Abstract
The transcription factor CYS3 of Neurospora crassa is a positive regulator of the sulfur regulatory circuit which contains many structural genes involved in sulfur metabolism. Expression and degradation of the CYS3 protein are precisely regulated in a sulfur-dependent manner. cys-3 expression was found to be fully repressed by high concentrations of methionine or inorganic sulfate present in the culture medium and to be derepressed when these favored sulfur sources were limited. cys-3 transcripts could be readily detected within 2 h after derepression, whereas the CYS3 protein was not found until after 4 h. CYS3 is stable, with a half-life greater than 4 h under low-sulfur conditions when it is required for cell growth. However, it is degraded relatively quickly when methionine or inorganic sulfate becomes available. Upon sulfur repression, cys-3 transcripts disappeared within 30 min with an estimated half-life of 5 min whereas CYS3 protein almost entirely disappeared in 1 h with a half-life of approximately 10 min. These results suggest that a selective elimination of CYS3 is a highly regulated process. Site-directed mutagenesis showed that Lys-105 of CYS3 is important for its instability. The change of this single residue from lysine to glutamine resulted in a prolonged half life of CYS3 and impaired responsiveness of CYS3 degradation to sulfur level changes.
Sulfur uptake and assimilation are subjected to sophisticated metabolic controls in the filamentous fungus Neurospora crassa. Sulfur-containing amino acids and inorganic sulfate are preferred sulfur sources for N. crassa. When the favored sulfur sources are missing or limited, other, less readily used sulfur compounds like choline-O-sulfate and aromatic sulfates can be utilized. The utilization of secondary sulfur sources requires the de novo synthesis of a set of catabolic enzymes including two sulfate permeases, a methionine-specific permease, aryl sulfatase, choline sulfatase, and an extracellular protease. Several regulatory genes, namely, cys-3, scon-1, and scon-2, form a hierarchical circuit, with cys-3 playing the decisive role to ensure that N. crassa cells have a steady supply of sulfur for growth (6, 12, 13, 16, 18–20). Of the three regulatory genes, cys-3 has been identified as the key positive regulator for the sulfur circuit. Expression of CYS3 protein under sulfur-limited conditions is a prerequisite for the expression of all of the structural genes in the sulfur circuit. A cys-3 null mutant is unable to express any of the sulfur catabolic enzymes and fails to grow on any of the secondary sulfur sources (13). scon-1 and scon-2 are identified as negative regulators for the sulfur circuit. Mutations in either gene lead to constitutive expression of all the catabolic enzymes regardless of the availability of sulfur sources (1). The CYS3 protein is expressed in these mutants in repression conditions, although a higher expression level still occurs upon derepression (20a). Heterokaryon studies showed that the function of scon-1 was intranuclear (1, 16). It appears that scon-1 prevents CYS3 expression and thus stops expression of the structural genes of the sulfur circuit during repression conditions (3, 18). The scon-2 gene, which encodes a protein with β-transducin repeats, also acts as a negative regulator for the expression of cys-3 (9, 18). Interestingly, recent studies showed that CYS3 protein acts positively for the expression of scon-2 via CYS3 binding sites located in scon-2 promoter region (9, 18).
As a basic region-leucine zipper (bZIP) transcription factor, CYS3 protein shares substantial homology to members of the bZIP protein family, especially Jun, Fos, and GCN4, and recognizes the consensus palindrome sequence ATGRYRYCAT (4, 5, 11). CYS3 binding sites have been found in the promoter regions of the cloned structural genes of the sulfur circuit, providing molecular evidence that links the expression of sulfur catabolic enzymes to CYS3 (9, 18). Promoter studies suggest that the expression of CYS3 is autoregulated via CYS3 binding sites in the promoter region of cys-3, which enables N. crassa to produce a large pool of CYS3 protein when cells encounter a shortage of sulfur (4). However, whether this CYS3 protein pool is eliminated in a timely fashion when the sulfur supply becomes abundant again is not known.
