Abstract
Recent work has shown that in Bacillus subtilis catabolite repression of several operons is mediated by a mechanism dependent on DNA-binding protein CcpA complexed to a seryl-phosphorylated derivative of HPr [HPr(Ser-P)], the small phosphocarrier protein of the phosphoenolpyruvate-sugar phosphotransferase system. In this study, it was found that a transposon insertional mutation resulted in the partial loss of gluconate (gnt) and xylose (xyl) operon catabolite repression by glucose, mannitol, and sucrose. The transposon insertion was localized to a gene, designated ccpB, encoding a protein 30% identical to CcpA, and relief from catabolite repression was shown to be due to the absence of CcpB rather than to the absence of a protein encoded by a downstream gene within the same operon. The relative intensities of CcpA- and CcpB-mediated catabolite repression depended on growth conditions. On solid media, and when cells were grown in liquid media with little agitation, CcpB and CcpA both proved to function in catabolite repression. However, when cells were grown in liquid media with much agitation, CcpA alone mediated catabolite repression. Like CcpA, CcpB appears to exert its catabolite-repressing effect by a mechanism dependent on the presence of HPr(Ser-P).
Carbon catabolite repression (CR) is a general phenomenon whereby the presence of a rapidly metabolizable carbon source in the growth medium of an organism inhibits the synthesis of enzymes involved in the utilization of other carbon sources. CR is mediated at the level of target gene transcription. Three distinct mechanisms of regulation have been well characterized, two in enteric bacteria and one in Bacillus subtilis. The first mechanism of CR, demonstrated in Escherichia coli, involves transcriptional activation (positive control) by the well-characterized cyclic AMP-binding cyclic AMP receptor protein (cyclic AMP-CRP) (20). The second mechanism of CR, also established in E. coli, involves transcriptional activation (positive control) mediated by the metabolite-binding catabolite repressor/activator (Cra) protein, previously called the fructose repressor, FruR (30, 34). Cyclic AMP-CRP and Cra together mediate catabolite repression of literally hundreds of genes in enteric bacteria. In B. subtilis, cyclic AMP is essentially lacking (19), suggesting that the cyclic AMP-CRP-dependent mechanism is nonoperative. Nevertheless, evidence for multiple CR mechanisms in this organism has been presented (reviewed in reference 26; 35). Recently, molecular evidence concerning one such mechanism involving transcriptional repression (negative control) has been forthcoming (12). This evidence can be summarized as follows. First, cis-acting sequences called catabolite-responsive elements (cre) were functionally identified in operons including the amyE, gntR, xylA, hutP, acsA, and acuA genes. These genes encode protein products involved in the utilization of amylose, gluconate, xylan, histidine, acetate, and acetoin, respectively (13, 40). Second, CR of over a dozen operons was shown to be affected by a trans-acting factor, catabolite control protein A (CcpA) (10, 12, 24). CcpA, which possesses a highly conserved helix-turn-helix DNA-binding motif, belongs to the LacI-GalR family of transcriptional repressors (10, 39). Finally, a second trans-acting factor, the small heat-stable protein HPr of the phosphoenol-pyruvate (PEP)-sugar phosphotransferase system (PTS), was shown to be involved in B. subtilis CR. HPr can be phosphorylated on histidyl residue 15 in a reaction which depends on PEP and enzyme I of the PTS, and this phosphorylation event is required for PTS-dependent sugar transport. Alternatively, HPr can be phosphorylated on seryl residue 46 [the product is designated HPr(Ser-P)] in a reaction which depends on a metabolite-activated, ATP-dependent protein kinase. This phosphorylation event appears to be important for CR of many operons (4, 15).
One molecular mechanism underlying cre-dependent B. subtilis CR involves an interaction between the two trans-acting factors, CcpA and HPr(Ser-P). Bacillus megaterium CcpA, which exhibits 75% amino acid identity with B. subtilis CcpA (11), specifically binds B. subtilis HPr(Ser-P) (3). Footprinting experiments have recently revealed that the CcpA protein protects cre from DNase I digestion (8, 14). However, it is not entirely clear whether CcpA alone or the complex of CcpA with HPr(Ser-P) binds to cre. The α-amylase cre was reported to be protected against DNase I digestion by CcpA alone (14), while the gluconate cre was protected against DNase I digestion only by the complex (8).
In order to further elucidate the signal transduction networks controlling B. subtilis CR, we initiated a genetic approach to identify genes whose products are important for gnt and xyl operon CR. We report the identification and characterization of a gene which we have designated ccpB. CcpB, the product of the ccpB gene, exhibits significant amino acid similarity with CcpA and may mediate CR in parallel to CcpA.
MATERIALS AND METHODS
Bacterial strains.
