Abstract
UMP kinase from Escherichia coli is one of the four regulatory enzymes involved in the de novo biosynthetic pathway of pyrimidine nucleotides. This homohexamer, with no counterpart in eukarya, might serve as a target for new antibacterial drugs. Although the bacterial enzyme does not show sequence similarity with any other known nucleoside monophosphate kinase, two segments between amino acids 35 to 78 and 145 to 194 exhibit 28% identity with phosphoglycerate kinase and 30% identity with aspartokinase, respectively. Based on these similarities, a number of residues of E. coli UMP kinase were selected for site-directed mutagenesis experiments. Biochemical, kinetic, and spectroscopic analysis of the modified proteins identified residues essential for catalysis (Asp146), binding of UMP (Asp174), and interaction with the allosteric effectors, GTP and UTP (Arg62 and Asp77).
Nucleoside monophosphate (NMP) kinases are ubiquitous enzymes in all forms of living cells, and they represent a homologous family of proteins with structural and catalytic properties similar to those of adenylate kinases (4, 7, 9). UMP kinase from Escherichia coli is a noticeable exception to this paradigm. The protein, encoded by the pyrH gene (16), does not show sequence similarity with any other known NMP kinase, has an oligomeric structure, and is subjected to complex regulatory mechanism in which UTP and GTP act as allosteric effectors (14). Another unique property of E. coli UMP kinase is a very low solubility at neutral pH (<0.1 mg/ml). UTP and/or alkaline pH increases the solubility of bacterial enzymes up to 50 times (15).
The fact that the pyrH gene product, with no counterpart in eukarya, is essential for cell growth and division makes it an attractive new target for antibacterial drugs. To properly use this property of UMP kinase, a number of questions concerning the architecture and mechanistic features of the protein should be answered. In the absence of high-resolution three-dimensional data for UMP kinase, other approaches such as chemical modification or site-directed mutagenesis in combination with spectroscopic techniques were considered.
Sequence comparison of E. coli UMP kinase with other known phosphotransferases allowed identification of a gap-free region (amino acids 145 to 194) exhibiting 28% identity and 51% similarity with aspartokinases. Another segment, situated between amino acids 35 and 78 of E. coli UMP kinase, exhibits 30% identity and 64% similarity with phosphoglycerate kinase from Haloarticula valismortis (Fig. 1). Based on these alignments, a number of residues of bacterial UMP kinase were selected for analysis by site-directed mutagenesis. The modified proteins were overproduced and purified by procedures similar to those used for the wild-type protein (14). Biochemical, kinetic, and spectroscopic analysis of the modified proteins revealed a number of residues essential for catalysis (Asp146), binding of UMP (Asp174), or interaction with effectors (Arg62 and Asp77). Two other modified forms of UMP kinase (D201N and D159N), described initially in previous papers (14, 15), were shown to play a role in the solubility of the protein at neutral pH.
FIG. 1.
Alignment of amino acid sequence of UMP kinase (UMPK) from E. coli with partial sequence of aspartokinase (AK) and phosphoglycerate kinase (PGK). Identical residues in UMP kinase and in the other proteins, expressed in one-letter code, are indicated in bold characters. Amino acid residues submitted to site-directed mutagenesis (R62H, D77N, D146N, D159N, D168N, D174N, and D201N) are marked by arrows. Ec, E. coli; Hv, H. valismortis; Sc, Saccharomyces cerevisiae.
MATERIALS AND METHODS
Chemicals.
Nucleotides, restriction enzymes, T4 DNA ligase, and coupling enzymes were from Boehringer Mannheim. T7 DNA polymerase and the four deoxynucleoside triphosphates used in sequencing reactions were from Pharmacia. Oligonucleotides were synthesized by the phosphoamidinate method, using a commercial DNA synthesizer (Cyclone; Biosearch). NDP kinase from Dictyostelium discoideum (2,000 U/mg of protein) was kindly provided by M. Véron.
Bacterial strains, plasmids, growth conditions, and DNA manipulations.
