Abstract
Conjugative transfer of the Enterococcus faecalis plasmid pPD1 is activated by cPD1, one of several peptide sex pheromones secreted by plasmid-free recipient cells, and is blocked by a donor-produced peptide inhibitor, iPD1. Using a tritiated pheromone, [3H]cPD1, we investigated how pPD1-harboring donor cells receive these peptide signals. Donor cells rapidly incorporated [3H]cPD1. The cell extract but not the membrane fraction of the donor strain exhibited significant [3H]cPD1-binding activity. On the basis of these data and those of tracer studies, it was demonstrated that cPD1 was internalized, where it bound to a high-molecular-weight compound. The cell extract of a strain carrying the traA-bearing multicopy plasmid (pDLHH21) also exhibited high [3H]cPD1-binding activity. A recombinant TraA exhibited a dissociation constant of 0.49 ± 0.08 nM against [3H]cPD1. iPD1 competitively inhibited [3H]cPD1 binding to TraA, whereas pheromones and inhibitors relating to other plasmid systems did not. These results show that TraA is a specific intracellular receptor for cPD1 and that iPD1 acts as an antagonist for TraA. A strain carrying the traC-bearing multicopy plasmid (pDLES23) exhibited significant [3H]cPD1-binding activity. A strain carrying traC-disrupted pPD1 (pAM351CM) exhibited lower [3H]cPD1-binding activity as well as lower sensitivity to cPD1 than a wild-type donor strain. Some of the other pheromones and inhibitors inhibited [3H]cPD1 binding to the traC transformant like cPD1 and iPD1 did. These results show that TraC, as an extracellular less-specific pheromone-binding protein, supports donor cells to receive cPD1.
Enterococcus faecalis produces a family of peptide signaling molecules designated as sex pheromones (6, 8, 11). Each pheromone triggers the conjugal transfer system of a particular plasmid such as the hemolysin plasmid pAD1, the bacteriocin plasmid pPD1, or the tetracycline resistance plasmid pCF10 (8, 9). Hosts carrying the plasmid shut off the activity of that pheromone by two functions encoded on the plasmid (29). One involves a reduction of the pheromone production, the so-called pheromone shutdown (1, 29, 36). The other is the production of a specific inhibitor competitive with the pheromone (21, 25, 30, 31). When the plasmid-containing donor bacteria are close to plasmid-free recipients and exposed to the pheromone secreted from the recipient, the conjugal transfer system encoded on the plasmid is activated, and a copy of the plasmid is transferred to the recipient. Synthesis of the aggregation substance is an important event in the pheromone-inducible conjugation system (13). The aggregation substance expressed on the donor cell surface leads to cell clumping between donor and recipient cells and facilitates the high-frequency transfer of the plasmid in liquid cultures (7, 13, 35). Five pheromones and their inhibitors have been identified as linear hepta- or octapeptides composed of protein amino acids (21–25, 28, 30, 31, 40). The pheromone and inhibitor corresponding to a certain plasmid, pX, are designated cX and iX, respectively. Pheromones exhibit clumping-inducing activity for donor strains at concentrations of approximately 0.1 to 0.01 nM. There is no cross-activity among these pheromones in the clumping-inducing bioassays. Moreover, the inhibitors specifically inhibited the mating response to the corresponding plasmid. These results suggest that those plasmids encode a system for peptide-specific pheromone signaling.
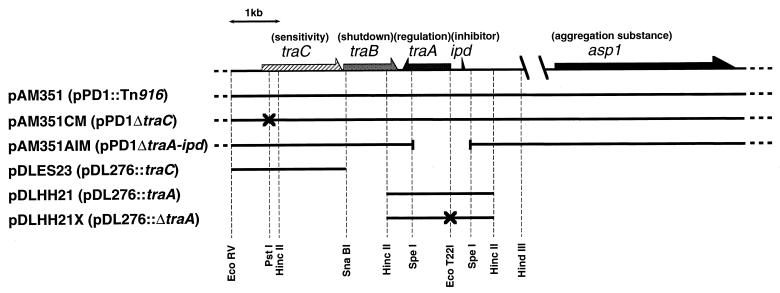
Bacteriocin plasmid pPD1 encodes a response to the octapeptide cPD1. A region of pPD1 involved in both pheromone response and pheromone shutdown has been sequenced, and tra genes have been characterized, as shown in Fig. 1 (12, 34, 36, 44). The traA and traC genes have been shown to contribute to pheromone sensing. The traA gene encodes a 38-kDa cytoplasmic protein. A strain carrying a disruption in traA constitutively clumped and transferred pPD1 without pheromone exposure. Thus, TraA is a negative regulator in the cPD1-inducible conjugation. The traC gene encodes a 61-kDa protein, TraC, with a putative signal sequence. The amino acid sequence of TraC is homologous to oligopeptide-binding proteins of other bacterial species (36), which is a part of a complex of an oligopeptide permease (Opp) (15, 18, 37, 38). A strain carrying a traC mutation (pAM351CM) required a fourfold-higher concentration of cPD1 than that needed by the wild-type strain for induction of sexual aggregation (36). These results suggest that TraC may contribute to pheromone sensitivity as a pheromone-binding protein.
FIG. 1.
Genetic organization of enterococcal plasmids related to this study. The arrows show the directions of transcription. The function attributed to each gene is indicated in parentheses above the gene. The genotype of each plasmid is shown in parentheses after the plasmid name. pAM351 is a derivative of pPD1 with an insertion of a tetracycline resistance transposon, Tn916. The insertion of Tn916 is located in the EcoRI-B fragment (not shown in this map); this insertion exhibited no effect on the phenotype relating to pheromone-inducible cell clumping and plasmid transfer. pDL276 is a multiple-copy E. coli-E. faecalis shuttle vector (10). The discontinuous region between the two slanted lines corresponds to a 2-kb segment (12). The vertical dashed lines indicate the restriction enzyme sites used for cloning, deletion, or site-directed mutagenesis. The crosses represent lesions of DNA which cause frameshift nonsense mutations. The discontinuous region between the two slanted lines represents a deleted region.
In this report, we describe a biochemical study on how donor cells receive the peptide-specific pheromone signal. Labeling of cPD1 has been difficult because modification or amino acid substitution greatly reduced its bioactivity. Thus, we designed and synthesized a radiolabeled cPD1 having the same chemical structure as native cPD1 except for replacement of some protons with tritium. Using the tritiated cPD1, we demonstrated that cPD1 permeates the cell wall with or without the aid of the pheromone-binding protein TraC and is internalized, where it binds to a specific receptor, TraA.
MATERIALS AND METHODS
Enterococcal plasmids, strains, and media.
