Abstract
The ability to predict a protein's three-dimensional conformation represents a crucial starting point for investigating evolutionary connections with other members of the corresponding protein family, examining interactions with other proteins, and potentially utilizing this knowledge for the purpose of rational drug design. In this work, we evaluated the feasibility of improving AlphaFold2’s three-dimensional protein predictions by developing a novel pipeline (AlphaMod) that incorporates AlphaFold2 with MODELLER, a template-based modeling program. Additionally, our tool can drive a comprehensive quality assessment of the tertiary protein structure by incorporating and comparing a set of different quality assessment tools. The outcomes of selected tools are combined into a composite score (BORDASCORE) that exhibits a meaningful correlation with GDT_TS and facilitates the selection of optimal models in the absence of a reference structure. To validate AlphaMod's results, we conducted evaluations using two distinct datasets summing up to 72 targets, previously used to independently assess AlphaFold2's performance. The generated models underwent evaluation through two methods: i) averaging the GDT_TS scores across all produced structures for a single target sequence, and ii) a pairwise comparison of the best structures generated by AlphaFold2 and AlphaMod. The latter, within the unsupervised setups, shows a rising accuracy of approximately 34% over AlphaFold2. While, when considering the supervised setup, AlphaMod surpasses AlphaFold2 in 18% of the instances. Finally, there is an 11% correspondence in outcomes between the diverse methodologies. Consequently, AlphaMod’s best-predicted tertiary structures in several cases exhibited a significant improvement in the accuracy of the predictions with respect to the best models obtained by AlphaFold2. This pipeline paves the way for the integration of additional data and AI-based algorithms to further improve the reliability of the predictions.
Keywords: Protein structure prediction, Deep learning, MODELLER, AlphaFold 2
Graphical Abstract

Highlights
-
•
We have combined AlphaFold2 with MODELLER into an integrated pipeline.
-
•
We have added a module that can perform the evaluation of the predicted structure.
-
•
Results show an improvement in the accuracy of the predictions made by AlphaFold2.
1. Introduction
Proteins play a crucial role in various biological processes, and their proper fold is essential for the correct functioning of biological systems. Understanding the three-dimensional (3D1) structure of a protein holds significant importance in elucidating its function, exploring evolutionary connections with other members of the same protein family [1], investigating interactions with other proteins and/or macromolecules both within and outside cells [2], and potentially applying this knowledge in diverse fields, including rational drug design [3]. Nevertheless, the determination of a protein structure by experimental methods is not at all straightforward, and each technique presents challenges that significantly increase the time and cost required to resolve structures [4]. Hence, until now, only a minute fraction of approximately 0.02% of the distinct entries in the UniProtKB database (a globally comprehensive and openly accessible resource of protein sequences and associated functional information) [5], has been linked to an experimentally determined 3D structure available in the wwPDB [6], the primary archive for 3D macromolecular structure data.
In the last decades, several methods were developed to predict the protein structures starting from their sequences. These methods can be divided into two main approaches. The ”template-based approach” needs to identify at least one known structure of a representative member of the structural family to which the protein of interest belongs. This structure is used as a template to build a model of the unknown structure, first by identifying structurally equivalent residues between the target sequence and the template, and then by creating the 3D coordinates of the unknown protein structure, following either of two main procedures: i. the assembly of rigid bodies, a sort of ”copy-and-paste” of the equivalent structural elements’ coordinates extracted from the template and mapped onto the target sequence, followed by relaxation of the assembly obtained; ii. the extraction of spatial restraints, in which the distances among residues in the template that are assumed to be spatially equivalent to the aligned residues in the target sequence are used as restraints, and the final model is obtained by minimizing the violations of all these restraints [7]. The most popular program that applies this last protocol is MODELLER [8]. In this program, the spatial relationships of distances and angles are expressed as conditional probability density functions (PDFs) and can be used directly as spatial restraints. The model is obtained by optimizing the objective function by employing methods of conjugate gradients and molecular dynamics with simulated annealing.
The second approach to model the unknown structure associated with a protein sequence is the “template-free modeling approach”. Most of the programs that deal with this approach assemble, with different strategies, fragments of known proteins and perform refinement of the assembled model in an iterative way [9]. The performances of these approaches remained stagnant until a few years ago, when the introduction of deep learning methods allowed to impressively improve the accuracy of the prediction of even hard targets. This led to the unexpected ”gigantic leap” of AlphaFold 2 (AF2) [10] at the 14th round of CASP, the biennial Critical Assessment of protein Structure Prediction, in which the best predictions made by the software developed by DeepMind reached a GDT_TS score [11] above 90%, meaning that their accuracy reached the level of experimental structures [12]. Less than 6 months after this terrific result, the authors of AF2 provided to the scientific community the predicted structures of more than 300,000 proteins, including 98.5% of the human proteome [13] plus the complete proteomes of other 20 model organisms [14]. And, one year later, the AlphaFold protein structure database (AlphaFoldDB, https://alphafold.ebi.ac.uk/), jointly developed by DeepMind and EMBL-EBI was released, which contains the structures of over 200 million proteins, i.e., nearly all the known sequences contained in the UniProt database at that date [15].
Subsequently, several research groups have worked on developing alternative protein structure predictors based on various deep learning approaches. Among these, particularly noteworthy achievements in performance have been observed with strategies grounded in natural language processing [16], [17]. Moreover, the possibility of refining the models obtained by AF2 predictions with other approaches has been explored, either by using traditional approaches such as molecular dynamics (MD) simulations, or by using a deep learning framework adding estimates of per-residue accuracy and residue-residue distances. The analysis of the results suggested that MD-based approaches perform best on the smallest targets, but for larger targets the addition of a deep learning framework outperforms the traditional, physically based approaches [18]. The motivation behind investigating the integration of traditional modeling procedures with deep learning-based strategies lies in the pursuit of further enhancing the predictive capabilities of protein structure prediction methods. Indeed, recent analyses showed that the AI-predicted structures performed constantly worse than experimental PDB structures in high-throughput docking experiments [19]. Therefore, while deep learning approaches have shown remarkable advancements in this field, it is essential to explore the potential synergistic effects that combining these techniques with more established methods might offer. Furthermore, the integration with these modeling procedures opens new possibilities for handling additional structural information that may not be easily integrated within a closed system like AF2.
For these reasons, we integrated AF2 with MODELLER, creating a sequential pipeline that we called AlphaMod. Furthermore, we designed an evaluation module for the pipeline, which integrates many different metrics into a unified scoring system. AlphaMod's efficacy was evaluated through its deployment on a curated test set from the CASP14 evaluation [20]. Moreover, our test extends to another independent protein dataset that was previously analyzed to enhance AF2's performance [21]. In both test sets, we evaluated our framework with the GDT_TS score serving as the principal metric for result assessment against AF2 and MODELLER, alone, and we provided an additional layer of analytical insight serving as an instrumental criterion in the selection of optimally folded templates.
In the Methods section (Section 2), we detail the techniques and methodologies employed to develop AlphaMod for predicting protein structures. The Results and discussion section (Section 3) presents our findings and comparisons over two well-known test sets. Furthermore, an in-depth analysis of the outcomes is shown. Finally, in the Conclusions section (Section 4), we summarize the key takeaways from our research and discuss potential future directions.
