Summary
Six infant human teeth and 112 animal tooth pendants from Borsuka Cave were identified as the oldest burial in Poland. However, uncertainties around the dating and the association of the teeth to the pendants have precluded their association with an Upper Palaeolithic archaeological industry. Using <67 mg per tooth, we combined dating and genetic analyses of two human teeth and six herbivore tooth pendants to address these questions. Our interdisciplinary approach yielded informative results despite limited sampling material, and high levels of degradation and contamination. Our results confirm the Palaeolithic origin of the human remains and herbivore pendants, and permit us to identify the infant as female and discuss the association of the assemblage with different Palaeolithic industries. This study exemplifies the progress that has been made toward minimally destructive methods and the benefits of integrating methods to maximize data retrieval from precious but highly degraded and contaminated prehistoric material.
Subject areas: Genetics, Paleogenetics
Graphical abstract

Highlights
-
•
Interdisciplinary analyses associate Borsuka Cave burial with the Upper Palaeolithic
-
•
Human remains and herbivore tooth pendants are directly dated to >30,000 years old
-
•
Genetic analyses demonstrate this is the earliest known female infant burial
-
•
Minimally invasive methods used <67 mg of tooth dentine for multiple analyses
Genetics; Paleogenetics
Introduction
Human remains from the Upper Palaeolithic (UP) in Eurasia are sparse, but the period is marked by the increasing visibility of human burials in the archaeological record, typically associated with the Gravettian culture (∼34 - 24 ka cal BP) and occasionally even earlier (∼39 - 34 ka cal BP). Eastern Central Europe has a rich record of Palaeolithic archaeology with several sites containing human remains (Figure 1), which are associated with different technological assemblages. Directly dated human remains at Mladeč Cave (Czech Republic),1 Oblazowa Cave (Poland),2,3,4,5,6 Peștera Muierii7 and Peștera Cioclovina8 in Romania, and Troisième Caverne of Goyet (individuals Q116-1 and Q376-3) in Belgium9,10 all fall within the ∼37 - 33 ka cal BP range and are tentatively associated with Aurignacian artifacts. The Kostenki-Borshchevo complex in Russia spans different periods of the UP and has yielded numerous human remains, including the burial of Kostenki 14 (∼39 - 37 ka cal BP), which is also associated with the Aurignacian.11,12 Isolated human remains from Buran Kaya III in Crimea13 are associated with Gravettian assemblages, but this is in contrast with their early direct age of ∼36.8–35.7 ka cal BP. In Central Europe, the Gravettian is split into the Early Gravettian (∼34 - 30 ka cal BP), Pavlovian (∼31 - 29 ka cal BP) and Late Gravettian/Willendorf Kostenkian (∼29 - 24 ka cal BP).14,15 The Pavlovian period is characterized by burials containing red ochre and grave goods such as seen at Dolní Věstonice-Pavlov16,17,18,19 and Předmostí14,20 in Czech Republic. Whilst UP child burials have been identified in the region, including the burial of infants at Krems-Wachtberg (Austria),21,22,23 they are extremely rare. Much more elaborate UP burials have been found at Sunghir (Russia), containing multiple individuals, abundant ochre, and an exceptionally rich assemblage of grave goods.24,25 Several attempts at directly dating these individuals produced vastly differing results, likely due to issues of contamination, thus contributing to uncertainty around the cultural association of the site. The most recent and rigorous compound specific dates place the burials at Sunghir between 35.8 and 32 ka cal BP.11,24,25,26,27,28,29,30
Figure 1.
Map of Upper Palaeolithic sites mentioned in the text from Central and Eastern Europe containing human remains and the archaeological complex with which they are currently associated
Sites number key: 1. Goyet Cave, 2. Krems-Wachtberg, 3. Dolní Věstonice, 4. Pavlov, 5. Mladeč, 6. Předmostí, 7. Oblazowa Cave, 8. Borsuka Cave, 9. Peștera Cioclovina, 10. Peștera Muierii, 11. Bacho Kiro Cave, 12. Buran Kaya III, 13. Kostenki, 14. Sunghir.
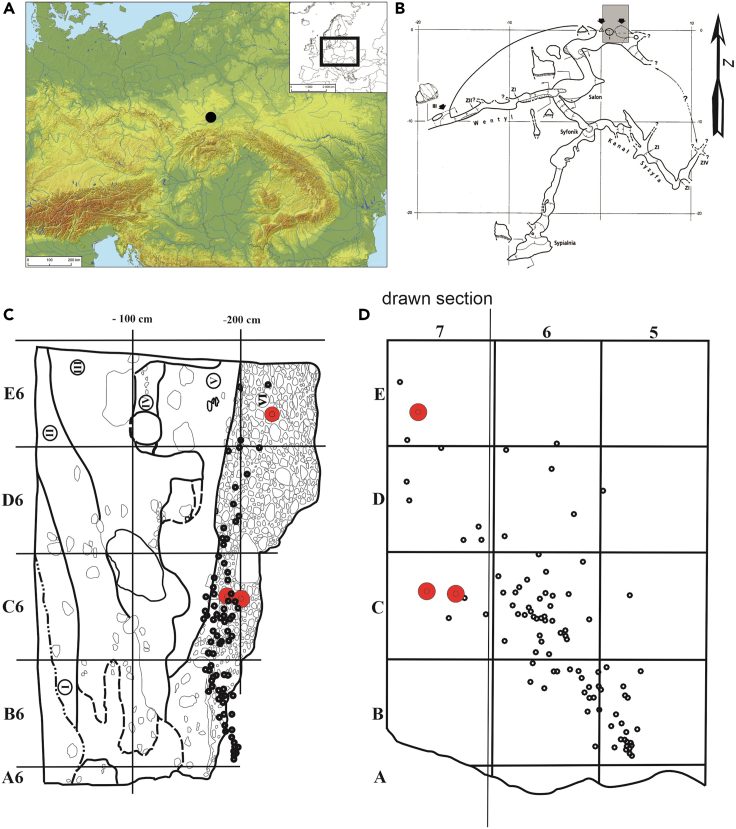
Borsuka Cave (50°9′53.94″N; 19°42′12.23″E), located in the Szklarka River valley in southern Poland (Figure 2A), adds important data to this discussion. Excavations next to the entrance of the cave in 2008–2010 (Figure 2B) uncovered six deciduous human teeth in association with 112 pierced teeth of Steppe wisent/aurochs (Bison priscus/Bos bison; n = 78) and European elk (Alces alces; n = 34), which have traces of ochre on them.31,32 Seven layers were excavated and separated into a lower Pleistocene unit (Layers VII - V) and an upper Holocene unit (Layers IV - I). At the base of the sequence, overlying bedrock, Layer VII was archaeologically sterile. In Layer VI (from 150 to 250 cm depth), the human remains and pierced teeth were found spread over several m2 in an NW linear distribution down the slope from the cave entrance. The pendants were concentrated by the southern wall of the trench at depths of 160–220 cm, with a few in the central and northern parts of the trench (Figure 2). The human teeth were located to the west of the concentration of pendants, in the northwest corner of the trench within squares C7, D7, and E7 (Figures 2C and 2D). These teeth were all identified as belonging to a child aged 12–18 months. Given the young age, it was not possible to determine the sex based on morphology, but the consistent phase of dentition indicated they likely belonged to one individual. Layer VI contained the human remains, perforated ungulate teeth, and one distal fragment of a flint blade but was otherwise archaeologically sterile. The overlying Layer V was also archaeologically and palaeontologically sterile but associated with Last Glacial Maximum (LGM) cooling. Layers IV-I contained Holocene assemblages spanning from the Mesolithic to modern (see31,33).
Figure 2.
Visuals of Borsuka Cave location, site plans, and stratigraphic profile
(A) Map showing the location of Borsuka Cave within Europe.
(B) Plan view of Borsuka Cave showing the 2008–2010 excavation trench (gray shaded rectangle).
(C) Stratigraphic profile with the location of ungulate tooth pendants (black circles) and human remains (red circles).
(D) Plan view of Layer VI showing the location of human remains and pendants. Note that three human teeth found in situ are plotted but three were found during wet-sieving so lack exact 3D coordinates. In C and D, each square is 1 m2.
In addition to the human remains, over 2,000 faunal skeletal fragments were excavated from Layer VI. The assemblage contained an unusual collection of fauna including cold steppe-tundra species, such as Rangifer tarandus (reindeer), Vulpes alopex (Arctic fox), Equus ferus (wild horse) and Coelodonta antiquitatis (woolly rhino), alongside taxa adapted to a warmer forested environment, including Alces alces (elk), Bos primigenius (aurochs), Martes martes (pine marten), Lynx lynx (Eurasian lynx), Castor fiber (beaver) and Meles meles (badger).33 The majority of the Pleistocene skeletal remains likely represent a natural accumulation, with signs of carnivore modifications on 1.7% of the mammalian and bird bones and no signs of anthropogenic modification aside from the pierced teeth.
Two of the herbivore tooth pendants were directly radiocarbon dated to 32,890 - 30,760 cal BP at 95.4% probability (Poz-32394: 27,350 ± 450 14C BP; 14C errors reported at 1σ) and 29,900–29,120 cal BP (Poz-38236: 25,150 ± 160 14C BP). A reindeer metatarsus from the same layer was dated to 31,060 - 30,270 cal BP (Poz-38237: 26,430 ± 180 14C BP).32,33 While the dates indicated a Mid-Upper Palaeolithic age for the Layer VI assemblage, the lack of agreement between the dates from the two pendants at the 95% range suggested that the pendants are not strictly contemporaneous. This, along with the distribution of the herbivore and human teeth, leaves open the question of the association between the pendants and the human remains, necessitating further chronological investigation. However, due to their small size, direct dating of the human teeth was initially determined to be impossible.
Although alternative theories have been discussed,32 the absence of associated “domestic” archaeology led to the interpretation that the assemblage represents an infant burial and grave goods that were disturbed post-deposition, resulting in the absence of a burial pit and their linear spread down the slope.32 The lack of any Palaeolithic archaeology necessitates relying on the dating and stylistic analysis of the pendants for determining the cultural association of the burial. The existing pendant 14C dates are roughly contemporaneous with Pavlovian burials ∼300-200 km away at Dolní Věstonice-Pavlov and Předmostí (Czech Republic)14,16,17,18,19,20 and infant burials ∼400 km away at Krems-Wachtberg (Austria).21,22,23 However, the typology of the pendants themselves are similar to those found at Mladeč32 and suggest a Late Aurignacian association. In addition, there have been numerous examples where human remains assumed to be Palaeolithic in origin in fact represent Holocene intrusions into Palaeolithic contexts.34,35,36,37,38,39 Given the small nature of fragmented deciduous human teeth and presence of Holocene archaeology in the upper layers, the possibility of this being the case at Borsuka Cave could not be ruled out.
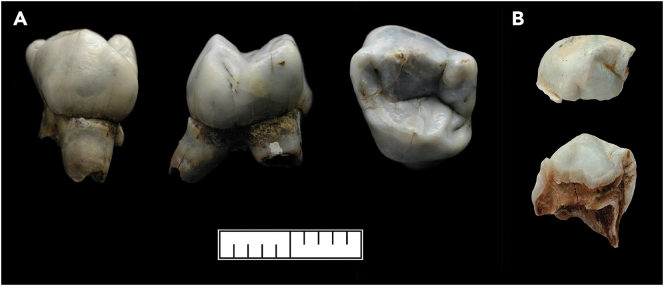
Significant advances have been made in recent years for the radiocarbon dating and genetic analysis of ancient and highly degraded skeletal material, including reducing sample sizes for dating,19,40,41,42 combining multi-method extractions from a single sample,43 non-destructive pre-screening of organic preservation,44 treatments for removing contaminant DNA and/or enriching for endogenous ancient DNA (aDNA),45,46,47,48,49 and non-destructive aDNA extraction from osseous artifacts.50 Such developments enable biomolecular analyses of material which were previously impossible. Considering the expected age and very limited amount of material available from the human and pierced herbivore teeth from Borsuka, we devised a multi-disciplinary study to investigate (i) if the human teeth originate from a single individual, (ii) the chronological relationship of the human teeth and pendants, and (iii) the association of the assemblage to different Palaeolithic cultures. In order to confirm the Palaeolithic origin of the teeth and ascertain if the human remains are contemporaneous with the pendants, we undertook radiocarbon dating of two deciduous human teeth (Figure 3) and six of the pierced herbivore teeth (Figure 4). From the human material, we selected two teeth (C7/675, C7/683) with partially intact roots to preserve the crown morphology and selected herbivore pendants from different squares to test across the scattered assemblage. Ancient DNA analysis was attempted on the same two human teeth to determine if they belonged to one individual and to see how this individual related to other Upper Palaeolithic individuals. The assemblage at Borsuka Cave represents a challenge in terms of the extremely limited material available, and the inherent issues of poor preservation and contamination of ancient samples for both 14C and aDNA. Nevertheless, we aimed to maximise data retrieval from extremely small samples to provide additional information about the Borsuka assemblage within the broader context of the UP.
