Abstract
CD47 and its receptor signal regulatory protein α (SIRPα) act as a dominant antiphagocytic, “don’t eat me” signal. Recent studies reveal CD24 as a novel target for cancer immunotherapy by macrophages in ovarian cancer and breast cancer. However, whether simultaneous blockade of CD47 and CD24 by a bispecific antibody may result in a potential synergy is still unclear. In the present study, we for the first time designed and developed a bispecific antibody fusion protein, PPAB001 for cotargeting CD47 and CD24. Data demonstrate that simultaneous blockade of CD47/SIRPα and CD24/Siglec-10 signaling by PPAB001 potently promoted macrophage phagocytosis of tumor cells. Compared to single CD47 or CD24 targeting agents, PPAB001 was more effective in inhibiting tumor growth in both mouse 4T-1 syngeneic and human SK-OV-3 xenogeneic tumor models. Mechanistically, we found that PPAB001 therapy markedly increased the proportion of tumor-infiltrating macrophages and upregulated interleukin-6 and tumor necrosis factor-α levels that were representative macrophage inflammatory cytokines. Notably, an increased ratio of M1/M2 in tumor-infiltrating macrophages in the mice treated with PPAB001 suggested that the dual blockade may promote the transition of macrophages from M2 to M1. Taken together, our data supported the development of PPAB001 as a novel immunotherapeutic in the treatment of CD47 and CD24 double-positive cancers.
Key words: CD47, CD24, macrophages, bispecific antibody, antitumor immunity
Graphical abstract
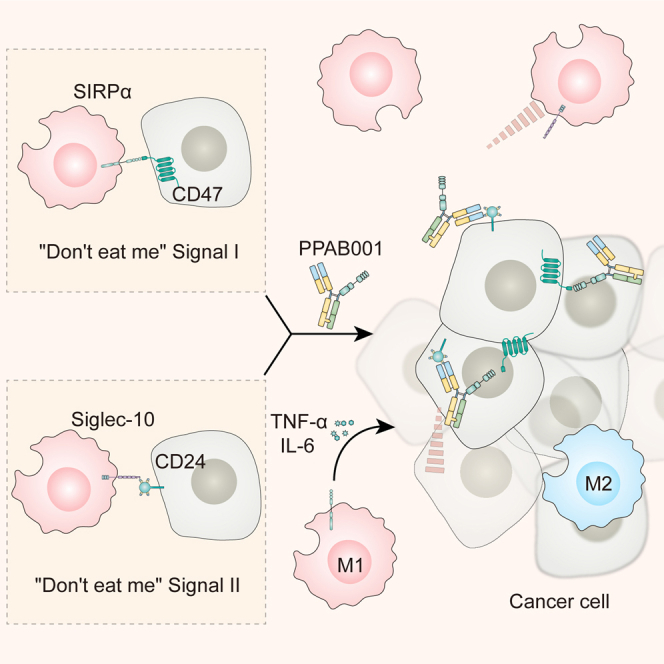
In this study, Wang and colleagues for the first time designed and developed a novel bispecific antibody fusion protein, PPAB001 for cotargeting CD47 and CD24. It will have great potential to provide a novel immunotherapy strategy by blocking dual “don’t eat me” signals for cancer patients.
Introduction
CD47, ubiquitously expressed on the membrane of normal cells, is a 50-kDa transmembrane protein known as integrin-associated protein.1 CD47 overexpression on various solid and hematopoietic tumors is always associated with poor prognosis in cancer patients.2,3,4 By interacting with its receptor signal regulatory protein α (SIRPα), it transmits a “don’t eat me” signal to phagocytic cells.5 CD47 blockade as a potent immunotherapy strategy has shown its efficacy in a variety of different cancers.4,6 Although anti-CD47 antibody monotherapy has exhibited benefits in preclinical studies and clinical trials, the synergetic advantages of simultaneous blocking CD47 and other receptors, including programmed cell death ligand 1 (PD-L1), human epidermal growth factor receptor 2 (HER2), or CD20, are being seen in cancer immunotherapy.7,8,9 Therefore, a few of the bispecific antibodies cotargeting CD47 and other tumor-specific antigens were designed to improve the efficacy of CD47 blockade.10,11,12 Although anti-CD47 antibody has shown efficacy in cancer immunotherapy, several cancers still show resistance to CD47 blockade that suggests the presence of additional, as yet unknown don’t eat me signals.13
CD24 is a glycosylphosphatidylinositol-anchored membrane protein and a ligand for P-selectin, an adhesion receptor expressed on normal endothelial cells and platelets.14,15 It is known as a tumor-associated marker, which is overexpressed in nearly 70% of human cancers.16,17 Recently, CD24 has been reported to release antiphagocytic signals through its interaction with sialic acid–binding immunoglobulin (Ig)-like lectin 10 (Siglec-10), which is located on the surface of macrophages, to compose a novel immune checkpoint.13,18 Barkal et al. demonstrated that combinatorial treatment with CD24- and CD47-blocking antibodies significantly promotes the phagocytic clearance of human cancer cells.13 Li et al. reported that ZBTB28, a tumor suppressor gene, regressed breast cancer by activating the interferon-α/β receptor (IFNAR) and combinatorial blockade of CD24 and CD47 to augment macrophage phagocytosis.19 Collectively, the findings above revealed CD24 as a promising therapeutic target in cancer immunotherapy.
Because both CD47 and CD24 are highly expressed on tumor cells and serve as critical dual immune checkpoints for macrophages, whether a potential synergy exists by simultaneously targeting the dual antigens using bispecific antibody was unknown. In this study, we for the first time generated a CD47/CD24 dual-targeting bispecific antibody fusion protein, PPAB001, that was designed by combining the CD47 receptor, the SIRPα high-affinity variant (CV1), and humanized anti-CD24 antibody. The elevated expression of CD47 and CD24 was detected in the HER2+ cancers by immunohistochemical (IHC) analysis. Moreover, CD47 and CD24 levels were positively associated with HER2 status. More important, the simultaneous blockade of CD24 and CD47 by PPAB001 markedly augmented the phagocytosis of HER2+ cancer cells by macrophages. The superior therapeutic efficacy of PPAB001 was also observed in both mouse 4T-1 syngeneic and human SK-OV-3 xenograft tumor models, which may be attributed to the increased proportion of tumor-infiltrating macrophages and the elevated ratio of the M1/M2 subtype. Collectively, these data pave the way for developing a novel immunotherapy strategy by blocking dual don’t eat me signals with PPAB001 for treating cancers.
