In this study, Zong et al. show that the loss of DNA helicase WRN in cancer cells with high microsatellite instability (MSI-H) induces successive accumulation of replication-associated DNA damage without CHK1 checkpoint activation that eventually results in cell death. They further show that cotargeting ATR, which is responsible for WRN and CHK1 phosphorylation, in instances of partial WRN deficiency could be an effective strategy to achieve complete inactivation of WRN, thus supporting the use of combinatorial therapy in MSI-H cancers.
Keywords: ATR, WRN, microsatellite instability, synthetic lethality
Abstract
Addiction to the WRN helicase is a unique vulnerability of human cancers with high levels of microsatellite instability (MSI-H). However, while prolonged loss of WRN ultimately leads to cell death, little is known about how MSI-H cancers initially respond to acute loss of WRN—knowledge that would be helpful for informing clinical development of WRN targeting therapy, predicting possible resistance mechanisms, and identifying useful biomarkers of successful WRN inhibition. Here, we report the construction of an inducible ligand-mediated degradation system in which the stability of endogenous WRN protein can be rapidly and specifically tuned, enabling us to track the complete sequence of cellular events elicited by acute loss of WRN function. We found that WRN degradation leads to immediate accrual of DNA damage in a replication-dependent manner that curiously did not robustly engage checkpoint mechanisms to halt DNA synthesis. As a result, WRN-degraded MSI-H cancer cells accumulate DNA damage across multiple replicative cycles and undergo successive rounds of increasingly aberrant mitoses, ultimately triggering cell death. Of potential therapeutic importance, we found no evidence of any generalized mechanism by which MSI-H cancers could adapt to near-complete loss of WRN. However, under conditions of partial WRN degradation, addition of low-dose ATR inhibitor significantly increased their combined efficacy to levels approaching full inactivation of WRN. Overall, our results provide the first comprehensive view of molecular events linking upstream inhibition of WRN to subsequent cell death and suggest that dual targeting of WRN and ATR might be a useful strategy for treating MSI-H cancers.
Microsatellite instability (MSI) is a pathological condition of pervasive hypermutation that occurs at short tandem repeat DNA sequences within the genome, caused by defects in cellular mismatch repair (MMR) (Olave and Graham 2022). MSI has been observed in many types of human cancers, most commonly in colorectal, endometrial, and gastric adenocarcinomas. Mismatch repair-deficient (MMRd) tumors with high levels of microsatellite instability (MSI-H) tend to respond well to immunotherapy (Jin and Sinicrope 2022; Taieb et al. 2022) but may be relatively less responsive to conventional chemotherapy. Nevertheless, chemotherapy remains the standard of care for patients with MSI-H cancers (Taieb et al. 2022). Moreover, only about half of all patients with MMRd tumors show durable responses to immunotherapy (Le et al. 2017; Westcott et al. 2023), underscoring an urgent need for additional means to combat these deadly diseases.
In the past 4 yr, multiple studies have identified a unique addiction of human MSI-H cancers to the RECQ family helicase WRN (Behan et al. 2019; Chan et al. 2019; Kategaya et al. 2019; Lieb et al. 2019; van Wietmarschen et al. 2020; Picco et al. 2021; Morales-Juarez and Jackson 2022), uncovering one of the most striking examples of synthetic lethality since PARP inhibition was shown to specifically kill homologous recombination-defective breast/ovarian cancers two decades ago (Bryant et al. 2005; Farmer et al. 2005). This has led to a huge push from the pharmaceutical industry to develop clinical-grade WRN inhibitors. However, despite the obvious benefit of targeting WRN, our understanding of how upstream WRN deficiency triggers downstream death of MSI-H cancer cells remains incomplete. We had shown earlier that MSI-H colorectal cancer cells contain unstable structure-forming TA repeats in their genome and that, in the absence of WRN, these become fragile and ultimately break (van Wietmarschen et al. 2020, 2021). Our study, along with many others, thoroughly documented the long-term effects of WRN deficiency in MSI-H cancers using a combination of siRNA, shRNA, and/or CRISPR-based approaches. However, due to the relatively slow and asynchronous nature of these reagents, it has not been possible so far to study the short-term, early effects of acute WRN loss in a manner that would mimic pharmacological inhibition.
To fill this gap in our knowledge and achieve a more complete elucidation of the cellular responses to WRN deficiency, we devised a strategy to engineer a degradable WRN protein encoded at its endogenous locus based on PROTAC technology. The modified WRN protein is functional and can be rapidly induced to degrade upon the addition of a small molecule ligand, dTAG-13 (Nabet et al. 2018). Using this model, we systemically mapped cell fates upon acute WRN loss. We show that the response of MSI-H cancer cells to WRN deficiency is biphasic and highly heterogeneous. In the acute phase, cells begin accruing replication-associated DNA damage almost as soon as WRN is degraded. Nevertheless, the majority of MSI-H cancer cells traverse two or more cell cycles before they die or become stably arrested (the chronic phase). Mechanistically, WRN degradation elicited a partially blunted DNA damage response (DDR) in MSI cancer cells, whereby repair factors such as 53BP1 and RAD51 were mobilized normally but a robust checkpoint response failed to initially engage, resulting in trans cell cycle propagation and amplification of DNA damage. Of therapeutic relevance, we found that MSI-H cancers do not readily adapt to loss of WRN, which is in contrast to the relative ease with which HR-defective cancers become resistant to PARP inhibitors. Finally, we found that ATR inhibition mimicked WRN inactivation in MSI-H cancers and that low doses of ATR inhibitor can compensate for incomplete WRN degradation. As ATR inhibitors are already approved clinically, our work thus supports the use of ATR inhibitors either alone (at higher doses) or in tandem (at lower doses) with WRN inhibition to combat human MSI-H cancers.
Results
An endogenous, rapidly inducible system for controlled degradation of WRN
Prolonged loss of WRN is extremely toxic to human MSI-H cancer cells, but the sequence of proximal events triggered upon acute WRN loss, particularly signaling upstream of eventual cell death, remains largely unexplored. We therefore sought to engineer a new system capable of inducing rapid and controlled tuning of WRN protein levels—properties that would allow us to probe both short-term and long-term consequences of WRN ablation.
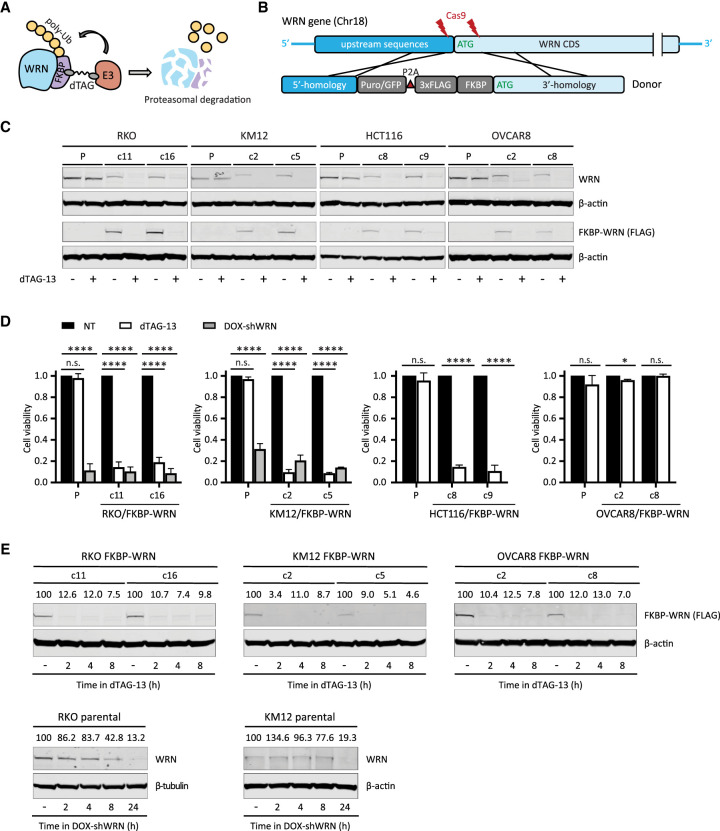
To this end, we adapted PROTAC and CRISPR technologies to fuse an FKBP12F36V degron domain to the N terminus of WRN at its endogenous locus in three MSI-H cell lines and one MSS cell line (Fig. 1A,B; Supplemental Fig. S1). We derived two types of clones: those that contained homozygous knock-in of FKBP-WRN and others that showed hemizygous knock-in with the second allele of WRN inactivated by a loss-of-function indel (deletion of ATG or nonsense mutation) (Supplemental Fig. S1B). In both cases, the FKBP-WRN protein was produced at a reduced level compared with unmodified WRN (Fig. 1C). This level of WRN protein was nonetheless sufficient to promote survival of human MSI-H cancer lines.
Figure 1.
Generation of human cancer cells expressing degradable FKBP-WRN. (A) Schematics of the FKBP-WRN degron system. (E3) Cellular E3 ubiquitin ligase (CRBN in the case of dTAG-13). (B) Targeting strategy to achieve knock-in of FKBP-WRN at the endogenous human WRN locus. (C) Immunoblotting confirming successful knock-in of FKBP-WRN in human cancer cell lines treated or not with 0.5 μM dTAG-13 for 24 h. One of two independent experiments is shown. (D) Viability of FKBP-WRN-expressing human cancer cell lines treated or not with 0.5 μM dTAG-13 for 6 d. Where indicated, shWRN expression was induced with 1 μg/mL doxycycline (DOX). Average (±SD) of three independent experiments is shown. (E) Immunoblotting comparing the kinetics of dTAG-induced FKBP-WRN degradation with that of shWRN-induced WRN depletion. One of two independent experiments is shown.