Figure 3.
The two human teeth from Borsuka cave analyzed in the study
(A) The deciduous molar C7/675 (uldm1) and (B) molar fragment C7/683. Scale bar is 1 cm.
Figure 4.
Four pendants made of large ungulate teeth from Borsuka Cave dated in the study
(A) European elk (Alces alces) left lower incisor, sample ID: C7/656; (B) European elk (Alces alces) left lower incisor, sample ID: B5/910; (C) Bos/Bison (Bos primigenius/Bos bison) right lower incisor, sample ID: B5/913; (D) Bos/Bison (Bos primigenius/Bos bison), sample ID: C7/658. Scale bar is 2 cm.
Results
Radiocarbon dating
The collagen yield of the two human teeth (4.3–8.5%) was within the range of the herbivore teeth (3.0–9.1%), consistent with degraded Palaeolithic material but all falling well above the ∼1% minimum required for dating. The elemental values (C%, N%, C/N) of all extracts fall within the commonly accepted ranges of well-preserved collagen (C%: ∼30–45%; N%: ∼11–16%; C/N: 2.9–3.6), indicating that the collagen extracts were suitable for radiocarbon dating51 (Table 1). However, several fall at the limit (3.5–3.6) of the accepted C/N range, indicating low levels of contamination may be present in the samples.52,53 For C7/675, C7/658, C6/494, sufficient collagen was available for additional quality analysis using Fourier Transform Infrared Spectroscopy (FTIR). All extracts had FTIR spectra characteristic of ancient bone collagen,54,55,56,57 but the higher intensity of the peak at ∼2900 cm−1 in the spectra of the human tooth C7/675 may be indicative of external contamination (Figure S1).
Table 1.
Pretreatment and radiocarbon dating of humans and herbivore pendants from Borsuka Cave
| Sample ID | Material | Sampled (mg) | Collagen (mg) | % Collagen | δ13C (‰) | δ15N (‰) | %C | %N | C/N | AMS lab number | combined BP | combined err | 95.4% cal BP | n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C7/675 | Human tooth - uldm1 | 41.1 | 3.5 | 8.5 | −20.3 | 14.9 | 42.9 | 13.7 | 3.6 | Aix-12047 | 25100 | 140 | 29860–29090 | 2 |
| C7/683 | Human tooth - dm | 23.4 | 1 | 4.3 | −20.1 | 13.6 | 39.7 | 13.3 | 3.5 | Aix-12058 | 26610 | 240 | 31160–30320 | 3 |
| C7/656 | Elk incisor pendant | 34.3 | 2.6 | 7.6 | −20.6 | 4.7 | 42.3 | 14.2 | 3.5 | Aix-12041 | 29310 | 240 | 34350–33260 | 2 |
| C7/658 | Bos/Bison incisor pendant | 48.5 | 4.4 | 9.1 | −20.3 | 7.4 | 42.7 | 15.2 | 3.3 | Aix-12042 | 30100 | 260 | 35190–34150 | 2 |
| C6/479 | Bos/Bison incisor pendant | 53.9 | 3.6 | 6.7 | −19.6 | 6.5 | 41.8 | 15 | 3.3 | Aix-12043 | 30080 | 260 | 35170–34130 | 2 |
| C6/494 | Elk incisor pendant | 67.4 | 4.5 | 6.7 | −19.8 | 2.3 | 42.7 | 15.1 | 3.3 | Aix-12044 | 30460 | 190 | 35290–34480 | 3 |
| B5/910 | Elk incisor pendant | 49.2 | 1.5 | 3 | −20.4 | 3.1 | 37.8 | 13.4 | 3.3 | Aix-12045 | 29310 | 370 | 34610–33860 | 3 |
| B5/913 | Bos/Bison incisor pendant | 49.9 | 2.9 | 5.8 | −20.1 | 6.3 | 40.4 | 14.5 | 3.3 | Aix-12046 | 30080 | 260 | 35170–34140 | 2 |
Combined dates of multiple replicates (n) are reported here with individual replicate dates and X2 values reported in Table S1.
The bulk collagen δ13C and δ15N values of the two human teeth fall within the range of stable isotopic values seen for other Mid-Upper Palaeolithic humans in Eurasia,19,23,58,59,60,61,62 with very high δ15N values that are well above the herbivorous signature of the Bos/Bison and elk teeth (Figure S2). Although the δ13C values of the two human teeth agree within instrumental error, the δ15N value of C7/675 is higher than C7/683 by 1.3‰. Given that the teeth are both deciduous molars with similar formation times, the difference is more likely to be indicative of contamination, or of separate individuals, rather than palaeodietary. The elevated δ15N values are at the highest end of the range seen in other Mid-UP humans and in the range of two trophic levels (trophic level increase: ∼3‰–5‰) above the herbivores. This is consistent with a breastfeeding signal given the age of the infant, but without comparative adult data and further chemical analyses it is not possible to elucidate this further.
Given the small size of the dentine samples (23.4–67.4 mg) and extracted collagen (1–4.5 mg), we followed a previously established approach for small samples.41,42 Multiple 14C dates were obtained from each collagen extract using both graphitization and CO2 gas ion source (GIS) methods, and subsequently combined. X2 tests were carried out to determine the statistical agreement between replicate dates from the same extract. All of the 14C dates obtained fall within the Mid-UP range, from 31,070 ± 770 14C BP to 24,830 ± 290 14C BP, or ∼35,290 to 29,090 cal BP (Figure 5; Table 1; Tables S1 and S2). For the herbivore pendants, all six pendants were dated to 35,290 - 33,260 cal BP (95.4% probability), much older than dates obtained previously on two of the pendants, overlapping with the Late Aurignacian/Early Gravettian periods. For the two human teeth, replicate measurements agree statistically for each collagen extract but the dates from C7/683 (Aix-12058 combined age: 26,610 ± 240 14C BP; 31,160 - 30,320 cal BP at 95.4% probability) are older than C7/675 (Aix-12047 combined age: 25,100 ± 140 14C BP; 29,860 - 29,090 cal BP at 95.4% probability) by ∼1500 14C years (Figure 5; Table S1). When the dates from Layer VI obtained thus far are modeled together in one phase in OxCal, the assemblage spans 6,290-4,770 years, between 36,650 and 27,620 cal BP (Figure 5; Table S3). No dates are available from the overlying and underlying layers to constrain the modeled range, due to a lack of material suitable for dating so none of the dates were identified as outliers. The radiocarbon data is included in Table S1, the background collagen measurements in Table S2 and the modeled ranges of the Borsuka data in Table S3.
Figure 5.
Single phase outlier model showing the posterior probability distributions of the 14C dates from Borsuka Cave Layer VI generated in this study (Aix-) and previously (Poz-)
The bracket beneath each distribution shows the 95% probability range. The outlier probability (O) is given in the format “[O: posterior outlier probability/prior outlier probability].” The ranges are given in Table S3.
Ancient DNA analysis
Two microsamples between 4.7 and 12.9 mg of dentine powder were drilled from each of the two human teeth (C7/675 and C7/683). The resulting four tooth powder sub-samples underwent DNA extraction, single-stranded library preparation and shotgun sequencing (see method details). Between 1,628,190 and 8,575,605 sequences were generated per library, of which between 2,495 and 37,436 mapped to the human reference genome (hg1963) for each library. While no ancient DNA was detected in C7/683, one of the sub-samples from C7/675 was identified as containing ancient DNA based on the observation of between 12.4% and 20.6% C-to-T substitutions on the terminal ends of recovered DNA (Table S4), which is characteristic of ancient DNA.64,65 The duplication rates, or number of times each DNA fragment was sequenced, for these libraries were all around 1 (Table S4), indicating that not all recovered DNA fragments were sequenced. Modern human DNA contamination for libraries produced from this subsample was estimated at 54–56% (Table S4) using the method AuthentiCT66 which calculates levels of contamination based on deamination patterns.
Despite not detecting ancient DNA in C7/683 to the limits of our resolution, libraries from both teeth were subsequently enriched for human mitochondrial (mt) DNA via hybridization capture,67 in an attempt to determine if both likely stem from the same individual (Table S5). Libraries from C7/683 were still included in hybridization captures as the shotgun sequencing was not exhaustive, and the content of nuclear and mitochondrial DNA has been shown to defer in ancient specimens,68 leaving the possibility that small amounts of endogenous DNA remained undetected. The enriched libraries contained between 243,078 and 938,984 sequences longer than 34 base pairs that mapped to the human mtDNA revised Cambridge Reference Sequence69 with elevated C-to-T substitutions (21.3–35.1% on the 5′ end and 14.5 to 24.6% on the 3′ end). As modern human contamination was detected in each library, ranging from 2.93% to 39.94%, all subsequent analyses were restricted to putatively deaminated fragments that contained a C-to-T substitution within the first three and/or last three terminal bases. Between 15,269 and 60,728 deaminated fragments (corresponding to mtDNA coverages of 46- and 223-fold, respectively) were used to reconstruct near-complete consensus mtDNA genomes. The resulting mtDNA haplotypes from each tooth were the same, consistent with the teeth either being from the same individuals or the same maternal lineage. However, as neither mtDNA genome was complete, we cannot exclude that individually discriminating variants were not detected. The more complete mtDNA genome from C7/675 was then used for tree building and molecular branch shortening (Tables S6 and S7). The resulting molecular date was estimated at 33,533 years BP (95% highest posterior density interval: 28,200 - 38,935 years BP) and the mtDNA genome falls within haplogroup U6 (Figure 6A). Haplogroup U6 is most commonly observed in Northern Africa in present-day humans,70,71 and has previously only been observed in Palaeolithic Europe in the specimens from the site of Peştera Muierii in Romania (∼34,000 years old).9,72,73 Similar to the Muierii 1 and 2 mtDNA genomes, which were subsequently determined to belong to the same individual,73 the Borsuka mtDNA genome is basal to the U6 haplogroup, sharing the 3348G, 10517A, and 16172C positions. Importantly, among the overlapping positions (6 missing positions) no differences are observed between the Muierii 2 and Borsuka mtDNA genomes. Even if both Borsuka teeth and Muierii 2 share an mtDNA genome, this only indicates that they likely lived within ∼2,500 years of each other (Figure S3, method details).
Figure 6.
The genetic analysis of the Borsuka C7/675 individual in relation to other modern and ancient humans
(A) The calculated allele sharing between the Borsuka individual and a selection of modern and ancient individuals (X) where warmer colors represent higher amounts of shared alleles. The f3 statistic f3(X, Borsuka; Mbuti) was calculated using 28,213 - 31,393 overlapping SNPs for modern populations and 6,697 - 28,642 SNPs for ancient individuals. Ancient individual key: 1- Goyet Q116, 2- EL Miron, 3- Dolní Věstonice16, 4- Bichon, 5- Oase1, 6- Bichon, 7- Mal’ta1, 8- Karelia, 9- Yana2, 10- Tianyuan, 11- Ust-Ishim, 12- Kostenki14, 13- Sunghir3.