The maps of the enterococcal plasmids used in this study are shown in Fig. 1. All enterococcal plasmids were expressed in strain OG1X (16) with the exception that strain 39-5Sα was used for the clumping-inducing bioassay (40). All E. faecalis strains were grown in Todd-Hewitt broth (Oxoid) at 37°C. pAM351 is a derivative of pPD1 with an insertion of a tetracycline resistance transposon, Tn916 (16). OG1X carrying pAM351 had the same phenotype relating to pheromone-inducible cell clumping and plasmid transfer as OG1X carrying pPD1. pAM351CM and pAM351AIM are mutant derivatives of pPD1, generated by site-directed mutagenesis. pAM351CM has a frameshift mutation proximal to the translation start site of traC (36). pAM351AIM has a deletion for most of traA and all of ipd (34). pDLES23 and pDLHH21 are chimeric plasmids of E. faecalis-Escherichia coli shuttle vector pDL276 (10) and a fragment containing traC or traA, respectively (34). pDLHH21 was digested with EcoT22I, blunted with T4 DNA polymerase, and then self-ligated. The resultant plasmid, designated pDLHH21X, had a 4-bp deletion at the EcoT22I site, generating a nonsense mutation proximal to the translation start site of traA.
Peptides.
All cold peptides used in this study and a precursor peptide for [3H]cPD1 were manually synthesized by the solid-phase method by using the Fmoc (9-fluorenylmethoxycarbonyl) strategy (Kokku-san peptide synthesis kit; Kokusan Kagaku) and purified by high-pressure liquid chromatography (HPLC) on a reverse-phase column (Pegasil octyldecylsilane [ODS]; Senshukagaku, Tokyo, Japan). The precursor, [4,5-dehydro-Leu2,6]cPD1 (1 mg), was dissolved in methanol and reduced with tritium gas in the presence of palladium black (10 mg) for 2 h (Tritium Labeling Service, Du Pont, Boston, Mass.) (14). The tritiated product ([4,5-3H-Leu2,6]cPD1, abbreviated [3H]cPD1), was purified by using a Pegasil ODS column (1 μg, 0.1% yield; 11.7 TBq/mmol).
Preparation of GST-TraA and rTraA.
The traA gene was amplified by Taq DNA polymerase by using the pHPEV plasmid (36) as a template and the following primers: 5′-GAGGATCCTGCATTTAAATGAATTAATG-3′ and 5′-GAGAATTCGTTAATCTATTTTTTTGTGG-3′. These primers create a BamHI site at the 5′ end of the gene and an EcoRI site at the 3′ end of the gene. The BamHI site was chosen to keep the traA gene in frame with the glutathione S-transferase (GST) gene upon cloning into pGEX-5X-1. The amplified fragment was cleaved to completion with EcoRI and BamHI and ligated into the pGEX-5X-1 vector cleaved with the same restriction endonucleases. This construct, called pGEX-traA, was transformed into E. coli JM109. The transformant was grown under appropriate antibiotic selection in 250 ml of Luria-Bertani broth to an optical density at 600 nm (OD600) of 1.6 at 26.5°C. The cells were induced to express GST-TraA by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.1 mM and grown for an additional 3 h at 26.5°C. All subsequent steps for the preparation of recombinant TraA (rTraA) and GST-TraA were done as described in the protocol of the Gene Fusion System (Pharmacia Biotech). rTraA was generated by digestion of GST-TraA bound to glutathione-Sepharose 4B (Pharmacia Biotech) with factor Xa (Danex Biotek, Mundelstrup, Denmark).
Preparation of spheroplasts, cell extract, and membrane fraction.
For the preparation of spheroplasts, the cell extract, and the membrane fraction, bacteria were grown to mid-log phase (OD660 = 0.5). For the preparation of spheroplasts, the cells were harvested by centrifugation, resuspended at 1/10 the original volume in STE (6.7% sucrose, 50 mM Tris-HCl, 1 mM EDTA [pH 8.0]) containing 1 mg of lysozyme per ml, and incubated for 30 min at 37°C. The spheroplasts obtained were washed once with STE and resuspended at 1/10 the original cultured volume in STE for the binding assay. The cell extract and the membrane were obtained by lysis of the protoplasts. For the preparation of protoplasts, the cells were harvested, resuspended at 1/100 of the original volume in STES (25% sucrose, 1 mM EDTA, 10 mM Tris-HCl, 0.1 N NaCl [pH 8.0]) containing 0.1 mg of lysozyme per ml, 0.25 mg of mutanolysin per ml, and 1 mM phenylmethylsulfonyl fluoride, and incubated for 30 min at 37°C. The protoplasts obtained were osmotically lysed by adding 10 times the volume of 1 mM MgCl2 containing 0.5 mg of DNase per ml. Unlysed cells were removed by centrifugation with a swing rotor at 780 × g for 20 min. The cell extract, which exhibited no significant dicyclohexylcarbodiimide-sensitive ATPase activity as a membrane enzyme marker (41) but significant inorganic pyrophosphatase activity as a cytoplasmic enzyme marker (17), was obtained by ultracentrifugation of the supernatant at 125,000 × g for 2 h. The membrane fraction, which exhibited an inorganic pyrophosphatase activity 1/100 that of the cell extract but a significant dicyclohexylcarbodiimide-sensitive ATPase activity, was obtained by washing the pellet with 1 mM MgCl2. The membrane fraction was resuspended at 1/10 the original culture volume in STE.
[3H]cPD1 binding assay.
For binding with intact cells, E. faecalis strains, except OG1X carrying pDLES23 or pDL276, were grown to mid-log phase (OD660 = 0.5), harvested by centrifugation, and resuspended at 1/10 the original volume in fresh medium. Strain OG1X carrying pDLES23 or pDL276 was grown to late log phase (OD660 = 1.0), harvested, and resuspended at 1/100 the original volume in fresh medium. After incubation with [3H]cPD1 at 37°C, the cells were harvested by centrifugation at 10,000 × g for 1 min and washed with an equal volume of fresh medium, and the radioactivity of the cells was measured. Except for the time course experiment, the incubation time for experiments was 15 min. For binding to spheroplasts, the spheroplasts were incubated with [3H]cPD1 for 30 min at 4°C, harvested by centrifugation at 600 × g for 5 min, and washed with an equal volume of STE, and the spheroplast-bound radioactivity was measured. Binding to the cell extract was measured by the equilibrium dialysis method (19) after incubation with 0.3 nM [3H]cPD1 at 4°C for 24 h. For binding to the membrane, the membrane fraction was incubated with [3H]cPD1 for 30 min at 4°C, harvested by ultracentrifugation at 125,000 × g for 30 min, and washed with an equal volume of STE, and the radioactivity of the membrane was measured. Binding to rTraA was determined by use of the equilibrium dialysis method (19) after incubation of 6.25 nM rTraA with [3H]cPD1 at 20°C for 48 h.