Strain BLI5 (14), used for overexpression of the pyrH gene, was derived from E. coli BL21(DE3) (Novagen Inc.). The strain expressed the lacI gene on plasmid pDIA17 (10) and the T7 RNA polymerase gene on the chromosome. Bacteria were grown in 2YT medium (11) supplemented with ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml). When the optical density at 600 nm reached 1.5, 1 mM isopropyl-β-d-thiogalactoside was added to the medium. The bacteria were harvested by centrifugation 3 h after induction. Site-directed mutagenesis was performed with the single-stranded DNA of phagemid pDIA5418 grown in strain CJ236 in the presence of plasmid pDIA17 and the helper phage M13KO7 (5). Sequences of oligonucleotides used for mutagenesis are given in Table 1. For each mutagenesis, the whole sequence of the pyrH gene was checked for the absence of any other mutation by the dideoxynucleotide sequencing method (12).
TABLE 1.
Amino acid substitutions and their consequences on the solubility and fluorescence properties of E. coli UMP kinase
| Amino acid substituted | Codon alteration | Sequence of mutagenic oligonucleotide | % of activity in the pelleta | Relative fluorescence intensityb
|
|
|---|---|---|---|---|---|
| −UTP | +UTP | ||||
| R62H | CGT→CAT | 5′CCAGCGCCATGGAACAGGTTA3′ | 83 | 89 | 203 |
| D77N | GAC→AAC | 5′CCCCATGTGGTTGCCCACAACG3′ | 84 | 112 | 108 |
| D146N | GAC→AAC | 5′GGAAGCTGCTGAGTTGGTGGT3′ | 84 | 178 | 220 |
| D159N | GAT→AAT | 5′CAGCACCACATTGGCTTCAATTTC3′ | 10 | 102 | 208 |
| D168N | GAC→AAC | 5′TAAACACGCCGTTAACTTTGGT3′ | 97 | 64 | 180 |
| D174N | GAT→AAT | 5′TCTTTCGCCGGATTAGCGGTA3′ | 98 | 67 | 164 |
| D201N | GAC→AAC | 5′GGCCGCCAGGTTCATGGACTTT3′ | 42 | 138 | 215 |
Sonicated bacterial extract in 50 mM Tris-HCl (pH 7.4) was centrifuged at 14,000 rpm for 4 min, and then the supernatant was separated from the pellet, which was resuspended at the original volume with 50 mM Tris-HCl (pH 7.4) or with 100 mM borate (pH 9). The activity of each fraction was determined at pH 7.4 with 1 mM ATP and 1 mM UMP as substrates.
The emission spectrum of purified UMP kinase (1 or 2 μM in terms of monomer) in 50 mM Tris-HCl (pH 7.4) was recorded between 305 and 400 nm. The fluorescence maxima were 332 nm for the wild-type enzyme and D146N mutant, 333 nm for the R62H and D159N mutants, 334 nm for the D168N and D174N mutants, and 335 nm for the D201N mutant. UTP when present did not shift the fluorescence maximum of UMP kinase. It was used at 10 μM for the wild type and D146N, D159N, D168N, D174N and D201N mutants and 300 μM for the R62H mutant. For the D77N mutant, up to 1 mM UTP no increase in fluorescence intensity was observed. The fluorescence intensity of the wild-type UMP kinase in the absence of UTP was considered 100%.
Purification of UMP kinase and activity assay.
Wild-type UMP kinase and several of the modified variants (D77N, R62H, D146N, D168N, and D174N) overproduced in the same bacteria were purified as described previously (14). The D159N and D201N variants were purified by ion-exchange and gel permeation chromatography (15). The UMP kinase activity was determined at 30°C and 0.5-ml final volume, using a coupled spectrophotometric assay (1) with a Beckman DU640 instrument. The reaction medium buffered with 50 mM Tris-acetate (pH 6) or with 50 mM Tris-HCl (pH 7.4 or 8) contained 50 mM KCl, 2 mM MgCl2, 1 mM phosphoenolpyruvate, 0.2 mM NADH, various concentrations of ATP and UMP, and 2 U each of pyruvate kinase, NDP kinase, and lactate dehydrogenase. The reaction was started with UMP kinase. One unit of the enzyme corresponds to 1 μmol of product formed per min.
Analytical procedures.