To validate the degron system, we administered a standard dose of 0.5 μM dTAG-13 to MSI-H cancer cells expressing FKBP-WRN, which engaged the cellular E3 ubiquitin ligase CRBN (Nabet et al. 2018) to induce WRN degradation (Fig. 1C). Since our RKO and KM12 cells contained a pre-existing doxycycline-inducible shWRN (DOX-shWRN) cassette (van Wietmarschen et al. 2020), direct comparison could be readily made between dTAG- and shRNA-mediated WRN depletion. Importantly, we found that dTAG-13 treatment impaired the viability of FKBP-WRN-expressing RKO and KM12 cells to an extent similar to that of DOX-shWRN, while their parental counterparts were eliminated by DOX-shWRN but not dTAG-13 (Fig. 1D). Similarly, the MSI-H cell line HCT116, but not the MSS OVCAR8, was also highly sensitive to dTAG-13 treatment (Fig. 1D), confirming the target specificity and MSI selectivity of our degron system. Detailed time course analysis in both MSI-H (RKO, KM12) and MSS (OVCAR8) cells revealed that ∼90% of WRN protein was degraded by 2 h, while depletion of WRN by DOX-shWRN was comparatively slower (Fig. 1E). On the other hand, dTAG-13 did not affect the stability of unmodified WRN in parental lines (Fig. 1C). Taken together, we concluded that our FKBP-WRN degron system can induce rapid and synchronous degradation of WRN protein and therefore is a suitable tool to dissect how MSI-H cancer cells sense and respond to acute loss of WRN function.
Acute loss of WRN triggers immediate DNA damage in cycling cells
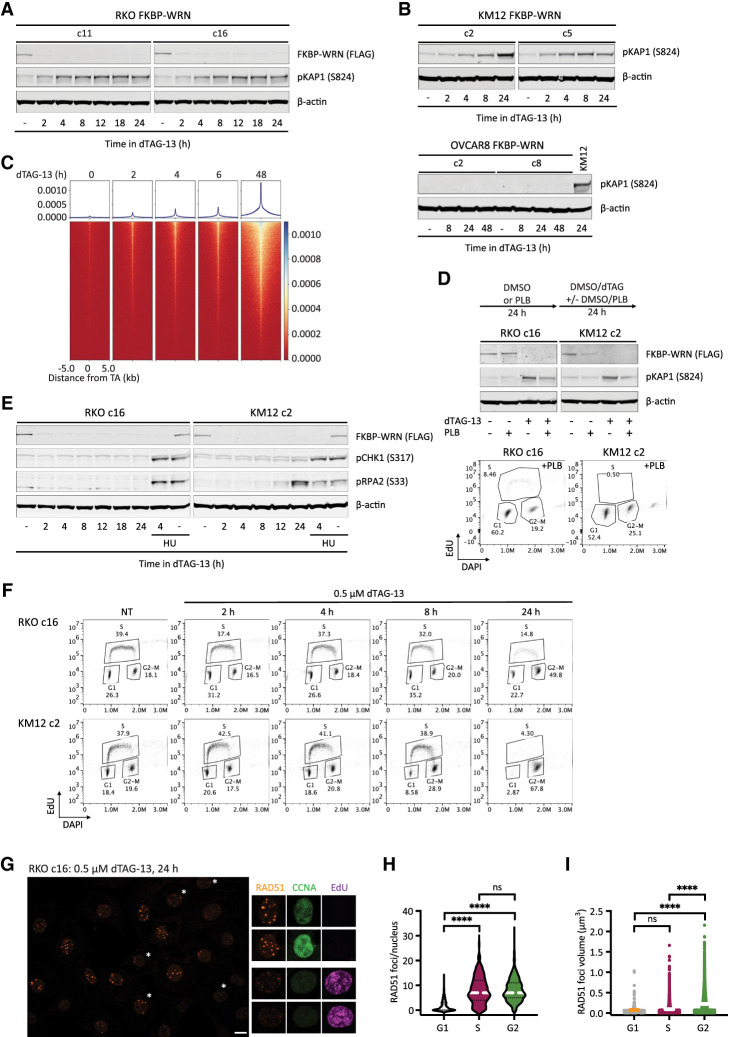
Previous studies using sh/siRNA and/or CRISPR have shown that DNA damage accumulates in MSI-H cancer cells several days after WRN ablation (Chan et al. 2019; Kategaya et al. 2019; Lieb et al. 2019; van Wietmarschen et al. 2020). However, these studies lacked sufficient temporal resolution to answer when and how DNA damage was first produced following WRN depletion. Using our inducible degron system, we found that acute degradation of FKBP-WRN led to measurable increases in phosphorylated KAP1 (pKAP1), a marker of DNA double-strand breaks, as soon as 2–4 h after the addition of dTAG-13 in RKO and KM12 cells (Fig. 2A,B). Consistent with this, double-stranded DNA breakage at sites of unstable TA repeats was also detected by END-seq within hours of dTAG-13 treatment in KM12 (Fig. 2C). Thus, MSI-H cells appeared to incur DNA damage almost immediately after FKBP-WRN had been degraded. In contrast, MSS OVCAR8 cells did not show induction of pKAP1 in response to WRN degradation, even days after dTAG-13 treatment (Fig. 2B). Next, we examined whether DNA damage induction upon acute WRN loss was dependent on cell cycle position. Notably, while dTAG-induced WRN degradation itself was cell cycle-independent, blocking S-phase entry with palbociclib (PLB) significantly reduced pKAP1 induction in RKO and KM12 cells as compared with their asynchronous counterparts (Fig. 2D). These observations suggest that DNA damage caused by loss of WRN depends on active proliferation.
Figure 2.
Acute degradation of WRN in MSI-H cancer cells causes replication-associated DNA damage without accompanying intra-S checkpoint activation. (A,B) Immunoblotting depicting the rapid induction of pKAP1, a marker of DNA double-strand breaks, by 0.5 μM dTAG-13 in RKO (A) and KM12 (B) clones expressing FKBP-WRN. In contrast, dTAG-13 did not induce pKAP1 in OVCAR8 clones expressing FKBP-WRN even after extended treatment. One of two independent experiments is shown. (C) END-seq showing the rapid appearance of DNA breakage at unstable expanded TA repeats in KM12 cells treated with 0.5 μM dTAG-13. (D, top panel) Immunoblotting showing that 0.5 μM dTAG-13 (24 h) induced significantly lower levels of pKAP1 in RKO and KM12 cells pretreated for 24 h with 5 μM palbociclib (PLB). (Bottom panel) Successful inhibition of S-phase entry by PLB was confirmed by FACS-based cell cycle analyses. One of three independent experiments is shown. (E) Immunoblotting demonstrating the lack of CHK1 (S317) and RPA2 (S33) phosphorylation in dTAG-treated RKO FKBP-WRN cells. CHK1/RPA2 phosphorylation was also undetectable in KM12 FKBP-WRN cells except at 24 h after dTAG-13 treatment. Where indicated, cells were treated with 3 mM hydroxyurea (HU) for 2 h, either alone or in combination with 0.5 μM dTAG-13 (2 h of dTAG-13 pretreatment followed by 2 h of HU + dTAG-13). Note that HU induced robust CHK1/RPA phosphorylation in both RKO and KM12 independently of WRN. One of two independent experiments is shown. (F) FACS-based cell cycle analyses showing that dTAG-treated RKO and KM12 cells failed to slow down or halt DNA synthesis during the first S phase after acute WRN degradation. Note that WRN-degraded cells eventually completed S phase and activated the G2 checkpoint. (G) Representative confocal images depicting RAD51 foci (maximum intensity projection) in RKO FKBP-WRN cells treated or not with 0.5 μM dTAG-13 for 24 h. Cyclin A (CCNA) and EdU positivity was used to distinguish cells in S phase from those in G2. Note that RAD51 foci appear to be larger in G2 cells (EdU-negative, CCNA-high), as compared with S-phase cells (EdU-positive, CCNA-low/intermediate). (H) Quantification of RAD51 number per nucleus as a function of cell cycle position. (I) Volumetric analysis of RAD51 foci as a function of cell cycle position. One of two independent experiments is shown.
WRN deficiency triggers a blunted ATR response in MSI-H cells
Eukaryotic cells typically respond to DNA damage by arresting cell cycle progression through the prompt activation of checkpoints, orchestrated by ATM–CHK2 and ATR–CHK1 (Waterman et al. 2020). In WRN-degraded MSI-H cancer cells, we found that the rapid elevation of pKAP1 was closely followed by the induction of CHK2 phosphorylation (Supplemental Fig. S2A). Therefore, the presence of DNA DSBs was clearly sensed by ATM. However, WRN degradation triggered little to no CHK1 phosphorylation, although the ATR–CHK1 pathway was intact in these cells and could be activated by hydroxyurea (Fig. 2E; Supplemental Fig. S2B).
In the absence of CHK1-enforced intra-S checkpoints, DNA replication proceeded unperturbed in WRN-degraded RKO and KM12 cells for at least 8 h (Fig. 2F). By 24 h, we detected a large accumulation of cells with 4N DNA content, indicating that most cells have completed S phase and entered G2, where CHK2 activity had presumably slowed down their progression toward mitosis. Interestingly, these G2 cells contained numerous prominent RAD51 foci that appeared qualitatively larger and brighter than RAD51 foci typically found in S-phase cells (Fig. 2G,H; Supplemental Fig. S2C). Detailed volumetric analyses subsequently confirmed that RAD51 foci present in G2 were indeed larger than those found in S phase (Fig. 2I). In contrast, ionizing irradiation-induced RAD51 foci were of similar sizes irrespective of cell cycle position in S or G2 (Supplemental Fig. S2C). Although the exact significance of this difference remains unclear, we speculate that the lack of robust checkpoints early on may limit the repair of replication-associated DNA damage in WRN-deficient MSI-H cancer cells and as such may continuously concentrate repair factors at DNA damage sites.
