SUMMARY
Variants of the RNA-editing enzyme ADAR1 cause Aicardi-Goutières syndrome (AGS), in which severe inflammation occurs in the brain due to innate immune activation. Here, we analyze the RNA-editing status and innate immune activation in an AGS mouse model that carries the Adar P195A mutation in the N terminus of the ADAR1 p150 isoform, the equivalent of the P193A human Zα variant causal for disease. This mutation alone can cause interferon-stimulated gene (ISG) expression in the brain, especially in the periventricular areas, reflecting the pathologic feature of AGS. However, in these mice, ISG expression does not correlate with an overall decrease in RNA editing. Rather, the enhanced ISG expression in the brain due to the P195A mutant is dose dependent. Our findings indicate that ADAR1 can regulate innate immune responses through Z-RNA binding without changing overall RNA editing.
Graphical abstract
In brief
Guo et al. analyze a mouse model carrying the ADAR1 Zα domain P195A mutation and find that this specific mutation does not affect the overall RNA-editing activity of ADAR1, while it results in activation of the MDA5-dependent RNA-sensing signaling pathway, leading to robust ISG expression in the brain.
INTRODUCTION
The adenosine-to-inosine (A-to-I) RNA-editing enzyme adenosine deaminase acting on RNA1 (ADAR1) modifies the sequence and structure of double-stranded cellular RNAs (dsRNAs), an essential process for innate immune homeostasis.1-5 Dysregulation of ADAR1-catalyzed RNA editing is associated with various autoimmune diseases.6-10 RNA editing is required for innate immune tolerance, as unedited cellular RNAs are recognized as “non-self” by retinoic acid-inducible gene I-like receptors (RLRs), especially melanoma differentiation-associated protein 5 (MDA5), which triggers interferon (IFN) pathway activation.1-4 Variants in the human ADAR1 coding gene ADAR (named Adar in mouse) have been linked to Aicardi-Goutières syndrome (AGS),6,8 a multiorgan inflammatory disease with severe brain injury and elevated type I IFN and IFN-stimulated gene (ISG) expression.11-13
ADAR1 is expressed as two isoforms. The p110 protein is constitutively expressed and the p150 form is induced by IFN. While p110 is localized to the nucleus, p150 undergoes nucleocytoplasmic shuffling. Both ADAR1 isoforms share a Zβ domain that is not known to bind nucleic acids, three downstream double-stranded RNA-binding motifs, and the C terminus deamination catalytic domain14-17 (Figure 1A). The Zα domain is present only in the N terminus of the ADAR1 p150 isoform and binds to both Z-DNA and Z-RNA with high affinity in a structure-dependent manner with no base-specific contacts.14,18-20 Among the ADAR variants identified in AGS patients, the most frequent is the loss of function (LOF) P193A variant in the Z-RNA-binding Zα domain of the IFN-induced ADAR p150 isoform.6,8,21 The frequency of the P193A variant is about 0.2% worldwide.21 A second, less-common LOF N173S variant in the Zα domain also causes AGS.21 Both N173S and P193A are causal for AGS when paired with a second LOF ADAR variant or a null p150 ADAR allele, a situation in which both the p110 alleles and the catalytic domain are wild type (WT).6,8,21 In contrast, other AGS causal ADAR variants alter the catalytic domain to reduce RNA-editing activity.4,8
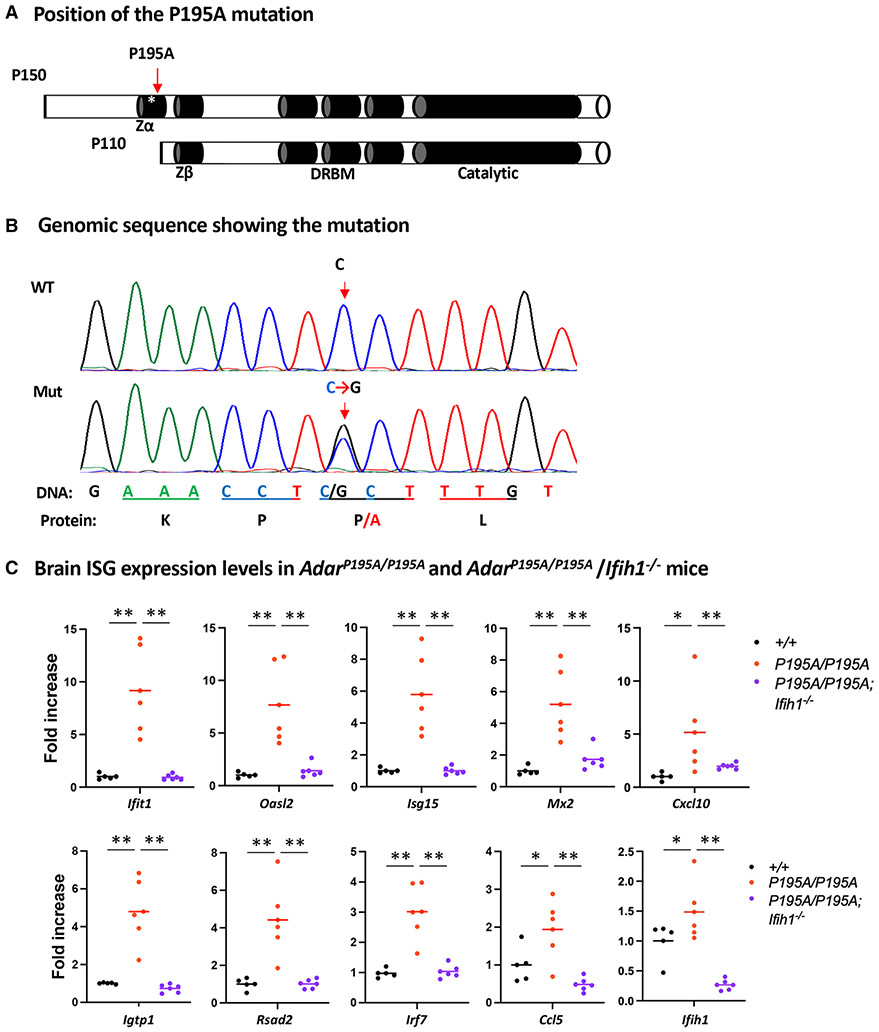
Figure 1. ADAR1 P195A mutation activates MDA5-dependent ISG expression in mouse brains.
(A) Two isoforms of ADAR1, p150 and p110, are expressed from the Adar1 gene. The p150 isoform contains the Zα domain at the N terminus; the Zβ domain, which is not known to bind nucleic acids, the three dsRNA-binding motifs (DRBMs), and the catalytic domain in the C terminus are shared by p150 and p110 isoforms. The asterisk marks the P195A site in the Zα domain; the Zα domain is not present in the p110 isoform.
(B) Mouse genomic sequences flanking the mutation site were confirmed using Sanger sequencing analysis. A single C>G nucleotide replacement that codes the P195A mutation is shown on the sequence histography indicated by the arrows, and the corresponding protein sequence is shown at the bottom.
(C) Mouse brain ISG expression was assessed at mRNA levels. Expression of a panel of 10 selected ISGs was measured using real-time RT-PCR, and the ISG expression levels in AdarP195A/P195A mice (n = 6) were compared with those of WT control (n = 5) and AdarP195A/P195A; Ifih1−/− mice (n = 6). Each dot shows the ISG level of a mouse, and the bars represent the means. The nonparametric Wilcoxon rank-sum test was used to test the differences between the two groups. *p < 0.05, **p < 0.01.
According to the current interpretation, the failure of immune regulation in AGS caused by ADAR variants is attributed to decreased editing of cellular dsRNAs.22-24 The view is supported by a number of mouse models showing that the RNA-editing activity of ADAR1 is essential for both innate immune homeostasis and embryonic development. Embryos with a catalytically dead Adar1E861A/E861A allele die around day 12 due to interferonopathy. Rescue to live birth can be achieved by deletion of MDA5 (encoded by Ifih1).2 Deletion of the downstream effector MAVS (encoded by Mavs) also rescues the ADAR1 knockout (KO) embryos to birth.4 Both Mavs and Ifih1 deletions prevent IFN pathway activation by dsRNA.2,4 Even partial ADAR1 RNA-editing activity loss leads to MDA5-dependent ISG expression, resulting in AGS-like neuroinflammatory pathogenesis.25-27
The human P193A variant results in partial loss of Z-binding activity.28 However, mice carrying homozygous P195A mutation, equivalent to human P193A, do not exhibit an abnormal phenotype.26,29,30 Recently, it was observed that postnatal death in P195A mutant mice occurred when a P195A mutant allele was paired with a p150-null allele.29 In that study, most Adar195A/p150− mice (Adar1p150− mice express the p110 but not the p150 isoform) died by 12 weeks of age. Further, most AdarP195A/− mice (the Adar null allele with deleted exons 7–9 [ΔE7–9] targets both p150 and p110 isoforms31) died within 5 weeks of birth.29 The protein kinase R (PKR)-dependent activation of the integrated stress response (ISR) signaling pathway was found responsible for the pathogenesis of the P195A mutation, and blockage of the ISR pathway rescued the lethality of AdarP195A/− mice, indicating a primary role for the Zα domain as a negative regulator of the ISR.29 In contrast, the studies in mice carrying a dual N173A/Y177A mutation that completely inactivated the Zα domain do not show PKR activation or an ISR response.24
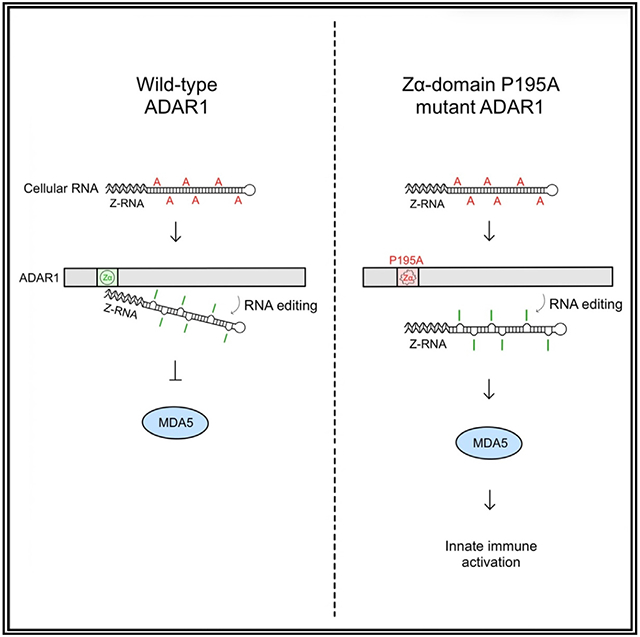
To investigate the function and mechanism of the P193A mutation in the Zα domain of ADAR1 in innate immune activation, we analyzed mice carrying the AdarP195A/P195A homozygous mutation and AdarP195A/− compound mutations using the Adar null allele (ΔE12-15).32 We fail to observe the previously reported lethal phenotype29 but do confirm that the effects of ADAR1 P195A mutation on ISG expression are MDA5 dependent. We demonstrate that the ADAR1 P195A mutation in an MDA5 null background does not affect the RNA-editing activity of ADAR1 at any of the tested well-known editing sites, nor at previously characterized p150 editing sites, nor does it alter the overall frequency of whole-transcriptome RNA editing. Furthermore, we find that ADAR1 suppressed the MDA5-dependent immune responses in a dose-dependent manner. Haploinsufficiency was only observed with the P195A mutant but not with the WT allele. Our results suggest an essential role for the binding activity of Z-RNA by the Zα domain of ADAR1 in regulating innate immune response, one that is likely independent of RNA-editing activity. Our in vivo findings show patterns of ISG expression in brains of P195A mutant mice that resemble those found in AGS patients.
