Abstract
We assessed the disease-causing potential of single nucleotide polymorphisms (SNPs) based on a simple set of sequence-based features. We focused on SNPs from the dbSNP database in G-protein-coupled receptors (GPCRs), a large class of important transmembrane (TM) proteins. Apart from the location of the SNP in the protein, we evaluated the predictive power of three major classes of features to differentiate between disease-causing mutations and neutral changes: (i) properties derived from amino-acid scales, such as volume and hydrophobicity; (ii) position-specific phylogenetic features reflecting evolutionary conservation, such as normalized site entropy, residue frequency and SIFT score; and (iii) substitution-matrix scores, such as those derived from the BLOSUM62, GRANTHAM and PHAT matrices. We validated our approach using a control dataset consisting of known disease-causing mutations and neutral variations. Logistic regression analyses indicated that position-specific phylogenetic features that describe the conservation of an amino acid at a specific site are the best discriminators of disease mutations versus neutral variations, and integration of all our features improves discrimination power. Overall, we identify 115 SNPs in GPCRs from dbSNP that are likely to be associated with disease and thus are good candidates for genotyping in association studies.
INTRODUCTION
G-protein-coupled receptors (GPCRs) are integral membrane proteins that include a large family of cell-surface receptors which are important in signal transduction processes. GPCRs recognize a wide range of extracellular ligands, such as nucleotides, peptides, amines and hormones. GPCRs transduce these extracellular signals through the interaction with guanine nucleotide-binding (G) proteins (1,2). This triggers changes in the levels of intracellular messengers, which set off a cascade of processes affecting a huge range of metabolic functions. Not surprisingly, they are important targets for the majority of prescription drugs, such as β-blockers for high blood pressure, β-adrenergic agonists for asthma and anti-histamine (H1 antagonist) for allergy (3,4). The main objective of this paper is to assess the disease-causing potential of single nucleotide polymorphisms (SNPs) in GPCRs from the public database dbSNP (5). SNPs are single base variations between genomes within a species. SNPs are defined as variations that occur at a frequency of at least 1% and are primarily used as markers for genome-wide mapping and study of disease genes. Additionally, it is also believed that these small genomic-level differences may be used to explain the differential drug–response behavior of individuals toward a drug and can be used to tailor drugs based on an individual's genetic makeup (6–8). The tremendous promise that SNPs hold has spurred a lot of research aimed at identifying SNPs. The publication of the human genome and the availability of more than 4 million SNPs in the public database dbSNP provides us with an opportunity to perform large-scale ‘in silico’ analysis of SNPs.
Given the important roles of GPCRs in many physiological processes and their pharmaceutical relevance as drug targets, understanding the role of sequence variations in GPCRs has potential implications for elucidating disease pathogenesis mechanisms and drug efficacy issues. To date, there has been only two published reports of a systematic study of SNPs in GPCRs (9,10). Small and co-workers (9) studied the variability in GPCR genes by sequencing 64 GPCR genes in an ethnically diverse group of 82 individuals. They reported that variability in GPCR genes were more than that observed in non-GPCR genes. Additionally, they found that ∼38% of SNPs were in transmembrane (TM) regions. Lee et al. (10) have analyzed coding variations in GPCR genes from various public sources. In particular, they studied the distribution of SNPs among the various domains of GPCRs, i.e. transmembrane, extracellular and intracellular regions. They found that disease-causing variations were overrepresented in TM regions. In contrast, non-disease-causing variations were underrepresented in TM regions.
With the explosion of data on the human genome and SNP discovery, it is essential to extract useful information from this deluge of data. Data mining of the public databases adds to the pool of useful information about disease genes. dbSNP has a heterogeneous collection of SNPs obtained by different methods and the quality of the SNP data is variable. It has been reported that ∼40% of SNPs from dbSNP were absent from a proprietary ‘genecentric’ database leading to the speculation that some of the SNPs in dbSNP may not be truly polymorphic (11). Another report estimates that 68% of nonsynonymous SNPs in GPCRs from dbSNP could be false positives based on the experimental verification of a subset of SNPs in GPCRs (12). Hence, there is a need for some kind of evaluation of SNPs from public databases to make them suitable targets for expensive association and genotyping studies.
While SNPs are widely used as markers, some of these SNPs may directly explain the pathogenesis of diseases. Nonsynonymous SNPs in coding regions may directly affect the function of the protein either by disrupting the 3D structure of the proteins dramatically or by subtle changes resulting in sub-optimal placement of important residues that affect active sites, ligand binding, etc. Several groups have studied the effect of SNPs on protein structure and function using both sequence and 3D structure-based analyses. Ng and Henikoff (13,14) have elegantly demonstrated the use of multiple sequence alignments (MSAs) to identify conserved amino acid positions that may be critical for protein function. They rationalized that an amino acid variation occurring in a conserved position is likely to affect the function of the protein. They developed an algorithm named SIFT to evaluate the effect of amino acid changes at any position based only on the sequence information.
Many other groups have assessed the effect of SNPs in soluble proteins on the basis of their location in the tertiary structure of protein. Chasman and Adams (15) predicted that ∼30% of nonsynonymous SNPs would affect protein function based on both sequence and structure-based criteria. Sunyaev and co-workers (16) estimate that ∼20% of nonsynonymous SNPs will have deleterious effects on protein structure based on the location of SNPs mapped onto 3D structures and comparative sequence homology analyses. In a very thorough study, Wang and Moult (17) developed a set of rules for predicting the effect of SNPs on protein function based on the results of in vitro studies of site-directed mutagenesis experiments in conjunction with data of known disease-causing mutations in the context of the 3D structures of proteins. They showed that SNPs resulting in deleterious amino acid changes predominantly affect the stability of proteins. Liang et al. mapped nonsynonymous SNPs from OMIM (18,19), a database consisting of human genetic disorders, on to the structural surfaces of proteins (20). Based on the geometric location of these structural sites, they showed that majority of disease-associated SNPs tend to be located in surface pockets or voids.
Although SNPs in soluble proteins have been evaluated computationally extensively based on the knowledge of 3D structure of proteins, a PubMed search for SNPs shows numerous reports of coding SNPs (21,22) as mere observations and few attempts to infer their effect on protein function. There has also been less emphasis on the systematic analysis of SNPs in membrane proteins by ‘in silico’ methods owing to the paucity of 3D structures for membrane proteins.
Mutations that are lethal to an organism are never observed. Fatal mutations are extremely low frequency changes and are by definition not included as polymorphisms. It is believed that there are common variants that contribute to disease (23). The goal of this study is therefore to correlate such SNPs and their potential to cause disease. It should be noted that correlating SNPs to a disease state is a very complex problem, and the in silico studies that have been discussed above are applicable only to monogenic disorders. The pathogenesis of many diseases has a very complex underlying mechanism involving several genes and pathways. Also, several SNPs that are mildly deleterious to a protein in isolation can be very deleterious to an organism when certain combinations of such SNPs occur together.
