phenotyped previously,36 showed no observable clustering based on age. Similarly, patients with variable age ranges clustered based on the type of mutation, but not on age, ruling out a simple age effect (Supplementary Fig. 5C and D). Finally, we confirmed that the four different network phenotypes that we observed here were not a result from changes in excitability due to alterations in cell viability or changes in AIS length (Supplementary Fig. 5E–H).
Febrile temperatures alter neuronal network organization
As all patients described in this study experienced febrile seizures, we examined the response of the neuronal networks to febrile temperatures (Fig. 4A). Increasing the temperature to 40°C led to an increase in NBR in control lines, which fully returned to baseline when the temperature was decreased to 37°C (Fig. 4B and C). Strikingly, all patient lines that displayed NBs with HFBs showed a significant increase in the number of HFBs at febrile temperatures (Fig. 4D and E). In basal conditions, the SCN1A family lines could not be distinguished, but after febrile temperature induction, only FAM001_FS and FAM001_GEFS neuronal network organization returned to baseline, while FAM001_DRAV neuronal networks did not (Fig. 4E). Interestingly, PAT001_DRAV showed a significantly higher increase in NBR than the control and other patient networks, signifying a higher sensitivity to febrile temperatures (Fig. 4G), which aggravated during development (Fig. 4H). Moreover, networks measured at febrile temperatures showed more distinguished clusters on the PCA based on mutation type (Supplementary Fig. 5I) than at basal temperatures (Fig. 3F). The response of the patient neuronal network to febrile temperatures resembled the earlier described response of the control network to proconvulsive compounds, with an increase in NBR and HFBs as the most prominent features, corresponding to Prediction Patterns 1 and 3, respectively (Fig. 1). To conclude, febrile temperatures alter neuronal network organization and aggravate the network phenotype associated with HFBs.
Anti-seizure medication affects GEFS+ neuronal networks in a clinically relevant matter
Although ASM prescribed for DS mostly works on the GABAergic system, we investigated whether ASM could also affect the excitatory network phenotype we described and if excitatory networks already give an indicative prediction of ASM non-responders versus responders. We therefore tested the response of the excitatory neuronal networks to the ASMs valproic acid, levetiracetam and topiramate, all of which have been prescribed to our patient cohort, and carbamazepine, which is a general contra-indicated ASM for DS (Supplementary material). Since the HFB was the main phenotypic driver in PAT001_GEFS, FAM001_GEFS and FAM001_DRAV, we compared the effect of the ASM on this parameter, as well as on the NBD in PAT001_DRAV. We did not see any significant changes in the number of HFBs in FAM001_DRAV or NBD in PAT001_DRAV, indicating that excitatory networks alone could not predict drug efficacy in DS patients (Fig. 5A). However, valproic acid significantly decreased the number of HFBs in FAM001_GEFS, while carbamazepine significantly increased the number of HFBs, in line with the clinical history. Indeed, carbamazepine also aggravated the HFB phenotype in PAT001_GEFS, whereas the neuronal network remained unresponsive to valproic acid, again in line with the clinical history of this patient (Fig. 5A and Table 2). While seizures in PAT001_GEFS were responsive to topiramate, it aggravated the neuronal network phenotype in vitro. This could be explained by the fact that topiramate most likely works by inhibiting Na+-channels in excitatory cultures and not by modulating GABAA receptor-mediated currents. Although we could not uncover differences between GEFS+ and DS in the basal phenotypes, the neuronal network remained unresponsive to ASM in DS patients. Indeed, PCA revealed clear clustering of ASM between treated and non-treated wells in GEFS+ patients but not in DS patients (Fig. 5B). To conclude, the response of the main phenotypic parameter in GEFS+ patients could be predicted in a clinical-relevant matter, further implicating excitatory neurons as a phenotypic contributor and HFBs as an important phenotypic driver.
Table 2.