Results
Design and characterization of PPAB001
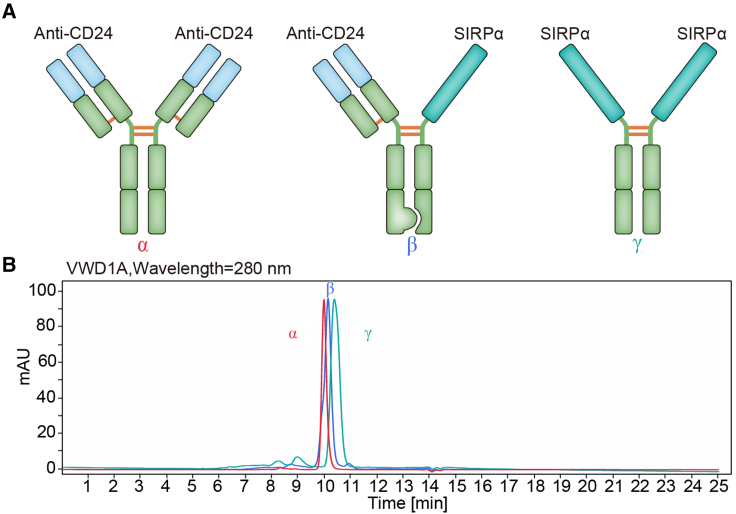
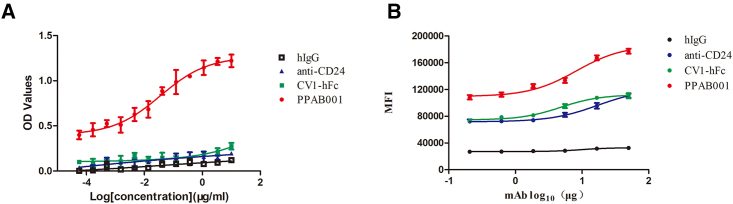
To assess whether CD47 and CD24 dual blockade may result in the enhanced antitumor effects, we developed PPAB001, which is a bispecific antibody fusion protein against CD47 and CD24 in an IgG1 format using knobs-into-holes (KIH) technology and a common light-chain architecture (Figure 1A). Size-exclusion chromatography (SEC) confirmed its purity and molecular weight (Figure 1B). PPAB001 was further demonstrated to bind to both CD47 and CD24 simultaneously by ELISA assay (Figure 2A). Moreover, PPAB001 also exhibits preferential binding activity on CD47 and CD24 in BT-474 cells that overexpressed CD47 and CD24 (Figures 2B and S1). To further verify the specificity of PPAB001, we also examined the binding activity of PPAB001 to HEK293T cells or the CD47 and CD24 knockout HEK293T cells by flow cytometry assay. As shown in Figure S2, the results showed that PPAB001 specifically bound to HEK293T cells while being hardly bound to CD47 and CD24 knockout HEK293T cells, which exhibited its specificity.
Figure 1.
Characterization of PPAB001
(A) Schematic representation of anti-CD24 antibody (α), PPAB001(β), and the engineered SIRPα variant-Fc (CV1-hFc) (γ). (B) Validation of purity and molecular size of PPAB001 by SEC chromatogram (SEC-ultra-performance liquid chromatography) analysis.
Figure 2.
Analysis of PPAB001 for simultaneous binding to CD47 and CD24
(A) Simultaneous binding of PPAB001 to recombinant CD47 and CD24 antigen by sandwich ELISA. (B) Flow cytometry of breast cancer BT-474 cells stained with PPAB001, anti-CD24, CV1-hFc, or hIgG at 10 μg/mL. Binding kinetics of PPAB001, anti-CD24, or CV1-hFc to membrane-associated CD47 and CD24 on BT-474 cells were shown. Data are shown as means ± SDs (n = 3).
Simultaneous blockade of CD24 and CD47 by PPAB001 synergistically enhances the phagocytosis of cancer cells
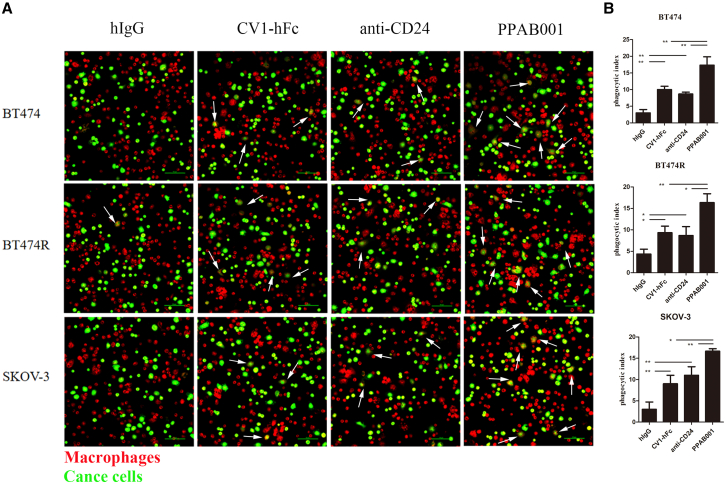
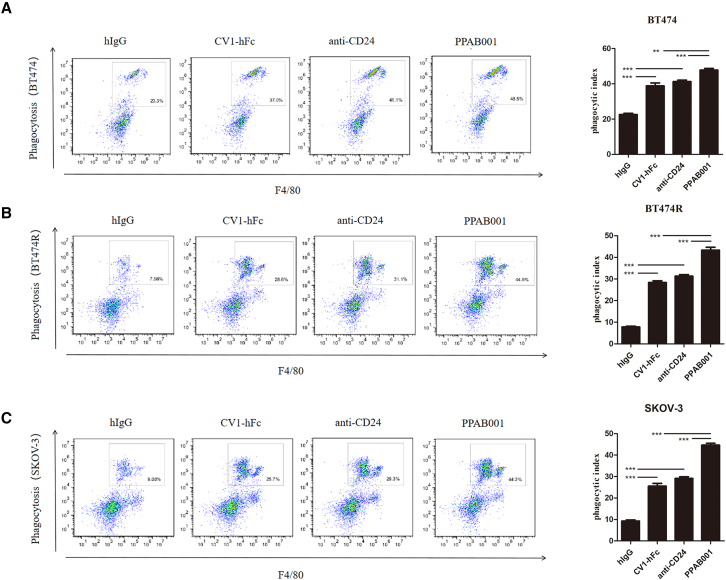
Candas-Green et al. demonstrate that CD47 is upregulated preferentially on HER2-expressing cells.8 In the present study, we also verified that CD47 and CD24 expression was overexpressed on HER2+ cancers, including BT-474, BT-474R, and SK-OV-3 (Figures S1A and S3). Moreover, CD47 and CD24 levels were positively associated with HER2 expression in 31 human tumor tissues from breast cancer patients by IHC staining (Figures S1B–S1D). To explore the impact of PPAB001 on the activity of macrophages to engulf CD47+ and CD24+ cancer cells, we cocultured macrophages with BT-474, BT-474R, and SK-OV-3 cells to investigate phagocytosis in vitro. Confocal microscopy visually exhibited that the blockade of CD24 and CD47 by PPAB001 significantly augmented the phagocytosis of BT-474, BT-474R, and SK-OV-3 cells by macrophages (Figures 3A and 3B). Furthermore, a flow cytometry assay further quantitatively showed that compared with anti-CD24 or SIRPα high-affinity variant-human Fc fragment (CV1-hFc), PPAB001 more effectively increased the phagocytic index of the cancer cells mentioned above by macrophages (Figures 4A–4C).
Figure 3.
Treatment with PPAB001 promotes phagocytosis of human cancer cells
(A) Representative phagocytosis images of macrophages engulfing BT-474, BT-474R, or SK-OV-3 cells with PPAB001, anti-CD24, CV1-hFc, or hIgG. Macrophages were stained red (CMTPX); cancer cells were labeled with green (CFSE). The white arrows indicate macrophages that engulfed cancer cells. (B) The extent of macrophage phagocytosis with PPAB001, anti-CD24, CV1-hFc, or hIgG was quantified for BT-474, BT-474R, or SK-OV-3 cells. Data are shown as means ± SDs (n = 3), and statistical significance was determined by a Student’s t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Figure 4.