GPCRs contain seven transmembrane regions separated by six loops: three extracellular and three intracellular, an extracellular N-terminus and an intracellular C-terminus. Several groups have attempted to model the tertiary structure of a GPCR of their interest based on the crystal structure of rhodopsin, the only available 3D structure for a GPCR (24–27). However, we have adopted a different approach in order to make it applicable to all membrane proteins. Given that there are very few high resolution 3D structures for membrane proteins, a general approach that will be applicable to all membrane proteins should be based on criteria independent of 3D structural information for the proteins. Moreover, the modeling of GPCRs based on rhodopsin itself presents some problems (28). Therefore, we have analyzed the SNPs in GPCRs from dbSNP primarily based on the properties of amino acids and the sequence-based tool SIFT to distinguish between disease-causing substitutions and neutral substitutions.
As 3D structural information is not available for most proteins, researchers have used several sequence-based and phylogenetic features to study the effect of amino acid variations on protein structure and function (16,29–37). These features are described in Table 1. Cai et al. (29) used several amino acid properties as features in their Bayesian approach for predicting pathogenic mutations. Of the several physicochemical properties of amino acids, they found that change in hydrophobicity was the only amino acid-based property that had a predictive value in conjunction with positional entropy. They also found that change in residue frequency was a good predictor in differentiating deleterious versus benign mutations. Saunders and Baker (36) used structural and evolutionary information to predict deleterious mutations. They clearly showed that a combination of just two features, SIFT score (a residue conservation index) and a solvent-accessibility term, were enough to differentiate between deleterious and neutral variations (13). Several studies have shown that substitutions at evolutionarily conserved sites are deleterious to the proteins (Table 1). Ferrer-Costa et al. (30) demonstrated that deleterious mutations are associated with extreme changes in sequence and structure-based features that relate to protein stability. Based on these results, we have included three major classes of features to study the pathogenic effect of SNPs in GPCRs:
Properties based on amino acid scale. We used changes in volume and hydrophobicity as simple physicochemical features describing an amino acid. In addition, we used an additional hydrophobicity feature, GES hydrophobicity scale, for TM regions, because it was specifically developed for helical TM regions and was shown to be better than several other hydrophobicity scales for TM helix prediction (38).
Position-specific phylogenetic features. We used SIFT scores, normalized site entropy and change in residue frequency at a given position as additional features. These features are calculated from MSAs.
Substitution matrix scores. We used BLOSUM62, GRANTHAM and PHAT substitution scores to assess amino acid changes and their potential to be deleterious to the protein. These are phylogenetic features that are not position-specific.
Table 1.
This table summarizes the different sequence-based features that have been used for identifying amino acid substitutions that could be deleterious to the protein and the results obtained from these studies
| Sequence-based features | Comment | Reference |
|---|---|---|
| Properties based on amino acid scale | ||
| Mass, volume, surface area, side-chain properties (charge, polarity), partial specific volume, hydrophobicity, alpha helix propensity, relative occurrence, percent buried, pKa. | The physicochemical properties were used as features in a Bayesian framework to predict the pathogenecity of an amino acid variation. Change in hydrophobicity coupled with low positional entropy was shown to be a good predictor. | (29) |
| Position-specific phylogenetic features | ||
| Positional entropy, modified Shannon entropy and normalized site entropy | Substitutions at evolutionarily conserved sites have been shown to be strongly correlated with disease-causing mutations. Conservation at a position in a protein sequence has been assessed using slightly modified versions of sequence entropy from MSAs. | (29,30,33–36) |
| Change in residue frequency | Residue frequency at a given amino acid position was calculated for both variants from multiple-sequence alignments. Change in residue frequency in conjunction with hydrophobicity correlated with the observed phenotype. | (29) |
| Conservation related to allele frequency | Absolutely conserved residues between at least three mammalian orthologs were identified and variations at these positions were shown to be underrepresented at high allele frequencies compared to variations at unconserved sites. | (31) |
| Degree of conservation using tree method | The number of substitutions at a given position in a sequence was estimated based on known phylogenetic relationships between species. Disease-associated mutations were more prevalent at conserved sites. | (32) |
| SIFT | Calculates a conservation index based on MSA. Normalized probabilities for all possible substitutions at a given amino acid position are obtained from the MSA and substitutions with probabilities below a certain cutoff are deemed intolerant to the protein. | (13,14) |
| Substitution matrices | ||
| BLOSUM, PAM and GRANTHAM | It was shown that ∼40% of disease-causing changes had highly unfavorable BLOSUM62 scores. Similar general trends were seen for PAM matrix scores (30). | (13,30–32,36) |
| A clear correlation between BLOSUM62 and allele frequency of nonsynonymous SNPs was not seen in a study of SNPs in membrane-transporter genes (31). | ||
| BLOSUM62 scores were able to distinguish tolerant from intolerant substitutions in a variety of proteins with total prediction accuracies ranging from 47 to 70% (13). | ||
| About 40% balanced classification error was reported by Saunders et al. (36) using BLOSUM62 scores as a predictive feature. | ||
| Miller et al. (32) showed that disease-causing amino acid changes are more radical than variation found among species using GRANTHAM scores. | ||
MATERIALS AND METHODS
Mapping SNPs on to GPCRs
SNPs from build 110 of dbSNP were used for this analysis. Sequences containing SNPs were downloaded from ftp://ftp.ncbi.nih.gov/snp/human. Homology matches to GPCRs were obtained by performing a six-frame translational BLAST (39) search of the sequences containing SNPs from dbSNP against the GPCRDB database (release 8) downloaded from www.gpcr.org (40,41). Matches which were at least 18 amino acids long with E-values <10−4 were considered as significant matches and for a given query sequence, the most significant match (i.e. the match with the smallest E-value) was chosen. Since the average length of a transmembrane helix is between 21 and 22 amino acids with a large variation around the mean (42,43), we used 18 amino acids as the minimum match length. Once the query sequences containing the SNPs were mapped on to GPCR proteins, sequences containing SNPs that lead to a change in amino acid, nonsynonymous SNPs, were extracted. At this stage, all matches to olfactory GPCR proteins were removed as it is known that nonsynonymous changes in olfactory receptors are predominantly owing to positive selection for a diverse olfactory repertoire (44,45). In addition, ∼60% of the complete olfactory subgenome are pseudogenes (46,47).
Domain information
The locations of nonsynonymous SNPs in the various domains (transmembrane, intracellular and extracellular) of the 7-TM GPCR proteins were elucidated based on the annotations from GPCRDB. In GPCRDB, TM helices were predicted using PredictProtein (48) and their positions were adjusted based on MSAs because it is hypothesized that the TMs must be aligned and of the same lengths for all the members of a receptor family/subfamily. The ends of Class A helices were determined from the alignment with bovine rhodopsin.
Validation datasets
Two control datasets were used to benchmark the predictive power of the sequence-based features to predict the disease-causing potential of an SNP.
Dataset containing disease mutations
Mutations in GPCRs that are associated with disease were compiled from SWISS-PROT (version 40.44) (49,50). All proteins containing disease mutations were extracted from SWISS-PROT. This list was cross-referenced with the protein IDs from GPCRDB to obtain disease-associated mutations in GPCRs.