B. subtilis strains used in this study are listed in Table 1. B. subtilis was transformed as described by Kunst et al. (17), and selection was carried out on Luria-Bertani (LB) plates (36) containing phleomycin (0.25 μg/ml), kanamycin (5 μg/ml), erythromycin (0.4 μg/ml), spectinomycin (100 μg/ml), and/or chloramphenicol (5 μg/ml). The Δ(spo0E) mutation and the xylA-lacZ fusion are associated with kanamycin resistance. The ptsH1 mutation, the Tn917lac insertion, and the gntRK′-′lacZ fusion are associated with chloramphenicol, erythromycin, and phleomycin resistance, respectively. E. coli K-12 strain TG1 [Δ(lac-proAB) thi supE hsdD5 (F′ traD36 proAB lacIq lacZΔM15)] (9) was used as a host to clone the mutated gene from B. subtilis. Standard procedures were used to transform E. coli (36), and selection was performed on LB plates supplemented with spectinomycin (150 μg/ml) or ampicillin (100 μg/ml).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotypea | Reference or sourceb |
|---|---|---|
| JH642 | trpC2 pheA1 | J. A. Hoch |
| JH12761 | trpC2 pheA1 Δ(spo0E) | 29 |
| GM1221 | trpC2 pheA1 Δ(bgalX) amyE::(gntRK′-′lacZ) (Camr) | 4 |
| GM1222 | trpC2 pheA1 Δ(bgalX) amyE::(gntRK′-′lacZ) ptsH1 (Camr) | 4 |
| WLN29 | aroG932 trpC2 ccpA::Tn917lac | 10 |
| MD164 | trpC2 amyE::(xylAp-CRE-spoVG-lacZ) | 2 |
| ST100 | trpC2 pheA1 Δ(bgalX) amyE::(gntRK′-′lacZ) Δ(spo0E) | JH12761 chromosomal DNA tf GM1221 |
| ST101 | trpC2 pheA1 Δ(bgalX) amyE::(gntRK′-′lacZ) Δ(spo0E) ccpA::Tn917lac | WLN29 chromosomal DNA tf ST100 |
| ST102 | trpC2 pheA1 Δ(bgalX) amyE::(gntRK′-′lacZ) Δ(spo0E) ccpB::Tn10 | This work |
| ST103 | trpC2 pheA1 Δ(bgalX) amyE::(gntRK′-′lacZ) Δ(spo0E) ccpA::Tn917lac ccpB::Tn10 | ST102 chromosomal DNA tf ST101 |
| ST104 | trpC2 pheA1 Δ(bgalX) Δ(spo0E) ccpA::Tn917lac | WLN29 chromosomal DNA tf ST100 |
| ST105 | trpC2 pheA1 Δ(bgalX) amyE::(gntRK′-′lacZ) Δ(spo0E) ptsH1 (Camr) | GM1222 chromosomal DNA tf St100 |
| ST121 | trpC2 pheA1 ccpA::Tn917lac | WLN29 chromosomal DNA tf JH642 |
| ST123 | trpC2 pheA1 ccpA::Tn917lac ccpB::Tn10 | ST102 chromosomal DNA tf ST121 |
| ST124 | trpC2 pheA1 ccpA::Tn917 | p917::Phl tf ST121 |
| ST125 | trpC2 pheA1 ccpA::Tn917 ccpB::Tn10 | p917::Phl tf ST123 |
| ST131 | trpC2 amyE::(xylAp-CRE-spoVG-lacZ) ccpA::Tn917 | ST124 chromosomal DNA tf MD164 |
| ST132 | trpC2 amyE::(xylAp-CRE-spoVG-lacZ) ccpB::Tn10 | ST125 chromosomal DNA tf MD164 |
| ST133 | trpC2 amyE::(xylAp-CRE-spoVG-lacZ) ccpA::Tn917 ccpB::Tn10 | ST125 chromosomal DNA tf MD164 |
| ST137 | trpC2 amyE::(xylAp-CRE-spoVG-lacZ) ccpB::pHT181 ccpA::Tn917 | pHT181 ΔccpB tf ST131 |
| ST138 | trpC2 amyE::(xylAp-CRE-spoVG-lacZ) orf126::pHT181 ccpA::Tn917 | pHT181 Δorf126 tf ST131 |
| ST139 | trpC2 amyE::(xylAp-CRE-spoVG-lacZ) orf184::pHT181 ccpA::Tn917 | pHT181 Δorf184 tf ST131 |
GM1221 and GM1222 contain a pJH101 derivative (conferring chloramphenicol [Cam] resistance) inserted downstream of ptsH1 in GM1222.
tf, the indicated chromosomal DNA was used to transform the indicated recipient strain.
Mutagenesis and cloning of mutated genes.
Mutagenesis of B. subtilis ST100 was performed by transposition with the pIC333 plasmid (37). pIC333 carries a mini-Tn10 containing a ColE1 origin and a spectinomycin resistance gene, an erythromycin resistance gene, a thermosensitive origin of replication for gram-positive hosts, and a Tn10 transposase gene. B. subtilis ST100(pIC333) transformants were obtained on LB plates containing erythromycin and spectinomycin incubated at 28°C. Transformants were grown in liquid LB supplemented with spectinomycin. The temperature was shifted from 28 to 37°C at the beginning of the exponential growth phase. Cells were grown for four additional hours at 37°C and then plated with 100 μl of 20-mg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) on 30-ml LB plates containing spectinomycin plus gluconate (1%) and glucose (1%). Plates were incubated at 37°C. The loss of plasmid pIC333 was identified by determining sensitivity to erythromycin. The gene carrying the Tn10 insertion was cloned in E. coli as described previously (37). Chromosomal DNA was extracted from the B. subtilis mutant and was cut with HindIII, which does not cut within the mini-Tn10 derivative sequence. The DNA fragments were self-ligated and used to transform E. coli. Selection of transformants was based on the spectinomycin resistance trait carried by the Tn10 insert.
DNA manipulations.
Chromosomal DNA was isolated from exponentially growing B. subtilis cells in Penassay antibiotic medium 3 (Difco) as described previously (25). A Qiagen kit (United States Biochemicals) was used to extract plasmids from E. coli. Restriction enzymes, Thermus aquaticus DNA polymerase, and T4 DNA ligase were used as recommended by the manufacturers. DNA adjacent to the Tn10 insertion was sequenced by using double-stranded templates with a Sequenase kit (United States Biochemicals) and two oligonucleotides, each hybridizing to a strand of an extremity of the mini-Tn10 derivative sequence. PCR amplification of double-stranded DNA was performed by using a thermal reactor (Hybaid Limited, Teddington, Middlesex, United Kingdom), with the following program: 99°C for 5 min, followed by 25 cycles, each consisting of 1 min at 55°C, 2 min at 72°C, and 1 min at 95°C, and ending with a 3-min incubation at 55°C and a 5-min incubation at 72°C. The nucleotide sequences of all cloned PCR products were verified by nucleotide sequencing (University of California at San Diego Core Facility).
