Abstract
Congenital insensitivity to pain (CIP) and hereditary sensory and autonomic neuropathies (HSAN) are clinically and genetically heterogeneous disorders exclusively or predominantly affecting the sensory and autonomic neurons. Due to the rarity of the diseases and findings based mainly on single case reports or small case series, knowledge about these disorders is limited.
Here, we describe the molecular workup of a large international cohort of CIP/HSAN patients including patients from normally under-represented countries. We identify 80 previously unreported pathogenic or likely pathogenic variants in a total of 73 families in the >20 known CIP/HSAN-associated genes. The data expand the spectrum of disease-relevant alterations in CIP/HSAN, including novel variants in previously rarely recognized entities such as ATL3-, FLVCR1- and NGF-associated neuropathies and previously under-recognized mutation types such as larger deletions. In silico predictions, heterologous expression studies, segregation analyses and metabolic tests helped to overcome limitations of current variant classification schemes that often fail to categorize a variant as disease-related or benign.
The study sheds light on the genetic causes and disease-relevant changes within individual genes in CIP/HSAN. This is becoming increasingly important with emerging clinical trials investigating subtype or gene-specific treatment strategies.
Keywords: neuropathies, CIP, HSAN, HSN, pain, genetics
Lischka et al. describe the molecular workup of a large cohort of patients with congenital insensitivity to pain/hereditary sensory and autonomic neuropathies. They identify 80 previously unreported pathogenic or likely pathogenic variants in 73 families, broadening the known mutational spectrum of these rare conditions.
Introduction
Complex genetic variability leads to individual differences in the perception of pain. In contrast to polygenic and environmental correlations, specific single nucleotide variants can have an effect such that the sensation of pain is absent from birth, or a progressive loss of pain sensitivity becomes apparent in the course of life. In these rare and monogenic diseases, there is usually a developmental disorder of pain-sensing neurons, neurodegeneration of peripheral nerves, or altered electrical activity of nociceptors. This heterogeneous group of genetic pain loss disorders includes congenital insensitivity to pain (CIP), hereditary sensory neuropathy (HSN) and, if autonomic nerves are involved, hereditary sensory and autonomic neuropathy (HSAN). The HSNs are assigned here to the group of HSAN diseases. The consequences of pain loss are recurrent injuries and fractures resulting in mutilation or amputation, often in combination with severely impaired wound healing. The sensation of itch, temperature and touch may also be impaired with negative impact on health. Affected patients can have marked autonomic dysfunction, such as anhidrosis, gastrointestinal and sexual dysfunction or blood pressure fluctuations. In some subtypes of CIP/HSAN, patients also show intellectual disability, muscle weakness, ataxia or other additional symptoms. To date, pathogenic variants in more than 20 genes are known to cause pain loss syndromes. Various cellular processes can be affected, including sodium channel activity,1,2 sphingolipid metabolism,3,4 membrane dynamics,5–7 axonal transport,8,9 neurotrophin signalling,10,11 epigenetic regulation12 or cytoskeletal architecture.13 Because of the rarity of CIP/HSAN disorders, knowledge of these conditions is limited and diagnosis is often delayed or incorrect resulting in a diagnostic odyssey. Moreover, except for few studies including larger patient numbers,14,15 the literature is often restricted to single case descriptions. Detailed studies on the genetic spectrum of the respective molecular subtypes have been largely lacking to date, but as the first therapeutic approaches for certain subtypes are in clinical trials, molecular classification is becoming increasingly important.16–18
In this retrospective study, we provide deeper insights into the clinical and genetic landscape of these rare conditions by sequencing of the as yet largest cohort of CIP/HSAN patients.
Materials and methods
Patient cohort
Genetic data were collected retrospectively from existing datasets from patients who had been either referred directly to the participating centres or whose blood samples and clinical information were sent to the participating centres. All patients showed clinical signs of HSAN or CIP (i.e. reduced sensation of pain, temperature and touch either congenital or developed later in life and/or clinical manifestations such as unnoticed injuries, skin ulcerations, amputations, osteomyelitis, painless fractures). Since neuropathic pain, especially in the initial stage, may be a sign of different forms of HSAN, patients with a combined phenotype and a variant in one of the HSAN-related genes were also included in the study. Patients with suspected other genetic disorders potentially mimicking the phenotype of HSAN, such as Charcot-Marie-Tooth disease (CMT) or e.g. Lesch-Nyhan syndrome as a typical example with self-mutilating behaviour, were not included in the study. Other possible underlying non-genetic causes of decreased pain sensitivity (e.g. toxic, metabolic or infectious causes of polyneuropathy) were queried by the respective clinical centres and resulted in exclusion from the study. The study was conducted in accordance with the Declaration of Helsinki and has been approved by the local ethics committees of the participating institutions. Prior to inclusion, written informed consent was obtained from patients or their legal guardians [Ethics approval Uniklinik RWTH Aachen: EK 086-20; Ethics approval London: 09/H0716/61 (‘CMT—A natural history study’); Ethics approvals University of Oxford: 12/LO/0017 (Painful Channelopathies Study, https://clinicaltrials.gov/ct2/show/NCT02696746), 18/SC/0263 (Pain in Peripheral Nerve Lesions), 13/EE/0325 (NIHR BioResource—Rare Diseases; Ethics approval of Antwerp and University Hospital of Antwerp: B300201422160 (V.T.) and B300201525715 (J.B.); Ethics approval University Hospital of Tübingen: 116/2015BO2].
Short-read next-generation sequencing
Project sites and collaborating research institutions provided genetic data of CIP/HSAN patients that had been analysed by short-read sequencing. Datasets were analysed with regard to novel variants in known CIP/HSAN genes [genes from the panel ‘pain syndromes’ v1.12, Genomics England Panel App were prioritized (https://panelapp.genomicsengland.co.uk/panels/288/)]. Variant calling was done by each research institute separately; protocols, consumables and pipelines used differed between the institutions. Detailed protocols can be provided upon request. Cases were considered if mono- or biallelic variants (for dominant and recessive disorders, respectively) were found in one of the core genes with no additional probably disease-associated variants detected by screening of the datasets. The MasterMind database was checked in June 2023 and variants were included if they (i) had not been published in the literature at all; (ii) had only been described in supplementary materials; or (iii) had only been reported in patients with a phenotype other than CIP/HSAN (Supplementary Table 1). If possible, segregation analyses within the families were performed using Sanger sequencing.