CD47/CD24 dual blockage potentiates the macrophage phagocytosis
(A) Representative flow cytometry plots depicting the phagocytosis of BT-474 cells treated with PPAB001, anti-CD24 mAb, or CV1-hFc, compared with the IgG control. Phagocytosis index is shown as a bar graph. (B) Representative flow cytometry plots depicting the phagocytosis of BT-474R cells treated with PPAB001, anti-CD24 mAb, or CV1-hFc, compared with the IgG control. Phagocytosis index is shown as a bar graph. (C) Representative flow cytometry plots depicting the phagocytosis of SK-OV-3 cells treated with PPAB001, anti-CD24 mAb, or CV1-hFc, compared with the IgG control. Phagocytosis index is shown as a bar graph. Data are shown as means ± SDs (n = 3), and statistical significance was determined by a Student’s t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
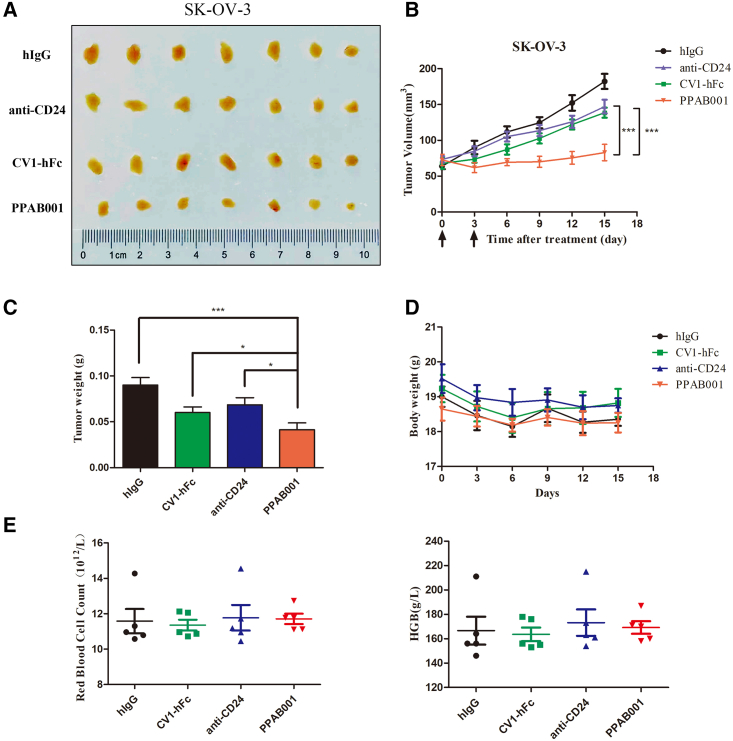
PPAB001 treatment showing superior efficacy in the SK-OV-3 tumor xenograft model
To determine the antitumor effects of PPAB001 in vivo, we established the SK-OV-3 xenogeneic mouse model. In this model, PPAB001 (5 mg/kg), anti-CD24 (5 mg/kg), or CV1-hFc (5 mg/kg) was administrated when tumors reached approximately 80 mm3 (Figure 5). Data confirmed that PPAB001 as a potent therapy significantly delayed the tumor growth compared to the anti-CD24 or the CV1-hFc treatment group in vivo (Figures 5A–5C). Moreover, no significant weight loss was observed and the counts of red blood cells (RBCs) and level of hemoglobin in these mice differed within 10%, which suggested that PPAB001 may be well tolerated and has a relatively low risk of hematological toxicities (Figures 5D and 5E). Meanwhile, we also conducted an agglutination assay with human RBCs (hRBCs) in vitro when incubated with anti-CD24, CV1-hFc, CV1-hFc plus anti-CD24, or PPAB001. Our data revealed that PPAB001 did not cause significant agglutination of hRBCs compared with CV1-hFc or CV1-hFc plus anti-CD24, which may be attributed to its lower binding activity to hRBCs (Figures S4A–S4C). Taken together, these results showed that PPAB001 may have both effective antitumor activity and a relatively safe profile.
Figure 5.
Antitumor activity of PPAB001 in immunodeficient SK-OV-3 xenogeneic tumor-bearing mice
(A) Images of excised subcutaneous tumors treated with PPAB001, anti-CD24, or CV1-hFc are shown. (B) BALB/c nude mice were subcutaneously transplanted with SK-OV-3 cells and treated with PPAB001, anti-CD24, CV1-hFc, or hIgG (5 mg/kg) (n = 6). (C) After xenograft tumors were removed, these tumor masses were weighted. (D) The body weights of tumor-bearing mice were determined in the whole period of therapy. Mice were weighed at regular intervals during the whole period to monitor unspecific toxicity. (E) The changes of RBCs and hemoglobin response to treatment with PPAB001, anti-CD24, CV1-hFc, or hIgG. Data are shown as means ± SDs (n = 6), and statistical significance was determined by a Student’s t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
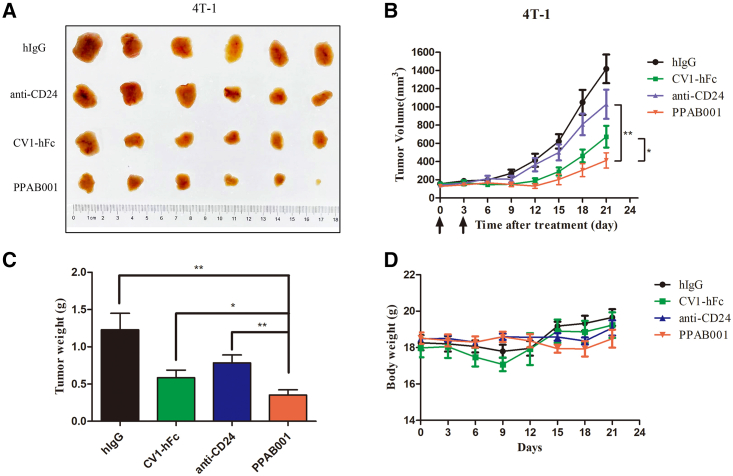
Therapeutic effect of PPAB001 in 4T-1 syngeneic tumor-bearing mice
To further examine whether PPAB001 functions in immunocompetent mice, C57BL/6 mice were inoculated with syngeneic 4T-1 cancer cells. PPAB001 has been verified to cross-react with murine CD47 and CD24 molecules (Figure S5). When tumors were well established, mice were intraperitoneally (i.p.) administered with PPAB001, anti-CD24, CV1-hFc, or human IgG (hIgG), and the tumor growth was monitored throughout the study period. Inconsistent with the results in SK-OV-3 xenografts, PPAB001 inhibited tumor development more effectively than single anti-CD24 or CV1-hFc treatment in the 4T-1 syngeneic model (Figures 6A–6C). Over the study period (approximately 21 days), these mice did not significantly lose body weight and behaved normally (Figure 6D). Thus, our data further clearly showed the therapeutic superiority of CD47 and CD24 dual blockade in an immunocompetent syngeneic host. Moreover, we also compared the therapeutic effects of PPAB001 with CV1-hFc plus anti-CD24 in 4T-1 tumor-bearing mice. Our results demonstrated that PPAB001 exhibited superior anti-tumor effects to CV1-hFc plus anti-CD24, which may be attributed to the synergistic phagocytosis-promoting effects caused by simultaneous CD47 and CD24 blocking action (Figures S6A–S6D).
Figure 6.
