Abstract
OBJECTIVE
Development of a systematic mutation detection assay strategy for denaturing high performance liquid chromatography (DHPLC).
DESIGN
Adaptation of Guanine and Cytosine (GC)-clamping from denaturing gradient gel electrophoresis (DGGE) to DHPLC.
METHODS
Three target sequences harboring known allelic variants were studied to develop a general DHPLC assay design strategy. These were exon 10 of the human RET (REarranged during Transfection) gene, exon 52 of the mouse Col1a2 gene, and exon 9 of the human FAS (APO-1, CD-95) gene. Available software was used to analyze melting curves and determine assay conditions. GC clamps of 20 bp or 36 bp were added to polymerase chain reaction (PCR) primers to introduce a high melting temperature (Tm) domain to each of the target molecules. DHPLC was performed under partially denaturing conditions.
RESULTS
DHPLC assays of PCR-amplified sequences can be developed using a personal computer. The following three steps allowed for mutation detection in all three targets.
The target sequence should have a uniform Tm
GC clamps of length sufficient to introduce a second melting domain with a Tm ≥ 8° above that of the target sequence should be appended to one of the primers.
The DHPLC assay should be performed at the highest temperature at which the target sequence is predicted to be ≥ 90% double stranded
CONCLUSION
Addition of GC-clamps to primers facilitates mutation detection by DHPLC.
The theoretical basis for this observation is identical to that underlying the utility of GC-clamps in DGGE.
Keywords: Heterozygote detection, Nucleic acid denaturation, Genetic screening, Polymerase chain reaction
INTRODUCTION
Efficient, robust mutation detection methods can potentially have a major impact on the diagnosis of genetic disorders and the identification of genetic contributions to multifactorial disorders. Many investigations depend on relating genotypic variations to specific phenotypes. Such assays should ideally possess several widely recognized performance characteristics.1,2
This report describes a simplified strategy for designing mutation detection assays by denaturing high performance liquid chromatography (DHPLC), based on DHPLC's ability to separate partially denatured double stranded DNA molecules. Particular attention is devoted to the use of GC clamps with this system, allowing assay design to be carried out in a nearly algorithmic fashion. This approach allows investigators to shift their efforts from mutation detection assay design to interpretation of the biological consequences of detected sequence variation.
MATERIALS AND METHODS
DNA preparation
Mouse and human DNA were prepared from tail and peripheral blood mononuclear cells using the Invitrogen (Carlsbad, CA) and Puregene (Minneapolis, MN) DNA extraction kits, respectively. Human subjects provided written informed consent under protocols approved by the Memorial Sloan-Kettering Cancer Center's IRB3,4 or the Hospital for Special Surgery's IRB.5–7
PCR
All amplifications were performed in 50 µL reactions including 1 U of Red Taq DNA Polymerase (Sigma, St. Louis, MO), 0.2 mM dNTP mix (Amersham Pharmacia, Piscataway, NJ), and 10 mM Tris-HCl (pH 8.3) containing 50 mM KCl, 1.5 mM MgCl2, 0.1% gelatin, and 20 to 50 ng of genomic DNA. Primers were designed using Primer 0.5 software.8 Reactions were performed in a PE Biosystems (Norwalk, CT) 2400 thermal cycler. Amplifications were carried out by an initial denaturation of 3 minutes at 95°C, followed by 35 cycles of 94°C x 30 seconds, Tannealing x 30 seconds, and 72°C x 30 seconds, and concluded by a final denaturation at 95°C x 5 minutes followed by slow cooling to room temperature. Primer sequences and annealing temperatures are as follows:
clamp 20 = GCGGCCCGCCGCCCCCGCCG clamp 36 = CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCG9, FOIM = GAAATGGCTTTCCTAGACCCCG, ROIM = AATGATTGTCTTGCCCCATTCA with Tannealing = 60°C, primers HFAS47 and HFAS495 and primers FRET10 and RRET1010 and their annealing temperatures were previously published.