Modified classification of pathogenicity
In accordance with the American College of Medical Genetics (ACMG) criteria,19 in a first step, only pathogenic or likely pathogenic variants were selected consistent with a very high probability of molecular diagnostic confirmation (Table 1). Subsequently, variants with formally unclear clinical significance (VUS) in the core genes were reassessed (Table 2). For this purpose, five additional objective criteria were established to cover variant features that support the pathogenicity of a variant but are not represented in the actual ACMG guidelines so far. Two novel criteria were classified as moderate and three as supporting (Supplementary Table 2). Additionally, we established the term of VUS+, defined as variants that do not meet the original criteria for likely pathogenic or pathogenic variants but fulfil at least one of the new criteria supporting their pathogenicity.
Table 1.
Novel (likely) pathogenic variants in CIP/HSAN genes
| Patient | Novel variant | Genotype | ACMG | Inh. | PP | Au | Mo | SL | F/M | ID |
|---|---|---|---|---|---|---|---|---|---|---|
| DST (NM_001374736) | ||||||||||
| 4 | c.4849C>T, p.(Arg1617*) | het + c.19942G>A (het) | LPV | AR | red | − | + | + | − | |
| 5 | c.22513C>T, p.(Arg7505*) | comp + c.19451A>T | LPV | AR | red | + | + | + | + | − |
| FLVCR1 (NM_014053) | ||||||||||
| 6 | c.139_151del, p.(Phe47Glyfs*62) | het + c.722C>T (het) | LPV | AR | red | − | + | − | + | + |
| 7 | c.868_871del, p.(Ile290*) | comp + c.655G>A | LPV | AR | abs | + | − | − | − | |
| 10 | c.1318_1321del, p.(Thr440Valfs*63) | comp + c.1317G>A | LPV | AR | red | + | − | |||
| 11 | c.1194C>A, p.(Tyr398*) | comp + c.1526-3C>T | LPV | AR | abs | − | + | + | − | |
| KIF1A (NM_001244008) | ||||||||||
| 12 | c.2839dup, p.(Leu947Profs*49) | hom° | LPV | AR | red | + | − | − | − | |
| NGF (NM_002506) | ||||||||||
| 13 | c.524_525del, p.(Phe175*) | hom° | LPV | AR | red | + | + | + | ||
| 14 | c.695_696del, p.(Val232Alafs*39) | hom° | LPV | AR | + | + | + | + | ||
| NTRK1 (NM_002529) | ||||||||||
| 15 | c.2T>A, p.(Met1?) | hom | LPV | AR | * | |||||
| 16 | c.145C>T, p.(Arg49*) | hom° | LPV | AR | abs | + | − | + | + | |
| 17 | c.213-1G>A, p.? | hom° | LPV | AR | abs | + | ||||
| 18 | c.228_229delGCinsTT, p.(Gln76_Gln77delinsHis*) | hom | LPV | AR | * | |||||
| 19 | c.287+2T>A, p.? | comp + known LPV | LPV | AR | * | |||||
| 22, 23 | c.605del, p.(Asn202Metfs*37) | het + known PV (het) | LPV | AR | red | + | − | + | + | + |
| 24 | c.717+1del, p.? | hom | LPV | AR | * | |||||
| 27 | c.850del, p.(Phe284Serfs*186) | hom | LPV | AR | * | |||||
| 28 | c.851-2A>G, p.? | hom° | LPV | AR | * | |||||
| 30 | c.1320del, p.(Asn440Lysfs*30) | hom | LPV | AR | abs | + | ||||
| 32, 33 | c.1865del, p.(Leu622Argfs*36) | hom° | LPV | AR | red | + | − | + | + | − |
| 34 | c.1953_1954insT, p.(Ala652Cysfs*17) | hom | LPV | AR | * | |||||
| PRDM12 (NM_021619) | ||||||||||
| 37 | c.575T>A, p.(Ile192Asn) | hom° | LPV | AR | abs | − | − | + | + | |
| 38 | c.788G>A, p.(Arg263His) | comp + known LPV | LPV | AR | abs | + | − | |||
| SCN9A (NM_002977) | ||||||||||
| 41 | c.116del, p.(Lys39Argfs*51) | hom | LPV | AR | * | |||||
| 42 | c.515T>G, p.(Leu172Arg) | hom° | LPV | AR | abs | + | − | + | − | |
| 43 | c.793C>T, p.(Gln265*) | hom | LPV | AR | * | |||||
| 44 | c.809_822del, p.(Asn270Metfs*7) | comp + c.1927C>T | LPV | AR | * | |||||
| 45 | c.954_955del, p.(Thr319Argfs*19) | hom | LPV | AR | * | |||||
| 46 | c.1368del, p.(Gly457Alafs*12) | hom | LPV | AR | * | |||||
| 47 | c.1449del, p.(Asn484Ilefs*81) | comp + known PV | PV | AR | * | |||||
| 48 | c.1602+2del, p.? | comp + known LPV | LPV | AR | abs | |||||
| 44 | c.1927C>T, p.(Gln643*) | comp + c.809_822del | LPV | AR | * | |||||
| 50 | c.2109G>A, p.(Trp703*) | comp + known PV | PV | AR | * | |||||
| 51, 52 | c.2362dup, p.(Asp788Glyfs*4) | hom° | LPV | AR | red | − | − | + | + | − |
| 54 | c.3309del, p.(Tyr1103*) | comp + c.5340del | LPV | AR | abs | − | − | + | − | |
| 55 | c.4331del, p.(Val1444Alafs*3) | hom | LPV | AR | red | − | − | + | − | − |
| 56 | c.4467del, p.(Asn1491Thrfs*10) | hom° | LPV | AR | * | |||||
| 57 | c.4470+1G>T, p.? | comp + known PV | PV | AR | * | |||||
| 59 | c.5118del, p.(Val1709Phefs*33) | hom | LPV | AR | * | |||||
| 54 | c.5340del, p.(Asp1781Metfs*6) | comp + c.3309del | LPV | AR | abs | − | − | + | − | |
| SPTLC1 (NM_006415) | ||||||||||
| 63 | c.397T>C, p.(Cys133Arg) | het | LPV | AD | * | |||||
| WNK1 (NM_001184985) | ||||||||||
| 73 | c.2159del, p.(Pro720Argfs*35) | hom° | LPV | AR | red | − | + | + | − | |
| 74, 75 | c.2392_2416del, p.(Ala798Profs*4) | hom | LPV | AR | red | − | − | + | + | − |
| 76 | c.2919_2920dup, p.(Pro974Hisfs*27) | hom° | LPV | AR | red | − | + | + | + | − |
| 77 | c.3071_3072del, p.(Asn1024Ilefs*28) | hom | LPV | AR | red | + | + | − | ||
| 78 | c.3909_3928del, p.(Gln1304Serfs*31) | comp + known PV | PV | AR | red | + | ||||
abs = absent (i.e. complete pain loss), ACMG = American College of Medical Genetics; AD = autosomal dominant; AR = autosomal recessive; Au = autonomic dysfunction; comp = compound heterozygosity, confirmed by segregation analyses; F/M = fractures and/or mutilations; het = heterozygous; hom = homozygosity, confirmed by segregation analyses; hom° = homozygosity, but no parental samples available for segregation analyses; Inh. = inheritance; ID = intellectual disability; LPV = likely pathogenic variant; Mo = motor dysfunction; red = reduced; PP = pain perception; PV = pathogenic variant; SL = skin lesions (including ulcerations).