2. Methods
In this section, the Section 2.1, Test sets, details the datasets used for evaluating our protein structure prediction tool, AlphaMod. Subsequently, Section 2.2, Pipeline description, outlines the design, execution and performance evaluation of the pipeline used to refine AF2's results with MODELLER, emphasizing a diverse comparison framework encompassing various supervised and unsupervised protein scores.
2.1. Test sets
AlphaMod underwent rigorous testing across two distinct datasets. The initial dataset (referred to as Test set A) was derived from carefully selected targets within the CASP14 competition, which initially presented approximately 115 potential target sequences for evaluation. However, this number was precluded from full consideration due to various factors: certain targets were withdrawn due to impending publication or because their structures consisted solely of a single helix, precluding tertiary structure evaluation, while others were dismissed as they lacked an accompanying structure within the PDB database. Further scrutiny reduced this pool significantly; 35 targets were excluded due to the absence of corresponding PDB files, and several others exhibited fragmented structures, exemplified by targets like T1027-D1 with non-continuous domains, thus failing to meet our continuous domain criterion for inclusion. Furthermore, an additional 23 targets were deemed incompatible with our assessment tools during the quality verification process, complicating the structural quality evaluation. Consequently, our refined Test set A comprised 47 target proteins, encompassing 9 TBM-easy, 13 TBM-hard, 15 FM, and 10 FM/TBM evaluation units (EUs), detailed exhaustively in Supplementary File 1, Table 1. Concurrently, a secondary dataset (Test set B) was employed, rooted in the single-chain protein analysis conducted by Terwilliger and colleagues [21], serving as a comparative platform for evaluating AlphaMod's proficiency against established benchmarks.
Furthermore, to identify classes of proteins on Test sets A and B, we clusterized the different targets at domain level using as criteria the percentage of secondary structures, calculated using the DSSP algorithm [22] (Supplementary File 1, Tables 2, 3). Through an in-house Perl script, it was possible to sum the percentage of secondary structures assigned by DSSP into 4 macro-classes: helices (sum of the percentage of codes H, G, and I as identified by DSSP), beta structures (percentage of code E), irregular structures (sum of the percentage of codes T, S, B) and coils (with no structure code assigned by DSSP). In parallel, each target was compared to all other targets using the SSAP algorithm [23] with parameters—slow-ssap-only and—max-score-to-slow-rerun = 75. The similarity matrix was built using the Ward method [24] and the SSAP score was used as a parameter to build a dendrogram that was used to represent the structural similarity among the selected targets (Supplementary Figs. 1 and 2).
2.2. Pipeline description
Our AlphaMod pipeline is composed of 5 sequential steps briefly described below and shown in Fig. 1. For more details, please refer to Supplementary File 2.
Fig. 1.
Scheme of the AlphaMod pipeline. AlphaMod is initialized by the Homolog Information Retrieval Engine (HIRE). The input selector S decides the entry data as either a single Fasta File or a group of Fasta Files, finding both the templates and Multiple Sequence Alignment (MSA). The Protein Backbone Construction Tool (PBCT), launches AlphaFold2 (AF2), using as input the MSAs and templates, producing 10 predictions as PDBs (5 relaxed and 5 unrelaxed). These predictions are analyzed by the Structure Model Assessment module (SMA), first by extracting the pLDDT from AF2, and second, calculating the QMEANDisCo score with a web-crawler; pLDDT and QMEANDisCo are used to compute BORDASCORE, all these results are stored in the Metrics Data Collector. Moreover, PBCT passes upon MODELLER the user criteria (OP1, OP2 and/or OP3), MODELLER will generate 5 new predictions based on the selected criteria. Each option is executed as follows: OP1 fetches the information stored in the Metrics Data Collector and selects the first and second best AF2 relaxed models by means of BORDASCORE. OP2 does not need any additional information and uses directly the first ranked predictions obtained from AF2. Finally, in OP3 (the test case when the ground truth is known), GDT_TS is calculated, the first and second models with the highest GDT_TS are given to MODELLER. Finally, the Comprehensive Model Quality Assessment module (CMQA) sequentially applies a series of unsupervised metrics, namely QMEANDisCo, PROCHECK, PROSA, MOLPROBITY, and DOPESCORE, to both AF2 and AlphaMod models. It is essential to highlight that the calculation of supervised metrics, specifically GDT_TS and RMSD, is exclusively enabled when the experimental structure is available and option OP3 (TEST MODE) is selected. In addition to the unsupervised metrics, all the previously mentioned supervised and unsupervised metrics are stored in the Metrics Data Collector for further analysis and evaluation.
2.2.1. Step 1: Homolog Information Retrieval Engine (HIRE)
HIRE's primary goal is to identify the templates and homologous sequences that most accurately represent a specified input protein. This is the default AF2’s protocol for searching for homologous sequences and templates for the target sequence, which includes JackHMMER v3.3 [25] on MGnify [26] and UniRef90 [27], followed by HHBlits v3.0-beta [28] on Uniclust30 [29] and BFD. Furthermore, the template search was done with HHSearch [30] on PDB70. Based on the scores of each search engine, AF2 creates two separated files: first, a Multiple Sequence Alignment (MSA) file, and second, a pickle file containing up to 20 templates classified in descending order being the first template the one with the highest similarity score compared with the target sequence. The following AF2's modeling step, keeps as reference only the top four templates. Sometimes, it is not possible to obtain at least four templates, despite this drawback AlphaFold2 is still able to make predictions.
2.2.2. Step 2: Protein Model Construction Tool (PMCT)—AF2 branch
PMCT fetches the MSA and pickle files in Step 1 and subsequently transfers them to AF2 for protein fold prediction using our local version downloaded from AF2 GitHub (AlphaFold v2.2.2, commit ab10514, with max_template_date=2020-05-14 and three recycling iterations as default). AF2 produces 10 PDB files divided into two groups, 5 relaxed and 5 unrelaxed. The objective of the relaxation process is to remove stereochemical violations [10] and is done by means of gradient descent in the Amber Force field without altering the accuracy measured by GDT_TS [11] or lDDT-Cα [31], the pLDDT score integrated in AF2 is used to rank relaxed models only. Hence, the naming convention follows an ascending order, where the model with the highest pLDDT score is denoted as ranked_0, while the model with the lowest pLDDT score is labeled as ranked_4.
2.2.3. Step 3: Structure Model Assessment (SMA)
In addition to pLDDT, we included QMEANDisCo [32], a composite scoring function assessing the major geometrical aspects of protein structures, computed on the relaxed models produced by AF2. The calculation of QMEANDisCo is performed by a web crawler using ad-hoc Python functions. In this context, both pLDDT and QMEANDisCo function as parameters for BORDASCORE evaluation [33] to determine the top-two relaxed performing models (See Fig. 1, SMA box). Finally, all the computed metrics are inputted to the Metrics Data Collector, also used in Step 4 and Step 5 (more information can be seen in Supplementary File 2).
2.2.4. Step 4: Protein Model Construction Tool (PMCT)—MODELLER’s branch
After the assessment of AF2’s models performed by SMA, the PMCT produces five new structural predictions by filtering and launching MODELLER with the best models. In detail, PMCT-MODELLER’s branch can be used with the option 1 (OP1—2best_unsupervised) to make predictions using as templates the best two models in agreement with BORDASCORE, or the option 2 (OP2—5best_unsupervised) to make predictions using as templates all the relaxed models produced by AF2. Finally, during the supervised testing phase, PMCT was initialized with option 3 (OP3—2best_supervised), which involves prior knowledge of Ground Truths of the three-dimensional protein structure taken as reference. Therefore, in OP3, we selected the best two models based on GDT_TS score. Subsequently, AlphaMod models, along with the information gathered by the Metrics Data Collector module are passed down to the Comprehensive Model Quality Assessment framework.