(B) The projection of 20 ancient individuals onto a principal component analysis of 1,267 modern individuals. Ancient individuals are colored based on their association with the Aurignacian (dark blue), Gravettian (orange), or IUP (light blue) archaeological assemblages or in green if their technological association is uncertain.
(C) A subset of the phylogenetic tree generated with BEAST2. Ancient humans are colored in blue (Borsuka in red). Nodes are labeled with their posterior probability. The x axis indicates calibrated years before present.
In order to determine the genetic affinity of the child buried in Borsuka Cave to other individuals from the Upper Palaeolithic, we enriched the libraries from C7/675 for ∼1.2 million single nucleotide polymorphisms (SNPs) in the human genome known to be informative for studying human population history.74,75 While the resulting libraries covered 513,237 SNPs, they were also identified as containing approximately 56% modern human contamination (Table S8). This required all downstream analysis to be restricted to the 46,286 putatively deaminated fragments, which were used to determine the sex of the child. As the ratio of autosome, X, Y chromosome SNPs in the 1240k does not follow conventional expectations due to the respective target size, we used an adjustment of the X and Y-rate as has been performed previously with this type of data.9 This resulted in an “X-rate” of 0.68 and “Y-rate” of 0.04, consistent with the individual being female (Table 2). When projected onto a principal component analysis (PCA) of modern-day individuals from West Eurasia, Central Asia and East Asia, the Borsuka individual clustered with previously published Upper Palaeolithic Western Eurasians and near modern day Western Eurasians (Figure 6B). The genetic sharing between the Borsuka individual and previously published modern and ancient individuals was calculated with f3-statistics, using present-day Mbuti individuals as an outgroup population (Figure 6A). Consistent with the PCA analysis, the greatest affinity among modern day populations was to Western Eurasians. Basal Eurasian ancestry was not detected in the Borsuka individual (Tables S9 and S10, method details). Among ancient individuals, the Borsuka individual shared the most alleles with the ∼35 ka cal BP Bacho Kiro Cave individual (BK1653), ∼34 ka cal BP Muierii 1, ∼31 ka cal BP Věstonice 16 and ∼34 ka cal BP Sunghir 3 individuals when tested with f3-statistics (Table S11). When directly comparing the genetic affinity of the Borsuka individual with other ancient individuals with D-statistics, the Borsuka female has a greater affinity to the Gravettian and Aurignacian individuals (Figures S4 and S5; Table S12). However, no significant difference was observed in the affinity of the Borsuka child to the Věstonice vs. Sunghir vs. BK1653 vs. Muierii 1 individuals, precluding the direct association of the Borsuka girl with one of these groups.
Table 2.
Sex determination based on coverage of nuclear SNPs in the '1240k′ array using putatively deaminated reads
| Sample ID | Material | Nyo (# bases covered on Y chromosome) | Nxo (# bases covered on X chromosome) | Nautoo (# bases covered on autosomal chromosomes) | Ny expected (targets on Y chromosome) | Nx expected (targets on X chromosome) | Nauto expected (targets on autosomal chromosomes) | X-ratio (Nxo/Nautoo)/(Nxe/Nautoe) | Y-ratio (Nyo/Nautoo)/(Nye/Nautoe) |
|---|---|---|---|---|---|---|---|---|---|
| C7/675 | Human tooth - left udm1 | 49 | 1,312 | 45,032 | 32,670 | 49,704 | 1,150,639 | 0.674 | 0.038 |
A Y-ratio <0.05 indicates a female and >0.2 indicates a male.
Discussion
The chronological and genetic data presented here confirms the Upper Palaeolithic origin of the infant remains from Borsuka Cave, making these remains currently the oldest female infant burial identified to date. While it is not possible to confirm that the teeth come from the same individual without nuclear DNA, the mitochondrial analysis is consistent with the two teeth belonging to the same individual or the same maternal lineage.
Radiocarbon dating is based on the exponential decay of 14C over time. As the concentration of 14C in modern carbon is therefore much larger than in ancient samples, any contamination with modern carbon will make 14C dates younger than the true age of the sample, with the effects getting progressively worse for more ancient material. Given the ubiquity of modern carbon introduced via the burial environment, handling, storage and analysis of artifacts, when inconsistent dating results are obtained from Palaeolithic material, older 14C ages are generally considered more accurate.29,76 The direct date from human tooth C7/683 is older than the date from C7/675. If the two teeth originate from the same individual, this implies that either 1) the older age of C7/683 is correct and the date from C7/675 is an under-estimation of the true age due to the presence of external carbon contamination, or 2) both dates are under-estimations.
Alternatively, the teeth could originate from two infants and both dates could be correct. In addition to the discrepancy in age between the direct human dates and the range of ages from the pendants, this may also be indicated by the difference in δ15N values between the teeth and the scatter of the assemblage over several square meters. Given the consistent form of pendant manufacture and that the mtDNA analysis indicates the teeth are at least from maternal relatives (if not from the same individual), a group from the same maternal lineage would need to have used the site for 4,000–6,000 years to account for the dating results in this scenario. The elevated C/N value and additional FTIR peak at ∼2900 cm−1 in the C7/675 extract do suggest contamination may have affected the date, and potentially the δ15N value. Thus, the most parsimonious interpretation of the results is that the two teeth originate from the same individual and are affected by contamination. If we consider the teeth as originating from one individual, the older date from C7/683 is more likely to be accurate than the younger date from C7/675 as contamination typically decreases the age. Given the small amount of material available for sampling and the C/N values from both teeth of 3.5–3.6, we consider the ∼31.2–30.3 ka cal BP date from C7/683 to be a minimum age for the human remains.
The older date from C7/683 is contemporaneous with the dates previously obtained from two Borsuka pendants and the ∼31 ka cal BP Pavlovian burials at Dolní Věstonice-Pavlov and Krems-Wachtberg, which would support the association of Borsuka with the Pavlovian culture in Central Europe (Figure 7). However, the new 14C dates on the herbivore tooth pendants range in age from 35,290 to 33,260 cal BP (95.4% probability). These new dates for the pendants are older than the ages obtained from the human infant teeth, but also much older than the dates previously obtained on two pendants and a reindeer bone from the same layer (Figure 5).
Figure 7.
Calibrated radiocarbon dates (95.4% range) from Borsuka Cave infant teeth and pendants compared to UP human burials and human remains in Central and Eastern Eurasia mentioned in the text
Human remains tentatively associated with Aurignacian assemblages are shown in blue and Gravettian assemblages in orange while uncertain cultural associations are shown in gray. The Borsuka infant dates are conservatively considered minimum ages.
It is possible that the burial assemblage dates to ∼31 - 30 ka cal BP, and the necklace was constructed from a collection of herbivore teeth of varying age, including some that were pierced and used for the construction of an ornament long after the death of the animal. The solitary lifestyle of European elk (Alces alces) implies that the elk teeth were unlikely to have been collected during one event. A recent study of the Sr isotopic compositions and trace element analysis of the enamel from four of the elk tooth pendants (at least 3 individuals) demonstrated a non-local origin of the teeth, indicating that the pendants were transported ∼250 km to Borsuka Cave from an area near the Austria/Slovakia border or northern Hungary, on the southern side of the Western Carpathians.77 This evidence for exchange or human regional mobility is supported by Borsuka Cave being the only site in the region containing evidence of the presence of elk ∼27 ka BP (beginning of MIS2) despite favorable environmental conditions.33,78,79,80,81 The presence of Trans-Carpathian raw stone materials in Central Europe also shows high human mobility around the Western Carpathian mountain region.80,82 This evidence suggests that the collection of the herbivore teeth over a period of time is feasible, however, it seems unlikely that the pendants represent animals varying in age by over 5,000 years. Alternatively, low levels of contamination may be affecting some of the 14C results with the ∼32 - 29 ka cal BP ages under-estimating the true age of the pendants and human teeth.
The older ∼35.3–33.3 ka cal BP range of dates from the pendants overlap in time with the ∼36.8–34.7 ka cal BP range of directly dated human remains from Mladeč Cave (Czech Republic)1 (Figure 7). It has been suggested that the relatively shallow human remains at Mladeč could be the result of intentional deposition,35,83,84 but the lack of contextual information means this hypothesis cannot be tested. Despite their uncertain context, based on the dates, the Mladeč humans have been associated with Aurignacian artifacts found elsewhere in the cave, including Mladeč-type bone points and pendants made from the teeth of large ungulates (e.g., elk, horse, bison/aurochs).85 In comparison to other UP perforated teeth, the Borsuka Cave pendants show close similarity to perforated teeth discovered at Mladeč and generally from Aurignacian sites.85,86,87 The entire roots of the perforated teeth were heavily scraped on both surfaces, remarkably reducing the root thickness and the perforations were made very close to the root apex by drilling from both sides. The only difference between pendants discovered at Borsuka and Mladeč can be observed in the larger range of species from which teeth were used at Mladeč. Thereby the assemblage from Borsuka cave represents the largest collection of pendants made of large ungulate teeth potentially associated with the Aurignacian culture. Currently, no aDNA is available from the Mladeč individuals to investigate this link further.
While the origin of the Gravettian has been widely debated,14 the relatively large collection of burials across Eurasia has provided a significant body of morphological and behavioral data on Gravettian humans across a wide geographical and chronological range. Recent aDNA studies10,88 have demonstrated a genetic distinction between the Gravettian cultures in Central Eastern Europe and Southwestern Europe. The oldest humans discovered to date with genetic continuity to Gravettians and modern West Eurasians are represented by three different genetic ancestry components from the ∼35,000-year-old GoyetQ116-1 individual,9 the ∼35,000-year-old individual from Bacho Kiro Cave (BK1653)89 and the ∼38,000-year-old Kostenki 149,.90 The BK1653 individual is related, but not identical, to the genetic ancestry of GoyetQ116-1 which contributes to the genetic ancestry of the Gravettian individuals in Southwestern Europe, while the former contributes ancestry to the Central European Gravettian individuals. Genetic ancestry from Kostenki 14 has been found in all individuals associated with the Gravettian culture. We aimed to clarify the genetic relationship of the Borsuka individual to individuals of different Palaeolithic cultures, however these associations were limited due to the low amounts of data recovered. The Borsuka individual shares a basal U6 mtDNA haplotype with Peştera Muierii 2, but there is not sufficient nuclear data to determine the full genetic relationship between these individuals in context with other ancient humans. The available data show that the Borsuka individual has the most nuclear genetic affinity to the Bacho Kiro 1653 individual (presumed to be associated with the Aurignacian), as well as individuals from Dolní Věstonice (Gravettian) and Sunghir (early UP). Interestingly, the Borsuka individual is significantly closer to the Bacho Kiro individual than they are to Kostenki 14. This is consistent with Borsuka being an UP Central European, but does not determine a closer affinity to Gravettian or Aurignacian individuals within Central Europe. The 14C dates provide a minimum age of 31.3–30.3 ka cal BP for the human remains, which agrees with the molecular date estimation of 33.5 ka that falls roughly at the border between the Late Aurignacian and Early Gravettian periods.
The Borsuka Cave assemblage represents an important data point in the catalog of Palaeolithic human remains for investigating the geographical and chronological range of UP burial practices. Of the very limited number of Upper Palaeolithic infant burials discovered and successfully analyzed with aDNA, this is the first female identified. The previously oldest confirmed female infant burial was found in Mesolithic Italy,91 at least 20,000 years after the burial of the Borsuka girl. This study therefore provides the first evidence that child burials incorporating grave goods were not limited to males during the UP. This study further highlights the challenges of working with limited and poorly preserved ancient materials, and emphasizes that employing an interdisciplinary, multi-method approach can still provide valuable insights to allow in-depth investigations of Palaeolithic contexts.
Limitations of the study
Palaeolithic human remains are rare, and the remains of UP infants even more so. The small size and partial fragmentation of the infant teeth presented an exceptionally limited amount of suitable material for analysis. We aimed to take as little material as possible and therefore limited destructive sampling to two of the six teeth to preserve the morphology of the crowns and ensure their preservation for future studies.