Studies conducted on cys-3 mutants and the downstream structural genes have suggested that the expression of cys-3 is precisely regulated by the availability of sulfur. Under sulfur-sufficient condition, expression of cys-3 as well as the sulfur circuit is fully repressed. As the sulfur level decreases, derepression of CYS3 synthesis takes place, which subsequently leads to expression of the entire set of sulfur catabolic enzymes (19). However, little is known about the transcriptional or translational regulation of CYS3 expression. In this report, we have examined regulation of cys-3 transcription and protein synthesis and degradation. Interestingly, we observed that the CYS3 protein is subjected to differential turnover, depending on the cellular sulfur supply. Site-directed mutagenesis plus analysis of N. crassa mutant strains have indicated that Lys-105 of CYS3 is important for its instability.
MATERIALS AND METHODS
N. crassa and E. coli strains.
The N. crassa wild-type strain 74-OR23-1A and cys-3 (allele P22) and cys-3 (NM27t) mutants were obtained from the Fungal Genetics Stock Center, University of Kansas Medical Center, Kansas City, Mo. The cys-3 revertant REV21 and cys-3 temperature-sensitive revertant REV65 were described before (13). Mycelia were cultured in Vogel’s liquid medium (2) with shaking. The amount of methionine supplement and the culture temperature are indicated for each experiment. Escherichia coli DH5α was used for plasmid propagation, E. coli CJ236 was used for single-stranded DNA template preparation, and E. coli BL21(DE3)(pLys) was used for protein expression.
To study the effect of derepression on cys-3 expression, N. crassa mycelia were initially grown overnight in 5 mM methionine as the sole sulfur source, then transferred to medium containing 0.25 mM methionine for derepression, and harvested at 1-h intervals for 8 h. For repression studies, mycelia were first cultured in a low-sulfur condition (0.25 mM methionine) overnight to allow the expression of CYS3. After methionine was increased to a repression concentration (5 mM), mycelia were harvested at 1-h intervals for 5 h.
Site-directed mutagenesis and CYS3 expression in E. coli.
Site-directed mutagenesis was used to generate a new cys-3 mutation, K105→Q105 (K105Q). A single-stranded DNA oligonucleotide containing the desired mutation (5′GCC GAG GAA GAC CAG CGA AAG CGC3′) was synthesized and annealed to the single-stranded cys-3 DNA template, which was generated by using E. coli CJ236. The daughter strand was synthesized in vitro, and the double-stranded product was used to transform E. coli. The mutant was selected and confirmed by DNA sequencing. The construct used for expressing wild-type CYS3 protein in E. coli was described before (4). The construct carrying cys-3 REV21 was generated by Kristin Coulter. The E. coli expression construct carrying the K105Q mutation was made by replacing the corresponding wild-type sequence in the coding region of cys-3 with the mutated sequence. CYS3 was expressed and purified as described previously (4).
Mobility shift assay.
DNA mobility shifts were carried out as described previously (4), with minor modifications. E. coli-expressed CYS3 proteins (0.1 to 1.0 μg) were used in the mobility shift assays. A 200-bp DNA fragment from the cys-14 promoter containing a strong CYS3 binding site was used as the probe. The mixtures of proteins and DNA were incubated at room temperature for 20 min in a total volume of 25 μl of binding buffer (12 mM HEPES, 4 mM Tris-HCl [pH 7.9], 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 0.3 mg of bovine serum albumin per ml, 10% glycerol) with 3 μg of poly(dI-dC) as a nonspecific competitor. Samples were separated on 4% polyacrylamide gels (acrylamide/bisacrylamide = 19:1) in 1/4× Tris-borate-EDTA buffer.
RNA preparation and Northern analysis.
Isolation of total RNA from N. crassa was done as described by Weaver et al. (22), with modifications. Mycelia were ground to a fine powder with a mortar and pestle in the presence of liquid nitrogen and suspended in lysis buffer (50 mM sodium acetate [pH 5.3], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS]) at a ratio of 1 g/5 ml. An equal volume of acidic phenol-chloroform (equilibrated with lysis buffer without SDS [pH 5.3], prewarmed at 65°C) was added to the lysate, and the reaction mixture was incubated at 65°C for 30 min with shaking. The aqueous phase was recovered by centrifugation, and this process was repeated several times until no protein interface could be seen. Total RNA was precipitated with 0.6 volume of isopropyl alcohol and then centrifuged. The RNA samples were fractionated in agarose gels and transferred to nitrocellulose filters.
cys-3 cDNA and β-tubulin cDNAs were labeled by random primer labeling using 32P-labeled dATP as probes for Northern analysis (GIBCO/BRL). The hybridization was carried out at 65°C in 1× hybridization buffer (0.5 M NaCl, 0.1 M NaPO4 [pH 7.0], 6 mM EDTA, 1% SDS). The membranes were washed at 65°C for 60 min with 1/4× hybridization buffer. Messages of cys-3 and β-tubulin were identified by their sizes.