During competitive inhibition analysis, binding was measured with the intact cells of OG1X carrying pDLES23, the spheroplasts of OG1X carrying pAM351, and GST-TraA. The spheroplasts of OG1X carrying pAM351 were prepared as described above. After incubation in STE containing 0.5 nM [3H]cPD1 and various concentrations of cold peptide for 30 min at 4°C, the spheroplasts were harvested by centrifugation and washed with an equal volume of STE, and the spheroplast-bound radioactivity was measured. OG1X carrying pDLES23 was cultured with 1 mM IPTG for 2 h from mid-log to late log phase. These cells were harvested by centrifugation and resuspended at 1/100 the original volume in fresh medium. After incubation with 0.5 nM [3H]cPD1 and various concentrations of cold peptide at 4°C for 30 min, the cells were harvested by centrifugation at 10,000 × g for 1 min and washed with an equal volume of fresh medium, and the radioactivity of the intact cells was measured. For binding to GST-TraA, 20 nM GST-TraA and 25 μl of a 75% slurry of glutathione-Sepharose 4B were added to 0.5 ml of STE containing 0.2 nM [3H]cPD1 and various concentrations of cold peptide. After incubation for 90 min at 4°C, the resin was collected by centrifugation and washed with an equal volume of STE, and the radioactivity of the resin was measured.
Gel filtration column chromatography.
Gel filtration column chromatography was done with a gel filtration HPLC column (Shodex KW-802.5, 6 mm [internal diameter] by 300 mm; Showadenko) equilibrated with 0.1 M potassium phosphate buffer (pH 7.0) containing 0.2 M NaCl. The elution was performed with the same buffer at a flow rate of 1 ml/min.
Tracer experiments of incorporated [3H]cPD1.
For the reverse-phase HPLC analysis, spheroplasts from 5 ml of culture of OG1X carrying pAM351 were incubated with 1.0 nM [3H]cPD1 at 37°C for 90 min. The spheroplasts were harvested by centrifugation, washed with STE, lysed with 100 μl of dimethyl sulfoxide, and then centrifuged at 15,000 × g. The supernatant was subjected to reverse-phase HPLC with the Pegasil ODS column and eluted with a linear gradient of 20 to 50% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 1.0 ml/min. For the gel filtration HPLC analysis, intact cells from 5 ml of culture of OG1X carrying pAM351 were incubated with 0.3 nM [3H]cPD1 at 37°C for 15 min. The cell extract was prepared by osmotic lysis after cell wall digestion as described above, and the lysate was applied to gel filtration HPLC analysis.
Tritium counting.
The tritium count was measured in scintillation vials containing 5 ml of aqueous counting scintillant (ACSII; Amersham) with a β-scintillation counter.
RESULTS
Synthesis of tritiated cPD1.
A tritiated cPD1, [3H]cPD1, was obtained by catalytic reduction of a precursor peptide, [4,5-dehydro-Leu2,6]cPD1, in the presence of tritium gas. [3H]cPD1 was isolated by reverse-phase HPLC. [3H]cPD1 was chromatographically identical to nonradioactive cPD1 and showed clumping-inducing activity at concentrations higher than 0.1 nM, which was comparable to that of the nonradioactive peptide. [3H]cPD1 had sufficiently high specific radioactivity (11.7 TBq/mmol) for the following binding experiments.
Time course of [3H]cPD1 binding to intact cells.
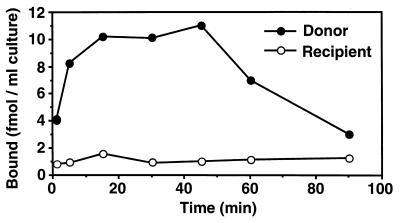
[3H]cPD1 was incubated with intact cells of E. faecalis, and the radioactivity associated with the cells was measured at various times (Fig. 2). The radioactivity of the donor cells increased immediately after the addition of [3H]cPD1 and reached a maximum within 30 min. This is not inconsistent with sexual aggregation starting at 30 to 45 min after pheromone exposure. On the other hand, no increase was detected in the case of the plasmid-free strain. These results implied that the donor cell specifically bound cPD1. After 45 min, the radioactivity associated with the donor cells gradually decreased.
FIG. 2.
Time course of [3H]cPD1 binding. [3H]cPD1 binding to intact cells of the recipient strain OG1X and the donor strain OG1X carrying pAM351 is shown. Intact cells prepared from the bacteria growing at mid-log phase (OD660 = 0.5) were resuspended at 1/10 of the original volume in fresh medium and incubated with 0.6 nM [3H]cPD1 at 37°C, and the radioactivity of the cells was then measured. The data represent average values from two incubations.
[3H]cPD1 binding to each fraction of donor cells.
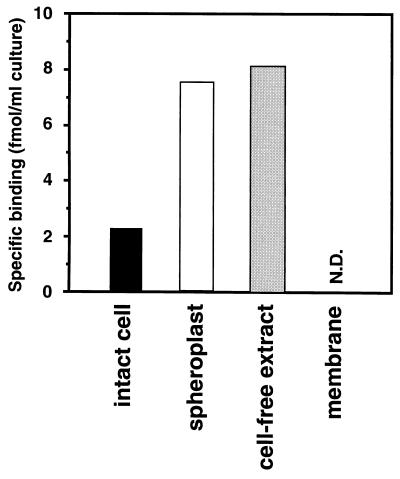
To survey the binding site for cPD1 on the donor cells, we examined the [3H]cPD1-binding activity to the intact cells, the spheroplasts, the cell extract, and the membrane fraction (Fig. 3). Spheroplasts, that is, lysozyme-treated cells, showed high [3H]cPD1-binding activity compared to that of intact cells. This implied that the cell wall is an obstacle to cPD1 approaching the cell membrane. The binding activity of the cell extract was determined in an assay using the equilibrium dialysis method with a membrane having a molecular size cutoff of 12,000 to 14,000 Da. The cell extract showed a binding activity as high as that of the spheroplasts. On the other hand, the membrane fraction showed no detectable specific [3H]cPD1-binding. On the basis of all these data, it was suggested that cPD1 is internalized into the cell and is then bound to a high-molecular-weight compound(s) existing in the cytosol.
FIG. 3.
[3H]cPD1 binding to intact cells, spheroplasts, cell extract, and membrane fraction of donor strain. Each sample was prepared from OG1X carrying pAM351 growing at mid-log phase (OD660 = 0.5) and incubated with 0.3 nM [3H]cPD1. Specific binding was calculated by subtracting the nonspecific binding determined in the presence of a 1,000-fold molar excess of cold cPD1 from the observed total binding. The data represent average values from two incubations. N.D., not detected (i.e., the nonspecific binding was higher than the total binding).
Tracer study of the internalized [3H]cPD1.