Summary of the clinical anti-seizure medication (ASM) response of each included patient, and the response of the patient-derived neuronal networks to the same ASM
| Sex | Clinical phenotype | Seizure control | Neuronal network response | |
|---|---|---|---|---|
| PAT001_DRAV | Female | Dravet syndrome | Levetiracetam, valproic acid. Seizures remain uncontrolled | No response |
| PAT001_GEFS | Female | GEFS+ | Seizures unresponsive to valproic acid. Seizure free with topiramate | Carbamazepine and topiramate |
| FAM001_GEFS | Female | GEFS+ | Seizure free with valproic acid | Valproic acid and carbamazepine |
| FAM001_DRAV | Male | Dravet syndrome | Seizure free with topiramate and valproic acid, seizures resumed after 5 years | No response |
Discussion
The contribution of excitatory neurons to the DS phenotype has been heavily debated. The observed excitatory phenotype in some studies, but not in others, might be attributed to differences in patient clinical phenotype or the type of mutation. Here, we provide further evidence for the involvement of excitatory neurons in DS, in line with previous reports,20,26,27 in four independent ways. First, we uncovered mutation-specific network phenotypes. Of interest, the neuronal network phenotypes, under basal conditions, did not differ between patients from one family with the same SCN1A mutation but with different clinical phenotypes. Rather, the neuronal network phenotype from this family clustered together with that of an independent patient line with a similar missense mutation in the voltage-sensing domain. Indeed, mutations in the voltage sensing domain were distinguished from mutations in the pore domain. Second, we showed that the patient-derived network phenotypes correspond to the response of naïve networks to pro-convulsive compounds. Third, we uncovered a previously unidentified excitatory neuronal network phenotype, HFBs, which were aggravated by clinical-relevant trigger febrile temperatures. Finally, the HFB phenotype in GEFS+ patient-derived neuronal networks responded to the ASMs valproic acid and carbamazepine in a clinically relevant manner.
We found no observable differences in neuronal network phenotypes between individuals from the same family that shared an identical mutation in SCN1A but presented different clinical phenotypes. As this family was diagnosed before whole exome sequencing was a routine clinical practice in the Netherlands, we cannot exclude that the family carries variants in epilepsy-related genes that could potentially modify the disease severity. More recently the view has shifted from rare variants with large effects on clinical phenotypes towards the presence of more common variants in modifier genes that might tip the balance over to a milder or more severe clinical phenotype.41 Further investigations into the expression of modifier genes in SCN1A families with different clinical presentations but identical mutations could help to underpin how severe or mild clinical phenotypes arise. Moreover, future studies should confirm the mutation-specific excitatory neuronal network phenotypes observed here, as this study is limited by the fact that we used a small cohort of patient lines.
While mutation-specific changes have not been described previously in SCN1A-deficient excitatory neuronal networks, previous research uncovered variant-specific alterations in Nav1.1 properties on a single cell level, in both excitatory and inhibitory neurons.42-45 The type or position of the mutation in the channel can predict if the mutation leads to Nav1.1 GoF or LoF to some degree.8 We observed a significant decrease in Na+-current density in DS-derived neurons with a missense mutation in the pore domain, matching the expected LoF properties of a pore-domain mutation.8 However, we could not detect significant changes in Na+-currents in neurons with mutations in the voltage sensing domain. We envisage three explanations. First, the elicited Na+-currents were affected by space clamp artifacts,27 caused by the expression of Na+-channels at sites electrically distant from the neuronal soma. This is in line with numerous other studies investigating Na+-currents in hiPSC-derived neurons.21-23,25-27 Second, the expression of Nav1.1 in excitatory neurons might be insufficient to properly detect more subtle changes in Nav1.1 currents, which could be overshadowed by other Na+-channels working in concert. Small changes in Nav1.1 currents might have a cumulative effect, explaining why we observed changes in activity on a neuronal network level. Finally, Na+-channels can function as a dimer and display coupled gating. Interaction with wild-type channels could therefore also influence channel kinetics.46 Taken together, true mechanistic insights into the biophysical properties of each variant should be measured carefully in expression systems with proper voltage control.