Protein concentration was measured by the method of Bradford (2). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (6). Fluorescence experiments were performed on a Shimadzu RF 5001 PC spectrofluorimeter at room temperature. Binding of UTP to UMP kinase was monitored from the nucleotide-induced enhancement of the protein fluorescence (14, 15).
RESULTS
Purification and molecular characterization of the modified forms of E. coli UMP kinase.
When E. coli overexpressing the wild-type pyrH gene was disrupted by sonication in 50 mM Tris-HCl (pH 7.4), over 90% of the enzyme activity was recovered in the pellet after centrifugation at 10,000 × g (15). The same was true for R62H, D77N, D146N, D168N, and D174N mutants (Table 1). These modified proteins were solubilized from the bacterial pellet with 100 mM sodium borate (pH 9). Overnight dialysis against 50 mM Tris-HCl (pH 7.4) yielded insoluble recombinant proteins. The precipitates were washed several times with the same buffer and gave homogeneous enzyme preparations as determined by SDS-PAGE. The enzymes were stored for several months at 4°C with no significant loss of activity. An exception was the D174N mutant, which lost two-thirds of its activity after 3 months at 4°C. Gel permeation chromatography on Sephacryl S-300 HR of pure modified enzymes in 50 mM Tris-HCl (pH 8.7) indicated that the active form (over 75% of total protein) corresponds, as in the case of the wild-type UMP kinase, to the hexamer (molecular mass, 150 ± 10 kDa).
The D159N and D201N mutants exhibited a much higher solubility than the wild-type protein at neutral pH (Table 1). These variants were purified by ion-exchange chromatography on DEAE-Sepharose CL-6B, using a linear gradient from 0 to 0.3 M of NaCl in 50 mM Tris-HCl (pH 7.4) or in 100 mM sodium borate (pH 9). Chromatography on Sephacryl S-300 HR showed that the active form of the D201N mutant, which exhibited a higher heterogeneity than expected, was the hexamer. It represented one-third of the total protein, in mixture with lower- or higher-molecular-mass oligomers, with no indication of an equilibrium between these forms. The active form of the D159N mutant represented over 80% of total protein (15).
Temperature and GdmCl denaturation of UMP kinase mutants.
Wild-type UMP kinase from E. coli showed a high stability against thermal (melting temperature [Tm] = 63°C) or chemical denaturation (14, 15). The R62H, D146N, D168N, and D174N mutants were half inactivated at temperatures between 59 and 62°C. Whereas the D159N mutant was particularly resistant against thermal denaturation (Tm = 73°C), the D77N and D201N mutants were half inactivated at 52 and 48°C, respectively. UTP, which at millimolar concentrations increased substantially the thermal stability of the wild-type enzyme (15), shifted the Tm of D201N mutant by almost 30°C. Denaturation of UMP kinase mutants by guanidinium hydrochloride (GdmCl) was monitored from the red shift of the Trp fluorescence maximum (15). Most of the variants obtained by site-directed mutagenesis behaved like the wild-type protein. The midpoint transition concentration of GdmCl was 1.8 M for the D146N and D201N mutants, 1.9 M for the wild type and the D77N and D174N mutants, and 2.0 M for the R62H and D168N mutants.
Kinetic properties of UMP kinase mutants.
Like many regulatory enzymes, E. coli UMP kinase exhibits a complex kinetic behavior, which varies with pH (15). At pH 6, the reaction rates at variable concentrations of one nucleotide in the presence of several fixed concentrations of cosubstrate yielded intersecting lines in double-reciprocal plots. Km values for both ATP and UMP and Vmax values at saturating concentrations of nucleotides are given in Table 2. ATP over 0.5 mM was inhibitory for the D77N and D201N variants, an effect independent of the concentration of Mg2+ ions. The reaction rates fitted by the equation v = Vmax. [ATP]/(KmATP + [ATP] + [ATP]2/KI) yielded KI values for these mutants of 0.64 and 0.94 mM, respectively. The Vmax of the D146N mutant represented only 0.14% of that of wild-type protein. This mutant was affected little or not at all in the Km for ATP and UMP or in its interaction with allosteric effectors, which suggests that Asp146 is involved primarily in catalysis. The D174N mutant, whose Vmax represented 30% of that of wild-type protein, exhibited a 22-fold increase in Km for UMP. The R62H mutant was affected in Vmax and Km for both substrates.