PPAB001 inhibits tumor growth in immunocompetent 4T-1 syngeneic tumor-bearing mice
(A) Photographs of excised tumors treated with hIgG, anti-CD24, CV1-hFc, and PPAB001. (B) C57BL/6J mice were subcutaneously transplanted with 4T-1 cells and treated with PPAB001, anti-CD24, CV1-hFc, or hIgG (5 mg/kg) (n = 6). (C) After xenograft tumors were removed, these tumor masses were weighted. (D) Body weight of tumor-bearing BALB/c mice on treatment with PPAB001, anti-CD24, CV1-hFc, or hIgG. Data are shown as means ± SEMs (n = 6), and statistical significance was determined by a Student’s t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
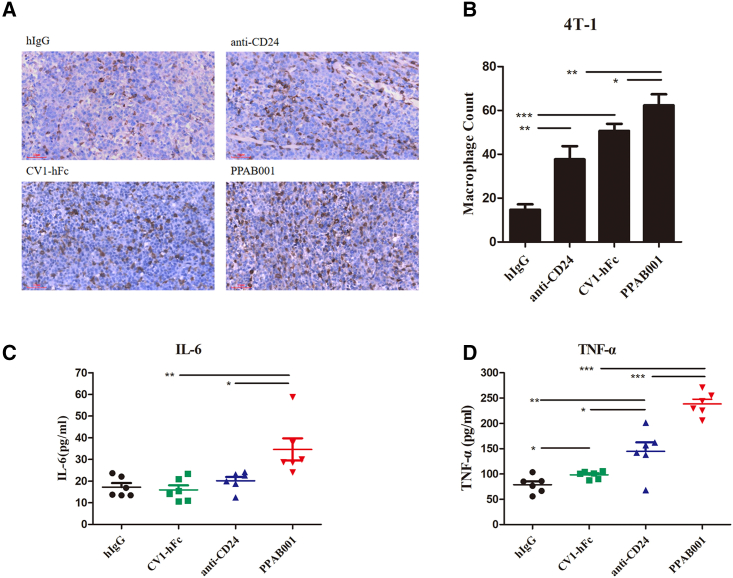
In vivo treatment with PPAB001 increased the proportion of tumor-associated macrophages and upregulated interleukin-6 and tumor necrosis factor-α
With PPAB001 therapy, we observed a significantly increased proportion of tumor-associated macrophages, which migrated into the 4T-1 tumor compared to anti-CD24 or CV1-hFc treatment (Figure 7A). Furthermore, the density of macrophages that appeared in the tumor with PPAB001 increased by approximately 1.65- and 1.23-fold, respectively, when compared to that of anti-CD24 or CV1-hFc (Figure 7B). These results suggest that CD47 and CD24 dual blockade leads to a significantly increased migration of macrophages into the cancer mass in vivo. In addition, we also evaluated the level of macrophage markers interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) in mice serum that hinted at the activation of macrophages. As shown in Figures 7C and 7D, PPAB001 treatment led to a marked upregulation of TNF-α and IL-6 compared to the anti-CD24- or the CV1-hFc-treated group. These findings indicated that the enhanced activity of PPAB001 may be correlated to a marked increase in and activation of the infiltrated macrophages.
Figure 7.
PPAB001 treatment promotes macrophage infiltration of 4T-1 tumors
(A) Representative F4/80 staining tumor images with hIgG, anti-CD24, CV1-hFc, and PPAB001 treatment (magnification ×20). (B) IHC analysis of tumor infiltrating F4/80+ cell counts. (C) The level of IL-6 in serum from tumor-bearing mice treated with different agents was measured by ELISA kit. Data are shown as means ± SEMs (n = 6). (D) The level of TNF-α in serum from tumor-bearing mice treated with different treatments was measured by ELISA kit. Data are shown as means ± SEMs (n = 6), and statistical significance was determined by a Student’s t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
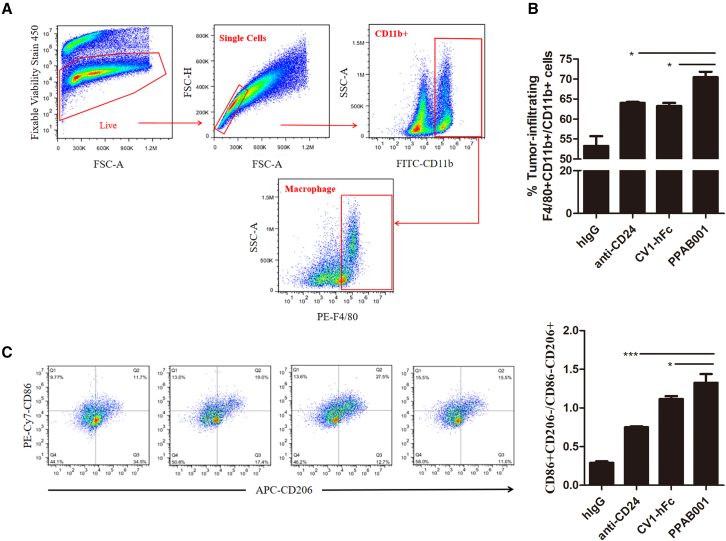
PPAB001 treatment promotes macrophages to polarize toward M1 phenotype and CD8+ T cell infiltration in the tumor microenvironment
Raising the ratio of M1/M2 is a promising strategy to ameliorate the tumor microenvironment toward tumor regression.20 In the present study, we investigated whether PPAB001 treatment in vivo mediated the macrophage polarization profile toward a favorable antitumorigenic microenvironment. Mice treated with PPAB001 were found to have a significantly increased number of total macrophages with markers CD11b+ F4/80+ in tumors relative to anti-CD24-, CV1-hFc-, or hIgG-treated mice (Figures 8A and 8B). Moreover, to further quantify and verify the phenotype of the macrophages, we examined M1 macrophage-related phenotypic marker CD86 and M2 macrophage-related phenotypic marker CD206. Results showing that the ratio of M1/M2 cell counts (CD86+/CD206− macrophage number) in the PPAB001-treated group increased by 1.77- and 1.19-fold, respectively, as compared to those in the anti-CD24 and CV1-hFc groups (Figure 8C). Therefore, these findings suggested that simultaneous disruption of the CD47-SIRPα and CD24-Siglec-10 axis may not only facilitate M1 macrophages infiltration into tumors but also convert M2 into the M1 phenotype, which may further explain the superior therapeutic effects of PPAB001.
Figure 8.
PPAB001 therapy increases the ratio of M1/M2 in tumor-infiltrating macrophages in the 4T-1 tumor model
(A) Flow cytometry gating strategy to identify tumor-infiltrating macrophages from CD11b+ immune cells; the CD11b+ immune cells were initially gated after the exclusion of dead cells and doublets. Intratumoral macrophages were defined as CD11b+ F4/80+. (B) Percentage of tumor-infiltrating macrophages among CD11b+ immune cells by FACS. (C) Representative FACS plots showing M1- and M2-subsets based on CD86 and CD206 staining. Bar chart depicting the M1/M2-macrophage ratio. Data are shown as means ± SDs (n = 3), and statistical significance was determined by a Student’s t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To explore how PPAB001 exerts effects on changing the secretion of cytokines and the ratio of M1/M2, we performed bulk RNA sequencing (RNA-seq) using tumor tissues from the 4T1 syngeneic model treated by PPAB001 or other agents. Comparing PPAB001 with other treatments, we observed that Tlr1, Tlr2, Tlr8, Cxcl10, Fcgr2b, Csf1r, Csf2ra, and Itgam were markedly upregulated in PPAB001-treated samples (Figures S7A and S7B). Then, these results revealed that the positive regulation of innate immune response, Toll-like receptor signaling pathway, and FcγR-mediated phagocytosis was significantly enriched in PPAB001-treated samples, which may partly explain the potential mechanism involved in the elevated secretion of cytokines and the ratio of M1/M2 (Figure S7C). In addition, we investigated the impact of PPAB001 on T cells around the tumor microenvironment and found that PPAB001 treatment significantly promotes CD8+ T cell infiltration in 4T-1 tumors (Figures S8A–S8C).
Discussion
It is worth noting that CD47 and CD24, two well-defined cell surface receptors with critical biological functions, were overexpressed in HER2+ breast cancer patients. Moreover, HER2+ tumors are predominantly positive for CD47 and CD24, suggesting that the expression level of CD47 and CD24 is tightly related. Previous studies also revealed that treatment with the anti-CD24 monoclonal antibody (mAb) promotes phagocytic clearance of CD47-directed antibody in human breast cancer cells.13 Collectively, these findings suggest that the blockade of both CD24 and CD47 by PPAB001 may further augment cellular engulfment.