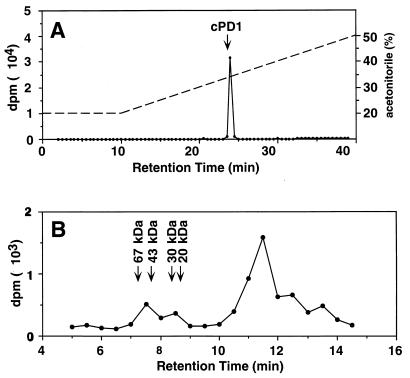
To investigate the fate of the internalized cPD1, we performed two kinds of tracer studies. One was to investigate proteolytic degradation of the internalized [3H]cPD1 (Fig. 4A). The donor spheroplasts were incubated with [3H]cPD1 and lysed with dimethyl sulfoxide, and the lysate was analyzed by reverse-phase HPLC. Dimethyl sulfoxide was used to rapidly lyse the spheroplasts. Eighty-eight percent of the incorporated radioactivity was recovered in a radioactive peak with the same retention time as that of cPD1. This result implied that the internalized cPD1 existed mostly in the intact form. This result did not negate our understanding that the internalized cPD1 was bound to a high-molecular-weight compound, because dimethyl sulfoxide and acetonitrile could dissociate cPD1 from the binding molecule. The other tracer study was to confirm the understanding that the internalized [3H]cPD1 was bound to a high-molecular-weight compound existing in the cytosol (Fig. 4B). The intact donor cells were incubated with [3H]cPD1, treated with lysozyme and mutanolysin, and osmotically lysed. Then, the cell extract was analyzed by gel filtration HPLC analysis. As expected, a radioactive peak was detected in the high-molecular-weight fractions.
FIG. 4.
Tracer studies of incorporated [3H]cPD1. (A) The spheroplasts from 5 ml of culture of OG1X carrying pAM351 were incubated with 1.0 nM [3H]cPD1 at 37°C for 90 min and then lysed with dimethyl sulfoxide. Ninety-seven percent of the incorporated radioactivity was recovered in the lysed fraction. The lysate was subjected to reverse-phase HPLC and eluted with a linear gradient of 20 to 50% (30 min) acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 1.0 ml/min. The radioactive peak at 24 min comprises 88% of the incorporated radioactivity. The arrow indicates the retention time of cPD1. The dotted line indicates the concentration of acetonitrile. (B) The intact cells from 5 ml of culture of OG1X carrying pAM351 were incubated with 0.3 nM [3H]cPD1 at 37°C for 15 min, treated with lysozyme and mutanolysin, and then osmotically lysed. The cell extract was subjected to gel filtration HPLC. Fractions were collected at 30-s intervals. Tritium counts were measured in each fraction.
Scatchard analysis of [3H]cPD1 binding to donor spheroplasts.
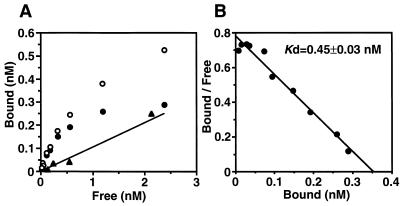
To characterize the nature of the binding molecule, Scatchard analysis of [3H]cPD1 binding was performed with the spheroplasts of the donor cells. The binding of [3H]cPD1 to the donor spheroplasts was saturable, as shown in Fig. 5A. Scatchard analysis of the binding data revealed one class of high-affinity binding sites (approximately 100 sites per cell), with a dissociation constant (Kd) of 0.45 ± 0.03 nM (mean ± standard deviation) (Fig. 5B). The Kd value is close to the minimum concentration (approximately 0.1 nM) required to induce cell clumping of the donor cells, suggesting that the binding molecule may function as a sensor for cPD1.
FIG. 5.
Equilibrium saturation binding (A) and Scatchard plot (B) of [3H]cPD1 binding to donor spheroplasts. The spheroplasts were prepared from OG1X carrying pAM351. The samples were run in duplicate. Specific binding (closed circles) was calculated by subtraction of the nonspecific binding (line [calculated from positions of closed triangles] determined for binding in the presence of a 1,000-fold molar excess of cold cPD1) from total binding (open circles).
[3H]cPD1 binding to tra mutants and tra transformants.
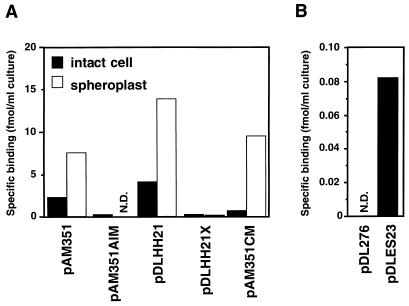
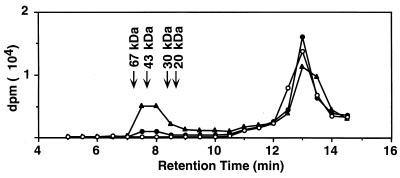
To investigate how the functions of TraA and TraC relate to cPD1 binding, we examined [3H]cPD1 binding to traA and traC mutants and traA and traC transformants (Fig. 6). Neither the intact cells nor spheroplasts of the strain carrying traA-disrupted pPD1 (pAM351AIM) showed significant binding activity. On the other hand, both the intact cells and spheroplasts of the strain carrying the traA-bearing multicopy plasmid (pDLHH21) exhibited very high binding activities. A frameshift mutation on the 5′ end of traA resulted in loss of the [3H]cPD1-binding activity, as shown in the strain carrying pDLHH21X. These observations implied that TraA was the binding molecule for cPD1 in the cytosol. This finding was confirmed by gel filtration analysis of the cell extract (Fig. 7). The cell extract was incubated with [3H]cPD1 and subjected to gel permeation chromatography. A radioactive peak was found in the large-molecular-size fractions as large as 40 kDa; traA encodes a 38-kDa protein. The radioactive peak of the strain carrying pDLHH21 was higher than that of the pAM351-carrying strain. The difference would depend on the copy number of the plasmids.
FIG. 6.
[3H]cPD1 binding to tra mutants and tra transformants. (A) Intact cells and spheroplasts prepared from plasmid-carrying OG1X cells growing at mid-log phase (OD660 = 0.5) were resuspended at 1/10 the original volume in THB medium and STE, respectively. (B) Intact cells prepared from plasmid-carrying OG1X cells growing at late log phase (OD660 = 1.0) were resuspended at 1/100 the original volume of THB medium. The intact cells and spheroplasts were incubated with 0.3 nM [3H]cPD1. Specific binding was calculated by subtraction of the nonspecific binding determined in the presence of a 1,000-fold molar excess of cold cPD1 from the observed total binding. The data represent average values from two incubations. N.D., not detected (i.e., the nonspecific binding was higher than the total binding).
FIG. 7.
Gel filtration analysis of cell extract incubated with [3H]cPD1. Cell extracts prepared from 1-ml cultures of strain OG1X (open circles), OG1X carrying pAM351 (closed circles), and OG1X carrying pDLHH21 (closed triangles) were incubated with 0.3 nM [3H]cPD1 at 4°C for 5 min and then subjected to gel filtration HPLC. Fractions were collected at 30-s intervals. Tritium counts were measured in each fraction.