Dataset consisting of neutral variations
For a dataset of neutral variations, homologs to all the GPCR proteins associated with disease were directly extracted using the multiple alignment files from GPCRDB. Amino acid variations between sequences >95% identical were considered as neutral variations similar to the approach used by Bork et al. (16). The logic behind this assumption is that variations in highly homologous sequences between species are generally neutral and are highly unlikely to be deleterious because deleterious changes will be selectively removed during the course of evolution. Nevertheless, it should be pointed out that in some instances, some of these changes may be functional changes important in one species, but not in the other. Paralogs with different functions could have high sequence similarity to homologs. To ensure that we do not include such functional variations as neutral changes in this dataset, we removed all paralogous homologs. This was accomplished in the following manner:
All homologs to the control dataset proteins containing disease mutations with >95% sequence identity were extracted from GPCRDB.
For each target disease protein, only one ortholog was chosen from each species based on the best match to the target protein. The sequence with higher percent identity to the target protein was chosen as the best match.
Distribution of mutations among the three domains of GPCRs
The partitioning of the mutations in the different datasets (the validation datasets and the dbSNP dataset) among the various domains of the GPCRs was assessed assuming a Poisson process to check whether the mutations within any dataset are distributed randomly in the transmembrane, intra and extracellular regions of the GPCRs. For example, in the case of the dataset containing the disease mutations, the occurrence of disease mutations in the three domains were modeled to fit a Poisson distribution using the following equation:
where m is the expected average number of disease mutations in a given domain obtained based on the density of disease mutations, y = 0, 1, 2, …, P(y) is the probability of random occurrences of ‘y’ number of disease mutations in that domain. The null hypothesis that we are testing is that disease mutations are randomly distributed in TM, extracellular and intracellular regions. Similar analyses were performed on the neutral variations and the SNP dataset.
For the dataset containing disease mutations, the average number of mutations in TM regions is calculated as follows:
where,
When the observed number of mutations is greater than the expected average number of mutations, we assessed the significance of this difference by calculating the sum of P(y) values for all values ≥y, where y is the observed number of mutations. Similarly, when the observed number of mutations is smaller than the expected average number of mutations, we calculated a cumulative P-value by adding P(y) values for all values ≤y. A small P-value (P < 0.05) indicates that the occurrence of ‘y’ number of mutations in a domain is not random.
Free energy changes
The changes in free energy of hydropathy, ΔΔG, owing to amino acid variations in transmembrane regions were evaluated using the GES hydrophobicity scale (38) as follows:
Here, ΔG refers to the transfer free energy of an amino acid from water to membrane. The various subscript notations on the right-hand side of the equations refer to the following:
For the dataset pertaining to disease mutations, ΔGvariant refers to the free energy value pertaining to amino acid causing disease and ΔGwild-type refers to the free energy value of the amino acid in the native protein.
For neutral variations, ‘variant’ refers to the neutral variation and ‘wild-type’ refers to the amino acid at that position in the native protein. For the SNPs from dbSNP, ‘variant’ refers to the altered amino acid as a result of an SNP.
Allele frequency information is not available for all variants in dbSNP. Therefore, for SNPs from dbSNP, the identity of the wild-type amino acid for a protein of interest was obtained directly from the amino acid sequence in GPCRDB and the other amino acid was designated as the ‘variant’ amino acid. In cases where both SNPs translated the codons to two different amino acids that differed from wild-type, they were considered as two variant amino acids and calculations were performed with respect to the wild-type amino acid from the parent sequence in GPCRDB. The absolute value of the free energy changes were used in the logistic regression analysis.
Volume calculations
For the volume calculations, changes in volumes, ΔV, were calculated. For this analysis, average residue volumes listed in Gerstein et al. (51) were used. These volumes were calculated according to the Richards's implementation of Voronoi method based on 118 structures from the PDB. The absolute value of the volume changes were used in the logistic regression analysis.
SIFT analysis
SIFT version 2.0 was used for the analyses (13,14). The default settings were used for executing SIFT. The proteins of interest were queried against SWISS-PROT (version 40.44) to extract sequences homologous to the query protein. The MSA sequence alignment used for calculating the conservation index was automatically generated by SIFT.
Change in hydrophobicity
Changes in hydrophobicity between two variants at a given amino acid position were evaluated using the Kyte Doolittle hydrophobicity scale (52). We calculated change in hydrophobicity using the same formalism that was used for change in free energy of hydropathy. Change in hydrophobicity as well as the absolute value of the hydrophobicity change were used in the initial stages of logistic regression analysis. Change in hydrophobicity was found to be a weak predictive feature and the absolute value of hydrophobicity difference performed better. Therefore, we only used the absolute value of hydrophobicity difference as a predictive feature for the various logistic regression analyses. The magnitude of change in hydrophobicity gives an estimate of how well the hydrophobic nature of a residue is conserved.
Normalized site entropy
Normalized site entropy for all the amino acid positions in the MSA was calculated using the software program AL2CO (53). The site entropy was calculated based on the entropy-based measure given as follows:
where Ce(i) is the entropy with the reverse sign at position i, fa(i) represents frequency of amino acid ‘a’ at ith position obtained from MSA generated by SIFT. The amino acid frequencies were estimated using an independent-count-based weighting scheme in order to correct for the masking effect of highly similar sequences over fewer divergent sequences in a MSA (54). The normalized site entropy was calculated by subtracting the mean site entropy from the site entropy and dividing by the standard deviation.
Change in residue frequency
The amino acid frequencies of the two amino acid variants at a given position were calculated directly from the alignments generated by SIFT. The change in residue frequency at a position was calculated using the same general formalism outlined above for the control datasets (disease and neutral) and the dbSNP dataset. The absolute value of change in residue frequency was used for the logistic regression analysis.
Logistic regression analysis
Logistic regression was used to discriminate disease-causing mutations from neutral ones. In the logistic regression model, the probability that a mutation is disease-causing is related to the weighted linear combination of scores for individual features in the following way:
| 1 |
where p is the probability that the mutation is disease-causing, and sj is the score of the jth feature for this mutation. To estimate the weights w0, w1, …, wM, a training set of N mutations is used where each mutation is known to be disease-causing or neutral. From the training set, the likelihood function, i.e. the probability of observing the data given the weights, is computed in the following way:
| 2 |
where for the ith mutation, pi is the probability that the mutation is diseasing-causing, computed from Equation 1. yi, the response variable, is equal to 1 if the ith mutation is disease-causing, and 0 if otherwise. Li, the likelihood of the logistic regression model given the ith mutation in the training set, is equal to pi if the mutation is disease-causing, and 1 − pi if the mutation is neutral. Finally, the weights w0, w1, …, wM are chosen such that the likelihood function L(w0, w1, …, wM) in Equation 2 is maximized.
Logistic regression analysis was performed using the Weka machine learning workbench (55). Error rates were calculated with 10-fold cross-validation.