2.2.5. Step 5: Comprehensive Model Quality Assessment (CMQA)
At this point in the execution of AlphaMod’s pipeline, the Metrics Data Collector module has stored on its local cache memory information of pLDDT, QMEANDisCo, BORDASCORE and GDT_TS, as explained in Step 3. The quality assessment of both AF2 and AlphaMod’s models is extended with CMQA. The latter is accomplished with five additional metrics including: (1) DOPESCORE (Discrete Optimized Protein Energy) [8], which is a statistical potential optimized for model assessment in MODELLER, (2) PROSA-Web Z-score [34] that measures the deviation of the total energy of the structure with respect to an energy distribution derived from random conformations, (3) MOLPROBITY score [35], a single number that results from the log-weighted combination of the clashscore, percentage of disallowed residues in the Ramachandran plot and percentage of bad side-chain rotamers, and (4) the classic evaluation of allowed and disallowed residues in the Ramachandran plot by means of PROCHECK [36]. PROSA, MOLPROBITY, and PROCHECK metrics are calculated with the web crawler, whereas DOPESCORE is acquired upon MODELLER’s execution. The fifth and last metric, (5) root mean square deviation (RMSD), is calculated only when PMCT-Modeller’s Branch is launched with OP3, as the ground truth template is required for comparison. Supplementary File 2 contains comprehensive details about the parameters employed to compute each metric.
3. Results and discussion
In this section, we present a comprehensive evaluation of the AlphaFold2 (AF2) tool alongside various configurations of AlphaMod (OP1, OP2, and OP3) for protein structure prediction. We evaluated the performance using a variety of quality assessment metrics, juxtaposed against the GDT_TS score. Initially, in Section 3.1, we delve into a comparison of average predictions on Test Set A, spotlighting the distinct output categories of AF2 and the enhancements brought about by integrating MODELLER. Then, we extended the analysis to Test Set B (Section 3.2). Section 3.3 extends our exploration to a pair-to-pair comparison of Test Set A and Test Set B, showing interesting insights and relative strengths of the prediction tools alone and combined in the AlphaMod pipeline. In detail, in Section 3.3.1, we provide a pair-to-pair comparison of the best models predicted. Concluding with Section 3.3.2, a meticulous statistical analysis is undertaken to discern the significance of the observed differences between the prediction tools and their scoring system. Finally, in Section 3.4, we present the computational costs associated with the full running of the AlphaMod pipeline.
3.1. Results on test set A—comparisons on averaged predictions
3.1.1. AF2
As delineated in Methods-Step 2, AF2 yields two categories of outputs: the "unrelaxed" models, which bypass the relaxation process, and the "relaxed" counterparts, which are subjected to AF2's subsequent refinement phase. Notably, the pLDDT score is assigned exclusively to the "relaxed" models, which are subsequently ranked based on this metric. We proceeded to determine the GDT_TS score for each model derived from AF2, with the aggregated results encapsulated in (Supplementary File 3, Table 1). The overarching mean GDT_TS score, computed across all the conclusive (ranked) models for each EU, stands at 81.01 ± 18.25, underscoring the predictor's robust reliability. Interestingly, we noted that several models classified as “unrelaxed” have a GDT_TS score higher with respect to the corresponding “ranked” models, indicating that the relaxing procedure of AF2 sometimes worsens, rather than improving, the results (see for more details Supplementary File 3, Table 2).
3.1.2. MODELLER
The combination in which MODELLER precedes AF2 in the pipeline was also considered during our research. In detail, after the execution of HIRE (see Methods-Step 1), the top-four templates were given as input to MODELLER. When less than four templates were found, we launched MODELLER with the available number of templates. MODELLER failed to deliver tertiary structures predictions with quality at least comparable to AF2 predictions in terms of GDT_TS. Therefore, this option was excluded from our pipeline The full pairwise comparisons using the top-ranked structures across different methodologies is shown in Supplementary File 3, Table 3.
3.1.3. AlphaMod-OP3
To assess MODELLER’s potential of improving AF2's predictions, we selected for each EU the two models predicted by AF2 with the highest GDT_TS, and launched AlphaMod with OP3 (see Methods, Step 4). We averaged the GDT_TS score for the five models obtained by AlphaMod-OP3 for each EU, as we did for AF2. The result presented a slight improvement (81.61 ± 18.28) with respect to AF2 (Supplementary File 3, Table 1). By making a pair-to-pair comparison of the average GDT_TS scores between AF2 and AlphaMod-OP3, the results indicated that AlphaMod-OP3’s models presented a higher average GDT_TS score than those obtained by AF2. The highest improvements were obtained for the targets T1038-D1 (more than 14 GDT_TS units, see Fig. 2), T1031-D1, T1037-D1, T1099-D1 (more than 4 GDT_TS units). These results proved that the addition of MODELLER has the potential to improve AF2’s predictions, if it is possible to select the two best models from which to model with MODELLER. However, all those targets whose models created by AF2 obtained a GDT_TS score <50, were not substantially improved by MODELLER.
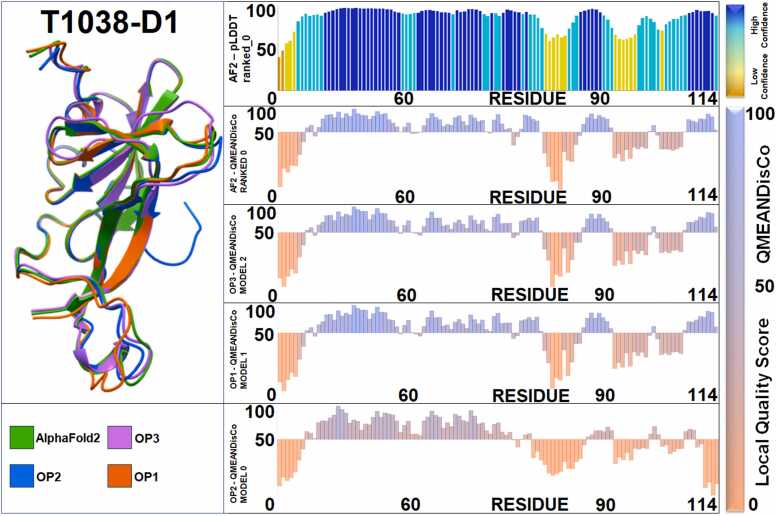
Fig. 2.
Illustration of four predictions of CASP14 Target T1038-D1. On the left side, the best models (i.e. those models with the highest overall GDT_TS score) produced with the different procedures: AF2 alone in green, OP1 in orange, OP2 in blue and OP3 in violet. On the right side, from top to bottom: 1st row, plot showing the confidence level of AlphaFold2’s prediction (pLDDT) residue by residue, rows 2nd to 5th illustrates a residue-by-residue assessment of the best models produced by the different procedures, utilizing the QMEANDisCo metric instead. In detail, 2nd row, AlphaFold2 ranked_0, 3rd row, AlphaMod’s OP3 model 2, 4th row, AlphaMod’s OP1 model 1, and 5th row, AlphaMod’s OP2 model 0. The bottom legend shows the number of residues, CASP14 Target T1038-D1 has a total of 114 residues.