Western analysis.
Mycelia were homogenized with a mortar and pestle in the presence of liquid nitrogen and suspended in 2 ml ice-cold lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 0.2% sodium azide, 100 μg of phenylmethylsulfonyl fluoride per ml, 1% Triton X-100, 2 mM EDTA). Cell debris was removed by centrifugation at 4°C for 5 min at full speed in a microcentrifuge, and the supernatant was subjected to Western analysis. The protein concentration of the cell lysate was determined by the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.). An equal amount of protein for each sample was run on SDS–15% polyacrylamide gels and transferred to a nitrocellulose filter by electroblotting in glycine buffer (25 mM Tris-HCl [pH 8.3], 192 mM glycine, 20% methanol) as described previously (7). After blotting, the filter was briefly rinsed with TBST (25 mM Tris-HCl [pH 8.0], 125 mM NaCl, 0.05% Tween 20) and was blocked by TBST containing 4% nonfat milk for 1 h at room temperature or at 4°C overnight. The filter was then incubated with rabbit anti-CYS3 antibodies for 1 h with shaking at room temperature (8). The filter was washed with TBST and then incubated with horseradish peroxidase-conjugated secondary antibodies (anti-immunoglobin) (1:7,000 dilution, in blocking buffer) for 45 min. After further washing, the antigen-antibody complex on the nitrocellulose filter was detected by the in situ chemiluminescence reaction (ECL system; Amersham), and the result was visualized on X-ray film.
Aryl sulfatase assay.
Mycelia were grown at 30°C in liquid Vogel’s minimum medium containing 0.25 mM methionine as the only sulfur source for 16 to 20 h. Cell lysates were prepared as described above, and protein concentrations were determined by the Bio-Rad protein assay. The aryl sulfatase enzyme assay was performed as described before with a few modifications (17). Cell lysate of 50 μl was mixed with 450 μl of the aryl sulfatase enzyme assay cocktail buffer (6.7 mM p-nitrophenol sulfate, 0.33 M Tris-HCl [pH 8.1]). The reaction mixtures were incubated at 30°C for 10 to 60 min, depending on the amount of activity, and 1 ml of stop solution (0.5 M NaOH, 90% ethyl alcohol) was added. For the time-zero control, 1 ml of stop solution was added to the mixture without incubation. The reaction mixtures were centrifuged in a microcentrifuge at full speed for 5 min to remove the precipitates. The absorbance at 405 nm of the supernatant was measured with a spectrophotometer.
RESULTS
Kinetics of CYS3 expression.
