Abstract
Heart failure is a leading cause of death and is often accompanied by activation of quiescent cardiac myofibroblasts, which results in cardiac fibrosis. In this study, we aimed to identify novel circular RNAs that regulate cardiac fibrosis. We applied transverse aortic constriction (TAC) for 1, 4, and 8 weeks in mice. RNA sequencing datasets were obtained from cardiac fibroblasts isolated by use of a Langendorff apparatus and then further processed by use of selection criteria such as differential expression and conservation in species. CircSMAD4 was upregulated by TAC in mice or by transforming growth factor (TGF)-β1 in primarily cultured human cardiac fibroblasts. Delivery of si-circSMAD4 attenuated myofibroblast activation and cardiac fibrosis in mice treated with isoproterenol (ISP). si-circSmad4 significantly reduced cardiac fibrosis and remodeling at 8 weeks. Mechanistically, circSMAD4 acted as a sponge against the microRNA miR-671-5p in a sequence-specific manner. miR-671-5p was downregulated during myofibroblast activation and its mimic form attenuated cardiac fibrosis. miR-671-5p mimic destabilized fibroblast growth factor receptor 2 (FGFR2) mRNA in a sequence-specific manner and interfered with the fibrotic action of FGFR2. The circSMAD4-miR-671-5p-FGFR2 pathway is involved in the differentiation of cardiac myofibroblasts and thereby the development of cardiac fibrosis.
Keywords: MT: Non-coding RNAs, circular RNA, transverse aortic constriction, cardiac fibrosis, circSMAD4, miR-671-5p, FGFR2
Graphical abstract

Kook and colleagues show that cardiac myofibroblast activation in heart failure leads to detrimental fibrosis. Our study highlights circSMAD4 as a key regulator of cardiac fibrosis, by its binding to miR-671-5p, subsequently activating FGFR2. Targeting this pathway reduces fibrosis and may offer therapeutic strategies for heart failure-related cardiac remodeling.
Introduction
Cardiovascular diseases (CVDs) are still a substantial public health problem and remain the leading cause of morbidity and mortality worldwide, contributing to 31% of all deaths.1 Myocardial fibrosis is a pathological process that occurs after injury to the heart, such as by myocardial infarction, and after heart failure.2 In the early stage of cardiac fibrosis, the fibrosis and subsequent scar formation are thought to be an adaptive and protective mechanism to injury to prevent cardiac rupture. In the late stage, however, excess fibrosis leads to pathological remodeling and impairs heart function.2 Cardiac fibrosis is characterized by activation of cardiac fibroblasts and differentiation of cardiac fibroblasts into cardiac myofibroblasts. This activation process adversely affects compliance and increases ventricular stiffness because functioning cardiomyocytes are replaced by non-functioning (non-contracting) fibroblasts. This remodeling ultimately leads to heart failure.3 Myofibroblast differentiation is accompanied by the secretion of extracellular matrix proteins and the expression of the contractile protein α-smooth muscle actin (αSMA).4 One way to prevent adverse cardiac remodeling is to regulate the late stage of myofibroblast activation and cardiac fibrosis. Thus, such regulation mechanisms are of great interest to both pharmaceutical companies and clinicians.
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs formed by backsplicing, which creates a covalently closed loop without a free 3′ or 5′ end.5,6 Compared with other non-coding RNAs, little is known about the working mechanism of the different circRNAs, although they do have some features in common. For example, multiple circRNA isoforms can be processed from one host gene.7 circRNAs are highly stable and are more resistant to digestion by exonuclease RNase R.8 The expression of circRNAs varies by cell type, tissue, and developmental stage.9 Although circRNAs are evolutionarily conserved in different species, many are specific to certain species without sequence homology.
In addition to their use as diagnostic or prognostic biomarkers because of their greater stability compared with linear RNAs, circRNAs function biologically by fine-tuning their associated gene targets or by generating proteins. Indeed, several major biological functions have been documented: (1) microRNA (miRNA) sponging, (2) protein sponging, (3) transcriptional regulation by splicing of host genes or scaffolding with other transcription factors, and (4) generation of proteins or peptides by translation of circRNAs.10 It is interesting that circRNAs remove miRNAs by binding to them directly in a sequence-specific manner, as exemplified by ciRS-7 and miR-7.11
Likewise, in the circRNA-mediated regulation of cardiac hypertrophy or diastolic dysfunction, heart-related circRNA (HRCR) was identified as an endogenous miR-223 sponge12 and circTTN was shown to work as an miR-432 sponge.13 In cardiac fibrosis-associated remodeling, some circRNAs have been reported to provoke cardiac fibrosis,12,14 whereas others block it.15,16 Transverse aortic constriction (TAC) is often used to study cardiac fibrosis and dysfunction. It should be noted, however, that most of these previous works obtained circRNA from whole hearts after TAC. Considering that the heart consists of several cellular components other than cardiomyocytes and that remodeling may alter the proportion of cellular constituents in the heart, it would be better to characterize cardiac fibroblast- or cardiomyocyte-specific circRNAs after isolation of those cells. This will enable us to more precisely understand the mechanisms of individual cell types and to develop more specific drugs targeting certain cell types. We were especially interested in cardiac fibroblast-specific circRNAs that may affect fibroblast differentiation and cardiac fibrosis. Thus, in the present study, using RNA sequencing (RNA-seq) analysis followed by diverse bioinformatics and biochemical studies, we identified and characterized a circRNA that may affect pathologic remodeling of the heart.
Results
Screening of circRNAs that are altered in cardiac fibroblasts after TAC-induced fibrosis
We first tried to identify the circRNAs that are responsible for the induction or maintenance of cardiac fibroblasts. We assumed that the time point of 1 week after TAC would reflect the initial triggering signal pathway of fibrosis, 4 weeks would represent cardiac fibrosis, and 8 weeks would represent established fibrosis.17,18,19 We performed total RNA-seq and analyzed as shown in Figure 1A. Detailed methods are described below. Our hierarchical clustering analysis showed that most of the replicate samples were clustered together (Figure S1). Cardiac fibrosis was accompanied by an increase in myofibroblast marker genes, such as actin alpha 2, smooth muscle, encodes αSMA, (Acta2), collagen type I alpha 1 chain (Col1a1), collagen type III alpha 1 chain (Col3a1), connective tissue growth factor (Ctgf), and fibronectin 1 (Fn1) (Table S1). After total RNA-seq followed by bioinformatics analysis, we identified 10 circRNAs that were upregulated in the relatively late phase of cardiac fibrosis in our experimental models (Figure 1B). The relative amount of each circRNA in the TAC model compared with the sham-operated one is shown in Table S2. To rule out circRNAs that did not share sequence homology with humans, we checked whether those circRNAs were properly expressed in human ventricular cardiac fibroblasts. Of the 10 circRNAs tested, only circNAV1, circSMAD4, and circBNC2 were highly expressed in human cardiac fibroblasts (Figure 1C) and the sequence homologies of those circRNAs between mouse and human were 88.7%, 85.0%, and 93.0%, respectively. We next checked the circularity of the candidate circRNAs by testing RNase R resistance. All three circRNAs (circNAV1, circSMAD4, and circBNC2) were resistant to RNase R treatment, whereas their corresponding mRNAs in linear forms were readily degraded (Figure 1D). To further confirm the circularity by checking the backsplicing junction, we performed Sanger sequencing with appropriate primers and found that all three circRNAs had an intact junction sequence as noted by the red arrows in Figure 1E.
Figure 1.
Screening of circRNAs in cardiac fibroblasts obtained after TAC for 1, 4, or 8 weeks in mouse
(A) Schematic diagram of screening flow. (B) Ten upregulated candidate circRNAs in cardiac fibroblasts. circRNAs with a p value less than 0.1 were selected as shown in Table S2. The values were expressed as fold changes in TAC groups relative to their matched sham controls. (C) Expression of circRNAs in human ventricular cardiac fibroblasts. One out of three samples shown. (D) Checking of circularity by RNase R treatment. Note that circRNAs were resistant to RNase R treatment, whereas linear host mRNAs were easily degraded. One out of three samples shown. (E) Sanger sequence to see the backsplice junctional sequence. Sequencing results of circNAV1, circSMAD4, and circBNC2 are shown on the left and the junctions are marked with a red arrow. The exon structures are shown on the right.
CircSmad4 activates cardiac fibroblasts
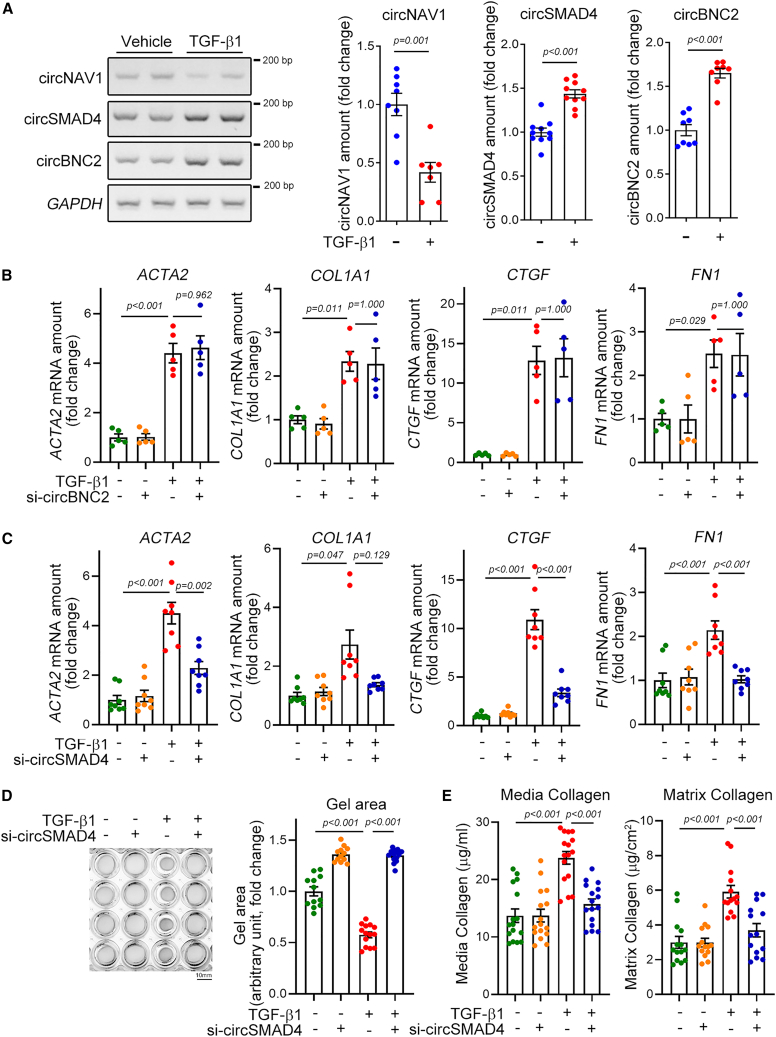
Cardiac fibrosis can be induced either actively by differentiation of cardiac myofibroblasts20 or passively by loss of intact cardiomyocytes.21 Thus, to check whether the circRNAs directly affect cardiac fibroblasts, we tested the effects of the candidate circRNAs on differentiation of cardiac myofibroblasts in primarily cultured human cardiac fibroblasts. Transforming growth factor (TGF)-β1 is an important inducer of fibroblast activation, and treatment with TGF-β1 mimics fibrosis in cellular models. Likewise, TGF-β1 induces cardiac fibrosis.22 We first checked whether the candidate circRNAs were upregulated in response to TGF-β1 in cellular models. TGF-β1 treatment significantly increased the amounts of circSMAD4 (second gel picture in left panel of Figure 2A) and of circBNC2 (third gel picture). However, TGF-β1 treatment failed to amplify the amount of circNAV1. In contrast, the amount of circNAV1 was significantly decreased in response to TGF-β1 (right bar graphs in Figure 2A), which was opposite of the in vivo results in Figure 1 and Table S3. We thus ruled out circNAV1 for further analysis.