WRN-degraded MSI-H cancer cells propagate and amplify DNA damage across multiple mitoses
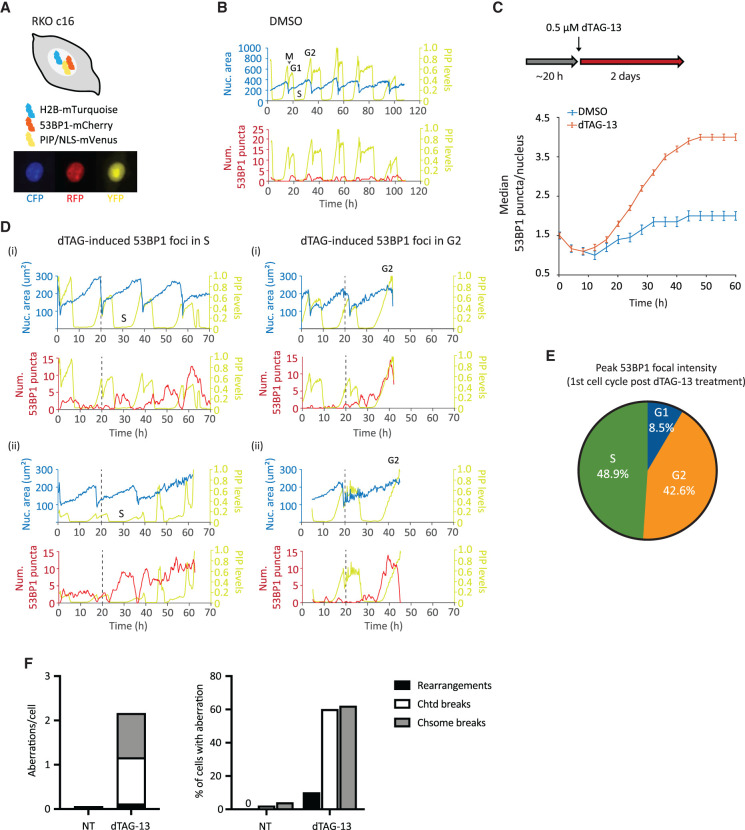
To gain additional insight into how MSI-H cancer cells respond to WRN ablation, we constructed an RKO FKBP-WRN cell line suitable for live-cell imaging (Fig. 3A,B). This system combined three separate reporters, enabling us to monitor chromatin (H2B-mTurquoise), cell cycle (PIP-NLS-mVenus), and DNA damage (53BP1-mCherry) simultaneously and in real time. We first confirmed that acute degradation of WRN indeed induced DNA damage, marked by 53BP1, very rapidly (Fig. 3C,D) and preferentially in S–G2 phases of the first cell cycle (Fig. 3D,E). The behavior of these 53BP1 foci was quite heterogeneous, with some being highly dynamic while others were relatively persistent. In accordance with previous studies (Nelson et al. 2009; Giunta et al. 2010), we observed that all 53BP1 foci dissolved in early mitosis. Nevertheless, we found that more than half of WRN-degraded cells harbored unrepaired DNA breaks (chromatid and/or chromosome type) in the first metaphase, and 10% additionally displayed complex rearrangements that were indicative of erroneous repair (Fig. 3F). Thus, cellular DNA repair machineries were not able to completely heal all WRN-associated lesions prior to cell division. Consistent with this idea, most WRN-degraded cells appeared to regain 53BP1 foci in the following G1, and the median 53BP1 foci per nucleus rose steadily over time (Fig. 3C).
Figure 3.
Induction and repair of DNA damage in WRN-degraded MSI-H cancer cells monitored by time-lapse microscopy. (A) Schematics of the RKO triple-reporter line used for live-cell imaging. An example of vehicle-treated cells is shown below in the CFP, RFP, and YFP channels. (B) Automated cell tracking was performed as described in the Materials and Methods. Example traces of a vehicle-treated reporter cell over time; different phases of the cell cycle are indicated. The birth of a cell at mitosis (M) coincides with a reduction in nuclear area (as defined by H2B-mTurquoise) as well as PIP sensor intensity. PIP intensity increases in G1 but is then abruptly lost as a cell enters S phase. Finally, PIP expression returns in G2. 53BP1 focus formation is also shown for this vehicle-treated cell. (C) Formation of 53BP1 puncta as a function of time. Cells were first imaged for ∼20 h without drug to establish baseline 53BP1 levels. Subsequent addition of dTAG-13 resulted in a rapid induction of 53BP1 puncta over the baseline, which steadily increased over time. One of two independent experiments is shown. (D) Example traces of four dTAG-treated reporter cells. The two cells in the left panels showed induction of 53BP1 puncta during the first S phase after WRN degradation. The two cells in the right panels had primarily induced 53BP1 in the first G2 after WRN degradation. Note, however, that these two cells also displayed a smaller induction of 53BP1 in the preceding S phase. (E) Quantification of 53BP1 punctum intensity during the first cell cycle following acute WRN degradation revealed that peak 53BP1 induction occur predominantly in S and G2. One of two independent experiments is shown. (F) Analyses of metaphase spreads showing that a majority of dTAG-treated RKO cells failed to completely repair DNA breaks incurred in S/G2 and carried residual damages into the first mitosis. One of two independent experiments is shown.
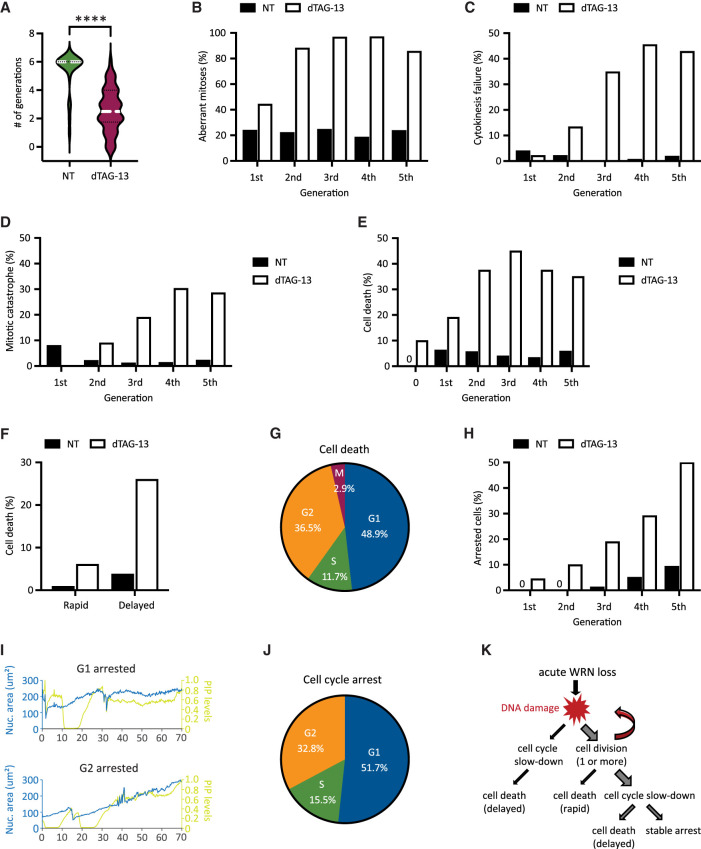
To examine how transmission of unresolved DNA damage may impact future cell division activities, we manually traced the lineage of a subset of RKO reporter cells over a span of 6 d. Overall, we found that most of the vehicle-treated RKO cells (n = 25) divided at least six times (after which accurate tracking became difficult due to cell movements and crowding), while WRN-degraded RKO cells (n = 50) divided on average two to three times (range zero to six) (Fig. 4A). Approximately 45% of first mitoses in dTAG-treated cells exhibited discernible abnormalities (Fig. 4B), such as chromosomal bridges and micronucleation. Subsequent mitoses in their progeny were even more error-prone, causing severe defects such as cytokinesis failures and mitotic catastrophe, in addition to bridges and micronuclei (Fig. 4B–D). In contrast, the frequencies of abnormal mitoses among vehicle-treated cells were significantly lower and stayed constant with each generation (Fig. 4B–D). Altogether, these observations support a model in which WRN-deficient MSI-H cells propagate and amplify DNA damage in a trans cell cycle manner due to a combination of inadequate repair and accrual of new lesions. As such, the massive genome fragility that we and others detected previously in MSI-H cancer cells after several days of prolonged WRN depletion via RNA interference likely reflects the culmination of DNA damage over multiple cell cycles rather than a single catastrophic event.
Figure 4.
Heterogeneity in the fate of WRN-degraded MSI-H cancer cells. RKO reporter cells were treated or not with 0.5 μM dTAG-13 and imaged by time-lapse microscopy over a span of 6 d. Manual tracking was performed on 25 vehicle-treated and 50 dTAG-treated cells. (A) Cell division activities. Most of vehicle-treated cells divided at least six times, producing 840 tracked progeny. In contrast, dTAG-treated cells divided less frequently (average two to three times, range zero to six) and produced less progeny (377 tracked). (B) Mitotic abnormalities as a function of cell division. A mitosis was scored as abnormal if it exhibited one or more of the following features: bridging, micronucleus formation, cytokinesis failure, and/or mitotic catastrophe. (C) Frequency of cytokinesis failure as a function of cell division. (D) Frequency of mitotic catastrophe as a function of cell division. Mitotic catastrophe was defined as any mitosis that resulted in complete failure to divide (failed mitosis), multipolar chromosome segregation, or rapid induction of mitotic cell death. (E) Frequency of cell death as a function of cell division. Note that 10% of dTAG-treated cells died without undergoing a single mitosis (denoted as generation 0). (F) Cell death as a function of time. A death event is defined as rapid if it occurred <10 h after the latest mitosis from which the cell was born; otherwise, it is defined as delayed. (G) Cell death as a function of cell cycle position. (H) Frequency of cell cycle arrest as a function of cell division. (I) Example traces of two dTAG-treated cells showing arrest in G1 (top panel) or G2 (bottom panel). (J) Cell cycle arrest as a function of cell cycle position. One of two independent experiments is shown. (K) Diagram depicting the various observed fates of RKO cells following acute WRN degradation.