TABLE 2.
Kinetic parameters of the modified forms of E. coli UMP kinase at two pH valuesa
| Enzyme | pH 6
|
pH 8
|
||||
|---|---|---|---|---|---|---|
| KmUMP (mM) | KmATP (mM) | VmaxUMP, ATP (μmol/min/mg of protein) | KmUMP (mM)b | KmATP (mM) | VmaxUMP, ATP (μmol/min/mg of protein) | |
| Wild typec | 0.17 ± 0.03 | 0.048 ± 0.004 | 105 ± 5 | 0.043c ± 0.002 | 0.12c ± 0.018 | 128 ± 11 |
| Mutants | ||||||
| R62H | 1.26 ± 0.21 | 0.24 ± 0.03 | 1.5 ± 0.03 | 0.32 ± 0.04 | 3.0 ± 0.2 | 4.4 ± 0.1 |
| D77N | 1.78 ± 0.30 | 0.12 ± 0.02 | 1.9 ± 0.20 | 0.47 ± 0.06 | 0.48 ± 0.12 | 8.2 ± 0.3 |
| D146N | 0.45 ± 0.009 | 0.03 ± 0.006 | 0.15 ± 0.01 | 0.059 ± 0.02 | 0.10 ± 0.03 | 0.32 ± 0.03 |
| D159N | 0.27 ± 0.06 | 0.095 ± 0.003 | 128 ± 13 | 0.052c ± 0.02 | 0.29c ± 003 | 153 ± 5 |
| D168N | 0.32 ± 0.06 | 0.14 ± 0.006 | 63 ± 4.2 | 0.06 ± 0.003 | 0.42 ± 0.01 | 86.6 ± 19 |
| D174N | 3.70 ± 0.36 | 0.17 ± 0.04 | 32 ± 3.5 | 0.31 ± 0.06 | 0.31 ± 0.06 | 45 ± 5 |
| D201N | 0.86 ± 0.16 | 0.20 ± 0.04 | 1.04 ± 0.20 | 0.22 ± 0.06 | 0.46 ± 0.09 | 8.2 ± 0.3 |
The reaction medium is described in Materials and Methods. The apparent Km for ATP and UMP was determined at a single, fixed concentration of cosubstrates. The VmaxUMP, ATP was obtained by extrapolating the reaction rates for infinite concentrations of ATP and UMP and by assuming that the concentration of one nucleotide substrate does not affect the apparent Km of the second nucleotide substrate. The mean values and the standard errors of the means are from three separate experiments.
At pH 8 (or 7.4), where inhibition by excess of UMP occurred, the reaction rates were fitted by the equation v = Vmax [UMP]/(KmUMP + [UMP] + [UMP]2/KI). The calculated KI values were 1.2 mM (wild type), 0.7 mM (D168N), 0.4 mM (D174N), and 0.65 mM (D159N). The other variants of E. coli UMP kinase did not exhibit inhibition by excess of UMP.
From reference 15, kinetic measurements being done at pH 7.4.
When measured at pH 8, the kinetic constants of the wild-type UMP kinase and those of the site-directed mutants revealed several differences from those determined at pH 6. UMP exerted an inhibitory effect on the wild type and the D146N, D159N, and D168N mutants. The KI (between 0.4 and 1.2 mM) was calculated by fitting reaction rates by the equation v = Vmax [UMP]/(KmUMP + [UMP] + [UMP]2/KI). On the other hand, the relative Vmax values of R62H, D77N, and D201N mutants, compared to that of the parent enzyme, were higher at alkaline pH than at acidic pH, and the increase of Km for ATP of the R62H mutant versus that of the wild-type protein was larger at pH 8 than at pH 6.