DHPLC conditions
Loading, elution and washing of the DHPLC column was carried out with varying combinations of three buffers injected at a flow rate of 0.9 ml/min: Buffer A contains 100 mM triethylamine acetate (TEAA), pH 7.0 and 0.025% acetonitrile, Buffer B contains 25% acetonitrile, 100 mM TEAA, pH 7.0, and 0.1 mM EDTA, and Buffer D contains 75% acetonitrile. Loading and elution buffers are combinations of buffers A and B, whose relative proportions form a gradient over a specified time interval. Buffer D is used to wash the column. DHPLC elution buffer gradients were generated by WAVEMAKER version 3.3.3 software (Transgenomic, Omaha, NE) and are reported as % buffer B at specified times. Assays were performed using the WAVE DNA fragment analysis system (Transgenomic, Omaha, NE). Oven temperature was determined from inspection of the melting profile, choosing the highest temperature at which the target sequence was predicted to be >90% duplex. Following the gradient elution, all remaining bound material was washed from the column for 36 seconds with buffer D and the column was re-equilibrated with the loading buffer for 156 seconds. Sample elution was monitored by absorbance at 260 nm.
DNA Sequencing
PCR products were purified by passage through Microcon microconcentrating centrifugal filter columns (Millipore, Bedford, MA) prior to sequencing. Sequencing reactions were performed using the Applied Biosystems Dye-Terminator Kit and analyzed on a ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA).
RESULTS
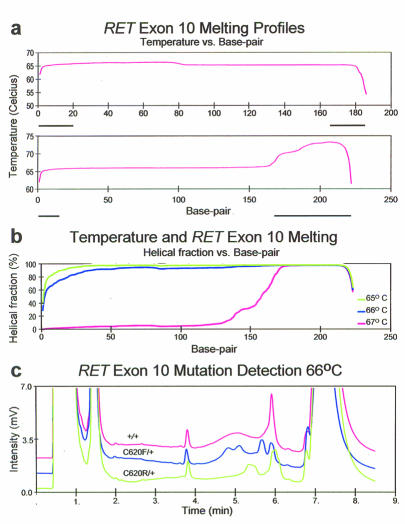
First, an established denaturing gradient gel electrophoresis (DGGE) assay to DHPLC was adapted, since both DGGE and DHPLC-based mutation detection operate on the principle of detecting differences in melting behavior among individual species in a mixed population of DNA molecules. Previously a GC-clamped DGGE assay was developed to detect pathogenic mutations in exon 10 of the RET protooncogene,3,4 this was used as the starting point in adapting GC-clamping to DHPLC. Without a GC clamp, the target sequence has a single, uniform melting domain, with a predicted Tm between 66°C (>90% double stranded) and 67°C (>80% single stranded). Addition of a 36 bp GC clamp introduces a second higher Tm melting domain (figures 1A and 1B). The target sequence therefore denatures under conditions in which the GC clamp remains double-stranded. The WAVEMAKER-generated elution profile was loading in 44% buffer B, 49% buffer B at 30 seconds, and 58% buffer B at 300 seconds. The software recommended 67°C for assay performance. RET exon 10 was amplified from subjects harboring the C620F and C620R mutations, suffering from familial medullary carcinoma of the thyroid and MEN 2A, respectively, and from unaffected family members.4 Mutations were detected at 66°C (figure 1C) and 67°C (not shown), but not at 65°C or 68°C (not shown) when the GC clamp was included. At 65°C a single sharp peak was seen, regardless of genotype, while at 68° all genotypes eluted as a broad, low intensity peak. Resolution of heteroduplex peaks was clearer at 66°C than at 67°C. These mutations were resolved at 60°C using a 20% to 60% gradient by DGGE.3 Mutations were not detected either by DHPLC or DGGE when the GC clamp was not used (not shown). At 65°C and 66°C, unclamped samples elute as a single sharp peak regardless of genotype, while at 67°C and 68°C they elute as a broad, low-amplitude peak regardless of genotype.
Figure 1.
A. Wavemaker-generated melting profiles for RET exon 10 without (top) and with (bottom) inclusion of a 36 base GC clamp on the 3′ amplification primer. The bold lines under the base pair axis indicate the extent of the primer sequences.
B. Predicted melting behavior of GC clamped RET10 amplicons at 65°C, 66°C, and 67°C. The GC clamp remains double stranded at all 3 temperatures. The target sequence is ≥ 90% double stranded at 65°C and 66°C, but not at 67°C.
C. CDHPLC elution profiles for 3 RET genotypes at 66°. Heterozygotes harboring the C620F and C620R mutations are readily distinguished from normals and from each other.