Interestingly, we report that SCN1A-deficiency and reduced Na+-current density did not result in reduced firing but rather increased excitability in SCN1A+/−-deficient neurons. This could be due to upregulation of other Nav channels. For example, homozygous null Scn1a−/− mice, showed specific upregulation of Nav1.3 in hippocampal interneurons.13 However, our data also point to a compensatory mechanism from Kv-channels. We uncovered significant changes in action potential decay kinetics and AHP in SCN1A+/−-deficient neurons. In addition, we observed a hyperpolarizing shift in ΔAHP in all neuronal lines that displayed HFBs, suggesting similar underlying mechanisms. Moreover, blocking Kv-channels by 4-AP and linopirdine resulted in a similar network phenotype to HFBs. Compensatory rebalancing in excitatory neurons was observed in previous work. In a Scn2a−/− mutant mouse model, the Kv-channel Kv8.2 functions as a genetic modifier, resulting in increased susceptibility to epilepsy in pyramidal cells.47 Additionally, in a mouse model with a GoF mutation in Scn2a, neocortical pyramidal cell hyperexcitability was explained by a reduction in hyperpolarizing K+-currents.48 Finally, in the Scn1b−/− DS mouse model, increased neuronal input resistance caused pyramidal cell hyperexcitability. This phenotype was rescued by the ASM retigabine, a Kv-channel opener that reduces neuronal input resistance.49 To conclude, compensatory rebalancing of Kv-channels might explain why SCN1A+/−-deficient excitatory neurons appear as paradoxically hyperactive.
Both this study and previous works fundamentally indicate that SCN1A mutations do not exclusively affect inhibitory neurons.20,26 However, the role of inhibitory neurons in the pathophysiology of DS remains indisputable, especially considering the broad group of ASMs prescribed for DS that target the GABAergic system. Nonetheless, we uncovered that in excitatory neurons, the main phenotypic driver, HFBs, responds to ASMs in a clinically relevant matter in GEFS+ patients, contrary to DS patient networks. On a clinical level, the DS patients in our cohort are not seizure free with ASM but rather show decreased seizure frequency. An excitatory-only neuronal network could therefore not predict changes in seizure frequency upon ASM administration. That we were nonetheless able to distinguish GEFS+ and DS patient responses to ASM further supports the role of excitatory neurons.
Through which mechanisms the ASM affects the HFB phenotype is still unclear. The working mechanisms of valproic acid are poorly understood and it predominantly functions through regional changes in GABA concentration. Previous research uncovered that valproic acid can also regulate pERK trough attenuation of the PKA system.50 Interestingly, proteomic signature analysis of Scn1a+/− mice revealed altered protein regulation in excitatory synapses specifically, implicating a dysregulation of the PKA pathway, before the seizure threshold.51 It could be that valproic acid alters excitatory neuronal networks through a similar mechanism. Moreover, topiramate acts as a positive allosteric modulator of GABAA receptor-mediated currents but also inhibits Nav channels. In our excitatory only networks, topiramate will clearly not affect GABAa-mediated currents. Therefore, the sole target of topiramate in our cultures are Nav channels, which is a contra-indication for DS. This could explain the aggravated or unaffected HFB phenotype that we observed. Future research should therefore focus on the incorporation of inhibitory neurons to the network,52 which might serve as a more ideal model for patient-specific drug response in the context of DS, especially considering the continuous improvement of methods that predict whether an SCN1A mutation will lead to a relatively mild or very severe clinical outcome.8 This could make an early, targeted pharmaceutical intervention possible. Since DS requires a different approach than GEFS+, it is of utmost interest to develop a predictable model system that allows patient-specific testing of ASMs.
Finally, in the human brain, SCN1A expression increases gradually from neonatal onset into adulthood,53 which is in line with the developmental expression in the hiPSC-derived neurons described here. During development, the expression ratio of SCN1A in excitatory versus inhibitory neurons decreases in the human brain, and other Na+-channels become predominantly expressed in excitatory neurons.54 There might be a critical time window in which mutations in SCN1A exert their pathogenic effect in excitatory neurons. Further investigations of how altered excitation contributes to different disease stages could be an important approach to further understanding the pathophysiology of DS.
Supplementary Material
Contributor Information
Eline J H van Hugte, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands; Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, The Netherlands; Department of Epileptology, ACE Kempenhaeghe, 5591 VE Heeze, The Netherlands.
Elly I Lewerissa, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands; Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, The Netherlands.
Ka Man Wu, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands.
Nicky Scheefhals, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands.
Giulia Parodi, Department of Informatics, Bioengineering, Robotics, and Systems Engineering (DIBRIS), University of Genova, 16145 GE Genova, Italy.