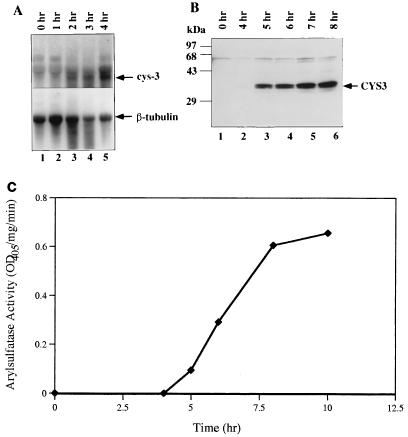
The expression of CYS3 and the catabolic enzymes of the sulfur circuit are precisely regulated by the availability of sulfur. Neither is expressed in the presence of a high concentration of methionine (5 mM) or inorganic sulfate (2 mM). However, when the sulfur level in the medium is limited (0.25 mM methionine or 0.02 mM inorganic sulfate), the expression of CYS3 and subsequently of the sulfur catabolic structural genes is derepressed. To determine the kinetics of cys-3 mRNA and protein upon derepression, Northern and Western analyses were carried out. Northern analysis using cys-3 cDNA as a probe showed that upon sulfur derepression, the synthesis of cys-3 mRNA occurs only slowly. No cys-3 transcripts were detected in RNA from cells under the repression condition or within 1 h after derepression (Fig. 1A, lanes 1 and 2). cys-3 mRNA first appeared at a low level 2 h after derepression and increased for 2 more h before leveling off (Fig. 1A, lanes 3 to 5). The bottom panel of Fig. 1A shows the same filter probed with β-tubulin gene as an internal control, indicating approximately equal loading of RNA in each lane. The expression of CYS3 protein is an even slower process. Western analysis using rabbit anti-CYS3 antibodies showed no detectable CYS3 protein accumulation within the first 3 h after depression. CYS3 protein first appeared at 4 h and kept increasing in the next 4 h before reaching a plateau (Fig. 1B). The CYS3 protein actually appears as two distinct bands (Fig. 1B), suggesting the possibility of posttranslational modification. Aryl sulfatase, a downstream structural gene regulated by CYS3, was chosen as an indicator for expression of the sulfur circuit. The enzyme activity was detected about 5 h after depression (Fig. 1C).
FIG. 1.
Derepression of cys-3. The derepression process was monitored by shifting wild-type N. crassa mycelia from medium with a repressing level of methionine (5 mM) to medium with a derepressing level of methionine (0.25 mM). Mycelia were harvested, and total RNA and protein extracts were prepared. The harvest time of each sample is indicated at the top. (A) RNA blot analysis of cys-3 transcript (1.6 kb) and β-tubulin (2.0 kb) control; (B) Western blot analysis of CYS3 protein, using rabbit anti-CYS3 antibodies; (C) time course of aryl sulfatase activity. OD405, optical density at 405 nm.
Kinetics of cys-3 mRNA and protein turnover.
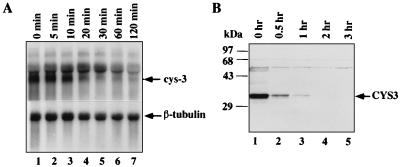
When cells growing under sulfur-limited conditions encounter an abundance of sulfur, the multiple sulfur-regulated permeases and enzymes are no longer needed. It was of interest to determine whether the sulfur circuit was shut down in response to repression conditions. We found that upon establishment of sulfur repression, the cys-3 mRNA and protein turn over in a rapid fashion. Northern analysis showed that the disappearance of cys-3 mRNA was very quick, and the transcript was undetectable after 20 min. The estimated half-life of mRNA at this transition stage was approximately 5 min (Fig. 2A). Western analysis indicated that the turnover of CYS3 protein is also a rapid process, with an estimated half-life of about 10 min (Fig. 2B). CYS3 nearly completely disappeared within 1 h after sulfur repression.
FIG. 2.
Turnover of cys-3 mRNA and protein. The repression kinetics was examined at the transition stage when the methionine level in culture was increased from low (0.25 mM) to high (5 mM). 74A mycelia were harvested at different times after the increase of Met in the medium and were used to prepare total RNA and protein extract. The half-lives of cys-3 mRNA and protein were estimated by using the SigmaGel program. (A) RNA blot analysis of cys-3 transcript; (B) immunoblot analysis of CYS3 protein.
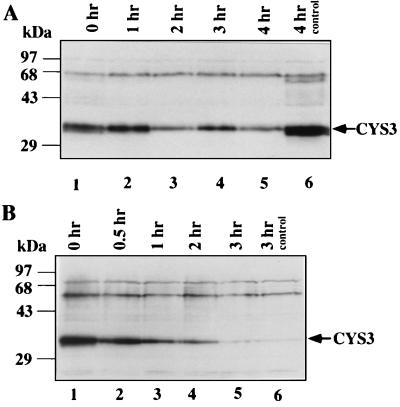
The stability of CYS3 protein during sulfur derepression conditions was also determined. Cycloheximide was added to the culture medium at 2 × 10−4 M to block de novo protein synthesis. Western analysis suggested that CYS3 was very stable under the low-sulfur condition, with an estimated half-life of about 4 h (Fig. 3A). A control experiment using the same concentration of cycloheximide to block the induction of Neurospora nitrate reductase was conducted at the same time. Nitrate reductase enzyme assay indicated that protein synthesis was completely inhibited under this condition (data not shown).