3.1.4. AlphaMod-OP2
When the reference structure of the predicted protein is not available, it is challenging to select the two best models produced by PMCT-AF2. An initial approach to face this issue was tested with AlphaMod-OP2, in which we used all AF2’s ranked models (five in total) for PMCT-MODELLER’s branch execution. Similarly, as with AlphaMod-OP3, we calculated the global average GDT_TS on the five models produced by PMCT-MODELLER’s branch (Supplementary File 3, Table 1). It is worth noting that the global average GDT_TS score for this procedure (80.77 ± 18.62) was slightly lower compared to the previous results obtained using AF2 alone.
3.1.5. AlphaMod-OP1
In light of AlphaMod-OP2’s results, our focus shifted to a way to classify and select the best top two templates like in AlphaMod-OP3 but in an unsupervised way, thus without requiring a reference structure (ground truth). Consequently, the development of AlphaMod-OP1 revolved around integrating and possibly improving the assessment made by AF2’s pLDDT score and MODELLER’s DOPESCORE [8], by shifting on traditional protein 3D-structure quality assessment measures, such as: QMEANDisCo [32], PROSA-Web Z-score [34], MOLPROBITY [35], PROCHECK [36]. This naturally leads to the question of which among these scores, or what combination thereof, best aligns with the representation of the EUs according to the GDT_TS supervised counterpart. Firstly, we analyzed the non-linear statistical correlation, arranging the predicted models into two groups: one that featured predicted models from both AF2 and AlphaMod with the highest-ranked GDT_TS and another that evaluated the average GDT_TS across each set of predictions (the latter, for the sake of thoroughness, is shown in the Supplementary Materials even if does not show relevant results (Supplementary File 4, Table 1). Meaningful correlations were identified only on pLDDT and QMEANDisCo each exhibiting a robust positive correlation with a Rho(p) value exceeding 0.75. Next, the Shapiro-Wilk test [37] is applied to confirm the non-normal nature of various measures, including GDT_TS, pLDDT, QMEANDisCo, PROSA, MOLPROBITY, DOPESCORE and PROCHECK, the p-values are detailed in (Supplementary File 4, Table 2). Following this, we implemented a normalization technique using robust scaling, which minimizes the impact of outliers by adjusting features based on median and interquartile ranges. After confirming the non-normality of the distributions, we utilized the Wilcoxon test [38], which yielded p-values of 8 × 10-3 for GDT_TS, 2 × 10-2 for QMEANDisCo, 7 × 10-4 for PROSA, followed by consistent values of 0.5 for both MOLPROBITY and DOPESCORE, finally, the least significant p-value was found for PROCHECK with a p-value of 0.7, (detailed information is available in Supplementary File 4, Table 3).
The p-values associated with GDT_TS and QMEANDisCo suggest that AlphaMod's results are statistically significant and can drive AlphaMod-OP1 evaluations. The average GDT_TS score recorded with the OP1 approach was (81.18 + 18.21), demonstrating a marginal enhancement compared to the scores achieved using only AF2, with comprehensive results detailed in Supplementary File 3, Table 1. A different perspective is illustrated at predictions made by AF2 for the whole proteins of targets T1024-D0 and T1038-D0, which gave a GDT_TS score significantly lower than those made by AlphaMod (Supplementary File 3, Table 4), whereas in the case of target T1053 the GDT_TS scores are similar. For the whole target T1030-D0, the results of AF2 and AlphaMod were similar when examining the average GDT_TS scores.
3.2. Results on test set B—comparisons on averaged predictions
To further validate our AlphaMod methodologies, we opted to extend their application to an independent Test Set B. This set comprises 25 single-domain proteins that were introduced following the CASP14 evaluation [21]. Notably, of these 25 targets, 12 possess more than 40% helical content in their secondary structures, while 17 exhibit over 40% irregular structures and coils. Only 3 targets feature more than 40% beta structures shown in Supplementary File 1, Table 3. Following the methodology applied to Test Set A, we began by generating model predictions with AF2, then proceeded with the implementation of the various AlphaMod-OPs. The full results of the analysis are shown in Supplementary File 3, Table 5. Within this dataset, the models produced by OP2 demonstrated an average GDT_TS score that closely matched those of AF2. In contrast, OP1 and OP3 improved slightly AF2 results. On closer inspection, AlphaMod-OP3 and AlphaMod-OP1 models have shown a higher GDT_TS score than AF2 models in 72% and 64% of the predicted models, respectively. Only one target obtained with AF2 showed a GDT_TS score lower than 50 (7MSW, it is formed of >40% irregular secondary structures and coils), and it was only slightly improved by AlphaMod. The average GDT_TS score for targets 7L6U, 7LV9, 7LX5 and 7M7B presented a gain higher than 4 units (reaching 13 units for 7LV9). Once again, apparently the presence of a particular secondary structure seems not related to the best performances of the predictors (Supplementary File 1, Table 3). Moreover, since Terwilliger et al. [21] utilized RMSD for comparisons with reference PDB structures, we calculated also the average RMSD of the models predicted with AF2 for each target and compared it with the average RMSD of the models predicted with our unsupervised methodologies OP1 and OP2. Complete findings are detailed in Supplementary File 3, Table 6 and summarized in Table 1. We completed this task using PHENYX [39], mirroring the software used by Terwilliger and colleagues. On average, the RMSD determined for all models derived through the OP1 process is less than that obtained for all models predicted by AF2. On the other hand, a one-to-one comparison reveals that 68% of the targets predicted by OP1 outperform those predicted by AF2. This result is better than the one obtained when evaluating the GDT_TS score, previously mentioned at 64%. Very interestingly, three targets were predicted very badly by AF2 (RMSD >10), but in two cases (7KU7 and 7LV9) AlphaMod-OP1 was able to rescue the final models to an RMSD value of about 5 Å. The comparison between the global average RMSD acquired through our AlphaMod procedures and those reported by Terwilliger et al. showed that our AlphaMod unsupervised procedures yield models with a lower RMSD values when compared to the reference PDB structure (see Table 1). We were very surprised to find that some targets predicted by Terwilliger et al. had a huge RMSD, while the same targets predicted by our AlphaMod procedure had an RMSD in line with that of the other targets. Since we do not have the structures predicted by Terwilliger et al., we are unable to explain the reason for this huge difference in performance. By excluding these outliers, both the Terwilliger’s and our AlphaMod procedures demonstrate comparable performance. Nevertheless, it is worth noting that AlphaMod’s approach exhibits greater robustness, featuring only one outlier compared to Terwilliger’s seven outliers within a sample of 25 targets (Supplementary File 3, Table 7).
Table 1.