Although trastuzumab has exhibited therapeutic effects for early-stage HER2+ breast cancer, the majority of advanced-stage HER2+ breast cancer patients who initially respond to trastuzumab developed resistance within 1 year of treatment.21 Impairments in trastuzumab-mediated antibody-dependent cell-mediated cytotoxicity (ADCC) also explain the cause of trastuzumab resistance.22,23 Upton et al. found that the combination of anti-CD47 antibody, Hu5F9-G4, and trastuzumab eliminated HER2+ breast cancer cells with increased potency due to the enhancement of antibody-dependent cellular phagocytosis by macrophages, even when the cancer cells were tolerant to trastuzumab-induced ADCC by natural killer cells.24 In our study, we also found that PPAB001 was effective in inducing phagocytosis against acquired trastuzumab-resistant breast cancer BT-474R cells, which suggests the potential of PPAB001 in overcoming trastuzumab resistance. Trastuzumab deruxtecan (DS-8201) was a novel HER2-targeting antibody-drug conjugate with topoisomerase I inhibitor payload that received accelerated approval by the US Food and Drug Administration for the treatment of patients with HER2+, unresectable breast cancer who have received at least 2 prior lines of anti-HER2-based treatments.25,26 Iwata et al. put forward that DS-8201 treatment benefited from combination with anti-PD-1 antibody, possibly due to increased T cell activity.27 Based on these studies, further study will need to be conducted to explore whether the combination of PPAB001 with HER2-targeted therapeutics such as trastuzumab or DS-8201 may exert synergistic effects in limiting tumor growth in HER2+ cancers.
Despite that CD47-targeted therapeutics have shown efficacy in eliminating tumors, the major concerns for the development of CD47/SIRPα-based immunotherapy are RBC toxicities and fast CD47-mediated clearance because of the broad expression profile of CD47 on normal cells.28,29 The development of bispecific antibodies against CD47 may be an attractive direction for addressing the issues.30,31 Wang et al. reported that 6MW3211, a novel bispecific antibody against CD47 and PD-L1, selectively binds to CD47 on tumor cells but not CD47 expression on RBCs, which suggests the good safety profile of 6MW3211 in nonhuman primate (NHP) studies and further human clinical trials.7 Weiskopf et al. indicated that the mice treated with 200 μg high-affinity SIRPα-Fc (CV1-hIgG4) resulted in the development of chronic anemia as a side effect.32 In our study, CV1-hFc treatment (approximately 100 μg per mouse) has not shown potential toxicity in RBCs, which suggested lower dosage and extended injection interval may reduce injury. Moreover, PPAB001 treatment did not lead to the significant reduction of the counts of RBCs and level of hemoglobin in xenografts. More important, the agglutination assay in our study revealed that PPAB001 did not cause significant agglutination of hRBCs compared with CV1-hFc or CV1-hFc plus anti-CD24, suggesting its relatively good safety profile. However, further safety studies on NHPs were still necessary.
As can be seen from our data, the effect of PPAB001 treatment against breast cancers is statistically significant but still not robust, which suggested that the cooperation of PPAB001 and other immune checkpoint inhibitors such as PD-L1 antagonists may be necessary to further augment antitumor immune responses and remodel the tumor microenvironment.
Macrophages of the M2 phenotype in carcinomas significantly promote tumor progression and metastasis, as opposed to the M1 phenotype that exhibits marked antitumor characteristics.33,34 Our results demonstrate that it is desirable to modulate the balance of the M1/M2 ratio toward M1 macrophages by dual blockade CD47 and CD24 for restoring antitumor immunity in malignant tumors. Then, we further investigated the signaling pathways involved in macrophage polarization in cancers upon PPAB001 treatment to elucidate the potential mechanism.
In summary, this study demonstrates that CD47 and CD24 functioning can coordinate the don’t eat me signal from macrophages to cancer cells. Targeting dual don’t eat me checkpoints using a bispecific approach has great potential to synergistically remodel the tumor microenvironment that contributes to the promotion of antitumor activity.
Materials and methods
Cell lines and human tumor samples
The human breast cancer cell line BT-474 and the ovarian cancer cell line SK-OV-3 were obtained from the American Type Culture Collection (ATCC). 4T-1 was a mouse breast cancer cell line obtained from the ATCC. Cells were cultured in DMEM (Hyclone, cat. no. SH30022.01), which was supplemented with 4.5 g/L glucose and 10% fetal bovine serum (FBS). Trastuzumab-resistant BT-474 (BT-474R) cancer cells were obtained according to the following procedures: DMEM medium with 10% FBS and trastuzumab (10 mg/mL) were added on top and replaced every 3 days. After 6 months, BT-474R cells were established and detected by a cell viability assay. CD47 and CD24 dual-knockout 293T cells were provided by the Guo lab. Primary human breast cancer tissues were obtained from the First Affiliated Hospital of Xinxiang Medical University. The pathological studies were approved by the institutional ethics committees of Xinxiang Medical University (approval notice no. XYLL-20230087), and informed consent was obtained from all of the subjects.
Reagents and antibodies
Reagents were purchased as follows: diaminobenzidine (DAB) (catalog no. ZLI-9018,ZSGB-BIO), thioglycolate medium (catalog no. C34552, Invitrogen), Cell-Tracker Red CMTPX (catalog no. C34552, Invitrogen), Cell-Tracker carboxyfluorescein diacetate succinimidyl ester (CFSE) (catalog no.70157, Sigma), Tetramethylbenzidine (TMB) Two-Component Substrate solution (for ELISA) (catalog no. PR1210, Solarbio), H&E Stain Kit (catalog no. G1120, Solarbio), CD24 Fusion Protein (catalog no. Ag11679, ProteinTech Group), Mouse TNF-α ELISA kit (catalog no. SEKM-0034, Solarbio), Mouse IL-6 ELISA kit (catalog no. SEKM-0007, Solarbio), TruStain FcX (anti-mouse CD16/32) Antibody (catalog no. 101319, BioLegend), fixation buffer (catalog no. 420801, BioLegend), Intracellular Staining Permeabilization Wash Buffer (10×) (catalog no. 421002, BioLegend), collagenase I (catalog no. 17100017, Invitrogen), hRBC isolation kit (catalog no. RBC2014TBD, Tbdscience), RNALater animal tissue RNA stable preservation solution (catalog no. R0118, Beyotime Biotechnology), antifluorescence quenching sealing solution (catalog no. P0133, Beyotime Biotechnology), and Tris-EDTA antigen repair solution (catalog no. P0084, Beyotime Biotechnology). Antibodies were used as follows: HER2 (1:1,000; catalog no. 54359; Cell Signaling Technology), CD47 (1:1,000; catalog no. ab284132; Abcam), CD24 (1:1,000; catalog no. ab278509; Abcam), F4/80 (1:2,000; catalog no. 28463-1-AP; ProteinTech Group), His-Tag monoclonal antibody (catalog no. HRP-66005; ProteinTech Group), fluorescein isothiocyanate (FITC)–conjugated Affinipure goat anti-mouse/rabbit/human IgG (H + L) (1:100; catalog no. SA00003-1 or SA00003-2 or SA00003-12; ProteinTech Group), horseradish peroxidase (HRP)–conjugated goat anti-mouse/rabbit secondary antibodies (1:5,000; catalog no. SA00001-1 or SA00001-2; ProteinTech Group), PE Rat Anti-Mouse F4/80 (catalog no. 565410; BD Pharmingen), FITC Rat Anti-CD11b(M1/70) (catalog no. 557396; BD Pharmingen), PE-Cy7 Rat Anti-Mouse CD86 (GL1) (catalog no. 560582; BD Pharmingen), Alexa Fluor 647 Rat Anti-Mouse CD206 (MR5D3) (catalog no. 565250; BD Pharmingen), Fixable Viability Stain 450 (catalog no. 562247; BD Pharmingen), and CD8α (2.43) Rat mAb (APC Conjugate) (catalog no. 64786S; Cell Signaling Technology).