Construction of ccpB, orf126, and orf184 knockout mutants.
Fragments of 313, 321, and 325 bp were amplified by PCR to clone the 5′ ends of ccpB, orf126, and orf184, respectively, without their initiation codons. The oligonucleotides used were 5′-ATGGGATCCATAAAAGAGATCGCAAGACT-3′ (BamHI site underlined) and 5′-ACAGAATTCAATATTTGATTTCAATATCG-3′ (EcoRI site underlined) for ccpB, 5′-TGCGGATCCAACATACAGCGGTCTGGGTC-3′ (BamHI) and 5′-TCAGAATTCCATAGTAGCCGTCTCCTGTT-3′ (EcoRI) for orf126, and 5′-TGAGGATCCAAAAAGAAAAAATGGCAGCC-3′ (BamHI) and 5′-GAGGAATTCCGGCCGTGTAGATATGTACA-3′ (EcoRI) for orf184. The PCR fragments were cloned into pHT181, a B. subtilis suicide plasmid which is unable to replicate in that host (18). The recombinant plasmids, pHT181ΔccpB, pHT181Δorf126 and pHT181Δorf184, were transformed into B. subtilis by selecting for erythromycin resistance and allowing integration into the chromosome by homologous recombination at either the ccpB, orf126, or orf184 locus through a Campbell-type mechanism. The correct insertion on the chromosome was checked by PCR with the universal primer hybridizing to pHT181 and the appropriate oligonucleotide hybridizing to the 3′ end of the cloned fragment. Thus, transformation into B. subtilis and selection for erythromycin resistance resulted in the integration of the recombinant plasmid into the chromosome by homologous recombination. The cloned fragment carried by the recombinant plasmid was therefore present in two copies on the chromosome—the original copy and the cloned copy brought in by the plasmid. The oligonucleotide hybridizes to the 3′ ends of both copies, but the hybridization to the original copy only gives a PCR product in the presence of the universal primer.
Construction of a ccpB′-′lacZ translational fusion.
Plasmids pAC7 and pJF751 are vectors allowing construction of translational fusions with codon 8 of the β-galactosidase gene (5, 41). Both plasmids cannot replicate in B. subtilis but can integrate into the chromosome via homologous recombination. pAC7 carries the promoterless lacZ gene, which lacks a ribosome binding site, as well as a chloramphenicol resistance determinant between two fragments of the B. subtilis amyE gene. pJF751 carries the promoterless lacZ gene and a kanamycin resistance determinant. The promoter region of ccpB was amplified by PCR on a 183-bp fragment with oligonucleotides 5′-ACAGAATTCTTGGATGGCGGATTGATTAT-3′ (EcoRI) and 5′-GCGGGATCCTTTATATTTGCCATCTCTTT-3′ (BamHI) and was cloned into pAC7 and pJF751. Recombinant plasmids pAC7B and pJF751B both carry a ccpB′-′lacZ translational fusion for which the point of fusion is the fifth codon of ccpB. pAC7B was linearized by using the unique ScaI site and was transformed into B. subtilis JH642 by chloramphenicol selection, allowing integration into the chromosome by homologous recombination at the amyE locus through a double-crossover event and disruption of the amyE gene by the translational gene fusion (α-amylase deficiency phenotype). pJF751B was transformed into B. subtilis JH642 by kanamycin selection, allowing integration into the chromosome by homologous recombination at the ccpB locus through a Campbell-type mechanism.
Enzyme assays.
B. subtilis cells from a single colony were grown overnight at 37°C in 5 ml of LB medium containing a 1% concentration of the desired sugar and the appropriate antibiotic(s). Cultures were grown either with a high level of agitation (approximately 275 rotations/min) or a low level of agitation (150 rotations/min). One milliliter of culture was harvested by centrifugation. Cells were suspended in 1 ml of Z buffer (21) supplemented with 40 μg of lysozyme and 6.25 μg of DNase per ml and were incubated for 10 min at 37°C. Cell debris was eliminated by centrifugation. β-Galactosidase activity was measured according to the method of Miller and was expressed as Miller units (21). Protein concentrations were measured with the Coomassie blue reagent supplied by Bio-Rad (1), with bovine serum albumin as a standard.
RESULTS
Isolation of mutants resistant to gnt operon catabolite repression.
The gnt operon of B. subtilis includes four open reading frames (ORFs): gntR, gntK, gntP, and gntZ, which respectively encode the transcriptional repressor of the operon, gluconate kinase, gluconate permease, and phosphogluconate dehydrogenase (6, 31). Expression of the gnt operon is subject to catabolite repression mediated by a mechanism involving (i) the cre sequence present in the coding region of gntR (22, 23), (ii) the CcpA protein (7), and (iii) HPr(Ser-P) (4). B. subtilis mutants resistant to the CR of gnt operon expression were generated with the pIC333 plasmid carrying a mini-Tn10 transposon (37). Strain GM1221 (Table 1) carries a gntRK′-′lacZ translational fusion inserted into the chromosomal amyE gene (4) and contains a chromosomal pJH101 derivative which shares homologous sequences with plasmid pIC333. To ensure that no homologous recombination between these sequences had occurred, the pJH101 derivative was removed by introducing a spo0E mutation, generating strain ST100 (Table 1). As for GM1221, expression of lacZ in ST100 is induced by gluconate (blue colonies on LB plates containing gluconate and X-Gal) and is repressed by glucose (white colonies on LB plates containing gluconate, glucose, and X-Gal). Transposon mutagenesis of 32 distinct ST100 (pIC333) transformants gave rise to 32 independently isolated mutants (blue colonies on LB plates containing gluconate, glucose, and X-Gal). According to their phenotypes, the 32 mutants, which were fully or partially resistant to glucose-promoted gnt operon CR, were grouped into 12 different classes. One of these classes was represented by strain ST102 (Table 1), which was isolated as a pale blue colony on an LB plate containing gluconate, glucose, and X-Gal.