TraC is homologous to oligopeptide-binding proteins of other bacterial species (36); therefore, it was likely that TraC has binding activity to cPD1. However, we could detect no significant [3H]cPD1 binding to the strain carrying pAM351AIM (TraA− TraC+). Therefore, we examined [3H]cPD1 binding to the strain carrying the traC-bearing multicopy plasmid (pDLES23). To detect the low binding activity, the cells were suspended at 1/100 of the original volume (i.e., concentrated 10 times more than the cells whose case is illustrated in Fig. 6A). As a result, the intact cells of the strain carrying pDLES23 exhibited significant [3H]cPD1-binding activity, while the intact cells of the strain carrying the vector (pDL276) exhibited no specific binding (Fig. 6B). This result indicates that TraC has binding ability for cPD1. However, the binding ability was very low compared with that of TraA. The intact cells of the strain carrying the traC-disrupted pPD1 (pAM351CM) (36) showed a [3H]cPD1-binding activity lower than that of the wild-type donor cells. On the other hand, the spheroplasts of the pAM351CM-carrying strain showed a [3H]cPD1-binding activity as high as that of the spheroplasts of wild-type donor cells, indicating that TraC did not contribute to the pheromone binding in spheroplasts.
cPD1-binding activity of TraA.
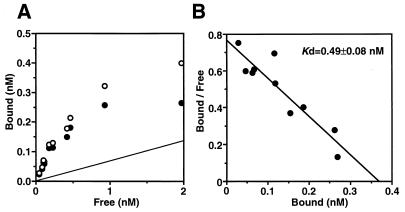
To elucidate the cPD1-binding activity of TraA, we prepared a recombinant protein, rTraA, and performed a Scatchard analysis of the [3H]cPD1 binding to rTraA (Fig. 8). GST-TraA, a fusion protein of GST and TraA, was expressed in E. coli, purified by glutathione-Sepharose affinity column chromatography, and digested with a site-specific protease, factor Xa. The sequence of the resultant protein, rTraA, should be the same as the deduced sequence of TraA except for three N-terminal amino acids. rTraA exhibited specific binding to [3H]cPD1. The binding of [3H]cPD1 to rTraA was saturable. The Kd value of 0.49 ± 0.08 nM is similar to that of spheroplasts of the donor cells, indicating that TraA is the binding molecule in the cytosol. The Kd value for maximum binding (0.37 nM) was much lower than the concentration of rTraA (6.25 nM). This discrepancy may be due to the denaturation of rTraA during the preparation or the incubation.
FIG. 8.
Equilibrium saturation binding (A) and Scatchard plot (B) of [3H]cPD1 binding to rTraA. Binding to rTraA was determined by the equilibrium dialysis method (19) after incubation of 6.25 nM rTraA with various concentrations of [3H]cPD1 at 20°C for 48 h. The samples were run in duplicate. Specific binding (closed circles) was calculated by subtraction of the nonspecific binding (line determined for binding in the presence of a 1,000-fold molar excess of cold cPD1) from total binding (open circles).
Competitive-inhibition analysis of [3H]cPD1 binding to TraA and TraC by cold peptides of cPD1, iPD1, and other pheromones and inhibitors.
There is no significant cross-activity among the five pheromones and their inhibitors thus far identified. To investigate the relationship between the peptide-specific pheromone signaling and the binding functions of TraA and TraC, we examined which of these pheromones and inhibitors could bind to TraA or TraC. The binding ability was examined by competitive-inhibition analysis of [3H]cPD1 binding with cold peptides (Table 1).
TABLE 1.
Competitive inhibition analysis with cold pheromones and inhibitors
| Pheromone or inhibitor | Sequence | Inhibition of [3H]cPD1 binding (IC50 [nM])a to:
|
cPD1 activitye (nM) | ||
|---|---|---|---|---|---|
| TraCb | Donor spheroplastc | GST-TraAd | |||
| cPD1 | H-Phe-Leu-Val-Met-Phe-Leu-Ser-Gly-OH | 34.2 | 1.49 | 0.50 | 0.1 |
| iPD1 | H-Ala-Leu-Ile-Leu-Thr-Leu-Val-Ser-OH | 78.5 | 0.45 | 0.33 | >350 |
| cAD1 | H-Leu-Phe-Ser-Leu-Val-Leu-Ala-Gly-OH | >1,000 | >1,000 | >1,000 | >350 |
| iAD1 | H-Leu-Phe-Val-Val-Thr-Leu-Val-Gly-OH | >1,000 | >1,000 | >1,000 | >350 |
| cCF10 | H-Leu-Val-Thr-Leu-Val-Phe-Val-OH | 126 | >1,000 | >1,000 | >1,000 |
| iCF10 | H-Ala-Ile-Thr-Leu-Ile-Phe-Ile-OH | 100 | >1,000 | >1,000 | >350 |
| cAM373 | H-Ala-Ile-Phe-Ile-Leu-Ala-Ser-OH | >1,000 | >1,000 | >1,000 | >350 |
| iAM373 | H-Ser-Ile-Phe-Thr-Leu-Val-Ala-OH | >1,000 | >1,000 | >1,000 | >350 |
| cOB1 | H-Val-Ala-Val-Leu-Val-Leu-Gly-Ala-OH | >1,000 | >1,000 | >1,000 | >350 |
| iOB1 | H-Ser-Leu-Thr-Leu-Ile-Leu-Ser-Ala-OH | 16.5 | >1,000 | >1,000 | >350 |
Concentrations of cold peptides that produce 50% inhibition of the specific binding (IC50) were estimated from the plots of [3H]cPD1 binding. The specific binding was calculated by subtraction of the nonspecific binding determined in the presence of 1 μM cold cPD1 from the maximum binding.
Binding to TraC was assayed with TraC-expressing cells (intact cells of OG1X carrying pDLES23) and 0.5 nM [3H]cPD1.
Binding to donor spheroplasts was assayed with spheroplasts of OG1X carrying pAM351 and 0.5 nM [3H]cPD1.
Binding to GST-TraA was assayed with 20 nM GST-TraA and 0.2 nM [3H]cPD1.
cPD1 activity represents the minimum concentration inducing cell clumping of a pPD1 donor strain (39-5Sα) in a microtiter bioassay (40).
The binding to TraC was detected on the strain carrying pDLES23 (Fig. 6B). The competitive inhibition of [3H]cPD1 binding to TraC was examined by measuring the radioactivity of the TraC-expressing cells after incubation with [3H]cPD1 and the cold peptide. iPD1 competitively inhibited the binding of [3H]cPD1 to TraC. The binding to TraC was also inhibited by cCF10, iCF10, and iOB1 at a concentration as low as that of cPD1, indicating that the binding function of TraC is not completely specific to cPD1 and iPD1.