Modern carbon contamination in a 14C sample produces an under-estimation of the true age of the material. As sample size decreases, the risk of contamination increases, which is clearly a consideration for the data presented. This is highlighted by the lack of agreement in dates from the two human teeth from the (potentially) same individual and their slightly elevated C/N values, indicating that low levels of C contamination may be present either due to incomplete removal of environmental contaminants during pretreatment or introduced in the laboratory. Given the ubiquity of carbon contaminants in the environment and laboratory, older ages produced from the same sample material are generally considered to be more accurate and reliable. Rigorous compound specific dating approaches can be employed to isolate endogenous hydroxyproline in bone collagen to overcome contamination issues.27,92,93 However, the small proportion of hydroxyproline in collagen means this approach requires large starting sample sizes, thus making the approach not feasible for this study. As contamination of the human dates is indicated we therefore consider them to be minimum ages. The molecular dating technique (calibrated using other directly radiocarbon dated human remains) indicates an age of ∼33.5 ka BP (albeit with a very large confidence interval), which supports the ∼31 ka cal BP being a minimum age. The new data presented nevertheless confirm the UP origin of the human remains. Given the challenges and the limited data available, we cannot conclude a definitive age for the burial assemblage or a more definitive association with a specific Palaeolithic culture. However, even using very small sample sizes for dating (in the order of aDNA sample sizes), we confirm the placement of the assemblage around the border of the Late Aurignacian/Early Gravettian, supported by the aDNA analysis and the typology of the pendants.
The high levels of modern human contamination and limited amount of endogenous DNA present in both infant teeth limited the power of determining different genetic affinities beyond broad population genetic relationships. This resulted in reduced coverage of both the mtDNA and nuclear genomes. As shown in Figures S6–S9, decreased amounts of data result in missing some genetic affinities. It is likely that with more data it would have been possible to deconvolute the association of the Borsuka girl to Bacho Kiro 1653, or the Věstonice and Sunghir clusters, but unfortunately the limited data prevented this.
The limitations arising due to contamination emphasize the importance of taking efforts to minimize the contamination of archaeological materials. This includes wearing gloves and face masks during excavation and handling of human remains and artifacts, appropriate storage and consideration of sampling prior to consolidants or glues being applied.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Osteological remain | This study | C7/675 |
| Osteological remain | This study | C7/683 |
| Osteological remain | This study | C7/656 |
| Osteological remain | This study | C7/658 |
| Osteological remain | This study | C6/479 |
| Osteological remain | This study | C6/494 |
| Osteological remain | This study | B5/910 |
| Osteological remain | This study | B5/913 |
| Chemicals, peptides, and recombinant proteins | ||
| 0.5M EDTA pH 8.0 | AppliChem | Cat.no. A4892 |
| 100% Tween 20 | Merck | Cat.no. P5927-100ML |
| 10 mg/mL Proteinase K | Sigma Aldrich | Cat.no. P6556 |
| H20, HPLC grade | Merck | Cat.no. 270733 |
| Guanidine hydrochloride | Sigma Aldrich | Cat.no. G3272 |
| Isopropanol | Merck | Cat.no. 109634 |
| 100% Ethanol | Merck | Cat.no. 100983 |
| PE Buffer | Qiagen | Cat.no. 19065 |
| 1 M Tris-HCl pH 8.0 | AppliChem | Cat.no. A4577 |
| 3M Sodium Acetate, pH 5.2 | Merck | Cat.no. S7899-500ML |
| 5 M NaCl | Merck | Cat.no. S5150-1L |
| 20% SDS | Thermo Fisher Scientific | Cat.no. AM9820 |
| 10x AmpliTaq Gold buffer (without Mg) | Life Technologies | Cat.no. 4379874 |
| 1M NaOH | Roth | Cat.no. 9062.3 |
| 20x SSC | Thermo Fisher Scientific | Cat.no. AM9763 |
| Silica magnetic beads | G-Biosciences, VWR | Cat.no. 786-915 |
| T4 RNA ligase reaction buffer including 50% (wt/vol) PEG 8000 | NEB | Cat.no. B0216L |
| ATP solution (100 mM) | Thermo Fisher Scientific | Cat.no. R0441 |
| T4 polynucleotide kinase 10 U uL−1 | Thermo Fisher Scientific | Cat.no. EK0031 |
| T4 DNA ligase high concentrated 30 U uL-1 | Thermo Fisher Scientific | Cat.no. EL0013 |
| Fast AP (1U/uL) | Thermo Fisher Scientific | Cat.no. EF0651 |
| Klenow fragment (10 U/uL), including 10X reaction buffer | Thermo Fisher Scientific | Cat.no. EP0052 |
| T4 DNA Ligase, 5 U/uL, including 10X reaction buffer and 50% (wt/vol) PEG-4000 | Thermo Fisher Scientific | Cat.no.EL0012 |
| Dynabeads MyOne Streptavidin C1 beads | Life Technologies | Cat.no. 65001 |
| PEG-8000 powder | Promega | Cat.no. V3011 |
| Sera-Mag SpeedBeads | Sigma Aldrich | Cat.no. GE6515-2105-050250 |
| UltraPure DNase/RNase-Free Distilled Water | Invitrogen | Cat.no. 10977049 |
| dNTPs 25mM each | Thermo Fisher Scientific | Cat.no. R1121 |
| Herculase II Fusion DNA polymerase, including 5X Herculase II reaction buffer | Agilent | Cat.no. 600679 |
| Human Cot-1 DNA | Thermo Fisher Scientific | Cat.no. 15279011 |
| 2x HI-RPM hybridization buffer, including Agilent blocking agent (dissolve in 1250uL water) | Agilent | Cat.no. 5188-5380_25 mL |
| Maxima Probe qPCR Master Mix (2x) | Thermo Fisher Scientific | Cat.no. K0261 |
| AccuPrime Pfx DNA polymerase, including 10x AccuPrime reaction mix | Thermo Fisher Scientific | Cat.no. 12344-024 |
| Maxima SYBR Green qPCR Master Mix (2X) | Thermo Fisher Scientific | Cat.no. K0253 |
| Ultrapure H2O (resistivity value: 18.2 MΩ·cm @ 25 °C; ≤ 5 ppb) | MilliporeSigma | MilliQ Element A10 Water Purification system |
| HCl 37% | Roth | Nr. 9277.1 |
| HCl (6N) | Roth | Nr. 0281.1 |
| NaOH (2N) | Roth | Nr. T135.1 |
| Ezee Filters 9ml | Elkay Labs | Cat.no. 127-3193-000 |
| Vivaspin Turbo 15 Ultrafilters PES (30 kDa MWCO) | Sartorius | Cat.no. VS15T01 |
| AMS standard reference material Oxalic acid | NIST | SRM 4990C |
| IAEA-CH-6 (EA-IRMS standard) | IAEA | https://nucleus.iaea.org/sites/ReferenceMaterials/Pages/IAEA-CH-6.aspx |
| IAEA-CH-7 (EA-IRMS standard) | IAEA | https://nucleus.iaea.org/sites/ReferenceMaterials/Pages/IAEA-CH-7.aspx |
| IAEA-N-1 (EA-IRMS standard) | IAEA | https://nucleus.iaea.org/sites/ReferenceMaterials/Pages/IAEA-N-1.aspx |
| IAEA-N-2 (EA-IRMS standard) | IAEA | https://nucleus.iaea.org/sites/ReferenceMaterials/Pages/IAEA-N-2.aspx |
| Methionine | Elemental Microanalysis, Okehampton, UK | B2100 |
| Critical commercial assays | ||
| Custom capture probes | Agilent | N/A |
| P5 8bp primer plate (384) | Eurogentec | N/A |
| P7 8bp primer plate (384) | Eurogentec | N/A |
| MinElute PCR Purification kit | Qiagen | Cat.no. 28006 |
| QIAquick Nucleotide Removal kit | Qiagen | Cat.no. 28306 |
| Deposited data | ||
| European Nucleotide Archive (nuclear DNA data) | This study | PRJEB66365 |
| Dryad (mtDNA genome) | This study | https://doi.org/10.5061/dryad.47d7wm3m4 |
| Oligonucleotides | ||
| (ddC, dideoxycytidine; TEG, triethylene glycol spacer; ∗, phosphothioate linkage; [N], 2′-O-methyl-RNA; {N}, locked nucleic acid (LNA); SpacerC12, 12-carbon spacer; FAM, fluorescein amidite; BHQ1, Black Hole Quencher-1) | ||
| TL 181 100uM (desalted), 1st adapter Phosphate-AGATCGGAAGAAA[A][A][A][A][A][A][A]-TEG-Biotin |
IDT | N/A |
| TL 159 100uM (desalted), splinter SpacerC12-[A][A][A]CTTCCGATCTNNNNNNNN[A]-AminoC6 |
Eurogentec | N/A |
| CL 128 100uM (HPLC), extension primer GTGACTGGAGTTCAGACGTGTGCTCTTCC∗G∗A∗T∗C∗T |
Eurogentec | N/A |
| CL 53 500uM (HPLC), 2nd adapter strand 1 CGACGCTCTTC-ddC |
Sigma-Adrich (Merck) | N/A |
| TL 178 500uM (desalted), 2nd adapter strand 2 Phosphate-GGAAGAGCGTCGTGTAGGGAAAGAGTGTA |
Eurogentec | N/A |
| CL 304 100uM (HPLC), control DNA Phosphate-ATTCAGCTCCGGTTCCCAACGATCAAGGC GAGTTACATGA-Phosphate |
Sigma-Adrich (Merck) | N/A |
| IS5 100uM (desalted), forward primer AATGATACGGCGACCACCGA |
Sigma-Adrich (Merck) | N/A |
| IS5 biotinylated 100uM (desalted), forward primer Biotin-AATGATACGGCGACCACCGA |
Sigma-Adrich (Merck) | N/A |
| IS6 100uM (desalted), reverse primer CAAGCAGAAGACGGCATACGA |
Sigma-Adrich (Merck) | N/A |
| CL105 100uM (desalted), forward primer ACACTCTTTCCCTACACGACGCTCTTCCTCGTCGTTT GGTATGGCTTC |
Sigma-Adrich (Merck) | N/A |
| CL106 100uM (desalted), reverse primer GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTC ATGTAACTCGCCTTGATCGT |
Sigma-Adrich (Merck) | N/A |
| IS7 100uM (HPLC), forward primer A&B ACACTCTTTCCCTACACGAC |
Sigma-Adrich (Merck) | N/A |
| IS8 100uM (HPLC), reverse primer A GTGACTGGAGTTCAGACGTGT |
Sigma-Adrich (Merck) | N/A |
| IS10 100uM (reverse phase - HPLC), probe A FAM-A{G}A{T}C{G}GAAGAGC{A}CAC-BHQ1 |
Eurogentec | N/A |
| CL107 100uM (HPLC), reverse primer B TCATGTAACTCGCCTTGATCGT |
Sigma-Adrich (Merck) | N/A |
| CL118 100uM (HPLC), probe B FAM-TTCAGCTCCGGTTCCCAACGAT-BHQ1 |
Sigma-Adrich (Merck) | N/A |
| Software and algorithms | ||
| leeHom | Renaud et al.94 | https://bioinf.eva.mpg.de/leehom/ |
| BWA version 0.5 | N/A | https://github.com/mpieva/network-aware-bwa |
| SAMtools version 1.3.1 | Li, H. et al.95 | http://www.htslib.org/doc/1.3.1/samtools.html |
| Bam-rmdup version 0.6.1 | NA | https://github.com/mpieva/biohazard-tools |
| MAFFT v.7 | Katoh, K. et al.96 | https://mafft.cbrc.jp/alignment/server/index.html |
| BEAST V2.6.6 | Bouckaert, R. et al.97 | http://www.beast2.org/ |
| MODEL_SELECTION | Leaché, A.D. et al.98 | https://beast.community/workshop_model_selection |
| Tracer v1.7 | Rambaut, A. et al.99 | https://beast.community/tracer |
| Bam-caller v0.1 | https://github.com/bodkan/bam-caller | |
| Smartpca | Patterson, N. et al., 2006 and Price, A.L. et al., 2006100,101 | https://reich.hms.harvard.edu/software |
| ADMIXTOOLS | Patterson, N. et al.102 | https://reich.hms.harvard.edu/software |
| Admixr v0.7.1 | Petr, M. et al.103 | https://github.com/bodkan/admixr |
| OxCal v4.4 | Bronk-Ramsey,104 | https://c14.arch.ox.ac.uk/oxcal.html |
| BATS v3.64 | Wacker et al.105 | |
| AGE3 software v4.666 | IonPlus AG | www.ionplus.ch |
| GIS software v4.15 | IonPlus AG | www.ionplus.ch |
| Other | ||
| Allen ancient DNA resource (aadr) for previously published ancient and modern DNA data | N/A | https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes- present-day-and-ancient-dna-data |
| IntCal20 Calibration curve | Reimer et al.106 | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Helen Fewlass (helen.fewlass@crick.ac.uk).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Nuclear DNA data have been deposited at the ENA (European Nucleotide Archive) repository: PRJEB66365 and mitochondrial DNA data at the Dryad repository: https://doi.org/10.5061/dryad.47d7wm3m4. Both are publicly available as of the date of publication. The accession number and DOI are also listed in the key resources table. This paper also analyses existing, publicly available data from the Allen Ancient DNA Resource. The link to the dataset is listed in the key resources table. The radiocarbon data is available in this paper’s supplemental information.