Figure 2.
CircSmad4 activates cardiac myofibroblasts
(A) Confirmation of changes of expression amounts of candidate circRNAs in TGF-β1-treated human cardiac fibroblasts. Band intensity was calculated after conventional RT-PCR as described in section “materials and methods” and the quantification results are shown in the bar graphs on the right. Note that circNAV1 was ruled out, because it was decreased, opposite to the initial screening after TAC. circNAV1: n = 7∼8 from four independent experimental sets with unpaired Student’s t test. circSMAD4: n = 10 from five sets with unpaired Student’s t test. circBNC2: n = 8 from four sets with Mann-Whitney’s U test. (B) Effects of si-circBNC2 on the expression of fibrosis-associated genes: ACTA2, actin alpha 2, smooth muscle, encodes α-smooth muscle actin (SMA); COL1A1, collagen type I alpha 1 chain; CTGF, connective tissue growth factor; FN1, fibronectin 1. Note that si-circBNC2 did not affect TGF-β1-induced myofibroblast differentiation. All four genes: n = 5 from two independent experimental sets. Each sample duplicated. ACTA2: one-way ANOVA with post hoc Tukey’s test. COL1A1, CTGF, and FN1: one-way ANOVA with post hoc Dunnett T3 test. (C) Effects of si-circSMAD4 on the expression of fibrosis-associated genes. All four genes: n = 8 from three sets. Each sample duplicated. ACTA2 and FN1: one-way ANOVA with post hoc Tukey’s test. COL1A1 and CTGF: one-way ANOVA with post hoc Dunnett’s T3 test. (D) Effects of si-circSMAD4 on TGF-β1-induced gel contraction. Left: representative gel picture. Right: quantification results. n = 12∼14 from four independent experimental sets. One-way ANOVA with post hoc Dunnett’s T3 test. Scale bar, 10 mm. (E) Effects of si-circSMAD4 on TGF-β1-induced releases of media collagen (left) and matrix collagen (right). Media collagen: n = 16 from four sets. One-way ANOVA with post hoc Tukey’s test. Matrix collagen: n = 14 from four sets. One-way ANOVA with post hoc Tukey’s test. Data are represented as mean ± SEM.
Next, we checked whether the candidate circRNAs could affect the TGF-β1-induced activation of cardiac myofibroblasts. circRNA can be readily knocked down by conventional small interfering RNAs (siRNAs). However, the siRNA should not affect the host mRNAs. To achieve this specificity, the siRNA should be designed to bind to the backsplice junctional nucleotides. Because of slight differences in the nucleotide sequence in the junctional area, we designed separate siRNAs for human and mouse, the sequences of which are shown in Table S3.
Of the two human si-circBNC2 designed, the knockdown efficiency of si-circBNC2-#2 was greater (upper gel in Figure S2A). The nucleotide sequence of si-circBNC2-#2 and its binding target at the junctional sequence (which is marked by a blue arrow) are shown in Figure S2B. It is noteworthy that BNC2 mRNA, which is a linear host gene, was not altered by this interference (the second gel in Figure S2A). Treatment with TGF-β1 increased the circBNC2 amounts, which was then successfully attenuated by si-circBNC2 (upper gel in Figure S2C). In contrast, the linear BNC2 mRNA level was not altered either by TGF-β1 or by si-circBNC2 (the second gel in Figure S2C). Treatment with TGF-β1 significantly increased the mRNA expression of marker genes such as ACTA2, COL1A1, CTGF, and FN1 (first bars versus third bars in four panels in Figure 2B). The increases in those four genes were also found in our total RNA-seq results in fibroblasts obtained from TAC-operated mouse hearts (Table S1). However, treatment with si-circBNC2 did not alter the TGF-β1-induced increase in those fibrosis marker genes (third bars versus fourth bars), which led us to rule out circBNC1 for further analysis.
Likewise, two si-circSMAD4s were designed and si-circSMAD4 #2 was more efficient to interfere with circSMAD4 (Figure S3A). Its binding target at the junctional sequence is shown in Figure S3B si-circSMAD4 successfully reduced the amount of circSMAD4 either in the presence or in the absence of TGFβ1 (upper gel in Figure S3C). However, it did not affect the linear SMAD4 mRNA (Figure S3C). It is noteworthy that TGF-β1 increased both circSMAD4 and SMAD4 mRNA. Next, we utilized this si-circSMAD4 for further experiments. In contrast, treatment with si-circSMAD4 significantly reduced the expression of ACTA2, CTGF, and FN1. Although it was not significant, si-circSMAD4 also reduced the amount of COL1A1 mRNA (Figure 2C).
When cardiac fibroblasts are activated and then differentiate, the cell contracts.23 In our experimental model, treatment with TGFβ1 significantly induced the contraction of gel-seeded cardiac myofibroblasts (first row versus third row in gel pictures of Figure 2D), as measured by the gel area (right bar graph in Figure 2D). Knocking down circSMAD4, however, prevented the gel contraction induced by TGF-β1 (third row versus fourth row in left panel and third bar versus fourth bar in right graph in Figure 2D). Activation of fibroblasts is accompanied by the release of either media collagen or matrix collagen. Thus, we measured both collagens using a kit. Treatment with TGF-β1 significantly increased the release of both media collagen and matrix collagen (first bars versus third bars in Figure 2E). However, these increases were completely abolished by simultaneous treatment with si-circSMAD4 (third bars versus fourth bars). These results suggest that circSMAD4, but not circBNC2 or circNAV1, is actively involved in regulating cardiac fibroblast activation.
si-circSmad4 prevents initiation of cardiac fibrosis but has limited effects on pre-established fibrosis
Prior to initiating the in vivo evaluation of circSmad4, we investigated its tissue-specific distribution. Notably, circSmad4 demonstrated pronounced expression in the heart, lung, and brain (Figure S4A). Intriguingly, circSmad4 expression was exclusive to cardiac fibroblasts, with an absence in cardiomyocytes (Figure S4B). To determine if circSmad4 is ubiquitously expressed in fibroblasts, we probed its expression in both ventricular and dermal fibroblasts, sourced from human postmortem specimens. It was discernible that circSmad4 was present in both types of fibroblasts (Figure S4C).
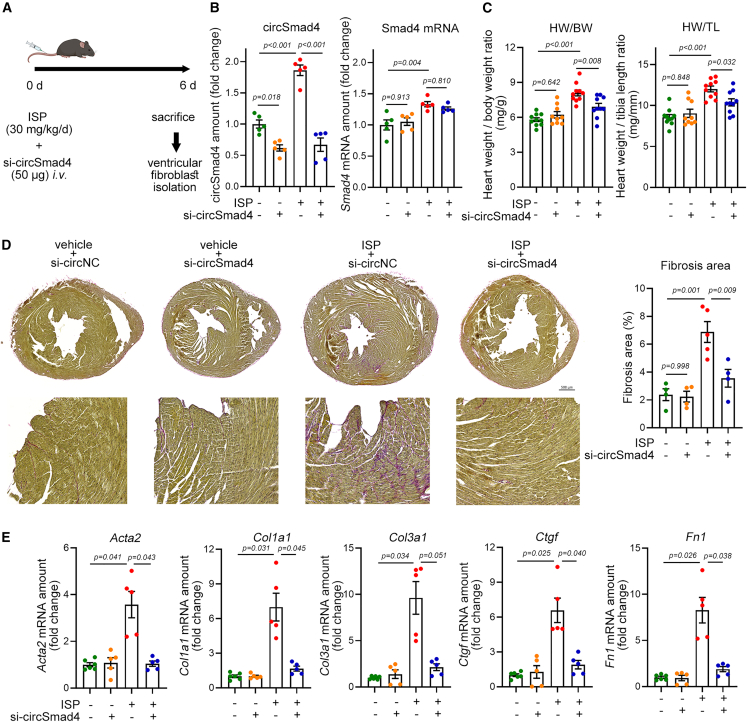
Daily administration of a high dose of isoproterenol (ISP), a β-adrenergic receptor agonist, is sufficient to induce both cardiac fibrosis and cardiac hypertrophy, even after a relatively short period of administration of less than 1 week.24 To check whether si-circSmad4 (mouse form) affects the induction of cardiac fibrosis in response to ISP, we administered a high dose of ISP to mice using an Alzet osmotic pump and injected a single dose of si-circSMAD4 (50 μg) through the tail vein on the starting day of ISP administration (Figure 3A). ISP significantly increased the amount of circSmad4 (first bar versus third bar in the left graph of Figure 3B), which mimicked the TAC-induced cardiac fibrosis (conditions that were shown in Figure 1). First, we checked the efficiency of in vivo delivery of si-circSmad4 to cardiac fibroblasts by measuring the endogenous circSmad4 amount. Intravenous injection of si-circSmad4 significantly blocked the expression of circSmad4 both in vehicle-treated (first bar versus second bar) and in ISP-administered (third bar versus fourth bar) mice (left graph of Figure 3B). The amount of Smad4 mRNA, a host gene of circSmad4 in a linear form, was also increased by ISP administration (first bar versus third bar in the right graph of Figure 3B). However, si-circSmad4 failed to reduce the Smad4 mRNA (right panel of Figure 3B). We investigated if si-circSmad4 could also diminish the levels of endogenous circSmad4 in the lung and brain, tissues wherein circSmad4 exhibits pronounced expression (Figure S4A). The administration of si-circSmad4 resulted in a notable reduction of circSmad4 in the lung and heart. However, such a reduction was not observed in the brain, potentially attributed to the inability of the siRNA to penetrate the blood-brain barrier (Figure S4D).
Figure 3.