Identification of the ccpB gene.
To ensure that only a single transposon insertion was present in ST102, a backcross was performed. ST100 was transformed with chromosomal DNA from ST102, and transformants were selected with spectinomycin. All of these transformants were blue on LB plates containing gluconate, glucose, and X-Gal, suggesting that the phenotype of resistance to CR by glucose was due to a single chromosomal transposon insertion. The mutated gene was cloned in E. coli and was sequenced from both extremities of the transposon. The nucleotide sequence of a 127-bp fragment was compared to sequences in the GenBank database and found to be identical to part of an ORF of unknown function identified by Ogasawara et al. (27). This ORF, previously designated yyaG, is now referred to as ccpB and is localized in the tetB (358°)-spo0J (359°) intergenic region. The putative gene product (CcpB) is 311 amino acids in length and is predicted to have a molecular mass of 34.8 kDa.
As reported by Ogasawara et al. (27), CcpB shows a high degree of similarity to proteins of the LacI-GalR family of transcriptional repressors (39). CcpB and B. subtilis CcpA exhibit 30% amino acid identity (Fig. 1). Like CcpA, CcpB possesses a highly conserved helix-turn-helix motif in its amino-terminal domain, suggesting that it is a DNA-binding protein. In the ST102 mutant, the CcpB gene was interrupted at codon 235 by the transposon insertion.
FIG. 1.
Sequence comparison of CcpB and CcpA of B. subtilis. Numbers preceding each line indicate the positions in the amino acid sequences. Identical residues are highlighted white on black. The predicted helix-turn-helix region is underlined. The position corresponding to the mini-Tn10 insertion in ccpB is indicated with a box. The CcpA and CcpB primary sequences are available from the SWISS-PROT database under accession numbers P25144 and P37517, respectively.
Structure of the putative ccpB operon.
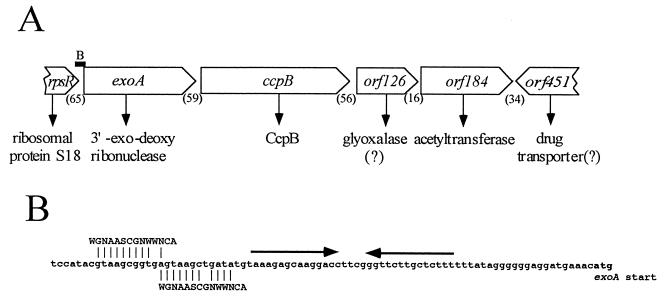
A ccpB′-′lacZ translational fusion was not expressed when only 183 nucleotides of upstream DNA were present (see below). This observation suggested that ccpB is not directly preceded by a promoter. An examination of the regions flanking ccpB suggested that ccpB is the second gene in a four-gene operon (Fig. 2). The first gene in this proposed operon is exoA, encoding 3′-exodeoxyribonuclease. Three genes encoding two ribosomal proteins and a single-stranded DNA-binding protein precede the exoA gene, but they are followed by a stem-loop structure that resembles a Rho-independent transcriptional terminator. We suggest that these three genes constitute a distinct operon. As shown in Fig. 2, two sequences resembling, but different from, the CcpA-binding cre consensus sequence precede the putative terminator. It is possible that these sequences act as CcpB binding sites although they precede the terminator of the upstream operon. It should be noted that it is not clear where transcription of the putative exoA-ccpB operon originates as there are no obvious candidate promoter sequences.
FIG. 2.
Proposed structure of the region of the B. subtilis chromosome that encompasses the ccpB gene. (A) The arrangement of genes surrounding the ccpB gene is depicted. The map shows the proposed gene and protein assignments, the direction of transcription (indicated by the open arrows), and the numbers of base pairs (in parentheses) in the intergenic regions. The solid bar labeled B indicates the position of the region shown in panel B. (B) The nucleotide sequence preceding the exoA gene. The sequence includes an imperfect palindrome that might serve as a Rho-independent terminator (indicated by the arrows) and two overlapping regions which show similarity with each other and the cre consensus sequence (13). See text for details.
Downstream of ccpB are two genes, orf126 and orf184. The product of orf126 is homologous but distantly related to glyoxalase I (R-S-lactoylglutathione methylglyoxal lyase [isomerizing]; also known as lactoylglutathione lyase, methylglyoxalase, aldoketomutase, and ketone-aldehyde mutase). Orf184 is homologous to numerous acetyltransferases, including the lacA gene product of E. coli. Downstream of orf184 is a set of genes with the opposite orientation on the chromosome. orf184 must therefore be the last gene in the putative operon that includes the ccpB gene.
Effects of ccpA and ccpB mutations on gntRK′-′lacZ expression.