-
•
All original code is available in this paper’s supplemental information.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Archaeological information
Three Elk and three Bos/Bison incisor tooth pendants were selected for dating from squares B5, C6 and C7 to represent the spread of the 112 pendants across the trench, each originating from a different animal based on tooth morphology. Five of the six teeth selected had pre-existing broken roots with only part of the drilled hole preserved. The two human teeth C7/675 (uldm1) and C7/683 (fragment of a cusp of a deciduous molar) were selected for analysis as they had the largest amount of dentine preserved and were microCT-scanned prior to the study to preserve their 3D morphology. Following excavation (2009-2010), the material was washed and wet-sieved in a local stream and handled without gloves prior to laboratory analysis, as per standard archaeological procedure. No consolidants were applied to the surface of the human or herbivore teeth.
Method details
Radiocarbon dating
Pretreatment for radiocarbon dating
Small samples of the dentine (human: 23.4 - 41.1 mg; herbivore: 34.4 - 67.4 mg) were removed using a dentist’s rotary drill. Dentine samples were pretreated in the Human Evolution department at the Max Planck Institute for Evolutionary Anthropology, Leipzig, using a previously published protocol for small samples.41 Briefly, sample chunks were demineralised in HCl 0.5M at 4°C until mechanically soft and CO2 effervescence stopped. Samples were treated with NaOH 0.1M to remove humic acid contamination and re-acidified in HCl 0.5M. The samples were gelatinised in HCl pH3 for 3-10 h at 70°C. The gelatin extracts were then filtered (Ezee-filters, Elkay labs, UK) to remove >80 μm particles and ultrafiltered to concentrate the >30 kDa weight fraction. Filters were pre-cleaned according to Bronk-Ramsey et al.107 Samples were rinsed to a neutral pH between each step. The extracts were freeze-dried for >48 hours and weighed immediately to determine the collagen yield. Small aliquots of bones dating beyond the 14C method (background) were pretreated and measured alongside the samples.
Collagen quality assessment
To assess the quality of the extracts, ∼0.4 mg collagen was weighed into a tin capsule and measured on a ThermoFinnigan Flash 2000 elemental analyser (EA) coupled to a Thermo Delta plus XP isotope ratio mass spectrometer (IRMS) via a Conflo III interface (Thermo Fisher Scientific, Germany) to determine the elemental (C%, N%, C/N) and stable isotopic values (δ13C, δ15N). Stable carbon isotope ratios were expressed relative to Vienna PeeDee Belemnite (VPDB) and stable nitrogen isotope ratios were measured relative to atmospheric N2 (AIR), using the delta notation (δ) in parts per thousand (‰). Stable isotope delta values were two-point scale normalised using international reference material IAEA-CH-6 (sucrose, δ13C = -10.449 ± 0.033 ‰), IAEA-CH-7 (polyethylene, δ13C = -32.151 ± 0.050 ‰), IAEA-N-1 (ammonium sulphate, δ15N = 0.4 ± 0.2 ‰) and IAEA-N-2 (ammonium sulphate, δ15N = 20.3 ± 0.2 ‰). Repeated analysis of internal and international standards indicates an analytical error of ± 0.2‰ (1σ). Two in-house quality control standards were used to quality check the scale normalisation and evaluate analytical precision: 1) EVA-0012 methionine (Elemental Microanalysis, Okehampton, UK), n = 12, δ13C = -28.0 ± 0.14 ‰ (1 s.d.), δ15N = -6.5 ± 0.12 ‰ (1 s.d.); and EVA MRG pig gelatin, n= 11, δ13C = -19.6 ± 0.24‰ (1 s.d.) and δ15N = 4.9 ± 0.11 ‰ (1 s.d.). These values compare well with the long-term average values of δ13C = -28.0 ± 0.1‰ (1 s.d.) and δ15N = -6.4 ± 0.1‰ (1 s.d.) for EVA-0012 and δ13C = -19.7 ± 0.3‰ (1 s.d.) and δ15N = 5.0 ± 0.1‰ (1 s.d.) for EVA MRG.
Sufficient collagen was available from C7/675, C7/658, C6/494 for additional quality checks using Fourier Transform Infrared Spectroscopy (FTIR).54,55,56 Roughly 0.3 mg collagen was homogenised and mixed with ∼40 mg of IR grade KBr powder in an agate mortar and pestle. The powder was pressed into a pellet using a manual hydraulic press (Wasserman) and analysed with an Agilent Technologies Cary FTIR with a DTGS detector. Spectra were recorded in transmission mode at 4 cm−1 resolution with averaging of 34 scans between 4000 and 400 cm−1 using Resolution Pro software (Agilent Technologies) and were compared to library spectra of well preserved collagen.
AMS dating
The extracts were dated on the AixMICADAS108 installed at CEREGE (Centre de Recherche et d'Enseignement de Geosciences de l'Environnement) in Aix-en-Provence, France. Each collagen extract was split and measured in several aliquots as per the protocol for small collagen extracts described in Fewlass et al.41,42 (Table S1). Where sufficient material was available, collagen (combustion weight ∼700 ug C) was weighed into aluminium cups and graphitized using the AGE 3109 (Automated Graphitisation Equipment) prior to dating on the AixMICADAS. Oxalic acid II standards and background collagen samples (Table S2) were measured in the same session and used in the age calculation of the archaeological samples in BATS.105 A relative error of 30% was applied to the blank value and an additional uncertainty of 1.6‰ was propagated in the F14C error calculation as per standard practice.
Gas measurements were performed using the protocol described in Tuna et al.110 and Fewlass et al.41 Small aliquots of collagen (combustion weight ∼60 ug C) were introduced into cleaned silver cups (800°C, 2 h) and combusted in an Elementar Vario MICRO cube EA (Elementar Analysensysteme GmbH, Germany) which was directly coupled to the gas ion source (GIS) of the AixMICADAS.111,112 Oxalic acid II NIST standards (from a gas canister) were measured to normalize and correct samples for fractionation. The age of each sample was corrected with background measurements from the same batch (Table S2). A relative error of 30% was applied to the blank value and an additional uncertainty of 4‰ was propagated in the F14C error.
Ancient DNA analysis
Data generation and shotgun sequencing
Sampling of the teeth for radiocarbon dating and genetic analysis was performed in a dedicated clean room at the Max Planck Institute for Evolutionary Anthropology. A microsampling approach was used for sample collection from the two of the infant teeth (C7/675 and C7/683) for downstream genetic analyses. Between 7.2 and 12.9 mg of dentine was collected from the two roots of C7/675 and two microsamples of 4.7 and 5.2 mg of dentine were drilled from the crown of C7/683 for a total of four samples. To extract DNA from the tooth powder, 500 uL of lysis buffer (for 5 mL lysis buffer: 4.5 mL 0.5M EDTA pH 8.0, 2.5 uL of Tween 20,125 uL of 10mg/mL proteinase K, 373 uL water) was added to each tooth powder aliquot for overnight lysis at 37°C. The resulting lysate was purified following the automated extraction protocol described,113 using binder buffer ‘D’. Each DNA extract was then converted into dual-indexed single-stranded libraries106 and sequenced on Illumina MiSeq or HiSeq2500 platforms in pools of 63-96 libraries that included libraries from other projects not discussed in this paper. Negative controls were included starting at both the DNA extraction and library preparation steps. Base calling was performed using Bustard (Illumina) and leeHom104 was used to trim adapters and merge overlapping forward and reverse sequences. The resulting sequences were mapped to the human reference genome (hg1963) using BWA version 0.5 (https://github.com/mpieva/network-aware-bwa) with ancient DNA parameters (-n 0.001 -o 2 -l 16500). Reads that were not a perfect match to expected index combinations, shorter than 35 base pairs, or had a mapping quality less than 25 were removed using SAMtools version 1.3.1104. Bam-rmdup version 0.6.1 (https://github.com/mpieva/biohazard-tools) was used to remove PCR duplicates. Each library (including controls) was evaluated for the presence of ancient human DNA based on the presence of elevated C-to-T substitutions on the terminal ends of DNA fragments (Table S4). Potential cross-contamination due to index swapping between libraries was evaluated using a previously published method67 that evaluates all sequenced index pairs in a sequencing run to estimate the number of reads assigned to a library that may originate from another library. All libraries were estimated to have less than 6 cross-contaminated reads. The negative controls were also examined for evidence of ancient DNA based on elevated C-to-T substitutions and no evidence of aDNA was identified in these libraries (Table S4).
Quantification and statistical analysis
Radiocarbon data analysis and reporting
The radiocarbon dates were calibrated and analysed using OxCal 4.4114 and the IntCal20 calibration curve.115 The dates were input into a one phase General t-type Outlier Model with a 5% prior likelihood of being an outlier, except C7/675 which was given an outlier likelihood of 100% due to the C/N value and FTIR analysis116 (Table S3). As per convention (e.g. Stuiver & Polach,94 Millard95), throughout the text uncalibrated 14C ages are reported with the abbreviation ‘BP’, meaning ‘radiocarbon years before AD 1950’ and are reported with 1σ errors, whereas calibrated ranges are denoted as ‘cal BP’ through the text and are given at the 95.4% (2σ) probability range. The agreement of the replicate dates from a single extract was tested using a X2 test using the R_Combine function. All previously reported dates have been re-calibrated with the IntCal20 curve.115
Mitochondrial DNA analysis
Six libraries (three from the samples and three negative controls) were enriched for human mitochondrial DNA via hybridization capture on an automated platform as described in67 and sequenced on Illumina’s MiSeq platform in pools of 32 to 76 libraries. Analysis of the resulting sequences was performed as described for the shotgun sequencing with the exception that libraries were mapped to the rCRS (Table S5). The reconstruction of mitochondrial genomes was performed on one library from each C7/675 and C7/683 where a coverage of at least 5-fold and support of at least 80% was required for calling a consensus base using only putatively deaminated reads.
No evidence of cross-contamination from index swapping between libraries (< 1 contaminating read per library) was identified in the sequencing runs. Reads with elevated C-to-T substitutions were also absent from the negative controls (Table S5). While the library negative controls had relatively few unique mtDNA reads (5 to 36) there were 680 unique mtDNA reads in the extraction negative control. Support for the variants observed in C7/675 and C7/683 were examined in the extraction negative control. The only shared variant was a G at position 73, which is a common variant across human populations,117,118 supporting that laboratory contamination did not impact downstream analyses.