RESULTS
Nonsynonymous SNPs in GPCRs, from the public database dbSNP, have been evaluated by ‘in silico’ methods in order to assess their pathogenic potential. Specifically, the effect of amino acid changes at a given position in a GPCR has been assessed using simple physicochemical indices of amino acids, position-specific phylogenetic features and substitution matrix scores. We used a dataset consisting of disease mutations and another comprising neutral variations in a set of GPCR proteins, as a training dataset in a logistic regression analysis to classify them as disease-causing and neutral variations. A correct prediction of ∼89% accuracy was obtained using a combination of all features. The model obtained from this training dataset was used to predict the pathogenecity of SNPs in GPCRs from dbSNP by logistic regression. A list of SNPs in GPCRs from dbSNP that would potentially affect the function of the proteins has been obtained using this methodology. The observed correlations of SNPs with the various features are discussed below.
Location of the amino acid variations
Of the 284 disease-causing mutations, 164 are found in transmembrane regions. Assuming that the mutations are distributed according to a Poisson process, the disease-causing changes are highly overrepresented in transmembrane regions as shown in Table 2. This is similar to the results obtained by Lee et al. (10) who used a different set of disease mutations. Among the mutations in the disease dataset, mutations in the extracellular and intracellular domains are underrepresented. This may imply that changes in TM regions are disease-causing presumably because such changes may directly affect either the structure or function of the receptor. Mutations in TM regions could abrogate or diminish the activity of the protein when a ligand-binding site is affected. On the other hand, a mutation in a TM region could compromise the protein's structural integrity owing to its effect on helix–helix packing interactions. Similar analyses of the dataset comprising neutral variations show a different trend. Here, the occurrence of neutral variations in the TM and extracellular regions appears to be random, whereas neutral variations are underrepresented in the intracellular regions. The SNPs in dbSNP are significantly underrepresented in TM regions and overrepresented in extracellular regions. The crude analysis at this level indicates that most of the SNPs in dbSNP are similar to neutral variations and are probably benign substitutions.
Table 2.
Distribution of the various amino acid changes among the TM, extracellular and intracellular regions for the disease-causing, neutral variations and SNPs from dbSNP
| Domain | Disease | Neutral | dbSNP |
|---|---|---|---|
| Transmembrane | 164 (93) P = 1.9 × 10−11 | 90 (86) P = 0.35 | 112 (158) P = 2.2 × 10−5 |
| Extracellular | 80 (111) P = 0.001 | 96 (82) P = 0.06 | 200 (159) P = 0.0009 |
| Intracellular | 40 (80) P = 5.5 × 10−7 | 61 (79) P = 0.019 | 152 (126) P = 0.056 |
The numbers in the parentheses is the expected number based on a Poisson distribution and the numbers left of the parentheses indicate the observed number of variations in the corresponding domain.
Distribution of scores based on different substitution matrices
The nature of amino acid changes were assessed in terms of scores using various substitution matrices. We used the BLOSUM62, GRANTHAM and PHAT substitution matrices. BLOSUM62 is a widely used robust substitution matrix (56). We also used GRANTHAM D values to evaluate the amino acid changes. In order to alleviate concerns about the suitability of BLOSUM matrices derived from a database of soluble proteins to TM proteins, we used the PHAT matrix for TM regions (57).
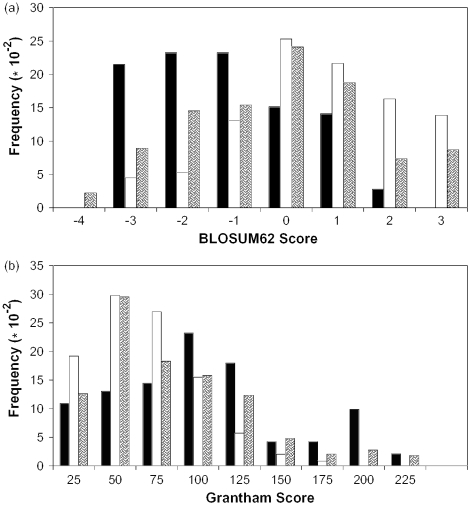
BLOSUM62 matrix: We assigned BLOSUM62 scores to the variations in all three datasets. Figure 1a is a histogram showing the distribution of BLOSUM62 scores for the disease, neutral and dbSNP variations. The distribution of scores for the disease and neutral variations is significantly different (χ2 = 141.07, P < 0.001, 6 degrees of freedom). A total of 44.7% of disease-causing mutations have scores <−1, whereas only 9.7% of neutral changes have scores <−1. For scores >1, only 2.8% are disease-causing, whereas 30.2% are neutral. For scores between −1 and 1, there is no way to discriminate between the two sets. Thus, extreme values of BLOSUM62 scores can be used to discriminate between disease-causing and neutral variations. Analyses of mutations in soluble proteins have yielded similar results (30). The correlation between BLOSUM62 scores and the deleterious nature of an amino acid substitution has been seen in some cases and not in others (13,30–32,36). For GPCRs, BLOSUM62 scores seem to be a fairly good predictor of deleterious substitutions. It is not obvious why this is the case. It is clear from Figure 1a that the distributions for the neutral and the dbSNP variations are extremely similar.
GRANTHAM matrix: GRANTHAM scores >100 are considered radical changes. Figure 1b depicts the distribution of GRANTHAM scores. The distribution of GRANTHAM scores for the disease and neutral variations are different (χ2 = 91.2, P < 0.001, 8 degrees of freedom). Variations with scores >100 are increasingly associated with disease-causing mutations. However, the distinction between disease-causing and neutral mutations is not as clear-cut as the BLOSUM62 results.
PHAT matrix: It has been previously reported that BLOSUM62 scores could not be used to discriminate deleterious mutations from benign changes in human membrane transporter genes (31). This could be owing to the fact that BLOSUM62 scores are derived primarily from soluble globular proteins. In the case of GPCRs, BLOSUM62 does seem to be a fairly good discriminator between disease-causing and neutral variations. Nevertheless, the variations in TM regions were assessed with PHAT, a transmembrane-specific substitution matrix. From Supplementary Figure 1S, it is very clear that PHAT scores <−1 are predominantly associated with disease-causing mutations. The distributions of PHAT scores for disease-causing and neutral changes in TM regions are significantly different (χ2 = 100.73, P < 0.001, 14 degrees of freedom). While 64.6% of disease mutations have PHAT scores <−1, only 5.6% of neutral variations have PHAT scores <−1. Thus, PHAT substitution scores <−1 is a very good discriminator for disease-causing and neutral variations in TM regions. A similar analysis of BLOSUM62 scores of amino acid changes in transmembrane regions shows that only 46% of disease mutations and 7.9% of neutral changes have BLOSUM62 scores <−1. This is depicted in Supplementary Figure 1S. Thus, PHAT scores are also a good discriminator of disease versus neutral amino acid changes in transmembrane regions similar to BLOSUM62 scores. Interestingly, logistic regression analysis (see Table 4) indicates that BLOSUM62 performs somewhat better than PHAT scores in TM regions.
Figure 1.