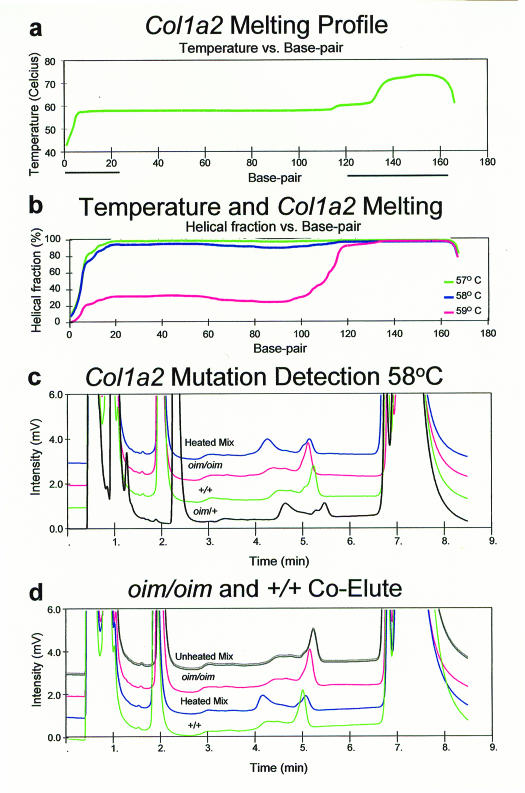
Next we designed a mutation detection assay for the mouse Col1a2oim mutation; a one-base pair deletion of a G residue in exon 52, encoding the c-terminal propeptide of the α2 chain of type 1 procollagen.11,12 The melting profile was determined to be uniform in the target region (figure 2A). The target sequence is >90% double stranded at 58°C but not at 59°C, so assays were conducted at 58°C (figure 2B). Gradient conditions were injection in 43% buffer B, 48% buffer B at 30 seconds, and 57% buffer B at 300 seconds. Heterozygotes were readily distinguished from either wild-type or mutant homozygotes when a 36 bp GC clamp was included (figure 2C), but not when it was omitted (not shown). As in the case of RET exon 10, the unclamped products all elute as single sharp peaks at 58°C, a temperature at which the molecule is predicted to be double stranded. We did not attempt to resolve this target at other temperatures. A 50:50 mix of wild-type and mutant homozygous samples eluted as a single sharp peak when unheated, but eluted as a pair of peaks when heated to 95°C and cooled slowly (figure 2D). This demonstrates that assay conditions are robust for detecting heterozygosity, but are not adequate for determining the allele present in homozygous samples. Moreover, the behavior of the unheated mixture reveals that there is no difference in the elution times of the wild-type and mutant products, as the peak is not broadened relative to the peak generated by homozygous samples. This finding indicates that decreased Tm at the site of mismatch is a critical element of mutation detection by DHPLC.
Figure 2.
A. Wavemaker-generated Col1a2 GC clamped melting profile. The bold lines under the base pair axis indicate the extent of the primer sequences.
B. The predicted melting behavior of Col1a2 amplicons at 57°C, 58°C, and 59°C.
C. Resolution of Col1a2 heterozygotes from homozygotes. A Col1a2oim/+ heterozygote is separated from either wild type or oim/oim homozygotes. A denatured and slowly reannealed mixture of the wild type and oim/oim PCR products produces an elution profile identical to that of the oim/+ heterozygote.
D. Inability to resolve +/+ homozygotes from oim/oim homozygotes. Both homozygotes and an unheated mixture of oim/oim and +/+ PCR products result in single elution peaks. However, heating the mixture to denature the DNA and allowing it to reanneal slowly allows heteroduplex formation and results in the appearance of a second elution peak, as for oim/+ heterozygotes.
Homozygous samples can be genotyped in a two-step assay.
The assay is performed on a pure sample
Samples found to be homozygous are mixed with a reference wild-type sample, denatured, reannealed and reanalyzed
Mutant homozygotes will yield a heterozygous pattern on mixing, while wild-type homozygotes will retain a homozygous pattern on mixing.
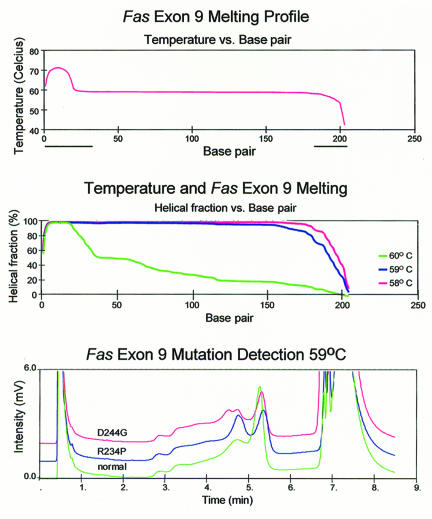
The same strategy was applied for the detection of the D244G and R234P missense mutations in exon 9 of the human FAS (APO-1, CD95) gene, which cause the Canale-Smith Syndrome5,7. The melting profile predicts that 59°C is the highest temperature at which the target sequence is >90% double stranded (figures 3A and 3B). DHPLC analysis at 59°C with injection at 45% buffer B and elution with a gradient profile of 50% buffer B at 30 seconds, and 59% at 300 seconds is shown in figure 3C. The mutations were readily detected with a 20 bp GC clamp, while omission of the GC clamp did not allow us to detect the mutations under these conditions, with all genotypes eluting as a single sharp peak (not shown).
