In this study, Hurd et al. characterize MYC lysine acetylation in human cancer cells and describe the functional significance of individual acetylated lysine (AcK) residues in regulating MYC stability and function. Specific MYC AcKs were found to be important for anchorage-dependent growth, apoptosis, cellular metabolism, and the induction of distinct transcriptional programs, all of which are deregulated in MYC overexpression transformed cells, thereby illustrating a novel avenue for phenotype-specific therapeutic targeting in cancer.
Keywords: MYC oncoprotein, lysine acetylation, malignant transformation
Abstract
The MYC oncogenic transcription factor is acetylated by the p300 and GCN5 histone acetyltransferases. The significance of MYC acetylation and the functions of specific acetylated lysine (AcK) residues have remained unclear. Here, we show that the major p300-acetylated K148(149) and K157(158) sites in human (or mouse) MYC and the main GCN5-acetylated K323 residue are reversibly acetylated in various malignant and nonmalignant cells. Oncogenic overexpression of MYC enhances its acetylation and alters the regulation of site-specific acetylation by proteasome and deacetylase inhibitors. Acetylation of MYC at different K residues differentially affects its stability in a cell type-dependent manner. Lysine-to-arginine substitutions indicate that although none of the AcK residues is required for MYC stimulation of adherent cell proliferation, individual AcK sites have gene-specific functions controlling select MYC-regulated processes in cell adhesion, contact inhibition, apoptosis, and/or metabolism and are required for the malignant cell transformation activity of MYC. Each AcK site is required for anchorage-independent growth of MYC-overexpressing cells in vitro, and both the AcK148(149) and AcK157(158) residues are also important for the tumorigenic activity of MYC transformed cells in vivo. The MYC AcK site-specific signaling pathways identified may offer new avenues for selective therapeutic targeting of MYC oncogenic activities.
The MYC proto-oncogene (also known as c-MYC) is amplified or activated in numerous human cancers, often leading to overexpression of the MYC oncoprotein, which correlates with increased tumor aggressiveness and poor prognosis (Vita and Henriksson 2006; Dang 2012; Schaub et al. 2018). MYC overexpression drives malignant transformation of various cell types in vitro and tumor initiation and maintenance in mouse models (Dang 2012; Gabay et al. 2014). In normal cells and tissues, MYC is tightly regulated by various upstream growth factor and mitogenic signaling pathways. MYC is essential for early development and for self-renewal and pluripotency of embryonic stem cells and plays a critical role in adult stem cell compartments of various regenerating tissues (Takahashi and Yamanaka 2006; Laurenti et al. 2009; Chappell and Dalton 2013). Indeed, MYC regulates many cellular processes in a context-dependent manner, including cell division, growth, apoptosis, metabolism, adhesion, and differentiation by regulating a large number of genes in mammalian cells (Grandori et al. 2000; Dang 2012; Tansey 2014).
MYC is a member of the MYC/MAX/MAD network of basic-helix–loop–helix/leucine zipper (bHLHZ) transcription regulators. MYC binds E-box (CACGTG or CANNTG) and more degenerate DNA elements, including non-E-box sequences (e.g., AACGTT) as an obligatory heterodimer with the bHLHZ partner MAX; the MYC:MAX heterodimer can also be recruited indirectly to promoters via interactions with other chromatin/DNA-binding proteins (Blackwood and Eisenman 1991; Amati et al. 1992, 1993; Grandori et al. 2000; Sabò and Amati 2014; Allevato et al. 2017; Pellanda et al. 2021). MYC (but not MAX) has a transcription regulatory domain at its N terminus that is unstructured and harbors the phylogenetically conserved MYC boxes (MBs) I–IV (MBI–IV). These MB amino acid sequences are important for MYC interaction with many cofactors, including histone acetyltransferases (HATs) and histone deacetylases (HDACs), as well as components of the general transcription machinery and transcription elongation factors, among others (Kalkat et al. 2018). Through these interactions, MYC is thought to regulate (activate or repress) transcription of specific genes or stimulate global transcription by all three RNA polymerases (I–III), leading to “amplification” of the transcriptional output of all active genes in specific cell types and contexts (Dang 2012; Lin et al. 2012; Nie et al. 2012; Tansey 2014; Walz et al. 2014; de Pretis et al. 2017; Lourenco et al. 2021).
The MYC protein is highly unstable in normal cells (with a half-life of ∼20 min) and is rapidly degraded by the ubiquitin-dependent proteasome system. MYC protein stability is regulated in response to growth signaling pathways by a variety of post-translational modifications (PTMs), including phosphorylation, O-linked glycosylation, SUMOylation, ubiquitination, and acetylation. Some of the PTMs of MYC have been shown to interplay to regulate not only MYC stability but also its subnuclear localization and/or transcriptional activity (Chou et al. 1995; Kim et al. 2003; von der Lehr et al. 2003; Hann 2006; Vervoorts et al. 2006; Farrell and Sears 2014; Sabò et al. 2014; González-Prieto et al. 2015; Jaenicke et al. 2016; Su et al. 2018; Sun et al. 2018).