Competitive inhibition of [3H]cPD1 binding to TraA was examined by the following two methods. One was to measure the radioactivity of the donor spheroplasts after incubation with [3H]cPD1 and cold peptide. As shown in Fig. 6A, the spheroplasts of the traA mutant strain (pAM351AIM) showed no [3H]cPD1-binding activity, indicating that the binding of [3H]cPD1 to the donor spheroplasts represents binding to TraA. The other was to measure the radioactivity bound to GST-TraA after incubation with [3H]cPD1 and the cold peptide. GST-TraA exhibited a dissociation constant against cPD1 (Kd = 1.03 ± 0.48 nM, data not shown) similar to that of rTraA, indicating that GST-TraA was useful for investigating the binding property of TraA. iPD1 inhibited [3H]cPD1 binding to both the donor spheroplasts and GST-TraA at a concentration as low as that of cold cPD1. These results indicate that iPD1 is internalized into the cell and blocks the cPD1 signal as an antagonist for TraA. Pheromones and inhibitors relating to other plasmids did not inhibit [3H]cPD1 binding to both the donor spheroplasts and GST-TraA at concentrations lower than 1 μM. This result implies that the binding function of TraA is completely specific to the related pheromone and inhibitor, cPD1 and iPD1.
DISCUSSION
In this study, we have investigated how a plasmid-harboring cell senses a pheromone signal by using the synthetic pheromone [3H]cPD1. The recipient bacteria produce 0.5 to 2.5 nM cPD1 in the culture broth (29, 40), and the minimum concentration required for the induction of mating aggregate is about 0.1 nM. The synthetic pheromone [3H]cPD1 had a sufficiently high specific radioactivity for [3H]cPD1 binding to the donor cells to be detected under these physiological concentrations. [3H]cPD1 binding to the donor cells was observed immediately after exposure to [3H]cPD1 (Fig. 2). [3H]cPD1-binding activity was detected in the cell extract from the donor strain (Fig. 3), suggesting the internalization of cPD1 and the existence of cytoplasmic binding molecules. The tracer study using reverse-phase HPLC (Fig. 4A) revealed that the internalized cPD1 existed mostly in the intact form. The tracer study using gel filtration chromatography (Fig. 4B) confirmed the finding that the internalized cPD1 was bound to a high-molecular-weight compound existing in the cytosol; this was subsequently identified as intracellular receptor TraA.
[3H]cPD1 binding to intact donor cells was lower than that to spheroplasts (Fig. 3), suggesting that the cell wall is an obstacle to cPD1 approaching the cell membrane. The mutation of traC further reduced the binding to intact cells, whereas it did not affect the binding ability of the spheroplasts (Fig. 6A, pAM351CM). It seems that TraC supports cPD1 permeation of the cell wall and helps donor cells sense the pheromone signal at the low concentration existing in nature. The binding experiment with the traC transformant provided direct evidence that TraC acts as the pheromone-binding protein (Fig. 6B). However, its affinity was very low compared with that of TraA. On the basis of all these data, it was suggested that TraC may act as a transporter or a carrier protein and may allow cPD1 to cross the cell wall. The precise function of TraC requires further investigation at the protein level, such as the localization or higher-order structure of TraC. It is noteworthy that [3H]cPD1 binding to intact cells or spheroplasts was observed only if the traA gene was transformed to the plasmid-free strain (Fig. 6A, pDLHH21). This indicates that cPD1 uptake requires no pPD1-encoded element other than TraA. Moreover, the disruption of traC abolished neither the [3H]cPD1-binding ability nor the pheromone response: the strain carrying pAM351CM exhibited one-third of the [3H]cPD1 binding found in the wild-type strain (Fig. 6A) and to respond to cPD1 required a concentration of it fourfold higher than that needed by the wild-type strain (36). A protein encoded by a chromosomal element, e.g., OppA (the lower-affinity peptide-binding protein described by Leonard et al. [20]), might act as another pheromone-binding protein.
The binding function of TraC was not completely specific to cPD1 and iPD1. The cold peptides of cCF10, iCF10, and iOB1, as well as those of cPD1 and iPD1, inhibited [3H]cPD1 binding to the TraC-expressing cell (Table 1). The pheromone-binding protein encoded by pCF10 (PrgZ) is highly homologous to TraC, with 86.6% identical amino acids (12, 36, 39). Especially, the N-terminal half (residues 1 to 300 of 545 amino acids) is extensively homologous, with 96.7% identical amino acids (95.4% identical nucleotides in the gene that encodes it). It seems that pPD1 is evolutionarily close to pCF10 in this region and that the pPD1-encoded TraC could bind both cCF10 and iCF10, which should be recognized by PrgZ. It is strange that there is less similarity among the structures of cPD1, iPD1, cCF10, and iCF10. On the other hand, iOB1 is similar to both cPD1 and iPD1; three leucines of iOB1 are common in iPD1. TraC seems to misrecognize iOB1 because of this structural similarity. In our previous genetic study, the function of pPD1 traC could be complemented by pAD1 traC (42), suggesting that pAD1-encoded TraC could bind cPD1 (34). However, cAD1 did not inhibit [3H]cPD1 binding to pPD1-encoded TraC (Table 1), indicating that cAD1 did not bind to pPD1-encoded TraC. On the contrary, the function of pPD1 traC could not be complemented by pCF10 prgZ (33), suggesting that pCF10-encoded PrgZ could not bind cPD1, although cCF10 can bind pPD1-encoded TraC.
The mechanism for membrane transport of a pheromone has been described by Leonard et al., who reported that inactivation of the chromosomal E. faecalis opp operon abolished the response at physiological concentrations of cCF10 (20). They proposed a model showing that cCF10 interacts with either PrgZ or OppA on the donor cell and is then transported into the cell via the Opp complex, which consists of two transmembrane proteins (OppB and OppC) and two membrane-associated cytoplasmic ATPases (OppD and OppF). To investigate whether cPD1 is imported via the Opp system, we prepared an oppD-disrupted E. coli and examined the uptake of [3H]cPD1. Unexpectedly, [3H]cPD1 was taken up by the oppB-negative E. coli expressed with GST-TraA, as occurred in wild-type E. coli expressed with GST-TraA (32). This suggests that cPD1 could permeate the bacterial cell membrane in a manner independent of the Opp system. Like TraC, the Opp system may help donor cells sense a pheromone at the low concentration existing in nature.