Bispecific antibody fusion protein generation
For anti-CD24 antibody generation, BALB/c mice were immunized with human CD24 recombinant protein. After that, hybridomas were generated and the supernatants were assayed for hCD24 binding by ELISA and fluorescence-activated cell sorting (FACS). The positive samples were subcloned followed by variable region (VH and VL) gene sequencing. Individual recombinant antibody was expressed in HEK293 cells and subsequently purified with protein A resin. Purified antibodies were then subjected to functional assays, and the top 10 candidates were humanized using complementarity determining region grafting strategy. After several rounds of heavy-chain and light-chain combinations, screening, engineering, and optimization, anti-CD24 antibody with common light chain was obtained. The amino acid sequence information of the VH and VL was shown as follows, respectively: [VKLVESGGGLVQPGGSLKLSCAASGDSVTSKSMSWVRQAPSGGLQWIGRTYPDNGDSSYTPSVRSRFIISRDNAKNTLYLQMRKVRSEDTGFYYCARQGDYWGQGTTLTVSS]; [DVVMTQTPLTLSLSPGQPASISCKSSQSGSIGRAKTYLNWYQQRPGQAPKLLIYDASKLDSGVPDRFTGSGSGTDFTLKISRVEAEDLGVYYCWQGTHFPYTFGGGTKLEIKR].
For CD47 receptor fusion protein generation, the engineered SIRPα variant (CV1) with high affinity was constructed and expressed as reported previously.32 CV1-hFc was generated by fusing the engineered SIRPα variant into hIgG1Fc. CD47/CD24 bispecific antibody PPAB001 was assembled as a full-length IgG1 subtype using the KIH technology, as reported.35 The knob arm used anti-CD24 heavy chain, and the hole arm used CV1-hFc heavy chain. PPAB001 was stably expressed in CHO cells. After production and purification, PPAB001 was characterized by SEC to show purity >95%.
RBC agglutination assay
Human whole blood collected from healthy donors isolated hRBCs using an hRBC isolation kit (Tbdscience) according to the manufacturer’s instructions. Then, RBCs were washed and resuspended with PBS. Approximately 5 × 106 RBCs in PBS were plated per well in polypropylene 96-well plates (LabServ) and incubated with serially diluted antibodies at room temperature. The hemagglutination of the plates was observed after 1 h under the microscope and photographed.
In vitro phagocytosis assay
Peritoneal macrophages were isolated following the injection of 1 mL 3% thioglycolate medium into the peritoneal cavity of 6-week-old BALB/c mice. After 3 days, a peritoneal wash was performed using 10 mL DMEM/F12 medium (10% FBS). The macrophage-containing medium was withdrawn, and the macrophages were cultured in DMEM/F12 medium.
For the imaging-based phagocytosis assay, macrophages were labeled with Cell Tracker Red CMTPX (Invitrogen), seeded onto a 6-well plate, and incubated in serum-free medium for 2 h at 37°C. Tumor cells were labeled with 2.5 μM CFSE according to the manufacturer’s protocol (Invitrogen), and CFSE-labeled tumor cells were subsequently added to macrophages. Then, hIgG, CV1-hFc, anti-CD24, or PPAB001 was added at a concentration of 10 μg/mL and incubated for 3 h at 37°C. Plates were washed with prewarmed culture medium after 3 h of coculture. Images were captured using a Nikon A1 with 200× magnification.
For the flow cytometry–based phagocytosis assays, CFSE-labeled tumor cells were co-incubated with macrophages in serum-free medium and antibody at 37°C for 3 h. Cells were washed 3 times with PBS, followed by incubation with PE Rat Anti-Mouse F4/80 antibody (BD Biosciences) in PBS for 30 min on ice (protected from light). Cells were washed 3 times with PBS and analyzed via flow cytometry. The phagocytic index denotes the ratio of F4/80+CFSE+ phagocytic macrophages to total F4/80+ macrophages.
Immunoblotting
Cells were lysed in radioimmunoprecipitation assay lysis buffer supplemented with 2 μL/mL protease inhibitor cocktail (Sigma) and 10 μL/mL phosphatase inhibitor cocktail (Sigma) for 10 min at 4°C. The protein concentration of the supernatants was measured by a QuantiPro bicinchoninic acid protein assay kit (Sigma-Aldrich). After denaturation, total cell lysates were separated using SDS-PAGE and immunoblotted with primary antibodies and HRP-conjugated secondary antibody, as mentioned above. After washing the membrane, image acquisition was performed using a ChemiDoc imaging system (Bio-Rad Laboratories) with the sensitive enhanced chemiluminescence reagent (Millipore).
ELISA
Secretions of TNF-α and IL-6 in in the blood of the tumor-bearing mice treated with the indicated agents were measured by ELISA kits according to the protocol provided by the manufacturers. TNF-α and IL-6 ELISA detection kits were purchased from Solarbio.
To determine the binding property of PPAB001, CD47 (0.5 μg/mL) antigen was coated on a 96-well enzyme-labeled plate, and the plate was sealed with skim milk powder. Antibodies (hIgG, anti-CD24, CV1-hFc, or PPAB001) diluted 3-fold by gradient from 10 μg/mL were then added, incubated at 37°C for 1 h, and washed 3 times with PBS 0.1% Tween 20 (PBST). His-tagged CD24 antigen (0.5 μg/mL) was added and incubated for 1 h, and then washed 3 times with PBST. HRP-conjugated anti-His antibody was added (diluted to the ratio recommended in the instructions) and incubated for 30 min. Then, TMB was added and incubated at 37°C for 15 min without light. The reaction was stopped by adding 1 M sulfuric acid. The absorbance (optical density) was measured at 450 nm.
FACS-based binding assays
Cells were incubated with antibodies for the indicated hours at 4°C in PBS. Antibody-bound cells were then washed and incubated with FITC-conjugated goat anti-hIgG secondary antibody for 45 min at 4°C. Cells were washed at least twice with PBS and analyzed via flow cytometry.
Animals
Five-week-old female nude mice and C57BL/6J mice (Skbex Laboratory Animal Co.) were maintained in a sterile environment. The animal research was conducted according to the Principle of Laboratory Animal Care (NIH Publication No. 85-23, revised in 1985). All of the experimental protocols were performed under the approval of the Animal Experimentation Ethics Committee of Xinxiang Medical University. The animals were treated in accordance with the guideline of the Animal Care and Use Committee of Xinxiang Medical University.
Xenogeneic tumor model
SK-OV-3 cells (1 × 107 per mouse) mixed with PBS were inoculated subcutaneously into the right flanks of nude mice. The mice were randomly divided into 4 groups of 6 mice each when tumor volumes reached an average of approximately 80 mm3. Each group was administered one of the following: hIgG, CV1-hFc, anti-CD24, or PPAB001 (5 mg/kg). These agents were administered i.p. twice per week for 1 week. The tumors were measured with digital calipers, and tumor volumes were calculated by the formula Volume = [Length × (Width)2]/2. Mice were euthanized with CO2 asphyxiation.
Syngeneic tumor model
The 4T-1 cells (2 × 106 per mouse) mixed with PBS were inoculated subcutaneously into the right flanks of C57BL/6J mice. When tumor volume reached 100 mm3, at approximately days 5–6 postinoculation, tumor-bearing mice were divided into 4 groups of 6 mice each. Each group was injected with indicated agents at a dose of 5 mg/kg. These agents were administered i.p. twice per week for 1 week. Tumors were measured with digital calipers, and tumor volumes were calculated by the formula Volume = [Length × (Width)2]/2. Mice were euthanized with CO2 asphyxiation. Body weight, maximum length of the major axis (L), and maximum length of the minor axis (W) of tumors were measured once every 3 days.