Table 2 summarizes the β-galactosidase activities of various B. subtilis gntRK′-′lacZ fusion strains in response to potential repressing sugars on solid media. The strains examined include the wild type (ST100), a ccpA mutant (ST101), a ccpB mutant (ST102), and a ccpA ccpB double mutant (ST103). In the presence of the inducer, gluconate, but in the absence of a repressing sugar, high levels of β-galactosidase were produced (colonies were dark blue). Glucose and mannitol were the most strongly repressing sugars, and essentially no β-galactosidase was produced by the wild-type strain in the presence of one of these sugars (colonies were white). The ccpA and ccpB mutants both showed less severe CR in the presence of one of these sugars, and the double mutant exhibited little or no CR. Repression by sucrose was less severe, but the relative responses of the mutants were similar to those observed when glucose or mannitol was the repressing sugar. Finally, glycerol did not exert a CR effect in any of the strains under the assay conditions employed.
TABLE 2.
Effects of ccpA and ccpB mutations on the expression of a gntRK′-′lacZ fusion by various sugars on solid medium containing X-Gala
| Repressing sugar | Response of
strainb:
|
||||
|---|---|---|---|---|---|
| ST100 | ST101 | ST102 | ST103 | ST105 | |
| None | ++ | ++ | ++ | ++ | ++ |
| Glucose | − | +/− | +/− | + | + |
| Mannitol | − | + | + | ++ | ++ |
| Sucrose | +/− | + | + | ++ | ++ |
| Glycerol | ++ | ++ | ++ | ++ | ++ |
Cells were streaked onto LB plates containing X-Gal and 1% gluconate as well as a 1% concentration of the repressing sugar indicated and were incubated overnight at 37°C. Responses were recorded as follows: ++, dark blue; +, blue; +/−, pale blue; −, white. In the absence of gluconate, all colonies were white.
Strain ST100 was the wild type; strains ST101, ST102, ST103, and ST105 carried defective genes ccpA, ccpB, ccpA and ccpB, and ptsH1, respectively.
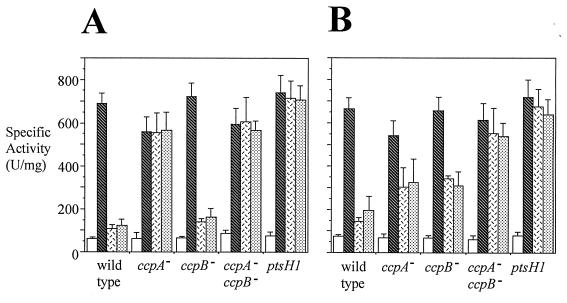
CR of gnt operon expression was quantified in strain ST100, the wild-type strain, as well as in strains ST101, ST102, ST103, and ST105, which are defective for ccpA, ccpB, ccpA and ccpB, and ptsH, respectively. The ptsH1 gene in strain ST105 codes for the HPr(Ser46Ala) mutant protein which cannot be phosphorylated at position 46 (33). In order to determine β-galactosidase production in the presence of the ccpA::Tn917lac mutation, ST104 was constructed. ST104 is isogenic with ST101 except that the gntRK′-′lacZ fusion was removed by congression (the simultaneous introduction of two unlinked markers by transformation [frequency around 1%]) (screening for phleomycin sensitivity). The β-galactosidase specific activity of ST104 was subtracted from all values obtained for ST101 and ST103. β-Galactosidase specific activities were determined with noninduced cells (absence of gluconate), induced cells (presence of gluconate), and repressed cells (presence of gluconate plus glucose or mannitol). Two growth conditions were used: cells were either grown with a high level of agitation (Fig. 3A) or with a low level of agitation (Fig. 3B).
FIG. 3.
Sensitivities of gnt operon expression to CR under conditions of high (A) and low (B) levels of culture agitation. Cell growth conditions, crude extract preparation, and β-galactosidase assays were as described in Materials and Methods. β-Galactosidase specific activity (units per milligram of protein) is given on the y axis, while the strains are indicated on the x axis as follows: wild type, ST100; ccpA−, ST101; ccpB−, ST102; ccpA− ccpB−, ST103; ptsH1, ST105. Open bars, LB medium; hatched bars, LB medium plus 1% gluconate; open bars with black dashes, LB medium plus 1% gluconate plus 1% glucose; stippled bars, LB medium plus 1% gluconate plus 1% mannitol. Error bars denote standard deviations.
When cells were grown with a high agitation rate (Fig. 3A), gnt operon expression in the wild-type strain (ST100) was strongly repressed by glucose or mannitol. The ccpA mutant and the ptsH1 mutant exhibited β-galactosidase specific activities in the presence of glucose or mannitol that approached those observed in the absence of a repressing sugar. These cells thus showed full resistance to CR. The ccpB mutant exhibited a specific activity similar to that of ST100 in the presence of glucose or mannitol, showing that the ccpB mutant was as sensitive to catabolite repression as the wild-type strain under these conditions. Indeed, the ccpA ccpB double mutant exhibited the same activity as the ccpA mutant.
When cells were grown with a low level of agitation (Fig. 3B), glucose or mannitol strongly repressed gnt operon expression in the wild-type strain (ST100). The ccpA mutant and the ccpB mutant exhibited β-galactosidase specific activities in the presence of glucose or mannitol that were approximately 50% of those observed in the absence of a repressing sugar. Under these conditions, and in contrast to what was found for cells grown with a high level of agitation, the ccpA mutant was only partially resistant to CR. In the presence of glucose or mannitol, the specific activity of the ccpA ccpB double mutant was significantly higher than that of either single mutant, showing that the two mutations had approximately additive effects. The ptsH1 mutant had the same specific activity as the ccpA ccpB double mutant, showing that the two strains were equally insensitive to CR. The results suggest that CcpB plays a significant role in CR under these conditions.
Effects of ccpA and ccpB mutations on xylA-lacZ expression.