Figure 3.
A. Wavemaker-generated FAS exon 9 GC clamped melting profile.
B. Temperature and FAS Exon 9 Melting. The predicted melting behavior of FAS amplicons at 58°C, 59°C and 60°C.
C. Resolution of R234P and D244G heterozygotes from a normal homozygote.
DISCUSSION
Sheffield and colleagues demonstrated the utility of GC clamps in designing DGGE assays,9 showing that the inclusion of a clamp allowed detection of mutations in the murine βmajor globin gene that were undetectable without clamping. The additional sensitivity arose from detection of mutations in the highest Tm domain of the target sequence. Subsequent sensitivity analyses of DGGE have consistently found that analyses performed with GC clamps have sensitivities of ∼95% and specificities approaching unity.13–16 The theoretical basis for this increase in sensitivity is that resolution of the various molecular species occurs primarily when individual molecules are partially denatured. Inclusion of a GC clamp provides an artificial, high Tm domain in the molecules being analyzed, allowing the target sequence to occur in the context of a low Tm domain. The data suggest that resolution of the various molecular species by DHPLC also occurs primarily when these have partially denatured, so that the principles guiding design of DGGE assays can be applied to DHPLC mutation detection.
Heteroduplexes include mismatches of one or more base pairs, resulting in early melting of the mismatched region. DHPLC allows resolution of heteroduplex from homoduplex DNA molecules based on differences in their retention time in the column under partially denaturing conditions, with heteroduplex molecules eluting earlier. This behavior mirrors heteroduplexes' lower Tm's and is similar to retardation of electrophoretic transport at different points along a denaturing gradient gel. In DGGE, mutations in the highest Tm domain are resolved poorly, if at all. It was determined that while the analytical technology differs in DHPLC, the principle of heteroduplex detection should be the same as in DGGE and that inclusion of GC clamps would facilitate mutation detection. The inability to detect sequence variants in the highest Tm domain may explain the anecdotally poor ability of DHPLC to detect mutations in short amplicons (K. Hecker, personal communication), which often include only a single melting domain.
A basic and general approach to designing new mutation screening assays by DHPLC was established. First, the target region's sequence is evaluated for uniformity of Tm. If this condition is not satisfied, alternative primers are chosen in order to generate a target sequence with a uniform Tm. Second, a GC clamp of sufficient length to create a high-Tm domain is appended to one of the primers. Once again, the predicted melting curve is inspected, to ensure that the clamp produces the desired high Tm domain, as illustrated in the “A” panels of the figures. Melting curves of the PCR product are calculated and the highest temperature at which the target sequence is >90% duplex is chosen for assay performance, as illustrated in the “B” panels of the figures. WAVEMAKER software, native to our DHPLC instrument, allowed both the calculation of melting profiles and generation of elution profiles. When using this software, inspection of the melting profiles is necessary. WAVEMAKER sometimes suggests an assay temperature that is too high for optimal resolution of sequence variation. Freely accessible programs perform equivalent calculations and give equivalent results.17–20
The trio of assays described here demonstrate that inclusion of a GC clamp allows detection of mutations by DHPLC under conditions that can be fully established prior to assay performance, using predicted melting curves. The experiments presented above do not exclude the development of assays without GC clamps, but rather show that the conditions identified in silico work well without further optimization when GC clamps are included. Unclamped primers for RET exon 10 were tested at four temperatures. The other two assays were tested at a single temperature. We did not attempt to optimize gradient conditions. It was expected that such optimization would likely have allowed mutation detection, even with unclamped molecules.
In the assays described here, the GC clamp had a melting point 8°C to 13°C higher than the target sequence, while the target sequence varied in Tm by no more than 3°C. Therefore, investigators should include GC clamp sequences sufficiently long to provide an 8°C increment in Tm relative to the target. The required length will vary according to the Tm of the target sequence, but it is straightforward to determine the minimum necessary length with melting profile software prior to ordering modified primers.