The functions of MYC acetylation have remained unclear. Intracellular MYC acetylation is induced by its coactivator HATs p300/CBP, GCN5/PCAF, and TIP60 and is maintained in check by continuous deacetylation by endogenous HDACs. Both MYC and MAX proteins are direct targets of acetylation by p300, while GCN5 only acetylates MYC, and TIP60 by itself does not directly acetylate MYC or MAX in vitro (Vervoorts 2003; Patel et al. 2004; Faiola et al. 2005, 2007; Zhang et al. 2014). Acetylation of the MYC:MAX complex requires the interaction of p300 and GCN5 with the N-terminal transcription regulatory domain of MYC (Faiola et al. 2005; Zhang et al. 2014). Although several MYC lysine residues acetylated by p300 and GCN5 have been identified, the predominant sites of acetylation by each HAT remain to be defined (Vervoorts et al. 2003, 2006; Patel et al. 2004; Faiola et al. 2005, 2007; Zhang et al. 2005).
The MYC protein is stabilized by interaction with its coactivator HATs p300/CBP, GCN5, and TIP60 via both acetylation-dependent and acetylation-independent mechanisms (Vervoorts 2003; Patel et al. 2004; Faiola et al. 2005). Intriguingly, p300-mediated acetylation of MYC has also been linked to its increased proteasomal degradation (Faiola et al. 2005; Lynch et al. 2013). Similarly, several different HDACs have been shown to deacetylate MYC and either stabilize or destabilize MYC (Marshall et al. 2011; Nascimento et al. 2011; Menssen et al. 2012; Garcia-Sanz et al. 2014). The reason for these discordant effects of acetylation on MYC stability is unclear.
Whether acetylation of MYC regulates its intrinsic transcriptional and biological functions is still largely unclear. Clinical HDAC inhibitors increased MYC acetylation at K323 and activated the TNFSF10 (TRAIL) gene and resulted in induction of apoptosis in AML cancer cells (Nebbioso et al. 2017). Intriguingly, a mutant of MYC having lysine (K)-to-arginine (R) substitutions at all but one K residue (i.e., K52) remained a target for efficient ubiquitin-dependent proteasomal degradation and retained overall functionality independently of acetylation, including the ability to activate transcription and stimulate mammary epithelial cell proliferation; this suggested that all K residues of MYC act redundantly in mediating its instability and function (Jaenicke et al. 2016). However, other activities of MYC, including cell transformation, were not addressed. Notably, K-to-R substitutions in mouse MYC at five acetylated K residues (K144, K149, K158, K317, and K323) or just two (K323 and K417) impaired cooperativity of HTLV-1 p30II with MYC in transformation/focus formation of rat myc−/− fibroblasts (Romeo et al. 2015). However, it has remained largely unclear whether acetylation of any of these K residues regulates the intrinsic biological or oncogenic activities of MYC.
In this study, we have defined the major MYC AcK sites preferentially acetylated by p300 and GCN5 and systematically analyzed their acetylation and roles in the regulation of MYC stability, MYC-induced apoptosis, metabolic remodeling, oncogenic transformation, and gene-specific regulation in different cell types. Our results suggest exquisitely specific mechanisms of MYC action and the possibility of therapeutic targeting of only select MYC functions important for its oncogenic activity.
Results
MYC is acetylated in diverse cancer cell lines at specific lysine residues differentially targeted by p300 and GCN5
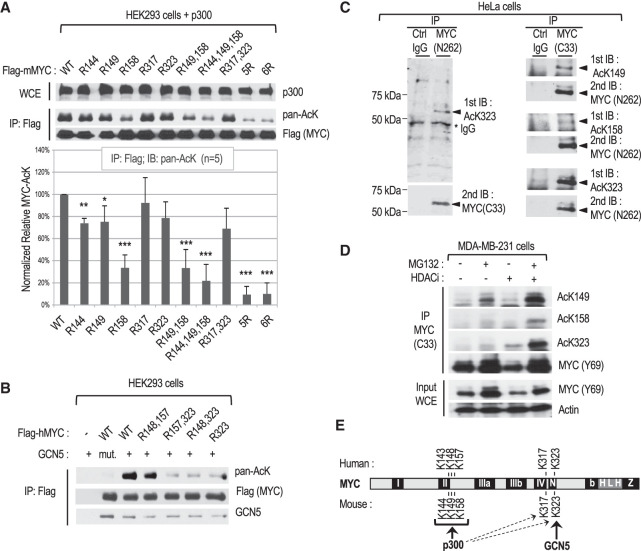
We previously found that p300 can directly acetylate mouse MYC in vitro and in cells at seven lysine residues (K144, K149, K158, K275, K317, K323 and K371) and that five of these residues (K144, K149, K158, K317, and K323) may encompass the major acetylation sites (Faiola et al. 2005; Zhang et al. 2005); note that we used the coordinates of the AUG-initiated 439-amino-acid MYC proteins (UniProt P01106-1 and P01108-2 for human and mouse MYC, respectively). Here we further investigated the preferred p300 target sites on MYC in HEK293 and HeLa cells transfected with either mouse MYC wild type (WT) or K-to-R substitution mutants (referred to here as R mutants). MYC immunoprecipitation (IP) and Western blot analyses with a pan-acetyl-lysine antibody indicated that p300 preferentially acetylates MYC at K144, K149, and K158 (K158 being the preferred site in this cell/assay system), while the K317 and K323 residues are weaker substrates (Fig. 1A, Supplemental Fig. S1A). Since the K-to-R substitutions did not influence MYC interaction with either p300 or MAX (Supplemental Fig. S1B), these results together with our previous in vitro results identify K144, K149, and K158 as the major MYC sites directly acetylated by p300.