Torben W van Voorst, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands; Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, The Netherlands.
Sofia Puvogel, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands.
Naoki Kogo, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands; Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, The Netherlands.
Jason M Keller, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands; Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, The Netherlands.
Monica Frega, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands; Department of Clinical Neurophysiology, University of Twente, 7522 NB Enschede, The Netherlands.
Dirk Schubert, Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, The Netherlands.
Helenius J Schelhaas, Department of Neurology, Stichting Epilepsie Instellingen Nederland (SEIN), 2103 SW Heemstede, The Netherlands.
Judith Verhoeven, Department of Epileptology, ACE Kempenhaeghe, 5591 VE Heeze, The Netherlands.
Marian Majoie, Department of Epileptology, ACE Kempenhaeghe, 5591 VE Heeze, The Netherlands.
Hans van Bokhoven, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands; Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, The Netherlands.
Nael Nadif Kasri, Department of Human Genetics, Radboudumc, 6500 HB Nijmegen, The Netherlands; Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, The Netherlands.
Data availability
All data and algorithms generated in this study can be requested from the corresponding author. Aside from the hiPSC lines, this study did not generate unique new reagents. All reagents, vectors or cell lines used in this study are available from the corresponding author upon request with a completed Materials Transfer Agreement.
Funding
This work was supported by the Netherlands Organisation for Health Research and Development ZonMw grant 91217055 (to H.v.B., N.N.K., M.M., J.V. and H.J.S.), the European Joint Programme on Rare Diseases JTC2020-SCN1A-up! (to N.N.K.), the EpilepsieNL WAR 18-02 (to N.N.K., J.V., M.M. and H.J.S), Simons Foundation (SFARI) Grant 890042 (to N.N.K.).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Dravet C. The core Dravet syndrome phenotype. Epilepsia. 2011;52:3–9. [DOI] [PubMed] [Google Scholar]
- 2. Deprez L, Jansen A, De Jonghe P. Genetics of epilepsy syndromes starting in the first year of life. Neurology. 2009;72:273–281. [DOI] [PubMed] [Google Scholar]
- 3. Wolff M, Cassé-Perrot C, Dravet C. Severe myoclonic epilepsy of infants (Dravet syndrome): natural history and neuropsychological findings. Epilepsia. 2006;47:45–48. [DOI] [PubMed] [Google Scholar]
- 4. Jansen FE, Sadleir LG, Harkin LA, et al. Severe myoclonic epilepsy of infancy (Dravet syndrome): recognition and diagnosis in adults. Neurology. 2006;67:2224–2226. [DOI] [PubMed] [Google Scholar]
- 5. Escayg A, Goldin AL. Sodium channel SCN1A and epilepsy: mutations and mechanisms. Epilepsia. 2010;51:1650–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SNC1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet 2001;68:1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Menezes LFS, Sabiá Júnior EF, Tibery DV, Carneiro LdA, Schwartz EF. Epilepsy-related voltage-gated sodium channelopathies: a review. Front Pharmacol. 2020;11:1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brunklaus A, Feng T, Brünger T, et al. Gene variant effects across sodium channelopathies predict function and guide precision therapy. Brain. 2022;145:4275–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Claes LRF, Deprez L, Suls A, et al. The SCN1A variant database: a novel research and diagnostic tool. Hum Mutat. 2009;30:E904–E920. [DOI] [PubMed] [Google Scholar]
- 10. Harkin LA, McMahon JM, Iona X, et al. The spectrum of SCN1A-related infantile epileptic encephalopathies. Brain. 2007;130:843–852. [DOI] [PubMed] [Google Scholar]
- 11. Zuberi SM, Birch R, Reavey E, Duncan J, Forbes GH. Genotype–phenotype associations in SCN1A -related epilepsies. Neurology 2011;76:594–600. [DOI] [PubMed] [Google Scholar]
- 12. Goldberg-Stern H, Aharoni S, Afawi Z, et al. Broad phenotypic heterogeneity due to a novel SCN1A mutation in a family with genetic epilepsy with febrile seizures plus. J Child Neurol. 2014;29:221–226. [DOI] [PubMed] [Google Scholar]