RESULTS
ADAR1 P195A mutation resulted in activation of the MDA5-dependent RNA-sensing signaling pathway in the brain
To determine the role and mechanism of ADAR1 P195A mutation in brain pathogenesis, we analyzed mice carrying the ADAR1 P195A mutation, equivalent to human ADAR1 P193A mutation, that we generated previously.26 We also crossed AdarP195A/P195A mice with MDA5 KO (Ifih1−/−) mice33 to test the role of the MDA5-dependent RNA-sensing signaling pathway in AGS development. First, we confirmed that the C>G nucleotide replacement that encodes the P195A mutation in the mouse genome was correctly made (Figure 1B). The sequences around this mutation were assessed by Sanger sequencing. Besides the single-nucleotide replacement as expected, no other genomic change was identified (Figure S1), excluding potential unintended mutations that could account for the phenotypes to be observed. Similar to previous reports,26,29,30 neither heterozygous (Adar+/P195A) nor homozygous (Adar195A/P195A) mutant mice exhibited an abnormal phenotype. No difference in development, growth rate, general health, motion, or behavior was observed between the mutant mice and the WT Adar+/+ control mice. Examining organ/tissue sections stained with hematoxylin and eosin dyes did not reveal noticeable pathologic changes. Considering that AGS predominantly causes brain injury accompanied by elevated ISG expression,11,12,34 we determined ISG mRNA levels in the brains of AdarP195A/P195A mice. A panel of selected ISGs was quantified by real-time RT-PCR.26,35 WT mice were used as controls. Among the 10 tested ISGs, expression of all was increased in AdarP195A/P195A mice compared with controls. In contrast, ISG expression was significantly decreased in the brains of AdarP195A/P195A; Ifih1−/− double-mutant mice, with transcript levels similar to or even lower than those found in controls (Figure 1C). These results indicate that the Adar P195A mutation alone can activate the MDA5-dependent RNA-sensing pathway in the brain even though the ADAR1 catalytic domain is intact. Previously, it was believed that ISG induction occurred only in a compound mutant Adar genotype in which the P195A variant was paired with an allele that has impaired editing activity.
AdarP195A/− compound mutation resulted in mouse growth retardation and excessive ISG expression in the brain
Like human P193A compound variants in AGS patients,6,8 the P195A mutation in mice was reported to severely affect newborn survival when the P195A mutant allele paired with either an ADAR1 KO allele (AdarP195/−) or ADAR1 p150 null allele (AdarP195A/p150−).29,30 Most AdarP195A/− mice died before 5 weeks of age, whereas AdarP195A/p150− mice, which only delete ADAR1 p150 without affecting p110 expression, survived up to 12 weeks of age.29 Moreover, it was found that the ISR pathway was activated in AdarP195A/p150− mice, which was responsible for the lethal phenotype.29 Attempting to replicate these findings, we crossed our AdarP195A/P195A mice with an Adar+/− mice32 to produce a compound AdarP195A/− mouse strain (Figure 2A). In the Adar KO mice used in this study, exons 12–15 of the Adar gene were deleted. The KO is complete; there is no detectable truncated protein produced with either the N-terminal Zα domain or the C-terminal domains.32,36 Among our AdarP195A/P195A and Adar+/− mouse breeding progenies, we found that all mice were healthy, including AdarP195A/− mice. The pups survived to adulthood, and their survival rate was not different from the controls (Figure 2B). We measured the expression levels of Hmox1, Cdkn1a, and Asns genes of the ISR pathway in our AdarP195A/− mice as these three genes were previously reported to be highly expressed in the liver and kidney, but not in the brain, of the AdarP195A/p150− mice.29 We assessed the mRNA levels in both the liver and brain but could not find a significantly increased expression of these genes in the liver, while less than 2-fold increases in Homox and Cdkn mRNA were observed in the brains of AdarP195A/− mice (Figure 2C).
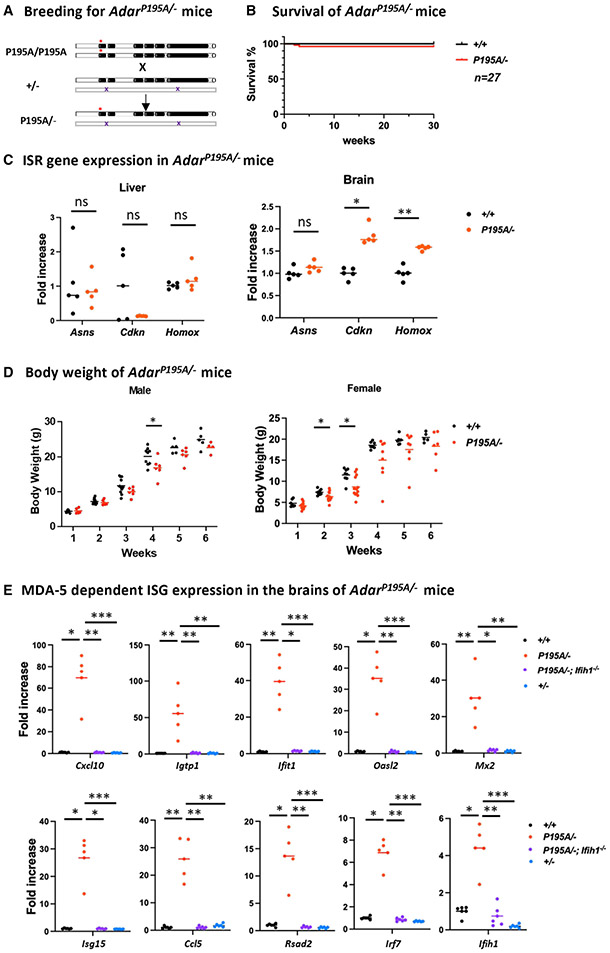
Figure 2. Phenotype and ISG expression in the brain of AdarP195A/− mice.
(A) AdarP195A/− mice were produced via breeding of AdarP195A/P195A mice and heterozygous ADAR1 KO Adar+/− mice. The ADAR1 KO allele did not produce detectable ADAR1 protein.
(B) AdarP195A/− mice were viable and healthy; the survival rate of AdarP195A/− mice was not significantly different from the WT control mice up to 30 weeks of age, as observed in 27 AdarP195A/− mice.
(C) Expression levels of ISR pathway genes Asns, Cdkn, and Homox in AdarP195A/− mice (n = 5) were not different from the expression levels in the livers of WT controls (n = 5). Cdkn and Homox expression in the brain of AdarP195A/− mice (n = 5) was increased by less than 2-fold from WT controls (n = 5), while Asns expression was not significantly different. The nonparametric Wilcoxon rank-sum test was used to test the differences between the two groups. *p < 0.05, **p < 0.01.
(D) The body weight of AdarP195A/− mice was less than that of the controls. In male mice, the body weight showed a statistically significant difference at 4 weeks of age, while the body weight of female mice showed more variation and was statistically different at 2 and 3 weeks of age. n = 5–11 (control male), n = 4–6 (AdarP195A/P195A male), n = 7–8 (control female), and n = 6–14 (AdarP195A/P195A female). The nonparametric Wilcoxon rank-sum test was used to test the differences between the two groups. *p < 0.05, **p < 0.01.
(E) The brain ISG expression levels of AdarP195A/− mice were compared with those of the controls and AdarP195A/−/Ifih1−/− mice. All 10 tested ISG mRNA levels were significantly increased in AdarP195A/− mice, which were significantly decreased in AdarP195A/−; Ifih1−/− mice. n = 5 (WT) and n = 6 (AdarP195A/P195A and AdarP195A/P195A; Ifih1−/−). The nonparametric Wilcoxon rank-sum test was used to test the differences between the two groups. **p < 0.01.
While we did not observe a lethal phenotype, we found that the growth of AdarP195A/− mice was retarded. The body weight of AdarP195A/− mice was significantly lower than that of controls, which was statistically significant at 4 weeks of age in male mice and at 2 and 3 weeks of age in female mice. The body weight of females varied dramatically. By 4–6 weeks of age, the differences were not statistically significant and likely due to the marked body weight variation and small mouse numbers (Figure 2D). We noticed that one of the female AdarP195A/− mice was significantly smaller than its littermates at the ages of 2–5 weeks; however, this mouse caught up quickly after 6 weeks of age, and its general health and survival were not affected.
Compared with the homozygous AdarP195A/P195 mice, the AdarP195A/− mice exhibited a haploinsufficiency effect due to the P195A mutation. To understand these outcomes further, we assessed ISG expression levels in the brains of AdarP195A/−, Adar+/+, and Adar+/− mice. We found the expression of all tested ISGs in AdarP195A/− mice was dramatically increased. The Cxcl10 expression was increased by 70-fold, whereas Igtp1, Ifit1, Ccl5, Oasl2, and Mx2 mRNAs were increased by 30- to 50-fold, and Isg15, Ccl5, Rsad2, Irf7, and Ifih1 were increased by 4- to 26-fold (Figure 2E). Expression of all these tested ISGs was significantly higher than that observed in AdarP195A/P195A mice (Figure 1C). We crossed AdarP195A/− with MDA5 KO (Ifih1−/−)mice33 and produced mice carrying AdarP195A/−; Ifih1−/− triple mutations. We tested ISG expression in the brains and found that the increased ISG expression in AdarP195A/− mice was dramatically reduced in the triplemutant mice, with expression at a level similar to those of the Adar+/+ control mice (Figure 2E). The results indicate that elevated ISG expression in AdarP195A/− mice is MDA5 dependent. Of note, in Adar+/− mice, no increased ISG expression was observed (Figure 2E), demonstrating that a single copy of the WT Adar allele is sufficient to maintain immune quiescence. In contrast, a single copy of the P195A mutant allele causes robust ISG expression. The P195A mutation produced robust ISG expression due to ADAR1 haploinsufficiency.