It has been reported that gene disruption of traA resulted in constitutive clumping and plasmid transfer of the host cells, indicating that TraA functions as a negative regulator in pheromone signaling (34, 44). This study revealed that TraA is the cytoplasmic cPD1-binding protein as well as the regulatory protein. The Kd value of rTraA was close to the minimum concentration needed to induce a mating response. On the basis of these data together, TraA is concluded to function as the receptor for cPD1. The Scatchard analysis with the donor spheroplasts (Fig. 5) showed approximately a hundred binding sites per cell; provided that TraA binds cPD1 at a ratio of 1:1, 100 molecules of TraA would exist in a donor cell. The data also showed that fewer than 10 molecules of cPD1 were taken up into the cell and bound to TraA in the presence of the minimum bioactive concentration (0.1 nM). iPD1 inhibited [3H]cPD1 binding to both GST-TraA and the donor spheroplasts at a concentration as low as that of cPD1 (Table 1). This coincides with the fact that iPD1 could inhibit the cPD1 activity at a pheromone/inhibitor ratio of about 1:1 (25). It was concluded that iPD1 is internalized into the cell and blocks the cPD1 signal as an antagonist for TraA (32). Pheromones and inhibitors relating to other plasmid systems did not inhibit [3H]cPD1 binding, indicating that the binding function of TraA is completely specific to cPD1 and iPD1. This coincides with the fact that there is a low homology among pPD1-encoded TraA, pAD1-encoded TraA, and pCF10-encoded PrgX. In conclusion, TraA plays a central role in the peptide-specific pheromone signaling as the specific receptor for cPD1.
We have found that TraA interacted with a DNA fragment including a promoter region of ipd, suggesting that TraA might function as a DNA-binding protein (27). A deletion of the promoter region resulted in the loss of the ability for pheromone response, suggesting that the induction for the aggregation substance gene expression requires the ipd region (34). These findings are coincident with a model of pAD1 where TraA functions as a DNA-binding protein and negatively regulates transcription of iad-traE1, which is involved in the positive regulation of expression of the aggregation substance gene (5, 26, 43). Recently, studies on pCF10 have shown that RNA molecules carried in the inhibitor and its proximal regions are involved in the positive regulation of the aggregation substance gene (2–4, 20). Considering these findings, TraA may bind a promoter region of ipd and repress expression of a transcript which might positively regulate for synthesis of the aggregation substance. When bound to cPD1, TraA may derepress the expression of the transcript, which might result in synthesis of the aggregation substance. However, considering that fewer than 10 molecules of cPD1 were internalized into the cell at the physiological concentration, it is estimated that more than 90% of TraA molecules were free-form. The cPD1 signal may be amplified through the TraA molecules, e.g., by autophosphorylation of TraA triggered by the cPD1 binding.
ACKNOWLEDGMENTS
We thank D. B. Clewell for the E. faecalis strains and his helpful comments on the manuscript, G. M. Dunny for the shuttle vector pDL276, and H. Kobayashi and F.-S. Che for their technical advice.
This work was supported in part by a Grant-in-Aid for Scientific Research (no. 09760112) from the Ministry of Education, Science, and Culture of Japan, by a grant from the Nissan Science Foundation, by a grant from the Naito Foundation, and by a grant from the Nougeikagaku Shoureikai.
REFERENCES
- 1.An F Y, Clewell D B. Characterization of the determinant (traB) encoding sex pheromone shutdown by the hemolysin/bacteriocin plasmid pAD1 in Enterococcus faecalis. Plasmid. 1994;31:215–221. doi: 10.1006/plas.1994.1023. [DOI] [PubMed] [Google Scholar]
- 2.Chung J W, Bensing B A, Dunny G M. Genetic analysis of a region of the Enterococcus faecalis plasmid pCF10 involved in positive regulation of conjugative transfer functions. J Bacteriol. 1995;177:2107–2117. doi: 10.1128/jb.177.8.2107-2117.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung J W, Dunny G M. Cis-acting, orientation-dependent, positive control system activates pheromone-inducible conjugation functions at distances greater than 10 kilobases upstream from its target in Enterococcus faecalis. Proc Natl Acad Sci USA. 1992;89:9020–9024. doi: 10.1073/pnas.89.19.9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung J W, Dunny G M. Transcriptional analysis of a region of the Enterococcus faecalis plasmid pCF10 involved in positive regulation of conjugative transfer functions. J Bacteriol. 1995;177:2118–2124. doi: 10.1128/jb.177.8.2118-2124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell D B. Bacterial sex pheromone-induced plasmid transfer. Cell. 1993;73:9–12. doi: 10.1016/0092-8674(93)90153-h. [DOI] [PubMed] [Google Scholar]
- 6.Clewell D B. Sex pheromones and the plasmid-encoded mating response in Enterococcus faecalis. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 349–367. [Google Scholar]
- 7.Dunny G M. Genetic functions and cell-cell interactions in the pheromone-inducible plasmid transfer system of Enterococcus faecalis. Mol Microbiol. 1990;4:689–696. doi: 10.1111/j.1365-2958.1990.tb00639.x. [DOI] [PubMed] [Google Scholar]
- 8.Dunny G M, Craig R A, Carron R L, Clewell D B. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid. 1979;2:454–465. doi: 10.1016/0147-619x(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 9.Dunny G M, Funk C, Adsit J. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid. 1981;6:270–278. doi: 10.1016/0147-619x(81)90035-4. [DOI] [PubMed] [Google Scholar]
- 10.Dunny G M, Lee L N, LeBlanc D J. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol. 1991;57:1194–1201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunny G M, Leonard B A B, Hedberg P J. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol. 1995;177:871–876. doi: 10.1128/jb.177.4.871-876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimoto S, Tomita H, Wakamatsu E, Tanimoto K, Ike Y. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J Bacteriol. 1995;177:5574–5581. doi: 10.1128/jb.177.19.5574-5581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galli D, Wirth R, Wanner G. Identification of aggregation substances of Enterococcus faecalis cells after induction by sex pheromones: an immunological and ultrastructural investigation. Arch Microbiol. 1989;151:486–490. doi: 10.1007/BF00454863. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa H, Shinohara Y, Baba S. Synthesis of [d-Ala2]Leu-enkephalin and [d-Ala2, d-Leu5]Leu-enkephalin with high specific tritiated activity in the leucine residue. J. Chem. Soc. Perkin Trans. I. 1990. pp. 2641–2644. [Google Scholar]