The resulting mtDNA genome from C7/675 was aligned to the rCRS with mtDNA genomes from one Neanderthal (Vindija33.19), 54 present day119 and 18 ancient humans (Table S6) with MAFFT96 for tree building and molecular dating using the program BEAST V2.6.697. Present-day individuals were set to 0 and the Borsuka individual was constrained to an age range of 0 to 60,000. A model using a strict clock and constant population was selected after using the path sampling approach with 40 path steps from the MODEL_SELECTION98 package in BEAST2 to evaluate different clock and tree models (Table S7). These models were selected because there was no significant difference identified between the models and therefore the simplest were selected. In this testing a chain length of 25 million iterations, 0.3 alpha parameter, pre-burn of 75,000 iterations, and an 80% whole chain burn-in were used. For both testing and the final analysis, clock models were set with a normal distribution around a mean mutation rate of 2.67E-08 per base per year120 with a sigma of 1.00E-10.121 The Tamura-Nei 193 (TN93)122 substitution model was used for all models. For the final tree building and molecular dating, 30 million iterations (1 million pre-burn iterations) of three Markov chain Monte Carlo runs with sampling every 2,000 trees were combined using Logcombiner2 from BEAST2. The tree annotator program from BEAST2 was used to identify a single tree from the combined tree file, which was visualised with Figtree v1.4.4. The Tracer v1.7119 program was used to examine the resulting molecular dates.
Estimating TMRCA for two mtDNA genomes with no differences
In order to estimate the potential time span between two mtDNA genomes that share an mtDNA haplotype, we followed a previously published approach100 to estimate the time to the most recent common ancestor (TMRCA) in this scenario. Following Skov et al., 2022,100 we used a probability density (see below), which combines equations 7, 8, and 9 from Tavare et al., 1996101 to determine the mean coalescence time for two sequences with 0 differences. To calculate Θ we used a mutation rate (μ) of 2.67e-8 mutations per base pair per year, a generation time of 29 years,102 length of 15,569, and set the effective population size to 1,000 individuals (Θ=2∗N∗μ∗L). As the resulting mean time (Figure S3) represents one branch length, the total branch length is then 2 ∗ 1,206 years, or 2,412 years.
Nuclear DNA analysis
Twelve libraries (six samples and six negative controls) were also enriched for ∼1.2 million SNPs (‘1240k array’) previously described in74 and75 via hybridization capture. They were sequenced on Illumina’s MiSeq and NextSeq platforms and analysed as described in the shotgun sequencing section (Table S8). For these libraries less than 0.001% of total reads (<15 reads) were estimated as originating from index swapping. The data from the two libraries from C7/675 identified as containing ancient DNA were merged for downstream analysis using samtools.123 Pseudo-haplotype genotyping was performed using random read sampling with bam-caller (https://github.com/bodkan/bam-caller; v0.1) after trimming the three termini bases of each fragment to minimise the effects of deamination on downstream analyses. These data were converted into EIGENSTRAT format and merged with previously published genotypes of ancient and modern humans previously published and compiled (https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes- present-day-and-ancient-dna-data). The principal component analysis was performed using smartpca103,124 on the 597,573 SNPs in the Affymetrix Human Origins array125 on 1,395 modern-day humans from West Eurasia, the Americas, Central Asia and Siberia, and East Asia. The Borsuka individual was then projected onto the PCA with 22 ancient individuals. The genetic relationship between the Borsuka individual and modern and ancient individuals was explored further with SNPs from the ‘1240k’ array using ADMIXTOOLS125 with the R package admixr v0.7.1.126 The modern populations used were from Simons Genome Diversity Project.127
The high amount of modern human contamination in the genetic data recovered from C7/675 required all downstream analysis to be restricted to putatively deaminated fragments only. This limited the data to only 46,286 SNPs in the ‘1240k’ panel. As this only represents 3.8% of the SNPs within the panel, this raises concern as to what conclusions are possible to draw with this limited amount of data. Therefore, comparisons between other ancient humans and the Borsuka individual were restricted only to individuals with at least 500,000 SNPs with one exception. The Věstonice complex includes multiple individuals with varying qualities of genetic data. Four of these individuals were included for analysis to serve as a proxy for Borsuka for the input of limited data on each analysis.
Basal Eurasian ancestry evaluation
D-statistics was used to explore how the Borsuka individual’s genetic affinities compared to other previously published ancient humans. It has previously been shown that the D statistic is ∼ 0 for both D(Kostenki14, X; Han/Dai, Mbuti) and D(W, Stuttgart; Han, Mbuti), when W/X is a European from ∼39,000 to 14,000 years ago without Basal Eurasian ancestry[9]. Calculating these D-statistics with Borsuka in the X or W position is consistent with an individual that also does not contain basal Eurasian ancestry (Tables S9 and S10).
Acknowledgments
This research was funded by the Max Planck Society. The AixMICADAS was acquired and operated in the framework of the EQUIPEX project ASTER-CEREGE (P.I.: E. Bard) with additional matching funds from the Collège de France. HF was partially funded by an EMBO post-doctoral fellowship (EMBO ALTF 590-2021). Elena I. Zavala’s work was partially funded by the Miller Institute for Basic Research in Science, University of California Berkeley. Genetic data was produced by the Ancient DNA Core Unit of the Max Planck Institute for Evolutionary Anthropology which is funded by the Max Planck Society. The authors would like to thank Laurits Skov for input in the TMRCA analysis.
Author contributions
This study was conceived by H.F., E.I.Z., and J.W. Data generation and analysis were performed by H.F., E.I.Z., Y.F., T.T., E.B., J-J.H. and M.H. The article was written by H.F., E.I.Z., and J.W. with input by all authors.
Declaration of interests
The authors declare no competing interests.
Published: October 23, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108283.
Contributor Information
Helen Fewlass, Email: helen.fewlass@crick.ac.uk.
Elena I. Zavala, Email: ezavala9@berkeley.edu.
Supplemental information
References
- 1.Wild E.M., Teschler-Nicola M., Kutschera W., Steier P., Trinkaus E., Wanek W. Direct dating of Early Upper Palaeolithic human remains from Mladeč. Nature. 2005;435:332–335. doi: 10.1038/nature03585. [DOI] [PubMed] [Google Scholar]
- 2.Valde-Nowak P., Nadachowski A., Wolsan M. Upper Palaeolithic boomerang made of a mammoth tusk in south Poland. Nature. 1987;329:436–438. [Google Scholar]
- 3.Hedges R.E.M., Housley R.A., Pettitt P.B., Bronk-Ramsey C., van Klinken G.J. Radiocarbon dates from the oxford AMS system: Archaeometry datelist 21. Archaeometry. 1996;38:181–207. [Google Scholar]
- 4.Trinkaus E., Haduch E., Valde-Nowak P.W., Wojtal P. The Obłazowa 1 early modern human pollical phalanx and Late Pleistocene distal thumb proportions. Homo. 2014;65:1–12. doi: 10.1016/j.jchb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Wilczyński J. In: Forgotten Times and Spaces. Sázelová S., Novák M., Mizerovápp A., editors. Brno & Masaryk University; 2015. The Gravettian and Epigravettian settlement of Poland; pp. 191–213. (Institute of Archaeology of the CAS). [Google Scholar]
- 6.Wilczyński J. In: The past societies Volume 1. Kabaciński J., editor. Institute of Archaeology and Ethnology, Polish Academy of Sciences; 2016. A new beginning: Modern humans in Poland. [Google Scholar]
- 7.Soficaru A., Doboş A., Trinkaus E. Early modern humans from the Peştera Muierii, Baia de Fier, Romania. Proc. Natl. Acad. Sci. USA. 2006;103:17196–17201. doi: 10.1073/pnas.0608443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soficaru A., Petrea C., Doboş A., Trinkaus E. The Human Cranium from the Peştera Cioclovina Uscată, Romania: Context, Age, Taphonomy, Morphology, and Paleopathology. Curr. Anthropol. 2007;48:611–619. [Google Scholar]
- 9.Fu Q., Posth C., Hajdinjak M., Petr M., Mallick S., Fernandes D., Furtwängler A., Haak W., Meyer M., Mittnik A., et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Posth C., Yu H., Ghalichi A., Rougier H., Crevecoeur I., Huang Y., Ringbauer H., Rohrlach A.B., Nägele K., Villalba-Mouco V., et al. Palaeogenomics of Upper Palaeolithic to Neolithic European hunter-gatherers. Nature. 2023;615:117–126. doi: 10.1038/s41586-023-05726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marom A., McCullagh J.S.O., Higham T.F.G., Sinitsyn A.A., Hedges R.E.M. Single amino acid radiocarbon dating of Upper Paleolithic modern humans. Proc. Natl. Acad. Sci. USA. 2012;109:6878–6881. doi: 10.1073/pnas.1116328109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinnis R., Bessudnov A., Reynolds N., Devièse T., Pate A., Sablin M., Sinitsyn A., Higham T. New data for the Early Upper Paleolithic of Kostenki (Russia) J. Hum. Evol. 2019;127:21–40. doi: 10.1016/j.jhevol.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Prat S., Péan S., Crépin L., Puaud S., Drucker D.G., Lázničková-Galetová M., van der Plicht J., Valladas H., Verna C., Patou-Mathis M., et al. The first anatomically modern humans from South-Eastern Europe. Contributions from the Buran-Kaya III site (Crimea) Bull. Mem. Soc. Anthropol. Paris. 2018;30:169–179. [Google Scholar]
- 14.Svoboda J. The Gravettian on the middle Danube. Paléo. 2007:203–220. [Google Scholar]
- 15.Wilczyński J., Goslar T., Wojtal P., Oliva M., Göhlich U.B., Antl-Weiser W., Šída P., Verpoorte A., Lengyel G. New Radiocarbon Dates for the Late Gravettian in Eastern Central Europe. Radiocarbon. 2020;62:243–259. [Google Scholar]
- 16.Sládek V., Trinkaus E., Hillson S.W., Holliday T.W., Svoboda J. Institute of Archaeology; 2000. The People of the Pavlovian: Skeletal Catalogue and Osteometrics of the Gravettian Fossil Hominids from Dolní Věstonice and Pavlov. Academy of Sciences of the Czech Republic. [Google Scholar]
- 17.Svoboda J. Institute of Archaeology; 2016. Dolní Věstonice II: Chronostratigraphy, Paleoethnology, Paleoanthropology. Academy of Sciences of the Czech Republic. [Google Scholar]
- 18.Svoboda J., Novák M., Sázelová S., Demek J. Pavlov I: A large Gravettian site in space and time. Quat. Int. 2016;406:95–105. [Google Scholar]
- 19.Fewlass H., Talamo S., Kromer B., Bard E., Tuna T., Fagault Y., Sponheimer M., Ryder C., Hublin J.-J., Perri A., et al. Direct radiocarbon dates of mid Upper Palaeolithic human remains from Dolní Věstonice II and Pavlov I, Czech Republic. J. Archaeol. Sci.: Reports. 2019;27 [Google Scholar]
- 20.A Svoboda J. The Upper Paleolithic burial area at Předmostí: ritual and taphonomy. J. Hum. Evol. 2008;54:15–33. doi: 10.1016/j.jhevol.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Einwögerer T., Friesinger H., Händel M., Neugebauer-Maresch C., Simon U., Teschler-Nicola M. Upper Palaeolithic infant burials. Nature. 2006;444:285. doi: 10.1038/444285a. [DOI] [PubMed] [Google Scholar]