WRN-degraded MSI-H cancer cells exhibit marked heterogeneity in their fate
Previously, it was reported that MSI-H cancer cells in which WRN was depleted by si/shRNA ultimately died in a manner consistent with induction of apoptosis (Chan et al. 2019; Kategaya et al. 2019). To validate and extend these findings, we conducted detailed manual cell fate mapping using our live imaging reporter system. We observed that 10% of dTAG-treated RKO cells died without ever dividing, whereas none of the vehicle-treated cells did so (Fig. 4E). Of those dTAG-treated cells that divided at least once, we carefully documented the fate of 377 progeny, of which 35% died within 6 d. In contrast, 4.9% of their vehicle-treated counterparts (840 tracked progeny) died during the same period. In concordance with the trans cell cycle propagation and amplification of DNA damage, we found that death following acute WRN loss tended to occur after at least two rounds of cell division, while the rate of death among vehicle-treated cells remained constant over time (Fig. 4E). Interestingly, the time elapsed between the birth of a given cell at mitosis and its subsequent demise was typically quite protracted (>10 h up to days) (Fig. 4F). During this time, the doomed cell might linger in the ensuing G1, attempt to replicate its DNA, or even enter G2 after completion of S phase (Fig. 4G). However, a minority of cells died rapidly (<10 h) after birth, either while still in M or shortly thereafter in G1 (Fig. 4F). Most of the observed cell death events exhibited chromatin condensation, consistent with the induction of apoptosis. However, we noted that in many cases the nucleus also seemed to “explode,” while in other instances the nucleus remained relatively intact (Supplemental Movie S1). Overall, these data suggest that WRN deficiency triggers cell death with significant temporal and morphological heterogeneity in MSI-H cancer cells, perhaps involving mechanisms in addition to classical apoptosis.
Of the cells that were still alive by the end of the experiment, we found that a significant proportion (15%–20% of all tracked progeny) appeared to have entered a state of stable arrest. As with cell death, the likelihood of cell cycle arrest also increased with each division (Fig. 4H). Similarly, CFSE dye dilution assay showed a progressive decline in cell division activities for RKO, KM12, and HCT116 cells following acute WRN degradation (Supplemental Fig. S3A), mirroring a steady rise in the levels of phosphorylated CHK2 over time (Supplemental Fig. S3B,C). We found that CHK1 activation remained undetectable in RKO FKBP-WRN clones even at late time points (Supplemental Fig. S3B), and accordingly, careful manual tracking of WRN-degraded RKO reporter cells showed that they arrested predominantly in G1 or G2, with only a minor fraction locked in S phase (Fig. 4I,J). Interestingly, WRN-degraded KM12 and HCT116 cells were able to activate CHK1 at late time points in addition to CHK2, which may explain why dTAG-13 treatment caused a stronger proliferative impairment in these two cell lines compared with RKO (Supplemental Fig. S3A,C). Altogether, these data highlight the heterogeneous fate that acute WRN loss imparts on MSI-H cancer cells (Fig. 4K).
MSI-H cancer cells do not readily adapt to WRN deficiency
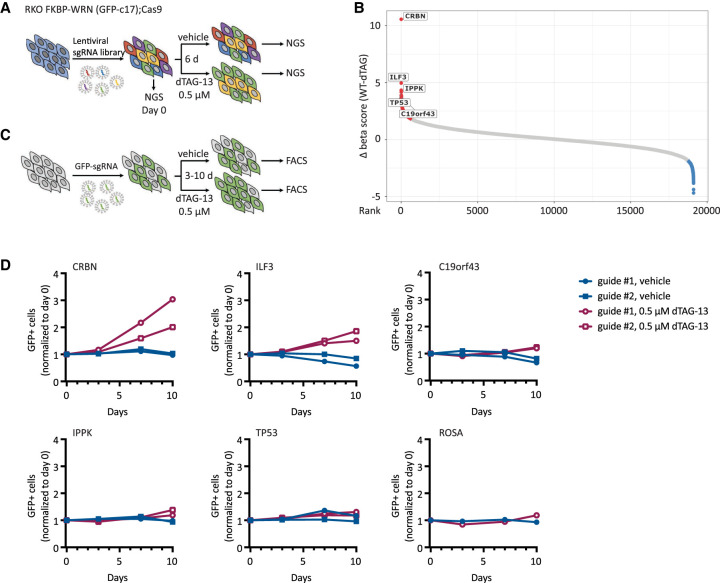
Given that WRN addiction is a striking and potentially actionable vulnerability in human MSI-H cancers, a key question that has not yet been addressed is whether WRN hypersensitivity can be circumvented by secondary inactivation of another cellular process. To test this possibility, we conducted a knockout CRISPR screen in a clone of Cas9-expressing RKO cells by transducing them with a genome-wide lentiviral library (Brunello) of single-guide RNAs (sgRNAs). The resultant pools of edited cells were either treated with vehicle or exposed to a near-lethal dose of dTAG-13 over 6 d (Fig. 5A). We used the model-based analysis of genome-wide CRISPR–Cas9 knockout (MAGeCK) algorithm to calculate gene enrichment or depletion (Fig. 5B; Supplemental Fig. S4; Li et al. 2014). Importantly, we identified CRBN, the gene encoding the ubiquitin E3 ligase engaged by dTAG-13, as the top hit whose deletion confers resistance to dTAG-13 (Fig. 5B), thereby confirming the validity of our approach.
Figure 5.
MSI-H cancer cells do not readily adapt to WRN deficiency. (A) Schematics of the CRISPR screening strategy used in RKO c17/Cas9 cells to identify genetic hits that could potentially confer resistance to WRN loss. (B) Candidate resistance genes identified in the screen. (C) Schematics of growth competition assays used to validate candidate resistance genes. (D) Growth competition assays confirmed that deletion of CRBN and, to a lesser degree, ILF3 rescued cytotoxicity caused by acute loss of WRN in RKO c17/Cas9 cells. Note that CRBN encodes the cellular E3 ubiquitin ligase that is engaged by dTAG-13 to promote FKBP-WRN degradation. One of two independent experiments is shown.
Apart from CRBN, we identified only a few additional candidate resistance genes, including ILF3, IPPK, TP53, and c19orf43 (Fig. 5B; Supplemental Table S1). Nevertheless, the sgRNAs targeting these genes showed much lower levels of enrichment than guides targeting CRBN. To validate potential hits, we transduced the RKO clone used in the original CRISPR screen with two individual guides targeting these candidate genes and performed growth competition assays in the presence or absence of dTAG-13 (Fig. 5C). We confirmed that deletion of CRBN completely abrogated dTAG-induced FKBP-WRN degradation, and as expected, CRBN-deficient RKO cells were significantly enriched after dTAG-13 treatment (Fig. 5D; Supplemental Fig. S5A,B). Deletion of ILF3 also conferred some level of resistance to dTAG-13 (Fig. 5D); however, cells deficient in ILF3 no longer appeared to degrade FKBP-WRN efficiently in response to dTAG-13 treatment (Supplemental Fig. S5B), thus ruling it out as a bona fide WRN resistance gene. Finally, the loss of IPPK, c19orf43, or TP53 had a modest to negligible impact on cell viability under conditions of WRN deficiency (Fig. 5D). These findings were recapitulated in a different clone of RKO and in KM12 (Supplemental Fig. S5C,D). Overall, we conclude that there is currently no evidence of any generalized mechanism of WRN resistance in human MSI-H cancer cells.
ATR inhibition sensitizes MSI-H cancer cells to partial loss of WRN
The apparent inability of human MSI-H cancer cells to adapt to WRN deficiency is highly encouraging and supports the therapeutic utility of WRN inhibitors. Clinically, it is often difficult to completely inactivate a target protein in vivo because of excessive toxicity and/or limited bioavailability. When we lowered the dose of dTAG-13 from 0.5 μM to the 2.5–10 nM range, we observed both higher levels of residual WRN protein and increased cell survival (Supplemental Fig. S6A–C), suggesting that low doses of dTAG-13 treatment can be used to simulate partial inhibition of WRN.
