Abstract
Translation is shown to be downregulated sharply between genes V and VII of IKe, a filamentous bacteriophage classed with the Ff group (phages f1, M13, and fd) but having only 55% DNA sequence identity to it. Genes V and VII encode the following proteins which are used in very different amounts: pV, used to coat the large number of viral DNA molecules prior to assembly, and pVII, used to serve as a cap with pIX in 3 to 5 copies on the end of the phage particle that emerges first from Escherichia coli. The genes are immediately adjacent to each other and are represented in the same amounts on the Ff and IKe mRNAs. Ff gene VII has an initiation site that lacks detectable intrinsic activity yet through coupling is translated at a level 10-fold lower than that of upstream gene V. The experiments reported reveal that by contrast, the IKe gene VII initiation site had detectable activity but was coupled only marginally to upstream translation. The IKe gene V and VII initiation sites both showed higher activities than the Ff sites, but the drop in translation at the IKe V-VII junction was unexpectedly severe, ∼75-fold. As a result, gene VII is translated at similarly low levels in IKe- and Ff-infected hosts, suggesting that selection to limit its expression has occurred.
In the circular genome of the closely related Ff filamentous phages f1, fd, and M13, genes V and VII are positioned as an adjacent pair bridging groups of genes that encode replication and structural proteins (29). The single-stranded DNA binding protein encoded by gene V (pV) coats progeny viral DNA molecules to generate the intracellular precursor for assembly of phage particles as they are extruded through the membranes of the Escherichia coli host. The small protein encoded by gene VII (pVII), together with the product of gene IX (pIX), is present in 3 to 5 copies on the end of the long thin virus particle that emerges first (36). Several findings make it likely that pVII and pIX have a role in initiating phage assembly. Phage production is nearly abolished if pVII or pIX is absent, indicating their involvement early in the process (23). Genetic evidence points to interactions between pVII and pIX, the packaging signal on viral DNA, and morphogenetic proteins. Deletions in the packaging signal can be compensated by mutations in pVII, pIX, and the morphogenetic protein pI (37). Immunoprecipitation data for membrane proteins extracted from infected cells also provide evidence for interactions of pVII with the major coat protein pVIII (12).
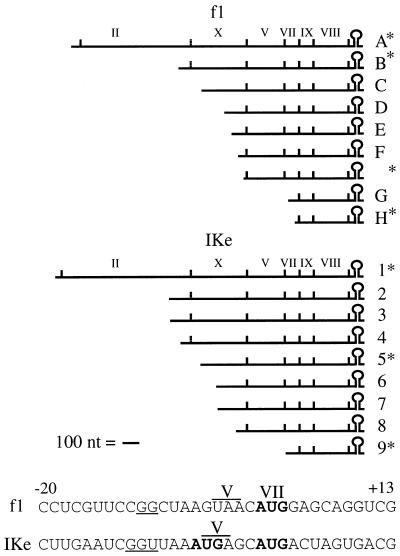
Expression of genes V and VII conforms to a general pattern that permits the phage to establish and maintain a persistent infection that does not kill or lyse the host. For this to occur, a balanced pattern of gene expression appears to be required, since overexpression of most of the phage genes and mutations in all but one are lethal to the host (6, 13a, 18, 22a, 29, 37a). The products of genes V and VII are found in the infected host in very different amounts. pV, needed in large quantities, is present in >105 copies per cell, whereas pVII, needed in few copies at one tip, is present in much lower amounts, despite the fact that a series of overlapping mRNAs maintains message for both genes at high levels (Fig. 1). About 10-fold-lower amounts of pVII are detected in infected cells (12), in agreement with studies of lacZ fusions suggesting that gene VII is translated about 10-fold less efficiently than gene V (3). These studies established that low-level synthesis of pVII was achieved by an unusual case of translational coupling. The gene VII initiation site was inactive when present by itself but via coupling acquired ∼10% of the translational activity from upstream gene V (19). The nature and distribution of mutations required for independent activity suggested that the gene VII site was intrinsically defective and throughout lacked sequences required for ribosome binding (20).
FIG. 1.
Major f1 and IKe mRNAs and junctions between genes V and VII. RNAs marked with an asterisk are primary transcripts; the others are the 3′ products of processing by host endonucleases. The f1 mRNA diagram is the work of many authors (29). The IKe mRNAs, as recently defined, range in length from 470 to ∼2,000 nt (49). Coding regions are indicated by vertical lines. In the sequences (17, 33), gene V stop codons are overlined and SD sequences are underlined. Known (42) or potential start codons for gene VII are shown in boldface type, with the 5′ nucleotide of the f1 initiator AUG denoted +1.
Although the properties of translational coupling between genes V and VII account reasonably well for the observed levels of the proteins, it is not known how tightly pVII synthesis must be regulated. Data of Endemann and Model (12) indicate that an excess of pVII is tolerated, since infected cells normally contain about 10 times more pVII and pIX than is incorporated into phage. However, since overexpression of gene VII is lethal, some degree of downregulation at the gene V-VII junction is probably required. It is also not known how general the translational coupling mechanism at the Ff gene V-VII junction is as a means of downregulating translation. An implication of the evidence is that by virtue of close spacing to a well-translated gene, even an intrinsically defective initiation site can transmit as much as 10% of upstream translation. We have addressed these issues by studying the homologous gene pair in a related filamentous phage for which the genomic sequence has been determined (33). Phage IKe, which infects E. coli bearing N pili, is classed with the Ff group based on structure, morphology, and life cycle. It has the same genes and gene order but only 55% sequence identity. Genes V and VII are immediately adjacent on a series of overlapping mRNAs (Fig. 1) that are very similar but not identical to the f1 mRNAs (49), making it likely that any control over pVII synthesis operates at the translational level. A previous study of IKe pV indicated that at least 105 copies are made per cell (32), but pVII levels were not determined. Compared to the f1 sequence, the sequence at the beginning of IKe gene VII (Fig. 1) appears potentially to provide a better initiation site. It has a somewhat better fit to several consensus sequences (20, 40, 48), a higher AU content (58% instead of 42%), and a slightly longer Shine-Dalgarno (SD) sequence. There are two in-frame AUG codons (33). The first overlaps the gene V stop codon (ATGA). This format is the most common overlap found in phage lambda (39) and E. coli (2) and is predicted to give efficient translational coupling (13). The second in-frame AUG is 2 nucleotides (nt) beyond the stop codon.
In this study, we examined translation from IKe genes V and VII by using lacZ fusions similar to those employed to study the f1 genes (3, 19). Site-directed mutagenesis, toeprinting, and N-terminal amino acid sequence analysis established which of the two in-frame AUGs serves as the start codon for gene VII. The effects of potentially interfering secondary structure were tested by making deletions extending into the gene VII initiation site, and the effects on coupled translation of varied spacing between the gene V stop codon and gene VII start codon were explored.
MATERIALS AND METHODS
Preparation of phage RFI DNA.