FIG. 3.
Differential elimination of CYS3 protein under different culture conditions. (A) The presence of CYS3 protein was examined after introduction of cycloheximide (2 × 10−4 M) to medium containing a derepressing amount of methionine (0.25 mM). Samples were harvested at different times after the addition of cycloheximide. In lane 6, a control sample grown without cycloheximide was harvested at the same time as the sample in lane 5, which was grown in cycloheximide for 4 h. (B) The presence of CYS3 protein was examined after shifting N. crassa mycelia from medium of derepression condition (0.25 mM methionine) to repression condition (5 mM methionine) and introducing cycloheximide to 2 × 10−4 M.
The 4-h half-life of CYS3 protein under the low-sulfur condition was confirmed by using a cys-3 temperature-sensitive mutant, REV65, which behaves as the wild type at 27°C but as a cys-3 null mutant at 37°C, when it fails to synthesize CYS3 as well as all the downstream sulfur catabolic enzymes. By shifting the culture temperature from 27 to 37°C, CYS3 protein synthesis is halted without altering the normal cell growth conditions. Western analysis showed that the preexisting CYS3 protein was very stable under the low-sulfur condition (data not shown). These results strongly suggest that a differential turnover of CYS3 occur under different growth conditions.
Since cycloheximide is known to selectively stabilize some highly unstable proteins, apparently due to a block of the synthesis of proteases, it was important to determine whether cycloheximide would stabilize CYS3 in cells transferred from low- to high-sulfur medium. When high-methionine medium and cycloheximide (2 × 10−4 M) were added to a culture at the same time, we found that cycloheximide did stabilize CYS3 somewhat, giving a longer half-life of about 30 min (Fig. 3B); however, the turnover of CYS3 was still much faster than under sulfur-limited conditions. The difference in the turnover rate of CYS3 under different growth conditions suggests that a specific degradation pathway is involved in CYS3 elimination when it is no longer needed.
Lysine 105 of CYS3 is required for rapid sulfur-regulated degradation.
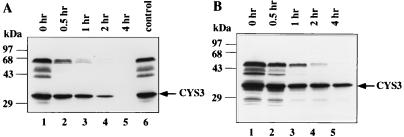
REV21, which was isolated as a revertant of the cys-3 null mutant, carries three changed amino acid residues, K105Q, R106Q, and F116Y (1a, 5). Analysis of expression of the downstream structural gene, aryl sulfatase, revealed that in REV21, the mutated CYS3 had an impaired transactivation function and retained about 10% of the activity of the wild-type protein (results not shown). However, Western analysis indicated that the CYS3 protein level was significantly elevated in REV21 compared to the wild-type strain under the derepression condition (data not shown). Since the expression of CYS3 protein is autoregulated, the CYS3 level in REV21 would be expected to be lower than in the wild type. Mobility shift analysis suggested that the reduction of transactivation activity was primarily due to a lower affinity of DNA binding, as shown below. One possible explanation for the elevated CYS3 protein level is that the stability of CYS3 protein in REV21 is changed. To examine this possibility, the stability of the mutant CYS3 protein was examined at the low-sulfur-to-high-sulfur transition stage. Indeed, the mutant CYS3 protein of REV21 was found to have a prolonged half-life of about 40 min, compared to 10 min for the wild type, and was still present after 2 h (Fig. 4). Thus, the amino acid substitutions appears to alter the turnover of the CYS3 of REV21 protein during sulfur repression, perhaps by partially hindering a sulfur-triggered degradation of CYS3.
FIG. 4.
Turnover of CYS3 mutant proteins upon sulfur repression. The turnover of CYS3 proteins was monitored after shifting N. crassa mycelia from low-sulfur medium to high-sulfur medium. The harvest time of each mycelial sample is indicated at the top. CYS3 proteins were detected by Western analyses. (A) Sulfur-regulated turnover of REV21 CYS3 protein; (B) sulfur-regulated degradation of K105Q CYS3 protein.