Theoretically, psoralen clamping provides an alternative to GC clamping by generating a covalent interstrand bond rather than by raising the Tm of a region of the target molecule.21 The covalent bond introduced by psoralenation and subsequent UV light exposure effectively clamps one end of the molecule regardless of temperature. The advantage of psoralen compared to GC clamping is that the clamp need not be changed from assay to assay; the disadvantage is that an additional UV crosslinking step must be added prior to analysis. However, experiments with psoralen clamps were not conducted, as a result it is not known if this approach would perform as well in practice as in theory.
Electrophoretic mutation detection assays are difficult to perform on a large scale, since casting and loading gels are tasks that are difficult to automate. Because DGGE and Single Strand Conformation Polymorphism (SSCP) suffer from these limitations, the DHPLC-based assay is more readily adaptable to even a medium-throughput setting. The WAVE system can load samples automatically from a 96-sample block. Each sample is processed in approximately 6 to 9 minutes. The automation provided by the WAVE system vs. the gel-based methods, allows investigators to perform mutation screens at higher throughput and with less hands-on time. Further improvements in throughput might be achieved by pooling samples prior to DHPLC analysis, as has been done for gel-based methods.22 These scale issues are most relevant to research laboratories engaged in mutation screening and genotype-phenotype correlation, and may become applicable to diagnostic clinical laboratories as well.
The assays we describe here scan relatively short target sequences and therefore cannot estimate the maximum length over which DHPLC based methods might work. High sensitivity and specificity have been reported for DHPLC in a growing body of analyses for sequences up to 400 bp in length.23–32 Our approach to primer design has a shorter practical limit of about 250 bp, imposed by our criterion that target sequences have a uniform Tm.
Other approaches may lead to further improvements in mutation scanning methodology. For example, two groups have recently described a purely optical assay strategy that obviates the need for physical separation of homoduplex and heteroduplex molecules.33,34 This approach may prove applicable to large-scale genotyping applications, but has yet to be widely validated.
The mutation detection strategy presented in this article leads to higher primer costs than other approaches, because of the need for either GC-clamped or psoralen-clamped primers, and shorter target sequences. At an estimated incremental cost of $0.70/base x 30 bases for “ordinary” scale primer synthesis, the extra primer costs for a typical length GC clamp is about $21. Psoralen clamps are more expensive than GC-clamps, requiring both an arbitrary primer adaptor arm and the psoralenation, thus adding about $40 to the cost of each clamped primer. Theoretically, psoralen primers could be reserved for the most GC-rich targets, in which GC clamps of practical length would not impact Tm. Costs arising from extra primer pairs needed as a result of studying shorter target sequences are less easily quantified and depend in part on the scope of the length of DNA to be scanned for mutations and the nature of its sequence. At an average cost of $50/GC clamped primer pair, addition of 10 additional primer pairs leads to a relatively small incremental cost. These are trivial costs compared with the time of technical personnel, whose salaries will typically exceed $10/hour. Therefore, even modest reductions in assay development time lead to net cost savings.
The greatest limitation on mutation scanning throughput, however, remains the time necessary to design mutation detection assays. Several previous reports addressed DHPLC assay design optimization.18,30,31,35,36 These authors all recommend performing assays at various temperatures to maximize sensitivity and some narrow the buffer gradient ranges, requiring a still greater number of temperatures. Another report37 suggests that a gradual decrease in Tm from the clamp to the free end of a molecule is the optimal configuration for mutation detection by heteroduplex-based methods. We have demonstrated that inclusion of GC clamps obviates the need for tedious assay optimization. GC clamps of sufficient length to create a domain with Tm 8°C greater than that of the target sequence allows an experimenter to develop a working assay in less than one hour. As genomic analysis moves from determination of additional sequence data to understanding the biological consequences of sequence variation, the ability to perform mutation screening efficiently will become ever more valuable. The algorithm for establishing DHPLC conditions described here will facilitate this.
CONCLUSION
Assay design principles adapted from those previously established for DGGE facilitate development of DHPLC mutation scanning strategies, as both methods resolve sequence variants by virtue of differences in the melting behavior of partially denatured DNA molecules. Use of GC clamps in DHPLC obviates the need for empirical optimization of new assays. The use of clamps is preferable to evaluation of a greater number of assay conditions for two reasons. First, unlike the situation with model assays such as those we have used for illustration, investigators performing mutation screens will not have positive controls available to test conditions. Second, the alternative strategy of varying conditions to increase sensitivity depends primarily on varying assay temperature. Heating and cooling the instrument oven is slow, and the time needed for temperature equilibration eliminates much of the time advantage arising from DHPLC.