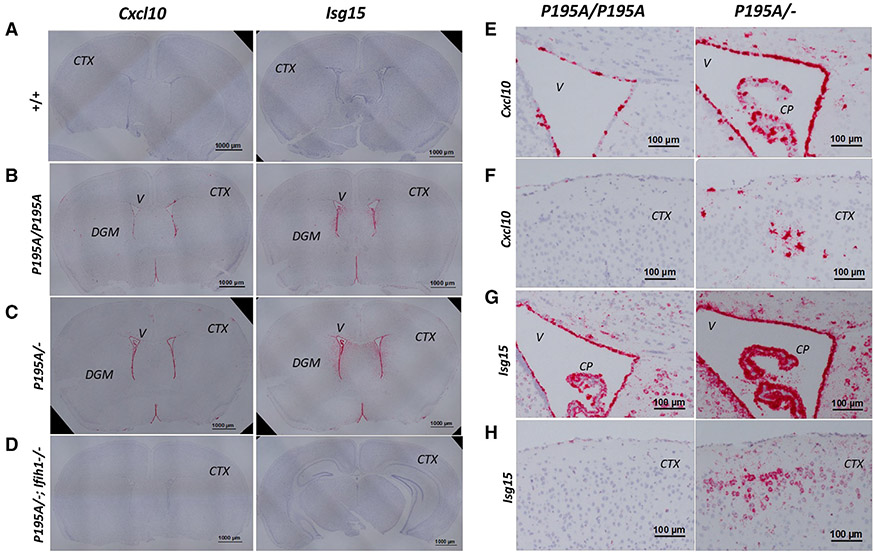
Our previous studies on other AGS-associated Adar mutations revealed that ISG expression in the brain was focally expressed in the cortex and deep gray-matter (DGM) areas and was cell-type specific.26,27,37 We performed RNA in situ hybridization (ISH) on brain sections of Adar195A/P195A and AdarP195A/− mice. The Isg15 and Cxcl10 probes showed intense staining in the periventricular areas, especially in the ependymal cells in both AdarP195A/P195A and AdarP195A/− mice (Figure 3), but no staining was observed in WT and AdarP195A/−;Ifih1−/− triple-mutant mice (Figures 3A and 3D). The Isg15 probe showed more intense staining in more widespread areas than the Cxcl10 probe. Fewer cells were stained for both the Cxcl10 and Isg15 probes in AdarP195A/P195A mouse brains than in AdarP195A/− mice (Figures 3E-3H). Scattered neurons and microglia were also stained in the cortex and DGM areas, whereas ISH with these probes did not detect ISG expression in most neurons and glial cells (Figure 3). These ISH studies confirmed MDA5-dependent ISG expression in the brains of ADAR1 P195A mutant mice, with the pattern of ISG expression matching the brain pathologic features of human AGS.
Figure 3. Distribution of ISG expression in AdarP195A/P195A and AdarP195A/− mouse brains determined by RNA ISH.
(A–D) Interferon-stimulated gene (ISG) expression in mouse brains was detected using RNA ISH on formalin-fixed paraffin-embedded sections of 8-week-old Adar+/+, AdarP195A/P195A, AdarP195A/−, and AdarP195A/−; Ifih1−/− mice. Shown are brain coronal sections stained with Cxcl10 or Isg15 probes. Both probes show ISG expression in the periventricular areas and on cell clusters scattered in the cortex and DGM areas of AdarP195A/P195A (B) and AdarP195A/− mice (C), while no staining was observed in Adar+/+ (A) and AdarP195A/−; Ifih1−/− triple-mutant mice (D). V, ventricle; CTX, cortex. Scale bar, 1,000 μm in (A)–(D).
(E) ISG expression in the periventricular areas detected with the CXCL10 probe in AdarP195A/P195A and AdarP195A/− mice. Ependymocytes and choroid plexus epithelial cells were highly stained on sections of AdarP195A/P195A and AdarP195A/− mice. Staining in AdarP195A/P195A mice was mainly on the ependymocytes, and more intense staining was observed in AdarP195A/− mice, including the adjacent cells in periventricular areas. V, ventricle; CP, choroid plexus. Scale bar, 100 μm.
(F) ISG expression in the brain surface cortical areas detected with the Cxcl10 probe in AdarP195A/P195A and AdarP195A/− mice. Scattered focuses were stained with the typical morphology of microglial cells in AdarP195A/− mice. Much fewer cells were stained in the AdarP195A/P195A mice. CTX, cortex. Scale bar, 100 μm.
(G) ISG expression detected with ISG-15 probe in the periventricular areas on brain sections of AdarP195A/P195A and AdarP195A/− mice. The staining pattern of the ISG-15 probe on ependymocytes and choroid plexus epithelial cells was similar to that of the CXCL-10 probe. The ISG-15 probe also stained cells in the periventricular areas showing neuron morphologic features. Staining in AAdarP195A/− mice was more intense than in AdarP195A/− mice. V, ventricle; CP, choroid plexus. Scale bar, 100 μm.
(H) ISG expression detected with Isg15 probe in the brain surface cortical areas on brain sections of AdarP195A/P195A and AdarP195A/− mice. Scattered areas with cells showing typical morphology of neurons were stained in AdarP195A/− mice, whereas much less staining was observed in AdarP195A/P195A mice. CTX, cortex. Scale bar, 100 μm.
Despite the dramatically increased ISG expression, AdarP195A/− mice did not exhibit noticeable neurological or behavioral abnormalities over 7 months of observation. The AdarP195A/− mice were fertile; however, we noticed that the pups of AdarP195A/− female mice grew slower than pups born from AdarP195A/P195A mothers, suggesting that increased ISG expression in AdarP195A/− mothers may affect nursing or feeding behavior, which affects postnatal growth.
P195A mutant ADAR1 protein exhibited a dose-dependent activity in innate immune suppression, while its RNA-editing activity was similar to WT ADAR1
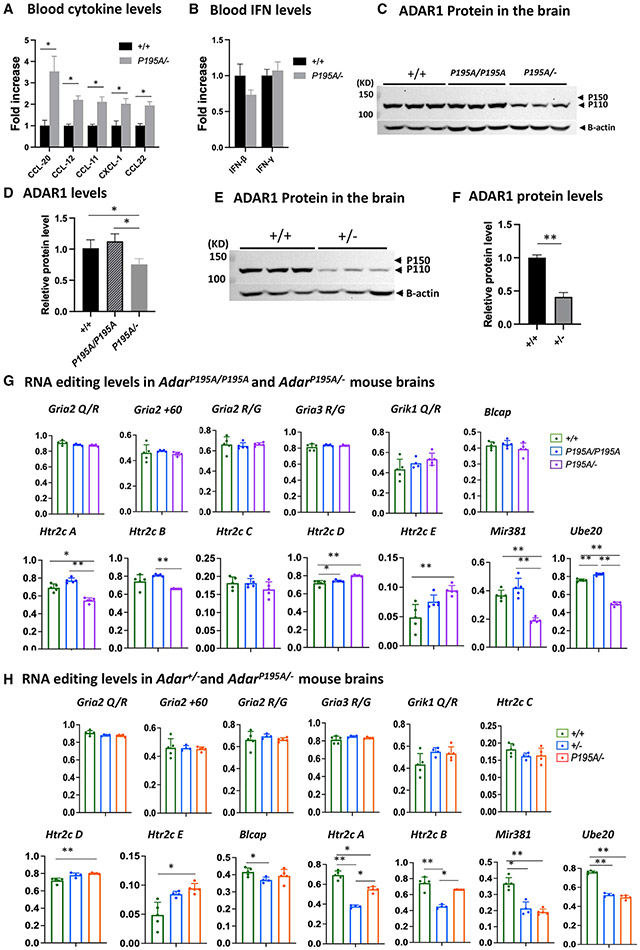
Next, we sought to determine the mechanism of ADAR1 P195A mutation in triggering ISG expression in the brain. We measured type I IFN levels in the blood of AdarP195A/− mice by Luminex assays with a complete panel of compatible antibodies and found that cytokine levels, including CCL-20(MIP-3a), CCL-12(MCP-5), CCL-11 (Eo-taxin), CXCL-1(KC) and CCL-22 (MDC), were significantly higher in AdarP195A/− mice than that in Adar+/+ control mice (Figure 4A and Table S1). In contrast, the IFN-β and IFN-γ levels did not differ from the controls (Figure 4B). This finding suggested that the increased ISG expression in the brain was probably not due to stimulation by blood IFNs. It was conceivable that the ADAR1 P195A mutation activated the brain’s intrinsic RNA-sensing pathway, leading to elevated ISG expression and inflammatory response in the brain. In addition, ISG levels in AdarP195A/− mice were dramatically higher than those in AdarP195A/P195A mice, suggesting that decreased ADAR1 protein might contribute to brain ISG expression. To determine whether the P195A mutation resulted in ADAR1 protein decrease, we quantified ADAR1 protein in the brains of the AdarP195A/P195A and AdarP195A/− mice by western blot and compared the results with those in Adar+/+ mice. Consistent with previous findings,23,38 the ADAR1 p110 isoform was abundantly expressed, while the p150 isoform was almost not detectable in the brains of the Adar+/+ mice (Figures 4C and 4D). ADAR1 protein in AdarP195A/P195A mice showed the same pattern as in the Adar+/+ mice, and the quantities were comparable, whereas ADAR1 protein in AdarP195A/− mice was significantly less than that in Adar+/+ and AdarP195A/P195A mice (Figures 4E and 4F). Thus, the robust ISG expression in AdarP195A/− mice was attributed to the protein decrease of the P195A mutant ADAR1. However, Adar+/− mice did not show excessive ISG expression even though one allele was also null (as shown in Figure 1E). It is possible that the WT Adar allele expressed more ADAR1 protein, which compensated for the null allele in Adar+/− mice, thus producing sufficient ADAR1 protein to suppress ISG expression. To test this possibility, we assessed the ADAR1 protein level in Adar+/− mice and found that ADAR1 protein was dramatically less than in the Adar+/+ mice, showing that, similar to the AdarP195A/− mice, there was no compensation in protein expression in Adar+/− mice (Figures 4E and 4F). Therefore, WT ADAR1 protein sufficiently suppressed the RNA-sensing pathway even when its quantity was dramatically decreased. In contrast, decreased P195A mutant ADAR1 protein resulted in robust ISG expression in AdarP195A/− mice. By comparison of the ISG levels in AdarP195A/P195A, AdarP195A/−, and Adar+/− mice, it is apparent that ADAR1 exhibits a dose-dependent effect in suppression of innate immune activation when the Z-nucleic-acid-binding activity is partially lost.