Since lysine is often modified for ubiquitin-controlled proteolysis, we constructed a new mutant, K105Q which has only one amino acid substitution, lysine 105, changed to glutamine. The K105Q mutant was transformed into the N. crassa cys-3 null mutant. Transformants carrying K105Q showed proper response to sulfur regulation, with about 20% transactivation activity comparing to wild-type CYS3 protein. The sulfur-regulated degradation of the CYS3 K105Q was examined at the low-sulfur-to-high-sulfur transition stage. Western analysis revealed that like REV21, CYS3 K105Q had a prolonged half life of about 40 min (Fig. 4B).
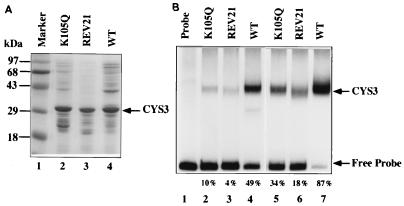
The DNA binding abilities of K105Q, REV21, and wild-type CYS3 proteins were examined. These three forms of CYS3 were expressed in E. coli and were partially purified (Fig. 5A). An identical amount of each CYS3 protein was used in a mobility shift analysis. As shown in Fig. 5B, analyses with two concentrations of proteins revealed that wild-type CYS3 protein has strong DNA binding ability, while the two mutant forms of CYS3 were significantly weaker in DNA binding. To measure the percentage shift of the probe, the density of each band was quantified with a phosphorimager; the percentage shift of the probe in each reaction is indicated at the bottom of Fig. 5B. The K105Q possesses about 20% of the wild-type protein’s binding affinity, while REV21 possesses only about 10%. This explains why these mutant proteins only poorly turn on the structural genes despite their presence in amounts greater than the wild-type level.
FIG. 5.
DNA mobility shift analysis of expressed CYS3 proteins. (A) Wild-type (WT) and mutants CYS3 proteins were expressed in E. coli and were partially purified. A Coomassie blue-stained SDS-polyacrylamide gel was used to confirm the presence of CYS3 proteins. (B) DNA mobility shift experiment. Lanes 2 to 4 contained 0.2 μg of protein; lanes 5 to 7 contained 0.6 μg of protein. A 200-bp DNA fragment taken from the cys-14 promoter containing a strong CYS3 binding site was end labeled by 32P and was used for the mobility shift assay. Free DNA fragment and CYS3-DNA complex are indicated by arrows. The percentage shift of the probe in each reaction as determined by a phosphorimager is indicated at the bottom.
DISCUSSION
The CYS3 protein plays the central role in controlling the expression of an entire family of structural genes which encode enzymes for acquisition of sulfur. CYS3 is composed of 236 amino acids and has a bZIP domain that confers sequence-specific DNA binding to elements with the consensus sequence ATGRYRYCAT. The cys-3 gene is itself highly regulated by the negative-acting scon genes and is not expressed, or is expressed only very weakly, during sulfur repression conditions. Thus, when cells growing with repressing levels of sulfur (i.e., 5 mM methionine) experience derepression conditions (i.e., 0.25 mM methionine), expression of cys-3 must occur before the various structural genes can be activated. Synthesis of aryl sulfatase occurs only approximately 5 h after wild-type cells are transferred from high-sulfur to low-sulfur conditions. It was of interest to determine the time course of cys-3 expression in cells undergoing the transition from sulfur repression to derepression conditions. We found that this is a very slow process, requiring approximately 2 h before cys-3 mRNA can be detected and an additional 2 to 3 h to reach a maximum level. It required at least 4 h following derepression to detect the CYS3 protein, which apparently explains the long delay in appearance of aryl sulfatase activity after cells are shifted from high to low levels of sulfur. Further reduction of the methionine concentration to 0.025 mM or use of a sulfur-free medium only slightly shortened the time period (to about 3 h) before the CYS3 protein could be detected by Western analysis (data not shown). The slow response of the sulfur regulatory circuit to the shift from sulfur repression to derepression conditions almost certainly is due to the presence of a substantial internal pool of sulfur compounds, which must be utilized before the cells actually experience a sulfur limitation. The ability to accumulate stored forms of sulfur, e.g., choline-O-sulfate, insures that Neurospora has sufficient intracellular sulfur for prolonged growth even in sulfur-poor environments before expression of the entire family of sulfur catabolic enzymes becomes necessary.