(a) Histogram of BLOSUM62 scores. (b) Histogram of GRANTHAM scores. Here, the black bars represent disease variations, white bars indicate neutral variations and the shaded bars are dbSNP variations.
Table 4.
Total error rate of misclassification of disease-causing and neutral variation when each feature was assessed by itself in the logistic regression analysis
| Feature | Error rate (%) |
|---|---|
| SIFT conservation score | 18.41 |
| Normalized site entropy | 18.60 |
| Change in residue frequency | 19.92 |
| BLOSUM62 score | 27.70 |
| GRANTHAM score | 31.31 |
| Change in volume | 34.91 |
| Change in hydrophobicity | 37.95 |
| Location of variation (i.e. TM or non-TM) | 39.47 |
| BLOSUM62 score (TM only) | 22.53 |
| PHAT (TM only) | 24.90 |
| Change in free energy of hydropathy (TM only) | 27.27 |
Here, phylogenetic features refer to SIFT score, normalized site entropy and change in residue frequency.
Free energy change of hydropathy associated with amino acid replacements in TM regions
Free energy changes associated with variations in TM regions were evaluated using the transfer free energies based on the GES hydrophobicity scale. Supplementary Figure 2S shows the frequency distribution of variations as a function of change in free energy of hydropathy. The change in free energy of hydropathy owing to neutral variations is small, varying predominantly between 0 and 2 kcal/mol. However, a substantial number of disease-causing variations also have similar destabilizing/stabilizing free energy changes. Therefore, small changes in free energy values do not allow the classification of an amino acid variation as either neutral or disease-causing. Substitutions that are highly destabilizing (>8 kcal/mol) are always associated with disease-causing variations, as seen in Supplementary Figure 2S. Overall, the dbSNPs in GPCR proteins have a similar distribution as neutral variations.
Change in side-chain volumes
The changes in the volume occupied by different side-chains were evaluated to see whether there was any correlation to disease-causing mutations versus neutral variations. Logistic regression analysis indicates that absolute volume change has a modest predictive value in differentiating between disease-causing and neutral variations (data shown in Table 4).
Change in hydrophobicity
The changes in hydrophobicity accompanying the substitution of one amino acid by another were evaluated to see whether it would be a useful feature to distinguish between disease-causing and neutral variations. Logistic regression analysis indicates that change in hydrophobicity also has a modest predictive value in differentiating between disease-causing and neutral variations (data shown in Table 4).
Change in residue frequency
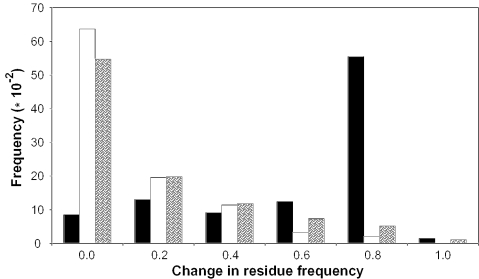
The amino acid frequencies of the two amino acid variants at a given position were calculated directly from the alignments generated by SIFT. Figure 2 shows the histogram of change in residue frequency for the two benchmark datasets and the dbSNP dataset. When the ‘change in residue frequency’ is small (values close to 0), the amino acid variations corresponding to these values tend to be neutral variations. In contrast, a large portion of disease-causing mutations are associated with big values of ‘change in residue frequency’. This distribution shows that SNPs in dbSNP are more similar to neutral SNPs than disease-causing mutations.
Figure 2.
Histogram of change in residue frequency for the disease-causing, neutral and dbSNP variation datasets. The absolute value of change in residue frequency is shown. The black bars represent disease variations, white bars indicate neutral variations and the shaded bars are dbSNP variations.
SIFT analysis
While all the above features used to evaluate amino acid variations are based on simple physicochemical parameters, we also analyzed the relationship between sequence conservation and the effect of variations in highly conserved positions using SIFT. Ng and Henikoff (13,14) have developed a tool called SIFT to identify conserved positions that may be critical for protein function using MSA.
SIFT scores were used to assess the two control datasets, disease-causing and neutral variations in GPCRs, Of the 284 disease-causing mutations, SIFT predicted 213 mutations to be deleterious. Thus, SIFT correctly identified 75% of disease-causing mutations as intolerant substitutions. In the case of neutral variations, the performance of SIFT was even better. SIFT predicts 94% of neutral variations to be tolerant substitutions. SIFT did not score 1 disease mutation and 3 neutral variations. SIFT was used to assess the dbSNPs in GPCRs. Based on SIFT scores, 74.8% of SNPs in GPCRs from the dbSNP database are neutral variations. Thus, only 25.2% of SNPs are predicted to be deleterious substitutions.
Normalized site entropy
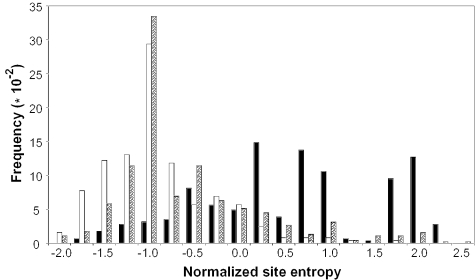
Figure 3 shows the distribution of normalized site entropy scores for disease mutations, neutral variations and SNPs in dbSNP. Clearly, the distribution of disease-causing mutations is different from neutral variations. Neutral variations are associated with a peak at a normalized site entropy value of −1, whereas the normalized site entropy values associated with disease mutations are spread over a range of values, most of which are >0.25. As with most other features described so far, the distribution of SNPs in dbSNP is very similar to neutral variations.
Figure 3.
Frequency distribution of normalized site entropy values for the disease-causing, neutral and dbSNP variation datasets. The black bars represent disease variations, white bars indicate neutral variations and the shaded bars are dbSNP variations.
Logistic regression analysis
It is clear that it is possible to use some of the above features to predict whether a SNP would be deleterious or neutral. Logistic regression analysis was performed to elucidate the best predictors and the relative contributions of the different features to a prediction. Logistic regression is a better alternative to linear regression when the response variable is dichotomous, which is true in our case: a mutation can be either disease-causing or neutral. We performed logistic regression analysis in several different ways. As the TM regions have more predictive features, the logistic regression was performed in two ways: (i) analysis of a dataset comprising all variations (TM and non-TM); and (ii) analysis of two datasets obtained by grouping the variations into TM and non-TM datasets.
In the first model, all variations were analyzed using the following features: BLOSUM, GRANTHAM, volume and hydrophobicity changes, location of the variation (TM or non-TM), SIFT scores, normalized site entropy and change in residue frequency. In the second model, variations in TM regions and non-TM regions were divided into two groups. For TM regions, two additional features were used: PHAT scores and change in free energy of hydropathy. The results of the logistic regression analyses are discussed below.