Routine blood examination
SK-OV-3 tumor-bearing mice were injected i.p. with hIgG, anti-CD24, CV1-hFc, or PPAB001. On day 3 after the last treatment, blood samples were withdrawn from the eyeballs and subjected to routine blood tests.
IHC
Paraffin-embedded tumor tissue sections (4 μm) were deparaffinized and subjected to heated-induced epitope retrieval using citrate buffer (pH 6.0). The tissues were incubated with the indicated antibody overnight at 4°C. Following these, the tissues were incubated with the corresponding secondary antibodies. Peroxidase activity was visualized with 3,3′-diaminobenzidine (DAB; Darko) and then counterstaining with H&E was performed.
Scoring of the immunostained tissue sections was performed in a blind fashion, and recorded as score 0 (no target protein staining), score 1 (low staining), score 2 (intermediate staining), and score 3 (high staining). Results were quantified by multiplying the percentage of positive cells by the staining intensity scores (0–3), with a maximum score of 300 (3 × 100).
Immunofluorescence
Tumor tissues were fixed in 4% formalin for 24 h, embedded in paraffin, and sectioned into 3-μm-thick sections. The antigen was repaired by microwave and the endogenous catalase activity was eliminated by 3% catalase. The sections were blocked with 10% goat serum and incubated with APC anti-mouse CD8α antibody at room temperature for 60 min. The slices were washed 3 times with PBS and then incubated with DAPI at room temperature for 5 min. The slices were washed with PBS 3 times, and then sealed with antifluorescence attenuation sealing solution. Photographs were taken using a confocal microscope, and 5 visual fields were selected for each slice.
Bulk RNA-seq assay
The bulk RNA-seq assay was performed by Shbio. Total RNA was extracted using the MJzol Animal RNA Isolation Kit (Majorivd) according to the standard operating procedures provided by the manufacturer. Purification was performed using the RNAClean XP Kit (Beckman Coulter) and the RNase-Free DNase Set (QIAGEN). RNA integrity was detected by the Agilent 2100 Bioanalyzer/Agilent 4200 TapeStation (Agilent Technologies). The total amount and purity of RNA were determined by the Qubit 2.0 fluorescence quantifier (Thermo Fisher Scientific) and the NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific).
The mRNA-seq library was constructed by mRNA separation, fragmentation, first-strand cDNA synthesis, second-strand cDNA synthesis, end repair, 3′ end A addition, splicing, enrichment, and other steps of the purified total RNA. The Qubit 2.0 fluorescence quantifier (Thermo Fisher Scientific) was used to detect the library concentration, and the Agilent 4200 TapeStation (Agilent Technologies) was used to detect the library fragment distribution. Then, sequencing was performed according to the effective concentration of the library and data production requirements. The sequencing platform used Illumina NovaSeq6000, and the sequencing mode used PE150 (pin-end 150 bp); that is, double-ended sequencing was conducted, with 150 bp measured at each end.
The protocol of high-throughput sequencing was fully according to the manufacturer’s instructions (Illumina). The raw reads were filtered by Seqtk before mapping to genome using Hisat2 (version 2.0.4).36 The fragments of genes were counted using stringtie (version 1.3.3b) followed by trimmed mean of M values normalization.37,38,39,40 Significant differentially expressed genes were identified as those with a false discovery rate value above the threshold (Q < 0.05) and fold-change >2 using edgeR software.40,41
Statistical analysis
Statistical analysis was performed with the SPSS 20.0 software (IBM SPSS Statistics) or GraphPad Prism version 5.0 (GraphPad). Data are shown as means ± SDs unless otherwise indicated. The Student’s t test was used to analyze the differences between two experimental groups. Differences were considered significant at p < 0.05 (∗). Nonlinear regression analyses were used to fit curves.
Data and code availability
The data that support the findings of this study are within the article. Additional data are available from the corresponding author on request.
Acknowledgments
We thank Shanghai Zhangjiang-Biotechnology and Mabwell (Shanghai) Bioscience. This work was supported by grants from the Program for Science and Technology Innovation Talents in Universities of Henan Province (22HASTIT049), the National Key R&D Program of China (2021YFC2501700), the Open Program of International Joint Research Laboratory for Recombinant Pharmaceutical Protein Expression System of Henan (KFKTYB202204), the Natural Science Foundation of Henan (222300420266), the Key Science and Technology Program of Henan Province (232102520027 and 232102310363), the Training Plan for Young Backbone Teachers in Universities of Henan Province (2020GGJS143), the Key Scientific Research Project of Higher Education of Henan Province (22A310007 and 22B310011), the National College Students Innovation and Entrepreneurship Training Program (202310472002), and the Innovation Project of Graduate in Xinxiang Medical University (YJSCX202130Y and YJSCX202218Y).
Author contributions
Yun Yang and T.W. designed the experiments, performed the data collection and analysis, and wrote the manuscript. Yun Yang, H.W., Y.K., Yan Yang, B.Z., J.Z., J.L., and C.G. carried out the experiments. H.G. and T.W. supervised and corrected the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2023.100747.
Supplemental information
References
- 1.Chen C., Wang R., Chen X., Hou Y., Jiang J. Targeting CD47 as a Novel Immunotherapy for Breast Cancer. Front. Oncol. 2022;12:924740. doi: 10.3389/fonc.2022.924740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majeti R., Chao M.P., Alizadeh A.A., Pang W.W., Jaiswal S., Gibbs K.D., Jr., van Rooijen N., Weissman I.L. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willingham S.B., Volkmer J.P., Gentles A.J., Sahoo D., Dalerba P., Mitra S.S., Wang J., Contreras-Trujillo H., Martin R., Cohen J.D., et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao M.P., Alizadeh A.A., Tang C., Myklebust J.H., Varghese B., Gill S., Jan M., Cha A.C., Chan C.K., Tan B.T., et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010;142:699–713. doi: 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okazawa H., Motegi S.i., Ohyama N., Ohnishi H., Tomizawa T., Kaneko Y., Oldenborg P.A., Ishikawa O., Matozaki T. Negative regulation of phagocytosis in macrophages by the CD47-SHPS-1 system. J. Immunol. 2005;174:2004–2011. doi: 10.4049/jimmunol.174.4.2004. [DOI] [PubMed] [Google Scholar]