Another significant change in environmental conditions occurs when cells growing under sulfur starvation conditions, with the sulfur circuit fully activated, suddenly encounter an abundance of sulfur. Under the new conditions of excess sulfur, the sulfur catabolic enzymes would no longer be required. We found that upon establishment of sulfur repression, synthesis of cys-3 mRNA apparently stopped immediately and the preexisting mRNA turned over rapidly and was completely gone within 20 min. Moreover, the CYS3 protein also turned over quickly, with a half-life of approximately 10 min. The rapid loss of cys-3 mRNA and protein ensures that the expression of the sulfur structural genes is turned off quickly upon sulfur repression.
A differential rate of turnover of the CYS3 protein was observed, depending on sulfur availability. The CYS3 protein was stable, with a half-life of about 4 h, in sulfur-derepressed cells. In contrast, during sulfur repression CYS3 turned over considerably more rapidly, with a half-life estimated to be approximately 10 min. The finding that cycloheximide partially stabilized the CYS3 protein suggested that its sulfur-dependent rapid turnover depended upon de novo protein synthesis, possibly of a proteolytic enzyme. The CYS3 protein with 236 amino acids, when expressed in E. coli, runs in SDS-polyacrylamide gels as a single species with a size of 31 kDa, whereas in Neurospora cells, CYS3 appears as a doublet running at approximately 35 kDa, suggesting that it may be subject to posttranslational modification. The faster-migrating form of the doublet of CYS3 from Neurospora cells is first observed when cells are derepressed, and it is the form that disappears first when cells are subject to sulfur repression. One interesting possibility is that CYS3 is controlled by phosphorylation and dephosphorylation. An experiment to determine whether CYS3 is phosphorylated revealed a doublet of 32P-labeled protein bands around of 35 kDa which was immunoprecipitated by anti-CYS3 antibody from cells grown under the sulfur derepression condition but not from cells grown under the sulfur repression condition, although a considerable background was present (result not shown).
In an attempt to address the mechanism involved in the selective degradation of the CYS3 protein, we found that amino acid residue lysine 105 is important in determining CYS3 instability. Both cys-3 REV21 and cys-3 K105Q mutant proteins, each with lysine 105 replaced with glutamine, showed an elevated cellular accumulation and prolonged half-life. It is well known that the ubiquitin-dependent proteolytic pathway, which involves the covalent attachment of ubiquitin to specific lysine residues of target proteins, is responsible for the selective degradation of proteins in eukaryotes. The bZIP protein c-Jun, which shares certain features with CYS3, is selectively degraded by the ubiquitin-dependent pathway (21). Our results that implicate lysine 105 as a determinant of CYS3 stability suggest that the ubiquitin pathway could be involved in CYS3 turnover.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM-23367 (to G.A.M.) from the National Institutes of Health.
We thank Kristin Coulter for providing the REV21 expression construct and all of our colleagues in our lab for their thoughtful discussions and suggestions.
REFERENCES
- 1.Burton E G, Metzenberg R L. Novel mutation causing derepression of several enzymes of sulfur metabolism Neurospora crassa. J Bacteriol. 1972;109:140–151. doi: 10.1128/jb.109.1.140-151.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Coulter, K., and G. A. Marzluf. Unpublished data.