Effects of si-circSmad4 on acute cardiac fibrosis induced by ISP administration
(A) Experimental scheme. si-circSmad4 was intravenously administered through a tail vein. (B) Efficiency of si-circSmad4. si-circSmad4 successfully reduced the expression of circSmad4 (left graph), whereas it failed to reduce the amount of linear mRNA of host Smad4 (right graph). Band intensity was calculated after conventional RT-PCR. n = 5 from two experimental sets. One-way ANOVA with post hoc Tukey’s test. (C) Effects of si-circSmad4 on the ISP-induced increase in heart weight. n = 5 from two experimental sets. Each sample duplicated. One-way ANOVA with post hoc Tukey’s test. (D) Picrosirius red staining and quantification of cardiac fibrosis. n = 4∼5. Each sample duplicated. One-way ANOVA with post hoc Bonferroni’s test. Scale bar, 500 µM. (E) Changes in cardiac fibrosis-associated genes. Col3a1, collagen type III alpha 1 chain. n = 5∼6 from two sets. Each sample duplicated. One-way ANOVA with post hoc Dunnett T3. Data are represented as mean ± SEM.
Treating mice with ISP for 6 days was sufficient to induce cardiac hypertrophy as analyzed by measuring heart weight (first bars versus third bars in both graphs of Figure 3C). Interestingly, the enlargement of heart was significantly reduced by simultaneous injection of si-circSmad4 (third bars versus fourth bars), which suggests that initiation of cardiac hypertrophy could be prevented by si-circSmad4.
ISP administration also induced cardiac fibrosis as shown by picrosirius red staining; the area in red was increased in the third heart section compared with the first one (Figure 3D). The increase was, however, reduced in si-circSmad4-treated mouse heart (fourth section). The anti-fibrotic effect of si-circSmad4 is quantified in the right bar graph of Figure 3D. The induction of cardiac fibrosis by ISP administration was further confirmed by increases in the mRNA amounts of fibrosis-associated genes (the third bars versus the first bars in Figure 3E). The increases in Acta2, Col1a1, Ctgf, and Fn1 were significantly attenuated by simultaneous treatment with si-circSmad4 (the fourth bars versus the third bars in Figure 3E). Although it was not statistically significant, the increase in the Col3a1 mRNA amount was also reduced by si-circSmad4. These results suggest that circSmad4 may play an important role as a triggering signal in the development of cardiac fibrosis.
Because the amount of circSmad4 was dramatically increased at 4 and 8 weeks, rather than at 1 week (Table S2), we assumed that circSmad4 plays an additional role in the relatively late phase of cardiac fibrosis, and we questioned whether si-circSmad4 could reverse the pre-established cardiac fibrosis. To examine this hypothesis, we established the experimental protocol as shown in Figure 4A; TAC was introduced to mice and si-circSmad4 was injected into the tail vein every 3 days from the fifth week after the TAC operation and cardiac fibroblasts were examined after isolation. We confirmed that cardiac fibrosis and cardiac hypertrophy were established at 4 weeks (Figure S5). TAC for 8 weeks further induced cardiac fibrosis with an increase in the expression of circSmad4 (first vs. third bars in Figure 4B left graph). The efficiency of knocking down circSmad4 in cardiac fibroblasts by si-circSmad4 was examined; si-circSmad4 significantly reduced the amounts of circSmad4 in cardiac fibroblasts from both sham-operated (left two bars) and TAC (right two bars in Figure 4B left graph) mice. It is noteworthy that TAC itself significantly increased the amount of both circSmad4 (left graph) and Smad4 mRNA, a linear host gene (right graph), compared with the sham-treated group (third bars versus first bars in both graphs). However, si-circSmad4 did not affect the expression of Smad4 mRNA (right graph).
Figure 4.
Effects of si-circSmad4 on the pre-established cardiac fibrosis induced by TAC for 8 weeks
(A) Experimental scheme. si-circSmad4 was intravenously administered every 3 days from 5 weeks after TAC (establishment of fibrosis after TAC for 4 weeks is shown in Figure S5). (B) Efficiency of circSmad4 knockdown. Band intensity was calculated after conventional RT-PCR. n = 6∼7. Each sample duplicated. One-way ANOVA with post hoc Dunnett’s T3 test. (C) Changes in heart weight. Unlike the effects seen with ISP administration (Figure 3), si-circSmad4 did not result in a significant reduction of heart weight, although there was a trend toward reduction (third bar vs. fourth bar). n = 14∼16. One-way ANOVA with post hoc Dunnett’s T3 test. (D) Representative one-dimensional echocardiogram. (E) Quantitative results of EF and FS. Note that TAC-induced reduction of contractility was recovered by si-circSmad4 administration. n = 10∼14. Both parameters: one-way ANOVA with post hoc Bonferroni’s test. (F) Changes in lung weight. si-circSmad4 significantly reduced the TAC-induced increase in lung weight (third bar vs. fourth bar). n = 8∼9. One-way ANOVA with post hoc Bonferroni’s test. (G) Effect of si-circSmad4 on TAC-induced cardiac fibrosis. Note that even pre-established fibrosis was reversed by knocking down circSmad4. n = 5∼14. Each sample duplicated. One-way ANOVA with post hoc Dunnett’s T3 test. Scale bar, 500 µM. (H) Changes in expression of fibrosis genes by si-circSmad4. n = 8∼9. Each sample duplicated. All analysis except Col3a1: one-way ANOVA with post hoc Dunnett’s T3 test. Col3a1: one-way ANOVA with post hoc Bonferroni’s test. Data are represented as mean ± SEM.
We first checked whether the cardiac hypertrophy could be reversed by administration of si-circSmad4 by measuring heart weight. The eventual increase in heart weight was reduced slightly by administration of si-circSmad4, but no statistical significance was determined (Figure 4C both graphs). We performed the echocardiographic analysis and observed the cardiac hypertrophy was developed (Figure 4D). The TAC-induced reduction of left ventricular internal dimension in both diastole and systole was significantly recovered by administration of si-circSmad4 (Figure S6). Cardiac output was examined by measuring ejection fraction (EF) and fractional shortening (FS), both of which are gold standard for the evaluation of cardiac contractility.25,26 TAC for 8 weeks significantly reduced both parameters (first and third bars in Figure 4E). However, these reductions were alleviated by administration of si-circSmad4 (third and fourth bars in Figure 4E). We next questioned whether the reduction of EF or FS resulted in the heart failure. Increase in the lung weight is one of standard parameters of heart failure, because it suggests the congestion of blood flow due to the reduced cardiac output.27,28 Thus, we measured lung weight. TAC for 8 weeks resulted in the increase in the lung weight (first and third bars in Figure 4F). However, this increase was significantly reduced by si-circSmad4 (third and fourth bars in Figure 4F).
The cardiac fibrosis was significantly reduced as determined by picrosirius red staining (Figure 4G). The quantification results are shown in the right bar graph in Figure 4G. The reduction of cardiac fibrosis was also accompanied by a significant reduction in mRNA amounts of fibrosis-associated genes such as Acta2, Col1a1, Col3a1, or Ctgf. Although it was not significant, TAC-mediated induction of Fn1 was reduced by si-circSmad4 injection (Figure 4H). In some mice, TAC for 8 weeks increased the appearance of severely deformed ventricular shapes as well as fibrosis, which suggests that cardiac remodeling had developed (Figure S7A). It is noteworthy that some mice mainly showed hypertrophic phenotypes (left two hearts in Figure S7A). However, in some mice, severe deformity accompanied with thinning of left ventricular free wall, which is remarkable feature of dilated cardiomyopathy, were observed (right six hearts in Figure S7A). This phenotypic diversity of TAC 8 weeks hearts might result in the relative wide range of values of heart weight (third bars in Figure 4C), EF and FS (third bars in Figure 4E), and fibrosis (third bar in Figure 4G). However, this remodeling was not seen when si-circSmad4 was administered (Figure S7B). These results suggest that circSmad4 plays a role in the relatively late phase of maintenance and further development of cardiac fibrosis and remodeling even after the establishment of initial cardiac fibrosis and remodeling.
circSMAD4 targets miR-671-5p and miR-671-5p attenuates myofibroblast activation
Recent advances in the study of non-coding RNA have elucidated that long non-coding RNAs modify chromosome structure, which results in the regulation of transcription of target genes in the nucleus.29,30 Nuclear circRNAs are involved in transcription through their binding to RNA-binding proteins (or transcription factors)31 or via the alternative splicing of exons.32 In contrast to nuclear circRNAs, cytoplasmic circRNAs have discrete roles. Although a new function of encoding small peptides33 has been reported, the most well-known and well-established function of cytoplasmic circRNAs is to sponge miRNA and thereby modulate the target genes of the miRNA.10 To further investigate this mechanism, we first analyzed the intracellular localization of circSMAD4. We found that circSMAD4 is highly expressed in the cytoplasm of human cardiac myofibroblasts (Figure S8A). We also noted that TGF-β1 treatment altered the distribution toward a cytoplasmic distribution (first bar and second bar in Figure S8B). This result likely suggests that transcriptional regulation or scaffold formation with transcription factors might not be the main function of circSMAD4.
Using bioinformatics analysis with two different bioinformatics databases, miRDB34 and miRNA_targets,35 we found that miR-512-3p, miR-671-5p, and miR-4425 contain complementary sequences to circSMAD4. However, by analysis with miRBase (http://www.mirbase.org), we found that miR-4425 was not identified in other species, which led us to rule it out for further study. As shown in Figure S9A, miR-512-3p has two possible binding sequences against circSMAD4 at 159–180 and 321–342. Treatment with TGF-β1 also significantly reduced the amount of miR-512-3p in human cardiac myofibroblasts (Figure S9B). We next checked whether miR-512-3p affected the expression of fibrosis-associated genes. Treatment with miR-512-3p mimic did not affect the TGF-β1-induced increase in the amount of ACTA2, COL1A1, CTGF, and FN1 (Figure S9C). miR-512-3p inhibitor also did not affect the TGF-β1-induced changes in those genes (Figure S9D). These results suggest that, although it may act in association with circSMAD4 and with TGF-β1, miR-512-3p itself does not modulate the development of fibrosis.