Average RMSD for the different types of models obtained (domains only) Test set B.
| Targets | RMSD (Å) Test set B | Average RMSD (Å) AF2 Ranked Models | Average RMSD (Å) AlphaMod OP1 | Average RMSD (Å) AlphaMod OP2 |
|---|---|---|---|---|
| 7BRM | 0.7* | 5.33 ± 2.20 | 4.20 ± 0.66 | 4.37 ± 0.78 |
| 7BXT | 0.8* | 1.60 ± 0.45 | 3.70 ± 1.34 | 2.61 ± 1.31 |
| 7C2K | 1.0* | 2.13 ± 0.65 | 1.49 ± 0.05 | 1.62 ± 0.01 |
| 7EDA | 21.6 | 2.26 ± 1.50 | 1.78 ± 0.53* | 3.43 ± 0.11 |
| 7EV9 | 0.5* | 1.61 ± 0.77 | 2.43 ± 0.66 | 1.76 ± 0.50 |
| 7KU7 | 1.7* | 12.52 ± 8.09 | 4.49 ± 0.69 | 18.31 ± 0.02 |
| 7KZZ | 1.4* | 2.50 ± 0.10 | 2.72 ± 0.44 | 2.59 ± 0.52 |
| 7L1K | 14.6 | 0.64 ± 0.02* | 0.64 ± 0.01* | 0.65 ± 0.02 |
| 7L6U | 1.3* | 2.31 ± 0.41 | 1.91 ± 0.04 | 2.34 ± 0.10 |
| 7LC6 | 10.2 | 0.82 ± 0.06 | 0.81 ± 0.02* | 0.83 ± 0.06 |
| 7LCI | 4.1 | 4.43 ± 0.34 | 4.09 ± 0.07* | 4.59 ± 0.11 |
| 7LS5 | 0.4* | 1.05 ± 0.04 | 1.08 ± 0.06 | 1.12 ± 0.05 |
| 7LSX | 23.7 | 1.36 ± 0.08* | 1.42 ± 0.03 | 1.39 ± 0.03 |
| 7LV9 | 16.4 | 10.52 ± 6.64 | 5.47 ± 0.21* | 8.72 ± 1.37 |
| 7LVR | 1.0* | 1.28 ± 0.05 | 1.23 ± 0.04 | 1.21 ± 0.02 |
| 7LX5 | 5.7 | 2.37 ± 2.28 | 1.16 ± 0.09* | 1.41 ± 0.20 |
| 7M7B | 2.9* | 3.56 ± 1.82 | 3.47 ± 0.52 | 3.06 ± 0.46 |
| 7M9C | 6.0 | 1.47 ± 0.05 | 1.49 ± 0.02 | 1.44 ± 0.03* |
| 7MBY | 19.5 | 1.54 ± 0.32* | 1.66 ± 0.59 | 1.55 ± 0.41 |
| 7ME0 | 0.4* | 0.77 ± 0.30 | 0.49 ± 0.02 | 0.85 ± 0.05 |
| 7MJS | 7.0 | 2.51 ± 0.39 | 2.21 ± 0.05* | 2.26 ± 0.11 |
| 7MLZ | 15.5 | 1.88 ± 0.22 | 1.71 ± 0.01* | 1.74 ± 0.07 |
| 7MSW | 17.0 | 16.55 ± 2.36 | 15.98 ± 0.11 | 15.45 ± 0.42* |
| 7N8I | 2.6 | 0.44 ± 0.01* | 0.46 ± 0.02 | 0.45 ± 0.02 |
| 7RB9 | 0.4* | 1.63 ± 0.05 | 1.60 ± 0.05 | 1.63 ± 0.04 |
| Global average RMSD | 7.06 ± 7.74 | 3.32 ± 4.43 | 2.71 ± 3.05* | 3.42 ± 4.37 |
Results in bold represent pair-to-pair models obtained with either AlphaMod procedure with a RMSD lower than the one obtained by AF2 alone. Results with * represent, for each target, the best result in terms of RMSD.
3.3. Results on test sets A and B
3.3.1. Pair-to-pair comparison of best models
In the preceding section, our discussion was centered on average scores. Moving forward, this section presents a comparison across AlphaMod’s OP methodologies and AF2, with a focus on the top-two best models predicted (in terms of GDT_TS). The selection of these premier models for OP1 is based on the BORDASCORE criterion, as outlined in the Method Section—Steps 3 and 4 (Supplementary File 5, Table 1). The obtained structures are listed in Table 2 together with their results for each methodology, further information is provided in Supplementary File 5, Table 2. The focus is on the average differences in GDT_TS scores and the variability of these scores when juxtaposed with the best methods predictions. Out of 72 predictions, AF2 emerges as the best method in approximately 38% of cases, a very low margin compared to OP1, OP2, and OP3, which excel in 21%, 13%, and 18% of cases, respectively. Qualitatively, at the differences in terms of GDT_TS, there are some instances when AF2 shows an uptick in performance, particularly in contrast with OP2, where there is an average increase of 0.11 points. Conversely, when juxtaposed with the other AlphaMod methods, its performance worsens. Interestingly, upon the exclusion of outliers, the performance decline is less pronounced, especially in the AF2 versus OP3 comparison. This indicates that these outliers significantly skew the general findings. In the OP1 versus AF2 setup, there are 15 targets considered, with an average improvement of 1.60 in the GDT_TS scores when OP1 leads. However, this improvement reduces to 0.67 when outliers are removed, indicating a substantial impact due to extreme cases. The standard deviation in OP1, in this setup, is 1.13, which tightens to 0.47 without outliers, further highlighting the influence of these extreme values. AF2 shows a standard deviation of 0.97 with outliers, and 0.69 without them. For OP2 versus AF2, out of 9 targets, there is an even more pronounced average improvement of 2.05 in GDT_TS scores (see Supplementary File 5, Table 3). From the above results, the most notable improvements are achieved on T1030-D0, 7KZZ-D1 and 7KU7-D1, with an increase on GDT_TS score of 14.65, 6.73 and 4.26 units, respectively (see Supplementary File 5, Table 4). This outcome shows particular significance as it underscores AlphaMod's potential protein quality enhancement. We investigated whether the success or failure of the different modeling procedures was related to the content in secondary structures of our dataset, to find if this procedure is sensitive towards these structural features. 20 of the targets belonging to our CASP14 dataset contain mainly helices as secondary structures (>40% of the total structures), whereas only 6 targets contain > 40% of beta structures; the targets particularly rich in irregular structures and coils (>40% of the total secondary structures) are 22 (Supplementary File 1, Table 2). Looking at the data, it seems that no relationship exists between the correctness of the predictions and the composition of the proteins in terms of secondary structures. Among those targets whose GDT_TS increases most with the addition of MODELLER after the AF2 predictions, T1038-D1 is formed mainly by beta and irregular structures, T1031-D1 by irregular structures, T1037-D1 and T1099-D1 mainly by helices. Those targets for which both AF2 and AlphaMod failed to reach a GDT_TS score of at least 50 generally contain, as expected, a high quantity of irregular structures and coils (T1029-D1, T1043-D1, T1047s1-D1, 7MSW-D1). Finally, in 11% of cases, the best models from both AF2 and AlphaMod present an equal GDT_TS score, rendering it challenging to determine a clear best-performing method. This underscores the fact that no single method consistently outperforms the others. When these insights are juxtaposed with the earlier analysis, a complex picture emerges. AF2, while often the frontrunner, does not maintain unchallenged dominance. OP1, OP2, and OP3, exhibit situational efficacy, sometimes rivaling or even surpassing AF2. The instances of a tie are particularly revealing, suggesting scenarios where the methods reach a performance plateau, making them indistinguishable in terms of efficacy. This variability in performance across the board reinforces the idea that predictive success is highly contextual. The data, rather than pointing towards an universally superior method, emphasizes the situational effectiveness of each. In essence, the quest for the best model is less about absolutes and more about understanding the conditional dynamics that play to the strengths of each method.
Table 2.