- 2.Davis R H, de Serres F J. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17A:79–143. [Google Scholar]
- 3.Dietrich P S, Metzenberg R L. Metabolic suppressors of a regulatory mutant in Neurospora. Biochem Genet. 1973;8:73–84. doi: 10.1007/BF00485558. [DOI] [PubMed] [Google Scholar]
- 4.Fu Y-H, Marzluf G A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. J Biol Chem. 1990;265:11942–11947. [PubMed] [Google Scholar]
- 5.Fu Y-H, Paietta J V, Mannix D G, Marzluf G A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a protein with a putative leucine zipper DNA-binding element. Mol Cell Biol. 1989;9:1120–1127. doi: 10.1128/mcb.9.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanson M A, Marzluf G A. Control of the synthesis of a single enzyme by multiple circuits in Neurospora crassa. Proc Natl Acad Sci USA. 1975;72:1240–1244. doi: 10.1073/pnas.72.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harlow E, Whyte P, Franza B R, Jr, Schley C. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanaan M, Marzluf G A. The positive-acting sulfur regulatory protein CYS3 of Neurospora crassa: nuclear localization, autogenous control, and regions required for transcriptional activation. Mol Gen Genet. 1993;239:334–344. doi: 10.1007/BF00276931. [DOI] [PubMed] [Google Scholar]
- 9.Ketter J S, Jarai G, Fu Y-H, Marzluf G A. Nucleotide sequence, messenger RNA stability, and DNA recognition elements of cys-14, the structural gene for sulfate permease II in Neurospora crassa. Biochemistry. 1991;30:1780–1787. doi: 10.1021/bi00221a008. [DOI] [PubMed] [Google Scholar]
- 10.Kumar A, Paietta J V. The sulfur controller-2 negative regulatory gene of Neurospora crassa encoded a protein with beta-transducin repeats. Proc Natl Acad Sci USA. 1995;92:3343–3347. doi: 10.1073/pnas.92.8.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Marzluf G A. Determination of the Neurospora crassa CYS3 sulfur regulatory protein consensus DNA-binding site: amino-acid substitutions in the CYS3 bZIP domain that alter DNA-binding specificity. Curr Genet. 1996;30:298–304. doi: 10.1007/s002940050136. [DOI] [PubMed] [Google Scholar]
- 12.Marzluf G A. Genetic and biochemical studies of distinct sulfate permease species in different developmental stage of Neurospora crassa. Arch Biochem Biophys. 1970;138:254–263. doi: 10.1016/0003-9861(70)90306-1. [DOI] [PubMed] [Google Scholar]
- 13.Marzluf G A. Genetics and molecular genetics of sulfur assimilation in fungi. Adv Genet. 1994;31:187–205. doi: 10.1016/s0065-2660(08)60398-3. [DOI] [PubMed] [Google Scholar]
- 14.Marzluf G A, Metzenberg R L. Positive control by the cys-3 locus in regulation of sulfur metabolism in Neurospora. J Mol Biol. 1968;33:423–437. doi: 10.1016/0022-2836(68)90199-x. [DOI] [PubMed] [Google Scholar]
- 15.Metzenberg R L, Ahigren S K. Structural and regulatory control of arylsulfatase in Neurospora: the use of interspecific differences in structural genes. Genetics. 1968;68:369–381. doi: 10.1093/genetics/68.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzenberg R L, Ahlgren S K. Structural and regulatory control of arylsulfatase in Neurospora: the use of interspecific differences in structural genes. Genetics. 1971;68:369–381. doi: 10.1093/genetics/68.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzenberg R L, Parson J W. Altered repression of some enzymes of sulfur utilization in a temperature-conditional lethal mutant of Neurospora. Proc Natl Acad Sci USA. 1966;55:629–635. doi: 10.1073/pnas.55.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paietta J V. Molecular cloning and analysis of the scon-2 negative regulatory gene of Neurospora crassa. Mol Cell Biol. 1990;10:5207–5214. doi: 10.1128/mcb.10.10.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paietta J V, Akins R A, Lambowitz A M, Marzluf G A. Molecular cloning and characterization of the cys-3 regulatory gene of Neurospora crassa. Mol Cell Biol. 1987;7:2506–2511. doi: 10.1128/mcb.7.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pall M L. Amino acid transport in Neuprspora crassa. II. Properties and regulation of a methionine transport system. Biochim Biophys Acta. 1971;233:201–214. doi: 10.1016/0005-2736(71)90372-5. [DOI] [PubMed] [Google Scholar]
- 20a.Tao, Y., and G. A. Marzluf. Unpublished data.
- 21.Treier M, Staszewski L, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the δ domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 22.Weaver P L, Sun C, Chang T-H. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol Cell Biol. 1997;17:1354–1365. doi: 10.1128/mcb.17.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]