Bacteriophage IKe and E. coli host strains bearing the N plasmid, JE2571/N3 (leu thr/N3) and P90C/N3 [ara Δ(gpt-lac)5 thi/N3] (50), were obtained from R. Webster (Duke University). Overnight cultures of host strains were grown at 37°C in LB broth supplemented with tetracycline (12.5 μg/ml) to maintain the N3 plasmid. Following dilution into LB broth and growth to a density of 2.0 × 108 to 2.5 × 108 cells/ml, shaking of the cultures was slowed from 200 to 50 rpm for 10 min. After infection (multiplicity of infection, 50), the culture was incubated without agitation for 5 min and then shaken at 50 rpm for 5 min and thereafter at 200 rpm. Bacteria were collected by centrifugation 2 h after infection and resuspended in 4 ml of a mixture of 50 mM glucose, 10 mM EDTA, 25 mM Tris-HCl (pH 8.0), and 4 mg of lysozyme per ml per 200-ml culture. IKe replicative-form (RFI) DNA was prepared by the method of Birnboim and Doly (1) with modifications described previously (49). Following resuspension in 2 ml of 10 mM Tris-HCl (pH 7.5)–1 mM EDTA, the DNA was digested with BamHI to linearize contaminating N3 DNA but leave RFI DNA intact. Samples were extracted with phenol, and the DNA was purified by cesium chloride-ethidium bromide centrifugation.
Plasmid construction.
Standard techniques were employed for plasmid construction (38). Plasmids were introduced into the lac deletion strain DS70 [F− Δ(lac) thi trpR λs] by transformation (14) or electroporation (10). Sequence junctions, point mutations, and deletion endpoints were verified by dideoxy DNA sequencing (51). PCRs employed Pfu DNA polymerase (Stratagene), primers (Table 1), and IKe RFI DNA or plasmid DNA as templates.
TABLE 1.
Primers
| Name | Sequencea | Target positions (nt) and restriction sites |
|---|---|---|
| 5′ 5-7 | 5′-ATGTAGATCTTTCTCAGCTATATCCTG-3′ | IKe (1181–1207), BglII, AluI |
| 5′ 7 | 5′-CGCTTTAAAGTACCGTTTACATCCTGC-3′ | IKe (1469–1495), DraI |
| 3′ 5-7,7 | 5′-ATTAGATCTTCACTAGTCATGCTCATT-3′ | IKe (1584–1558), BglII |
| 3′ ATG→CTG | 5′-ATTAGATCTTCACTAGTCAT/GGCTCAT/GTTAA-3′ | IKe (1584–1555), BglII |
| 3′ stop codon | 5′-ATTAGATCTTCACTAGTCATGCC/TC/TATTTAA-3′ | IKe (1584–1555), BglII |
| 3′ TGG | 5′-ATTAGATCTTCACTAGTCATGCCCATTTAA-3′ | IKe (1584–1555), BglII |
| 3′ CAG | 5′-ATTAGATCTTCTGTAGTCATGCTCATTTAA-3′ | IKe (1584–1555), BglII |
| 5′ SD mutant | 5′-CTCAGCTATATCCTGAATGGTATGTT-3′ | IKe (1193–1218), AluI |
| 3′ SD mutant | 5′-cgAGATCTTCACTAGTCATGCTCATTCCTCC-3′ | IKe (1581–1553), BglII |
| 5′ EcoRI | 5′-GTATCACGAGGCCCT-3′ | pKC8 |
| 3′ KpnI | 5′-GACGAATTCCCCGGGGTA--CCAC-3′ | pKC8, EcoRI, SmaI, KpnI |
| 3′ lacZ | 5′-GGCGATTAAGTTGGGTAACGCC-3′ | pKC8 |
| 5′ 7 BglII | 5′-GAGTGGAATTCCAGATCTCCGTTTACATC-3′ | pSM7, BglII |
| 3′ 7 MstII | 5′-ACAGTATCGGCCTCAGGAAGATCGCACT-3′ | pSM7 |
| IKe toeprint | 5′-CATCGGAACACTCCAGCAG-3′ | IKe (1656–1638) |
Positions in oligonucleotides that differ from target sequences are indicated as follows: changes to generate restriction sites are in boldface type; point mutations are underlined; two nucleotides present in an equal mixture during synthesis are separated by a slash; nucleotides complementary to vector sequences are in lowercase letters; a deletion in the primer relative to the target is indicated by a dashed line.
Plasmids (Table 2) derive from pKC8, a pBR322 derivative containing the lac operon structural genes under control of the lacIq promoter (8). pSM5 contains in the SmaI site a 149-bp DdeI/HhaI fragment from IKe RFI DNA (nt 1194 to 1342) made blunt by repair synthesis. pSM5-7, pSM7, and derivatives were constructed by PCR amplification of the IKe sequence by using the primers indicated (Table 1). The DNA used to construct pSM5-7, after digestion with BglII, was ligated into pKC8 digested with BamHI. DNA used to construct pSM7, after digestion with DraI and BglII, was ligated into pKC8 digested with SmaI and BamHI. Derivatives were constructed in the same way by using primers with the desired mutations. Deletions extending into the VII site were made by using pSM5-7 so that V site deletions could be made at the same time. The EcoRI-SmaI fragment of pSM5-7 so that V site deletions could be made at the same time. The EcoRI-SmaI fragment of pSM5-7 containing the lacIq promoter was replaced with one containing a KpnI restriction site generated by PCR amplification of the same region of pKC8. Unidirectional deletions in the resulting plasmid were generated by exonuclease III digestion. DNA digested with KpnI and SmaI was resuspended at 0.5 mg/ml in 66 mM Tris-HCl (pH 7.6)–0.66 mM MgCl2–1 mM β-mercaptoethanol. Nucleotides were removed from the SmaI end by treatment with exonuclease III (2 U/μg of DNA) for 3 to 4 min at 30°C. After digestion with S1 nuclease and repair synthesis with the DNA polymerase I Klenow fragment, the plasmids were circularized with DNA ligase.
TABLE 2.
Plasmids
| Plasmid(s) | Description | Source or reference |
|---|---|---|
| pKC8 | pMC1403 with lacIq promoter in EcoRI site upstream from lac sequences | 8 |
| pSM5 | pKC8 with IKe gene V initiation site and first 15 codons in frame with lacZ coding region | This work |
| pSM7 | pKC8 with the IKe gene VII initiation site and first 4 codons in frame with the lacZ coding region | This work |
| pSM5-7 | pKC8 with IKe gene VII-lacZ fusion as in pSM7, but preceded by IKe gene V | This work |
| pSM7AC, pSM7CA, pSM7CC | pSM7 with second or first ATG or both changed to CTG | This work |
| pSM5-7AC, pSM5-7CA, pSM5-7CC | pSM5-7 with second or first ATG or both changed to CTG | This work |
| pSM5-7TAG, pSM5-7TAA | pSM5-7 with gene V TGA changed to TAG or TAA | This work |
| pSM5-7CAG | pSM5-7 with out-of-frame GTG in gene VII changed to CAG | This work |
| pGI73 | pSP73 with 335-bp Sau3A/Csp451 fragment from IKe (nt 1404 to 1738) inserted downstream from SP6 promoter | 49 |
| pSM7.87, etc.a | pSM7 deletion derivatives with the indicated number of nucleotides remaining upstream from gene VII initiator ATG | This work |
| pSM5-7TGG | pSM5-7 with gene V stop codon (TGA) changed to TGG | This work |
| pSMΔ5-7 | pSM5-7 with 135-nt in-frame internal deletion in gene V | This work |
| pSMΔ5-7 +54, +15, −2 | pSMΔ5-7 with the distal portion of gene V in alternate reading frames to place a stop codon at the indicated positions before or after gene VII initiator ATG | This work |
| pM506, pM565, pM563, pJ32 | M13 vectors carrying lacZ coding region segments | 25 |
For details on these plasmids, see the legend to Fig. 6.