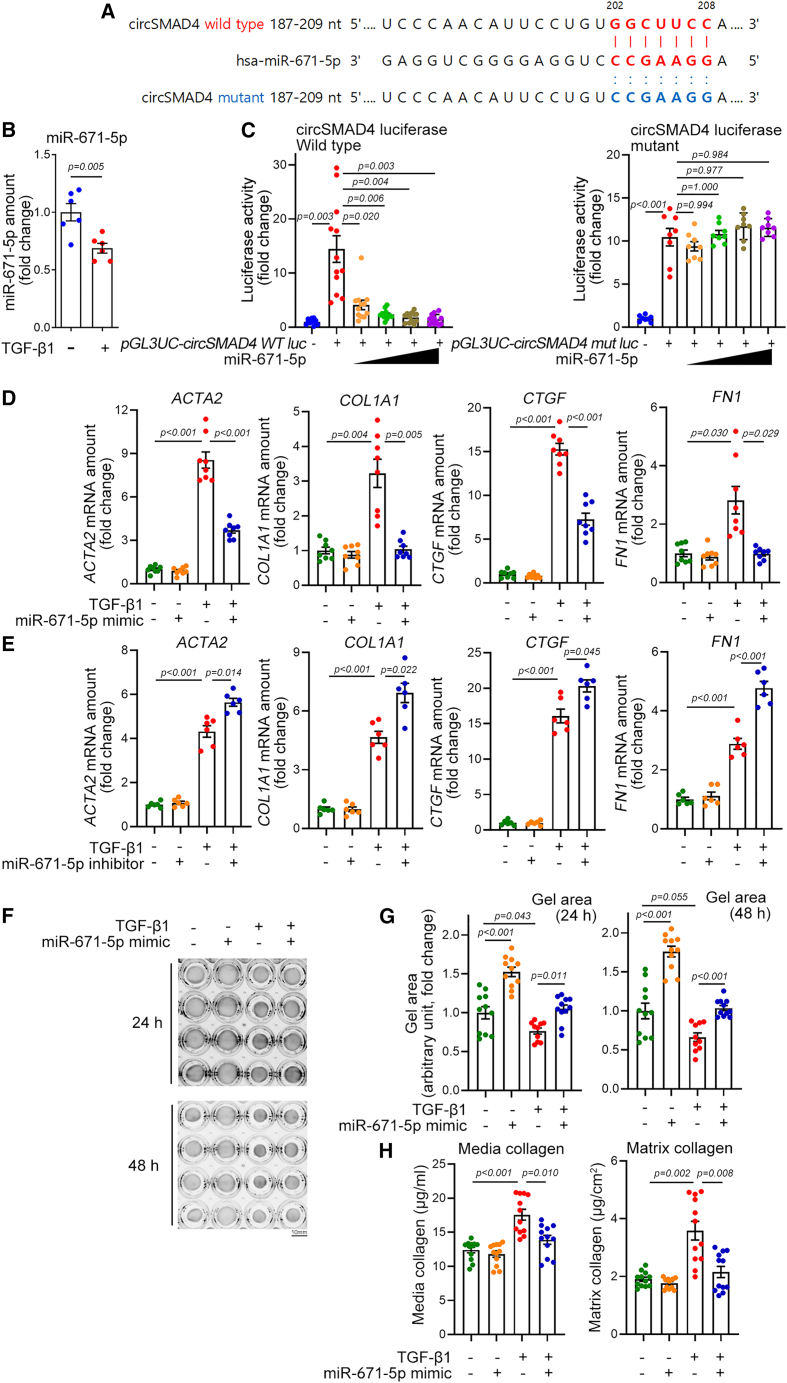
Thus, we investigated whether miR-671-5p is a target of circSMAD4 and whether miR-671-5p modulates fibroblast activation. The possible binding sequence between circSMAD4 and has-miR-671-5p is shown in Figure 5A. As for miR-512-3p, the amount of miR-671-5p was also significantly reduced by TGF-β1 in human cardiac fibroblasts (Figure 5B) or by TAC 8 weeks in mouse cardiac fibroblasts (Figure S10). To examine whether circSMAD4 can act as a sponge against miR-671-5p, we used a luciferase reporter assay. We inserted the circSMAD4 downstream behind the luciferase coding sequence to locate as a 3′ UTR and co-transfected this luciferase construct together with miR-671-5p. Transfection of miR-671-5p dramatically reduced the luciferase activity of circSMAD4 wild type in a dose-dependent fashion (left graph of Figure 5C). To check whether this reduction of luciferase activity was circSMAD4 sequence specific, we generated a binding sequence-disrupted mutant luciferase construct by use of site-directed mutagenesis. The sequence is shown in Figure 5A. miR-671-5p failed to reduce the luciferase activity of mutant circSMAD4 (right graph of Figure 5C).
Figure 5.
circSmad4 targets miR-671-5p
(A) Sequence alignment of human circSMAD4 with has-miR-671-5p. Binding sequences are shown in red. The mutant sequence in which the binding element is disrupted is shown in blue. (B) TGF-β1 reduced the amount of miR-671-5p. n = 6 from three independent experimental sets. Each sample duplicated. Unpaired Student’s t test. (C) Effects of miR-671-5p on circSMAD4-luciferase activity. Note that circSMAD4 wild-type luciferase was inhibited in a dose-dependent fashion (left panel, miRNA-671-mimic, 10–60 nM), whereas circSMAD4 mutant was not (right panel, miRNA-671-mimic, 10–60 nM). Wild type: n = 12 from three sets. One-way ANOVA with post hoc Dunnett’s T3 test. Mutant: n = 8 from two sets. One-way ANOVA with post hoc Dunnett’s T3 test. (D) miR-671-5p mimic attenuated the TGF-β1-induced increase in fibroblast genes. n = 8 from three sets. Each sample duplicated. One-way ANOVA with post hoc Dunnett’s T3 test. (E) miR-671-5p inhibitor potentiated the TGF-β1 effects. n = 6 from three sets. Each sample duplicated. ACTA2, COL1A1, and CTGF: one-way ANOVA with post hoc Dunnett’s T3 test. FN1: one-way ANOVA with post hoc Tukey’s test. (F and G) Representative gel pictures to show effects of miR-671-5p mimic on TGF-β1-induced gel contraction (F). Scale bar, 10 mm. Quantification results (G). Twenty-four hours: n = 10∼11 from three sets. One-way ANOVA with post hoc Bonferroni’s test. Forty-eight hours: n = 10∼11 from three sets. One-way ANOVA with post hoc Dunnett’s T3 test. (H) Effects of miR-671-5p mimic on TGF-β1-induced releases of media collagen (left) and matrix collagen (right). n = 12 from three sets. Both: one-way ANOVA with post hoc Dunnett’s T3 test. Data are represented as mean ± SEM.
We next checked whether miR-671-5p modulated fibroblast differentiation. Transfection of miR-671-5p mimic significantly reduced TGF-β1-mediated induction of ACTA2, COL1A1, CTGF, and FN1 (Figure 5D). In contrast, miR-671-5p inhibitor potentiated the TGF-β1-mediated increase in the expression of these genes (Figure 5E). To confirm the role of miR-671-5p in fibroblast differentiation, a gel contraction assay was performed. The TGF-β1-mediated reduction in gel area was recovered by addition of miR-671-5p mimic (Figure 5F). The quantification results are shown in Figure 5G. The TGF-β1-mediated gel contraction was accompanied by an increase in collagen in media (left graph) or in matrix (right graph in Figure 5H). However, these increases were blocked when miR-671-5p was transfected (Figure 5H).
miR-671-5p mimic blocks circSMAD4 overexpression-induced fibroblast differentiation
We next checked whether overexpression of circSMAD4 induces fibroblast differentiation into myofibroblasts and whether miR-671-5p mimic attenuates the effect of circSMAD4. To do this, we first constructed pcDNA3.1-zkscan1-circSMAD4 to ensure a circular structure. Transfection of zkscan1-circSMAD4 increased the expression of myofibroblast genes of ACTA2, COL1A1, CTGF, and FN1 (first vs. second bars). These increases, however, were blocked by co-transfection of miR-671-5p mimic (second vs. third bars in Figure 6A). We also checked whether miR-671-5p mimic could act on myofibroblast contraction, which is an opposite action against circSMAD4 (Figure 2D). Transfection of pcDNA3.1-zkscan1-circSMAD4 itself for 24 h induced contraction of the gel (left picture in Figure 6B), which appears as a reduction in the gel area (left graph in Figure 6C). The reduction, however, was abolished by co-transfection of miR-671-5p (left graph and picture in Figures 6B and 6C). Transfection of miR-671-5p also attenuated the gel contraction induced by transfection of circSMAD4 for 48 h (right picture and graph in Figures 6B and 6C). The results in Figures 5 and 6 suggest that circSMAD4 is pro-fibrotic mediator that is at least in part mediated by its antagonizing effect on anti-fibrotic miR-671-5p.
Figure 6.
Effects of circSmad4 overexpression and miR-671-5p on fibroblast differentiation
(A) Transfection of pcDNA3.1-zkscan1-circSMAD4 increased the expression of fibrosis genes, whereas co-transfection of miR-671-5p mimic blunted it. n = 9∼11 from approximately three to five independent experimental sets. Each sample duplicated. ACTA and FN1: one-way ANOVA with post hoc Bonferroni’s test. COL1A1 and CTGF: one-way ANOVA with post hoc Dunnett’s T3 test. (B) Representative gel pictures. Scale bar, 10 mm. (C) Quantification results of gel contraction. Transfection of zkscan1-circSMAD4 reduced the gel area, which represents contraction of the gel. However, co-transfection of miR-671-5p relieved the contraction. n = 9∼11 from three sets. Each sample duplicated. One-way ANOVA with post hoc Bonferroni’s test. Data are represented as mean ± SEM.
miR-671-5p targets FGFR2
To identify genes that are related to the cardiac fibroblast activation in our experimental models, we screened target candidates of miR-671-5p in silico. miRNAs inhibit their target gene mRNA by complementary sequence-specific binding to the 3′ UTR, which reduces the stability of the target mRNA.36 We narrowed the candidates related to fibrosis by doing a literature search. These approaches led us to focus on two candidates: disheveled 3 (DVL3) and fibroblast growth factor receptor 2 (FGFR2).
The DVL family, including DVL3, are key regulators in Wnt signaling and are known to induce fibrosis by inducing the nuclear translocation of β-catenin.37 Although a role of DVL3 in cardiac fibrosis has not been reported, we postulated that it may share a common pathway in cardiac fibroblasts. FGFR2 and its ligand fibroblast growth factor (FGF) are well known to induce fibrosis in lung38 and kidney.39 Likewise, its pro-fibrotic function in cardiac fibrosis and remodeling has been documented. FGFR2 is a target of an alternate miRNA, miR-338-3p, in cardiac fibrosis.40
The DVL3 3′ UTR has four putative binding sites at 646–652, 907–914, 1,579–1,585, and 2,210–2,215. The sequence alignments are shown in Figure S11A. We first generated a pGL3UC-DVL3 luciferase construct by inserting DVL3 3′ UTR. Transfection of miR-671-5p mimic dose-dependently decreased the luciferase activity (Figure S11B). Next, we tested whether pro-fibrotic stresses could induce an increase in DVL3. In human cardiac fibroblasts, treatment with TGF-β1 mildly but significantly increased the DVL3 mRNA amount (Figure S11C). In in vivo models, however, either ISP for 6 days (Figure S11D) or TAC for 8 weeks (Figure S11E) failed to increase the amount of DVL3 mRNA. Considering that these in vivo conditions were sufficient to induce cardiac fibrosis, which was accompanied by a significant increase in the myofibroblast marker genes (Figures 3 and 4), DVL3 per se might not be involved in fibrosis.
We next focused on FGFR2 as a target of miR-671-5p. The 1,112–1,118 region of the 3′ UTR of FGFR2 was predicted to bind to miR-671-5p (Figure 7A). Transfection of miR-671-5p reduced the 3′ UTR luciferase activity of FGFR2 (left graph in Figure 7B). To check the sequence specificity, we constructed a mutant FGFR2 3′ UTR luciferase construct (blue letters in Figure 7A). Transfection of miR-671-5p failed to reduce the luciferase activity (right graph in Figure 7B).