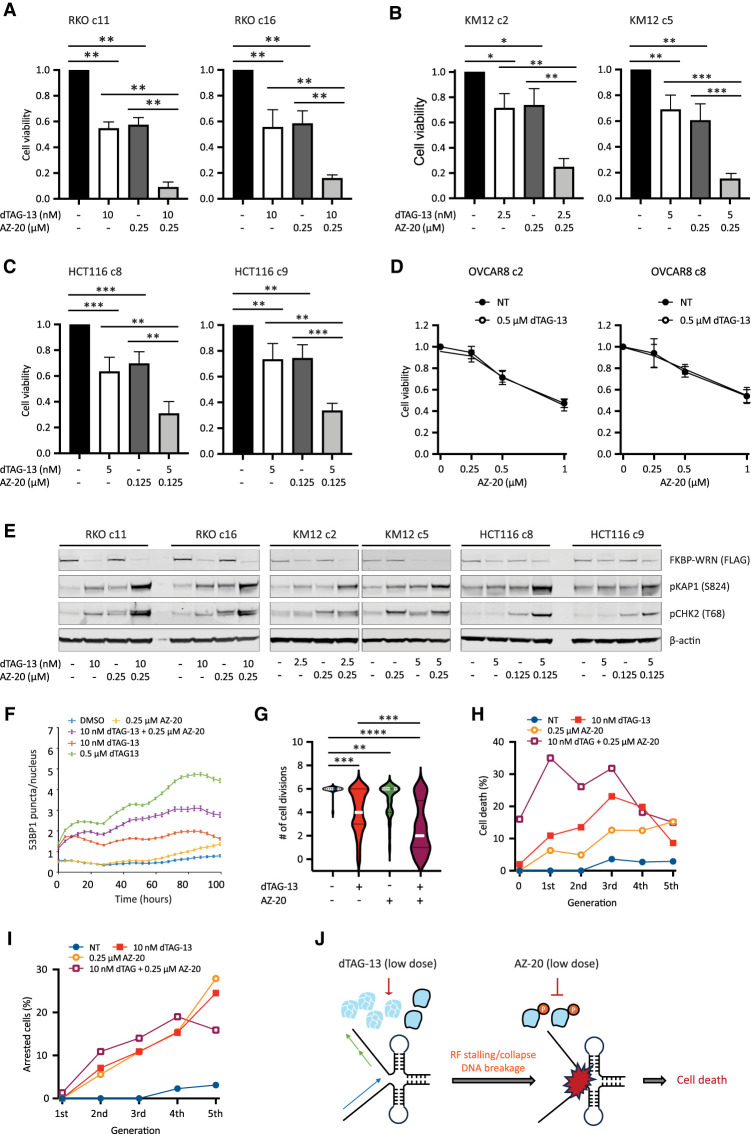
Next, we explored ways to augment the efficacy of low-dose dTAG-13 treatment. Previous studies from our laboratory have shown that WRN phosphorylation by ATR is required for its genome-protective functions in MSI-H cancer cells (van Wietmarschen et al. 2020). We therefore tested whether ATR inhibition could complement low-dose dTAG-13 treatment, potentially by impairing the function of residual active WRN. However, we found that MSI-H cancer cells were highly sensitive to the ATR inhibitor AZ-20 even under WRN-proficient conditions, especially at doses of ≥0.5 μM (Supplemental Fig. S6D,E; data not shown). This notion was further supported by data extracted from several genome-wide screens and public drug sensitivity data sets (Supplemental Fig. S6F,G). Although microsatellite instability clearly confers hypersensitivity to ATR inhibition, it is possible that MMR deficiency in of itself could also play a role (Wang et al. 2023). While these results support the potential use of ATR inhibitor monotherapy in MSI-H human cancers, the inherent hypersensitivity of MSI-H cells to ATR inhibitors made it difficult to interpret combinatorial effects of dual WRN/ATR inhibition. To circumvent this issue, we decided to combine low doses of dTAG-13 with concentrations of AZ-20 that were lower than those used in our single-agent studies. Notably, we found that the combination regimen imparted significantly greater losses in the viability of RKO, KM12, and HCT116 cells than what was achievable with either agent alone (Fig. 6A–C; Supplemental Fig. S7A,B). On the contrary, the combination regimen yielded an effect comparable with that of high-dose ATR inhibition alone in MSS OVCAR8 cells (Fig. 6D). Mechanistically, the combination regimen led to increased DNA damage load, as shown by elevated KAP1 phosphorylation (Fig. 6E) and breakage at TA repeat sites (Supplemental Fig. S7C). Corroborating these findings, live-cell imaging showed that 53BP1 focus formation was significantly enhanced in RKO reporter cells treated with the combination regimen as compared with their single-agent-treated counterparts (Fig. 6F), which likely contributed to decreased cell proliferation, augmented cell cycle arrest, and more rapid induction of cell death (Fig. 6G–I). Taken together, these data provide a reasonable rationale for the further development and testing of combination regimens of ATR and WRN inhibitors to treat human MSI-H cancers.
Figure 6.
ATR inhibition can compensate for incomplete WRN degradation. (A–C) Human cancer cells expressing FKBP-WRN were treated as indicated for 6 d. When used in combination, the ATR inhibitor AZ-20 was always added 2 h after the start of dTAG-13 treatment. Viability assays showed that the combination regimen produced significantly superior cytotoxicity as compared with single-agent treatments in MSI-H RKO (A), KM12 (B), and HCT116 (C) clones but not in MSS OVCAR8 (D). Average (±SD) of three (OVCAR8 and KM12 c2) or four (RKO, HCT116, and KM12 c5) independent experiments is shown. (E) MSI-H cancer cells expressing FKBP-WRN were treated as indicated for 24 h. Immunoblotting showing that the combination regimen resulted in markedly elevated levels of pKAP1 as compared with either single agent alone. One of two independent experiments is shown. (F) RKO reporter cells were treated as indicated and subjected to live-cell imaging. Automated tracking of 53BP1 puncta showed that the combination regimen induced more 53BP1 than either agent alone. (G–I) Manual fate mapping of RKO reporter cells: vehicle (n = 25; 1094 tracked progeny), 10 nM dTAG-13 (n = 50; 992 tracked progeny), 0.25 μM AZ-20 (n = 40; 1074 tracked progeny), and dTAG-13/AZ-20 (n = 50; 497 tracked progeny). Cells treated with the combination regimen exhibited reduced cell division activities (G) accompanied by increases in both cell death (H) and cell cycle arrest (I). For F–I, one of two independent experiments is shown. (J) Proposed model of how ATR inhibition compensates for incomplete WRN degradation.
Discussion
Successful targeting of tumor-specific vulnerabilities is the ultimate goal of modern precision anticancer therapy. Recently, the RECQ helicase WRN has emerged as a crucial survival factor in human MMRd MSI-H cancers (Behan et al. 2019; Chan et al. 2019; Kategaya et al. 2019; Lieb et al. 2019; van Wietmarschen et al. 2020; Picco et al. 2021; Morales-Juarez and Jackson 2022). The unique specificity of WRN addiction in MSI-H cancer cells and the extreme toxicity unleashed by WRN loss put it on par with PARP inhibition in BRCA1/2-deficient tumors (van Wietmarschen et al. 2021; Morales-Juarez and Jackson 2022). Despite holding considerable promise, however, clinical-grade WRN inhibitors are not currently available. To get a sense of how MSI-H cancer cells might respond to pharmacological WRN inactivation in both the short and long term, we engineered a rapidly inducible degron system and probed it with state-of-the art reporter-based live-cell imaging. We discovered that accrual of replication-associated DNA double-strand breakage is a conserved immediate early consequence of acute WRN loss, while cell death and cell cycle arrest occur relatively late in a more heterogeneous manner.
The lesions that accumulate in the genomes of WRN-degraded MSI-H cancer cells are quickly sensed by ATM, leading to the mobilization of many canonical signaling and repair factors including pKAP1, pCHK2, 53BP1, RPA, and RAD51. Surprisingly, however, MSI-H cancer cells fail to robustly activate CHK1 during the first S phase after acute WRN loss, thus allowing replication to continue unabated and eventually be completed in the presence of DNA damage. Through real-time monitoring of 53BP1 foci, we also found that a substantial fraction of WRN-degraded RKO cells appears to incur additional DNA damage in G2 (Fig. 3D), possibly the result of nucleolytic processing of unresolved aberrant replication intermediates. In this manner, the lack of a viable intra-S checkpoint early on could trigger a cascade of damage accumulation as cells progress through S/G2, which ultimately overwhelms the cellular DNA repair machineries. Moreover, without activated CHK1 to support pCHK2, the G2 checkpoint is likely to become more vulnerable to slippage. Indeed, examination of metaphase chromosomes of RKO cells entering the first mitosis after WRN degradation revealed both unresolved DNA breaks and genomic rearrangements associated with misrepair. Thus, the inability of MSI-H cancer cells to halt DNA synthesis immediately upon WRN loss leads to transmission of DNA damage to the next cell cycle, which could be a major cause of subsequent cytotoxicity. We further suggest that these effects should be recapitulated by acute pharmacological inhibition of WRN.
By carefully tracking the fate of WRN-degraded RKO reporter cells, we found that they typically progress through two or more cell cycles before succumbing. In addition, some cells apparently entered a particular cell cycle phase from which they did not emerge, though it is not yet clear whether this leads to a permanent exit from the cell cycle (i.e., senescence) or whether it is simply a prolonged transitional state after which cells either die or recover. Previously, it was reported that WRN-deficient MSI-H cancer cells undergo apoptosis, a form of cell death that is nonimmunogenic. However, based on morphological characteristics and a relative lack of PARP1 cleavage (data not shown), our data raise the possibility that cell death triggered by WRN deficiency in MSI-H cancer cells may be biochemically distinct from classical apoptosis. If WRN inhibition indeed induces potentially immunogenic forms of cell death as well as senescence in MSI-H tumors, combining it with immunotherapy may prove to be even more beneficial.
As with any targeted therapy, the clinical benefit of prospective WRN inhibitors is potentially limited by the emergence of resistance mechanisms. In this regard, a recent study reported that inactivation of TP53 completely abrogated apoptosis induced by WRN depletion in MSI-H colorectal cancer cells (Hao et al. 2022). However, TP53 did not behave as a bona fide WRN resistance gene in our system. In addition, KM12 cells are known to be p53 mutated yet respond strongly to WRN depletion by either dTAG-13 or shRNA. Moreover, both we and others (Lieb et al. 2019) have observed that p53-deficient HCT116 cells were just as (hyper)sensitive as their WT counterparts to si/shWRN (data not shown). Overall, aside from a few potential mild and clone-specific modifiers, we found no evidence of any generalized mechanisms that could confer resistance to WRN deficiency in MSI-H cancer cells. Nevertheless, we cannot rule out the possibility that such resistance might be obtained through screening additional MSI-H cellular models from diverse tissues, overexpression of certain cellular factors, and/or gain-of-function mutations. In addition, pre-existing cancer subclones harboring relatively limited repeat expansions could potentially emerge under the pressure of sustained WRN inhibition. We also note that our WRN degradation model may not perfectly recapitulate the action of a pharmacological agent that impairs WRN protein function without causing its degradation. It would therefore be important to revisit the topic of resistance mechanisms once clinical-grade WRN inhibitors become available.