Determination of lac-specific mRNA.
Plasmid-bearing strains grown to early log phase in M9 medium were labelled for 3 min with [5,6-3H]uridine (37 Ci/mmol, 20 μCi/ml; ICN). The nucleic acids were isolated by rapid lysis of 1-ml cultures at 65°C in 10 mM Tris-HCl (pH 7.5)–10 mM EDTA–0.5% sodium dodecyl sulfate (SDS), followed by immediate extraction with phenol equilibrated at 65°C. The aqueous phase was extracted with phenol-chloroform (1:1 [vol/vol]), and the nucleic acids were collected by precipitation from 0.3 M sodium acetate (pH 7.0) with 2 volumes of ethanol. The dried precipitate was dissolved in 0.4 ml of 200 mM Tris-HCl (pH 7.5) and precipitated with 2 volumes of ethanol. The nucleic acids were hybridized as described previously (19) to an excess of single-stranded phage DNA prepared from the following M13 vectors carrying the indicated nucleotides of the lacZ gene (25): pM605, nt 1725 to 2122; pM565, nt 2349 to 3238; pM563, nt 3239 to 3947; and pJ32, nt 3948 to 4310. RNA concentrations over a fourfold range were tested to ensure that conditions of DNA excess were met. Radioactivity recovered from hybridization of each RNA sample with the four probes was compared to ensure that the RNA detected represented full-length transcripts. Values for the four probes were averaged and corrected for background radioactivity and represent at least two independent RNA preparations.
β-Galactosidase assays.
β-Galactosidase assays were done as described by Miller (27), with cultures that had been grown to mid-log phase in LB broth containing 0.4% (vol/vol) glycerol. Chilled cells were permeabilized by adding 2 drops of chloroform and 1 drop of 0.1% (wt/vol) SDS to each milliliter of culture diluted in assay buffer and then vortexing vigorously for 10 to 15 s.
Toeprint analysis.
Toeprinting was done as described previously (15), with RNA generated by runoff transcription in vitro. Purified E. coli 30S ribosomal subunits were gifts of L. Spremulli (University of North Carolina) and P. Wollenzien (North Carolina State University). Plasmid pGI73, a derivative of pSP73 (Promega) containing the IKe sequence of interest, was linearized at the PvuII site and transcribed with SP6 RNA polymerase. Reactions were carried out for 30 to 60 min at 40°C in 50 μl of 40 mM Tris-HCl (pH 7.5)–6 mM MgCl2–2 mM spermidine–10 mM NaCl–10 mM dithiothreitol–500 μM each nucleoside triphosphate (NTP)–50 U of RNasin (Promega)–20 U of SP6 RNA polymerase (Promega)–2 to 5 μg of template DNA. The 405-nt transcript was purified on sequencing gels. The primer was end labelled with [γ-32P]ATP (4,500 Ci/mmol; ICN) and T4 polynucleotide kinase and annealed to the RNA. Toeprint reaction mixtures (10 μl) contained 0.4 pmol of primer, 0.2 pmol of RNA, 6.5 or 11.5 pmol of purified 30S ribosomal subunits, 60 mM NH4Cl, 10 mM Tris-acetate (pH 7.4), 6 mM β-mercaptoethanol, 10 mM magnesium acetate, and 400 to 800 μM each dNTP. Reaction mixtures contained 100 pmol tRNAfMet (Subriden RNA), when included. Preincubations were for 10 min at 37 or 42°C prior to the addition of 4 U of avian myeloblastosis virus reverse transcriptase (Amersham Corp.), and incubations were for 15 min. Sequencing reaction mixtures omitted tRNA and 30S ribosomes and contained a ddNTP at 200 μM. Reactions were stopped by the addition of an equal volume of buffered formamide dye (45 mM Tris-borate [pH 8.3], 2 mM EDTA, 80% formamide, 0.1% bromophenol blue, 0.1% xylene cyanol FF) and placed at 0°C. After heating to 95°C for 3 min, the reaction mixtures were fractionated on 8% sequencing gels.
Purification of the pVII–β-galactosidase fusion protein.
DS70 bearing a pSM5-7 derivative with 16 bp of IKe sequence upstream from the gene V AUG was used for higher yields of the pVII–β-galactosidase fusion protein. Overnight cultures were grown in LB broth supplemented with ampicillin (50 μg/ml). Following a 1:400 dilution into 200 ml of LB broth containing ampicillin, the culture was grown to a density of 2 × 108 cells/ml. After the bacteria were collected by centrifugation, cell extracts were prepared and chromatographed on an anti-β-galactosidase antibody affinity column (Promega) as described in the manufacturer’s instructions. A sample of the eluate was mixed with an equal volume of 3 mM Tris-HCl (pH 7.5)–0.07 mM β-mercaptoethanol–0.5% SDS–5% glycerol–0.05% bromophenol blue and analyzed on SDS–6% polyacrylamide gels (21). Following concentration by precipitation (53) and resuspension in 10 mM Tris-HCl (pH 7.5)–1 mM EDTA, samples were electrophoresed on an SDS–6% polyacrylamide gel and transferred electrophoretically to Immobilon-P membranes (Millipore Corp.). N-terminal sequence analysis was performed on an Applied Biosystems 477A gas-phase sequence analyzer.
RESULTS
IKe conserves differential translation of genes V and VII.
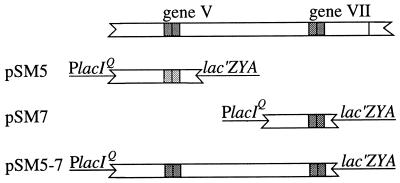
To measure translation of genes V and VII, lacZ fusions similar to those used to study the f1 initiation sites (3) were generated. Each contained a small IKe DNA fragment placed between the lacIq promoter and lacZ coding region such that β-galactosidase synthesis depended on initiation at the gene V or gene VII start (Fig. 2). The fusions included the N-terminal 15 codons of gene V or the first 6 triplets in the gene VII reading frame. The VII site-lacZ fusions in pSM7 and pSM5-7 were identical, but pSM5-7 contained gene V as well. pSM7 and pSM5-7 were used to assay gene VII translation in the absence or presence of upstream translation, and pSM5 was used to determine the upstream translation level (Table 3). The gene VII site showed low but detectable activity (260 U) in the absence of upstream translation, but with gene V translated upstream, activity increased only 2.5-fold (630 U). The activity value of 46,400 U for pSM5 confirmed earlier indications that pV is abundant in IKe-infected hosts (32). The substantial differences in β-galactosidase activity reflected translational effects and were not explained by differences in steady-state mRNA levels, which varied less than threefold (Table 3). Comparison with the results for the f1 initiation sites (3) revealed that the IKe gene VII initiation site differed in two ways from the inactive f1 gene VII site (<2 U). The IKe gene VII site had detectable independent activity and showed only limited activation by upstream translation. The IKe gene V initiation site gave nearly sixfold more activity than the f1 gene V site (8,000 U), suggesting that the IKe gene is expressed at a higher level than the f1 gene. However, because the decrease in translation across the IKe gene V-VII junction was more severe than that for the f1 gene V-VII junction, ∼75-fold instead of 10-fold, the level of IKe gene VII translation (630 U) was very similar to that observed for f1 gene VII (880 U).