Figure 7.
miR-671-5p targets FGFR2
(A) Sequence alignment of miR-671-5p and wild-type or mutant FGFR2 3′ UTR. (B) miR-671-5p dose dependently reduced the 3′ UTR luciferase activity (left, miRNA-671-mimic, 10–60 nM). In contrast, it failed to inhibit the mutant luciferase (right, miRNA-671-mimic, 10–60 nM). n = 8 from two independent experimental sets. Wild type: one-way ANOVA with post hoc Dunnett’s T3 test. Mutant: one-way ANOVA with post hoc Tukey’s test. (C and D) Effect of miR-671-5p mimic (C) or inhibitor (D) on TGF-β1-induced increase in FGFR2 mRNA expression. Mimic (C): n = 7 from three sets. One-way ANOVA with post hoc Dunnett’s T3 test. Inhibitor (D): n = 6 from two sets. One-way ANOVA with post hoc Dunnett’s T3 test. (E) Effect of si-circSMAD4 on TGF-β1-induced increase in FGFR2 mRNA expression. n = 8∼13 from approximately three to five sets. One-way ANOVA with post hoc Dunnett’s T3 test. (F and G) Effect of intravenous injection of si-circSmad4 on the increase in Fgfr2 expression induced by ISP for 6 days (F) or by TAC for 8 weeks (G). The experimental schemes of those animal models are identical to Figures 3A and 4A. ISP: n = 5∼9 from two ∼ three sets. One-way ANOVA with post hoc Bonferroni’s test. TAC: n = 9∼11 from two sets. One-way ANOVA with post hoc Bonferroni’s test. (H) Schematic diagram of working mechanism. Data are represented as mean ± SEM.
We next questioned whether fibrosis-provoking stresses can increase the FGFR2 mRNA level and whether these increases can be affected by si-circSMAD4, which would suggest that circSMAD4 may participate in the induction of FGFR2 expression in cardiac fibrosis. In human cardiac fibroblasts, treatment with TGF-β1 increased the expression of FGFR2, which was then blocked by simultaneous treatment with miR-671-5p mimic (third vs. fourth bar in Figure 7C). Although not significant, transfection of miR-671-5p inhibitor tended to increase the TGF-β1-induced increase in FGFR2 mRNA (third vs. fourth bar in Figure 7D).
We next wanted to link the circSMAD4 and FGFR2. Transfection of si-circSMAD4 attenuated the TGF-β1-induced increase in FGFR2 mRNA in human cardiac fibroblasts (Figure 7E). Prevention of the TGF-β1-induced increase in FGFR2 mRNA was also observed in si-circSmad4-administered ISP (Figure 7F) or TAC (Figure 7G) mouse hearts.
Discussion
Almost all etiologies of heart disease can produce cardiac fibrosis, which adversely contributes to cardiac remodeling and then reduced compliance and function in the diseased hearts. Thus, cardiac fibrosis is a high-impact health problem worldwide. In the current study, we identified a novel signal pathway involving circSMAD4-miR-671-5p-FGFR2, which mediates myofibroblast differentiation in cardiac fibroblasts. Knocking down circSMAD4 by si-circSMAD4 blocked cardiac fibrosis in mouse hearts that had been administered ISP for 6 days. Notably, it reversed the pre-established cardiac fibrosis in the mice subjected to TAC for 8 weeks. In human cardiac fibroblasts, circSMAD4 provoked cardiac fibroblast activation, while miR-671-5p, a novel anti-fibrotic miRNA, prevented fibrosis. Mechanistically, we showed that circSMAD4 squelches out miR-671-5p by direct binding in a sequence-specific manner. This reduction of miR-671-5p induces upregulation of FGFR2, which then induces myofibroblast differentiation and fibrosis (Figure 7H).
RNA-targeting therapeutics for human diseases have emerged as an important platform in recent years. Like other non-coding RNAs, circRNAs have been implicated as new modulators in many diseases.41,42 Because of their stable structure, circRNAs have greater potential over other RNAs. Indeed, circRNAs are believed to act as mediators between distant cells and organs43 and some circRNAs have drawn much attention from clinicians and industry as reliable biomarkers. Indeed, in previous studies in our laboratory, we showed that two circRNAs, circSAMD4a and circSMOC1-2, are potential targets for treatment of vascular calcification.44,45 Moreover, circRNA expression patterns tell us that circRNAs are quite tissue specific and have many isoforms that may help to increase specific actions in target organs and thereby to reduce unwanted effects.46,47 It is noteworthy that circSMAD4 also has different isoforms with different sets of exons and backsplice regions. Our circSMAD4 consists of exons 5 to 8 (circSMAD4-III, exon 5 to 8). In our initial screening with RNA-seq data, we noticed that cardiomyocytes have different isoforms of circSmad4, such as circSmad4-I (exon 2 to 4), circSmad4-II (exon 1 to 7), and circSmad4-III (exon 5 to 8). Their expressions, however, were not detected in cardiac fibroblasts. Considering this isoform expression pattern, circSMAD4 (exon 5 to 10), which we studied in the current work with cardiac fibroblasts, may have specificity against cardiac fibrosis.
circSMAD4 has been shown in previous studies to attenuate inflammation by targeting miR-377-3p.48 In contrast, its opposite action in inflammation has also been reported in a study showing that circSMAD4 provokes experimental colitis through miR-135a-5p.49 Considering its roles in inflammation, it is also plausible that circSMAD4 plays a role in cardiac fibrosis and remodeling, because local inflammation is a crucial factor for the initiation of cardiac fibrosis and remodeling.50 We did not address the involvement of inflammation in the function of circSMAD4/miR-135-5p in the current work, however, and this may need further study. Our results suggest, at least in part, that circSMAD4 directly regulates cardiac fibrosis by inhibiting miR-671-5p. It is also noteworthy that, although the circSmad4 amount was dramatically increased in the relatively late phase (8 weeks) after TAC in our experimental models, si-circSmad4 successfully attenuated the initial phase of acute cardiac fibrosis induced by ISP (Figure 3). In addition, although it failed to eventually reverse the cardiac hypertrophy in the late phase of TAC (8 weeks), it could effectively reduce the pre-established cardiac fibrosis (Figure 4). These wide-ranging effects raise the possible involvement of multiple mechanisms or multiple cell types, including cardiomyocytes.
Recent studies have suggested that many circRNAs are involved in cardiac fibrosis. Like our report, other reports have suggested sponging of miRNAs as a working mechanism of circRNAs; some examples are circ_0036176,51 circSnx12,52 and circHIPK3.53 Although C2C12 skeletal myoblasts were used, one study showed that circHIPK3 actively mediates myoblast differentiation.54 Notably, other research has shown that circYap attenuates cardiac fibrosis by its direct binding to proteins, tropomyosin-4, and gamma-actin.19 Alternatively, circNlgn generates the protein Nlgn173, which then activates H2AX to induce other proteins, including interleukins, and thereby prevent doxorubicin-induced cardiac fibrosis.55 It should be mentioned, however, that those studies used whole hearts for the initial screening and identification of candidate circRNAs. Not only the active differentiation of fibroblasts into myofibroblasts56 but also the loss of functionally intact cardiomyocytes57,58 can provoke overall fibrosis and substitution of functional cardiomyocytes. Thus, using specific cell types for screening and identification may help to delineate the mechanism of action and thereby reduce unwanted effects when the mechanism is applied for future therapeutics. Thus, in the present study, we used the Langendorff isolation system to first obtain cardiac fibroblasts after TAC. Compared with other previous reports in which whole hearts were used after TAC, the isolation of cardiac fibroblasts after TAC is a strength of our study that allowed us to focus on cardiac fibroblast-specific circRNAs.
In TAC 8 weeks mice, si-circSmad4 administration significantly improved cardiac manifestations, including dilatation, fibrosis, and cardiac output (Figures 4 and S7). However, it did not substantially alleviate cardiac hypertrophy (Figure 4C). Considering that cardiomyocyte enlargement predominantly drives cardiac hypertrophy in response to exogenous stressors, and given that si-circSmad4 primarily targets cardiac fibroblasts due to the exclusive expression of circSmad4 in these cells (Figure S4B), its limited efficacy in curbing hypertrophy during the late stages of TAC might result from its narrowed effect on cardiac myofibroblasts and scant influence on cardiomyocytes. However, this specificity could be advantageous, allowing si-circSmad4 to target cardiac fibroblasts without impacting cardiomyocytes during fibrosis-related remodeling in the advanced stages of heart failure. Given circSmad4’s pronounced expression in the lungs, a compelling question arises: might si-circSmad4 mitigate interstitial lung fibrosis? This remains an avenue for future exploration.
In the current work, we did not consider mechanisms other than miRNA sponging. Although active internal ribosome entry site (IRES) was not found (IRESpy; https://irespy.shinyapps.io/IRESpy/), and other possible proteins were not predicted to be translated, protein translation or direct binding to other proteins may still participate in the pro-fibrotic mechanism of circSMAD4, which requires further study.
SMAD4 is a host gene of circSMAD4 and both genes are simultaneously upregulated in hypertrophic stresses in in vivo conditions (Figures 3 and 4). Still, it is curious what the biological function of circSMAD4 in normal physiology is and what the relationship between circSMAD4 and SMAD4 mRNA is. Although not likely, whether SMAD4 has other functions mediated by protein generation or direct binding to other DNA or protein would be of interest, too. In addition, the distribution of isoforms and their functions remain unknown. Nonetheless, this work has merit due to our discovery of a novel signal pathway involving circSMAD4/miR-671-5p/FGFR2, which can be considered for future therapeutic platforms in cardiac fibrosis.
Materials and methods
Bioinformatics
RNA-seq and circRNA selection strategy
For the RNA-seq analysis, total RNA was treated with the Ribo-Zero Gold rRNA Removal Kit (Illumina) to remove ribosomal RNAs and applied to the TruSeq Stranded Total RNA Kit (Illumina) to construct an RNA-seq library. The library was sequenced by HiSeq 2500 (Illumina) in the paired-end mode with 100 sequencing cycles. The FastQC was used for the initial quality check of the sequencing results,59 and the reads with low quality were removed with Trimmomatic.59 The expression counts of circRNAs were calculated using the STAR aligner60 and DCC algorithm61 and normalized by the total counts of circRNAs in each sample. Among the total of 19,737 circRNAs with at least two reads count in the 24 samples, we selected circRNAs with a mean read count of at least 1 across all samples, resulting in 1,717 circRNAs passing this criterion. Detailed schemes are shown in Figure 1A.
miRNA prediction
Prediction of miRNAs that bind to circRNA was performed by utilization of two websites of miRDB34 and miRNA_targets.35 The sequences of selected circRNAs were provided as input in the prediction tool of the miRDB (http://mirdb.org/index.html) web tool and TargetScan software to identify miRNAs potentially interacting with circRNA. The list of miRNAs expressed in lung fibroblast was obtained from a previous publication (GEO: GSE125183). After retrieving the list of miRNAs with binding sites in circRNAs, the expression of these miRNAs was identified by miRNA assay using human cardiac fibroblast samples with fibrosis induced by TGF-β1. The miRNAs with significant expression changes after treatment of cardiac fibroblasts with TGF- β1 were selected as final candidates.