Finally, just how much residual WRN is sufficient to promote survival of MSI-H cancers is a key question that should be considered when developing future WRN inhibitors. Using our FKBP-WRN degron system, we observed the most efficient cell killing at doses of 0.04–0.5 μM dTAG-13, which degraded ∼90% of WRN protein, while dTAG-13 doses below this range resulted in significantly less cytotoxicity. Because this applied to both homozygous and hemizygous clones, we estimate that ∼95% of functional WRN protein would need to be inhibited for optimal outcome. In other words, very little WRN may be able to sustain the viability of MSI-H cancer cells. Interestingly, previous findings have shown WRN to be functionally regulated by ATR-mediated phosphorylation (Pichierri et al. 2003; Ammazzalorso et al. 2010; van Wietmarschen et al. 2020), suggesting that a combination approach might promote stronger suppression of WRN activity. Indeed, we were able to significantly enhance cytotoxicity in MSI-H cancer cells by combining low doses of dTAG-13 and relatively low concentrations of the ATR inhibitor AZ-20, with the efficacy of the combination regimen approaching that of high-dose dTAG-13. We hypothesize that the combination works by augmenting partial degradation of WRN with functional inactivation of the remaining proteins, effectively achieving near-complete inhibition of WRN (Fig. 6J). The inhibition of ATR may subvert additional critical genome surveillance mechanisms, such as protection against excessive nucleolytic activity at stalled replication forks (RFs) (Couch et al. 2013; Matos et al. 2020), compounding the damage leading to replication catastrophe (Fig. 6J). Whatever the precise mechanism, our data suggest that dual targeting of WRN and ATR could be a productive strategy to explore for the treatment of human MSI-H cancers.
Materials and methods
Cell lines and culture conditions
RKO, KM12, SW48, NCIH508, SW620, and SW837 cells containing a doxycycline-inducible shWRN cassette were generated as previously described (Chan et al. 2019; van Wietmarschen et al. 2020). HCT116, LS180, and OVCAR8 cells were obtained from commercial sources. RKO and LS180 were grown under standard conditions in EMEM (ATCC). KM12, SW48, NCIH508, SW837, and OVCAR8 cells were cultured in RPMI-1640, while HCT116 and SW620 cells were maintained in McCoy 5A and Leibovitz L-15 media, respectively. RPMI-1640, McCoy 5A, and L-15 were from Thermo Fisher Scientific. All media were supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gemini Bio-Products) and 1% antibiotic (penicillin and streptomycin; Gibco). All cell lines were confirmed free of mycoplasma.
Construction of the FKBP-WRN donor and the WRN targeting vector
DNA encoding the 5′ and 3′ homology arms of WRN was obtained from Twist Biosciences and subsequently cloned into pGMC00018, along with the PCR product encoding PuromycinR-2A-3X-FLAG-FKBP12 or eGFP-2A-3X-FLAG-FKBP12, using isothermal assembly. Guide RNAs targeting the N terminus of WRN (ENST00000298139.6) were designed using sgRNA Scorer 2.0 (Chari et al. 2017). Candidates 3537 (TGACACCTAGGTCCAAGCATAGG; PAM in bold) and 3539 (TGGAAACAACTGCACAGCAGCGG) were chosen based on superior cutting efficiency in initial test experiments. Oligonucleotides corresponding to these two guide sequences were subsequently annealed and ligated into the Cas9-containing pDG458 vector (Adikusuma et al. 2017) using golden gate assembly; pDG458 was a gift from Dr. Paul Thomas (Addgene plasmid 100900, RRID: Addgene_100900). The FKBP-WRN donor plasmid construct and WRN targeting vector were verified using Sanger sequencing (ACGT DNA sequencing services).
Generation of FKBP-WRN cell lines
Recipient cells were transiently transfected with both the FKBP-WRN donor and the WRN targeting vector using X-tremeGENE 9 (Sigma) as per the manufacturer's instructions. After 48 h, cells were either subjected to puromycin selection (if PuromycinR-2A-3X-FLAG-FKBP12 was used as the donor, as in RKO and KM12) or FACS-sorted (if eGFP-2A-3X-FLAG-FKBP12 was used as the donor, as in HCT116 and RKO c17). Single-cell-derived candidate clones were harvested for DNA and protein. The targeted region was amplified by PCR (MyTaq Red mix, Bioline) from 100 ng of genomic DNA. Successful knock-in of FKBP-WRN was confirmed by Sanger sequencing and by immunoblotting (for both WRN and FLAG).
To generate the FKBP-WRN triple-reporter cell line, RKO clone #16 was stably transduced with lentiviral vectors encoding H2B-mTurquoise, PIP/NLS-mVenus, and 53BP1-mCherry. H2B-mTurquoise (Spencer et al. 2013) and PIP/NLS-mVenus (Grant et al. 2018) have been described previously. 53BP1-mCherry was excised from mCherry-BP1-2 pLPC-Puro (Addgene plasmid 19835) (Dimitrova et al. 2008) and inserted into H53-LV (a gift from Dr. Michael Ward) using isothermal assembly. Triple-positive cells were FACS-sorted (Sony MA900) and expanded for future use.
Chemicals
dTAG-13 was purchased from Tocris Bioscience. Doxycycline, hydroxyurea, and CFSE (carboxyfluorescein diacetate N-succinimidyl ester) were from Sigma. Palbociclib (PLB) and AZ-20 were from Selleck Chemicals. EdU (5-ethynyl-2′-deoxyuridine) was from Invitrogen. Formaldehyde and paraformaldehyde were obtained from Sigma and Electron Microscopy Sciences, respectively. DAPI (4′,6-diamidino-2-phenylindole) was from Thermo Fisher Scientific. All other chemical reagents were from Sigma.
Cell viability assays
Cells were seeded in six-well plates (10,000–20,000 cells per well) overnight and treated with drugs the following day. DOX-shWRN expression was induced with 1 μg/mL doxycycline. The standard dosage of both dTAG-13 and AZ-20 was 0.5 μM, with DMSO serving as the vehicle. Where indicated, lower doses of either or both were also used. When used in combination, AZ-20 was always added 2 h after dTAG-13. Cells were treated continuously for 6 d. Viability was determined using the CellTiter-Glo luminescent cell viability assay (Promega) as per the manufacturer's instructions.
Live-cell imaging
RKO FKBP-WRN triple-reporter cells were seeded in a 24-well plate (Ibidi 89626) ∼18 h prior to imaging. Cells were plated such that cell density remained subconfluent until the end of the imaging period (3–6 d). Depending on the goal of the experiment, different drug treatment and imaging schedules were used. To examine 53BP1 induction, cells were first imaged for ∼20 h in drug-free media to establish baseline 53BP1 level and behavior, after which 0.5 μM dTAG-13 or vehicle (DMSO) was quickly added and imaging resumed for an additional 3 d. To conduct long-term cell fate mapping, cells were treated with DMSO or 0.5 μM dTAG-13 and imaged over a span of 6 d. To explore the effect of WRN/ATR single inhibition or coinhibition, cells were first treated with vehicle or 10 nM dTAG-13, followed by the addition of vehicle or 0.25 μM AZ-20 2 h later before commencing image acquisition (6 d). In all cases, time-lapse images were taken in CFP, YFP, and RFP channels every 12 min on a Nikon Ti2-E inverted microscope (Nikon) with a 20× 0.45 NA objective. NIS Elements (Nikon v5.11.00) software was used for controlling image acquisition. Total light exposure time was kept under 600 msec for each time point. The microscope was housed within an environmental chamber to maintain cells at 37°C with 5% humidified CO2.
Image processing and single-cell tracking
All image analyses were performed with custom Matlab scripts as previously described (Cappell et al. 2016). Cells were segmented for their nuclei based on H2B-mTurquoise. Nuclear PIP sensor activity was calculated as median nuclear intensity and then normalized to obtain a range of values between 0 and 1. 53BP1-mCherry puncta corresponding to pixel locations within the nuclear mask were quantified by applying a TopHat filter followed by segmentation. The number of unique segmented objects corresponding to the mCherry channel was counted to obtain the number of 53BP1 puncta, and the pixel intensity of each spot was quantified to obtain 53BP1 punctum intensity.
Segmentation and automated tracking of cells from time-lapse microscopy films were performed using a previously published Matlab pipeline (Cappell et al. 2016; code is available at https://github.com/scappell/cell_tracking). Briefly, cell tracks were linked by screening the nearest future neighbor using segmented cell nuclei in adjacent frames. When the imaging plate was removed from and put back on the microscope stage (for addition of drugs), the plate jitter was calculated by registering images of the nucleus-stained channel and corrected prior to tracking.
Fate mapping of a subset of cells was performed by manual analyses of time-lapse microscopy films using Fiji (Schindelin et al. 2012). Only cells that were present in the first time-lapse frame and not already showing overt signs of abnormalities were included in the analyses. Care was also taken to exclude cells that rapidly moved out of frame. Cell cycle positions were assigned based on manual examination of PIP sensor intensity.
Immunoblotting
Western blotting was performed as previously described (Zong et al. 2015). Briefly, cells were washed in PBS and lysed in an extraction buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5% Tween-20, 2% Igepal, 2 mM PMSF, and 2.5 mM b-glycerophosphate (all from Sigma) and supplemented with protease and phosphatase inhibitor tablets (both from Roche Diagnostics). Equal amounts of protein were loaded into precast 4%–12% Bis-Tris minigels (Invitrogen) and resolved by SDS-PAGE. Thereafter, proteins were transferred onto nitrocellulose membranes, blocked in a TBS-based blocking agent (LiCor Biosciences), and incubated with the corresponding primary antibodies recognizing the following proteins: WRN (1:1000; Novus Biologicals), FLAG (1:1000; Sigma), pKAP1 (S824; 1:1000; Bethyl Laboratories), pATR (T1989; 1:500; Cell Signaling Technologies), pCHK1 (S345; 1:500; Cell Signaling Technologies), pCHK1 (S317; 1:500; Novus Biologicals), pCHK1 (S296; 1:1000; a gift from Dr. Lee Jung-Min), CHK1 (1:1000; Cell Signaling Technologies), pRPA2 (S33; 1:1000; Bethyl Laboratories), RPA2 (1:2000; Cell Signaling Technologies), pCHK2 (T68; 1:1000; Cell Signaling Technologies), CHK2 (1:1000; Cell Signaling Technologies), β-actin (1:5000; Sigma), and Tubulin (1:5000; Sigma). Fluorescently labeled secondary antibodies were used at a dilution of 1:10,000 (Li-Cor Biosciences). Visualization and quantification of protein bands were achieved by fluorescence imaging using a Li-Cor Odyssey CLx imaging system (Li-Cor Biosciences).