FIG. 2.
IKe gene-lacZ fusions used to measure translation of genes V and VII. The IKe sequences shown are as follows: pSM5, 103 bp upstream from gene V ATG and 15 codons; pSM7, 89 bp before the first in-frame ATG for gene VII and 5 additional triplets; pSM5-7, 110 bp upstream from the gene V ATG, gene V, and the gene VII site-lacZ fusion present in pSM7. Transcription from these and all other lacZ fusions reported was directed by the lacIq promoter in parent vector pKC8. pKC8 contains the lacIq promoter and 11 bp of lacI sequence downstream from the transcription start site, followed by restriction sites (EcoRI, SmaI, and BamHI) and the lacZ coding region starting at codon 9 (8). lacI and lacZ sequences in the vector (continuous lines), the IKe gene sequences (open bars), and initiator regions (shaded bars; with ATG position indicated by a vertical line) are shown.
TABLE 3.
Activities of the IKe gene V and gene VII initiation sites
| Plasmida | β-Galactosidase activity
|
Relative lacZ-specific mRNA level (%)d | |
|---|---|---|---|
| Ub | %c | ||
| pSM5 | 46,400 | 100.0 | 100 |
| pSM7 | 260 | 0.6 | 40 |
| pSM5-7 | 630 | 1.3 | 60 |
o-Nitrophenyl absorbancy units at A420/min per A600 unit of cells (27). Background values for DS70 bearing the parent vector lacking an IKe insert (pKC8) have been subtracted. The values are the average of at least four determinations.
Expressed relative to that of pSM5.
Levels of [3H]uridine-labelled lacZ-specific RNA, determined as described in Materials and Methods, are expressed relative to that of pSM5.
Mutational analysis of potential gene VII start codons.
As a first step in determining whether one or both of the ATGs serve as initiator codons, they were eliminated by base substitution (Fig. 3). The location of the gene V stop codon (ATGA; stop codon underlined) required that the first position of the ATG codons be changed, and the fact that ATG, GTG, and TTG all function as initiators left CTG as the only substitution possible. This was not ideal, since C is the least frequent nucleotide at most positions of initiation sites and is generally a down mutation (47). On the other hand, none of the changes was predicted to alter the basic base-pairing potential of the region containing the gene VII initiation site (57). Parallel sets of pSM7 and pSM5-7 derivatives were made to assess the effects of the mutations in the absence or presence of upstream translation. Additional constructs (pSM5-7TAA and pSM5-7TAG) changed the first ATG to ATA, also changing the identity but not the position of the gene V stop codon. pSM5-7CAG eliminated an out-of-frame GTG to determine whether ribosome binding to the out-of-frame GTG interfered with in-frame initiation.
FIG. 3.
Sequences and β-galactosidase activities of wild-type VII site-lacZ fusions and mutants eliminating ATG codons in the gene VII reading frame. The SD sequence is underlined, and ATG and mutant CTG codons are in boldface type. The gene V stop codon is shaded in the sequences of plasmids in which gene V is present. The out-of-frame GTG changed to CAG is indicated by a wavy line.
When both ATGs were eliminated, activity dropped to the background level in the absence or presence of upstream translation, indicating that one or both of the ATGs do function as the gene VII start. The fact that activity did not increase when the out-of-frame GTG was eliminated supports the argument that it does not interfere with in-frame initiation. Substituting the first ATG with CTG had effects, decreasing activity of the pSM7 derivative fivefold and that of the pSM5-7 derivative twofold. However, eliminating the second ATG reduced activity of the pSM7 derivative to the background level and reduced activity of the pSM5-7 derivative fivefold. Eliminating the second ATG was thus associated with more loss of function. The decreased activity observed for mutants with the change in the first ATG could have been due to either inactivating a functional start codon or introducing a down mutation within the initiation site defined by the second ATG. Since the constructs which eliminated the first ATG by changing the gene V stop codon (pSM5-7TAA and pSM5-7TAG) both showed wild-type or higher levels of activity, the latter appeared more likely. Thus, differences in the activities of the various constructs probably reflected the nature of the nucleotide substitution (47) and not the loss of a functional start codon. pSM5-7TAA and pSM5-7TAG also provided evidence that the gene VII initiation site is as efficient with just the second ATG present as it is when both are present. The results suggest that the second ATG is the gene VII initiator codon.
30S ribosomes toeprint to the second AUG.
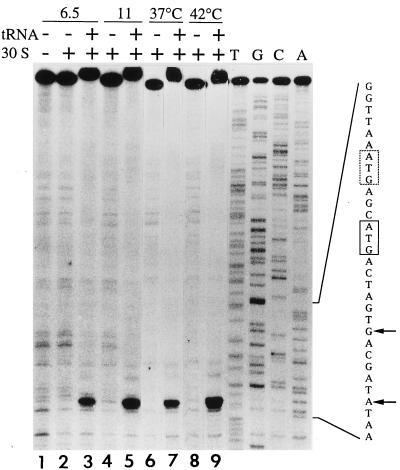
Two factors suggested the need for directly localizing ribosomes bound to mRNA containing the VII site. The 3-nt spacing between the SD sequence and the first AUG was less common than the 9-nt spacing to the second AUG, but use of the first was not ruled out based on known initiation sites (47). Also, because abolishing the second AUG did not reduce activity to the background level when gene V was being translated, it remained possible that some pVII arises from initiation at the first AUG. The position(s) of 30S ribosome binding was localized by use of the toeprinting assay (15). The mRNA template was a 405-nt in vitro transcript containing the 3′ half of gene V and the gene VII initiator region. An end-labelled primer complementary to the 3′ end of gene VII was annealed to the transcript, and the position at which reverse transcriptase stopped primer extension in the presence of purified 30S subunits was identified on sequencing gels by using dideoxy sequencing products as size standards. When initiator tRNA is present in the ribosomal P site, primer extension stops at +16 relative to the 5′ nucleotide (+1) of the initiator codon (15). The results revealed a single strong stop dependent on the presence of 30S subunits and tRNAfMet which mapped to position +16 relative to the second AUG (Fig. 4, lanes 1 to 3). If a higher concentration of 30S subunits was used to enhance detection of weaker ribosome binding sites (Fig. 4, lanes 4 and 5), an additional stop was not detected. If the reaction mixtures were incubated at 42°C to favor any unfolding of the mRNA (Fig. 4; compare lanes 6 and 7 to lanes 8 and 9), binding to the second AUG increased somewhat, but binding to the first AUG was not detected. The results indicated that ribosomes bind to the gene VII initiation site with the second AUG occupying the P site and provided no evidence for ribosomes positioned with the first AUG in the P site.
FIG. 4.