*Suspected clinical diagnosis of HSAN, no further clinical information available. For clinical data, ‘+’ indicates the presence and ‘–’ the absence of symptoms in the respective category.
Table 2.
Novel variants in CIP/HSAN genes classified as likely pathogenic or VUS+ after reclassification
| Patient | Novel variant | Genotype | ACMG | Reclass. | Inh. | PP | Au | Mo | SL | F/M | ID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ATL3 (NM_015459) | |||||||||||
| 1 | c.544G>A, p.(Asp182Asn) | het | VUS | VUS+ | AD | NP | + | + | + | − | |
| 2 | c.1027A>G, p.(Met343Val) | het | VUS | VUS+ | AD | red | − | ||||
| 3 | c.1053C>A, p.(Asn351Lys) | het | VUS | VUS+ | AD | red | + | + | + | − | |
| DST (NM_001374736) | |||||||||||
| 5 | c.19451A>T, p.(Gln6484Leu) | comp + c.22513C>T | VUS | LPV | AR | red | + | + | + | + | − |
| 4 | c.19942G>A, p.(Val6648Ile) | het + c.4849C>T (het) | VUS | VUS+ | AR | red | − | + | + | − | |
| FLVCR1 (NM_014053) | |||||||||||
| 7 | c.655G>A, p.(Gly219Ser) | comp + c.868_871del | VUS | LPV | AR | abs | + | − | − | − | |
| 6 | c.722C>T, p.(Ala241Val) | het + c.139_151del (het) | VUS | VUS+ | AR | red | − | + | − | + | + |
| 8 | c.758T>A, p.(Phe253Tyr) | het + c.1369G>A (het) | VUS | LPV | AR | abs | + | + | |||
| 9 | c.1034C>G, p.(Thr345Ser) | hom° | VUS | VUS+ | AR | red | + | + | + | ||
| 10 | c.1317G>A, p.(Met439Ile) | comp + c.1318_1321del | VUS | LPV | AR | red | + | − | |||
| 8 | c.1369G>A, p.(Glu457Lys) | het + c.758T>A (het) | VUS | VUS+ | AR | abs | + | + | |||
| 11 | c.1526-3C>T, p.? | comp + c.1194C>A | VUS | VUS+ | AR | abs | − | + | + | − | |
| NTRK1 (NM_002529) | |||||||||||
| 20, 21 | c.287+5G>A, p.? | hom | VUS | VUS+ | AR | abs | + | + | + | ||
| 25, 26 | c.717+4A>T, p.? | comp + known PV/LPV | VUS | LPV | AR | * | |||||
| 29 | c.1136T>A, p.(Met379Lys) | hom | VUS | LPV | AR | red | + | + | + | + | + |
| 31 | c.1514T>A, p.(Ile505Asn) | hom | VUS | VUS+ | AR | * | |||||
| PRDM12 (NM_021619) | |||||||||||
| 36 | c.131_139del, p.(Val44_Gly46del) | hom | VUS | LPV | AR | red | + | ||||
| RAB7A (NM_004637) | |||||||||||
| 39, 40 | c.467C>T, p.(Ala156Val) | het | VUS | VUS+ | AD | red | + | − | + | − | |
| SCN9A (NM_002977) | |||||||||||
| 49 | c.1650C>G, p.(Ser550Arg) | comp + c.1660C>A | VUS | VUS+ | AR | * | |||||
| 49 | c.1660C>A, p.(Leu554Ile) | comp + c.1650C>G | VUS | VUS+ | AR | * | |||||
| 53 | c.2689_2691del, p.(Trp897del) | hom° | VUS | LPV | AR | * | |||||
| 58 | c.5059G>C, p.(Ala1687Pro) | comp + known PV | VUS | LPV | AR | * | |||||
| SPTLC1 (NM_006415) | |||||||||||
| 64 | c.1037C>T, p.(Ala346Val) | het | VUS | VUS+ | AD | * | |||||
| SPTLC2 (NM_004863) | |||||||||||
| 65 | c.302A>G, p.(His101Arg) | het | VUS | VUS+ | AD | * | |||||
| 66 | c.359A>G, p.(Asn120Ser) | het | VUS | VUS+ | AD | * | |||||
| 67 | c.430G>A, p.(Ala144Thr) | het | VUS | VUS+ | AD | * | |||||
| 68 | c.707G>T, p.(Gly236Val) | het | VUS | VUS+ | AD | * | |||||
| 69, 70 | c.1276A>T, p.(Ile426Phe) | het | VUS | VUS+ | AD | NP | + | + | − | − | − |
| 71 | c.1304G>T, p.(Gly435Val) | het | VUS | VUS+ | AD | * | |||||
| 72 | c.1513G>A, p.(Glu505Lys) | het | VUS | VUS+ | AD | red | + | + | + | − | |
abs = absent (i.e. complete pain loss); AD = autosomal dominant; AR = autosomal recessive; Au = autonomic dysfunction; comp = compound heterozygosity, confirmed by segregation analyses; F/M = fractures and/or mutilations; het = heterozygous; hom = homozygosity, confirmed by segregation analyses; hom° = homozygosity, but no parental samples available for segregation analyses; Inh. = inheritance; ID = intellectual disability; LPV = likely pathogenic variant; Mo = motor dysfunction; NP = neuropathic pain; red = reduced; PP = pain perception; PV = pathogenic variant; Reclass. = reclassification; SL = skin lesions (including ulcerations); VUS = variant of uncertain significance.
*Suspected clinical diagnosis of HSAN, no further clinical information available. For clinical data, ‘+’ indicates the presence and ‘–’ the absence of symptoms in the respective category.
Additional methods
Detailed information about long-read next-generation sequencing (NGS), sphingolipid profiling and electrophysiology is provided in the Supplementary material.