Target mRNA prediction
The functions of circRNAs were regulated by the genes targeted in the circRNA-miRNA-mRNA network. The miRDB (http://mirdb.org/index.html) and TargetScan (http://targetscan.org/vert_72/) were used for analysis of the target mRNAs that can potentially bind with miRNAs. Target mRNAs were predicted by using miRNA name or sequence. After searching a list of mRNAs that might interact with miRNAs, mRNAs related to fibrosis were selected. The mRNAs having a negative correlation with miRNA in their expression change during fibrosis were selected as final targets.
Sequence data deposition
We deposited the RNA-seq dataset, including raw sequencing reads and processed data, at GEO with the accession number GSE240332.
Cell culture, vector construct, transfection, and luciferase assay
Cell culture
Primarily cultured human cardiac fibroblasts (LONZA, #CC-2904, Lonza, Basel, Switzerland) were cultured in fibroblast basal medium (FBM, #CC-3131, Lonza, Basel, Switzerland) supplemented with FGM-3 Bullet Kit (#CC-4526, Lonza) at 37°C in a humidified incubator with 5% CO2. Cells were cultured from passages 6 to 10. To induce fibroblast activation, primary human ventricular cardiac fibroblasts were starved in FBM for 16 h followed by treatment with human TGF-β1 (#PHG9214, Gibco, MA, USA) for 24 h.
Vector constructs
The whole length of circRNA was cloned into the overexpression vector of pcDNA3.1(+). Zkscan1 MCS exon vector (Addgene, plasmid #69901) was used for overexpression of circSMAD4. The vectors were digested and ligated with the PCR product using the EZ-Fusion Cloning Kit (Enzynomics) following the manufacturer’s instructions. The putative binding site mutants were inserted into the MCS of the pcDNA3.1(+) Zkscan1 vector (Cosmogentech, Seoul, Korea). Primers for mutant construct for luciferase assay are described in Table S3. After cloning, all constructs were confirmed by sequencing (Macrogen, Korea).
Transfection
For circRNA overexpression, human cardiac fibroblasts were seeded at a density of 2 × 105 cells. When confluent, cardiac fibroblasts were transfected with pZkscan1-circ-SMAD4, using Lipofectamine 3000 (#L3000001, Thermo), as per the manufacturer’s instruction. For circRNA knockdown, cardiac fibroblasts were treated with siRNA, using Lipofectamine RNAiMax (#13778075, Thermo), according to the manufacturer’s protocol.
Luciferase assay
Luciferase reporter pGL3UC vector, which was constructed by inserting CMV promoter sequences and multiple cloning sites into pGL3 basic (Promega) was used. To detect the interaction between circSMAD4 and miRNAs, the sequence of circSMAD4-wt and the sequence of circSMAD4-mutant were cloned into the multiple cloning sites at the 3′ UTR of pGL3UC (Cosmogentech). Human cardiac fibroblasts were seeded into 24-well plates. When the cell confluency reached around 70%–80%, cells were co-transfected with either 100 ng of empty pGL3UC vector or pGL3UC vector with circSMAD4-wild type and circSMAD4-mutant, along with miRNA mimics using Lipofectamine 3000.
To verify the interaction between miRNA and mRNA, the sequence of the mRNA 3′ UTR was cloned into the multiple cloning sites of pGL3UC vector. Human cardiac fibroblasts were seeded into 24-well plates. When the cell confluency reached about 70%–80%, cells were co-transfected with either 100 ng of empty pGL3UC vector or pGL3UC vector with mRNA 3′ UTR sequence and mRNA 3′ UTR mutant sequence, along with miRNA mimics using Lipofectamine 3000.
Luciferase and galactosidase activities were measured 24 h after transfection with the dual luciferase reporter assay system (#E397A, Promega, Madison, WI, USA) following the manufacturer’s protocol. Luciferase activity was normalized by β-galactosidase luciferase activity.
Sircol collagen assay
The manufacturer’s instruction for Sircol collagen assay kit (#S1000, Biocolor Assays, Ireland) was followed. Briefly, to extract matrix collagen, cells were incubated with 0.1 mg/mL pepsin (#P7012, Sigma-Aldrich, USA) in 0.5 M acetic acid at 4°C with gentle agitation overnight. Then, acid-neutralizing reagent was added to the cell extracts. To measure contents of soluble collagens in the conditioned medium and matrix, medium and cell extracts sample were added to Sircol dye reagent and agitated for 30 min at room temperature followed by centrifugation at 12,000 rpm for 10 min. Then, the pellet was dissolved in alkali reagent. The absorbance was measured at 555 nm using a Spectramax Abs Plus (Molecular Devices). Soluble collagen contents were attained with a standard graph of supplied collagen.
Collagen gel assay
The contractile property of cardiac fibroblasts was determined by collagen gel contraction assay. Confluent cultures of cardiac fibroblasts were detached by trypsin and suspended in serum-free FBM. The cell number was then counted with a hemocytometer. Amounts of 2.5 105 cells were resuspended with medium containing 1 mg/mL Collagen type I (#354231, Corning Life Sciences, NY, USA). Following this, 500 μL of the mixture was mixed with 1 M NaOH to prepare a collagen lattice and then immediately transferred to the 24-well plate. The mixture was then allowed to polymerize at room temperature for 40 min. After polymerization, the gels were covered with serum-free medium with TGF-β1 or vehicle and were gently detached with a sterilized spatula. The area of gels was measured daily with the ENDURO GDS gel documentation system (Labnet International, Iselin, NJ, USA). The gel area was determined by using ImageJ software.
RNA studies
RNA isolation, cDNA synthesis, conventional PCR, and qRT-PCR
TRI Reagent (#TR118, Molecular Research Center, OH, USA) was used to isolate total RNA in accordance with the manufacturer’s protocol. DNase I (#2270A, Tokyo, Takara) was used to remove residual DNA in the samples. Total RNAs (1 μg, 20 μL) were reverse transcribed using a RevertAid reverse transcriptase (#EP0442, Thermo Scientific) and random hexamers (#SO142, Thermo Scientific) to obtain cDNA. cDNAs were then subjected to conventional PCR and real-time qRT-PCR. qRT-PCR was performed to measure myofibroblast marker gene expression using QuantiTect SYBR Green PCR Kit (#204143, Qiagen, Germantown, MD, USA) on a Rotor-Gene Q real-time PCR cycler (Qiagen). The relative expressions of genes were calculated on the cycle threshold values using the 2−ΔΔCt method. GAPDH was used as a control gene for normalization. The primers used are listed in Table S1.
To validate circRNA expression, conventional PCR was performed using the nTaq (Mg2+Plus) kit (#P050A, Enzynomics, Deajeon, Korea) with specifically designed primers for circRNA amplification. Divergent primers were designed across the backsplicing junction to amplify circRNA transcripts. Used primers are listed in Table S3. Amplification of circRNA transcripts from agarose gel electrophoresis was quantified by measuring the band intensity with ImageJ software.
The cDNA of miR-671-5p and miR-512-3p were synthesized by adding a poly(A) tail to the 3′ end and ligating an adapter to the 5′ end of miRNA followed by reverse transcription with universal primer using the TaqMan Advanced miRNA cDNA Synthesis Kit (A28007, Applied Biosystems, Thermo Fisher Scientific). The cDNAs were analyzed by qPCR using a QuantiTect SYBR Green PCR Kit with gene-specific primers for miR-671-5p, miR-512-3p, and 18S rRNA (483035_mir, 478971_mir, and 4333760F, Applied Biosystems).
RNase R digestion
The RNase R treatment assay is a common method used to remove linear RNAs. Total RNA samples were mixed with RNase R (#RNR07250, Lucigen, Middleton, WI, USA), while the control samples were without RNase R. RNase R digestion was conducted at 37°C for 20 min and inactivated at 95°C for 3 min following the manufacturer’s protocol.
siRNAs
The siRNAs for each circRNA at the head-to-tail junctions were designed using the siDesign center in Horizon Discovery (https://horizondiscovery.com/en/products/tools/siDESIGN-Center) or the i-Score designer (https://www.med.nagoya-u.ac.jp/neurogenetics/iScore/iscore.html). The siRNAs were synthesized by Bioneer (Daejeon, Korea). siRNAs for FGFR2, DVL3, and AccuTarget negative-control siRNA (#SN-1003) were obtained from Bioneer (Daejeon, Korea). siRNA sequences were provided in Table S3.
Animal models
Adult male C57BL/6J mice (8–10 weeks old) were purchased from Damul Science (Damul Science, Dajeon, Korea). All animal experiments as well as the use of Avertin were approved by the Institutional Review Board of Chonnam National University Medical School Research Institutional Animal Care and Use Committee (CNU-IACUC-H-2021-39 and CNU-IACUC-H-2022-11) and followed the Guide for the Care and Use of Laboratory Animals (eighth edition, National academies Press, Washington, DC, USA, 2011). As used in other papers,62,63 Avertin was chosen for anesthesia because it is known to have fewer side effects on cardiovascular and hemodynamic stability.64,65,66 In the current study, Avertin was used for temporary induction of anesthesia but not for obtaining of results from the live animals.
Osmotic pump implantation for ISP administration
The animal model of cardiac fibrosis was induced by implantation of an osmotic pump containing 30 mg/kg/day ISP for 6 days, as previously described.67 Pumps were dorsally implanted in the mice for 6 days. For the control group, age-matched normal control mice received the equivalent volume of PBS. After infusion, mice were sacrificed and heart tissues were harvested for further analysis.
TAC surgery
TAC surgery was described in previous reports.68,69 Briefly, 8-week old male C57BL/6J mice were anesthetized with 2,2,2-tribromoethanol (Avertin, #T48402, Sigma, St. Louis, MO, USA). The mice were then ventilated and the chest was opened to expose the thymus and aortic arch. A 7-0 silk suture was placed between the innominate and left carotid arteries. A 27G needle was placed parallel to the transverse aorta and the suture was tied. After ligation, the needle was removed and the rib cage was closed using 4-0 silk suture. As a control, sham mice were subjected to the same operation without ligation.
Isolation of mouse cardiac fibroblasts from the heart
The Langendorff perfusion system was used to isolate cardiac fibroblasts from mouse hearts as described in Yoon et al.70 After anesthesia, the hearts were immediately excised and were instantly perfused with Ca2+-free Tyrode buffer (137 mM NaCl, 5.4 mM KCl, 10 mM HEPES pH 7.4, 1 mM MgCl2, 5 mM taurine, 10 mM glucose, and 10 mM BDM) for 7 min followed by perfusion with enzyme solution (0.1 mg/mL hyaluronidase and 0.35 U/mL collagenase type B in Ca2+-free Tyrode buffer) for an additional 25 min. Then, hearts were dissociated into small pieces with fine forceps, resuspended in 5% BSA in Tyrode buffer for 10 min, and then passed through a 100-μm cell strainer (#352360, Falcon, NY, USA). The suspension was allowed to settle by centrifugation at 50 × g for 2 min to purify cardiomyocytes. The supernatants were centrifugated to obtain cardiac fibroblasts.