Figure 4. Decreased ADAR1 protein and RNA-editing levels in AdarP195A/− and Adar+/− mice.
(A) Luminex assay was used to measure the blood cytokine/chemokine levels of AdarP195A/− mice. Among the tested cytokines of the available panels, levels of CCL-20, CCL12, CCL-11, CXCL-1, and CCL22 were significantly increased in AdarP195A/− mice in comparison to the WT mice. n = 5 for both control and AdarP195A/− groups. The nonparametric Wilcoxon rank-sum test was used to test the differences between the two groups. *p < 0.05. The complete panel is listed in Table S1.
(B) IFN-β and IFN-γ levels in the blood were assessed in the Luminex panels. n = 5 for both control and AdarP195A/− groups. Their levels were not different between AdarP195A/− and WT mice.
(C) Brain ADAR1 protein levels in AdarP195A/P195A and AdarP195A/− mice were analyzed by western blotting and compared with WT mice. ADAR1 p110 isoform was abundantly expressed in the brains of the WT and AdarP195A/P195A mice, while no obvious expression of ADAR1 p150 isoform was observed. ADAR1 protein in AdarP195A/P195A mice was not significantly different from that in WT mice. However, much less ADAR1 protein was observed in AdarP195A/− mouse brains.
(D) Quantification of ADAR1 protein shown in (C). There is no significant difference between WT and AdarP195A/P195A mice, while the ADAR1 protein level in AdarP195A/− mice is significantly lower than in WT and AdarP195A/P195A mice. n = 3, *p < 0.05 (nonparametric Wilcoxon rank-sum test).
(E) To determine whether a single copy of WT Adar allele also causes ADAR1 protein level decrease, ADAR1 in Adar+/− mice was assessed and compared with WT mice. Significantly less ADAR1 was observed in Adar+/− mice.
(F) ADAR1 protein levels in Adar+/− and Adar+/+ mice were quantified and compared. The ADAR1 protein level in Adar+/− mice was significantly less than that in Adar+/+ mice. n = 3, *p < 0.05 (nonparametric Wilcoxon rank-sum test).
(G) Brain RNAs were isolated from AdarP195A/P195A and AdarP195A/− mouse brains, and A-to-I RNA-editing levels were assessed via Sanger sequencing analysis. RNA editing at the known editing sites in defined neural RNA-editing substrates, including editing sites in the Gira2, Rria3, Grik1, and Htr2c neuron receptor, as well as the editing sites in mRNAs for Blcap, Ube2o, and the miR381, were compared with the WT control mice and between AdarP195A/P195A and AdarP195A/− mice. At all tested sites, RNA-editing levels in AdarP195A/P195A mice were the same or even higher than the WT control mice, while, in AdarP195A/− mice, only editing in miR381 and Ube2o mRNAs was significantly lower than that in AdarP195A/P195A and WT control mice.
(H) RNA-editing activities in the brain of Adar+/− and AdarP195A/− mice were assessed by Sanger sequencing analysis to determine whether the P195A mutation affects the RNA-editing activity of ADAR1 at a low protein dose. As shown in (G), the RNA-editing levels at the defined editing sites were assessed and compared with the WT control mice. Except for Htr2c A and B sites and in miR381 and Ube2o mRNA, RNA-editing levels in Adar+/− and AdarP195A/− mice were the same or even higher than those in the WT control mice. RNA-editing levels at Htr2c A and B sites and in miR381 and Ube2o mRNA were significantly lower in Adar+/− and AdarP195A/− mice. However, editing levels in Htr2c and Ube20 mRNAs in AdarP195A/− mice were not different from those in Adar+/− mice and were even higher than those in Adar+/− mice at the Htr2c A and B sites. In (G) and (H), the average editing levels and SDs are shown. n = 5 (WT) and n = 6 (AdarP195A/P195A and AdarP195A/P195A/Ifih1−/−). The nonparametric Wilcoxon rank-sum test was used to test the differences between the two groups. The significant differences are indicated. *p < 0.05, **p < 0.01.
RNA editing on cellular RNAs by ADAR1 is an essential process for immune homeostasis, with the engagement of unedited dsRNAs by MDA5 leading to ISG expression. To test whether ADAR1 P195A mutant results in decreased RNA editing of cellular RNAs, we assessed RNA-editing levels in AdarP195A/P195A and AdarP195A/− mice. We then correlated RNA-editing levels in various RNA substrates with ISG expression to test whether diminished editing by the P195A mutation caused immune activation. We first examined RNA editing at well-defined editing sites in cellular RNAs by Sanger sequencing analysis of cDNAs from mouse brains. In particular, we analyzed editing of the glutamate ionotropic receptor (encoded by Gria2) and the serotonin 5-HT receptor 2c (encoded by Htr2c),32,39-41 as well as Blcap, Ube2o, and miR381 RNAs.42 We included ADAR2 editing sites to control how well we quantitated editing. We found that the AdarP195A/P195A homozygous genotype did not affect the RNA-editing activity in the brains. In all the sites tested, editing levels in the AdarP195A/P195A mice remained the same as in the Adar+/+ mice, with the sole exceptions of Ube2o and Htr2c D sites, where editing was slightly increased (Figure 4G). Thus, the increased ISG expression in AdarP195A/P195A mice was not associated with decreased RNA-editing levels at these known editing sites. In AdarP195A/− mice, where ADAR1 protein was decreased as only a single-mutant allele was expressed, the editing at Htr2c D and E sites, the known ADAR2 editing sites, was higher than that in Adar+/+ mice, whereas there was a marked decrease in the editing at the Htr2c A and B sites and in miR381 and Ube2o mRNA (Figure 4G). We also compared the editing levels in AdarP195A/− mice with Adar+/− mice. At the Htr2c D and E sites, Adar+/− and AdarP195A/− mice showed higher editing levels than Adar+/+ mice. At the Htr2c A and B sites, editing in both AdarP195A/− and Adar+/− mice was diminished, but, surprisingly, editing in AdarP195A/− mice was higher than that in Adar+/− mice. In miR381 and Ube2o mRNA, there was no difference between AdarP195A/− and Adar+/− mice. Only Adar+/−, but not AdarP195A/−, mice showed a lower editing level at the Blcap editing site (Figure 4H).
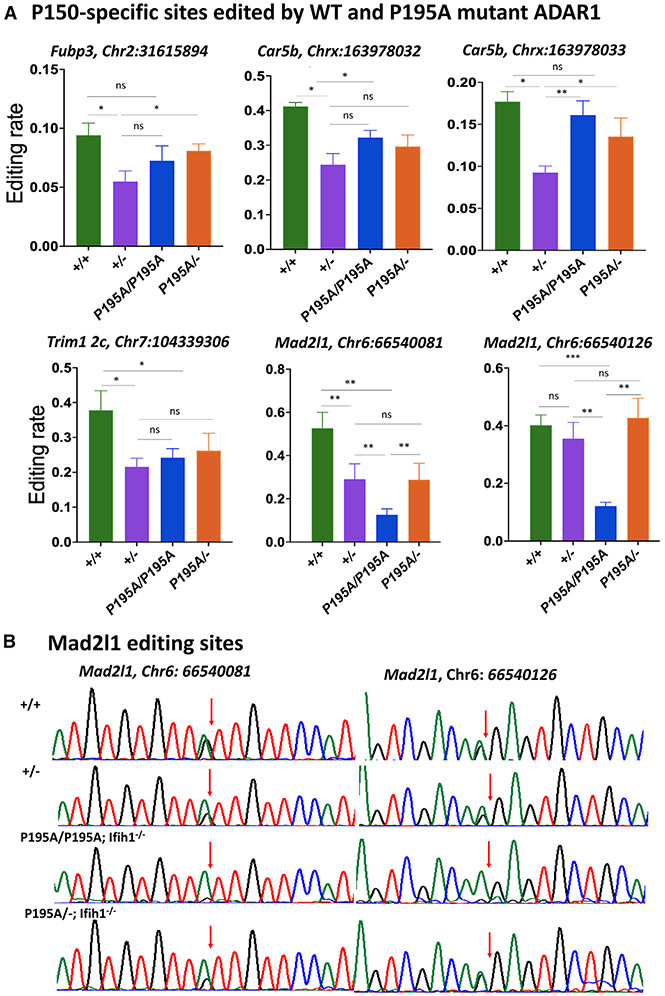
It was recently reported that a small number of cellular RNAs was specifically edited by ADAR1 p150 isoform in the brains of mice in which both ADAR2 and ADAR1 p110 were deleted.38 The mRNAs of Fubp3, Mad2l1, Trim2c, and Car5b were the most significantly edited targets. We performed a Sanger sequencing analysis to assess whether the ADAR1 P195A mutation affected editing in these specific RNAs. The potential effects of MDA5-induced ISG activation on editing levels were minimized by assessing ADAR1 P195A RNA editing in AdarP195A/P195A; Ifih1−/− and AdarP195A/−; Ifih1−/− mice where MDA5 was absent. Results were compared with RNA-editing levels in WT and Adar+/− mice. The RNA-editing levels were significantly decreased in Adar+/− mice compared with Adar+/+ mice, reflecting the lower levels of ADAR1 protein present in mice with a null allele. However, the editing levels in AdarP195A/P195A; Ifih1−/− and AdarP195A/−; Ifih1−/− mice were similar to, or even higher than those in Adar+/− mice. The only exception was the editing in Mad2l1 mRNAs, in which editing of both sites was lower in AdarP195A/P195A; Ifih1−/− than in Adar+/− mice. Nevertheless, the editing level of Mad2l1 was not associated with the P195A mutation, as the editing levels in AdarP195A/−; Ifih1−/− mice were unaffected (Figure 5A). Repeated Sanger sequencing analysis confirmed these editing levels in this specific cellular RNA (Figure 5B). Overall, a single copy of the ADAR1 P195A mutation was not associated with decreased ADAR1 editing of these ADAR1 p150-specific RNA substrates compared with a single copy of the WT allele. Therefore, the increased ISG expression in AdarP195A/− mice could not be reasonably attributed to decreased editing in these ADAR1 p150-specific RNA substrates.
Figure 5. RNA editing at the P150-specific sites in AdarP195A/− and Adar1+/− mice.