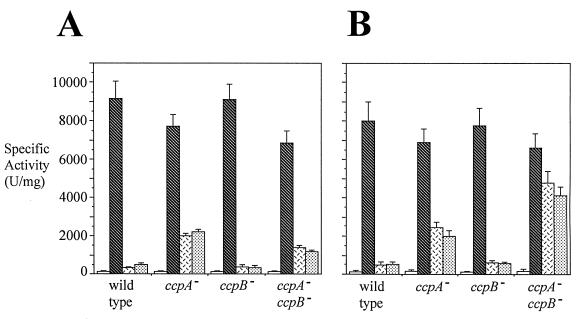
In order to determine if the ccpB knockout mutation exerts an effect on CR of the xyl operon of B. subtilis, the ccpA and ccpB mutants and the ccpA ccpB double mutant were constructed in the genetic background of a xylA-lacZ transcriptional fusion. β-Galactosidase production in the presence of the ccpA::Tn917lac mutation was eliminated by replacing the lacZ- erythromycin resistance region with phleomycin resistance by using plasmid p917::Phl as described by Steinmetz and Richter (38). The resulting strains, ST124 and ST125, were used to construct ST131, ST132, and ST133 carrying the ccpA mutation, the ccpB mutation, and both the ccpA and ccpB mutations, respectively (Table 1). These isogenic strains, together with the wild type, MD164, were used to quantify CR of xyl operon expression in liquid media under conditions of high and low levels of agitation (Fig. 4A and B, respectively). When cells were grown with a high level of agitation (Fig. 4A), xyl operon expression in the wild-type strain and the ccpB mutant was strongly repressed by glucose or sucrose. In addition, the ccpA ccpB double mutant exhibited a specific activity similar to that of the ccpA mutant in the presence of glucose or sucrose, showing that CcpB is not involved in CR of xyl operon expression under these conditions. When cells were grown with a low level of agitation (Fig. 4B), xyl operon expression in wild-type and ccpB mutant strains was strongly repressed by glucose or sucrose. The β-galactosidase specific activity of the ccpA ccpB double mutant in the presence of glucose or sucrose was significantly higher than that of the ccpA single mutant, showing that CcpB is involved in the CR of xyl operon expression under these conditions. β-Galactosidase production on solid plates not only confirmed the results observed for cells grown with a low level of agitation but also showed that mannitol had the same effect as glucose or sucrose (Table 3).
FIG. 4.
Sensitivities of xyl operon expression to CR under conditions of high (A) and low (B) levels of culture agitation. The experimental design and format of presentation are as described in the legend to Fig. 3 with the following differences: wild type, MD164; ccpA−, ST131; ccpB−, ST132; ccpA− ccpB−, ST133; hatched bars, LB medium plus 1% xylose; white bars with black dashes, LB medium plus 1% xylose plus 1% glucose; stippled bars, LB medium plus 1% xylose plus 1% sucrose. Error bars denote standard deviations.
TABLE 3.
Effects of ccpA and ccpB mutations on the expression of a xylA-lacZ fusion by various sugars on solid medium containing X-Gala
| Repressing sugar | Response of
strainb:
|
||||||
|---|---|---|---|---|---|---|---|
| MD164 | ST131 | ST132 | ST133 | ST137 | ST138 | ST139 | |
| None | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Glucose | − | + | − | ++ | ++ | + | + |
| Mannitol | +/− | + | +/− | ++ | ++ | + | + |
| Sucrose | − | + | − | ++ | ++ | + | + |
Cells were streaked onto LB plates containing X-Gal and 1% xylose as well as a 1% concentration of the repressing sugar indicated and were incubated overnight at 37°C. Responses were recorded as follows: ++, dark blue; +, blue; +/−, pale blue; −, white. In the absence of xylose, all colonies were white.
Strain MD164 was the wild type; the other strains carried defective genes as follows: ST131, ccpA; ST132, ccpB; ST133, ccpA and ccpB; ST137, ccpA and ccpB; ST138, ccpA and orf126; ST139, ccpA and orf184.
Null mutations in genes of the ccpB operon.
Since ccpB, orf126, and orf184 are apparently included in a single operon, the phenotype resulting from the mini-Tn10 mutation in ccpB could have been due to loss of function of either CcpB or the product of one of the downstream genes. In order to distinguish between these three possibilities, null mutations were constructed in each of these three genes by inserting an erythromycin resistance cartridge into each of them. Three strains, ST137 (ccpB mutant), ST138 (orf126 mutant), and ST139 (orf184 mutant) were thus generated in a ccpA mutant genetic background in the presence of the xylA-lacZ fusion (Table 1). These strains were assayed for β-galactosidase activities both on solid media (Table 3) and in liquid media (data not shown). The plate phenotype of ST137 proved to be identical to that of strain ST133, while the plate phenotype of ST138 and ST139 proved to be identical to that of ST131. The same conclusion resulted when β-galactosidase activity was assayed after growth in liquid media with a low level of agitation as described in the legend to Fig. 4. In the presence of the ccpA mutation, the ccpB mutation partially relieved CR of xyl operon expression (from 70% repression to 35% repression) although the orf126 and orf184 mutations did not (70% repression for strains ST138 and ST139 as well as for strain ST131; data not shown). These results establish that CcpB and not the product of one of the downstream genes is responsible for the CR-resistant phenotype described above.
Constitutivity of ccpB expression.
A ccpB′-′lacZ translational fusion was inserted into the chromosome at either the amyE locus or the native ccpB locus. The resulting strains were examined for β-galactosidase production by the in vitro assay described in Materials and Methods. When the fusion was inserted at the amyE locus, essentially no β-galactosidase activity (<20 U/mg of protein) was detected, showing that the fragment amplified by PCR did not bear a promoter. This observation suggested that ccpB may be in an operon with the upstream gene, which encodes a protein homologous to 3′-exodeoxyribonuclease.