Toeprint analysis of a gene VII transcript. Assays were carried out as described in Materials and Methods, and samples of identical sizes from the reaction mixtures were electrophoresed on an 8% sequencing gel. In lanes containing tRNA, the full-length cDNA product reproducibly showed decreased mobility. Symbols at the top of the lanes indicate the presence (+) or absence (−) of tRNAfMet and 30S ribosomal subunits, the amounts of 30S subunits present (6.5 and 11 pmol), and incubation temperatures (37 and 42°C). Where not otherwise indicated, reaction mixtures contained 6.5 pmol of 30S subunits and were incubated at 37°C. Sequencing reactions were performed in the absence of tRNA and 30S ribosomes, with the indicated ddNTP present at 200 μM. The sequence highlights the two in-frame ATGs (boxed) and +16 positions (arrows) corresponding to each.
N-terminal sequencing of the pVII–β-galactosidase fusion protein.
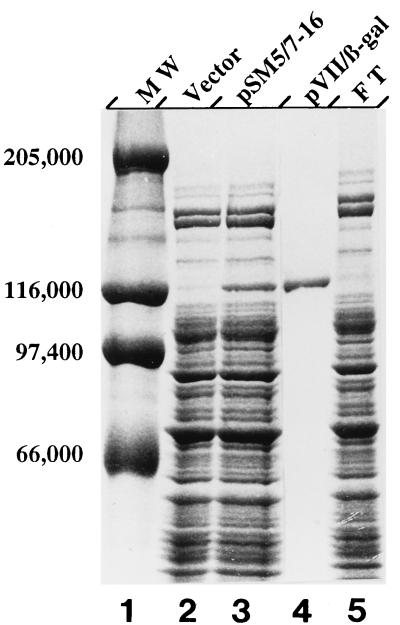
With the N-terminal amino acid sequence of IKe pVII available in the pVII–β-galactosidase fusion protein, it was possible to determine whether the sequence reflected a single start or a mixture of two. A protein initiated at the first AUG should have the sequence Met-Ser-Met-Thr-Ser-Glu-Asp-Pro, whereas one initiated at the second AUG should have the sequence Met-Thr-Ser-Glu-Asp-Pro-Val-Val. The pVII–β-galactosidase fusion protein was isolated from a pSM5-7 construct so that the gene VII site-lacZ fusion would be expressed as it normally is, with gene V translated upstream. Purification was as described in Materials and Methods with an anti-β-galactosidase antibody affinity column. Crude lysates from the strain bearing the construct (Fig. 5, lane 3) contained substantial amounts of a protein not seen in the lysate from strains carrying the vector (Fig. 5, lane 2). The protein comigrated with purified β-galactosidase (Fig. 5, lane 1) (116,000 Da) and was absent from the flowthrough material (Fig. 5, lane 5). The material eluted from the column (Fig. 5, lane 4) appeared to be nearly homogeneous. Sequence analysis showed that ≥95% of the sequenceable protein arose from initiation at the second AUG. The sequence was identical to that predicted, with the N-terminal methionine retained. Virtually no material with a sequence beginning Met-Ser-Met-Thr and yielding Met in cycle 3 was detectable. The results confirmed that the second AUG is the gene VII start codon.
FIG. 5.
Purification of the pVII–β-galactosidase fusion protein. Affinity purification was as described in Materials and Methods. Samples of crude lysates from strains bearing the vector (lane 2), the pSM5-7 clone (lane 3), and flowthrough material from the column (lane 5) contained 5 μg of protein as determined by the Bradford dye binding assay (5). The sample of affinity-purified fusion protein (lane 4) contained 0.5 μg of protein. Samples were fractionated on an SDS–6% polyacrylamide gel and stained with Coomassie brilliant blue. The sizes of molecular mass standards (lane 1) used are indicated on the left in daltons.
An unlikely role for secondary structure in inhibiting gene VII translation.
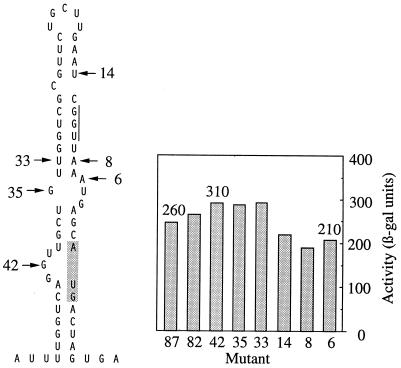
To determine whether the limited translation of gene VII was due to structural occlusion of an otherwise efficient ribosome binding site, deletions extending into the gene VII initiation site were made. Successively removing sequences that form structure usually increases initiation efficiency substantially and hence is diagnostic of interfering RNA structure (9, 31, 41). The 5′→3′ deletions were made in pSM7 without gene V upstream. They were designated by the number of nucleotides of IKe sequence remaining before the initiator AUG and are shown (Fig. 6) superimposed on the most stable theoretical structure for the VII site (57). The activities determined for pSM7.87 and pSM7.82, predicted to leave the base-pairing scheme intact, were similar to that of the wild-type parent (260 U). The activity of pSM7.42 increased slightly as the deletion extended into the ascending arm of the stem. However, more extensive deletions, to 35 or 33 nt upstream from the initiator AUG, did not give further increases in activity, even though the predicted ΔG dropped by more than half, from −12.8 to −5.5 kcal/mol. Deletions up to positions 14, 8, and 6 nt upstream from the AUG extended into the ribosome binding site and, as expected, decreased activity. Thus, although a structure was predicted to sequester the SD sequence and AUG, little evidence that RNA structure inhibits initiation significantly was obtained from lowering the base-pairing potential. The activities of the wild-type parent and the deletion mutants were more consistent with the simple alternative that gene VII has a weak initiation site.
FIG. 6.
A predicted structure for the IKe gene V-VII sequence and β-galactosidase activities of deletion mutants (mutant 87 is, e.g., pSM7.87). The most stable secondary structure predicted for the gene VII initiation site (57) is shown on the left. The 5′→3′ deletion mutants generated from pSM7 are designated by the number of nucleotides in the IKe sequence remaining upstream from the initiator AUG. The AUG is shaded, and the SD sequence is indicated by a vertical line.
Coupled translation and spacing at the gene V-VII junction.