(A) RNA-editing levels at typical ADAR1 p150 isoform-specific editing sites in Adar+/+, Adar+/−, AdarP195A/P195A; Ifih1−/−, and AdarP195A/−; Ifih1−/− mice were assessed, including the editing sites in the mRNAs of Fub3 (Chr2: 31615894), Trim12c (Chr7: 104339306), Car5b (Chrx: 163978032, 163978033), and Mad2l1 (Chr6: 66540081, 66540126). Except for the site of Mad2l1 (Chr6: 66540126), editing levels in in Adar+/− mice were significantly lower than those in Adar+/+ mice, whereas editing levels in AdarP195A/P195A; Ifih1−/− and AdarP195A/−; Ifih1−/− mice were the same as those in Adar+/− mice or between Adar+/+ mice and Adar+/− mice. Editing at the two sites in Mad2l1 mRNA (Chr6: 66540081, 66540126) were the only sites with significantly lower editing levels in AdarP195A/P195A; Ifih1−/− mice than those in Adar+/+, Adar+/-, and AdarP195A/−; Ifih1−/− mice, while editing at these two sites in AdarP195A/−; Ifih1−/− mice was not significantly different from Adar+/− and Adar+/+ mice. Shown in the figure are the average editing levels with SD values. n = 5 for all the groups. The nonparametric Wilcoxon rank-sum test was used to test the differences between the two groups. *p < 0.05, **p < 0.01.
(B) Different RNA-editing levels at the two specific sites in Mad2l1 mRNA (Chr6: 66540081, 66540126) in Adar+/+, Adar+/−, AdarP195A/P195A; Ifih1−/−, and AdarP195A/−; Ifih1−/− mice were shown by Sanger sequencing analysis. Shown here are the representative chromatograms of the editing sites. The arrows indicate the editing sites, and the editing level was calculated by the A and G peak values (G/(A + G)). Significantly decreased editing levels were observed in AdarP195A/P195A; Ifih1−/− mice.
ADAR1 P195A mutation did not change whole-transcriptome-wide RNA-editing levels, although RNA-editing patterns were highly variable
We also conducted a high-throughput RNA sequencing (RNA-seq) analysis of editing in ADAR1 P195A mutant mice. First, we isolated RNAs from the brain tissues of three mice of each genotype, including AdarP195A/P195A; Ifih1−/−, AdarP195A/−; Ifih1−/−, Adar+/+, and Adar+/− mice. Deep sequencing with an average of 80 million paired-end reads was collected for each RNA sample. Both A>G mismatches in (+) strands and T>C mismatches in (−) strands with the reference genome sequence were recorded. Single-nucleotide polymorphism (SNP) sites defined by the dbSNP database were removed, along with any difference with an editing rate of either 100% or 50% that was likely due to an unannotated SNP. Only sites with more than two A>G or T>C mismatch reads and total coverage greater than 10 were considered reliable editing sites for further analysis. There were 100,941, 112,138, 107,749, and 100,453 editing sites found in Adar+/+, Adar+/−, AdarP195A/P195A; Ifih1−/−, and AdarP195A/−; Ifih1−/− mice, respectively (Tables S2-S5). We evaluated our RNA-seq data by comparing outcomes with our Sanger sequencing results and found that the editing levels at all RNA sites assessed by Sanger sequencing analysis were consistent with those from RNA-seq analysis (Figure S2), indicating that our RNA-seq data were reliable.
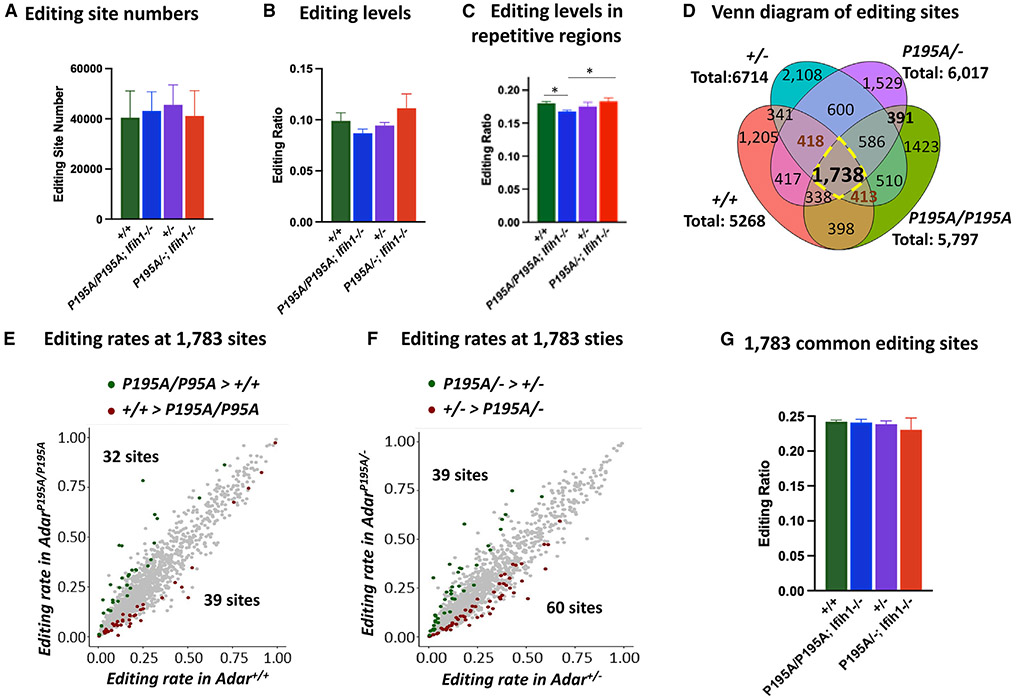
The numbers of editing sites detected were highly variable between mice in each group (Figure S3). This outcome was expected because of the stochastic nature of editing when many adenosines in the short interspersed nuclear elements (SINEs) are modified to a small extent.43 After pooling the editing sites for individual mice in each group, there was no significant difference in the numbers of editing sites and average editing rates between different groups (Figures 6A and 6B).
Figure 6. RNA editing assessed at whole-transcriptome level.
(A) High-throughput RNA sequencing (RNA-seq) study was performed with brain RNAs from Adar+/+, Adar+/−, AdarP195A/P195A; Ifih1−/−, and AdarP195A/−; Ifih1−/− mice. The numbers of RNA-editing sites identified in three mice of each genotype were compared. n = 3 for each group. Welch two-sample t test was used to test the differences between the two groups. No significant difference was observed in these four groups of mice.
(B) The average editing rates of all the editing sites in three mice of each genotype were compared, which showed no significant difference.
(C) The average editing rates of all the editing sites in repetitive sequences in three mice of each genotype were compared. The average editing rate in AdarP195A/P195A; Ifih1−/− mice was significantly lower than that in Adar+/+ and AdarP195A/−; Ifih1−/− mice but not significant compared with Adar+/− mice. For(A)–(C), the bars show the means of the three mice of each genotype, and the error bars are the corresponding SD values. n = 3 for each group. *p < 0.05, **<0.01 (Welch two-sample t test).
(D) The editing sites found in three mice of each genotype are compared in the Venn diagram. The total sites shown for each genotype are the shared editing sites found in all three mice of each genotype. Each genotype shows a unique selection of editing sites, with 1,738 editing sites shared by all the analyzed mice. Subgroups of the editing sites differ between the genotypes.
(E) Comparison of the editing rates of the 1,738 shared editing sites in AdarP195A/P195A; Ifih1−/− mice versus Adar+/+ mice; 32 sites were edited at significantly higher levels in AdarP195A/P195A; Ifih1−/− mice and 39 sites were edited significantly more in Adar+/+ mice.
(F) Comparison of the editing rates of the 1,738 shared editing sites in AdarP195A/−; Ifih1−/− mice versus Adar+/− mice; 39 sites were edited at significantly higher level in AdarP195A/−; Ifih1−/− mice and 60 sites were editing significantly more in Adar+/− mice.
(G) The average editing rates of the 1,783 editing sites in the Adar+/+, Adar+/−, AdarP195A/P195A; Ifih1−/−, and AdarP195A/−; Ifih1−/− mice were compared. The bars show the means of three mice of each genotype, and the error bars are the corresponding SD values. n = 3 for each genotype. Welch two-sample t test was used to test the differences between the two groups. No significant difference was observed in these four groups of mice.
We further compared the editing ratio detected only in repetitive genomic regions such as SINEs where A-to-I editing preferentially occurs, examining how this measure was affected by the ADAR1 P195A mutation. Editing of repetitive elements was lower in AdarP195A/P195A; Ifih1−/− mice compared with either WT or AdarP195A/−; Ifih1−/− mice. Notably, editing levels did not significantly differ between Adar+/+, Adar+/− and AdarP195A/−; Ifih1−/− genotypes (Figure 6C). There was no correlation between the number of RNA-editing sites and average editing levels in AdarP195A/P195A; Ifih1−/− and Adar+/−; Ifih1−/− mice that might account for the increased ISG expression.
Next, we surveyed the high-throughput RNA-seq to evaluate RNA-editing levels at the sites reported as specific for the ADAR1 p150 isoform. These included the 36 editing sites in 3′ UTRs and the 17 editing sites in introns.38 No significant difference was found in AdarP195A/P195A; Ifih1−/− mice versus Adar+/+ mice and in AdarP195A/−; Ifih1−/− versus Adar+/− mice (Figures S4 and S5).
We also compared the whole-transcriptome editing levels in Adar+/+, Adar+/−, AdarP195A/P195A; Ifih1−/−, and AdarP195A/−; Ifih1−/− mice. The editing sites shared by the mice of different genotypes varied significantly (Figure 6D). The overlap of sites for AdarP195A/P195A and AdarP195A/− mice differed dramatically from those shared by Adar+/+ and Adar+/− mice, with only 1,738 sites common to all four genotypes. Among these common editing sites, only 39 and 60 sites were edited higher in Adar+/+ and Adar+/− mice than in AdarP195A/P195A; Ifih1−/− and AdarP195A/−; Ifih1−/− mice respectively, and 32 and 39 sites were edited higher in AdarP195A/P195A; Ifih1−/− and AdarP195A/−; Ifih1−/− mice than in Adar+/+ and Adar+/− mice (Figures 6E and 6F and Table S6). These common sites’ average editing levels did not differ between genotypes (Figure 6G).