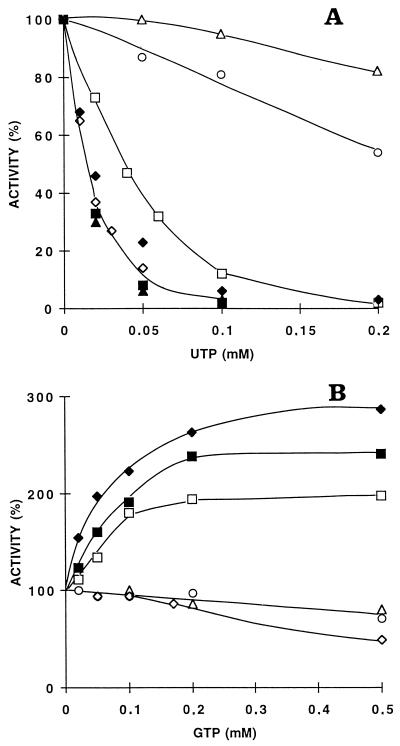
The sensitivity of the UMP kinase variants to UTP and GTP was also examined under different conditions. The effect of these nucleotides on UMP kinase depends not only on the pH but also on the concentration of UMP. Thus, at pH 8 and a low (0.1 mM) UMP concentration, the inhibitory effect of UTP is maximal. On the other hand, the activating effect of GTP is most visible at the same pH but in the presence of a high (1 mM) concentration of UMP (15). The wild type and the D159N, D168N, D174N, and D201N mutants were half inhibited at less than 25 μM UTP, whereas the 50% inhibitory concentration for the D146N mutant was 40 μM. At these concentrations of nucleotide, the D77N mutant was fully active and the R62H mutant was only slightly inhibited. The R62H and D77N mutants were insensitive to the activation by GTP, whereas the D201N mutant was inhibited by GTP over 0.2 mM. The other variants of UMP kinase as well as the wild-type enzyme were activated by GTP by a factor of between 2 and 4 (Fig. 2).
FIG. 2.
Dependence of the activities of various mutants of E. coli UMP kinase on UTP (A) and GTP (B). The reaction medium buffered with Tris-HCl (pH 8) is described in Materials and Methods. The concentrations of ATP (1 mM) and UMP (0.1 mM [A] and 1 mM [B]) were kept constant; 100% corresponds to the activity of each mutant in the absence of effectors. ○, R62H; ▵, D77N; □, D146N; ⧫, D159N; ▪, D168N; ▴, D174N; ◊, D201N.
Fluorescence analysis of UMP kinase variants.
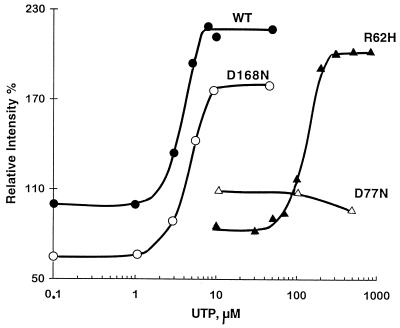
Binding of UTP to E. coli UMP kinase is a cooperative process. It is accompanied by a 2.2-fold enhancement in the intrinsic fluorescence of the wild-type protein (14, 15). The modified forms of UMP kinase exhibiting sensitivity to inhibition by UTP similar to that of the wild-type protein behaved very similarly to the parent molecule with respect to fluorescence. The fluorescence of the R62H mutant was sensitive to the addition of UTP, but as expected from inhibition studies, much higher concentrations of nucleotide (>75 μM) were required to enhance its fluorescence than in the case of the wild-type enzyme. The D146N variant of UMP kinase exhibited an intrinsic fluorescence 1.8-fold higher than that of the wild-type protein. The protein was still sensitive to the addition of UTP, and the overall increase in fluorescence upon addition of UTP was similar to that of the wild-type protein. The fluorescence of the D77N mutant was insensitive to the enhancing effect of UTP in the range of concentration used in our experiments (Table 1 and Fig. 3).
FIG. 3.
Fluorescence analysis of the interaction between the modified forms of UMP kinase and UTP. UMP kinase (1 μM in terms of monomer) in 50 mM Tris-HCl (pH 7.4) and 100 mM NaCl was titrated at 332 nm (wild type), 333 nm (R62H and D77N), or 334 nm (D168N) with UTP (between 0.1 and 700 μM). The fluorescence intensity of wild-type UMP kinase in the absence of UTP was considered 100%.