END-seq
Detailed protocol for END-seq has been described elsewhere (Canela et al. 2016; Wong et al. 2021). Briefly, 10 million KM12 or HCT116 cells along with the spike-in control (20%) were carefully resuspended in cell suspension buffer (Bio-Rad). The resultant homogeneous single-cell suspensions were embedded in agarose and transferred into plug molds (Bio-Rad CHEF mammalian genomic DNA plug kit). Plugs were allowed to solidify at 4°C and were then incubated with proteinase K (Qiagen) for 1 h at 50°C and then overnight at 37°C. The deproteinized plugs were sequentially washed in wash buffer (10 mM Tris at pH 8.0, 50 mM EDTA) and TE buffer (10 mM Tris at pH 8.0, 1 mM EDTA), treated with RNase A (Qiagen), and washed again in wash buffer. Blunting, A-tailing, and ligation of biotinylated hairpin adaptor 1 (END-seq-adaptor-1, 5′-Phos-GATCGGAAGAGCGTCGTGTAGGGAAAGAGTGUU[Biotin-dT]U[Biotin-dT]UUACACTCTTT CCCTACACGACGCTCTTCCGATC*T-3′ [* indicates phosphorothioate bond]) were performed in the plug. Thereafter, DNA was recovered by first melting the agarose plugs and then digesting the agarose with GELase (Epicenter) as per the manufacturer's recommendations. The recovered DNA was then sheared to a length between 150 and 200 bp by sonication (Covaris), followed by capture of biotinylated DNA fragments using streptavidin beads (MyOne C1, Invitrogen). The second end of the captured DNA was repaired using an enzymatic cocktail (15 U of T4 DNA polymerase, 5 U of Klenow fragment, 15 U of T4 polynucleotide kinase), A-tailed with 15 U of Klenow exo(−), and ligated to hairpin adaptor 2 (END-seq-adaptor-2, 5′-Phos-GATCGGAAGAGCACACGTCUUUUUUUUAGACGTGTGCTCTTCCGATC*T-3′ [* indicates phosphorothioate bond]) (quick ligation kit, NEB). To prepare libraries for sequencing, the hairpins on both END-seq adapters were digested with USER enzyme (NEB) and PCR-amplified for 16 cycles using TruSeq index adapters. All libraries were quantified using Picogreen or qPCR. Sequencing was performed on an Illumina NextSeq500 (75-bp single-end reads). Alignment of reads was performed as previously described (van Wietmarschen et al. 2020). The bamtobed function in the bedtools package was used to convert the bam files to bed files. The genomecov function in the bedtools package was used to convert the bed files into bedGraph files, scaled to normalize to spike-in control. The bedGraphToBigWig function in the UCSC package was used to convert the bedGraph files to bigWig files. The computeMatrix and plotHeatmap functions in the deeptools package were used to generate heat maps of double-strand break intensity around sites of TA breaks (as defined in van Wietmarschen et al. 2020) by reference point with –missingDataAsZero.
Immunofluorescence
Cells were seeded on 12-mm round glass coverslips overnight and treated with DMSO or 0.5 μM dTAG-13 the following day for 24 h. Cells were additionally incubated with 10 μM EdU during the last 30 min. Depending on the application, cells were either fixed immediately with 4% paraformaldehyde for 10 min or first pre-extracted (20 mM HEPES, 50 mM NaCl, 3 mM MgCl2, 0.3 M sucrose, 0.2% Triton X-100) for 5 min on ice to remove soluble nuclear proteins prior to fixation. Samples were then permeabilized with 0.5% Triton X-100 for 5 min and blocked in 2% BSA/PBS. Incubation of primary antibodies recognizing RAD51 (1:250; Millipore), Cyclin A (1:200; Santa Cruz Biotechnology), and/or RPA2 (1:2000; Cell Signaling Technologies) was followed by appropriate fluorochrome-conjugated secondary antibodies (Invitrogen). Next, Click-IT chemistry was performed as per the manufacturer's instructions (Thermo Fisher Scientific), and DNA was counterstained with DAPI. Wide-field fluorescence images were captured at 40× magnification on a Lionheart LX automated microscope (BioTek Instruments). Quantification of nuclear foci was performed using Gen5 spot analysis software (BioTek Instruments). Confocal z-stacks were acquired using a Nikon SoRa spinning disk microscope equipped with a 60× apo TIRF oil immersion objective lens (1.49 NA) and Photometrics BSI sCMOS camera. Z-stacks were collected with a 0.2-μm step size and 0.110-μm X–Y pixel size. A denoise.ai algorithm was applied to the images in the data set using Nikon Elements (v.5.41) image analysis software. The denoised images were imported into Imaris image analysis software (Oxford Instruments), and the ImarisCell module was used to segment the individual nuclei and measure both the number of RAD51 foci per nucleus and the volume of each focus.
Flow cytometry
Vehicle- and dTAG-13-treated cells were incubated with 10 μM EdU for 30 min at 37°C, fixed with 1% formaldehyde for 10 min, and stained using the Click-IT EdU Alexa fluor 647 flow cytometry assay kit (Thermo Fisher Scientific) as per the manufacturer's instructions. DNA content was assessed by incorporation of 1 μg/mL DAPI. Where indicated, cells were first pulse-labeled with 5 μM CFSE for 10 min at 37°C and treated or not with 0.5 μM dTAG-13 the following day. At 24–72 h after treatment, live cells were collected for FACS. Data were acquired on a Cytoflex tabletop flow cytometer (Beckman Coulter) and analyzed using FlowJo (BD Biosciences).
Preparation and analysis of metaphase spreads
Cells were treated with DMSO or 0.5 μM dTAG-13 for 18 h and subsequently arrested at mitosis with 0.1 mg/mL colcemid (Roche) for 6 h (total 24 h). Metaphase chromosome spreads were prepared as previously described (Zong et al. 2015, 2019). First, cells were induced to swell in a prewarmed hypotonic solution containing 0.75 M KCl (Sigma) for 20 min at 37°C. Next, suspensions of single cells were fixed with a solution mixture containing methanol and glacial acetic acid at a 3:1 ratio. Thereafter, cells were further washed extensively with the same fixative solution and dropped onto slides in a humidified chamber (Thermotron Industries). To visualize metaphase chromosomes by fluorescence in situ hybridization (FISH), samples immobilized on slides were sequentially treated with 5–10 μg/mL pepsin in 0.01 N HCl for 5 min at 37°C, washed, dehydrated with ethanol, briefly heat-denatured for 1.25 min at 80°C in a slide moat (Boekel Scientific), and incubated with a commercially available Cy3-labeled (CCCTAA)3 peptide nucleic acid probe (PNA Bio) recognizing mammalian telomere sequences. After extensive washes, DNA was counterstained with DAPI. Images were acquired at 63× magnification using the Metafer automated scanning and imaging platform (MetaSystems). Fifty individual metaphases were scored for the presence of chromosomal aberrations.
CRISPR–Cas9 screen
CRISPR–Cas9 screen was performed using the whole-genome human Brunello CRISPR knockout pooled library (Addgene 73178) (Doench et al. 2016). RKO FKBP-WRN (GFP clone #17) cells stably expressing Cas9 were transduced at a multiplicity of infection (MOI) of 0.2 and 200-fold coverage of the library. The following day, cells were selected with 0.5 μg/mL puromycin for 3 d and further expanded for 4 d. Aliquots of day 0 cells were collected, and the remaining cells were split into triplicates for drug treatment (DMSO vs. 0.5 μM dTAG-13), maintaining 200-fold coverage for a further 6 d. Surviving cells from each condition were collected, and genomic DNA was isolated (Blood and Cell Culture DNA midi kit, Qiagen). DNA was PCR-amplified with Illumina-compatible primers (P7 and P5 mix), gel-purified, and processed for Illumina sequencing. Genes enriched or depleted in the dTAG-13-treated samples were determined with MAGeCK software package version 0.5.9.5.
Growth competition assay
Sequences for sgRNAs of interest were cloned into LRG2.1-GFP (Addgene 108098) (Tarumoto et al. 2018) as described previously (Girish and Sheltzer 2020) and Sanger-sequenced to confirm correct insertion. RKO FKBP-WRN (GFP clone #17, Puro clone #16) and KM12 FKBP-WRN (Puro clone #5) stably expressing Cas9 were seeded and transduced with LRG2.1-GFP containing the sgRNA of interest (MOI ∼0.5). After 3 d, cells were split into control and dTAG-13 (0.5 μM), and an aliquot was run on flow cytometry to establish GFP expression at baseline. On the indicated days, an aliquot of cells was collected for flow cytometry analysis, and the remaining cells were split and maintained in dTAG-13 as needed. To verify knockdown of target genes, cells were treated or not with 0.5 μM dTAG-13 for 24 h before collection. RNA was extracted (RNeasy mini kit, Qiagen), and 1 μg was used to make cDNA (SuperScript VILO cDNA synthesis kit, Invitrogen). Quantitative RT-PCR was performed with iTAQ SYBR Green using 1/40th of the cDNA reaction per sample and the appropriate primers. Western blotting was performed as described above.