We then asked whether there were differences in editing levels of shared sites attributable to a particular genotype. We examined 418 unique sites in AdarP195A/−; Ifih1−/− mice shared by Adar+/+ and Adar+/− mice (Figure S6A and Table S7), and the 413 unique sites in AdarP195A/P195A; Ifih1−/− mice shared by Adar+/+ and Adar+/− mice (Figure S7A and Table S8). Similar to the 1,738 sites common to all groups, the average editing levels at sites in ADAR1 P195A mutant mice were the same as in Adar+/+ mice. Only a small number of editing sites with either higher or lower editing levels were found in ADAR1 P195A mutant mice compared with Adar+/+ and Adar+/− mice. Editing sites with increased and decreased editing levels were detected in the P195A mutant mice (Figures S6A, S6B, S7A, and S7B).
DISCUSSION
The P193A variant in the Zα domain of the human ADAR1 p150 isoform is the most frequent in AGS patients,6,8,21 and it triggers innate immune activation in the brain when paired with a p150 null allele or another LOF Adar allele.6,8,14 The P193A variant reduces the Z-DNA/RNA-binding affinity28,44; how it modulates innate immune activation is the question we addressed here. In this study, we crossed the AdarP195A/P195A mutant mice (referred to as ADAR1 P193A mice previously26) with Adar+/− (ΔE12-15) mice to generate an AdarP195A/− mouse model, the equivalent of a haploid AdarP195A genotype. Although AdarP195A/P195A homozygous mice did not show apparent abnormality, we detected expression of ISGs in the brain triggered by the MDA5-dependent dsRNA-sensing pathway. This finding demonstrated that the P195A mutation alone can cause increased ISG expression, even though the ADAR1 catalytic domain is intact and protein expression is unaffected. Furthermore, we found that the level of ISGs in the haploid AdarP195A/− genotype was dramatically higher than that in the AdarP195A/P195A homozygotes. In contrast, ISG expression was not induced in haploid WT Adar+/− mice. A single copy of WT ADAR1 was sufficient to prevent innate immune activation, whereas two copies of the P195A Adar mutant could not entirely abolish ISG expression.
How ADAR1 mutations cause innate immune activation has yet to be fully understood, although there has been much recent progress. Mutations affecting the catalytic domain of ADAR1 show that RNA-editing activity is essential for preventing innate immune activation.2,4,5,45,46 Mice carrying the E861A mutation, which abolishes the RNA-editing activity of ADAR1, die at an early embryonic stage, while deletion of the dsRNA sensor MDA5 rescues the mice from intrauterine death.2 Even partial loss of RNA-editing activity in mice carrying the catalytic domain K999N and D1113H mutations leads to MDA5-dependent dsRNA-sensing signaling pathway activation.25-27 These results support the hypothesis that unedited endogenous RNAs trigger the MDA5 RNA sensor, leading to the initiation of IFN responses.
In mice with an ADAR1 Zα domain N175A/Y179A mutation with a WT Ifih1 gene, IFN responses in multiple organs were spontaneously activated. A-to-I edits are decreased in transposable elements.22 The mice showed increased resistance to influenza virus infection. A similar finding was made in another study of an N175A/Y179A mutant mouse. In this case, resistance to the encephalomyocarditis virus was increased. Interestingly, cultures of fibroblasts developed ISG expression after 7 days of culture, indicating that negative regulation of the response was defective.24 Similarly, mice with the ADAR1 Zα domain W197A mutation and a KO of the related Adar2 gene showed severe disease due to a failure to turn off the MDA5-induced response.23 In these mice, RNA editing was decreased at a subset of, but not all, ADAR1 target sites, while other sites showed an increase, indicating the effects of the absence of Zα targeting of ADAR1. This finding indicates that the ADAR1 Zα domain is required to edit a subset of RNA substrates after MDA5 activation to suppress the MDA5 positive-feedback loop.
There may be more than one way in which p150 acts to restrain an innate immune response. Any cellular Z-RNA can also activate the Z-RNA sensor, Z binding protein 1 (ZBP-1), which is capable of inducing inflammatory cell death.47 The activation of ZBP1 can be suppressed by the ADAR1 Zα domain.48-50 Indeed, ADAR1 depletion or mutation resulted in Z-RNA accumulation and activation of ZBP-1.47 Further, the innate immune response of mice with ADAR1 KO or with Zα mutations, including N175A/Y179A,50 N175D/Y179A,48 and P195A/p150 null49 genotypes, was rescued by ZBP1 KO. The most likely mechanism is through competition by ADAR1 for Z-RNA substrates, although ADAR1 will also diminish the formation of Z-RNA by editing dsRNA.
In this study, we found that the overall RNA-editing levels did not differ in ADAR1 P195A mutant mice in an MDA5 null background, either in the total number of edits or by the average level of editing. Even at the editing sites specific for the ADAR1 p150 isoform,38 we did not find a difference between the WT and mutant mice. However, we found a dosage effect that affected editing levels at specific sites, including the 5-HTR2c A and B sites, with reduced editing in miR381 and Ube2o mRNA. Nevertheless, the editing levels in AdarP195A/− mice were not different from or were even higher than Adar+/− mice. We noticed variations in the editing sites in RNA-seq data analysis between mice of the same genotype, most likely reflecting the stochastic nature and the variable editing of adenosines in repeat elements.43 Jiao et al. recently reported a considerable loss of editing sites in Adar1mZa/− (N175D/Y179A) mice, and the majority of the editing was found in SINE RNAs.48 de Reuver et al. observed an overall reduction in the quality of repeat editing sites in Adar1Za/− (N175A/Y179A) mice.50 The mutations at the N175 and Y179 sites more severely affect the Z nucleic acid-binding activity of ADAR1 than the mutation at the P195 site,21 which may account for why the mZa mutations reduce the RNA editing while the P195A mutation does not.
Discrepancies were also found in the mouse models carrying the same ADAR1 P195A mutation. First, the postnatal viability of different P195A mutant mice was different. The AdarP195A/− (ΔE7-9) mice in a previous study did not survive beyond 5 weeks after birth.29 In contrast, we followed 27 mice until they reached 7 months of age, and only two mice died within 2 weeks after birth, with no significant difference from the controls. Furthermore, activation of the ISR pathway was reported in AdarP195A/p150− (ΔE7-9) mice, with significantly increased expression of the ISR pathway’s genes Hmox1, Cdkn1a, and Asns in the liver and kidney; blocking the ISR pathway rescued AdarP195A/p150− mice from postnatal death.29 However, in this study, expression of the ISR genes was not upregulated in the livers of our AdarP195A/− (ΔE12-15) mice. In the brain, the expression levels of Cdkn and Homox genes increased less than 2-fold. The significance of this change is unknown, as it did not affect the survival and general health of the mutant mice. Very recently, another independent AdarP195A/− (ΔE2-13) mouse model was reported in which P195A mutation showed partial penetrance. About 45% of AdarP195A/− mice showed normal phenotype and long-term survival, while another half showed runted phenotype and a shortened life span.30 An editing-dead p150 allele rescued the phenotypes of the AdarP195A allele, indicating that it was not a defect in RNA editing that primarily caused the ISG induction. The restricted PKR activation and ISR signature were not seen in this mouse model, even in the runted mice.30 It is unknown whether the ISR pathway activation and the early-death phenotype were associated with the Adar null allele used in the previous study. The Adar ΔE7-9 allele used in the previous study29 produced an unstable truncated ADAR1 protein with the Zα domain intact,45 while in our study the Adar ΔE12-15 allele completely abolished ADAR1 protein production.32,36 Curiously, the previous study obtained a similar result with another allele that expressed only the p110 isoform but not the p150 isoform, suggesting that this allele requires further investigation. We noticed that the genetic background of the mice used in our study differed from other studies, which might contribute to the phenotype differences. Considering the significant variation of the genetic background of AGS patients, we intentionally kept our mice in the mixed background of C57/BL6J and DBA2 strains; meanwhile, other studies used mice in pure C57/BL6 background. However, we carefully checked the genomic sequence of our mutant mice and confirmed that only the single G>C mutation presents in the genome without an extra genetic modification. In the mixed background of C57/BL6J and DBA2, the compound mutation of AdarP195A/− at the least did not affect postnatal survival in our study. It is likely that undefined factors exist that modify the activity of P195A mutation in innate immune activation, which needs further investigations.
Limitations of the study
This study focuses on the molecular changes in the brain tissue, which does not exclude potential differences in RNA editing and ISG expression in other tissues, especially those that express higher levels of the P150 isoform of ADAR1, such as the spleen and thymus. The Adar P195A mutation does not alter the overall RNA-editing level, which does not exclude the possibility of a higher or lower editing level in a few specific RNA molecules that remain to be identified, while it is difficult to confirm that a change in a few RNA molecules is the cause of the innate immune response. The mouse model used in this study shows a high-level ISG expression in the periventricular regions, a pattern like a typical brain pathologic change in AGS patients; however, the high-level ISG expression does not cause apparent brain functional lesions that lead to neurologic phenotypes as observed in AGS patients.The roles of the ADAR1 Z-RNA binding activity and the MDA5 helicase in inducing Z-RNA formation are not assessed in this study. The finding from this study has not been confirmed in human neural cells.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Qingde Wang (wangqd@pitt.edu).
Materials availability
Mouse lines generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ADAR1 Antibody (15.8.6) | Santa Cruz Biotechnology, Inc. | Cat#: sc-73408 |
| β-Actin Antibody (C4) | Santa Cruz Biotechnology, Inc. | Cat#: sc-47778 |
| Critical commercial assays | ||
| MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel | Millipore Sigma | Cat#: MCYTOMAG-70K |
| MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel II | Millipore Sigma | Cat#: MECY2MAG-73K |
| Deposited data | ||
| RNA-seq data | NCBI Gene Expression Omnibus | accession ID GSE229884 |
| Experimental models: Organisms/strains | ||
| Mouse:AdarP195A/P195A | Guo et al. 202126 | NA |
| Mouse:AdarP195A/− | This paper | NA |
| Mouse:Adar +/− | Wang et al. 200436 | NA |
| Mouse: Ifih1−/− | The Jackson Laboratory | Stock#: 015812 |
| Oligonucleotides | ||
| See Table S9. | This paper | N/A |
| Software and algorithms | ||
| Trimmomatic | Bolger et al. 201459 | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR | Dobin et al. 201360 | https://github.com/alexdobin/STAR |
| bcftools | Li et al. 200961 | https://samtools.github.io/bcftools/bcftools.html |
| snpSift | Cingolani et al. 201262 | https://pcingola.github.io/SnpEff/ |
EXPERIMENTAL MODEL AND SUBJECT PARTICIPANT DETAILS
Mouse models
P195A mutant mice were prepared using CRISPR/Cas9 technology as described previously.26 ADAR1 knockout Adar+/− mice were prepared via homologous recombination that deleted Exon 12–15 as described previously.32 Mice used in this study, including AdarP195A/P195A, AdarP195A/P195A/Ifih1−/−, AdarP195A/−, AdarP195A/−/Ifih1−/−, and Adar+/− mice were in C57/BL6J and DBA2 mixed background. Ifih1 −/− mice were purchased from Jackson lab, strain #: 015812.