Figure 1.
Characterization of site-specific MYC acetylation in different cell lines. (A) Mapping of the main MYC lysine (K) residues acetylated by p300 in HEK293 cells transfected with p300 and FLAG-tagged mouse MYC or the indicated K-to-R substitution mutants. (Top) Representative Western blot with the indicated antibodies of whole-cell extracts (WCEs) and immunoprecipitated FLAG-mMYC. (Bottom) Chart showing the quantitation of acetylated MYC signals obtained with the pan-acetyl-lysine antibody from five independent experiments. (B) Mapping of the main human MYC lysine residue acetylated by GCN5 in transfected cells as above. (mut.) Acetylation-defective GCN5 HAT mutant. (C) Endogenous MYC in HeLa cervical cancer cells was immunoprecipitated with the indicated antibodies and analyzed by Western blot by immunoblotting (IB) with MYC AcK site-specific antibodies, followed by stripping and reprobing with a MYC antibody. (D) Analysis by IP and Western blot of endogenous site-specific acetylated MYC in MDA-MB-231 mammary cancer cells with or without MG132 and/or HDAC inhibitor (HDACi) treatment for 2 h before lysis. (E) Summary of MYC K site(s) preferentially acetylated by p300 and GCN5.
Human and mouse MYC proteins were shown to be directly acetylated by GCN5 (and the purified STAGA complex) in vitro, and two lysine residues (K323 and K417) were found acetylated in mouse MYC (Patel et al. 2004; Faiola et al. 2005). In human MYC, which lacks K417, the R323 mutation inhibited most GCN5-mediated acetylation and did not affect MYC interaction with GCN5 (Fig. 1B; Supplemental Fig. S1B). These results show that GCN5 does not target the K residues acetylated by p300 but specifically acetylates MYC at the K323 site. Cell fractionation studies further showed that acetylated MYC and the MYC–GCN5 complex localize in the nucleus of HeLa cells (Supplemental Fig. S1C).
To investigate endogenous MYC acetylation in cancer cells, we performed liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) analyses of native MYC:MAX complexes immunoprecipitated from HeLa cells. LC-MS/MS identified an acetylated human MYC peptide carrying an acetyl group at lysine K148, which is the equivalent of K149 in mouse MYC (Supplemental Fig. S1D). We further investigated MYC acetylation using antibodies specific for human MYC acetyl-lysine (AcK) residues AcK148(149), AcK157(158), and AcK323 (homologous mouse residues are in parentheses) (Supplemental Fig. S1E). These AcK antibodies recognized p300-acetylated MYC WT but not the corresponding site-specific R mutants (R149, R158, or R323) by Western blot (Supplemental Fig. S1F) and preferentially immunoprecipitated acetylated MYC WT relative to the corresponding R mutant proteins expressed in HEK293 cells (Supplemental Fig. S1G). Western blot analyses with these site-specific AcK antibodies confirmed that p300 can acetylate MYC at all three residues, while GCN5 selectively acetylates K323 (Supplemental Fig. S1H). All three MYC AcK antibodies recognized endogenous MYC in HeLa cells by Western blot (Fig. 1C) and immunoprecipitated endogenous MYC (Supplemental Fig. S1I). Endogenous MYC was also acetylated at all three residues in the human breast cancer cell line MDA-MB-231, and the levels of MYC acetylation at K148(149) increased most significantly (e.g., compared with AcK323) after short (2-h) proteasome inhibition with MG132 (Fig. 1D). This suggested that MYC acetylated at different residues may have different stabilities (see also below). Similarly, MYC acetylation at these three K sites was observed in the human P493-6 lymphoblastoid cell line, a model of Burkitt's lymphoma with doxycycline-regulated MYC expression (Supplemental Fig. S1J).
Collectively, these results establish that MYC acetylation at K148(149), K157(158), and K323 (mouse MYC coordinates in parentheses) is conserved in diverse human cancer cell lines and that these residues are differentially targeted by p300 and GCN5: p300 targets all three sites but preferentially acetylates K148(149) and K157(158), while GCN5 selectively acetylates K323 (Fig. 1E; Supplemental Fig. S1K). Here, our analyses focus on the p300-specific sites K148(149) and K157(158) and the preferred GCN5 target site (K323).