Results
The cohort studied was composed of patients who presented at the participating centres or whose findings were referred from peripheral hospitals or treating physicians. The inclusion of patients from countries or regions with limited resources led to variable availability of clinical information (Supplementary Fig. 1). Inclusion criteria for the genetic test were the suspicion of CIP/HSAN due to a decreasing or absent pain sensation and written consent to participate in the study. Cases were excluded if a secondary cause of insensitivity to pain such as leprosy or abusive injury was confirmed or suspected. The predefined inclusion and exclusion criteria are detailed in the ‘Materials and methods’ section. A cohort of 78 patients from 73 families with the suspected diagnosis of CIP/HSAN had been analysed by NGS using gene panels, whole exome (WES) or whole genome sequencing (WGS). The core genes included in the analysis were the following 22 genes: ATL1, ATL3, DST, ELP1, GLA, KIF1A, NGF, NTRK1, PRDM12, RAB7A, RETREG1/FAM134B, SCN9A, SCN11A, SPTLC1, SPTLC2, TTR, WNK1, MPV17, NAGLU, CLTCL1, FAAHP1 and FLVCR1.
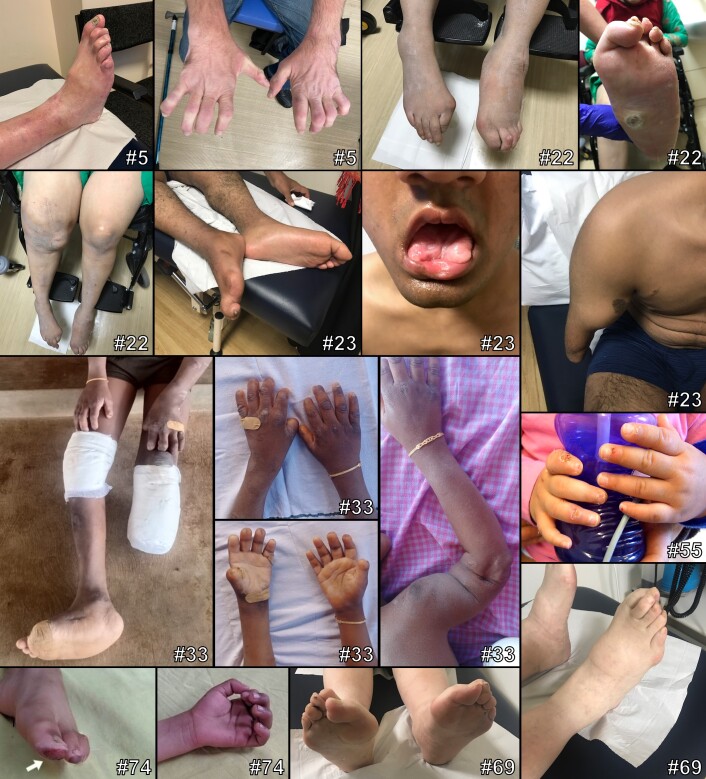
By applying the ACMG criteria and our additional criteria for pathogenicity, 80 novel CIP/HSAN-related variants were identified in 78 patients (Tables 1 and 2 and Supplementary Table 3). Complete clinical and genetic details are given in Supplementary Table 1 and clinical images for a subset of patients are shown in Fig. 1. In some patients with recessive conditions, the novel variant occurred in compound heterozygosity with a second, previously described pathogenic or likely pathogenic variant as indicated in Tables 1 and 2. The mutation spectrum across all genes included missense variants, non-frameshift variants, frameshift variants and stop-gains whereby for some genes, exclusively one mutation type was found (e.g. missense variants in SPTLC1 and SPTLC2) (Fig. 2). For most genes, no mutational hot spot was identified and the variants showed a distribution pattern across the entire coding regions, for some genes also including adjacent splice sites. An exception was the WNK1 gene, for which a known clustering of variants was confirmed in the HSN2 exon known to code for part of the neuron-specific isoform. One of these variants, however, affected the pan-isoform of WNK1 in compound heterozygosity with an HSN2-specific variant.
Figure 1.
Phenotypic findings in CIP/HSAN patients. The pictures show exemplary findings in patients with novel (likely) pathogenic variants in DST (Patient 5), NTRK1 (Patients 22, 23 and 33), SCN9A (Patient 55), SPTLC2 (Patient 69), and WNK1 (Patient 74). Additional clinical details are provided in Supplementary Table 1. Number sign indicates patient ID.
Figure 2.
Novel variants in known CIP/HSAN genes. The location within the respective genes is shown for all variants identified in this study. Green = missense/non-frameshift; yellow = splicing; red = truncating. For WNK1, the neuron-specific exon HSN2 is highlighted, in which the majority of the known pathogenic variants (to date) is located.
Novel CIP/HSAN-related variants were identified in 12 different CIP/HSAN genes, namely in ATL3 (n = 3), DST (n = 2), KIF1A (n = 1), NGF (n = 2), NTRK1 (n = 21), PRDM12 (n = 3), RAB7A (n = 2), SCN9A (n = 22), SPTLC1 (n = 2), SPTLC2 (n = 8), WNK1 (n = 6) and FLVCR1 (n = 6) (Supplementary Fig. 2), where ‘n’ corresponds to the number of patients per gene.
Genomic data revealed a larger intragenic deletion of 1.3 kb (Patient 35) in NTRK1 and two larger intragenic deletions of 3.8 kb (Patient 60) and 3.4 kb (Patient 61) in SCN9A, respectively. Patient 62 showed a 322 kb deletion spanning the entire SCN9A locus. In these patients, the deletion was in a compound heterozygous situation with a single nucleotide variant on the other allele. For the 3.4 kb deletion (Patient 61) spanning exon 20 of SCN9A, the DNA quality was sufficient to determine the exact size of the maternally inherited deletion by long-read sequencing (Oxford Nanopore Technologies, ONT) (Supplementary Fig. 3A). The deletion had a size of 3421 bp [chr2:166,235,764-166,239,185 (hg38)] and was further confirmed by quantitative PCR (qPCR) (Supplementary Fig. 3B). The father is a carrier of the single base pair deletion c.5318del, confirming compound heterozygosity in the index patient. For eight patients carrying SPTLC1 and SPTLC2 variants, plasma samples were available and 1-deoxy-sphingolipids (1-deoxySL) levels were elevated in line with assumed pathogenicity of the variant (Patients 63–68 and Patients 71–72) (Supplementary Fig. 4). For the homozygous missense variant p.(Leu172Arg) in Nav1.7 (SCN9A) in transmembrane segment 2 of channel domain I (D1), in silico predictions regarding a role in CIP were inconsistent, so functional studies were performed (Patient 42). The respective variant was electrophysiologically analysed upon heterologous expression in HEK293 cells and showed a complete loss-of-function in line with pathogenicity (Supplementary Fig. 5).