In vivo siRNA delivery
siRNA of circSmad4 was delivered into C57BL/6J mice using in vivo-jetPEI (#101000040, Polyplus, Illkirch, France), following the manufacturer’s instruction. The experimental scheme is described in Figures 3A and 4A. Briefly, in vivo-jetPEI reagent and siRNA were diluted in 5% glucose solution, followed by incubation for 15 min at room temperature. The siRNA mixture was intravenously injected through the tail vein.
Echocardiography
Echocardiographic studies were performed using a Vivid S5 echocardiography system (GE Healthcare, Chicago, IL, USA) equipped with 13-MHz linear array transducer at 8 weeks after TAC by an expert who was not notified about experimental conditions to exclude bias. Two-dimensional guided M mode of the left ventricle (LV) was obtained from the parasternal view. LV cavity dimension and LV free and interventricular septum wall thickness were measured, and percentage change in LV dimension (FS, LV%FS) was calculated as LV%FS = [(LVDd − LVDs)/LVDd] × 100, where LVDd is LV dimension at end-diastole and LVDs is LV dimension at end systole. LV% EF was calculated as LV%EF = [(EDVESV)/EDV] × 100, where EDV is LV volume at end-diastole and ESV is LV volume at end systole. LV volume was estimated by the Teicholz method.
The LV myocardial performance index (MPI) was calculated from mitral valve inflow and aortic valve outflow Doppler images. The time from cessation to the beginning of mitral inflow (a) and the LV ejection time (b) were measured. The LV MPI was calculated as follows: LV MPI = (ICT + IRT)/ejection time = (a − b)/b where ICT is the LV isovolumic contraction time and IRT is the isovolumic relaxation time. Doppler-derived intervals were measured offline from stored data.
Histochemical and Sirius red staining
Mouse heart tissues for heart histology were fixed in 4% paraformaldehyde. The fixed heart tissues were generally dehydrated and paraffin embedded. The paraffin blocks were cut into sections with a thickness of 15 μm. Heart sections were then processed for Sirius red staining. Sirius red staining was performed on paraffin sections of mouse hearts to detect collagen deposition.
Statistical analysis
Analyses were conducted using IBM SPSS Statistics 26 (IBM, Chicago, IL). We first verified data normality through the Shapiro-Wilk test (because all numbers in group were less than 50). For data confirmed as normally distributed, we applied a parametric test—either a two-tailed Student’s t test or ANOVA with subsequent post hoc analysis—dependent on the number of groups being either two or more. In case of evaluating more than two groups, following ANOVA, we examined variance homogeneity between groups with Levene’s test. Absence of homogeneity necessitated the use of Dunnett’s T3 as the post hoc test. When homogeneity was evident, the choice between the post hoc Tukey’s honestly significant difference test or Bonferroni’s test was determined based on whether the number of cases within groups was equal. For non-normally distributed data, which is determined by Shapiro-Wilk test, we employed non-parametric methods: the Kruskal-Wallis test with a Bonferroni adjustment or the Mann-Whitney U test, predicated on whether there were more than two groups in consideration. Significance was confirmed for a p value of less than 0.05 and each p value is noted in the figures.
Data and code availability
All data from this study are included in this article, the associated source data file, and the supplemental information. The data that support the findings of this study are available on request from the corresponding author.
Acknowledgments
This research was supported by funding from Basic Science Research Program of the National Research Foundation of Korea (NRF) by Ministry of Science, ICT & Future Planning (NRF-2021R1A2B5B02001501, RS-2023-00208097, and NRF-2022R1A4A2000767). The authors would like to thank Jennifer Holmes at Medical Editing Services for language editing and careful reading of the manuscript.
Author contributions
A.J. drafted the manuscript. A.J. and Y.L. executed most in vivo and in vitro experiments. D.-H.K. undertook Langendorff isolation in the preliminary screening. T.K. established the TAC operation for initial analysis. Y.K.C. conducted and interpreted mouse echocardiography. J.R. initiated the analysis of circRNA candidates. Y.G.L. was responsible for histological preparations. S.S. handled in vitro cell preparation and the luciferase assay. N.C. mainly worked on RNA preparation. H.J.C. assisted with histological preparation. J.C.K. oversaw animal operations. Y.C. delved into bioinformatic analysis, particularly with the cDNA microarray. S.-J.L. and H.-S.K. procured and analyzed human postmortem samples. H.J.K. and M.H.J. supplied the mouse echocardiography equipment. K.I.N. contributed to histology. Y.S.K and Y.A. offered insights into the conceptualization of cardiac fibrosis. W.J.P. shaped the study’s conceptual framework and assisted with Langendorff and TAC processes. Y.-K.K. engaged in RNA-related tasks, the bioinformatic analysis of total RNA-seq, and furthered the study’s conceptualization. H.K. orchestrated the study design, conceptualization, and secured funding.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.102071.
Contributor Information
Young-Kook Kim, Email: ykk@jnu.ac.kr.
Hyun Kook, Email: kookhyun@jnu.ac.kr.
Supplemental information
References
- 1.Hinderer S., Schenke-Layland K. Cardiac fibrosis - A short review of causes and therapeutic strategies. Adv. Drug Deliv. Rev. 2019;146:77–82. doi: 10.1016/j.addr.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Liu T., Song D., Dong J., Zhu P., Liu J., Liu W., Ma X., Zhao L., Ling S. Current Understanding of the Pathophysiology of Myocardial Fibrosis and Its Quantitative Assessment in Heart Failure. Front. Physiol. 2017;8:238. doi: 10.3389/fphys.2017.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nwabuo C.C., Vasan R.S. Pathophysiology of Hypertensive Heart Disease: Beyond Left Ventricular Hypertrophy. Curr. Hypertens. Rep. 2020;22:11. doi: 10.1007/s11906-020-1017-9. [DOI] [PubMed] [Google Scholar]
- 4.Leask A. Getting to the heart of the matter: new insights into cardiac fibrosis. Circ. Res. 2015;116:1269–1276. doi: 10.1161/CIRCRESAHA.116.305381. [DOI] [PubMed] [Google Scholar]
- 5.Hentze M.W., Preiss T. Circular RNAs: splicing's enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X.O., Dong R., Zhang Y., Zhang J.L., Luo Z., Zhang J., Chen L.L., Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu T., Wu J., Han P., Zhao Z., Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genom. 2017;18:680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng S., Zhang X., Odame E., Xu X., Chen Y., Ye J., Zhou H., Dai D., Kyei B., Zhan S., et al. CircRNA-Protein Interactions in Muscle Development and Diseases. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22063262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 12.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T., et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 13.Wang X., Cao X., Dong D., Shen X., Cheng J., Jiang R., Yang Z., Peng S., Huang Y., Lan X., et al. Circular RNA TTN Acts As a miR-432 Sponge to Facilitate Proliferation and Differentiation of Myoblasts via the IGF2/PI3K/AKT Signaling Pathway. Mol. Ther. Nucleic Acids. 2019;18:966–980. doi: 10.1016/j.omtn.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Xu J.D., Fang X.H., Zhu J.N., Yang J., Pan R., Yuan S.J., Zeng N., Yang Z.Z., Yang H., et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc. Res. 2020;116:1323–1334. doi: 10.1093/cvr/cvz215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu N., Li C., Xu B., Xiang Y., Jia X., Yuan Z., Wu L., Zhong L., Li Y. Circular RNA mmu_circ_0005019 inhibits fibrosis of cardiac fibroblasts and reverses electrical remodeling of cardiomyocytes. BMC Cardiovasc. Disord. 2021;21:308. doi: 10.1186/s12872-021-02128-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mester-Tonczar J., Winkler J., Einzinger P., Hasimbegovic E., Kastner N., Lukovic D., Zlabinger K., Spannbauer A., Traxler D., Batkai S., et al. Association between Circular RNA CDR1as and Post-Infarction Cardiac Function in Pig Ischemic Heart Failure: Influence of the Anti-Fibrotic Natural Compounds Bufalin and Lycorine. Biomolecules. 2020;10 doi: 10.3390/biom10081180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia Y., Lee K., Li N., Corbett D., Mendoza L., Frangogiannis N.G. Characterization of the inflammatory and fibrotic response in a mouse model of cardiac pressure overload. Histochem. Cell Biol. 2009;131:471–481. doi: 10.1007/s00418-008-0541-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B., Zhang P., Tan Y., Feng P., Zhang Z., Liang H., Duan W., Jin Z., Wang X., Liu J., et al. C1q-TNF-related protein-3 attenuates pressure overload-induced cardiac hypertrophy by suppressing the p38/CREB pathway and p38-induced ER stress. Cell Death Dis. 2019;10:520. doi: 10.1038/s41419-019-1749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu N., Xu J., Du W.W., Li X., Awan F.M., Li F., Misir S., Eshaghi E., Lyu J., Zhou L., et al. YAP Circular RNA, circYap, Attenuates Cardiac Fibrosis via Binding with Tropomyosin-4 and Gamma-Actin Decreasing Actin Polymerization. Mol. Ther. 2021;29:1138–1150. doi: 10.1016/j.ymthe.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M., López de Juan Abad B., Cheng K. Cardiac fibrosis: Myofibroblast-mediated pathological regulation and drug delivery strategies. Adv. Drug Deliv. Rev. 2021;173:504–519. doi: 10.1016/j.addr.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber K.T., Sun Y., Bhattacharya S.K., Ahokas R.A., Gerling I.C. Myofibroblast-mediated mechanisms of pathological remodelling of the heart. Nat. Rev. Cardiol. 2013;10:15–26. doi: 10.1038/nrcardio.2012.158. [DOI] [PubMed] [Google Scholar]
- 22.Frangogiannis N.G. Cardiac fibrosis. Cardiovasc. Res. 2021;117:1450–1488. doi: 10.1093/cvr/cvaa324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czubryt M.P. Cardiac Fibroblast to Myofibroblast Phenotype Conversion-An Unexploited Therapeutic Target. J. Cardiovasc. Dev. Dis. 2019;6 doi: 10.3390/jcdd6030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang S.C., Ren S., Rau C.D., Wang J.J. Isoproterenol-Induced Heart Failure Mouse Model Using Osmotic Pump Implantation. Methods Mol. Biol. 2018;1816:207–220. doi: 10.1007/978-1-4939-8597-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill J.A., Karimi M., Kutschke W., Davisson R.L., Zimmerman K., Wang Z., Kerber R.E., Weiss R.M. Cardiac hypertrophy is not a required compensatory response to short-term pressure overload. Circulation. 2000;101:2863–2869. doi: 10.1161/01.cir.101.24.2863. [DOI] [PubMed] [Google Scholar]