Pairwise comparison of the top-ranked predicted targets across different methodologies including: AF2, OP1, OP2 and OP3.
| Target | AF2 |
AFM-OP1 |
AFM-OP2 |
AFM-OP3 |
Target | AF2 |
AFM-OP1 |
AFM-OP2 |
AFM-OP3 |
Target | AF2 |
AFM-OP1 |
AFM-OP2 |
AFM-OP3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GDT_TS | GDT_TS | GDT_TS | GDT_TS | GDT_TS | GDT_TS | GDT_TS | GDT_TS | GDT_TS | GDT_TS | GDT_TS | GDT_TS | |||
| 7BRM-D1 | 83.82 | 86.24* | 81.49 | 84.69 | 7RB9-D1 | 85.59 | 85.46 | 85.39 | 85.12 | T1042-D1 | 61.32 | 59.78 | 59.78 | 60.60 |
| 7BXT-D1 | 93.99 | 90.38 | 94.71 | 90.63 | T1024-D0 | 85.68 | 87.02 | 65.73 | 87.02 | T1043-D1 | 25.51 | 25.84 | 25.34 | 27.70 |
| 7C2K-D1 | 93.70 | 93.29 | 91.14 | 93.48 | T1024-D1 | 90.29 | 89.90 | 90.16 | 89.90 | T1045s1-D1 | 95.62 | 96.10 | 96.27 | 96.59 |
| 7EDA-D1 | 98.36 | 92.01 | 90.00 | 95.07 | T1024-D2 | 91.18 | 91.30 | 91.30 | 91.79 | T1045s2-D1 | 93.83 | 92.02 | 92.62 | 93.37 |
| 7EV9-D1 | 97.00 | 89.69 | 90.60 | 89.69 | T1025-D1 | 98.35 | 98.15 | 98.25 | 98.35 | T1046s1-D1 | 97.57 | 96.87 | 97.22 | 97.92 |
| 7KU7-D1 | 79.63 | 80.46 | 83.89* | 80.56 | T1026-D1 | 92.64 | 92.81 | 92.98 | 92.47 | T1046s2-D1 | 98.76 | 98.58 | 98.40 | 98.76 |
| 7KZZ-D1 | 71.90 | 75.89 | 78.63* | 75.89 | T1028-D1 | 92.98 | 93.15 | 92.38 | 93.15 | T1047s1-D1 | 47.87 | 46.68 | 46.45 | 47.99 |
| 7L1K-D1 | 97.83 | 97.83 | 97.67 | 97.83 | T1029-D1 | 46.20 | 46.20 | 45.40 | 46.20 | T1047s2-D1 | 89.12 | 88.61 | 87.08 | 87.58 |
| 7L6U-D1 | 91.35 | 90.22 | 86.38 | 90.22 | T1030-D0 | 53.48 | 68.13* | 67.12 | 62.64 | T1047s2-D3 | 50.43 | 50.22 | 49.78 | 50.22 |
| 7LC6-D1 | 97.49 | 97.31 | 97.27 | 97.18 | T1030-D1 | 71.10 | 68.34 | 69.64 | 70.94 | T1049-D1 | 93.28 | 92.91 | 92.72 | 92.91 |
| 7LCI-D1 | 67.83 | 67.70 | 61.87 | 69.54* | T1030-D2 | 89.50 | 85.92 | 82.14 | 85.29 | T1053-D0 | 88.72 | 87.76 | 88.24 | 88.28 |
| 7LS5-D1 | 96.52 | 96.31 | 95.80 | 96.21 | T1031-D1 | 94.47 | 94.47 | 90.00 | 94.74 | T1053-D1 | 89.01 | 89.01 | 89.26 | 89.26 |
| 7LSX-D1 | 94.72 | 93.80 | 93.90 | 94.92 | T1032-D1 | 65.59 | 65.29 | 65.00 | 64.85 | T1053-D2 | 84.65 | 82.46 | 82.31 | 85.96 |
| 7LV9-D1 | 73.47 | 69.64 | 57.40 | 69.64 | T1033-D1 | 52.75 | 42.00 | 50.00 | 46.00 | T1054-D1 | 89.34 | 88.99 | 88.46 | 88.99 |
| 7LVR-D1 | 94.29 | 94.12 | 94.00 | 94.29 | T1034-D1 | 95.51 | 95.19 | 95.35 | 95.35 | T1055-D1 | 87.91 | 88.73 | 87.91 | 88.73 |
| 7LX5-D1 | 93.91 | 93.66 | 92.64 | 94.04 | T1035-D1 | 87.75 | 88.24 | 87.01 | 88.24 | T1056-D1 | 97.48 | 95.71 | 95.41 | 96.01 |
| 7M7B-D1 | 90.12 | 90.36 | 89.41 | 90.36 | T1036s1-D1 | 83.17 | 74.80 | 63.20 | 74.80 | T1065s1-D1 | 92.86 | 93.07 | 93.91 | 93.07 |
| 7M9C-D1 | 88.28 | 86.92 | 87.69 | 87.98 | T1037-D1 | 84.71 | 84.03 | 86.32* | 86.08 | T1065s2-D1 | 97.96 | 98.47 | 98.21 | 98.47 |
| 7MBY-D1 | 90.96 | 92.06 | 91.18 | 91.91 | T1038-D0 | 86.84 | 85.13 | 41.84 | 87.24 | T1074-D1 | 93.18 | 93.37 | 91.67 | 92.42 |
| 7ME0-D1 | 99.28 | 99.28 | 96.55 | 99.28 | T1038-D1 | 85.53 | 81.36 | 82.89 | 86.40 | T1076-D1 | 98.98 | 99.12 | 99.07 | 99.12 |
| 7MJS-D1 | 88.53 | 88.16 | 88.35 | 88.53 | T1038-D2 | 91.78 | 90.79 | 90.46 | 91.12 | T1078-D1 | 94.96 | 94.96 | 94.57 | 94.38 |
| 7MLZ-D1 | 82.49 | 83.12 | 82.36 | 83.12 | T1039-D1 | 85.25 | 83.70 | 80.59 | 82.92 | T1082-D1 | 91.67 | 90.33 | 90.33 | 90.33 |
| 7MSW-D1 | 45.01 | 44.58 | 41.94 | 46.15 | T1040-D1 | 56.54 | 55.96 | 55.58 | 57.12 | T1090-D1 | 89.53 | 89.53 | 88.87 | 89.79 |
| 7N8I-D1 | 98.83 | 98.83 | 98.83 | 99.07 | T1041-D1 | 84.71 | 85.54 | 85.12 | 85.12 | T1099-D1 | 79.35 | 80.76 | 82.16 | 80.62 |
Results in bold represent pair-to-pair models obtained with either AlphaMod procedure with a GDT_TS score higher than the one obtained by AF2 alone. Results with * represent, for each target, a notable quality increase in terms of GDT_TS score. Results in italic represent pair-to-pair models obtained with either AlphaMod procedure with a GDT_TS score equal to those obtained by AF2 alone.