Acetylation of MYC is enhanced by overexpression and regulates MYC turnover in an AcK site-specific manner but is dispensable for adherent cell proliferation
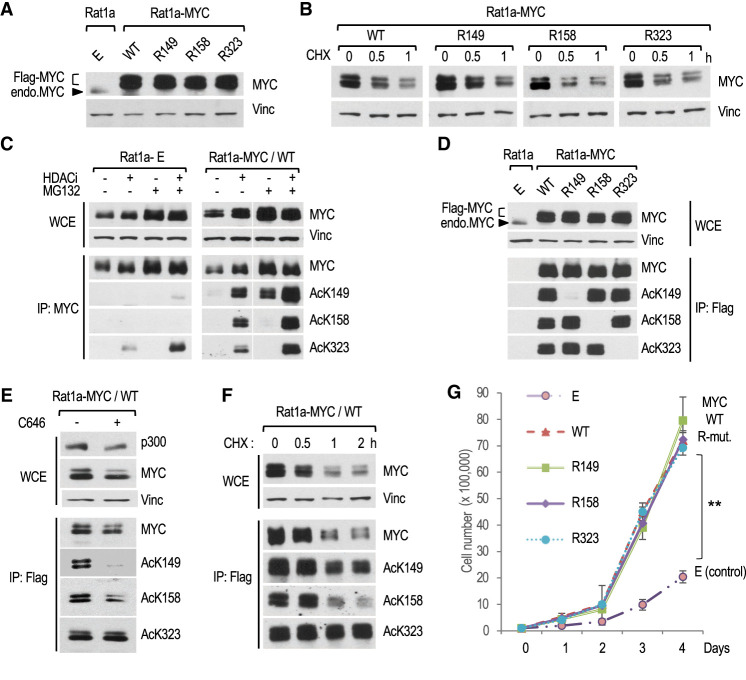
To investigate the functions of MYC acetylation, we overexpressed mouse FLAG-tagged MYC WT and the mutants R149, R158, and R323 in Rat1a fibroblasts. Rat1a cells are immortal but nontumorigenic and have been extensively used to characterize the normal and oncogenic functions of MYC (Stone et al. 1987). Cells were stably transfected with expression vectors for either MYC WT or the different R mutants (or the empty [E] vector control) and pools of stable transfectants (i.e., Rat1a-E [empty vector] and Rat1a-MYC WT, R149, R158, and R323) (Supplemental Fig. S2A) were analyzed at equivalent passage numbers. The ectopic FLAG-tagged MYC WT and R mutants were overexpressed to similar levels (Fig. 2A), accumulated in the nucleus (Supplemental Fig. S2B), and had a similar short (∼30-min) half-life (Fig. 2B). Thus, substitution of K149, K158, or K323 by arginine did not affect the subcellular localization or rapid turnover of the bulk of overexpressed MYC.
Figure 2.
Characterization of MYC acetylation, stability, and stimulation of cell proliferation in MYC transformed Rat1a fibroblasts. (A) Western blot analysis of total MYC in WCEs of control (E) and the indicated MYC-overexpressing Rat1a cell lines. (Vinc) Vinculin. Ectopic FLAG-MYC and endogenous MYC are indicated. (B) Rat1a-MYC cell lines were treated with cycloheximide (CHX) for the indicated times, and WCEs were analyzed by Western blot. (C) Rat1a-E (control) and RAT1a-MYC/WT cell lines were treated for 2 h with the indicated inhibitors before WCE and IP analyses by Western blot with MYC and MYC AcK site-specific antibodies. (D) Western blot analyses of WCEs and anti-FLAG immunoprecipitated material from the indicated cell lines. (E) C646 inhibition of MYC acetylation analyzed by Western blot, as above. (F) Western blot analyses of the stability of total and site-specific acetylated MYC in Rat1a-MYC/WT cells treated with HDAC inhibitors for 2 h, followed by CHX (in the presence of HDAC inhibitors) for the indicated times. (G) Proliferation of Rat1a-E and Rat1a-MYC (WT or R mutants) cell lines on normal adherent cell culture plates.
Western blot analyses using the site-specific MYC AcK antibodies indicated that endogenous MYC in Rat1a-E control cells was acetylated at K323 and weakly at K149 but not at K158 (Fig. 2C, left). In contrast, overexpressed MYC in Rat1a-MYC WT cells was acetylated at all three residues (Fig. 2C, right). As expected, acetylation of these lysine residues was not detected in the corresponding R mutant cell lines (Fig. 2D). In addition, mutation of a specific acetylated K residue to R did not alter acetylation at the other two K sites (Fig. 2D). These results indicate that the overall and relative levels of acetylation at these three specific sites are differentially enhanced by MYC overexpression. We also noted that the levels of MYC acetylated at each site, but most prominently at K149, increased upon proteasomal inhibition (Fig. 2C, right, +MG132). Moreover, the p300-specific HAT inhibitor C646 (Bowers et al. 2010) inhibited acetylation of MYC selectively at K149 and K158 but not at K323 (Fig. 2E). Hence, these results suggest that MYC overexpression stimulates p300-mediated acetylation at K149 and K158, while the K323 residue could be acetylated by a different endogenous HAT(s), possibly GCN5 (and/or PCAF) as in HEK293 cells (Fig. 1).