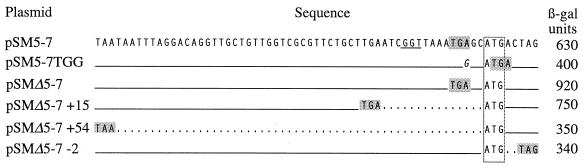
To determine how the spatial arrangement of genes V and VII governs coupled translation from gene VII, the effects of varying the stop-start distance were explored. This approach often provides an indication of how the distal gene in a pair is regulated by revealing how dependent activity is on proximity to the upstream gene and whether downstream translation is hindered or enhanced by local RNA structure. For the f1 gene pair, the sharp decrease in gene VII translation that resulted from a small increase in spacing from gene V (1 to 5 nt) was key in suggesting that the gene VII initiation site was incapable of binding ribosomes de novo (19). Sequences at the IKe gene V-VII junction in the pSM5-7 derivatives are shown in Fig. 7. In the pSM5-7TGG sequence, the gene V TGA was changed to TGG to overlap the functional stop codon with the gene VII ATG. The modest decrease in activity observed suggested that minimizing the spacing does not improve gene VII translation. Alternatively, the A→G change could represent a down mutation. To avoid this complication, the spacing in other constructs was varied without changing the sequence at the V-VII junction. This was done by using deletions of slightly different lengths within gene V to place the distal segment in different reading frames. In the pSMΔ5-7 sequence, the 0 frame and wild-type 2-nt spacing was maintained. In pSMΔ5-7 +15 and pSMΔ5-7 +54, the spacing was increased to 15 or 54 nt, and in pSMΔ5-7 −2, the stop codon was moved 2 nt beyond the gene VII ATG (−2). Increasing the spacing from 2 to 15 nt reduced activity by only ∼20%. Increasing it to 54 nt decreased activity to a value not appreciably higher than the activity observed for the gene VII site in the absence of upstream translation. Activity was also low if the stop codon was 2 nt beyond the gene VII ATG. The results are not atypical for translationally coupled genes. Generally, activity decreases if the stop codon for the upstream gene is placed beyond the start of the distal gene, is relatively insensitive to small increases in spacing, and drops to the independent level when the spacing is increased further. Modest increases in spacing frequently also unmask substantial activity from occluded initiation sites by positioning the terminating ribosome on a sequence that would otherwise assume structure. That they did not do this at the IKe V-VII junction concurs with indications from deletions extending into the VII site that interfering RNA structure is probably not the basis for inefficient translation.
FIG. 7.
Effect of spacing on gene VII translation. Diagrams show the V-VII junctions in IKe gene-lacZ fusions with the spacing varied between the gene V stop codon and the gene VII start codon. The sequence of the parent plasmid, pSM5-7, is shown at the top, and the change in the sequence of pSM5-7TGG is indicated in italics. Other fusions had the wild-type sequence at the V-VII junction but were derivatives of pSMΔ5-7, which contained an internal deletion in gene V that removed about half of the coding region. Derivatives are designated by the number of nucleotides between the functional stop codon for gene V and the gene VII ATG, with spacings represented by dotted lines. The gene VII SD sequence is underlined, functional stop codons are shaded, and the gene VII ATG is boxed.
The activities observed for pSM5-7 and pSMΔ5-7 eliminated another explanation for inefficient translation of gene VII under conditions in which gene V is translated. Phage f1 pV is known to repress translation of genes II and X by binding to the f1 mRNAs (28, 55). While IKe pV appears not to repress translation of gene II (56), it is not clear whether the protein has lost function entirely as a translational repressor. Deleting about half of gene V in pSMΔ5-7, including the amino acids important for DNA binding and dimer formation (26), did not substantially increase activity from the VII site. This ruled out the possibility that pV functions in trans as a repressor of gene VII translation.
DISCUSSION
Translation of IKe genes V and VII has been examined, and several basic features of the gene VII initiation site have been defined. The genes are represented in the same amounts on the phage mRNAs, but the products are needed in different amounts to produce phage. The lacZ fusions driven by the respective initiation sites showed that differences are brought about during translation, and the expression levels were more disparate than would have been expected. Gene V is translated at a very high level, consistent with reports that pV is abundant in IKe-infected hosts (32). Gene VII is translated at low levels from an initiation site defined by a single AUG. Unlike the Ff gene VII initiation site, the IKe gene VII site has detectable intrinsic activity and is coupled only marginally to upstream translation. These properties suggest that IKe has evolved a distinct mechanism for lowering translation at the gene V-VII junction. This and the fact that expression is downregulated between genes V and VII to similar levels by the two distantly related phages suggest that selection has occurred to maintain expression of pVII at a low level. While that level is not set precisely to the amount incorporated into phage (12), the functions or properties of the protein appear to require that its synthesis be limited. Neither the independent activities of the IKe gene V and VII initiation sites nor the close spacing between the genes would have predicted a level of gene VII translation so similar to that seen in Ff phage infections.
The fact that translation on the IKe mRNAs is sharply downregulated at the gene V-VII junction increases the likelihood that the other coat protein involved in initiating phage assembly (pIX) is translated at similarly low levels. On the Ff mRNAs, efficient binding of ribosomes to the gene IX initiation site requires removing a structure 15 nt upstream from the AUG (3). From this result and evidence that amber mutations in gene VII are polar on gene IX expression (42, 43), it was suggested (19) that inefficient coupling at the gene V-VII junction determines the number of ribosomes transmitted into gene VII. Ribosomes traversing gene VII are then needed to unfold the structure masking the gene IX start. Since the IKe sequence at the junction of genes VII and IX (33) has a format identical to that of the gene V-VII junction, a substantial predicted structure (57) 15 nt upstream from overlapping stop and start codons, control of gene IX translation may be achieved by the same mechanism.
The sixfold-higher activity of the IKe gene V initiation site compared with that of the f1 gene V initiation site is notable in two respects. Certainly, the basis for its efficiency is not obvious. The f1 V site (Fig. 8A), based on direct binding and the activity of the lacZ fusion (8,000 U) (3) compared to that of fusions for other initiation sites (8, 19, 20), is a strong ribosome binding site. The IKe V site in our standard lacZ fusion gives the highest value we have encountered (46,400 U), yet the sequence has only modest SD complementarity (GAG) and, at the initiator AUG, lacks the optimal context AUGA (52). However, the V site contains regions similar to U-rich sequences recognized in a site-specific manner by ribosomal protein S1 and proposed to serve as determinants for recognition (4). It also includes two 9-nt stretches nearly identical to the epsilon translational enhancer (30) and a 14-nt sequence that at 11 positions is identical to the downstream box (44). Present evidence (46) indicates that these elements are important in some but not other RNAs (34, 45). How they function is not yet clear. They have complements in 16S rRNA, but the involvement of the 16S rRNA sequences in base-pairing has not been demonstrated. Nevertheless, the possibility that the IKe gene V initiation site derives its efficiency from interactions with the ribosome by using these or other as-yet-unidentified elements merits further study.
FIG. 8.
Sequences of the f1 and IKe gene V initiation sites (A) and SD-AUG spacings in the IKe gene VII initiation site (B). SD sequences are underlined, the start codons are shown in boldface type, and the gene V stop codon is overlined. (A) Boxed regions in which 7 of 9 nt are identical to the epsilon translational enhancer are shown (30). The sequence with similarity to the downstream box (UCAcGaUUCuCAAG; identical nucleotides are capitalized) occurs at positions +18 to +31 downstream from the AUG, included in all of the V site-lacZ fusions. (B) Spacing is defined with reference to the 5′ A of the ASD sequence. Aligned spacing between the SD sequence and the first or second AUG in the gene VII initiation site is indicated by dots beyond the 5′-terminal position of the ASD sequence.