DISCUSSION
UMP kinase from E. coli is in many respects a unique member of the NMP kinase family. The homohexameric protein, whose activity is regulated by GTP and UTP, uses UMP as single phosphate acceptor and ATP or dATP as donors. UMP kinase genes have been identified by gene sequencing in many other bacteria (Haemophilus influenzae, Mycobacterium tuberculosis, Mycoplasma pneumoniae, Chlamydia trachomatis, and Synechocystis sp.). The E. coli UMP kinase gene is identical with the smbA gene, originally recognized as involved in proper chromosomal positioning during cell division (17). The homology of E. coli UMP kinase with the pyrH gene product from other organisms or with other nucleotide binding enzymes served to target the putative catalytic or regulatory residues. However, conserved and essential residues are not synonymous, and loss of a specific function by single amino acid substitution might result from local or propagated conformational effects. To minimize such undesired effects, the six Asp residues selected for site-directed mutagenesis were substituted by Asn as the most conservative replacement with respect to the size and the polar effect of this side chain. With these inherent limitations of mutational analysis in mind, we shall concentrate on those results that help to decipher a protein whose function in the bacterial cell is not well understood.
(i) From the seven amino acids of UMP kinase substituted by site-directed mutagenesis, two (D146 and D174) showed significant alteration of a single kinetic parameter, the Vmax and the KmUMP, respectively. The activation by GTP or the inhibition by UTP, the solubility, or the resistance against denaturation of these variants were similar to that of the wild-type protein. One might therefore conclude that Asp146 is involved in catalysis, whereas Asp174 is primarily responsible for binding of UMP. The specific role of these residues can be tentatively assigned by comparing UMP kinase with enzymes whose structure in complex with inhibitors, substrates, or reaction products is known at high resolution (8, 13, 18). It is conceivable that Asp174 interacts with the hydroxyl group(s) of the ribose ring in UMP. Similarly, we suppose that Asp146 in UMP kinase, like Asp84 in E. coli adenylate kinase or Asp89 in D. discoideum UMP kinase, stabilizes the transition state (by ca. 3.9 kcal/mol) by interaction with MgATP.
(ii) Although affected in several kinetic parameters, the major characteristic of D77N and R62H mutants of bacterial UMP kinase is the loss of activation by GTP and a substantial decrease in affinity for UTP. For this reason, we assigned Arg62 and Asp77 as regulatory residues. Their location in the N-terminal third of the polypeptide chain, far from the catalytic residues Asp146 and Asp174, might indicate that the regulatory and active sites of UMP kinase belong to separate domains. Deletion mutagenesis, which might give an answer to this question, was not conclusive. Even short deletions (up to 25 amino acid residues) from the N-terminal end of UMP kinase yielded essentially insoluble and inactive proteins.
(iii) Among E. coli smbA mutants responsible for the altered morphological phenotype under nonpermissive conditions, cold-sensitive growth, and hypersensitivity to SDS, two resulted from substitution of Arg62 with His (smbA9) and of Asp201 with Asn (smbA2) (17). The two modified UMP kinases have as common properties a low catalytic activity and loss of activation by GTP. They differ markedly in thermal stability and sensitivity to inhibition by UTP. Assuming that participation of UMP kinase in cell proliferation depends solely on its role in the synthesis of UTP, the relationship between the altered catalytic properties and the pleiotropic phenotype of smbA2 and smbA9 mutants is straightforward. As a building block for synthesis of different forms of RNAs including mRNA or of various uridinyl sugars, UTP is essential for cell growth and division. This is possibly an oversimplified view which does not consider the complex structure of UMP kinase. It is therefore legitimate to hypothesize another function of the pyrH/smbA gene product, independent of its catalytic activity. Charlier et al. (3) suggested a direct role of UMP kinase in the regulation of the upstream, pyrimidine-specific promoter P1 of the carAB operon. These authors identified a pyrH mutant in which the modified enzyme (A94E) has a quasi-normal catalytic activity. Overexpression of the mutant pyrH allele results in UMP kinase levels that far exceed that of the wild-type strain but does not restore normal pyridimine-mediated repression.