Notably, the levels of endogenous rat MYC protein (and Myc mRNA) (see below) were suppressed by overexpressed MYC WT and each of the R mutant proteins (Fig. 2A,D, arrowhead). This down-regulation of the endogenous Myc gene, which may be related to the transcriptional autoinhibitory activity of MYC (Facchini et al. 1997), is therefore independent of MYC acetylation at these sites.
Although the K-to-R substitutions did not impact overall MYC stability, we tested more directly the stability of MYC acetylated at K149, K158, or K323 by using the AcK site-specific antibodies. Interestingly, MYC acetylated at K149 or K158 retained a short half-life like the bulk of total MYC (∼30 min), while MYC acetylated at K323 was more stable, with a half-life of >2 h (Fig. 2F). We note that a fraction (∼10%) of total MYC had a half-life >60 min (Fig. 2F, CHX 1- and 2-h time points). Thus, the fraction of MYC acetylated at K149 and K158 (p300 sites) remains relatively unstable, while MYC acetylated at K323 (GCN5 site) has an increased stability. These results are reminiscent of previous findings indicating the existence of distinct cellular pools of stable and unstable MYC (Tworkowski et al. 2002).
We further investigated the functions of the different AcK residues for MYC stimulation of cell proliferation during the logarithmic growth phase. Cells transformed with MYC WT or the three R mutants proliferated faster than the control Rat1a-E cells. However, there was no difference in the proliferation rates between the MYC WT and the different R mutant cell lines (Fig. 2G). No significant cell death was observed under these conditions (data not shown). These results demonstrate that MYC acetylation at K149, K158, or K323 is dispensable for stimulation of Rat1a cell division/proliferation under typical adherent cell culture conditions.
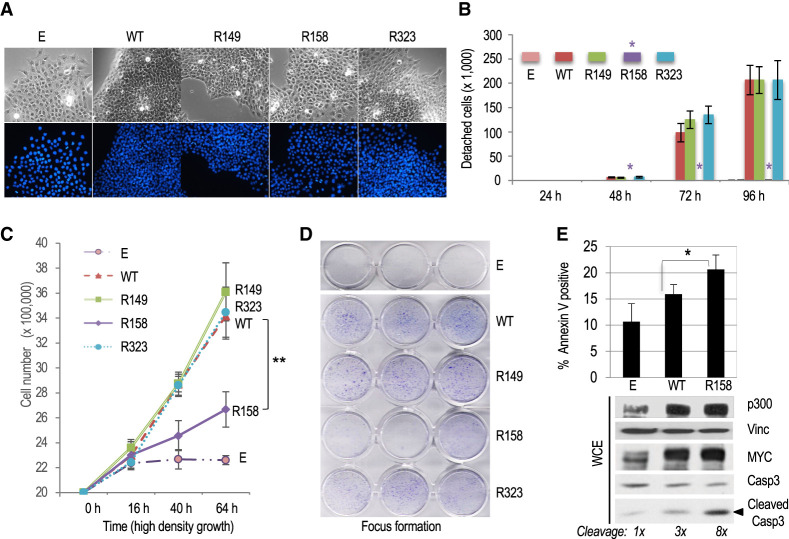
Substitution of MYC-acetylated lysine residue K158 to arginine (R158) increases cell adhesion, contact inhibition, and apoptosis of MYC transformed Rat1a cells
Cells overexpressing MYC WT had a characteristic transformed appearance by phase contrast microscopy. Compared with control Rat1a-E cells, MYC transformed cells were smaller and more refringent and grew in tight clusters. This was also evident for cells transformed with the MYC R149 and R323 mutants. In contrast, cells transformed with MYC R158 were larger and grew in a more spread manner, like control Rat1a-E cells (Fig. 3A; Supplemental Fig. S2B). Another characteristic of the R158 cells was their increased adherence to the plates compared with cells transformed with MYC WT or the R149 and R323 mutants. The adherence of R158 cells was very similar to untransformed Rat1a-E control cells (Fig. 3B; Supplemental Fig. S3A,B). Interestingly, the proliferation of R158 cells was partially impaired upon reaching confluence, whereas cells transformed with MYC WT or the R149 and R323 mutants continued to proliferate. As expected, nontransformed Rat1a-E cells stopped proliferating after reaching confluence (Fig. 3C). In accord with the above results, cells transformed with MYC R158 formed fewer foci in confluent cultures compared with cells transformed with MYC WT or the R149 and R323 mutants (Fig. 3D). These results suggest that the ability of MYC transformed Rat1a cells to bypass cell contact-dependent growth inhibition in dense cultures is dependent specifically (or predominantly) on the K158 residue of MYC.
Figure 3.