- 13. Yu FH, Mantegazza M, Westenbroek RE, et al. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9:1142–1149. [DOI] [PubMed] [Google Scholar]
- 14. Cheah CS, Yu FH, Westenbroek RE. et al. et al. Specific deletion of NaV1.1 sodium channels in inhibitory interneurons causes seizures and premature death in a mouse model of Dravet syndrome. Proc Natl Acad Sci U S A. 2012;109:14646–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liautard C, Scalmani P, Carriero G, de Curtis M, Franceschetti S, Mantegazza M. Hippocampal hyperexcitability and specific epileptiform activity in a mouse model of Dravet syndrome. Epilepsia. 2013;54:1251–1261. [DOI] [PubMed] [Google Scholar]
- 16. Ogiwara I, Miyamoto H, Morita N, et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tai C, Abe Y, Westenbroek RE, Scheuer T, Catterall WA. Impaired excitability of somatostatin- and parvalbumin-expressing cortical interneurons in a mouse model of Dravet syndrome. Proc Natl Acad Sci U S A. 2014;111:3139–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martin MS, Dutt K, Papale LA, et al. Altered function of the SCN1A voltage-gated sodium channel leads to γ-aminobutyric acid-ergic (GABAergic) interneuron abnormalities. J Biol Chem. 2010;285:9823–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chopra R, Isom LL. Untangling the Dravet syndrome seizure network: the changing face of a rare genetic epilepsy. Epilepsy Curr. 2014;14:86–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mistry AM, Thompson CH, Miller AR, Vanoye CG, George AL Jr, Kearney JA. Strain- and age-dependent hippocampal neuron sodium currents correlate with epilepsy severity in Dravet syndrome mice. Neurobiol Dis. 2014;65:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun Y, Paşca SP, Portmann T, et al. A deleterious Na v 1.1 mutation selectively impairs telencephalic inhibitory neurons derived from Dravet syndrome patients. Elife 2016;5:e13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J, Gao C, Chen W, et al. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry. 2016;6:e703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim HW, Quan Z, Kim YB, et al. Differential effects on sodium current impairments by distinct SCN1A mutations in GABAergic neurons derived from Dravet syndrome patients. Brain Dev. 2018;40:287–298. [DOI] [PubMed] [Google Scholar]
- 24. Higurashi N, Uchida T, Lossin C, et al. A human Dravet syndrome model from patient induced pluripotent stem cells. Mol Brain. 2013;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiao J, Yang Y, Shi Y, et al. Modeling Dravet syndrome using induced pluripotent stem cells (iPSCs) and directly converted neurons. Hum Mol Genet. 2013;22:4241–4252. [DOI] [PubMed] [Google Scholar]
- 26. Liu Y, Lopez-Santiago LF, Yuan Y, et al. Dravet syndrome patient-derived neurons suggest a novel epilepsy mechanism. Ann Neurol. 2013;74:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie Y, Ng NN, Safrina OS, et al. Comparisons of dual isogenic human iPSC pairs identify functional alterations directly caused by an epilepsy associated. Neurobiol Dis. 2019;134:104627. [DOI] [PubMed] [Google Scholar]
- 28. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 29. Mandegar MA, Huebsch N, Frolov EB, et al. CRISPR Interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016;18:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frega M, Linda K, Keller JM, et al. Neuronal network dysfunction in a model for kleefstra syndrome mediated by enhanced NMDAR signaling. Nat Commun. 2019;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y, Pak C, Han Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frega M, van Gestel SHC, Linda K, et al. Rapid neuronal differentiation of induced pluripotent stem cells for measuring network activity on micro-electrode arrays. J Vis Exp. 2017;119:54900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mossink B, Verboven AHA, van Hugte EJH, et al. Human neuronal networks on micro-electrode arrays are a highly robust tool to study disease-specific genotype-phenotype correlations in vitro. Stem Cell Reports. 2021;16:2182–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mendis GDC, Morrisroe E, Petrou S, Halgamuge SK. Use of adaptive network burst detection methods for multielectrode array data and the generation of artificial spike patterns for method evaluation. J Neural Eng. 2016;13:026009. [DOI] [PubMed] [Google Scholar]