Discussion
This retrospective cross-sectional study was aimed at molecular characterization of patients with CIP/HSAN and to our knowledge, includes the largest number of molecularly resolved cases to date. The study has its main limitation in that in some cases the inclusion criteria were met, but detailed clinical data were not available. This was because data were collected from multiple sites, some with only limited clinical research and documentation capabilities. This is a frequently observed difficulty in ultrarare diseases, as access to patients is already a major hurdle. In addition, the total number of cases investigated at each centre over the years is variable, making it difficult to accurately determine detection rates for CIP/HSAN. The strength and focus of this study were therefore on molecular characterization and careful assessment of the pathogenicity of variants in CIP/HSAN-associated genes. Thus, a large cohort of patients with these extremely rare diseases could be studied in a collaborative network and patients from normally under-represented countries could also be included.
Overall, the study supports that SCN9A and NTRK1 are the most frequently mutated genes in congenital painlessness. Whereas CIP/HSAN-related pathogenic SCN9A variants frequently lead to anosmia as a secondary symptom, NTRK1-related neuropathy is accompanied by lack of sweat gland innervation with anhidrosis and sometimes life-threatening hyperthermia. Intellectual disability was not observed in SCN9A-associated neuropathy and was present with a variable degree in NTRK1-related disease. In adult onset HSN/HSAN, pathogenic variants are most frequently found in the enzymes of the sphingolipid metabolism pathway (SPTLC1/2). The study has a bias at this point: previously, more pathogenic variants were described for SPTLC1 than for SPTLC2. We report more cases with SPTLC2 variants in this study, but this is due to the fact that we solely report new disease-relevant variants. In the UK, for example, there is a high frequency of SPTLC1-related patients due to the p.(Cys133Trp) founder mutation.
In addition, to address these more frequently mutated genes, we were able to substantiate the role of genes that have so far only very rarely been described as the cause of CIP/HSAN. To date, only three causal variants have been reported in NGF, two of which are missense variants.10,20,21 The study expands the mutation spectrum to include two homozygous loss-of-function variants and further corroborates a clinical presentation broadly equivalent to that of pathogenic variants in the NGF receptor encoding gene, NTRK1.
For ATL3, only two causal missense variants have been described to date.6,22 As for the previous changes, the here identified heterozygous variants are missense changes located at very highly conserved residues of the protein. The data further support that a dominant-negative effect of missense variants is likely the central mechanism of ATL3-associated disease. A recent report of an early stop-gain variant in ATL3, p.(Arg6Ter)23 would argue against this assumption, but proof of pathogenicity of this variant is pending. FLVCR1 variants have predominantly been reported in cases of autosomal-recessive posterior column ataxia with retinitis pigmentosa (PCARP).24 Since the first description of FLVCR1 variants as cause of HSAN, only a few pathogenic variants have been reported, including missense and loss-of-function variants.25–27 The findings in this study based on six additional patients show that the clinical transitions between PCARP and HSAN are fluid and often result in a complex phenotype with overlapping symptoms.
In several patients, the underlying genetic variants are immediately classifiable as likely pathogenic or pathogenic according to ACMG criteria. A greater difficulty arises with missense variants that often have to be classified as VUS. Here, further parameters such as homozygosity/compound heterozygosity with a likely pathogenic or very rare variant for recessive disorders, occurrence in more than one affected individual, a highly specific phenotype, detailed evaluation of the functionally critical domains and amino acids of the protein, or the lack of evidence of another genetic cause by broad genetic screening using WES or WGS were used to prove the pathogenicity of the suspicious variants. For all variants listed in this study, those that were formally classified as VUS according to ACMG criteria were considered as VUS+ (i.e. assumed to be likely pathogenic) or likely pathogenic based on such additional criteria. This critical review of variants was performed to exclude false positives as much as possible and to provide reliable genetic counselling to patients and their families.
Further functional assessment of VUS in SPTLC1 or SPTLC2 was achieved by detecting toxic sphingolipid species in the serum of patients by mass spectrometry. Both genes encode key enzymes of the de novo sphingolipid synthesis pathway, the so-called serine palmitoyltransferases (SPT). Pathogenic gain-of-function variants in SPTLC1 and SPTLC2 lead to the increased formation of toxic 1-deoxySL, which have been measured in patient’s plasma to further assess suspicious VUS. Heterologous expression studies with functional measures were additionally performed in selected cases to corroborate pathogenicity of a VUS. As an additional example, we show whole-cell voltage-clamp recordings of HEK293 cells transiently expressing the SCN9A missense variant p.(Leu172Arg), confirming a complete loss-of-function of this variant.
The analysis of larger deletions has long been a difficulty in NGS-based diagnostics, but with increasingly better bioinformatic algorithms and new sequencing methods, this type of genetic alteration can more frequently be detected in CIP/HSAN. In rare cases, large deletions in NTRK1 have already been reported in HSAN.28 Our results show another case of an NTRK1 deletion and, in addition, three patients with larger deletions in SCN9A confirming their expected relevance in CIP/HSAN. Determination of copy number variants (CNV) from genomic data is therefore generally recommended in the case of an underlying loss-of-function mechanism. Long-read sequencing technologies, such as nanopore sequencing, have proven useful for rapidly determining the size and position of deletions with base pair precision,29 as we also exemplify in a case of a 3.4 kb SCN9A deletion.
Another feature in CIP/HSAN concerns isoform-specific pathogenic variants. HSAN-relevant recessive variants in WNK1 cluster in a neuron-specific alternatively spliced exon (HSN2 exon) of the gene, whereas biallelic pan-WNK1 loss is most likely lethal. We report here one of the rare cases of a compound heterozygosity for a mutation in the neuron-specific exon in trans with a loss-of-function mutation affecting the pan-isoform of WNK1, similarly to a previous report.30
In conclusion, our results broaden the mutational spectrum of CIP/HSAN and the cohort provides a framework for natural history studies and improvement of care in these rare debilitating conditions.
Supplementary Material
Acknowledgements
We would like to thank the patients and their relatives who have supported our research activity for years.
Contributor Information
Annette Lischka, Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Katja Eggermann, Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Christopher J Record, Department of Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Maike F Dohrn, Department of Neurology, Medical Faculty of the RWTH Aachen University, 52074 Aachen, Germany; Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Petra Laššuthová, Department of Paediatric Neurology, 2nd Faculty of Medicine, Charles University in Prague and Motol University Hospital, 150 06 Praha, Czechia.