Cell adhesion, contact inhibition and apoptosis of MYC transformed Rat1a fibroblasts are regulated by the acetylated K158 residue of MYC. (A) Phase contrast (top) and DAPI fluorescence microscopy (bottom) of Rat1a-E and RAT1a-MYC WT and R mutant cell lines. (B) Detachment assay of logarithmically growing cell lines at different times after seeding on regular cell culture plates. The asterisk indicates the lack of significant detachment of Rat1a-MYC/R158 mutant cells (Rat1a-E control cells do not detach). (C) High-density (postconfluency) proliferation assay. Cell lines were confluent at 16 h after plating on regular cell culture plates. (D) Postconfluency growth and formation of dense foci visualized by crystal violet staining. (E) The indicated Rat1a-E or Rat1a-MYC (WT and R158) cell lines were serum-starved, and the fraction of apoptotic cells was determined by annexin V staining or by Western blot analysis of cleaved caspase-3. Cleaved caspase-3 signals were normalized to vinculin and are relative to control (E) cells.
We further analyzed the proapoptotic activity of MYC WT and R mutants in cells deprived of serum growth factors. Compared with cells overexpressing MYC WT or the R149 and R323 mutants, the R158 cells had a reduced viability after serum starvation (Supplemental Fig. S3C) that correlated with increased caspase-3 cleavage and apoptotic cell death (Fig. 3E; Supplemental Fig. S3D). We note that p300 protein levels in serum-starved cells were markedly higher in cells overexpressing MYC relative to control cells, suggesting that MYC overexpression sustains p300-dependent pathways in quiescent cells. The above R158 phenotypes were observed in two different stably transfected Rat1a cell line pools and in cells transiently transduced with retroviral expression vectors (Fig. 3E; Supplemental Fig. S3D). Altogether, these results suggest that the p300-acetylated K158 residue of MYC inhibits cell adhesion and dampens contact inhibition and apoptosis of MYC-overexpressing Rat1a cells.
MYC-acetylated lysine residues are important for malignant transformation of both fibroblastic and epithelial cell types
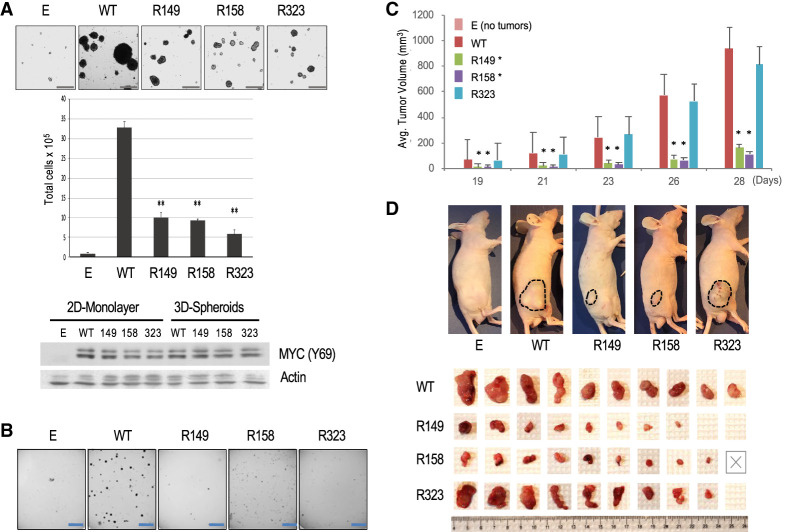
To analyze the malignant phenotype of Rat1a fibroblasts overexpressing MYC WT or the different MYC R mutants, we first tested their anchorage-independent growth as suspension spheroids (Fig. 4A) and ability to form colonies in soft agar (Fig. 4B). Spontaneous spheroid formation is a characteristic of tumor-initiating “stem cell-like” cancer cells and has been associated with MYC overexpression (Ishiguro et al. 2017; Poli et al. 2018), while colony formation in semisolid soft agar is a well-established characteristic of malignant cells (Shin et al. 1975). As expected, the control Rat1a-E cells did not significantly grow under nonadherent conditions, while MYC WT transformed cells formed many suspension spheroids (Fig. 4A) and soft agar colonies (Fig. 4B). Notably, compared with MYC WT transformed cells, cells overexpressing each of the MYC R mutants had a significant proliferation defect in both the tumor spheroid growth assays (Fig. 4A, Supplemental Fig.S4A,B) and the soft agar colony formation assay (Fig. 4B). This was observed with two independent pools of stably transfected cells and with cells transduced with retroviral vectors that did not undergo antibiotic selection (Supplemental Fig. S4A). Expression levels of MYC WT and R mutant proteins were similar in cells grown as suspension tumor spheroids (Fig. 4A; Supplemental Fig. S4A) and there was no significant cell death observed, suggesting that spheroid growth rather than survival was affected (data not shown). Notably, a similar anchorage-independent growth defect was observed when Rat1a cells were transduced with MYC mutants with lysine-to-glutamine (Q) substitutions at the K149 or K158 sites (i.e., Q149 and Q158) (Supplemental Fig. S4C), indicating that the simple removal of the positive charge at these K sites is not sufficient to reconstitute function.
Figure 4.