- 26.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W., et al. Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohammed S.F., Storlie J.R., Oehler E.A., Bowen L.A., Korinek J., Lam C.S.P., Simari R.D., Burnett J.C., Jr., Redfield M.M. Variable phenotype in murine transverse aortic constriction. Cardiovasc. Pathol. 2012;21:188–198. doi: 10.1016/j.carpath.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards D.A., Aronovitz M.J., Calamaras T.D., Tam K., Martin G.L., Liu P., Bowditch H.K., Zhang P., Huggins G.S., Blanton R.M. Distinct Phenotypes Induced by Three Degrees of Transverse Aortic Constriction in Mice. Sci. Rep. 2019;9:5844. doi: 10.1038/s41598-019-42209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han P., Li W., Lin C.H., Yang J., Shang C., Nuernberg S.T., Jin K.K., Xu W., Lin C.Y., Lin C.J., et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon G., Lim Y.H., Jo D., Ryu J., Song J., Kim Y.K. Obesity-linked circular RNA circTshz2-2 regulates the neuronal cell cycle and spatial memory in the brain. Mol. Psychiatr. 2021;26:6350–6364. doi: 10.1038/s41380-021-01303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 33.Li F., Cai Y., Deng S., Yang L., Liu N., Chang X., Jing L., Zhou Y., Li H. A peptide CORO1C-47aa encoded by the circular noncoding RNA circ-0000437 functions as a negative regulator in endometrium tumor angiogenesis. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong N., Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar A., Wong A.K.L., Tizard M.L., Moore R.J., Lefèvre C. miRNA_Targets: a database for miRNA target predictions in coding and non-coding regions of mRNAs. Genomics. 2012;100:352–356. doi: 10.1016/j.ygeno.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang D., Li S., Duan X., Ren J., Liang S., Yakoumatos L., Kang Y., Uriarte S.M., Shang J., Li W., Wang H. TLR4 induced Wnt3a-Dvl3 restrains the intensity of inflammation and protects against endotoxin-driven organ failure through GSK3beta/beta-catenin signaling. Mol. Immunol. 2020;118:153–164. doi: 10.1016/j.molimm.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L., Zhang S., Wei L., Wang Z., Ma W., Liu F., Qian Y. FGF2 and FGFR2 in patients with idiopathic pulmonary fibrosis and lung cancer. Oncol. Lett. 2018;16:2490–2494. doi: 10.3892/ol.2018.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Z., Dai C. Ablation of FGFR2 in Fibroblasts Ameliorates Kidney Fibrosis after Ischemia/Reperfusion Injury in Mice. Kidney Dis. 2017;3:160–170. doi: 10.1159/000484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang C., Wang R., Lu J., He Y., Wu Y., Ma W., Xu J., Wu Z., Feng Z., Wu M. MicroRNA-338-3p as a therapeutic target in cardiac fibrosis through FGFR2 suppression. J. Clin. Lab. Anal. 2022;36 doi: 10.1002/jcla.24584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Q., Li F., He A.T., Yang B.B. Circular RNAs: Expression, localization, and therapeutic potentials. Mol. Ther. 2021;29:1683–1702. doi: 10.1016/j.ymthe.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y.K. RNA therapy: rich history, various applications and unlimited future prospects. Exp. Mol. Med. 2022;54:455–465. doi: 10.1038/s12276-022-00757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M., Ding W., Liu G., Wang J. Extracellular Circular RNAs Act as Novel First Messengers Mediating Cell Cross-Talk in Ischemic Cardiac Injury and Myocardial Remodeling. J. Cardiovasc. Transl. Res. 2022;15:444–455. doi: 10.1007/s12265-022-10219-y. [DOI] [PubMed] [Google Scholar]
- 44.Ryu J., Kwon D.H., Choe N., Shin S., Jeong G., Lim Y.H., Kim J., Park W.J., Kook H., Kim Y.K. Characterization of Circular RNAs in Vascular Smooth Muscle Cells with Vascular Calcification. Mol. Ther. Nucleic Acids. 2020;19:31–41. doi: 10.1016/j.omtn.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu J., Choe N., Kwon D.H., Shin S., Lim Y.H., Yoon G., Kim J.H., Kim H.S., Lee I.K., Ahn Y., et al. Circular RNA circSmoc1-2 regulates vascular calcification by acting as a miR-874-3p sponge in vascular smooth muscle cells. Mol. Ther. Nucleic Acids. 2022;27:645–655. doi: 10.1016/j.omtn.2021.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patop I.L., Wüst S., Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38 doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu R., Niu Z., Ren G., Ruan L., Sun L. CircSMAD4 alleviates high glucose-induced inflammation, extracellular matrix deposition and apoptosis in mouse glomerulus mesangial cells by relieving miR-377-3p-mediated BMP7 inhibition. Diabetol. Metab. Syndrome. 2021;13:137. doi: 10.1186/s13098-021-00753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J., Lin Z., Ying P., Zhao Z., Yang H., Qian J., Gong Y., Zhou Y., Dai Y., Jiao Y., et al. circSMAD4 promotes experimental colitis and impairs intestinal barrier functions by targeting JAK2 through sponging miR-135a-5p. J. Crohns Colitis. 2023;17:593–613. doi: 10.1093/ecco-jcc/jjac154. [DOI] [PubMed] [Google Scholar]
- 50.Smolgovsky S., Ibeh U., Tamayo T.P., Alcaide P. Adding insult to injury - Inflammation at the heart of cardiac fibrosis. Cell. Signal. 2021;77 doi: 10.1016/j.cellsig.2020.109828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo J., Chen L.W., Huang Z.Q., Guo J.S., Li H., Shan Y., Chen Z.R., Yan Y.M., Zhu J.N., Guo H.M., et al. Suppression of the Inhibitory Effect of circ_0036176-Translated Myo9a-208 on Cardiac Fibroblast Proliferation by miR-218-5p. J. Cardiovasc. Transl. Res. 2022;15:548–559. doi: 10.1007/s12265-022-10228-x. [DOI] [PubMed] [Google Scholar]
- 52.Zheng H., Shi L., Tong C., Liu Y., Hou M. circSnx12 Is Involved in Ferroptosis During Heart Failure by Targeting miR-224-5p. Front. Cardiovasc. Med. 2021;8 doi: 10.3389/fcvm.2021.656093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ni H., Li W., Zhuge Y., Xu S., Wang Y., Chen Y., Shen G., Wang F. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int. J. Cardiol. 2019;292:188–196. doi: 10.1016/j.ijcard.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 54.Yao R., Yao Y., Li C., Li X., Ni W., Quan R., Liu L., Li H., Xu Y., Zhang M., et al. Circ-HIPK3 plays an active role in regulating myoblast differentiation. Int. J. Biol. Macromol. 2020;155:1432–1439. doi: 10.1016/j.ijbiomac.2019.11.119. [DOI] [PubMed] [Google Scholar]
- 55.Xu J., Du W.W., Wu N., Li F., Li X., Xie Y., Wang S., Yang B.B. The circular RNA circNlgnmediates doxorubicin-inducedcardiac remodeling and fibrosis. Mol. Ther. Nucleic Acids. 2022;28:175–189. doi: 10.1016/j.omtn.2022.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vallée A., Lecarpentier Y. TGF-beta in fibrosis by acting as a conductor for contractile properties of myofibroblasts. Cell Biosci. 2019;9:98. doi: 10.1186/s13578-019-0362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kong P., Christia P., Frangogiannis N.G. The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci. 2014;71:549–574. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piek A., de Boer R.A., Silljé H.H.W. The fibrosis-cell death axis in heart failure. Heart Fail. Rev. 2016;21:199–211. doi: 10.1007/s10741-016-9536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng J., Metge F., Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2016;32:1094–1096. doi: 10.1093/bioinformatics/btv656. [DOI] [PubMed] [Google Scholar]
- 62.de Lucia C., Grisanti L.A., Borghetti G., Piedepalumbo M., Ibetti J., Lucchese A.M., Barr E.W., Roy R., Okyere A.D., Murphy H.C., et al. G protein-coupled receptor kinase 5 (GRK5) contributes to impaired cardiac function and immune cell recruitment in post-ischemic heart failure. Cardiovasc. Res. 2022;118:169–183. doi: 10.1093/cvr/cvab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ge W., Guo X., Song X., Pang J., Zou X., Liu Y., Niu Y., Li Z., Zhao H., Gao R., Wang J. The role of immunoglobulin E and mast cells in hypertension. Cardiovasc. Res. 2022;118:2985–2999. doi: 10.1093/cvr/cvac010. [DOI] [PubMed] [Google Scholar]
- 64.Cope D.K., Impastato W.K., Cohen M.V., Downey J.M. Volatile anesthetics protect the ischemic rabbit myocardium from infarction. Anesthesiology. 1997;86:699–709. doi: 10.1097/00000542-199703000-00023. [DOI] [PubMed] [Google Scholar]
- 65.Headrick J.P., See Hoe L.E., Du Toit E.F., Peart J.N. Opioid receptors and cardioprotection - 'opioidergic conditioning' of the heart. Br. J. Pharmacol. 2015;172:2026–2050. doi: 10.1111/bph.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molojavyi A., Preckel B., Comfère T., Müllenheim J., Thämer V., Schlack W. Effects of ketamine and its isomers on ischemic preconditioning in the isolated rat heart. Anesthesiology. 2001;94:623–629. doi: 10.1097/00000542-200104000-00016. [DOI] [PubMed] [Google Scholar]
- 67.Eom G.H., Cho Y.K., Ko J.H., Shin S., Choe N., Kim Y., Joung H., Kim H.S., Nam K.I., Kee H.J., Kook H. Casein kinase-2alpha1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation. 2011;123:2392–2403. doi: 10.1161/CIRCULATIONAHA.110.003665. [DOI] [PubMed] [Google Scholar]
- 68.Yoon S., Kook T., Min H.K., Kwon D.H., Cho Y.K., Kim M., Shin S., Joung H., Jeong S.H., Lee S., et al. Author Correction: PP2A negatively regulates the hypertrophic response by dephosphorylating HDAC2 S394 in the heart. Exp. Mol. Med. 2019;51:1. doi: 10.1038/s12276-019-0222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gorski P.A., Jang S.P., Jeong D., Lee A., Lee P., Oh J.G., Chepurko V., Yang D.K., Kwak T.H., Eom S.H., et al. Role of SIRT1 in Modulating Acetylation of the Sarco-Endoplasmic Reticulum Ca(2+)-ATPase in Heart Failure. Circ. Res. 2019;124:e63–e80. doi: 10.1161/CIRCRESAHA.118.313865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoon S., Kim M., Min H.K., Lee Y.U., Kwon D.H., Lee M., Lee S., Kook T., Joung H., Nam K.I., et al. Inhibition of heat shock protein 70 blocks the development of cardiac hypertrophy by modulating the phosphorylation of histone deacetylase 2. Cardiovasc. Res. 2019;115:1850–1860. doi: 10.1093/cvr/cvy317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from this study are included in this article, the associated source data file, and the supplemental information. The data that support the findings of this study are available on request from the corresponding author.