Florian Kraft, Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Matthias Begemann, Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Daniela Dey, Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Thomas Eggermann, Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Danique Beijer, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Jana Šoukalová, Department of Medical Genetics, University Hospital Brno, 625 00 Brno, Czechia.
Matilde Laura, Department of Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Alexander M Rossor, Department of Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Radim Mazanec, Department of Neurology, Faculty of Medicine, Charles University in Prague and Motol University Hospital, 150 06 Prague, Czechia.
Jonas Van Lent, Peripheral Neuropathy Research Group, Department of Biomedical Sciences, Institute Born Bunge, University of Antwerp, 2160 Antwerp, Belgium.
Pedro J Tomaselli, Department of Neurosciences and Behaviour Sciences, Clinical Hospital of Ribeirão Preto, University of São Paulo, Ribeirão Preto, 14015-130, Brazil.
Martin Ungelenk, Institute of Human Genetics, University Hospital Jena, 07747 Jena, Germany.
Karlien Y Debus, Center for Molecular Biomedicine Institute for Biophysics, Friedrich-Schiller Universität Jena, 07745 Jena, Germany.
Shawna M E Feely, Department of Neurology, University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA; Division of Pediatric Neurology, Seattle Children’s Hospital, University of Washington School of Medicine, Seattle, WA 98105, USA.
Dieter Gläser, Center for Human Genetics, Genetikum®, 89231 Neu-Ulm, Germany.
Sujatha Jagadeesh, Department of Clinical Genetics and Genetic Counselling, Mediscan Systems, Chennai 600032, Tamilnadu, India.
Madelena Martin, Davis and Davis Children's Hospital, University of California, Sacramento, CA 95817, USA.
Geeta M Govindaraj, Department of Pediatrics, Government Medical College, Kozhikode, Kerala 673 008, India.
Pratibha Singhi, Pediatric Neurology and Neurodevelopment, Medanta, The Medicity, Gurgaon, Haryana 122 001, India.
Revanth Baineni, Department of Pediatrics, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry 605 006, India.
Niranjan Biswal, Department of Pediatrics, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry 605 006, India.
Marisol Ibarra-Ramírez, Genetics Department, Hospital Universitario Dr. José Eleuterio González Universidad Autónoma de Nuevo León, 64460 Monterrey, Nuevo León, México.
Maryse Bonduelle, Centre for Medical Genetics, Universitair Ziekenhuis Brussel, 1090 Jette, Brussels, Belgium.
Burkhard Gess, Department of Neurology, Medical Faculty of the RWTH Aachen University, 52074 Aachen, Germany; Department of Neurology, University Hospital, Evangelisches Klinikum Bethel, University of Bielefeld, 33617 Bielefeld, Germany.
Juan Romero Sánchez, Pediatría, Clínica Premium, 29601 Marbella, Spain.
Renu Suthar, Pediatric Neurology and Neurodevelopment Unit, Department of Pediatrics, Advanced Pediatric Centre, Post Graduate Institute of Medical Education and Research (PGIMER), Chandigarh 160 012, India.
Vrajesh Udani, Department of Child Neurology, PD Hinduja Hospital and Medical Research Centre, Mumbai, Maharashtra 400 016, India.
Atchayaram Nalini, Department of Neurology, National Institute of Mental Health and Neurosciences, Bengaluru 560 029, India.
Gopikrishnan Unnikrishnan, Department of Neurology, National Institute of Mental Health and Neurosciences, Bengaluru 560 029, India.
Wilson Marques, Junior, Department of Neurosciences and Behaviour Sciences, Clinical Hospital of Ribeirão Preto, University of São Paulo, Ribeirão Preto, 14015-130, Brazil.
Sandra Mercier, CHU Nantes, Service de Génétique Médicale, Centre de Référence des Maladies Neuromusculaires AOC, 44000 Nantes, France.
Vincent Procaccio, Department of Biochemistry and Genetics, MitoVasc Institute, UMR CNRS 6015- INSERM U1083, CHU Angers, 49055 Angers, France.
Céline Bris, Department of Biochemistry and Genetics, MitoVasc Institute, UMR CNRS 6015- INSERM U1083, CHU Angers, 49055 Angers, France.
Beena Suresh, Department of Clinical Genetics and Genetic Counselling, Mediscan Systems, Chennai 600032, Tamilnadu, India.
Vaishnavi Reddy, Department of Clinical Genetics and Genetic Counselling, Mediscan Systems, Chennai 600032, Tamilnadu, India.
Mariola Skorupinska, Department of Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Nathalie Bonello-Palot, INSERM, MMG, U 1251, Marseille, France, Aix Marseille Univ, 13385 Marseille, France.
Fanny Mochel, Genetics Department, Sorbonne Université, Paris Brain Institute, APHP, INSERM, CNRS, 75013 Paris, France.
Georg Dahl, Pediatric Neurology, Children’s Hospital of the King’s Daughters in Norfolk, Norfolk, VA 23507, USA.
Karthika Sasidharan, Department of Pediatrics, Government Medical College, Kozhikode, Kerala 673 008, India.
Fiji M Devassikutty, Department of Pediatrics, Government Medical College, Kozhikode, Kerala 673 008, India.
Sheela Nampoothiri, Department of Pediatric Genetics, Amrita Institute of Medical Sciences and Research Center, Cochin, Kerala 682 041, India.
Maria J Rodovalho Doriqui, Department of Genetics, Hospital Infantil Doutor Juvêncio Mattos, São Luis, Maranhão 65015-460, Brazil.
Wolfgang Müller-Felber, Department of Neuropediatrics, Developmental Neurology and Social Pediatrics, LMU Campus Innenstadt, University of Munich, 80337 Munich, Germany.
Katharina Vill, Department of Pediatric Neurology and Developmental Medicine, Dr. von Hauner Children's Hospital, University Hospital, LMU Munich, 80337 Munich, Germany; Institute of Human Genetics, School of Medicine, Technical University of Munich, 81675 Munich, Germany.
Tobias B Haack, Institute of Medical Genetics and Applied Genomics, University of Tübingen, 72076 Tübingen, Germany.
Andreas Dufke, Institute of Medical Genetics and Applied Genomics, University of Tübingen, 72076 Tübingen, Germany.
Michael Abele, Neurologie, Praxis für Neurologie und Schlafmedizin, 53359 Rheinbach, Germany.
Rolf Stucka, Friedrich Baur Institute at the Department of Neurology, LMU University Hospital, LMU Munich, 80336 Munich, Germany.
Saima Siddiqi, Genomics Group, Institute of Biomedical and Genetic Engineering (IBGE), Islamabad 44000, Pakistan.
Noor Ullah, Institute for Paramedical Sciences, Khyber Medical University, Peshawar, KPK 25100, Pakistan.