Analysis of malignant transformation of Rat1a fibroblasts overexpressing MYC WT or individual K-to-R mutants. (A) Spontaneous spheroid formation assay with the indicated Rat1a cell lines. (Top) Microscopic images of spheroids. Scale bar, 250 μm. (Middle) Total cell counts after spheroid trypsinization. (Bottom) Western blot with equal amounts of WCE protein from cell lines grown as two-dimensional (2D) monolayers or 3D spheroids (with MYC Y69 and β-actin antibodies). (B) Representative images of soft agar colony formation assays with the indicated Rat1a cell lines. Scale bar, 5 mm. (C,D) Xenograft assays in nude mice injected with the indicated Rat1a cell lines. (C) Volume of tumors at different times after injection. (D) Images of tumors in mice before and after surgical excision (Rat1a-E cells did not generate tumors). The “X” indicates a mouse that died early.
We further tested the in vivo oncogenic activity of the MYC transformed Rat1a cells in subcutaneous xenograft assays in nude mice. Control Rat1a-E cells did not form tumors in any of the 10 injected mice, while large tumors rapidly developed in all mice injected with cells transformed with MYC WT. Notably, cells transformed with the MYC mutant R149 or R158 generated slow-growing tumors that remained significantly smaller than MYC WT tumors during the 4-wk period (Fig. 4C,D). The R323 mutant cells formed tumors in nine out of 10 mice that were similar in size to MYC WT tumors; however, the R323 tumors were often more vascularized, less compact, and more tightly attached to the underlying muscle tissue, making them more difficult to dissect (Fig. 4C,D; data not shown). These results demonstrate an important role of all three MYC-acetylated residues (AcK149, AcK158, and AcK323) in supporting anchorage-independent growth and indicate that the AcK149 and AcK158 residues (but not the AcK323 site) are also important for the in vivo tumorigenic activity of MYC transformed Rat1a fibroblasts.
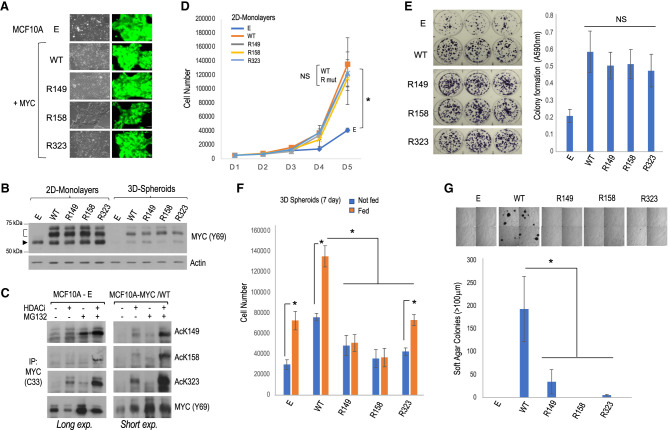
To extend these analyses to a different cell type, we tested the importance of the AcK residues in MYC transformation of human MCF10A mammary epithelial cells, which are immortal but nonmalignant. Retroviral MYC-IRES-GFP bicistronic vectors for MYC WT or R mutants (or empty vector [E]) were transduced into MCF10A cells, and GFP-positive polyclonal cell lines were obtained that overexpressed MYC WT or the R mutants to similar extents (Fig. 5A,B). Note that MCF10A cells have a focal amplification of the MYC gene locus (Kadota et al. 2010), and endogenous MYC expression is not inhibited by overexpressed ectopic MYC in these cells (Fig. 5B), as shown previously (Lourenco et al. 2019). Both endogenous MYC and overexpressed MYC were acetylated at all three K sites, and overexpression of MYC enhanced its acetylation and altered the response to HDACs and proteasome inhibitors at the AcK149 site (Fig. 5C). Furthermore, MYC acetylation at the different K sites was regulated differently by different classes of HDAC inhibitors (Supplemental Fig. S5A). The MYC AcK149 and AcK158 marks were most sensitive to nicotinamide (NAM), which inhibits NAD+-dependent class III HDACs (sirtuins), while the AcK323 mark was only sensitive to trichostatin A (TSA), which inhibits zinc-dependent class I, II, and IV HDACs. Note, however, that treatment of cells with both NAM and TSA inhibitors further increased the levels of acetylation at all sites, suggesting both direct and indirect effects of these HDAC inhibitors (Supplemental Fig. S5A).
Figure 5.
Analysis of malignant transformation of human MCF10A mammary epithelial cells overexpressing MYC WT or individual K-to-R mutants. (A) Phase contrast and GFP fluorescent images of the cell lines. (B) Western blot analysis of MYC expression levels for cell lines grown as monolayers and as spheroids. The arrowhead indicates endogenous MYC, and the bracket indicates ectopic FLAG-tagged MYC. (C) Analysis of MYC acetylation by IP and Western blot with the indicated antibodies in control (MCF10A-E) and MYC WT transformed (MCF10A-MYC/WT) cell lines; long and short exposures, respectively, are shown. (D) Proliferation of the different cell lines over a 5-d period in regular adherent culture plates. (E) Colony formation assays on regular adherent culture plates and quantitation of crystal violet staining. (NS) No significant difference between MYC WT and the R mutant cell lines. (F) Spheroid growth assays. Total number of cells at the end of the 7-d growth period without (blue bars) or with (orange bars) fresh growth medium feeding is indicated. (G) Soft agar colony formation assays. (Top) Representative microscopic images of four quadrants of each plate are shown. Scale bar, 250 μm. (Bottom) Plot showing the number of colonies ≥100 μm.