- 22.Einwögerer T., Händel M., Neugebauer-Maresch C., Simon U., Steier P., Teschler-Nicola M., Wild E.M. 14C Dating of the Upper Paleolithic Site at Krems-Wachtberg, Austria. Radiocarbon. 2009;51:847–855. [Google Scholar]
- 23.Teschler-Nicola M., Fernandes D., Händel M., Einwögerer T., Simon U., Neugebauer-Maresch C., Tangl S., Heimel P., Dobsak T., Retzmann A., et al. Ancient DNA reveals monozygotic newborn twins from the Upper Palaeolithic. Commun. Biol. 2020;3:650. doi: 10.1038/s42003-020-01372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettitt P.B., Bader N.O. Direct AMS Radiocarbon dates for the Sungir mid Upper Palaeolithic burials. Antiquity. 2000;74:269–270. [Google Scholar]
- 25.Dobrovolskaya M., Richards M.P., Trinkaus E. Direct radiocarbon dates for the Mid Upper Paleolithic (eastern Gravettian) burials from Sunghir, Russia. Bull. Mém. Soc. Anthropol. 2012;24:96–102. [Google Scholar]
- 26.Kuzmin Y., Burr G.S., Jull A.J.T., Sulerzhitsky L.D. AMS 14C age of the Upper Palaeolithic skeletons from Sungir site, Central Russian Plain. Nucl. Instrum. Methods Phys. Res. B. 2004;223–224:731–734. [Google Scholar]
- 27.Nalawade-Chavan S., McCullagh J., Hedges R. New hydroxyproline radiocarbon dates from Sungir, Russia, confirm early Mid Upper Palaeolithic burials in Eurasia. PLoS One. 2014;9 doi: 10.1371/journal.pone.0076896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds N., Dinnis R., Bessudnov A.A., Devièse T., Higham T. The Kostënki 18 child burial and the cultural and funerary landscape of Mid Upper Palaeolithic European Russia. Antiquity. 2017;91:1435–1450. [Google Scholar]
- 29.Higham T.F. Removing contaminants: a restatement of the value of isolating single compounds for AMS dating. Antiquity. 2019;93:1072–1075. [Google Scholar]
- 30.Soldatova T. Spatial Distribution and Problems in the Interpretation of Radiocarbon Dates of the Sungir Site, Russia. Radiocarbon. 2019;61:e1–e15. [Google Scholar]
- 31.Wilczyński J., Wojenka M., Wojtal P., Szczepanek A., Sobieraj D. Human occupation of the Borsuka Cave (Southern Poland) from Upper Paleolithic to the Post-Mediaeval Period. Eurasian Prehistory. 2012;9:77–91. [Google Scholar]
- 32.Wilczyński J., Szczepanek A., Wojtal P., Diakowski M., Wojenka M., Sobieraj D. A mid upper Palaeolithic child burial from Borsuka Cave (southern Poland) Int. J. Osteoarchaeol. 2016;26:151–162. [Google Scholar]
- 33.Wilczyński J., Miękina B., Lipecki G., Lõugas L., Marciszak A., Rzebik-Kowalska B., Stworzewicz E., Szyndlar Z., Wertz K. Faunal remains from Borsuka Cave – an example of local climate variability during Late Pleistocene in southern Poland. Acta Zool. Cracov. 2012;55:131–155. [Google Scholar]
- 34.Tillier A.-M., Mester Z., Bocherens H., Henry-Gambier D., Pap I. Direct dating of the “Gravettian” Balla child’s skeleton from Bükk Mountains (Hungary): unexpected results. J. Hum. Evol. 2009;56:209–212. doi: 10.1016/j.jhevol.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Svoboda J.A., van der Plicht J., Kuželka V. Upper Palaeolithic and Mesolithic human fossils from Moravia and Bohemia (Czech Republic): some new 14C dates. Antiquity. 2002;76:957–962. [Google Scholar]
- 36.Trinkaus E., Pettitt P.B. The Krems-Hundssteig “Gravettian” human remains are Holocene. Homo. 2000;51:258–260. [Google Scholar]
- 37.Douka K., Slon V., Stringer C., Potts R., Hübner A., Meyer M., Spoor F., Pääbo S., Higham T. Direct radiocarbon dating and DNA analysis of the Darra-i-Kur (Afghanistan) human temporal bone. J. Hum. Evol. 2017;107:86–93. doi: 10.1016/j.jhevol.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Talamo S., Hajdinjak M., Mannino M.A., Fasani L., Welker F., Martini F., Romagnoli F., Zorzin R., Meyer M., Hublin J.-J. Direct radiocarbon dating and genetic analyses on the purported Neanderthal mandible from the Monti Lessini (Italy) Sci. Rep. 2016;6 doi: 10.1038/srep29144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benazzi S., Peresani M., Talamo S., Fu Q., Mannino M.A., Richards M.P., Hublin J.-J. A reassessment of the presumed Neandertal remains from San Bernardino Cave, Italy. J. Hum. Evol. 2014;66:89–94. doi: 10.1016/j.jhevol.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Cersoy S., Zazzo A., Rofes J., Tresset A., Zirah S., Gauthier C., Kaltnecker E., Thil F., Tisnerat-Laborde N. Radiocarbon dating minute amounts of bone (3–60 mg) with ECHoMICADAS. Sci. Rep. 2017;7:7141. doi: 10.1038/s41598-017-07645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fewlass H., Tuna T., Fagault Y., Hublin J.-J., Kromer B., Bard E., Talamo S. Pretreatment and gaseous radiocarbon dating of 40–100 mg archaeological bone. Sci. Rep. 2019;9:5342. doi: 10.1038/s41598-019-41557-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fewlass H., Talamo S., Tuna T., Fagault Y., Kromer B., Hoffmann H., Pangrazzi C., Hublin J.-J., Bard E. Size Matters: Radiocarbon Dates of <200 μg Ancient Collagen Samples with AixMICADAS and Its Gas Ion Source. Radiocarbon. 2017;60:425–439. [Google Scholar]
- 43.Korlević P., Talamo S., Meyer M. A combined method for DNA analysis and radiocarbon dating from a single sample. Sci. Rep. 2018;8:4127. doi: 10.1038/s41598-018-22472-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sponheimer M., Ryder C.M., Fewlass H., Smith E.K., Pestle W.J., Talamo S. Saving Old Bones: a non-destructive method for bone collagen prescreening. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-50443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korlević P., Meyer M. Pretreatment: Removing DNA Contamination from Ancient Bones and Teeth Using Sodium Hypochlorite and Phosphate. Methods Mol. Biol. 2019:15–19. doi: 10.1007/978-1-4939-9176-1_2. [DOI] [PubMed] [Google Scholar]
- 46.Damgaard P.B., Margaryan A., Schroeder H., Orlando L., Willerslev E., Allentoft M.E. Improving access to endogenous DNA in ancient bones and teeth. Sci. Rep. 2015;5 doi: 10.1038/srep11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boessenkool S., Hanghøj K., Nistelberger H.M., Der Sarkissian C., Gondek A.T., Orlando L., Barrett J.H., Star B. Combining bleach and mild predigestion improves ancient DNA recovery from bones. Mol. Ecol. Resour. 2017;17:742–751. doi: 10.1111/1755-0998.12623. [DOI] [PubMed] [Google Scholar]
- 48.Schroeder H., de Barros Damgaard P., Allentoft M.E. Pretreatment: Improving Endogenous Ancient DNA Yields Using a Simple Enzymatic Predigestion Step. Methods Mol. Biol. 2019;1963:21–24. doi: 10.1007/978-1-4939-9176-1_3. [DOI] [PubMed] [Google Scholar]
- 49.Essel E., Korlević P., Meyer M. A method for the temperature-controlled extraction of DNA from ancient bones. Biotechniques. 2021;71:382–386. doi: 10.2144/btn-2021-0025. [DOI] [PubMed] [Google Scholar]
- 50.Essel E., Zavala E.I., Schulz-Kornas E., Kozlikin M.B., Fewlass H., Vernot B., Shunkov M.V., Derevianko A.P., Douka K., Barnes I., et al. Ancient human DNA recovered from a Palaeolithic pendant. Nature. 2023;618:328–332. doi: 10.1038/s41586-023-06035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Klinken G.J. Bone Collagen Quality Indicators for Palaeodietary and Radiocarbon Measurements. J. Archaeol. Sci. 1999;26:687–695. [Google Scholar]
- 52.Guiry E.J., Szpak P. Quality control for modern bone collagen stable carbon and nitrogen isotope measurements. Methods Ecol. Evol. 2020;11:1049–1060. [Google Scholar]
- 53.Schwarcz H.P., Nahal H. Theoretical and observed C/N ratios in human bone collagen. J. Archaeol. Sci. 2021;131 [Google Scholar]
- 54.DeNiro M.J., Weiner S. Chemical, enzymatic and spectroscopic characterization of “collagen” and other organic fractions from prehistoric bones. Geochem. Cosmochim. Acta. 1988;52:2197–2206. [Google Scholar]
- 55.Yizhaq M., Mintz G., Cohen I., Khalaily H., Weiner S., Boaretto E. Quality Controlled Radiocarbon Dating of Bones and Charcoal from the Early Pre-Pottery Neolithic B (PPNB) of Motza (Israel) Radiocarbon. 2005;47:193–206. [Google Scholar]
- 56.D’Elia M., Gianfrate G., Quarta G., Giotta L., Giancane G., Calcagnile L. Evaluation of Possible Contamination Sources in the 14C Analysis of Bone Samples by FTIR Spectroscopy. Radiocarbon. 2007;49:201–210. [Google Scholar]
- 57.Talamo S., Fewlass H., Maria R., Jaouen K. “Here we go again”: the inspection of collagen extraction protocols for 14C dating and palaeodietary analysis. Sci. Technol. Archaeol. Res. 2021;7:62–77. doi: 10.1080/20548923.2021.1944479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richards M.P., Pettitt P.B., Stiner M.C., Trinkaus E. Stable isotope evidence for increasing dietary breadth in the European mid-Upper Paleolithic. Proc. Natl. Acad. Sci. USA. 2001;98:6528–6532. doi: 10.1073/pnas.111155298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettitt P.B., Richards M., Maggi R., Formicola V. The Gravettian burial known as the Prince (“Il Principe”): new evidence for his age and diet. Antiquity. 2003;77:15–19. [Google Scholar]
- 60.Richards M.P., Trinkaus E. Isotopic evidence for the diets of European Neanderthals and early modern humans. Proc. Natl. Acad. Sci. USA. 2009;106:16034–16039. doi: 10.1073/pnas.0903821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bocherens H., Drucker D.G., Germonpré M., Lázničková-Galetová M., Naito Y.I., Wissing C., Brůžek J., Oliva M. Reconstruction of the Gravettian food-web at Předmostí I using multi-isotopic tracking (13C, 15N, 34S) of bone collagen. Quat. Int. 2015;359–360:211–228. [Google Scholar]
- 62.Wißing C., Rougier H., Baumann C., Comeyne A., Crevecoeur I., Drucker D.G., Gaudzinski-Windheuser S., Germonpré M., Gómez-Olivencia A., Krause J., et al. Stable isotopes reveal patterns of diet and mobility in the last Neandertals and first modern humans in Europe. Sci. Rep. 2019;9:4433. doi: 10.1038/s41598-019-41033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Church D.M., Schneider V.A., Graves T., Auger K., Cunningham F., Bouk N., Chen H.-C., Agarwala R., McLaren W.M., Ritchie G.R.S., et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 65.Briggs A.W., Stenzel U., Johnson P.L.F., Green R.E., Kelso J., Prüfer K., Meyer M., Krause J., Ronan M.T., Lachmann M., Pääbo S. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. USA. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peyrégne S., Peter B.M. AuthentiCT: a model of ancient DNA damage to estimate the proportion of present-day DNA contamination. Genome Biol. 2020;21:246. doi: 10.1186/s13059-020-02123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zavala E.I., Aximu-Petri A., Richter J., Nickel B., Vernot B., Meyer M. Quantifying and reducing cross-contamination in single- and multiplex hybridization capture of ancient DNA. Mol. Ecol. Resour. 2022;22:2196–2207. doi: 10.1111/1755-0998.13607. [DOI] [PubMed] [Google Scholar]
- 68.Furtwängler A., Reiter E., Neumann G.U., Siebke I., Steuri N., Hafner A., Lösch S., Anthes N., Schuenemann V.J., Krause J. Ratio of mitochondrial to nuclear DNA affects contamination estimates in ancient DNA analysis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 70.Secher B., Fregel R., Larruga J.M., Cabrera V.M., Endicott P., Pestano J.J., González A.M. The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents. BMC Evol. Biol. 2014;14:109. doi: 10.1186/1471-2148-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pereira L., Silva N.M., Franco-Duarte R., Fernandes V., Pereira J.B., Costa M.D., Martins H., Soares P., Behar D.M., Richards M.B., Macaulay V. Population expansion in the North African late Pleistocene signalled by mitochondrial DNA haplogroup U6. BMC Evol. Biol. 2010;10:390. doi: 10.1186/1471-2148-10-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hervella M., Svensson E.M., Alberdi A., Günther T., Izagirre N., Munters A.R., Alonso S., Ioana M., Ridiche F., Soficaru A., et al. The mitogenome of a 35,000-year-old Homo sapiens from Europe supports a Palaeolithic back-migration to Africa. Sci. Rep. 2016;6 doi: 10.1038/srep25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Svensson E., Günther T., Hoischen A., Hervella M., Munters A.R., Ioana M., Ridiche F., Edlund H., van Deuren R.C., Soficaru A., et al. Genome of Peştera Muierii skull shows high diversity and low mutational load in pre-glacial Europe. Curr. Biol. 2021;31:2973–2983.e9. doi: 10.1016/j.cub.2021.04.045. [DOI] [PubMed] [Google Scholar]