The substantially higher level at which IKe pV is synthesized relative to Ff pV also has implications for DNA replication and phage production. Ff pV molecules have two known functions: they bind single strands (SS) displaced by rolling-circle replication during the switch from replicative-form (RF) to SS synthesis and, when in excess over DNA, bind operators on the mRNAs to repress translation of genes II and X (29). An increase in pV synthesis over normal levels is believed to compromise phage production in several ways. It shortens the period of RF production before the switch, limits the number of RF molecules to serve as templates for SS synthesis and transcription, and prematurely represses synthesis of pII needed for RF production. In IKe-infected hosts, the higher level of pV synthesis should similarly limit RF levels, which may in part explain the weaker infection generally observed. On the other hand, IKe pV differs from Ff pV by not functioning as a translational repressor for gene II (56). This may help the host tolerate the high level of pV synthesis by ensuring that some level of continuous RF production maintains the infection.
Certain properties of the IKe gene VII initiation site are readily understood. The detectable but low independent activity is probably not unexpected. The bias toward an AU-rich sequence (58%) may be closer than the atypical f1 VII site to the range seen in ribosome binding sites, but other features are only marginally better (Fig. 1). In the region of direct ribosome contact (−20 to +13), the fit to a consensus sequence derived from independent initiation sites is 36%, while that for f1 is 33% (20). By comparison, the values for the respective gene V initiation sites are much higher (55%). The SD sequence in the IKe VII site is 1 nt longer (Fig. 1), but since a 3-bp interaction with 16S rRNA would involve the 5′ end of the anti-SD (ASD) sequence rather than the 3′ end used most often (47), the 2-bp interaction is more likely (Fig. 8B). The SD-ASD interaction also rationalizes selective use of the second AUG. Spacing to the first AUG is only 3 nt if the SD sequence pairs with the 5′ end of the ASD sequence, and if base pairs involve the more frequently used 3′ end, there is no space between the 5′ nucleotide of the ASD sequence and the position that would be occupied by the initiator tRNA anticodon. The equivalent spacings to the second AUG are 9 and 6 nt, respectively. Recent studies using the notion of alignment (11) to explore the constraints on SD-AUG spacing indicate that a spacing minimum is required for translation, and spacings less than 4 nt sharply decrease activity (7, 22, 35). Thus, the fact that ribosomes terminating at the end of gene V ignore an AUG overlapping the stop codon in favor of a second AUG with better alignment contributes further evidence that the SD-AUG spacing in mRNA is constrained to allow simultaneous interactions with the ASD region of 16S rRNA and initiator tRNA in the P site.
The most puzzling findings of the work presented here are clearly the severity of the downregulation at the IKe gene V-VII junction and the limited extent to which gene VII is coupled to gene V translation. The genes have an arrangement typical of translationally coupled gene pairs, coding regions separated by only 2 nt and the start for the distal gene located just after the upstream stop. Based on mutational and comparative analysis of highly expressed genes, the sequence requirements for coupled translation are less stringent than those for de novo initiation (20, 24). The f1 gene VII initiation site illustrates this by extreme example, giving coupled translation at 10% of the upstream translation level despite no detectable independent activity and a sequence that lacks the characteristic features of ribosome binding sites (19, 20). Thus, since both initiation sites in the IKe gene pair showed higher activities than the f1 sites, a higher level of gene VII translation was expected when gene V was being translated upstream. Instead, translation occurred at a much lower level, ∼1% of gene V activity. The most prevalent means of decreasing or preventing coupling, interference by RNA secondary structure (16, 31, 54), appears an unlikely explanation. The effects of lowering the potential for structure were modest, and varying the spacing at the V-VII junction did not unmask substantially higher activity. Moreover, ribosomes terminating 2 nt before the gene VII AUG presumably have unfolded any local structure. Competing signals do not appear to have significant effects, since removal of the upstream AUG or an out-of-frame GUG did not increase translation from the gene VII AUG. In addition, little indication that the identity of the termination codon or limiting release factors is the basis of the defect in translational coupling was provided by the modest effects of varying the gene V stop codon (Fig. 3). In view of the unexpected nature of our findings, studies designed to identify situations that increase gene VII translation are under way to determine the basis for the low efficiency of translational coupling between genes V and VII.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grant GM33349 to D.A.S. S.M.-A. was supported in part by National Institute of General Medical Sciences Predoctoral Traineeship GM07184.
We thank L. Spremulli and P. Wollenzien for 30S subunits and J. Abernethy and T. Vanaman for help in interpreting the protein sequencing data.
REFERENCES
- 1.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1976;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Blumer K J, Ivey M R, Steege D A. Translational control of phage f1 gene expression by differential activities of the gene V, VII, IX, and VIII initiation sites. J Mol Biol. 1987;197:439–451. doi: 10.1016/0022-2836(87)90557-2. [DOI] [PubMed] [Google Scholar]
- 4.Boni I V, Isaeva D M, Musychenko M L, Tzareva N V. Ribosome-messenger recognition: mRNA targets sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brissette J L, Russel M. Secretion and membrane integration of a filamentous phage-encoded morphogenetic protein. J Mol Biol. 1990;211:565–580. doi: 10.1016/0022-2836(90)90266-O. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;23:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cone K C, Steege D A. Functional analysis of lac repressor restart sites in translational initiation and reinitiation. J Mol Biol. 1985;186:733–742. doi: 10.1016/0022-2836(85)90393-6. [DOI] [PubMed] [Google Scholar]
- 9.de Smit M H, van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]
- 10.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn J J, Studier F W. Nucleotide sequence from the genetic left end of bacteriophage T7 DNA to the beginning of gene 4. J Mol Biol. 1981;148:303–330. doi: 10.1016/0022-2836(81)90178-9. [DOI] [PubMed] [Google Scholar]
- 12.Endemann H, Model P. Location of filamentous phage minor coat proteins in phage and in infected cells. J Mol Biol. 1995;250:496–506. doi: 10.1006/jmbi.1995.0393. [DOI] [PubMed] [Google Scholar]
- 13.Gold L. Posttranscriptional regulatory mechanisms in E. coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 13a.Haigh, N., and R. E. Webster. Personal communication.
- 14.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Hartz D, McPheeters D S, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 16.Hellmuth K, Rex G, Surin B, Zinck R, McCarthy J E G. Translational coupling varying in efficiency between different pairs of genes in the central region of the atp operon of Escherichia coli. Mol Microbiol. 1991;5:813–824. doi: 10.1111/j.1365-2958.1991.tb00754.x. [DOI] [PubMed] [Google Scholar]
- 17.Hill D F, Petersen G B. Nucleotide sequence of bacteriophage f1 DNA. J Virol. 1982;44:32–46. doi: 10.1128/jvi.44.1.32-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horabin J I, Webster R E. Morphogenesis of f1 filamentous bacteriophage: increased expression of gene I inhibits bacterial growth. J Mol Biol. 1986;188:403–413. doi: 10.1016/0022-2836(86)90164-6. [DOI] [PubMed] [Google Scholar]
- 19.Ivey-Hoyle M, Steege D A. Translation of phage f1 gene VII occurs from an inherently defective initiation site made functional by coupling. J Mol Biol. 1989;208:233–244. doi: 10.1016/0022-2836(89)90385-9. [DOI] [PubMed] [Google Scholar]
- 20.Ivey-Hoyle M, Steege D A. Mutational analysis of an inherently defective translation initiation site. J Mol Biol. 1992;224:1039–1054. doi: 10.1016/0022-2836(92)90468-y. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lee K, Holland-Staley C A, Cunningham P R. Genetic analysis of the Shine-Dalgarno interaction: selection of alternative functional mRNA-rRNA combinations. RNA. 1996;2:1270–1285. [PMC free article] [PubMed] [Google Scholar]
- 22a.Locklear, J., and D. A. Steege. Unpublished results.