- 38. Tukker AM, Wijnolts FMJ, de Groot A, Westerink RHS. Applicability of hiPSC-derived neuronal cocultures and rodent primary cortical cultures for in vitro seizure liability assessment. Toxicol Sci. 2020;178:71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bradley JA, Luithardt HH, Metea MR, Strock CJ. In vitro screening for seizure liability using microelectrode array technology. Toxicol Sci. 2018;163:240–253. [DOI] [PubMed] [Google Scholar]
- 40. Raghuram V, Werginz P, Fried SI. Scaling of the AIS and somatodendritic compartments in α S RGCs. Front Cell Neurosci. 2019;13:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Lange IM, Mulder F, Slot R, et al. Modifier genes in SCN1A -related epilepsy syndromes. Mol Genet Genomic Med. 2020;8:e1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Volkers L, Kahlig KM, Verbeek NE, et al. Na v1.1 dysfunction in genetic epilepsy with febrile seizures-plus or Dravet syndrome. Eur J Neurosci. 2011;34:1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Volkers L, Kahlig KM, Das JHG, van Kempen MJA, Lindhout D, Koeleman BPC, Rook MB. Febrile temperatures unmask biophysical defects in Nav1.1 epilepsy mutations supportive of seizure initiation. J Gen Physiol. 2013;142:641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rhodes TH, Lossin C, Vanoye CG, Wang DW, George AL. Noninactivating voltage-gated sodium channels in severe myoclonic epilepsy of infancy. Proc Natl Acad Sci U S A. 2004;101:11147–11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sugawara T, Tsurubuchi Y, Fujiwara T, Inoue Y, Yamakawa K. Nav1.1 channels with mutations of severe myoclonic epilepsy in infancy display attenuated currents. Epilepsy Res. 2003;54:201–207. [DOI] [PubMed] [Google Scholar]
- 46. Clatot J, Hoshi M, Wan X, et al. Voltage-gated sodium channels assemble and gate as dimers. Nat Commun. 2017;8:2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jorge BS, Campbell CM, Miller AR, et al. Voltage-gated potassium channel KCNV2 (Kv8.2) contributes to epilepsy susceptibility. Proc Natl Acad Sci U S A. 2011;108:5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spratt PWE, Alexander RPD, Ben-Shalom R, et al. Paradoxical hyperexcitability from NaV1.2 sodium channel loss in neocortical pyramidal cells. Cell Rep. 2021;36:109483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reid CA, Leaw B, Richards KL, et al. Reduced dendritic arborization and hyperexcitability of pyramidal neurons in a Scn1b-based model of Dravet syndrome. Brain. 2014;137:1701–1715. [DOI] [PubMed] [Google Scholar]
- 50. Boeckeler K, Adley K, Xu X, Jenkins A, Jin T, Williams RSB. The neuroprotective agent, valproic acid, regulates the mitogen-activated protein kinase pathway through modulation of protein kinase A signalling in dictyostelium discoideum. Eur J Cell Biol. 2006;85:1047–1057. [DOI] [PubMed] [Google Scholar]
- 51. Miljanovic N, Hauck SM, van Dijk RM, Di Liberto V, Rezaei A, Potschka H. Proteomic signature of the Dravet syndrome in the genetic Scn1a-A1783V mouse model. Neurobiol Dis. 2021;157:105423. [DOI] [PubMed] [Google Scholar]
- 52. Mossink B, van Rhijn J-R, Wang S, et al. Cadherin-13 is a critical regulator of GABAergic modulation in human stem-cell-derived neuronal networks. Mol Psychiatry. 2021;27:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heighway J, Sedo A, Garg A, et al. Sodium channel expression and transcript variation in the developing brain of human, rhesus monkey, and mouse. Neurobiol Dis. 2022;164:105622. [DOI] [PubMed] [Google Scholar]
- 54. Du J, Simmons S, Brunklaus A, et al. Differential excitatory vs inhibitory SCN expression at single cell level regulates brain sodium channel function in neurodevelopmental disorders. Eur J Paediatr Neurol. 2020;24:129–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and algorithms generated in this study can be requested from the corresponding author. Aside from the hiPSC lines, this study did not generate unique new reagents. All reagents, vectors or cell lines used in this study are available from the corresponding author upon request with a completed Materials Transfer Agreement.