Stephanie Spranger, MVZ Humangenetik Bremen, Limbach Genetics, 28209 Bremen, Germany.
Deborah Chiabrando, Department of Molecular Biotechnology and Health Sciences, Molecular Biotechnology Center ‘Guido Tarone’, University of Torino, 10124 Turin, Italy.
Behiye S Bolgül, Department of Pedodontics, Faculty of Dentistry, Dicle University, 21200 Diyarbakir, Turkey.
Yesim Parman, Neuromuscular Unit, Department of Neurology, Istanbul Faculty of Medicine, Istanbul University, 34093 Istanbul, Turkey.
Pavel Seeman, Department of Paediatric Neurology, 2nd Faculty of Medicine, Charles University in Prague and Motol University Hospital, 150 06 Praha, Czechia.
Angelika Lampert, Institute of Neurophysiology, Medical Faculty, Uniklinik RWTH Aachen University, 52074 Aachen, Germany.
Jörg B Schulz, Department of Neurology, Medical Faculty of the RWTH Aachen University, 52074 Aachen, Germany; JARA-BRAIN Institute Molecular Neuroscience and Neuroimaging, Research Centre Jülich GmbH, and RWTH Aachen University, 52056 Aachen, Germany.
John N Wood, Molecular Nociception Group, Wolfson Institute for Biomedical Research, University College London, London WC1E 6BT, UK.
James J Cox, Molecular Nociception Group, Wolfson Institute for Biomedical Research, University College London, London WC1E 6BT, UK.
Michaela Auer-Grumbach, Department of Orthopedics and Trauma Surgery, Medical University of Vienna, 1090 Vienna, Austria.
Vincent Timmerman, Peripheral Neuropathy Research Group, Department of Biomedical Sciences, Institute Born Bunge, University of Antwerp, 2160 Antwerp, Belgium.
Jonathan de Winter, Translational Neurosciences and Institute Born Bunge, Faculty of Medicine and Health Sciences, University of Antwerp, 2610 Antwerp, Belgium; Neuromuscular Reference Centre, Department of Neurology, Antwerp University Hospital, 2610 Antwerp, Belgium.
Andreas C Themistocleous, Nuffield Department of Clinical Neuroscience, University of Oxford, Oxford OX3 9DU, UK.
Michael Shy, Department of Neurology, University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA.
David L Bennett, Nuffield Department of Clinical Neuroscience, University of Oxford, Oxford OX3 9DU, UK.
Jonathan Baets, Translational Neurosciences and Institute Born Bunge, Faculty of Medicine and Health Sciences, University of Antwerp, 2610 Antwerp, Belgium; Neuromuscular Reference Centre, Department of Neurology, Antwerp University Hospital, 2610 Antwerp, Belgium.
Christian A Hübner, Institute of Human Genetics, University Hospital Jena, 07747 Jena, Germany.
Enrico Leipold, Department of Anesthesiology and Intensive Care and CBBM—Center of Brain, Behavior and Metabolism, University of Luebeck, 23562 Luebeck, Germany.
Stephan Züchner, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Miriam Elbracht, Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Arman Çakar, Neuromuscular Unit, Department of Neurology, Istanbul Faculty of Medicine, Istanbul University, 34093 Istanbul, Turkey.
Jan Senderek, Friedrich Baur Institute at the Department of Neurology, LMU University Hospital, LMU Munich, 80336 Munich, Germany.
Thorsten Hornemann, Department of Clinical Chemistry, University Hospital Zurich, University of Zurich, 8006 Zurich, Switzerland.
C Geoffrey Woods, Cambridge Institute for Medical Research, Keith Peters Building, Cambridge Biomedical Campus, Cambridge CB2 0XY, UK.
Mary M Reilly, Department of Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Ingo Kurth, Institute for Human Genetics and Genomic Medicine, Medical Faculty, RWTH Aachen University Hospital, 52074 Aachen, Germany.
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
Funding
The ‘European Network on Inherited Sensory Neuropathies and Insensitivity to Pain (ENISNIP)’ is supported by the Deutsche Forschungsgemeinschaft (DFG) (I.K. and J.S. KU 1587/6-1, SE 1839/2-1), the Ministry of Education, Youth and Sports (MEYS), Czech Republic (P.L.), the Austrian Science Fund (FWF) (M.A.-G.), the Swiss National Science Foundation (SNSF) (T.H.), the Scientific and Technological Research Council of Turkey (TUBITAK) (A.C., Y.P.), under the frame of EJP RD, the European Joint Programme on Rare Diseases. In addition, the ENISNIP project has received funding from the European Union's Horizon 2020 research and innovation programme under the EJP RD COFUND-EJP N° 825575. The study was supported by a grant from the Interdisciplinary Centre for Clinical Research within the faculty of Medicine at the RWTH Aachen University (A.La., A.Li., I.K. and M.F.D.: IZKF TN1-1/IA 532001, IZKF TN1-2/IA 532002, IZKF TN1-9/IA 532009) and the BMBF consortium ‘Precision2Treat’ (German Federal Ministry of Education and Research/Bundesministerium für Bildung und Forschung, BMBF, contract number 13GW0656). A.La. has a research contract Grünenthal and receives consulting fees from Grünenthal, which do not affect the work presented here. J.J.C. is supported by the Medical Research Council (MR/R011737/1) and the Wellcome Trust (200183/Z/15/Z). J.N.W. is supported by the Wellcome trust (200183/Z/15/Z) and Versus Arthritis (21950). C.G.W.: This research was supported by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health & Social Care. This work was supported by the Association Belge contre les Maladies neuro-Musculaires (ABMM to J.B. and V.T.). J.B. is supported by a Senior Clinical Researcher mandate of the Research Foundation Flanders (FWO) under grant agreement number 1805021N. J.V.L. is supported by a DOC-PR04 PhD fellowship from the University of Antwerp. Several authors of this publication are member of the European Reference Network for Rare Neuromuscular Diseases (ERN EURO-NMD) and of the European Reference Network for Rare Neurological Diseases (ERN-RND). V.T. and J.B. are partners in the Solve-RD EU project Horizon 2020 under grant agreement N° 779257. J.B. and V.T. are members of the µNEURO Research Centre of Excellence of the University of Antwerp. D.L.H.B., and A.C.T are members of: the DOLORisk consortium funded by the European Commission Horizon 2020 (ID633491), which received funding from European Union Seventh Framework Program (FP7/2007-2013); the International Diabetic Neuropathy Consortium (IDNC) research programme, which is supported by a Novo Nordisk Foundation Challenge Programme grant (Grant number NNF14OC0011633); and PAINSTORM consortium funded by UKRI and Versus Arthritis (MR/W002388/1). E.L. was supported by the German Research Foundation, DFG (LE-2338/3-1). A.C.T is supported by the Academy of Medical Sciences (Starter Grant SGL022\1086), and is a Honorary Senior Research Fellow and Carnegie-Wits Diaspora Fellow at the Brain Function Research Group, School of Physiology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa. D.C. is supported by the Fondazione Telethon ETS (GMR22T1076). P.J.T. was supported by an MRC strategic award to establish an International Centre for Genomic Medicine in Neuromuscular Diseases (ICGNMD) MR/S005021/1.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Cox JJ, Reimann F, Nicholas AK, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leipold E, Liebmann L, Korenke GC, et al. A de novo gain-of-function mutation in SCN11A causes loss of pain perception. Nat Genet. 2013;45:1399–1404. [DOI] [PubMed] [Google Scholar]