When the fusion was inserted into the native chromosomal ccpB gene, a high level of β-galactosidase activity was observed (about 250 U/mg of protein). This activity was the same when cells were grown in LB medium with or without glucose. Moreover, this activity did not change when the cultures were maintained with either a low or high level of agitation. These results suggest that CcpB, like CcpA (24), is regulated at the posttranslational level.
DISCUSSION
CR in B. subtilis is a complex and still poorly understood phenomenon. Evidence has been presented suggesting that one mechanism of CR depends on a cre DNA sequence, the CcpA protein, and HPr(Ser-P) (see above). This mechanism is postulated to be sensitive to cytoplasmic sugar catabolite concentrations by virtue of the fact that the ATP-dependent HPr(Ser)-kinase is allosterically activated by metabolites of sugar catabolism such as fructose-1,6-bisphosphate, gluconate-6-phosphate, and 2-phosphoglycerate (3, 4, 32).
In the present study, we report the identification of a novel transcription factor, CcpB, which apparently mediates CR of the B. subtilis gnt and xyl operons in parallel with CcpA. The ccpB mutant was initially isolated in a gntRK′-′lacZ translational fusion host strain as a pale blue colony on LB plates containing gluconate, glucose, and X-Gal. The fact that gnt operon expression exhibited normal inducibility suggested that the ccpB mutation confers partial resistance of gnt operon expression to CR by glucose. In vivo β-galactosidase assays indeed showed that the ccpB mutation partially relieves the sensitivity of gnt operon expression to glucose, mannitol, and sucrose CR. Similarly, this mutation partially relieves the sensitivity of xyl operon expression to glucose, mannitol, and sucrose CR. The possibility that the product of a gene downstream of the ccpB gene was responsible for the phenotype was eliminated. These observations clearly suggest that CcpB, like CcpA, may function as a general mediator of CR in B. subtilis. The mechanism of CcpB action remains unknown. Whether it acts directly on the control regions of target operons or acts indirectly by regulating expression of a gene whose product influences CR has yet to be determined.
A CcpB dependency was observed when cells were grown on solid media or when the liquid culture agitation rate was low, but not when the agitation rate was high. These observations indicate that environmental conditions apparently affect the relative intensities of CcpA- and CcpB-mediated CR. Dependence of B. subtilis gene expression on growth conditions has been reported previously. Regulation of the citrulline biosynthetic operon, argC-F (28), and of the levansucrase gene, sacB (16), is different for cells grown on solid versus liquid media. The growth conditions which are likely to give rise to differential gene expression may include (i) oxygen availability, (ii) cell density, (iii) the growth phase, and (iv) the concentrations of diffusible substances which accumulate in or are removed from the media. Our attempts to identify which of these conditions determine the sensitivity of CR to CcpA versus CcpB action have as yet been inconclusive (unpublished results).
CcpB displays 30% identity to CcpA, and like CcpA, it bears a highly conserved amino-terminal helix-turn-helix motif, suggesting that it is a DNA-binding protein. The effects of the loss of each of the two proteins, CcpA and CcpB, on CR are apparently additive, and loss of both proteins is quantitatively equivalent to the loss of serine 46 in HPr, due to the ptsH1 mutation. Both proteins appear to function in CR by mechanisms that involve posttranslational regulation. Our observations lead to the following mechanistic proposal. Both CcpA and CcpB function in CR by similar mechanisms. That is, both proteins bind to cre in the DNA, both exert comparable repressive effects on gnt and xyl operon expression, and both are probably activated for DNA binding by HPr(Ser-P). Since expression of both the ccpA and ccpB genes is apparently unaffected by the growth conditions that determine relative sensitivities of B. subtilis to CcpA- and CcpB-mediated CR, we suggest that either the activities or the stabilities of these two transcription factors are reciprocally regulated by growth conditions. Experiments are currently in progress to determine the causative agents that determine the relative sensitivities to CcpA and CcpB action and to establish the mechanistic details of CcpB-mediated CR.
ACKNOWLEDGMENTS
Ian Paulsen was supported by a C. J. Martin fellowship from the National Health and Medical Research Council (Australia). This work was supported by USPHS grant 9RO1 GM55434 from the National Institute of General Medical Sciences.