Table 3 shows the results obtained from a logistic regression analysis of all variations (disease and neutral changes) using only the features common to both TM and non-TM regions. It can be seen that the overall error rate drops from 18.41 to 11.20% when SIFT is complemented with other features. To assess the predictive power of each feature, logistic regression analyses were performed using each feature individually for the classification. The total error rates obtained from this analysis are shown in Table 4. The error rates are reported for the analysis on the training dataset including all variations (TM and non TM) in all cases, except for the last three features in the row (PHAT, BLOSUM62 and change in free energy of hydropathy). For those three features, the error rates are reported for the dataset comprising variations only in the TM regions. It is clear from Table 4 that the top three best discriminators of disease versus neutral variations are the position-specific phylogenetic features that describe evolutionary conservation. All three features, change in residue frequency, SIFT score and normalized site entropy, have individual prediction error rates ∼18–20%. In the absence of these three features, the error rate is 26.38%. The error rate drops to 11.95% when the three position-specific phylogenetic features are used together for the logistic regression analysis. The addition of other features lowers the error rate even further to 11.20%.
Table 3.
The results of logistic regression analysis of all variations using the features common to both TM and non-TM regions
| All features (excluding position-specific phylogenetic features) | SIFT onlya | Position-specific phylogenetic features only | All features | |||||
|---|---|---|---|---|---|---|---|---|
| Disease | Neutral | Disease | Neutral | Disease | Neutral | Disease | Neutral | |
| Correct classification | 221 | 167 | 257 | 173 | 247 | 217 | 249 | 219 |
| Wrong classification | 62 | 77 | 26 | 71 | 36 | 27 | 34 | 25 |
| Total number of errors | 139 (26.38%) | 97 (18.41%) | 63 (11.95%) | 59 (11.20%) | ||||
Here, phylogenetic features refer to SIFT score, normalized site entropy and change in residue frequency.
aThe classification obtained by logistic regression analysis using only the SIFT score as the determining feature.
Tables 5 and 6 summarize the results obtained from a logistic regression analysis of the variations in the control datasets sub-grouped into two sets: one consisting of variations only in TM domains and the other comprising variations in non-TM domains. For variations in non-TM regions, the error rate was almost twice that of the error rate in TM regions (Table 6). It is seen that predictions for the TM regions are more accurate than the non-TM regions. In all the cases, the combination of all three position-specific phylogenetic features, SIFT score, normalized site entropy and change in residue frequency, significantly improves the overall prediction accuracy. This underscores the importance of position-specific phylogenetic features in the assessment of disease-causing potential of an amino acid substitution at a particular site in a protein.
Table 5.
The results of logistic regression analysis of variations in TM regions
| All features excluding position-specific phylogenetic features | SIFT only | Position-specific phylogenetic features only | All features | |||||
|---|---|---|---|---|---|---|---|---|
| Disease | Neutral | Disease | Neutral | Disease | Neutral | Disease | Neutral | |
| Correct classification | 143 | 58 | 157 | 68 | 155 | 71 | 155 | 80 |
| Wrong classification | 21 | 31 | 7 | 21 | 9 | 18 | 9 | 9 |
| Total number of errors | 52 (20.55%) | 28 (11.07%) | 27 (10.67%) | 18 (7.11%) | ||||
Here, phylogenetic features refer to SIFT score, normalized site entropy and change in residue frequency.
Table 6.
The results of logistic regression analysis of variations in non-TM regions
| All features excluding position-specific phylogenetic features | SIFT only | Position-specific phylogenetic features only | All features | |||||
|---|---|---|---|---|---|---|---|---|
| Disease | Neutral | Disease | Neutral | Disease | Neutral | Disease | Neutral | |
| Correct classification | 77 | 117 | 100 | 114 | 93 | 142 | 94 | 143 |
| Wrong classification | 42 | 38 | 19 | 41 | 26 | 13 | 25 | 12 |
| Total number of errors | 80 (29.20%) | 60 (21.90%) | 39 (14.23%) | 37 (13.50%) | ||||
Here, phylogenetic features refer to SIFT score, normalized site entropy and change in residue frequency.
It is clear from Tables 3–6 that in all the cases the position-specific phylogenetic features perform the best. On the other hand, in the absence of the phylogenetic features, the other features can still be used with a prediction accuracy of ∼70%.
Logistic regression was also performed to classify all the variations as disease-causing or neutral using each phylogenetic feature individually. The prediction error rates for this analysis are shown in Table 7. Of the three phylogenetic features, SIFT scores perform better in TM regions than in non-TM regions. For the other two features, their predictive power is not significantly different for TM versus non-TM regions.
Table 7.
The error rate of misclassification of disease-causing and neutral variations using the SIFT score, normalized site entropy and change in residue frequency individually as predictors in the logistic regression analysis
| Dataset | SIFT score (%) | Normalized site entropy (%) | Change in residue frequency (%) | Combining all three features (%) |
|---|---|---|---|---|
| All variations | 18.41 | 18.60 | 19.92 | 11.95 |
| TM only | 11.07 | 19.37 | 19.37 | 10.67 |
| Non-TM only | 21.90 | 19.71 | 20.07 | 14.23 |
From the above analyses, it is clear that position-specific phylogenetic features that describe the conservation of amino acid residue at a specific site are the best predictors for discriminating disease-causing versus neutral variation. When SIFT is used with its default settings, substitutions with SIFT scores <0.05 are predicted to be intolerant substitutions. This is a very conservative cutoff. It can be seen that SIFT combined with other features can be used to predict a higher number of disease-causing mutations correctly by logistic regression analysis. Of the 283 disease-causing mutations, 213 are predicted to be intolerant substitutions using the default SIFT setting. However, logistic regression analysis using SIFT score in conjunction with the other features classifies 249 of them to be disease-causing (Table 3). Using the regression coefficients for the model obtained from Table 3, 115 SNPs in GPCRs from dbSNP are predicted to be deleterious. A list of the 464 SNPs in GPCRs from dbSNP including the features used in the logistic regression model can be downloaded from http://www.gersteinlab.org/proj/gpcrsnp. The log odds ratio as calculated by Equation 1 is also included for each SNP and the list is ordered according to the score. Thus, the SNPs that are likely to be deleterious are shown in the top rows of the table.
DISCUSSION
We have evaluated the disease-causing potential of nonsynonymous coding SNPs in GPCRs by assessing the nature of the amino acid change using a variety of features, such as BLOSUM62, GRANTHAM and PHAT substitution score matrices, free energy change of hydropathy associated with a substitution and changes in side-chain volume of residues and hydrophobicity changes. In addition, we used three different position-specific phylogenetic features, SIFT score, normalized site entropy and change in residue frequency, to evaluate the impact of an amino acid variation caused by a nonsynonymous coding SNP.
Two control datasets were used to assess the relationship between the above mentioned features and amino acid variations. The disease dataset has a preponderance of mutations in transmembrane regions, whereas the neutral variations are randomly distributed. Extreme values of BLOSUM62 can be used to distinguish between disease-causing and neutral variation. BLOSUM62 scores <−1 are predominantly associated with disease mutations and scores >1 are associated with neutral variations. GRANTHAM scores cannot be used to clearly differentiate between the two datasets. PHAT scores <−1 are associated with disease mutations and scores >+2 are associated with neutral variations. In all the cases, the distribution of dbSNPs in GPCRs is more similar to the neutral variations than disease mutations. This indicates that most of the dbSNPs in GPCRs are neutral variations and will not severely affect the function of the protein.