- 3. Dawkins JL, Hulme DJ, Brahmbhatt SB, Auer-Grumbach M, Nicholson GA. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat Genet. 2001;27:309–312. [DOI] [PubMed] [Google Scholar]
- 4. Rotthier A, Auer-Grumbach M, Janssens K, et al. Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. Am J Hum Genet. 2010;87:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guelly C, Zhu PP, Leonardis L, et al. Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I. Am J Hum Genet. 2011;88:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kornak U, Mademan I, Schinke M, et al. Sensory neuropathy with bone destruction due to a mutation in the membrane-shaping atlastin GTPase 3. Brain. 2014;137(Pt 3):683–692. [DOI] [PubMed] [Google Scholar]
- 7. Kurth I, Pamminger T, Hennings JC, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. 2009;41:1179–1181. [DOI] [PubMed] [Google Scholar]
- 8. Rivière JB, Ramalingam S, Lavastre V, et al. KIF1A, An axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet. 2011;89:219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verhoeven K, De Jonghe P, Coen K, et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003;72:722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Einarsdottir E, Carlsson A, Minde J, et al. A mutation in the nerve growth factor beta gene (NGFB) causes loss of pain perception. Hum Mol Genet. 2004;13:799–805. [DOI] [PubMed] [Google Scholar]
- 11. Indo Y, Tsuruta M, Hayashida Y, et al. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat Genet. 1996;13:485–488. [DOI] [PubMed] [Google Scholar]
- 12. Baets J, Duan X, Wu Y, et al. Defects of mutant DNMT1 are linked to a spectrum of neurological disorders. Brain. 2015;138(Pt 4):845–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edvardson S, Cinnamon Y, Jalas C, et al. Hereditary sensory autonomic neuropathy caused by a mutation in dystonin. Ann Neurol. 2012;71:569–572. [DOI] [PubMed] [Google Scholar]
- 14. Palma JA, Yadav R, Gao D, Norcliffe-Kaufmann L, Slaugenhaupt S, Kaufmann H. Expanding the genotypic spectrum of congenital sensory and autonomic neuropathies using whole-exome sequencing. Neurol Genet. 2021;7:e568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan JH, Higuchi Y, Ando M, et al. Multi-type RFC1 repeat expansions as the most common cause of hereditary sensory and autonomic neuropathy. Front Neurol. 2022;13:986504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garofalo K, Penno A, Schmidt BP, et al. Oral L-serine supplementation reduces production of neurotoxic deoxysphingolipids in mice and humans with hereditary sensory autonomic neuropathy type 1. J Clin Invest. 2011;121:4735–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fridman V, Suriyanarayanan S, Novak P, et al. Randomized trial of l-serine in patients with hereditary sensory and autonomic neuropathy type 1. Neurology. 2019;92:e359–e370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Auranen M, Toppila J, Suriyanarayanan S, et al. Clinical and metabolic consequences of L-serine supplementation in hereditary sensory and autonomic neuropathy type 1C. Cold Spring Harb Mol Case Stud. 2017;3:a002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carvalho OP, Thornton GK, Hertecant J, et al. A novel NGF mutation clarifies the molecular mechanism and extends the phenotypic spectrum of the HSAN5 neuropathy. J Med Genet. 2011;48:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shaikh SS, Nahorski MS, Woods CG. A third HSAN5 mutation disrupts the nerve growth factor furin cleavage site. Mol Pain. 2018;14:1744806918809223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fischer D, Schabhuttl M, Wieland T, Windhager R, Strom TM, Auer-Grumbach M. A novel missense mutation confirms ATL3 as a gene for hereditary sensory neuropathy type 1. Brain. 2014;137(Pt 7):e286. [DOI] [PubMed] [Google Scholar]
- 23. Mohammadi S, Jafari Khamirani H, Baneshi M, et al. A novel nonsense variant in the ATL3 gene is associated with disturbed pain sensitivity, numbness of distal limbs and muscle weakness. Ann Hum Genet. 2023;87:147–157. [DOI] [PubMed] [Google Scholar]
- 24. Rajadhyaksha AM, Elemento O, Puffenberger EG, et al. Mutations in FLVCR1 cause posterior column ataxia and retinitis pigmentosa. Am J Hum Genet. 2010;87:643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiabrando D, Castori M, di Rocco M, et al. Mutations in the heme exporter FLVCR1 cause sensory neurodegeneration with loss of pain perception. PLoS Genet. 2016;12:e1006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castori M, Morlino S, Ungelenk M, et al. Posterior column ataxia with retinitis pigmentosa coexisting with sensory-autonomic neuropathy and leukemia due to the homozygous p.Pro221Ser FLVCR1 mutation. Am J Med Genet B Neuropsychiatr Genet. 2017;174:732–739. [DOI] [PubMed] [Google Scholar]
- 27. Bertino F, Firestone K, Bellacchio E, et al. Heme and sensory neuropathy: Insights from novel mutations in the heme exporter feline leukemia virus subgroup C receptor 1. Pain. 2019;160:2766–2775. [DOI] [PubMed] [Google Scholar]
- 28. Li L, Jia C, Tang Y, Kong Y, Xia Y, Ma L. Novel gross deletion mutations in NTRK1 gene associated with congenital insensitivity to pain with anhidrosis. Front Pediatr. 2021;9:638190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kraft F, Kurth I. Long-read sequencing to understand genome biology and cell function. Int J Biochem Cell Biol. 2020;126:105799. [DOI] [PubMed] [Google Scholar]
- 30. Shekarabi M, Girard N, Riviere JB, et al. Mutations in the nervous system–specific HSN2 exon of WNK1 cause hereditary sensory neuropathy type II. J Clin Invest. 2008;118:2496–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.