METHOD DETAILS
ISG expression quantification
RNA isolation was performed with RNeasy Plus Mini Kit (Qiagen Cat # 74134) following the manufacturer’s instructions. Quantitative RT-PCR was performed using the iTaqTM Universal SYBR Green One-Sep Kit (Bio-Rad cat #1725151). Assayed genes comprised Cxcl10, Igtp1, Ifit1, Ccl5, Oasl2, Mx2, Isg15, Rsad2, Irf7, and Ifih1. Gadph and Hprt were used as internal controls. The specificity of PCR amplifications was confirmed by the melting curve and by electrophoresis analysis of the final PCR products. The quantification of the mRNA levels was calculated by the Ct values using ΔΔt method with internal references of the average value of the Hprt and Gadph measurements as described previously.26,35
RNA editing essays
Total brain RNA was isolated from undissected whole brains, and reverse transcription PCR was performed with the total RNA samples. The PCR products of the entire brain RNA pool were subjected to Sanger sequencing analysis. The relative quantities of inosine (read as guanosine) and adenosine at each editing site were determined on the chromatographs by the ratio of the G and A peaks. The primer sequences used for the PCR amplifications of the editing sites were 5′-cactgaggaatttgaagatgga-3′ and 5′-agcaggcatg gaatgatagg-3’ (for the Gria2 Q/R site and the intron hot spots), 5′-cttgcgacaccatgaaagtg-3′ and 5′-gccagaaatgtgggtaaagg-3’ (for the Gria2 R/G site), 5′-agcagagaaagccgtgtgat-3′ and 5′-agaacaccacatccatgcaa-3’ (for the Gria3 R/G site), 5′-acccgtgcaaccct gact-l 3′ and 5′-ttgcaggaaattttgtccagt-3’ (for the Grik1 Q/R site), 5′-attatgtctggcctttacctagatat-3′ and 5′-ataggaactgaaactcctattga tattgc-3’ (for the editing sites A to E in 5-HT 2cR mRNA). RNA-editing efficiency was assessed on the editing sites in Gria2, Gria3, and Grik1 mRNAs for the Q/R and R/G sites and on the A-E sites in Htr2c receptor mRNA by calculation of the relative ratio of the average of G peak to A peak on each of the Sanger sequencing chromatograms for samples from mutant and wild-type groups.
ADAR protein analysis
Brain tissues were collected and immediately frozen in liquid nitrogen until analysis. Homogenization was performed in RIPA buffer with the addition of protease inhibitor cocktail. ADAR1 protein was detected in the brain tissues using Western blot as described previously.18 In brief, 30ug of protein extract was loaded to each lane and separated on 8% polyacrylamide gel with 0.1% SDS. ADAR1 was detected with ADAR1 antibody clone 15.8.6 (Santa Cruz sc-73408) at 1:1,000 dilution.
Cytokine and chemokine luminex assay
Blood cytokine levels in Adar+/+ and AdarP195A/− mice were measured by Luminex assay. Forty-five (45) cytokines and chemokines were measured utilizing Luminex xMAP technology. The multiplexing analysis was performed using the Luminex 200 system (Luminex, Austin, TX, USA) by Eve Technologies Corp. (Calgary, Alberta, Canada). Forty-five markers were simultaneously measured in the samples using a MILLIPLEX Mouse Cytokine/Chemokine 32-plex kit and a MILLIPLEX Mouse Cytokine/Chemokine 13-plex kit (Millipore, St. Charles, MO, USA) according to the manufacturer’s protocol. The 45-plex consisted of Eotaxin, Erythropoietin, 6Ckine, Fractalkine, G-CSF, GM-CSF, IFNB1, IFNγ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-11, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-16, IL-17, IL-20, IP-10, KC, LIF, LIX, MCP-1, MCP-5, M-CSF, MDC, MIG, MIP-1α, MIP-1β, MIP-2, MIP-3α, MIP-3B, RANTES, TARC, TIMP-1, TNFα, and VEGF. The assay sensitivities of these markers range from 0.3–30.6 pg/mL.
RNA in situ hybridization (ISH)
ISH studies were performed on FFPE tissue sections using two commercial RNAscope Target Probes (Advanced Cell Diagnostics, Hayward, CA) catalog # 559271 and 408921 complementary to sequences 2–561 of Isg15 and 11–1012 of Cxcl10, respectively. Pretreatment, hybridization, and detection techniques (RNAscope 2.5HD) were performed according to the manufacturer’s protocols and as previously described.51
High throughput RNA sequencing (RNA-seq) and RNA editing site identification
Three mice of each genotype including Adar+/+, Adar+/−, AdarP195A/P195A;Ifih1−/−, and AdarP195A/−;Ifih1−/− mice were used for RNA-seq study. The whole brain was homogenized for RNA isolation, and 100ng total RNA of each mouse was used for the RNA-seq study. Twelve independent RNA libraries were prepared using the KAPA RNA HyperPrep Kit with the removal of ribosome RNAs by RiboErase (Kapa Biosystems) according to the manufacturer’s protocol, and the libraries were sequenced using the NovaSeq6000 platform (Illumina) to an average of 80M 101PE reads. Illumina adapter sequences and low-quality reads were filtered using the tool Trimmomatic.52 Then, surviving reads were aligned to the mouse reference genome mm10 by STAR aligner.53 Then variant calling was performed by the bcftools mpileup function.54,55 The candidate A-to-I editing sites were defined by A-to-G alteration on the positive strand or T-to-C alteration on the negative strand and were further annotated by repetitive regions and SNP by RepeatMasker database (Smit, AFA, Hubley, R& Green, P. RepeatMasker Open-4.0.2013–2015. http://www.repeatmasker.org) and snpSift software.56 Eventually, the valid RNA editing sites per library were defined by (1) non-SNP sites; (2) total aligned reads (sum of aligned reference reads and alternative reads) higher than 10; (3) the number of altered reads covering the site >2 (4) editing ratio (alternative reads over total aligned reads) not equivalent to 0%, 50% nor 100%. These criteria were chosen to eliminate variations caused by rare sequencing error events. For the three mice of each genotype, their common editing sites were defined as the valid alternative sites in all three repeats. Differential editing analysis was performed comparing AdarP195A/P195A;Ifih1−/− over Adar+/+, and AdarP195A/−;Ifih1−/− over Adar+/−. Per comparison, the Welch Two Sample t test on the log10 values of the calculated editing rate was performed, and the significant differential editing sites were defined by p value <0.05, as the method described by de Reuver et al.24
QUANTIFICATION AND STATISTICS ANALYSIS
The nonparametric Wilcoxon rank-sum test was used to test for differences between two groups of continuous data. The nonparametric Kruskal–Wallis test by ranks, followed by Dunn’s test, was used for multiple pairwise comparisons. All tests used were two-tailed. pP-values of less than 0.05 indicate statistical significance. Stata-17, Stata Corporation, Collage Station TX, USA, and Prism 9 GraphPad Software, Inc. were used in the analysis.
Supplementary Material
Highlights.
The ADAR1 P195A mutation alone causes brain innate immune response in AdarP195A/P195A mice
Haploinsufficiency of ADAR1 P195A mutation dramatically enhances ISG expression in the brain
ISGs are highly expressed in the periventricular areas in the brains of P195A mutant mice
The ADAR1 P195A mutation does not affect the overall RNA-editing levels in the brain
ACKNOWLEDGMENTS
We thank Hanna Wang and Christine Burr for editing the manuscript, Richard J. Bodnar for his microscopic imaging assistance, and Julia J. Dahm for assistance in figure preparation. We thank the University of Pittsburgh Biospecimen Core for brain tissue pathologic processing.