- 15.Hiles I D, Gallagher M P, Jamieson D J, Higgins C F. Molecular characterization of the oligopeptide permease of Salmonella typhimurium. J Mol Biol. 1987;195:125–142. doi: 10.1016/0022-2836(87)90332-9. [DOI] [PubMed] [Google Scholar]
- 16.Ike Y, Craig R A, White B A, Yagi Y, Clewell D B. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc Natl Acad Sci USA. 1983;80:5369–5373. doi: 10.1073/pnas.80.17.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josse J. Constitutive inorganic pyrophosphatase of Escherichia coli. J Biol Chem. 1966;241:1938–1947. [PubMed] [Google Scholar]
- 18.Kashiwagi K, Yamaguchi Y, Sakai Y, Kobayashi H, Igarashi K. Identification of the polyamine-induced protein as a periplasmic oligopeptide-binding protein. J Biol Chem. 1990;265:8387–8391. [PubMed] [Google Scholar]
- 19.Kim H S, Nihira T, Tada H, Yanagimoto M, Yamada Y. Identification of binding protein of virginiae butanolide C, an autoregulator in virginiamycin production, from Streptomyces virginiae. J Antibiot. 1989;42:769–778. doi: 10.7164/antibiotics.42.769. [DOI] [PubMed] [Google Scholar]
- 20.Leonard B A B, Podbielski A, Hedberg P J, Dunny G M. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci USA. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori M, Isogai A, Sakagami Y, Fujino M, Kitada C, Clewell D B, Suzuki A. Isolation and structure of the Streptococcus faecalis sex pheromone inhibitor, iAD1, that is excreted by the donor strain harboring plasmid pAD1. Agric Biol Chem. 1986;50:539–541. [Google Scholar]
- 22.Mori M, Sakagami Y, Ishii Y, Isogai A, Kitada C, Fujino M, Adsit J C, Dunny G M, Clewell D B. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J Biol Chem. 1988;263:14574–14578. [PubMed] [Google Scholar]
- 23.Mori M, Sakagami Y, Narita M, Isogai A, Fujino M, Kitada C, Craig R A, Clewell D B, Suzuki A. Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett. 1984;178:97–100. doi: 10.1016/0014-5793(84)81248-x. [DOI] [PubMed] [Google Scholar]
- 24.Mori M, Tanaka H, Sakagami Y, Isogai A, Fujino M, Kitada C, White B A, An F Y, Clewell D B, Suzuki A. Isolation and structure of the Streptococcus faecalis sex pheromone, cAM373. FEBS Lett. 1986;206:69–72. doi: 10.1016/0014-5793(86)81342-4. [DOI] [PubMed] [Google Scholar]
- 25.Mori M, Tanaka H, Sakagami Y, Isogai A, Fujino M, Kitada C, Clewell D B, Suzuki A. Isolation and structure of the sex pheromone inhibitor, iPD1, excreted by Streptococcus faecalis donor strains harboring plasmid pPD1. J Bacteriol. 1987;169:1747–1749. doi: 10.1128/jb.169.4.1747-1749.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muscholl A, Galli D, Wanner G, Wirth R. Sex pheromone plasmid pAD1-encoded aggregation substance of Enterococcus faecalis is positively regulated in trans by traE1. Eur J Biochem. 1993;214:333–338. doi: 10.1111/j.1432-1033.1993.tb17928.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama, J. Unpublished data.
- 28.Nakayama J, Abe Y, Isogai A, Suzuki A. Isolation and structure of the Enterococcus faecalis sex pheromone, cOB1, that induces conjugal transfer of the hemolysin/bacteriocin plasmids, pOB1 and pYI1. Biosci Biotechnol Biochem. 1995;59:703–705. doi: 10.1271/bbb.59.703. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama J, Dunny G M, Clewell D B, Suzuki A. Quantitative analysis for pheromone inhibitor and pheromone shutdown in Enterococcus faecalis. Dev Biol Stand. 1995;85:35–38. [PubMed] [Google Scholar]
- 30.Nakayama J, Ono Y, Isogai A, Clewell D B, Suzuki A. Isolation and structure of the sex pheromone inhibitor, iAM373, of Enterococcus faecalis. Biosci Biotechnol Biochem. 1995;59:1358–1359. doi: 10.1271/bbb.59.1358. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama J, Ruhfel R E, Dunny G M, Isogai A, Suzuki A. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid, pCF10, encodes a peptide inhibitor, iCF10. J Bacteriol. 1994;176:7405–7408. doi: 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama, J., A. Horii, and A. Suzuki. Bacterial peptide pheromone is imported where it directly binds to an intracellular receptor. In Proceedings of the 1st International Peptide Symposium 1997, in press. Kluwer Academic Publishers, B.V., Dordrecht, The Netherlands.
- 33.Nakayama, J., and A. Suzuki. Unpublished data.
- 34.Nakayama J, Suzuki A. Genetic analysis of plasmid-specific pheromone signaling encoded by pPD1 in Enterococcus faecalis. Biosci Biotechnol Biochem. 1997;61:1796–1799. doi: 10.1271/bbb.61.1796. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama J, Watarai H, Nagasawa H, Isogai A, Clewell D B, Suzuki A. Immunological characterization of pheromone-induced proteins associated with sexual aggregation in Enterococcus faecalis. Biosci Biotechnol Biochem. 1992;56:264–269. [Google Scholar]
- 36.Nakayama J, Yoshida K, Kobayashi H, Isogai A, Clewell D B, Suzuki A. Cloning and characterization of a region of Enterococcus faecalis plasmid pPD1 encoding pheromone inhibitor (ipd), pheromone sensitivity (traC), and pheromone shutdown (traB) genes. J Bacteriol. 1995;177:5567–5573. doi: 10.1128/jb.177.19.5567-5573.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perego M, Higgins C F, Pearce S R, Gallagher M P, Hoch J A. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol Microbiol. 1991;5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- 38.Rudner Z D, Ledeaux J R, Ireton K, Grossman A D. The spoOK locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J Bacteriol. 1991;173:1388–1398. doi: 10.1128/jb.173.4.1388-1398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruhfel R E, Manias D A, Dunny G M. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone binding function. J Bacteriol. 1993;175:5253–5259. doi: 10.1128/jb.175.16.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki A, Mori M, Sakagami Y, Isogai A, Fujino M, Kitada C, Craig R A, Clewell D B. Isolation and structure of the bacterial sex pheromone, cPD1. Science. 1984;226:849–850. doi: 10.1126/science.6436978. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki T, Umemoto T, Kobayashi H. Novel streptococcal mutants defective in the regulation of H+-ATPase biosynthesis and FO complex. J Biol Chem. 1988;263:11840–11843. [PubMed] [Google Scholar]
- 42.Tanimoto K, An F Y, Clewell D B. Characterization of the traC determinant in the Enterococcus faecalis hemolysin/bacteriocin plasmid pAD1: binding of sex pheromone. J Bacteriol. 1993;175:5260–5264. doi: 10.1128/jb.175.16.5260-5264.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanimoto K, Clewell D B. Regulation of the pAD1-encoded sex pheromone response in Enterococcus faecalis: expression of the positive regulator TraE1. J Bacteriol. 1993;175:1008–1018. doi: 10.1128/jb.175.4.1008-1018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanimoto K, Tomita H, Ike Y. The traA gene of the Enterococcus faecalis conjugative plasmid pPD1 encodes a negative regulator for the pheromone response. Plasmid. 1996;36:55–61. doi: 10.1006/plas.1996.0032. [DOI] [PubMed] [Google Scholar]