- 74.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K., et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu Q., Hajdinjak M., Moldovan O.T., Constantin S., Mallick S., Skoglund P., Patterson N., Rohland N., Lazaridis I., Nickel B., et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Higham T. European Middle and Upper Palaeolithic radiocarbon dates are often older than they look: problems with previous dates and some remedies. Antiquity. 2011;85:235–249. [Google Scholar]
- 77.Kowalik N., Anczkiewicz R., Wilczyński J., Wojtal P., Müller W., Bondioli L., Nava A., Gasparik M. Tracing human mobility in central Europe during the Upper Paleolithic using sub-seasonally resolved Sr isotope records in ornaments. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-67017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willis K., Vanandel T. Trees or no trees? The environments of central and eastern Europe during the Last Glaciation. Quat. Sci. Rev. 2004;23:2369–2387. [Google Scholar]
- 79.Sommer R.S., Nadachowski A. Glacial refugia of mammals in Europe: evidence from fossil records. Mamm Rev. 2006;36:251–265. [Google Scholar]
- 80.Přichystal A. Masaryk University Press; 2014. Lithic Raw Materials in Prehistoric Times of Eastern Central Europe. [Google Scholar]
- 81.Stefaniak K., Pawłowska K., Ratajczak U., Roblíčková M., Gumiński W., Wojtal P. Annales Societatis Geologorum Poloniae. Polskie Towarzystwo Geologiczne; 2014. Middle and Late Pleistocene elks (Cervalces Scott, 1855 and Alces Gray, 1821) from Poland: palaeoenvironmental and palaeogeographic implications; pp. 341–362. [Google Scholar]
- 82.Kozłowski J.K. The Lithic Raw Material Sources and the Interregional Human Contacts in the Northern Carpathian Regions. Polska Akademia Umiejętności; 2013. Raw materials procurement in the Late Gravettian of the Carpathian Basin. 63–85. [Google Scholar]
- 83.Svoboda J. The depositional context of the Early Upper Paleolithic human fossils from the Koněprusy (Zlatý kůň) and Mladeč Caves, Czech Republic. J. Hum. Evol. 2000;38:523–536. doi: 10.1006/jhev.1999.0361. [DOI] [PubMed] [Google Scholar]
- 84.Svoboda J.A. In: Early Modern Humans at the Moravian Gate: The Mladeč Caves and their Remains. Teschler-Nicola M., editor. Springer Vienna; 2006. The Structure of the Cave, Stratigraphy, and Depositional Context; pp. 27–40. [Google Scholar]
- 85.Oliva M. In: Early Modern Humans at the Moravian Gate: The Mladeč Caves and their Remains. Teschler-Nicola M., editor. Springer Vienna; 2006. The Upper Paleolithic Finds from the Mladeč Cave; pp. 41–74. [Google Scholar]
- 86.Vanhaeren M., d’Errico F. Aurignacian ethno-linguistic geography of Europe revealed by personal ornaments. J. Archaeol. Sci. 2006;33:1105–1128. [Google Scholar]
- 87.White R., Normand C. Early and archaic Aurignacian personal ornaments from Isturitz cave: Technological and regional perspectives. Palethnologie. 2015;7 doi: 10.4000/palethnologie.789. [DOI] [Google Scholar]
- 88.Villalba-Mouco V., van de Loosdrecht M.S., Rohrlach A.B., Fewlass H., Talamo S., Yu H., Aron F., Lalueza-Fox C., Cabello L., Cantalejo Duarte P., et al. A 23,000-year-old southern Iberian individual links human groups that lived in Western Europe before and after the Last Glacial Maximum. Nat. Ecol. Evol. 2023;7:597–609. doi: 10.1038/s41559-023-01987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hajdinjak M., Mafessoni F., Skov L., Vernot B., Hübner A., Fu Q., Essel E., Nagel S., Nickel B., Richter J., et al. Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry. Nature. 2021;592:253–257. doi: 10.1038/s41586-021-03335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seguin-Orlando A., Korneliussen T.S., Sikora M., Malaspinas A.-S., Manica A., Moltke I., Albrechtsen A., Ko A., Margaryan A., Moiseyev V., et al. Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346:1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 91.Hodgkins J., Orr C.M., Gravel-Miguel C., Riel-Salvatore J., Miller C.E., Bondioli L., Nava A., Lugli F., Talamo S., Hajdinjak M., et al. An infant burial from Arma Veirana in northwestern Italy provides insights into funerary practices and female personhood in early Mesolithic Europe. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-02804-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Devièse T., Massilani D., Yi S., Comeskey D., Nagel S., Nickel B., Ribechini E., Lee J., Tseveendorj D., Gunchinsuren B., et al. Compound-specific radiocarbon dating and mitochondrial DNA analysis of the Pleistocene hominin from Salkhit Mongolia. Nat. Commun. 2019;10:274. doi: 10.1038/s41467-018-08018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Devièse T., Abrams G., Hajdinjak M., Pirson S., De Groote I., Di Modica K., Toussaint M., Fischer V., Comeskey D., Spindler L., et al. Reevaluating the timing of Neanderthal disappearance in Northwest Europe. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2022466118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stuiver M., Polach H.A. Discussion Reporting of 14C Data. Radiocarbon. 1977;19:355–363. [Google Scholar]
- 95.Millard A.R. Conventions for Reporting Radiocarbon Determinations. Radiocarbon. 2014;56:555–559. [Google Scholar]
- 96.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N., et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leaché A.D., Fujita M.K., Minin V.N., Bouckaert R.R. Species delimitation using genome-wide SNP data. Syst. Biol. 2014;63:534–542. doi: 10.1093/sysbio/syu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Skov L., Peyrégne S., Popli D., Iasi L.N.M., Devièse T., Slon V., Zavala E.I., Hajdinjak M., Sümer A.P., Grote S., et al. Genetic insights into the social organization of Neanderthals. Nature. 2022;610:519–525. doi: 10.1038/s41586-022-05283-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tavaré S., Balding D.J., Griffiths R.C., Donnelly P. Inferring coalescence times from DNA sequence data. Genetics. 1997;145:505–518. doi: 10.1093/genetics/145.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fenner J.N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 103.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Renaud G., Stenzel U., Kelso J. leeHom: adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 2014;42:e141. doi: 10.1093/nar/gku699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wacker L., Christl M., Synal H.-A. Bats: A new tool for AMS data reduction. Nucl. Instrum. Methods Phys. Res. B. 2010;268:976–979. [Google Scholar]
- 106.Gansauge M.-T., Aximu-Petri A., Nagel S., Meyer M. Manual and automated preparation of single-stranded DNA libraries for the sequencing of DNA from ancient biological remains and other sources of highly degraded DNA. Nat. Protoc. 2020;15:2279–2300. doi: 10.1038/s41596-020-0338-0. [DOI] [PubMed] [Google Scholar]
- 107.Bronk-Ramsey C., Higham T., Bowles A., Hedges R. Improvements to the Pretreatment of Bone at Oxford. Radiocarbon. 2004;46:155–163. [Google Scholar]
- 108.Bard E., Tuna T., Fagault Y., Bonvalot L., Wacker L., Fahrni S., Synal H.-A. AixMICADAS, the accelerator mass spectrometer dedicated to 14C recently installed in Aix-en-Provence, France. Nucl. Instrum. Methods Phys. Res. B. 2015;361:80–86. [Google Scholar]
- 109.Wacker L., Němec M., Bourquin J. A revolutionary graphitisation system: Fully automated, compact and simple. Nucl. Instrum. Methods Phys. Res. B. 2010;268:931–934. [Google Scholar]
- 110.Tuna T., Fagault Y., Bonvalot L., Capano M., Bard E. Development of small CO2 gas measurements with AixMICADAS. Nucl. Instrum. Methods Phys. Res. B. 2018;437:93–97. [Google Scholar]
- 111.Ruff M., Szidat S., Gäggeler H., Suter M., Synal H.-A., Wacker L. Gaseous radiocarbon measurements of small samples. Nucl. Instrum. Methods Phys. Res. B. 2010;268:790–794. [Google Scholar]
- 112.Wacker L., Fahrni S.M., Hajdas I., Molnar M., Synal H.-A., Szidat S., Zhang Y.L. A versatile gas interface for routine radiocarbon analysis with a gas ion source. Nucl. Instrum. Methods Phys. Res. B. 2013;294:315–319. [Google Scholar]
- 113.Rohland N., Glocke I., Aximu-Petri A., Meyer M. Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc. 2018;13:2447–2461. doi: 10.1038/s41596-018-0050-5. [DOI] [PubMed] [Google Scholar]
- 114.Bronk-Ramsey C. Bayesian Analysis of Radiocarbon Dates. Radiocarbon. 2009;51:337–360. [Google Scholar]
- 115.Reimer P.J., Austin W.E.N., Bard E., Bayliss A., Blackwell P.G., Bronk-Ramsey C., Butzin M., Cheng H., Edwards R.L., Friedrich M., et al. The IntCal20 Northern Hemisphere Radiocarbon Age Calibration Curve (0–55 cal kBP) Radiocarbon. 2020;62:725–757. [Google Scholar]
- 116.Bronk-Ramsey C. Dealing with Outliers and Offsets in Radiocarbon Dating. Radiocarbon. 2009;51:1023–1045. [Google Scholar]
- 117.Weissensteiner H., Pacher D., Kloss-Brandstätter A., Forer L., Specht G., Bandelt H.-J., Kronenberg F., Salas A., Schönherr S. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–W63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dür A., Huber N., Parson W. Fine-Tuning Phylogenetic Alignment and Haplogrouping of mtDNA Sequences. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22115747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ingman M., Kaessmann H., Pääbo S., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 120.Fu Q., Li H., Moorjani P., Jay F., Slepchenko S.M., Bondarev A.A., Johnson P.L.F., Aximu-Petri A., Prüfer K., de Filippo C., et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fu Q., Mittnik A., Johnson P.L.F., Bos K., Lari M., Bollongino R., Sun C., Giemsch L., Schmitz R., Burger J., et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 123.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 125.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Petr M., Vernot B., Kelso J. admixr—R package for reproducible analyses using ADMIXTOOLS. Bioinformatics. 2019;35:3194–3195. doi: 10.1093/bioinformatics/btz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A., et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Nuclear DNA data have been deposited at the ENA (European Nucleotide Archive) repository: PRJEB66365 and mitochondrial DNA data at the Dryad repository: https://doi.org/10.5061/dryad.47d7wm3m4. Both are publicly available as of the date of publication. The accession number and DOI are also listed in the key resources table. This paper also analyses existing, publicly available data from the Allen Ancient DNA Resource. The link to the dataset is listed in the key resources table. The radiocarbon data is available in this paper’s supplemental information.
-
•
All original code is available in this paper’s supplemental information.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.