Acknowledgments
The authors thank Dr. Nancy Camacho for provision of DNA samples and Drs. Adele Boskey, Nancy Camacho, Paul Fogle, Karl Hecker and Misi Robinson for critical review and editorial consultation of the manuscript.
Contributor Information
Robert J. Wurzburger, Research Division, Hospital for Special Surgery, New York, New York.
Rajarsi Gupta, Laboratory for Fluorescence Dynamics, Physics Department, University of Illinois, Urbana-Champaign, Urbana, Illinois.
Andrew P. Parnassa, Department of Microbiology and Immunology, Weill Medical College, Cornell University, New York, New York.
Sargam Jain, State University of New York at Stony Brook, School of Medicine, Stony Brook, New York.
Jason A. Wexler, Division of Endocrinology, Clinical Nutrition and Vascular Medicine, University of California, Davis Medical Center, Sacramento, California.
Jia Li Chu, Division of Rheumatology, University of Washington, Seattle, Washington.
Keith B. Elkon, Division of Rheumatology, University of Washington, Seattle, Washington.
Robert D. Blank, Section of Endocrinology, Department of Medicine, University of Wisconsin-Madison, Madison, Wisconsin and Geriatrics Research, Education and Clinical Center, William S. Middleton Veterans Hospital, Madison, Wisconsin.
References
- 1.Grompe M. The rapid detection of unknown mutations in nucleic acids. Nat Genet. 1993;5:111–117. doi: 10.1038/ng1093-111. [DOI] [PubMed] [Google Scholar]
- 2.Cotton RG. Slowly but surely towards better scanning for mutations. Trends Genet. 1997;13:43–46. doi: 10.1016/s0168-9525(97)01011-1. [DOI] [PubMed] [Google Scholar]
- 3.Blank RD, Sklar CA, Martin ML. Denaturing gradient gel electrophoresis to diagnose multiple endocrine neoplasia type 2. Clin Chem. 1996;42:598–603. [PubMed] [Google Scholar]
- 4.Blank RD, Sklar CA, Dimich AB, LaQuaglia MP, Brennan MF. Clinical presentations and RET protooncogene mutations in seven multiple endocrine neoplasia type 2 kindreds. Cancer. 1996;78:1996–2003. [PubMed] [Google Scholar]
- 5.Drappa J, Vaishnaw AK, Sullivan KE, Chu JL, Elkon KB. Fas gene mutations in the Canale-Smith syndrome, an inherited lymphoproliferative disorder associated with autoimmunity. N Engl J Med. 1996;335:1643–1649. doi: 10.1056/NEJM199611283352204. [DOI] [PubMed] [Google Scholar]
- 6.Vaishnaw AK, Toubi E, Ohsako S, Drappa J, Buys S, Estrada J, Sitarz A, Zemel L, Chu JL, Elkon KB. The spectrum of apoptotic defects and clinical manifestations, including systemic lupus erythematosus, in humans with CD95 (Fas/APO-1) mutations. Arthritis Rheum. 1999;42:1833–1842. doi: 10.1002/1529-0131(199909)42:9<1833::AID-ANR7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 7.Vaishnaw AK, Orlinick JR, Chu JL, Krammer PH, Chao MV, Elkon KB. The molecular basis for apoptotic defects in patients with CD95 (Fas/Apo-1) mutations. J Clin Invest. 1999;103:355–363. doi: 10.1172/JCI5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lincoln SE, Daly MJ, Lander ES. Primer: a computer program for automatically selecting PCR primers. Cambridge, MA: Whitehead Institute for Biomedical Research; 1991. [Google Scholar]
- 9.Sheffield VC, Cox DR, Lerman LS, Myers RM. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donis-Keller H, Dou S, Chi D, Carlson KM, Toshima K, Lairmore TC, Howe JR, Moley JF, Goodfellow P, Wells SA., Jr Mutations in the RET proto-oncogene are associated with MEN 2A and FMTC. Hum Mol Genet. 1993;2:851–856. doi: 10.1093/hmg/2.7.851. [DOI] [PubMed] [Google Scholar]