- 23.Lopez J, Webster R E. Morphogenesis of filamentous bacteriophage f1: orientation of extrusion and production of polyphage. Virology. 1983;127:177–193. doi: 10.1016/0042-6822(83)90382-3. [DOI] [PubMed] [Google Scholar]
- 24.McCarthy J E G. Post-transcriptional control in the polycistronic operon environment: studies of the atp operon of Escherichia coli. Mol Microbiol. 1990;4:1233–1240. doi: 10.1111/j.1365-2958.1990.tb00702.x. [DOI] [PubMed] [Google Scholar]
- 25.McCormick J R, Zengel J M, Lindahl L. Intermediates in the degradation of mRNA from the lactose operon of Escherichia coli. Nucleic Acids Res. 1991;19:2767–2776. doi: 10.1093/nar/19.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPherson A, Brayer G D. The gene 5 protein and its molecular complexes. In: Jurnak F A, McPherson A, editors. Biological molecules and assemblies. II. Nucleic acids and interactive proteins. New York, N.Y: John Wiley & Sons, Inc.; 1985. pp. 325–392. [Google Scholar]
- 27.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 28.Model P, McGill C, Mazur B, Fulford W D. The replication of bacteriophage f1: gene V protein regulates the synthesis of gene II protein. Cell. 1982;29:329–335. doi: 10.1016/0092-8674(82)90149-0. [DOI] [PubMed] [Google Scholar]
- 29.Model P, Russel R. Filamentous bacteriophage. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 375–456. [Google Scholar]
- 30.Olins P O, Rangwala S H. A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in Escherichia coli. J Biol Chem. 1989;264:16973–16976. [PubMed] [Google Scholar]
- 31.Pati S, DiSilvestre D, Brusilow W. Regulation of the Escherichia coli uncH gene by mRNA secondary structure and translational coupling. Mol Microbiol. 1992;6:3559–3566. doi: 10.1111/j.1365-2958.1992.tb01791.x. [DOI] [PubMed] [Google Scholar]
- 32.Peeters B P H, Konings R N H, Schoenmakers J G G. Characterization of the DNA binding protein encoded by the N-specific filamentous Escherichia coli phage IKe. Binding properties of the protein and nucleotide sequence of the gene. J Mol Biol. 1983;169:197–215. doi: 10.1016/s0022-2836(83)80180-6. [DOI] [PubMed] [Google Scholar]
- 33.Peeters B P H, Peters R M, Schoenmakers J G G, Konings R N H. Nucleotide sequence and genetic organization of the genome of the N-specific filamentous bacteriophage IKe. Comparison with the genome of the F-specific filamentous phages M13, fd, and f1. J Mol Biol. 1985;181:27–39. doi: 10.1016/0022-2836(85)90322-5. [DOI] [PubMed] [Google Scholar]
- 34.Resch A, Tedin K, Grundling A, Mundlein A, Blasi U. Downstream box-anti-downstream box interactions are dispensable for translation initiation of leaderless mRNAs. EMBO J. 1996;15:4740–4748. [PMC free article] [PubMed] [Google Scholar]
- 35.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo G D, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 36.Russel M. Moving through the membrane with filamentous phages. Trends Microbiol. 1995;3:223–228. doi: 10.1016/s0966-842x(00)88929-5. [DOI] [PubMed] [Google Scholar]
- 37.Russel M, Model P. Genetic analysis of the filamentous bacteriophage packaging signal and of the proteins that interact with it. J Virol. 1989;63:3284–3295. doi: 10.1128/jvi.63.8.3284-3295.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Russel, M. Personal communication.
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sanger F, Coulson A R, Hong G F, Hill D F. Nucleotide sequence of bacteriophage λ DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 40.Scherer G F E, Walkinshaw M D, Arnott S, Moore D J. The ribosome binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 1980;8:3895–3907. doi: 10.1093/nar/8.17.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt B F, Berkhout B, Overbeek G P, van Strien A, van Duin J. Determination of the RNA secondary structure that regulates lysis gene expression in the bacteriophage MS2. J Mol Biol. 1987;195:505–516. doi: 10.1016/0022-2836(87)90179-3. [DOI] [PubMed] [Google Scholar]
- 42.Simons G F M, Konings R N H, Schoenmakers J G G. Genes VI, VII, and IX of phage M13 code for minor capsid proteins of the virion. Proc Natl Acad Sci USA. 1981;78:4194–4198. doi: 10.1073/pnas.78.7.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simons G F M, Veeneman G H, Konings R N H, Van Boom J H, Schoenmakers J G G. Oligonucleotide-directed mutagenesis of gene IX of bacteriophage M13. Nucleic Acids Res. 1982;10:821–832. doi: 10.1093/nar/10.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sprengart M L, Fatscher H P, Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16S rRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990;18:1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sprengart M L, Fuchs E, Porter A G. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 46.Sprengart M L, Porter A G. Functional importance of RNA interactions in selection of translation initiation codons. Mol Microbiol. 1997;24:19–28. doi: 10.1046/j.1365-2958.1997.3161684.x. [DOI] [PubMed] [Google Scholar]
- 47.Stormo G D. Translation initiation. In: Reznikoff W, Gold L, editors. Maximizing gene expression. Stoneham, Mass: Butterworth Publishers; 1986. pp. 195–224. [Google Scholar]
- 48.Stormo G D, Schneider T D, Gold L M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stump M D, Madison-Antenucci S, Kokoska R J, Steege D A. Filamentous phage IKe mRNAs conserve form and function despite divergence in regulatory elements. J Mol Biol. 1997;266:51–65. doi: 10.1006/jmbi.1996.0766. [DOI] [PubMed] [Google Scholar]
- 50.Sun T-P, Webster R E. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J Bacteriol. 1986;165:107–115. doi: 10.1128/jb.165.1.107-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabor S, Richardson C C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taniguchi T, Weissmann C. Site-directed mutations in the initiator region of the bacteriophage Qβ coat cistron and their effect on ribosome binding. J Mol Biol. 1978;118:533–565. [Google Scholar]
- 53.Wessel D, Flügge U I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 54.Wikström P M, Lind L K, Berg D E, Björk G R. Importance of mRNA folding and start codon accessibility in the expression of genes in a ribosomal protein operon of Escherichia coli. J Mol Biol. 1992;224:949–966. doi: 10.1016/0022-2836(92)90462-s. [DOI] [PubMed] [Google Scholar]
- 55.Yen T S B, Webster R E. Translational control of bacteriophage f1 gene II and gene X proteins by gene V protein. Cell. 1982;29:337–345. doi: 10.1016/0092-8674(82)90150-7. [DOI] [PubMed] [Google Scholar]
- 56.Zaman G J, Kaan A M, Schoenmakers J G, Konings R N. Gene V protein-mediated translational regulation of the synthesis of gene II protein of the filamentous bacteriophage M13: a dispensable function of the filamentous-phage genome. J Bacteriol. 1992;174:595–600. doi: 10.1128/jb.174.2.595-600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]