This study was supported by the National Institutes of Health grant R01AI139544, P30 DK120531 and US Department of Veterans Affairs grant I01RX001455, as well as the University of Pittsburgh Brain Institute Assault on Alzheimer’s seed grant program. This study was supported in part by the University of Pittsburgh Center for Research Computing with use of the HTC cluster, which is supported by NIH award S10OD028483.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112733.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Heraud-Farlow JE, and Walkley CR (2016). The role of RNA editing by ADAR1 in prevention of innate immune sensing of self-RNA. J. Mol. Med 94, 1095–1102. 10.1007/s00109-016-1416-1. [DOI] [PubMed] [Google Scholar]
- 2.Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, and Walkley CR (2015). RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120. 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, and Stetson DB (2015). Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity 43, 933–944. 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. (2014). The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 9, 1482–1494. 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang S, Deng P, Zhu Z, Zhu J, Wang G, Zhang L, Chen AF, Wang T, Sarkar SN, Billiar TR, and Wang Q (2014). Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs. J. Immunol 193, 3436–3445. 10.4049/jimmunol.1401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice GI, Kitabayashi N, Barth M, Briggs TA, Burton ACE, Carpanelli ML, Cerisola AM, Colson C, Dale RC, Danti FR, et al. (2017). Genetic, Phenotypic, and Interferon Biomarker Status in ADAR1-Related Neurological Disease. Neuropediatrics 48, 166–184. 10.1055/s-0037-1601449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livingston JH, Lin JP, Dale RC, Gill D, Brogan P, Munnich A, Kurian MA, Gonzalez-Martinez V, De Goede CGEL, Falconer A, et al. (2014).A type I interferon signature identifies bilateral striatal necrosis due to mutations in ADAR1. J. Med. Genet 51, 76–82. 10.1136/jmedgenet-2013-102038. [DOI] [PubMed] [Google Scholar]
- 8.Rice GI, Kasher PR, Forte GMA, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. (2012). Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet 44, 1243–1248. 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson S, Steiner A, Harapas CR, and Masters SL (2018). An Update on Autoinflammatory Diseases: Interferonopathies. Curr. Rheumatol. Rep 20, 38. 10.1007/s11926-018-0748-y. [DOI] [PubMed] [Google Scholar]
- 10.Maroofian R, Sedmík J, Mazaheri N, Scala M, Zaki MS, Keegan LP, Azizimalamiri R, Issa M, Shariati G, Sedaghat A, et al. (2021). Biallelic variants in ADARB1, encoding a dsRNA-specific adenosine deaminase, cause a severe developmental and epileptic encephalopathy. J. Med. Genet 58, 495–504. 10.1136/jmedgenet-2020-107048(2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow YJ, and Stetson DB (2022). The type I interferonopathies: 10 years on. Nat. Rev. Immunol 22, 471–483. 10.1038/s41577-021-00633-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crow YJ, and Manel N (2015). Aicardi-Goutieres syndrome and the type I interferonopathies. Nat. Rev. Immunol 15, 429–440. 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 13.Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, et al. (2007). Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am. J. Hum. Genet 81, 713–725. 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, and Rich A (1997). A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. USA 94, 8421–8426. 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai F, Drakas R, and Nishikura K (1995). Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem 270, 17098–17105. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, George CX, Patterson JB, and Samuel CE (1997). Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem 272, 4419–4428. [DOI] [PubMed] [Google Scholar]
- 17.Patterson JB, Thomis DC, Hans SL, and Samuel CE (1995). Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 210, 508–511. 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- 18.Placido D, Brown BA 2nd, Lowenhaupt K, Rich A, and Athanasiadis A (2007). A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure 15, 395–04. 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert A, Schade M, Lowenhaupt K, Alfken J, Schwartz T, Shlyakhtenko LS, Lyubchenko YL, and Rich A (1998). The Zalpha domain from human ADAR1 binds to the Z-DNA conformer of many different sequences. Nucleic Acids Res. 26, 3486–3493. 10.1093/nar/26.15.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown BA 2nd, Lowenhaupt K, Wilbert CM, Hanlon EB, and Rich A (2000). The zalpha domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl. Acad. Sci. USA 97, 13532–13536. 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbert A (2020). Mendelian disease caused by variants affecting recognition of Z-DNA and Z-RNA by the Zalpha domain of the double-stranded RNA editing enzyme ADAR. Eur. J. Hum. Genet 28, 114–117. 10.1038/s41431-019-0458-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Q, Rigby RE, Young GR, Hvidt AK, Davis T, Tan TK, Bridgeman A, Townsend AR, Kassiotis G, and Rehwinkel J (2021). Adenosine-to-inosine editing of endogenous Z-form RNA by the deaminase ADAR1 prevents spontaneous MAVS-dependent type I interferon responses. Immunity 54, 1961–1975.e5. 10.1016/j.immuni.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakahama T, Kato Y, Shibuya T, Inoue M, Kim JI, Vongpipatana T, Todo H, Xing Y, and Kawahara Y (2021). Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutieres-syndrome-like encephalopathy. Immunity 54, 1976–1988.e7. 10.1016/j.immuni.2021.08.022. [DOI] [PubMed] [Google Scholar]
- 24.de Reuver R, Dierick E, Wiernicki B, Staes K, Seys L, De Meester E, Muyldermans T, Botzki A, Lambrecht BN, Van Nieuwerburgh F, et al. (2021). ADAR1 interaction with Z-RNA promotes editing of endogenous double-stranded RNA and prevents MDA5-dependent immune activation. Cell Rep. 36, 109500. 10.1016/j.celrep.2021.109500. [DOI] [PubMed] [Google Scholar]
- 25.Inoue M, Nakahama T, Yamasaki R, Shibuya T, Kim JI, Todo H, Xing Y, Kato Y, Morii E, and Kawahara Y (2021). An Aicardi-Goutieres Syndrome-Causative Point Mutation in Adar1 Gene Invokes Multiorgan Inflammation and Late-Onset Encephalopathy in Mice. J. Immunol 207, 3016–3027. 10.4049/jimmunol.2100526. [DOI] [PubMed] [Google Scholar]
- 26.Guo X, Wiley CA, Steinman RA, Sheng Y, Ji B, Wang J, Zhang L, Wang T, Zenatai M, Billiar TR, and Wang Q (2021). Aicardi-Goutieres syndrome-associated mutation at ADAR1 gene locus activates innate immune response in mouse brain. J. Neuroinflammation 18, 169. 10.1186/s12974-021-02217-9(2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo X, Steinman RA, Sheng Y, Cao G, Wiley CA, and Wang Q (2022). An AGS-associated mutation in ADAR1 catalytic domain results in early-onset and MDA5-dependent encephalopathy with IFN pathway activation in the brain. J. Neuroinflammation 19, 285. 10.1186/s12974-022-02646-0(2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schade M, Turner CJ, Lowenhaupt K, Rich A, and Herbert A (1999). Structure-function analysis of the Z-DNA-binding domain Zalpha of dsRNA adenosine deaminase type I reveals similarity to the (alpha + beta) family of helix-turn-helix proteins. EMBO J. 18, 470–79. 10.1093/emboj/18.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurano M, Snyder JM, Connelly C, Henao-Mejia J, Sidrauski C, and Stetson DB (2021). Protein kinase R and the integrated stress response drive immunopathology caused by mutations in the RNA deaminase ADAR1. Immunity 54, 1948–1960.e5. 10.1016/j.immuni.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Z, Chalk AM, Taylor S, Goradia A, Heraud-Farlow JE, and Walkley CR (2023). The phenotype of the most common human ADAR1p150 Zalpha mutation P193A in mice is partially penetrant. EMBO Rep. 24, e55835. 10.15252/embr.202255835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartner JC, Schmittwolf C, Kispert A, Müller AM, Higuchi M, and Seeburg PH (2004). Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem 279, 4894–4902. 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, and Nishikura K (2004). Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem 279, 4952–4961. 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 33.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, and Colonna M (2006). Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyo-carditis picornavirus. Proc. Natl. Acad. Sci. USA 103, 8459–8464. 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GMA, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, et al. (2015). Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet 167A, 296–312. 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Liu S, Yan R, Nguyen V, Zenati M, Billiar TR, and Wang Q (2022). ADAR1 RNA editing regulates endothelial cell functions via the MDA-5 RNA sensing signaling pathway. Life Sci. Alliance 5, e202101191. 10.26508/lsa.202101191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinman RA, and Wang Q (2011). ADAR1 isoform involvement in embryonic lethality. Proc. Natl. Acad. Sci. USA 108, E199, author reply E200; author reply E200. 10.1073/pnas.1105004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiley CA, Steinman RA, and Wang Q (2023). Innate immune activation without immune cell infiltration in brains of murine models of Aicardi-Goutieres Syndrome. Brain Pathol. 33, e13118. 10.1111/bpa.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JI, Nakahama T, Yamasaki R, Costa Cruz PH, Vongpipatana T, Inoue M, Kanou N, Xing Y, Todo H, Shibuya T, et al. (2021). RNA editing at a limited number of sites is sufficient to prevent MDA5 activation in the mouse brain. PLoS Genet. 17, e1009516. 10.1371/journal.pgen.1009516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, and Seeburg PH (2000). Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81. 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 40.Seeburg PH, Higuchi M, and Sprengel R (1998). RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res. Brain Res. Rev 26, 217–229. 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 41.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, and Emeson RB (1997). Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387, 303–308. [DOI] [PubMed] [Google Scholar]
- 42.Costa Cruz PH, Kato Y, Nakahama T, Shibuya T, and Kawahara Y (2020). A comparative analysis of ADAR mutant mice reveals site-specific regulation of RNA editing. RNA 26, 454–469. 10.1261/rna.072728.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth SH, Levanon EY, and Eisenberg E (2019). Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods 16, 1131–1138. 10.1038/s41592-019-0610-9. [DOI] [PubMed] [Google Scholar]
- 44.Langeberg CJ, Nichols PJ, Henen MA, Vicens Q, and Vögeli B (2023). Differential Structural Features of Two Mutant ADAR1p150 Zalpha Domains Associated with Aicardi-Goutieres Syndrome. J. Mol. Biol 435, 168040. 10.1016/j.jmb.2023.168040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bajad P, Ebner F, Amman F, Szabó B, Kapoor U, Manjali G, Hildebrandt A, Janisiw MP, and Jantsch MF (2020). An internal deletion of ADAR rescued by MAVS deficiency leads to a minute phenotype. Nucleic Acids Res. 48, 3286–3303. 10.1093/nar/gkaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartner JC, Walkley CR, Lu J, and Orkin SH (2009). ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol 10, 109–115. 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang T, Yin C, Fedorov A, Qiao L, Bao H, Beknazarov N, Wang S, Gautam A, Williams RM, Crawford JC, et al. (2022). ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 606, 594–602. 10.1038/s41586-022-04753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiao H, Wachsmuth L, Wolf S, Lohmann J, Nagata M, Kaya GG, Oikonomou N, Kondylis V, Rogg M, Diebold M, et al. (2022). ADAR1 averts fatal type I interferon induction by ZBP1. Nature 607, 776–783. 10.1038/s41586-022-04878-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hubbard NW, Ames JM, Maurano M, Chu LH, Somfleth KY, Gokhale NS, Werner M, Snyder JM, Lichauco K, Savan R, et al. (2022). ADAR1 mutation causes ZBP1-dependent immunopathology. Nature 607, 769–775. 10.1038/s41586-022-04896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Reuver R, Verdonck S, Dierick E, Nemegeer J, Hessmann E, Ahmad S, Jans M, Blancke G, Van Nieuwerburgh F, Botzki A, et al. (2022). ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature 607, 784–789. 10.1038/s41586-022-04974-w(2022. [DOI] [PubMed] [Google Scholar]
- 51.Brien JD, Uhrlaub JL, Hirsch A, Wiley CA, and Nikolich-Zugich J (2009). Key role of T cell defects in age-related vulnerability to West Nile virus. J. Exp. Med 206, 2735–2745. 10.1084/jem.20090222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup(2009).The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, and Lu X (2012). Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front. Genet 3, 35. 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ADAR1 Antibody (15.8.6) | Santa Cruz Biotechnology, Inc. | Cat#: sc-73408 |
| β-Actin Antibody (C4) | Santa Cruz Biotechnology, Inc. | Cat#: sc-47778 |
| Critical commercial assays | ||
| MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel | Millipore Sigma | Cat#: MCYTOMAG-70K |
| MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel II | Millipore Sigma | Cat#: MECY2MAG-73K |
| Deposited data | ||
| RNA-seq data | NCBI Gene Expression Omnibus | accession ID GSE229884 |
| Experimental models: Organisms/strains | ||
| Mouse:AdarP195A/P195A | Guo et al. 202126 | NA |
| Mouse:AdarP195A/− | This paper | NA |
| Mouse:Adar +/− | Wang et al. 200436 | NA |
| Mouse: Ifih1−/− | The Jackson Laboratory | Stock#: 015812 |
| Oligonucleotides | ||
| See Table S9. | This paper | N/A |
| Software and algorithms | ||
| Trimmomatic | Bolger et al. 201459 | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR | Dobin et al. 201360 | https://github.com/alexdobin/STAR |
| bcftools | Li et al. 200961 | https://samtools.github.io/bcftools/bcftools.html |
| snpSift | Cingolani et al. 201262 | https://pcingola.github.io/SnpEff/ |