ACKNOWLEDGMENTS
We are grateful to H. Sakamoto for valuable help in proposing several UMP kinase variants, D. Charlier for inspiring discussion, and M. Ferrand for excellent secretarial assistance. S. Landais is deeply indebted to J.-C. Mazié for continuous support. N. Bucurenci is grateful to M. Simionescu for help with spectrofluorimetric experiments.
This work was supported by grants from the Centre National de la Recherche Scientifique (URA 1129), the Institut Pasteur, France, and the Ministry for Research and Technology, Romania.
REFERENCES
- 1.Blondin C, Serina L, Wiesmüller L, Gilles A-M, Bârzu O. Improved spectrophotometric assay of nucleoside monophosphate kinase activity using the pyruvate kinase/lactate dehydrogenase coupling system. Anal Biochem. 1994;220:219–221. doi: 10.1006/abio.1994.1326. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Charlier D, Kholti A, Gigot M, Roovers M, Huysveld N, Glansdorff N. Abstracts of the 24th FEBS Meeting. Heidelberg, Germany: Springer International; 1996. p. PS4.4-09. [Google Scholar]
- 4.Konrad M. Cloning and expression of the essential gene for guanylate kinase from yeast. J Biol Chem. 1992;267:25652–25655. [PubMed] [Google Scholar]
- 5.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1985;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 6.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 7.Liljelund P, Sanni A, Priesen J D, Lacroute F. Primary structure of the S. cerevisiae gene encoding uridine monophosphokinase. Biochem Biophys Res Commun. 1989;165:464–473. doi: 10.1016/0006-291x(89)91093-0. [DOI] [PubMed] [Google Scholar]
- 8.Müller C W, Schulz G E. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 Å resolution. A model for a catalytic transition state. J Mol Biol. 1992;224:159–177. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]
- 9.Müller-Dieckmann H-J, Schulz G E. The structure of uridylate kinase with its substrates, showing the transition state geometry. J Mol Biol. 1994;236:361–367. doi: 10.1006/jmbi.1994.1140. [DOI] [PubMed] [Google Scholar]
- 10.Munier H, Gilles A-M, Glaser P, Krin E, Danchin A, Sarfati R S, Bârzu O. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur J Biochem. 1991;196:469–474. doi: 10.1111/j.1432-1033.1991.tb15838.x. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 12.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheffzek K, Kliche W, Wiesmüller L, Reinstein J. Crystal structure of the complex of UMP/CMP kinase from Dictyostelium discoideum and the bisubstrate inhibitor P1-(5′-adenosyl)P5-(5′-uridyl) pentaphosphate (UP5A) and Mg2+ at 2.2 Å: implications for water-mediated specificity. Biochemistry. 1996;35:9716–9727. doi: 10.1021/bi960642s. [DOI] [PubMed] [Google Scholar]
- 14.Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, Gilles A-M, Bârzu O. Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine nucleotides and UTP. Biochemistry. 1995;34:5066–5074. doi: 10.1021/bi00015a018. [DOI] [PubMed] [Google Scholar]
- 15.Serina L, Bucurenci N, Gilles A-M, Surewicz W K, Fabian H, Mantsch H H, Takahashi M, Petrescu I, Batelier G, Bârzu O. Structural properties of UMP-kinase from Escherichia coli: modulation of protein solubility by pH and UTP. Biochemistry. 1996;35:7003–7011. doi: 10.1021/bi960062v. [DOI] [PubMed] [Google Scholar]
- 16.Smallshaw J, Kelln R A. Cloning, nucleotide sequence and expression of the Escherichia coli K-12 pyrH gene encoding UMP kinase. Genetics (Life Sci Adv) 1992;11:59–65. [Google Scholar]
- 17.Yamanaka K, Ogura T, Niki H, Hiraga S. Identification and characterization of the smbA gene, a suppressor of the mukB null mutant of Escherichia coli. J Bacteriol. 1992;174:7517–7526. doi: 10.1128/jb.174.23.7517-7526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan H, Tsai M-D. Mechanism of adenylate kinase. Demonstration of a functional relationship between aspartate 93 and Mg2+ by site-directed mutagenesis and proton, phosphorus-31 and magnesium-25 NMR. Biochemistry. 1991;30:5539–5546. doi: 10.1021/bi00236a029. [DOI] [PubMed] [Google Scholar]