Statistical analyses
Unless otherwise indicated, statistical significance was evaluated with unpaired Student's t-test or Mann–Whitney nonparametric test. The resultant P-values (represented by asterisks) are indicated in the figures (P > 0.05 [ns], P < 0.05 [*], P < 0.01 [**], P < 0.001 [***], and P < 0.0001[****]) and were calculated using GraphPad Prism. The total number of replicates and the mean and error bars are explained in the figure legends.
Supplementary Material
Acknowledgments
We thank Dr. Eros Lazzerini Denchi, Dr. Sergio Ruiz, Dr. Michael Lichten, Dr. Sam John, and members of the Laboratory of Genome Integrity for discussions and advice; Dr. Ferenc Livak, Dr. Subhadra Banerjee, and Dr. Shafiuddin Siddiqui at the Center for Cancer Research (CCR)/Laboratory of Genetic Integrity Flow Cytometry Core for cell sorting; and the National Cancer Institute (NCI)/CCR Genomics Core for help with sequencing. The computational resources of the National Institutes of Health (NIH) High-Performance Computing Biowulf cluster were used for data analyses. This work is supported by the Intramural Research Program of the NIH, funded in part with federal funds from the NCI under contract HHSN2612015000031. The A.N. laboratory is also supported by an Ellison Medical Foundation Senior Scholar in Aging Award (AG-SS-2633-11), two Department of Defense Awards (W81XWH-16-1-599 and W81XWH-19-1-0652), and the Alex's Lemonade Stand Foundation Award. The S.D.C. laboratory is supported by the NIH Intramural Research Program (ZIA BC 011830).
Author contributions: D.Z. and A.N. initiated the project. D.Z. and N.C.K. carried out most of the experiments with assistance from N.W. Live-cell imaging and confocal microscopy were performed by J.A.C. and M.J.K., respectively, with assistance from D.Z.; A.V.P. and N.C.K. were responsible for computational analyses. R.C. designed and constructed vectors used for FKBP-WRN knock-in. D.Z. and A.N. wrote the manuscript with input from N.C.K., J.A.C., and S.D.C.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.351085.123.
Competing interest statement
The authors declare no competing interests.
References
- Adikusuma F, Pfitzner C, Thomas PQ. 2017. Versatile single-step-assembly CRISPR/Cas9 vectors for dual gRNA expression. PLoS One 12: e0187236. 10.1371/journal.pone.0187236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. 2010. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J 29: 3156–3169. 10.1038/emboj.2010.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, et al. 2019. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568: 511–516. 10.1038/s41586-019-1103-9 [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917. 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, Nussenzweig A. 2016. DNA breaks and end resection measured genome-wide by end sequencing. Mol Cell 63: 898–911. 10.1016/j.molcel.2016.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappell SD, Chung M, Jaimovich A, Spencer SL, Meyer T. 2016. Irreversible APCCdh1 inactivation underlies the point of no return for cell-cycle entry. Cell 166: 167–180. 10.1016/j.cell.2016.05.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Shibue T, McFarland JM, Gaeta B, Ghandi M, Dumont N, Gonzalez A, McPartlan JS, Li T, Zhang Y, et al. 2019. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568: 551–556. 10.1038/s41586-019-1102-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R, Yeo NC, Chavez A, Church GM. 2017. sgRNA Scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth Biol 6: 902–904. 10.1021/acssynbio.6b00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Bétous R, Carroll CM, Jung SY, Qin J, Cimprich KA, et al. 2013. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev 27: 1610–1623. 10.1101/gad.214080.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528. 10.1038/nature07433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34: 184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921. 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- Girish V, Sheltzer JM. 2020. A CRISPR competition assay to identify cancer genetic dependencies. Bio Protoc 10: e3682. 10.21769/BioProtoc.3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S, Belotserkovskaya R, Jackson SP. 2010. DNA damage signaling in response to double-strand breaks during mitosis. J Cell Biol 190: 197–207. 10.1083/jcb.200911156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GD, Kedziora KM, Limas JC, Cook JG, Purvis JE. 2018. Accurate delineation of cell cycle phase transitions in living cells with PIP-FUCCI. Cell Cycle 17: 2496–2516. 10.1080/15384101.2018.1547001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Tong J, Jha A, Risnik D, Lizardo D, Lu X, Goel A, Opresko PL, Yu J, Zhang L. 2022. Synthetical lethality of werner helicase and mismatch repair deficiency is mediated by p53 and PUMA in colon cancer. Proc Natl Acad Sci 119: e2211775119. 10.1073/pnas.2211775119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Sinicrope FA. 2022. Mismatch repair-deficient colorectal cancer: building on checkpoint blockade. J Clin Oncol 40: 2735–2750. 10.1200/JCO.21.02691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kategaya L, Perumal SK, Hager JH, Belmont LD. 2019. Werner syndrome helicase is required for the survival of cancer cells with microsatellite instability. iScience 13: 488–497. 10.1016/j.isci.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. 2017. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357: 409–413. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15: 554. 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb S, Blaha-Ostermann S, Kamper E, Rippka J, Schwarz C, Ehrenhöfer-Wölfer K, Schlattl A, Wernitznig A, Lipp JJ, Nagasaka K, et al. 2019. Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells. Elife 8: e43333. 10.7554/eLife.43333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos DA, Zhang JM, Ouyang J, Nguyen HD, Genois MM, Zou L. 2020. ATR protects the genome against R loops through a MUS81-triggered feedback loop. Mol Cell 77: 514–527.e4. 10.1016/j.molcel.2019.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Juarez DA, Jackson SP. 2022. Clinical prospects of WRN inhibition as a treatment for MSI tumours. NPJ Precis Oncol 6: 85. 10.1038/s41698-022-00319-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, et al. 2018. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol 14: 431–441. 10.1038/s41589-018-0021-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Buhmann M, von Zglinicki T. 2009. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle 8: 3379–3383. 10.4161/cc.8.20.9857 [DOI] [PubMed] [Google Scholar]
- Olave MC, Graham RP. 2022. Mismatch repair deficiency: the what, how and why it is important. Genes Chromosomes Cancer 61: 314–321. 10.1002/gcc.23015 [DOI] [PubMed] [Google Scholar]
- Picco G, Cattaneo CM, van Vliet EJ, Crisafulli G, Rospo G, Consonni S, Vieira SF, Rodríguez IS, Cancelliere C, Banerjee R, et al. 2021. Werner helicase is a synthetic-lethal vulnerability in mismatch repair-deficient colorectal cancer refractory to targeted therapies, chemotherapy, and immunotherapy. Cancer Discov 11: 1923–1937. 10.1158/2159-8290.CD-20-1508 [DOI] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F, Franchitto A. 2003. Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene 22: 1491–1500. 10.1038/sj.onc.1206169 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T. 2013. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155: 369–383. 10.1016/j.cell.2013.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb J, Svrcek M, Cohen R, Basile D, Tougeron D, Phelip JM. 2022. Deficient mismatch repair/microsatellite unstable colorectal cancer: diagnosis, prognosis and treatment. Eur J Cancer 175: 136–157. 10.1016/j.ejca.2022.07.020 [DOI] [PubMed] [Google Scholar]
- Tarumoto Y, Lu B, Somerville TDD, Huang YH, Milazzo JP, Wu XS, Klingbeil O, El Demerdash O, Shi J, Vakoc CR. 2018. LKB1, Salt-inducible kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol Cell 69: 1017–1027.e6. 10.1016/j.molcel.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wietmarschen N, Sridharan S, Nathan WJ, Tubbs A, Chan EM, Callen E, Wu W, Belinky F, Tripathi V, Wong N, et al. 2020. Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature 586: 292–298. 10.1038/s41586-020-2769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wietmarschen N, Nathan WJ, Nussenzweig A. 2021. The WRN helicase: resolving a new target in microsatellite unstable cancers. Curr Opin Genet Dev 71: 34–38. 10.1016/j.gde.2021.06.014 [DOI] [PubMed] [Google Scholar]
- Wang M, Ran X, Leung W, Kawale A, Saxena S, Ouyang J, Patel P, Dong Y, Yin T, Shu J, et al. 2023. ATR inhibition induces synthetic lethality in mismatch repair-deficient cells and augments immunotherapy. Genes Dev (this issue). 10.1101/gad.351084.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman DP, Haber JE, Smolka MB. 2020. Checkpoint responses to DNA double-strand breaks. Annu Rev Biochem 89: 103–133. 10.1146/annurev-biochem-011520-104722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PMK, Muyas F, Hauck H, Smith OC, Sacks NJ, Ely ZA, Jaeger AM, Rideout WM III, Zhang D, Bhutkar A, et al. 2023. Mismatch repair deficiency is not sufficient to elicit tumor immunogenicity. Nat Genet 55: 1686–1695. 10.1038/s41588-023-01499-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, John S, Nussenzweig A, Canela A. 2021. END-seq: an unbiased, high-resolution, and genome-wide approach to map DNA double-strand breaks and resection in human cells. Methods Mol Biol 2153: 9–31. 10.1007/978-1-0716-0644-5_2 [DOI] [PubMed] [Google Scholar]
- Zong D, Callén E, Pegoraro G, Lukas C, Lukas J, Nussenzweig A. 2015. Ectopic expression of RNF168 and 53BP1 increases mutagenic but not physiological non-homologous end joining. Nucleic Acids Res 43: 4950–4961. 10.1093/nar/gkv336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong D, Adam S, Wang Y, Sasanuma H, Callén E, Murga M, Day A, Kruhlak MJ, Wong N, Munro M, et al. 2019. BRCA1 haploinsufficiency is masked by RNF168-mediated chromatin ubiquitylation. Mol Cell 73: 1267–1281.e7. 10.1016/j.molcel.2018.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.