- 11.Chipman SD, Sweet HO, McBride DJ, Jr, Davisson MT, Marks SC, Jr, Shuldiner AR, Wenstrup RJ, Rowe DW, Shapiro JR. Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: a model of human osteogenesis imperfecta. Proc Natl Acad Sci U S A. 1993;90:1701–1705. doi: 10.1073/pnas.90.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride DJ, Jr, Shapiro JR. Confirmation of a G nucleotide deletion in the Cola-2 gene of mice with the osteogenesis imperfecta mutation. Genomics. 1994;20:135–137. doi: 10.1006/geno.1994.1141. [DOI] [PubMed] [Google Scholar]
- 13.Guldberg P, Henriksen KF, Guttler F. Molecular analysis of phenylketonuria in Denmark: 99% of the mutations detected by denaturing gradient gel electrophoresis. Genomics. 1993;17:141–146. doi: 10.1006/geno.1993.1295. [DOI] [PubMed] [Google Scholar]
- 14.Nissen H, Petersen NE, Mustajoki S, Hansen TS, Mustajoki P, Kauppinen R, Horder M. Diagnostic strategy, genetic diagnosis and identification of new mutations in intermittent porphyria by denaturing gradient gel electrophoresis. Hum Mutat. 1997;9:122–130. doi: 10.1002/(SICI)1098-1004(1997)9:2<122::AID-HUMU4>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 15.Macek M, Jr, Mercier B, Mackova A, Miller PW, Hamosh A, Ferec C, Cutting GR. Sensitivity of the denaturing gradient gel electrophoresis technique in detection of known mutations and novel Asian mutations in the CFTR gene. Hum Mutat. 1997;9:136–147. doi: 10.1002/(SICI)1098-1004(1997)9:2<136::AID-HUMU6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Gejman PV, Cao Q, Guedj F, Sommer S. The sensitivity of denaturing gradient gel electrophoresis: a blinded analysis. Mutat Res. 1998;382:109–114. doi: 10.1016/s1383-5726(98)00002-8. [DOI] [PubMed] [Google Scholar]
- 17.Oefner PJ, Underhill PA. Current Protocols in Human Genetics. New York: Wiley & Sons; 1998. DNA mutation detection using denaturing high-performance liquid chromatography (DHPLC) pp. 7.10.1–7.10.12. [DOI] [PubMed] [Google Scholar]
- 18.Jones AC, Austin J, Hansen N, Hoogendoorn B, Oefner PJ, Cheadle JP, O'Donovan MC. Optimal temperature selection for mutation detection by denaturing HPLC and comparison to single-stranded conformation polymorphism and heteroduplex analysis. Clin Chem. 1999;45:1133–1140. [PubMed] [Google Scholar]
- 19.Hansen NF, Oefner PJ. DHPLC Melt. Stanford Genome Technology Center; 2002. [Google Scholar]
- 20.Lerman LS, Silverstein K, Fripp W, Sauer P, Dresselhaus C. Melt94. Cambridge, MA: MIT; 1994. [Google Scholar]
- 21.Costes B, Girodon E, Ghanem N, Chassignol M, Thuong NT, Dupret D, Goossens M. Psoralen-modified oligonucleotide primers improve detection of mutations by denaturing gradient gel electrophoresis and provide an alternative to GC-clamping. Hum Mol Genet. 1993;2:393–397. doi: 10.1093/hmg/2.4.393. [DOI] [PubMed] [Google Scholar]
- 22.Zarbl H, Aragaki C, Zhao LP. An efficient protocol for rare mutation genotyping in a large population. Genet Test. 1998;2:315–321. doi: 10.1089/gte.1998.2.315. [DOI] [PubMed] [Google Scholar]
- 23.Liu WO, Oefner PJ, Qian C, Odom RS, Francke U. Denaturing HPLC-identified novel FBN1 mutations, polymorphisms, and sequence variants in Marfan syndrome and related connective tissue disorders. Genet Test. 1997–98;1:237–242. doi: 10.1089/gte.1997.1.237. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Smith DI, Rechtzigel KJ, Thibodeau SN, James CD. Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acids Res. 1998;26:1396–1400. doi: 10.1093/nar/26.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Donovan MC, Oefner PJ, Roberts SC, Austin J, Hoogendoorn B, Guy C, Speight G, Upadhyaya M, Sommer SS, McGuffin P. Blind analysis of denaturing high-performance liquid chromatography as a tool for mutation detection. Genomics. 1998;52:44–49. doi: 10.1006/geno.1998.5411. [DOI] [PubMed] [Google Scholar]