Logistic regression analyses of the predictions show that the position-specific phylogenetic features are the best predictors of the effect of amino acid variation at a particular position on the function of a protein. This is because these features quantify how well conserved a given amino acid is at a specific position in a protein. Substitution scores, such as BLOSUM62, are also phylogenetic features but are not position-specific. Therefore, variations involving two amino acids are given the same weight irrespective of their context in the protein in substitution matrices. But features, such as SIFT scores, change in residue frequency and normalized site entropy describe the conservation of an amino acid at a specific position in a sequence. Thus these position-specific phylogenetic features, elucidated from multiple sequence alignments, describe the strong evolutionary constraints placed on the specific amino acids necessary for the protein's function. Therefore, they are better discriminators of disease-causing versus neutral variations. Hence, position-specific phylogenetic features can be used as the most powerful tools for the evaluation of SNPs and amino acid variations.
Conservation indices based on MSA cannot be used for species-specific sequences, i.e. those proteins that do not have homologs in other organisms. In addition, some SIFT predictions are labeled ‘low confidence’ predictions. This occurs either when there are few sequences homologous to the query sequence or when the homologous sequences are closely related and not very diverse. In such cases, the simple physicochemical parameters of amino acids can be used to get an estimate of the effect of an amino acid variation on protein function. Thus, simple sequence features based on the properties of amino acids can be useful to evaluate sequence variations for those sequences which have no homologs (species-specific SNPs), have few homologs or are not very divergent, albeit with lower prediction accuracy.
Logistic regression analyses using all the features described above indicate that 115 SNPs in GPCRs in dbSNP could be deleterious to the protein. This subset of SNPs from dbSNP in GPCRs are the best candidate SNPs for further genotyping and in-depth experimental analyses to evaluate their effect on the protein's structure and function and thus their pathogenecity. Based on our analysis of the assessment of the amino acid variations using phylogenetic features in conjunction with substitution matrix scores and other simple amino acid features, it is clear that the majority of dbSNPs in GPCRs are neutral variations.
In an analysis of variations in amino acid membrane transporter genes, it was seen that the amino acid diversity in TM regions was less than that of the extracellular and intracellular loop regions (31). From a phylogenetic analysis of TM proteins, Li and Tourasse (58) found that non-TM regions accumulate twice the number of changes as their corresponding TM regions. This study on the 7-TM GPCRs also shows similar trends. It is of interest to note that the SNPs in GPCRS from dbSNP are significantly underrepresented in TM regions compared with the loop regions. Similar observations were reported by Lee et al. (10). This indicates that TM regions are less variable than the soluble extra and intracellular loops. Presumably, this is owing to the general sequence constraints in membrane proteins.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
M.G. thanks NIH for support (P01 GM54160). Y.X. is a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. S.B. thanks Florence Horn and Gert Vriend for answering all queries regarding GPCRDB and making available additional files that were requested. Specifically, S.B. thanks Florence Horn for providing several files on very short notice. S.B. thanks Pauline Ng for providing the source code for SIFT analysis as well as responding to all our questions regarding SIFT. The authors thank Rajkumar Sasidharan for comments on the paper. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rana B.K., Shiina T., Insel P.A. Genetic variations and polymorphisms of G protein-coupled receptors: functional and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2001;41:593–624. doi: 10.1146/annurev.pharmtox.41.1.593. [DOI] [PubMed] [Google Scholar]
- 2.Wess J. Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol. Ther. 1998;80:231–264. doi: 10.1016/s0163-7258(98)00030-8. [DOI] [PubMed] [Google Scholar]
- 3.Dorn G.W., II, Tepe N.M., Wu G., Yatani A., Liggett S.B. Mechanisms of impaired beta-adrenergic receptor signaling in G(alphaq)-mediated cardiac hypertrophy and ventricular dysfunction. Mol. Pharmacol. 2000;57:278–287. [PubMed] [Google Scholar]
- 4.Liggett S.B. The pharmacogenetics of beta2-adrenergic receptors: relevance to asthma. J. Allergy Clin. Immunol. 2000;105:S487–S492. doi: 10.1016/s0091-6749(00)90048-4. [DOI] [PubMed] [Google Scholar]
- 5.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drysdale C.M., McGraw D.W., Stack C.B., Stephens J.C., Judson R.S., Nandabalan K., Arnold K., Ruano G., Liggett S.B. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl Acad. Sci. USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips K.A., Veenstra D.L., Oren E., Lee J.K., Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286:2270–2279. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 8.Roses A.D. Pharmacogenetics and drug development: the path to safer and more effective drugs. Nature Rev. Genet. 2004;5:645–656. doi: 10.1038/nrg1432. [DOI] [PubMed] [Google Scholar]
- 9.Small K.M., Tanguay D.A., Nandabalan K., Zhan P., Stephens J.C., Liggett S.B. Gene and protein domain-specific patterns of genetic variability within the G-protein coupled receptor superfamily. Am. J. Pharmacogenomics. 2003;3:65–71. doi: 10.2165/00129785-200303010-00008. [DOI] [PubMed] [Google Scholar]
- 10.Lee A., Rana B.K., Schiffer H.H., Schork N.J., Brann M.R., Insel P.A., Weiner D.M. Distribution analysis of nonsynonymous polymorphisms within the G-protein-coupled receptor gene family. Genomics. 2003;81:245–248. doi: 10.1016/s0888-7543(03)00009-0. [DOI] [PubMed] [Google Scholar]
- 11.Jiang R., Duan J., Windemuth A., Stephens J.C., Judson R., Xu C. Genome-wide evaluation of the public SNP databases. Pharmacogenomics. 2003;4:779–789. doi: 10.1517/phgs.4.6.779.22821. [DOI] [PubMed] [Google Scholar]
- 12.Small K.M., Seman C.A., Castator A., Brown K.M., Liggett S.B. False positive non-synonymous polymorphisms of G-protein coupled receptor genes. FEBS Lett. 2002;516:253–256. doi: 10.1016/s0014-5793(02)02564-4. [DOI] [PubMed] [Google Scholar]
- 13.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng P.C., Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chasman D., Adams R.M. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- 16.Sunyaev S., Ramensky V., Koch I., Lathe W., III, Kondrashov A.S., Bork P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Moult J. SNPs, protein structure, and disease. Hum. Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 18.Hamosh A., Scott A.F., Amberger J., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM) Hum. Mutat. 2000;15:57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 19.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stitziel N.O., Tseng Y.Y., Pervouchine D., Goddeau D., Kasif S., Liang J. Structural location of disease-associated single-nucleotide polymorphisms. J. Mol. Biol. 2003;327:1021–1030. doi: 10.1016/s0022-2836(03)00240-7. [DOI] [PubMed] [Google Scholar]