- 6.Iribarren K., Buque A., Mondragon L., Xie W., Lévesque S., Pol J., Zitvogel L., Kepp O., Kroemer G. Anticancer effects of anti-CD47 immunotherapy in vivo. Oncoimmunology. 2019;8:1550619. doi: 10.1080/2162402X.2018.1550619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R., Zhang C., Cao Y., Wang J., Jiao S., Zhang J., Wang M., Tang P., Ouyang Z., Liang W., et al. Blockade of dual immune checkpoint inhibitory signals with a CD47/PD-L1 bispecific antibody for cancer treatment. Theranostics. 2023;13:148–160. doi: 10.7150/thno.79367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candas-Green D., Xie B., Huang J., Fan M., Wang A., Menaa C., Zhang Y., Zhang L., Jing D., Azghadi S., et al. Dual blockade of CD47 and HER2 eliminates radioresistant breast cancer cells. Nat. Commun. 2020;11:4591. doi: 10.1038/s41467-020-18245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piccione E.C., Juarez S., Liu J., Tseng S., Ryan C.E., Narayanan C., Wang L., Weiskopf K., Majeti R. A bispecific antibody targeting CD47 and CD20 selectively binds and eliminates dual antigen expressing lymphoma cells. mAbs. 2015;7:946–956. doi: 10.1080/19420862.2015.1062192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Ni H., Zhou S., He K., Gao Y., Wu W., Wu M., Wu Z., Qiu X., Zhou Y., et al. Tumor-selective blockade of CD47 signaling with a CD47/PD-L1 bispecific antibody for enhanced anti-tumor activity and limited toxicity. Cancer Immunol. Immunother. 2021;70:365–376. doi: 10.1007/s00262-020-02679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y., Guo R., Chen Q., Liu Y., Zhang P., Zhang Z., Chen X., Wang T. A novel bispecific antibody fusion protein co-targeting EGFR and CD47 with enhanced therapeutic index. Biotechnol. Lett. 2018;40:789–795. doi: 10.1007/s10529-018-2535-2. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y., Yang Z., Yang Y. Potential Role of CD47-Directed Bispecific Antibodies in Cancer Immunotherapy. Front. Immunol. 2021;12:686031. doi: 10.3389/fimmu.2021.686031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barkal A.A., Brewer R.E., Markovic M., Kowarsky M., Barkal S.A., Zaro B.W., Krishnan V., Hatakeyama J., Dorigo O., Barkal L.J., Weissman I.L. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. 2019;572:392–396. doi: 10.1038/s41586-019-1456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aigner S., Sthoeger Z.M., Fogel M., Weber E., Zarn J., Ruppert M., Zeller Y., Vestweber D., Stahel R., Sammar M., Altevogt P. CD24, a mucin-type glycoprotein, is a ligand for P-selectin on human tumor cells. Blood. 1997;89:3385–3395. [PubMed] [Google Scholar]
- 15.Hosonaga M., Arima Y., Sugihara E., Kohno N., Saya H. Expression of CD24 is associated with HER2 expression and supports HER2-Akt signaling in HER2-positive breast cancer cells. Cancer Sci. 2014;105:779–787. doi: 10.1111/cas.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith S.C., Oxford G., Wu Z., Nitz M.D., Conaway M., Frierson H.F., Hampton G., Theodorescu D. The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res. 2006;66:1917–1922. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 17.Kristiansen G., Sammar M., Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J. Mol. Histol. 2004;35:255–262. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 18.Freile J.Á., Ustyanovska Avtenyuk N., Corrales M.G., Lourens H.J., Huls G., van Meerten T., Cendrowicz E., Bremer E. CD24 Is a Potential Immunotherapeutic Target for Mantle Cell Lymphoma. Biomedicines. 2022;10:1175. doi: 10.3390/biomedicines10051175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Gong Y., Tang J., Yan C., Li L., Peng W., Cheng Z., Yu R., Xiang Q., Deng C., et al. ZBTB28 inhibits breast cancer by activating IFNAR and dual blocking CD24 and CD47 to enhance macrophages phagocytosis. Cell. Mol. Life Sci. 2022;79:83. doi: 10.1007/s00018-021-04124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao C.C., Han Q.J., Ying H.Y., Gu X.X., Yang N., Li L.Y., Zhang Q.Z. TNFSF15 facilitates differentiation and polarization of macrophages toward M1 phenotype to inhibit tumor growth. Oncoimmunology. 2022;11:2032918. doi: 10.1080/2162402X.2022.2032918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y. The root cause of drug resistance in HER2-positive breast cancer and the therapeutic approaches to overcoming the resistance. Pharmacol. Ther. 2021;218:107677. doi: 10.1016/j.pharmthera.2020.107677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Shao B., Yan Y., Song G., Liu X., Wang J., Liang X. Efficacy and safety of trastuzumab combined with chemotherapy for first-line treatment and beyond progression of HER2-overexpressing advanced breast cancer. Chin. J. Cancer Res. 2016;28:330–338. doi: 10.21147/j.issn.1000-9604.2016.03.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilken J.A., Maihle N.J. Primary trastuzumab resistance: new tricks for an old drug. Ann. N. Y. Acad. Sci. 2010;1210:53–65. doi: 10.1111/j.1749-6632.2010.05782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Upton R., Banuelos A., Feng D., Biswas T., Kao K., McKenna K., Willingham S., Ho P.Y., Rosental B., Tal M.C., et al. Combining CD47 blockade with trastuzumab eliminates HER2-positive breast cancer cells and overcomes trastuzumab tolerance. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2026849118. e2026849118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunte S., Abraham J., Montero A.J. Novel HER2-targeted therapies for HER2-positive metastatic breast cancer. Cancer. 2020;126:4278–4288. doi: 10.1002/cncr.33102. [DOI] [PubMed] [Google Scholar]
- 26.Doi T., Shitara K., Naito Y., Shimomura A., Fujiwara Y., Yonemori K., Shimizu C., Shimoi T., Kuboki Y., Matsubara N., et al. Safety, pharmacokinetics, and antitumour activity of trastuzumab deruxtecan (DS-8201), a HER2-targeting antibody-drug conjugate, in patients with advanced breast and gastric or gastro-oesophageal tumours: a phase 1 dose-escalation study. Lancet Oncol. 2017;18:1512–1522. doi: 10.1016/S1470-2045(17)30604-6. [DOI] [PubMed] [Google Scholar]
- 27.Iwata T.N., Ishii C., Ishida S., Ogitani Y., Wada T., Agatsuma T. A HER2-Targeting Antibody-Drug Conjugate, Trastuzumab Deruxtecan (DS-8201a), Enhances Antitumor Immunity in a Mouse Model. Mol. Cancer Ther. 2018;17:1494–1503. doi: 10.1158/1535-7163.MCT-17-0749. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W., Huang Q., Xiao W., Zhao Y., Pi J., Xu H., Zhao H., Xu J., Evans C.E., Jin H. Advances in Anti-Tumor Treatments Targeting the CD47/SIRPalpha Axis. Front. Immunol. 2020;11:18. doi: 10.3389/fimmu.2020.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayat S.M.G., Bianconi V., Pirro M., Jaafari M.R., Hatamipour M., Sahebkar A. CD47: role in the immune system and application to cancer therapy. Cell. Oncol. 2020;43:19–30. doi: 10.1007/s13402-019-00469-5. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B., Li W., Fan D., Tian W., Zhou J., Ji Z., Song Y. Advances in the study of CD47-based bispecific antibody in cancer immunotherapy. Immunology. 2022;167:15–27. doi: 10.1111/imm.13498. [DOI] [PubMed] [Google Scholar]
- 31.Buatois V., Johnson Z., Salgado-Pires S., Papaioannou A., Hatterer E., Chauchet X., Richard F., Barba L., Daubeuf B., Cons L., et al. Preclinical Development of a Bispecific Antibody that Safely and Effectively Targets CD19 and CD47 for the Treatment of B-Cell Lymphoma and Leukemia. Mol. Cancer Ther. 2018;17:1739–1751. doi: 10.1158/1535-7163.MCT-17-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiskopf K., Ring A.M., Ho C.C.M., Volkmer J.P., Levin A.M., Volkmer A.K., Ozkan E., Fernhoff N.B., van de Rijn M., Weissman I.L., Garcia K.C. Engineered SIRPalpha variants as immunotherapeutic adjuvants to anticancer antibodies. Science. 2013;341:88–91. doi: 10.1126/science.1238856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chittezhath M., Dhillon M.K., Lim J.Y., Laoui D., Shalova I.N., Teo Y.L., Chen J., Kamaraj R., Raman L., Lum J., et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity. 2014;41:815–829. doi: 10.1016/j.immuni.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atwell S., Ridgway J.B., Wells J.A., Carter P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J. Mol. Biol. 1997;270:26–35. doi: 10.1006/jmbi.1997.1116. [DOI] [PubMed] [Google Scholar]
- 36.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 38.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are within the article. Additional data are available from the corresponding author on request.