3.3.2. Statistical analysis
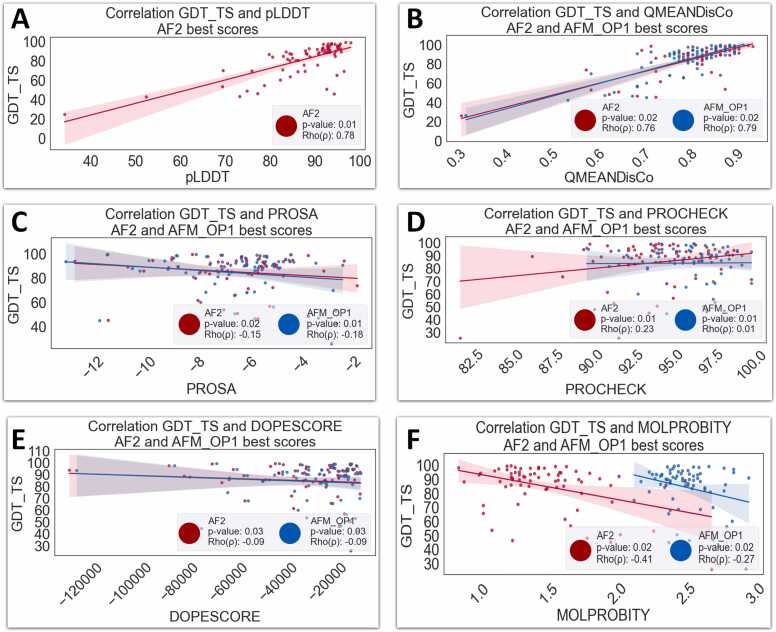
Analogous to the procedure outlined in Section 3.1.4, a linear correlation analysis has been executed and is visually presented in Fig. 3. In this case, we focus only on the top-performing models. Particularly, meaningful p-values (<0.02) and correlations Rho(p) > 0.78 were found only on pLDDT and QMEANDisCo. However, despite this linear correlation, a more significant non-linear relationship between GDT_TS and QMEANDisCo is found. A permutation test involving the QMEANDisCo score in connection with GDT_TS was designed to evaluate the statistical hypothesis about the relevance of the relationship between GDT_TS and QMEANDisCo (scoring bivariates). More precisely, we investigated whether their original bivariate distribution significantly deviates from what would be expected by chance as evidenced by the random permutation of the scoring bivariates in the AF2 model and the AlphaMod models in their various configurations: OP1, OP2, and OP3. To achieve this, we selected the best top-two ranked GDT_TS and its QMEANDisCo over Test Set A and B associated as bivariate Beta distributions, incorporating variable parameters α_1 and α_2. These parameters were selected recursively via a grid search method that iterates from 0.1 to 2.0 by 0.1 steps, with the constraint that the sum of α_1 and α_2 equals to one. The scores associated with each model are treated as distinct statistical distributions, we utilized the symmetrical Kullback-Leibler (KL) divergence [40] to evaluate the statistical separation between them. This method allows us to quantify the divergence between the probability distributions of AF2 and each OP configuration (OP1, OP2, and OP3). The KL divergence is particularly suitable for this purpose as it measures the difference between two probability distributions, providing a statistical basis for comparison. The core of this analysis lies in the calculation of p-values, which serve as indicators of the statistical significance of the observed differences. Specifically, these p-values assess whether the disparities in the distributions are substantial enough to not be attributed to random chance. Our findings present p-values for the comparisons between AF2 and each OP configuration, with the values being 0.021, 0.019, 0.019, and 0.022 for AF2, OP1, OP2, and OP3, respectively (detailed results are available in Supplementary File 4, Table 4). For a fair comparison in the permutation tests the seed is fixed. These figures suggest that the differences observed are statistically significant, particularly for the comparisons involving OP1 and OP2, underscoring the importance of the configurations in the models' performance.
Fig. 3.
Correlation between best scores of supervised metric GDT_TS and unsupervised metrics: pLDDT, QMEANDisCo, MOLPROBITY, PROSA, DOPESCORE and PROCHECK. AF2 is represented in red and AFM-OP1 in blue. Panel A: relationship between GDT_TS and AF2’s pLDDT, (p-value=0.01, Rho(ρ) = 0.78). Panel B: relationship GDT_TS and QMEAN, AF2: (p-value=0.02, Rho(ρ) = 0.76), AFM-OP1: (p-value=0.02, Rho(ρ)= 0.79). Panel C: relationship GDT_TS and PROSA, AF2: (p-value=0.02, Rho(ρ) = −0.15), AFM-OP1: (p-value=0.01, Rho(ρ) = −0.18). Panel D: relationship GDT_TS and PROCHECK, AF2: (p-value=0.01, Rho(ρ) = 0.23), AFM-OP1: (p-value=0.01, Rho(ρ) = 0.01). Panel E: relationship GDT_TS and DOPESCORE, AF2: (p-value=0.03, Rho(ρ) = −0.09), AFM-OP1: (p-value=0.03, Rho(ρ) = −0.09). Panel F: relationship GDT_TS and PROCHECK, AF2: (p-value=0.02, Rho(ρ) = −0.41), AFM-OP1: (p-value=0.02, Rho(ρ) = −0.27).
3.4. Computational cost
In assessing the computational costs tied to our methodology, we separately monitored the runtimes for AF2 and MODELLER. AF2 typically takes an average of 4 h to generate predictions for each of the chosen 72 targets. Conversely, MODELLER's predictions take an average of around 45 s per target when run on MARCONI100 in Bologna (IT). However, on a more budget-constrained machine equipped with a CPU-I7 and an Nvidia GPU GTX1070, the runtime roughly doubles. Overall, the introduction of AlphaMod does not significantly increase the time complexity compared to AF2, as detailed in Supplementary File 4, Table 5.
4. Conclusions
While AF2 has achieved remarkable accuracy in predicting protein structure, our study has highlighted the potential for further improvement. We have demonstrated that, in principle, by combining this cutting-edge deep learning tool with traditional modeling strategies, it is possible to achieve a substantial improvement in the quality of a protein’s tertiary structure, especially in terms of GDT_TS. Only where AF2 fails to achieve high quality results on average and top-two best comparisons over these targets: T1029-D1, T1043-D1, T1047s1-D1, 7MSW-D1, our AlphaMod procedures cannot significantly improve prediction accuracy.
Furthermore, as described in Section 2 and Supplementary File 4, Tables 1–4, large-scale protein predictions can be effectively applied, thanks to the automation integrated into the AlphaMod pipeline, spanning from data retrieval to automatic processing. Finally, our pipeline provides a unified platform for comprehensive protein structural quality assessment, encompassing several metrics. This addresses the current challenge where these tools are dispersed across multiple service providers. AlphaMod, on the other hand, offers an integrated solution by centralizing all these quality assessment tools within a single, easily accessible platform.
The current pipeline is only the first brick for the development of a tool that will also handle heterogeneous information, in addition to sequence-related features, to perform better predictions for selected subsets of proteins, with non-common structural features. According to our research, the addition of supplementary data has the potential to improve the predictive accuracy in most of the predicted models.
Moreover, in future research it would be of great interest to study the feasibility of jointly using Supplementary data and AI-based integration models to improve predictions in situations where AF2’s performance level is below 50%.
Funding
This work was supported by University of Salerno, [grant numbers ORSA208455, ORSA219407, and ORSA229241]; by MIUR, [grant FFABR2017 and PRIN 2017 program, grant number: 2017483NH8]; and by BANCA D’ITALIA (NDA, AM).
CRediT authorship contribution statement
Fabio Hernan Gil Zuluaga: Software, Resources, Data Curation, Investigation, Validation, Writing – original draft. Nancy D’Arminio: Data Curation, Validation, Resources. Francesco Bardozzo: Conceptualization, Software, Formal analysis, Writing – review & editing, Supervision. Roberto Tagliaferri: Conceptualization, Formal analysis, Writing – review & editing, Supervision, Project administration, Funding acquisition. Anna Marabotti: Conceptualization, Formal analysis, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work used MARCONI100 (https://www.hpc.cineca.it/hardware/marconi100) at HPC@CINECA, which is provided under ELIXIR, the research infrastructure for life-science data.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.10.056.