- 21.Iida A., Saito S., Sekine A., Kataoka Y., Tabei W., Nakamura Y. Catalog of 300 SNPs in 23 genes encoding G-protein coupled receptors. J. Hum. Genet. 2004;49:194–208. doi: 10.1007/s10038-004-0133-8. [DOI] [PubMed] [Google Scholar]
- 22.Saito S., Iida A., Sekine A., Kawauchi S., Higuchi S., Ogawa C., Nakamura Y. Catalog of 178 variations in the Japanese population among eight human genes encoding G protein-coupled receptors (GPCRs) J. Hum. Genet. 2003;48:461–468. doi: 10.1007/s10038-003-0062-y. [DOI] [PubMed] [Google Scholar]
- 23.Smith D.J., Lusis A.J. The allelic structure of common disease. Hum. Mol. Genet. 2002;11:2455–2461. doi: 10.1093/hmg/11.20.2455. [DOI] [PubMed] [Google Scholar]
- 24.Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H., Fox B.A., Le Trong I., Teller D.C., Okada T., Stenkamp R.E., et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 25.Stenkamp R.E., Filipek S., Driessen C.A., Teller D.C., Palczewski K. Crystal structure of rhodopsin: a template for cone visual pigments and other G protein-coupled receptors. Biochim. Biophys. Acta. 2002;1565:168–182. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- 26.Bissantz C., Bernard P., Hibert M., Rognan D. Protein-based virtual screening of chemical databases. II. Are homology models of G-protein coupled receptors suitable targets? Proteins. 2003;50:5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 27.Bissantz C., Logean A., Rognan D. High-throughput modeling of human G-protein coupled receptors: amino acid sequence alignment, three-dimensional model building, and receptor library screening. J. Chem. Inf. Comput. Sci. 2004;44:1162–1176. doi: 10.1021/ci034181a. [DOI] [PubMed] [Google Scholar]
- 28.Becker O.M., Shacham S., Marantz Y., Noiman S. Modeling the 3D structure of GPCRs: advances and application to drug discovery. Curr. Opin. Drug Discov. Devel. 2003;6:353–361. [PubMed] [Google Scholar]
- 29.Cai Z., Tsung E.F., Marinescu V.D., Ramoni M.F., Riva A., Kohane I.S. Bayesian approach to discovering pathogenic SNPs in conserved protein domains. Hum. Mutat. 2004;24:178–184. doi: 10.1002/humu.20063. [DOI] [PubMed] [Google Scholar]
- 30.Ferrer-Costa C., Orozco M., de la Cruz X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol. 2002;315:771–786. doi: 10.1006/jmbi.2001.5255. [DOI] [PubMed] [Google Scholar]
- 31.Leabman M.K., Huang C.C., DeYoung J., Carlson E.J., Taylor T.R., de la Cruz M., Johns S.J., Stryke D., Kawamoto M., Urban T.J., et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc. Natl Acad. Sci. USA. 2003;100:5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller M.P., Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum. Mol. Genet. 2001;10:2319–2328. doi: 10.1093/hmg/10.21.2319. [DOI] [PubMed] [Google Scholar]
- 33.Mooney S.D., Klein T.E. The functional importance of disease-associated mutation. BMC Bioinformatics. 2002;3:24. doi: 10.1186/1471-2105-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mooney S.D., Klein T.E., Altman R.B., Trifiro M.A., Gottlieb B. A functional analysis of disease-associated mutations in the androgen receptor gene. Nucleic Acids Res. 2003;31:e42. doi: 10.1093/nar/gng042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mooney S.D., Altman R.B. MutDB: annotating human variation with functionally relevant data. Bioinformatics. 2003;19:1858–1860. doi: 10.1093/bioinformatics/btg241. [DOI] [PubMed] [Google Scholar]
- 36.Saunders C.T., Baker D. Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J. Mol. Biol. 2002;322:891–901. doi: 10.1016/s0022-2836(02)00813-6. [DOI] [PubMed] [Google Scholar]
- 37.Sunyaev S., Ramensky V., Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 38.Engelman D.M., Steitz T.A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 39.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horn F., Weare J., Beukers M.W., Horsch S., Bairoch A., Chen W., Edvardsen O., Campagne F., Vriend G. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 1998;26:275–279. doi: 10.1093/nar/26.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horn F., Bettler E., Oliveira L., Campagne F., Cohen F.E., Vriend G. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arkin I.T., Brunger A.T. Statistical analysis of predicted transmembrane alpha-helices. Biochim. Biophys. Acta. 1998;1429:113–128. doi: 10.1016/s0167-4838(98)00225-8. [DOI] [PubMed] [Google Scholar]
- 43.Hildebrand P.W., Preissner R., Frommel C. Structural features of transmembrane helices. FEBS Lett. 2004;559:145–151. doi: 10.1016/S0014-5793(04)00061-4. [DOI] [PubMed] [Google Scholar]
- 44.Sharon D., Gilad Y., Glusman G., Khen M., Lancet D., Kalush F. Identification and characterization of coding single-nucleotide polymorphisms within a human olfactory receptor gene cluster. Gene. 2000;260:87–94. doi: 10.1016/s0378-1119(00)00467-4. [DOI] [PubMed] [Google Scholar]
- 45.Gilad Y., Segre D., Skorecki K., Nachman M.W., Lancet D., Sharon D. Dichotomy of single-nucleotide polymorphism haplotypes in olfactory receptor genes and pseudogenes. Nature Genet. 2000;26:221–224. doi: 10.1038/79957. [DOI] [PubMed] [Google Scholar]
- 46.Glusman G., Yanai I., Rubin I., Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 47.Fuchs T., Glusman G., Horn-Saban S., Lancet D., Pilpel Y. The human olfactory subgenome: from sequence to structure and evolution. Hum. Genet. 2001;108:1–13. doi: 10.1007/s004390000436. [DOI] [PubMed] [Google Scholar]
- 48.Rost B., Yachdav G., Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Donovan C., Martin M.J., Gattiker A., Gasteiger E., Bairoch A., Apweiler R. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief. Bioinformatics. 2002;3:275–284. doi: 10.1093/bib/3.3.275. [DOI] [PubMed] [Google Scholar]
- 51.Gerstein M., Sonnhammer E.L., Chothia C. Volume changes in protein evolution. J. Mol. Biol. 1994;236:1067–1078. doi: 10.1016/0022-2836(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 52.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 53.Pei J., Grishin N.V. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 54.Sunyaev S.R., Eisenhaber F., Rodchenkov I.V., Eisenhaber B., Tumanyan V.G., Kuznetsov E.N. PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
- 55.Frank E., Hall M., Trigg L., Holmes G., Witten I.H. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 56.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng P.C., Henikoff J.G., Henikoff S. PHAT: a transmembrane-specific substitution matrix. Predicted hydrophobic and transmembrane. Bioinformatics. 2000;16:760–766. doi: 10.1093/bioinformatics/16.9.760. [DOI] [PubMed] [Google Scholar]
- 58.Tourasse N.J., Li W.H. Selective constraints, amino acid composition, and the rate of protein evolution. Mol. Biol. Evol. 2000;17:656–664. doi: 10.1093/oxfordjournals.molbev.a026344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.