- 26.Choy YS, Dabora SL, Hall F, Ramesh V, Niida Y, Franz D, Kasprzyk-Obara J, Reeve MP, Kwiatkowski DJ. Superiority of denaturing high performance liquid chromatography over single-stranded conformation and conformation-sensitive gel electrophoresis for mutation detection in TSC2. Ann Hum Genet. 1999;63:383–391. doi: 10.1046/j.1469-1809.1999.6350383.x. [DOI] [PubMed] [Google Scholar]
- 27.Gross E, Arnold N, Goette J, Schwarz-Boeger U, Kiechle M. A comparison of BRCA1 mutation analysis by direct sequencing, SSCP and DHPLC. Hum Genet. 1999;105(1–2):72–78. doi: 10.1007/s004399900092. [DOI] [PubMed] [Google Scholar]
- 28.Wagner T, Stoppa-Lyonnet D, Fleischmann E, Muhr D, Pages S, Sandberg T, Caux V, Moeslinger R, Langbauer G, Borg A, Oefner P. Denaturing high-performance liquid chromatography detects reliably BRCA1 and BRCA2 mutations. Genomics. 1999;62:369–376. doi: 10.1006/geno.1999.6026. [DOI] [PubMed] [Google Scholar]
- 29.Dobson-Stone C, Cox RD, Lonie L, Southam L, Fraser M, Wise C, Bernier F, Hodgson S, Porter DE, Simpson AH, Monaco AP. Comparison of fluorescent single-strand conformation polymorphism analysis and denaturing high-performance liquid chromatography for detection of EXT1 and EXT2 mutations in hereditary multiple exostoses. Eur J Hum Genet. 2000;8:24–32. doi: 10.1038/sj.ejhg.5200409. [DOI] [PubMed] [Google Scholar]
- 30.Buyse IM, Fang P, Hoon KT, Amir RE, Zoghbi HY, Roa BB. Diagnostic testing for Rett syndrome by DHPLC and direct sequencing analysis of the MECP2 gene: identification of several novel mutations and polymorphisms. Am J Hum Genet. 2000;67:1428–1436. doi: 10.1086/316913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takashima H, Boerkoel CF, Lupski JR. Screening for mutations in a genetically heterogeneous disorder: DHPLC versus DNA sequence for mutation detection in multiple genes causing Charcot-Marie-Tooth neuropathy. Genet Med. 2001;3:335–342. doi: 10.1097/00125817-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Genet Med 2001 Sep–Oct;3(5):335–42 Denaturing high-performance liquid chromatography (DHPLC) is a highly sensitive, semi-automated method for identifying mutations in the TSC1 gene. J Biochem Biophys Methods. 2001;47:33–37. doi: 10.1016/s0165-022x(00)00149-4. [DOI] [PubMed] [Google Scholar]
- 33.Lipsky RH, Mazzanti CM, Rudolph JG, Xu K, Vyas G, Bozak D, Radel MQ, Goldman D. DNA melting analysis for detection of single nucleotide polymorphisms. Clin Chem. 2001;47:635–644. [PubMed] [Google Scholar]
- 34.Akey JM, Sosnoski D, Parra E, Dios S, Hiester K, Su B, Bonilla C, Jin L, Shriver MD. Melting curve analysis of SNPs (McSNP): a gel-free and inexpensive approach for SNP genotyping. Biotechniques. 2001;30:358, 362, 364, 366–367. doi: 10.2144/01302tt05. [DOI] [PubMed] [Google Scholar]
- 35.Erlandson A, Stibler H, Kristiansson B, Wahlstrom J, Martinsson T. Denaturing high-performance liquid chromatography is a suitable method for PMM2 mutation screening in carbohydrate-deficient glycoprotein syndrome type IA patients. Genet Test. 2000;4:293–297. doi: 10.1089/10906570050501533. [DOI] [PubMed] [Google Scholar]
- 36.Antonarakis ES, Sampson JR, Cheadle JP. Temperature modulation of DHPLC analysis for detection of coexisting constitutional and mosaic sequence variants in TSC2. J Biochem Biophys Methods. 2002;51:161–164. doi: 10.1016/s0165-022x(02)00011-8. [DOI] [PubMed] [Google Scholar]
- 37.Gille C, Gille A. TGGE-STAR: primer design for melting analysis using PCR gradient gel electrophoresis. Biotechniques. 2002;32:264, 266, 268. doi: 10.2144/02322bm06. [DOI] [PubMed] [Google Scholar]