We thank Mary Beth Hiller for expert assistance in the preparation of this manuscript.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Dahl M K, Hillen W. Contribution of XylR, CcpA and HPr to catabolite repression of the xyl operon in Bacillus subtilis. FEMS Microbiol Lett. 1995;132:79–83. [Google Scholar]
- 3.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrari F, Trach K, Hoch J A. Sequence analysis of the spo0Blocus reveals a polycistronic transcription unit. J Bacteriol. 1985;161:556–562. doi: 10.1128/jb.161.2.556-562.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita Y, Fujita T, Miwa Y, Nihashi J, Aratani Y. Organization and transcription of the gluconate operon, gnt, of Bacillus subtilis. J Biol Chem. 1986;261:13744–13753. [PubMed] [Google Scholar]
- 7.Fujita Y, Miwa Y. Catabolite repression of the Bacillus subtilis gntoperon mediated by the CcpA protein. J Bacteriol. 1994;176:511–513. doi: 10.1128/jb.176.2.511-513.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 9.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge, England: University of Cambridge; 1984. [Google Scholar]
- 10.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galRrepressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 11.Hueck C, Kraus A, Hillen W. Sequences of ccpA and two downstream Bacillus megaterium genes with homology to the motAB operon from Bacillus subtilis. Gene. 1994;143:147–148. doi: 10.1016/0378-1119(94)90621-1. [DOI] [PubMed] [Google Scholar]
- 12.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 13.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 14.Kim J H, Guvener Z T, Cho J Y, Chung K-C, Chambliss G H. Specificity of DNA binding activity of the Bacillus subtiliscatabolite control protein CcpA. J Bacteriol. 1995;177:5129–5134. doi: 10.1128/jb.177.17.5129-5134.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krüger S, Stülke J, Hecker M. Catabolite repression of β-glucanase synthesis in Bacillus subtilis. J Gen Microbiol. 1993;139:2047–2054. doi: 10.1099/00221287-139-9-2047. [DOI] [PubMed] [Google Scholar]
- 16.Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst F, Msadek T, Rapoport G. Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis. In: Piggot P J, Moran C P Jr, Youngman P, editors. Regulation of bacterial differentiation. Washington, D.C: ASM Press; 1995. pp. 1–20. [Google Scholar]
- 18.Lereclus D, Arantes O. spbAlocus ensures the segregational stability of pHT1030, a novel type of Gram-positive replicon. Mol Microbiol. 1992;6:35–46. doi: 10.1111/j.1365-2958.1992.tb00835.x. [DOI] [PubMed] [Google Scholar]
- 19.Mach H, Hecker M, Mach F. Evidence for the presence of cyclic adenosine monophosphate in Bacillus subtilis. FEMS Microbiol Lett. 1984;22:27–30. [Google Scholar]
- 20.Magasanik B, Neidhardt F C. Regulation of carbon and nitrogen utilization. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1318–1325. [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 22.Miwa Y, Fujita Y. Determination of the cis sequence involved in catabolite repression of the Bacillus subtilis gnt operon; implication of a consensus sequence in catabolite repression in the genus Bacillus. Nucleic Acids Res. 1990;18:7049–7053. doi: 10.1093/nar/18.23.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miwa Y, Fujita Y. Promoter-independent catabolite repression of the Bacillus subtilis gntoperon. J Biochem. 1993;113:665–671. doi: 10.1093/oxfordjournals.jbchem.a124100. [DOI] [PubMed] [Google Scholar]
- 24.Miwa Y, Saikawa M, Fujita Y. Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology. 1994;140:2567–2575. doi: 10.1099/00221287-140-10-2567. [DOI] [PubMed] [Google Scholar]
- 25.Msadek T, Kunst F, Henner D, Klier A, Rapoport G, Dedonder R. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J Bacteriol. 1990;172:824–834. doi: 10.1128/jb.172.2.824-834.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nihashi J-I, Fujita Y. Catabolite repression of inositol dehydrogenase and gluconate kinase syntheses in Bacillus subtilis. Biochim Biophys Acta. 1994;798:88–95. doi: 10.1016/0304-4165(84)90014-x. [DOI] [PubMed] [Google Scholar]
- 27.Ogasawara N, Nakai S, Yoshikawa H. Systematic sequencing of the 180 kilobase region of the Bacillus subtilischromosome containing the replication origin. DNA Res. 1994;1:1–14. doi: 10.1093/dnares/1.1.1. [DOI] [PubMed] [Google Scholar]
- 28.O’Reilly M, Woodson K, Dowds B C A, Devine K M. The citrulline biosynthetic operon, argC-F, and a ribose transport operon, rbs, from Bacillus subtilisare negatively regulated by Spo0A. Mol Microbiol. 1994;11:87–98. doi: 10.1111/j.1365-2958.1994.tb00292.x. [DOI] [PubMed] [Google Scholar]
- 29.Perego M, Hoch J A. Negative regulation of Bacillus subtilis sporulation by the spo0Egene product. J Bacteriol. 1991;173:2514–2520. doi: 10.1128/jb.173.8.2514-2520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramseier T M, Bledig S, Michotey V, Feghali R, Saier M H., Jr The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol Microbiol. 1995;16:1157–1169. doi: 10.1111/j.1365-2958.1995.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 31.Reizer A, Deutscher J, Saier M H, Jr, Reizer J. Analysis of the gluconate (gnt) operon of Bacillus subtilis. Mol Microbiol. 1991;5:1081–1089. doi: 10.1111/j.1365-2958.1991.tb01880.x. [DOI] [PubMed] [Google Scholar]
- 32.Reizer J, Novotny M J, Stuiver I, Saier M H., Jr Regulation of glycerol uptake by the phosphoenolpyruvate-sugar phosphotransferase system in Bacillus subtilis. J Bacteriol. 1984;159:243–250. doi: 10.1128/jb.159.1.243-250.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reizer J, Sutrina S L, Saier M H, Jr, Stewart G C, Peterkofsky A, Reddy P. Mechanistic and physiological consequences of HPr(ser) phosphorylation on the activities of the phosphoenolpyruvate:sugar phosphotransferase system in Gram-positive bacteria: studies in site-specific mutants of HPr. EMBO J. 1989;8:2111–2120. doi: 10.1002/j.1460-2075.1989.tb03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saier M H, Jr, Ramseier T M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saier M H, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J-J. Protein phosphorylation and regulation of carbon metabolism in Gram-negative versus Gram-positive bacteria. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Steinmetz M, Richter R. Easy cloning of mini-Tn10 insertions from the Bacillus subtilischromosome. J Bacteriol. 1994;176:1761–1763. doi: 10.1128/jb.176.6.1761-1763.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinmetz M, Richter R. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivorecombination. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 39.Weickert M J, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 40.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinrauch Y, Msadek T, Kunst F, Dubnau D. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J Bacteriol. 1991;173:5685–5693. doi: 10.1128/jb.173.18.5685-5693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]