3d: three-dimensional; AF2: AlphaFold 2; TBM: template-based modeling; FM: free-modeling; EU: evaluation units
Contributor Information
Roberto Tagliaferri, Email: robtag@unisa.it.
Anna Marabotti, Email: amarabotti@unisa.it.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Data availability
The data underlying this article are available at the following GitHub repository https://github.com/Fabio-Gil-Z/AlphaMod, in the article and in its online supplementary material.
References
- 1.Orengo C.A., Todd A.E., Thornton J.M. From protein structure to function. Curr Opin Struct Biol. 1999;9:374–382. doi: 10.1016/S0959-440X(99)80051-7. [DOI] [PubMed] [Google Scholar]
- 2.Russell R.B., Alber F., Aloy P., Davis F.P., Korkin D., Pichaud M., Topf M., Sali A. A structural perspective on protein–protein interactions. Curr Opin Struct Biol. 2004;14:313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Batool M., Ahmad B., Choi S. A structure-based drug discovery paradigm. Int J Mol Sci. 2019;20:2783. doi: 10.3390/ijms20112783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seffernick J.T., Lindert S. Hybrid methods for combined experimental and computational determination of protein structure. J Chem Phys. 2020;153 doi: 10.1063/5.0026025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The UniProt Consortium. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res 2023;51:D523–31.
- 6.wwPDB consortium. Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res 2019;47:D520–8.
- 7.Fiser A. Template-based protein structure modeling. Methods Mol Biol. 2010;673:73–94. doi: 10.1007/978-1-60761-842-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sali A., Blundell T.L. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 9.Dhingra S., Sowdhamini R., Cadet F., Offmann B. A glance into the evolution of template-free protein structure prediction methodologies. Biochimie. 2020;175:85–92. doi: 10.1016/j.biochi.2020.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kryshtafovych A., Schwede T., Topf M., Fidelis K., Moult J. Critical assessment of methods of protein structure prediction (CASP)—round XIV. Proteins. 2021;89:1607–1617. doi: 10.1002/prot.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., Velankar S., Kleywegt G.J., Bateman A., Evans R., Pritzel A., Figurnov M., Ronneberger O., Bates R., Kohl S.A.A., Potapenko A., Ballard A.J., Romera-Paredes B., Nikolov S., Jain R., Clancy E., Reiman D., Petersen S., Senior A.W., Kavukcuoglu K., Birney E., Kohli P., Jumper J., Hassabis D. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callaway E. DeepMind’s AI predicts structures for a vast trove of proteins. Nature. 2021;596:635. [Google Scholar]
- 15.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., Žídek A., Green T., Tunyasuvunakool K., Petersen S., Jumper J., Clancy E., Green R., Vora A., Lutfi M., Figurnov M., Cowie A., Hobbs N., Kohli P., Kleywegt G., Birney E., Hassabis D., Velankar S. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhury R., Bouatta N., Biswas S., Floristean C., Kharkar A., Roy K., Rochereau C., Ahdritz G., Zhang J., Church G.M., Sorger P.K., AlQuraishi M. Single-sequence protein structure prediction using a language model and deep learning. Nat Biotechnol. 2022;40(11):1617–1623. doi: 10.1038/s41587-022-01432-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Z., Akin H., Rao R., Hie B., Zhu Z., Lu W., Smetanin N., Verkuil R., Kabeli O., Shmueli Y., Dos Santos Costa A., Fazel-Zarandi M., Sercu T., Candido S., Rives A. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science. 2023;379(6637):1123–1130. doi: 10.1126/science.ade2574. [DOI] [PubMed] [Google Scholar]
- 18.Simpkin A.J., Sánchez Rodríguez F., Mesdaghi S., Kryshtafovych A., Rigden D.J. Evaluation of model refinement in CASP14. Proteins. 2021;89(12):1852–1869. doi: 10.1002/prot.26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scardino V., Di Filippo J.I., Cavasotto C.N. How good are AlphaFold models for docking-based virtual screening? iScience. 2022;26(1) doi: 10.1016/j.isci.2022.105920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinch L.N., Schaeffer R.D., Kryshtafovych A., Grishin N.V. Target classification in the 14th round of the critical assessment of protein structure prediction (CASP14) Proteins. 2021;89:1618–1632. doi: 10.1002/prot.26202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terwilliger T.C., Poon B.K., Afonine P.V., Schlicksup C.J., Croll T.I., Millán C., Richardson J.S., Read R.J., Adams P.D. Improved AlphaFold modeling with implicit experimental information. Nat Methods. 2022;19:1376–1382. doi: 10.1038/s41592-022-01645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 23.Orengo C.A., Taylor W.R. SSAP: sequential structure alignment program for protein structure comparison. Methods Enzym. 1996;266:617–635. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- 24.Ward J.H., Jr. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 25.Johnson L.S., Eddy S.R., Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinforma. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., Crusoe M.R., Kale V., Potter S.C., Richardson L.J., Sakharova E., Scheremetjew M., Korobeynikov A., Shlemov A., Kunyavskaya O., Lapidus A., Finn R.D. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res. 2020 8;48(D1):D570–D578. doi: 10.1093/nar/gkz1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H. UniProt Consortium. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31(6):926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remmert M., Biegert A., Hauser A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9(2):173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 29.Mirdita M., von den Driesch L., Galiez C., Martin M.J., Söding J., Steinegger M. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 2017;45(D1):D170–D176. doi: 10.1093/nar/gkw1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinegger M., Meier M., Mirdita M., Vöhringer H., Haunsberger S.J., Söding J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinforma. 2019;20(1):473. doi: 10.1186/s12859-019-3019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariani V., Biasini M., Barbato A., Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29(21):2722–2728. doi: 10.1093/bioinformatics/btt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studer G., Rempfer C., Waterhouse A.M., Gumienny G., Haas J., Schwede T. QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics. 2020;36:1765–1771. doi: 10.1093/bioinformatics/btz828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Erp M., Schomaker L. Variants of the Borda count method for combining ranked classifier hypotheses. In 7th International Workshop on frontiers in handwriting recognition, pages 443–452. International Unipen Foundation, 2000.
- 34.Sippl M.J. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 35.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., Jain S., Lewis S.M., Arendall W.B., 3rd, Snoeyink J., Adams P.D., Lovell S.C., Richardson J.S., Richardson D.C., Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., Jain S., Lewis S.M. Arendall WB 3rd, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 37.Shapiro S.S., Wilk M.B. An analysis of variance test for normality (complete samples) Biometrika. 1965;52(3/4):591–611. [Google Scholar]
- 38.Wilcoxon F. Individual comparisons by ranking methods. In Breakthroughs in statistics: methodology and distribution. Springer; New York, NY: 1992. pp. 196–202. [Google Scholar]
- 39.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J., Richardson J.S., Richardson D.C., Sammito M.D., Sobolev O.V., Stockwell D.H., TerwilligerTC, Urzhumtsev A.G., Videau L.L., Williams C.J., Adams P.D. Macromolecular structure determination using x-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr D Struct Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyd S., Vandenberghe L. Convex optimization. Cambridge University Press; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
The data underlying this article are available at the following GitHub repository https://github.com/Fabio-Gil-Z/AlphaMod, in the article and in its online supplementary material.