Consistent with our observation of p300 preferentially targeting K149 and K158 in HEK293 and Rat1a cells, the p300-specific inhibitor C646 inhibited MYC acetylation at K149 and K158 but not at K323 in transformed MCF10A cells (Supplemental Fig. S5B). Like in Rat1a cells, MCF10A cells overexpressing MYC WT or each of the R mutants had an increased proliferation rate compared with control MCF10A-E cells, and the K-to-R substitutions did not affect MYC stimulation of cell proliferation (Fig. 5D) or colony formation (Fig. 5E) under adherent cell culture conditions. Notably, as seen in Rat1a cells, MCF10A cells overexpressing the MYC R158 mutant (but not MYC WT or the other R mutants) were unable to bypass contact inhibition; moreover, the ability of MYC WT transformed cells to grow beyond confluency was inhibited by the p300-specific inhibitor C646 (Supplemental Fig. S5C). Thus, p300 HAT activity and the MYC K158 site are required to overcome contact inhibition.
Importantly, MCF10A cells transformed with each of the MYC R mutants had a reduced ability to proliferate under anchorage-independent conditions as 3D suspension spheroids (Fig. 5F) and in soft agar (Fig. 5G). We note that culture medium replenishment during the 7-d spheroid assay stimulated growth of MCF10A-E control cells and cells transformed with MYC WT or R323 but not cells transformed with the MYC R149 or R158 mutants (Fig. 5F; Supplemental Fig. S5D); this suggests a more essential role for the MYC AcK149 and AcK158 sites, compared with AcK323, in sustaining tumor spheroid growth. Altogether, these results establish essential contributions of the MYC AcK149 and AcK158 residues in MYC-driven malignant transformation of fibroblastic (Rat1a) and epithelial (MCF10A) cell types and suggest that the transforming function of the MYC AcK323 site is more context-dependent.
Different MYC-acetylated lysine residues play distinct cell type-specific functions in the regulation of MYC stability and remodeling of cellular metabolism
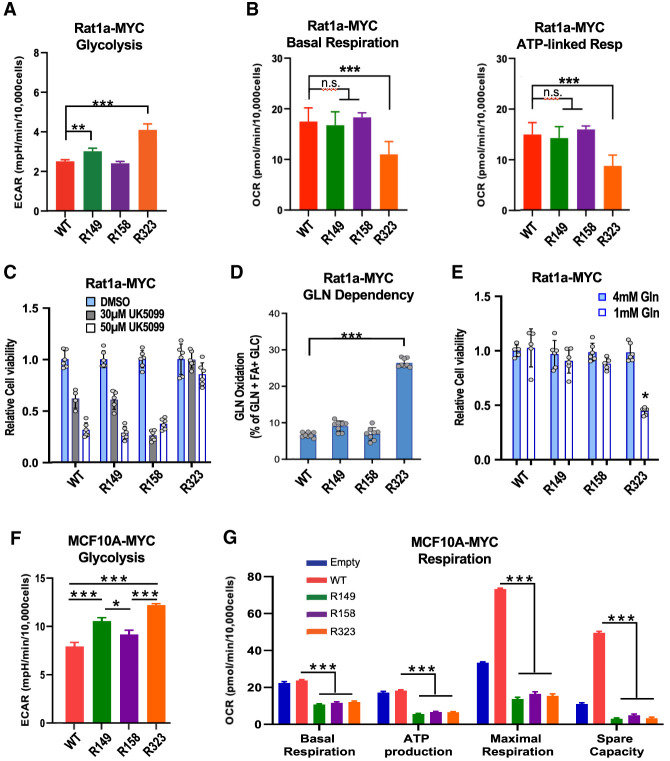
Given the importance of the MYC AcK sites for cell transformation and evidence linking MYC activity to its lysine ubiquitination-dependent proteasomal degradation (see above), we tested whether the increased stability of oncogenic MYC acetylated at K323 observed in Rat1a cells (Fig. 2F) was conserved in MYC transformed MCF10A cells. Consistent with the results in Rat1a cells, MYC WT and R mutants had the same overall instability in MCF10A cells (Supplemental Fig. S5E). Analysis of each acetylated form of MYC revealed that MYC acetylated at either K149, K158, or K323 had only a moderately increased half-life (37, 30, and 29 min, respectively) compared with the ∼24-min half-life of the bulk of total MYC (Supplemental Fig. S5F). This was in stark contrast to the higher stability (half-life >2 h) of MYC acetylated selectively at K323 in Rat1a cells (Fig. 2F). Thus, site-specific acetylation impacts MYC turnover differently in Rat1a and MCF10A cells and does not correlate with the requirement of each AcK site in MYC-dependent transformation of both cell types.
We further analyzed the influence of the MYC AcK sites in MYC remodeling of metabolism. In MYC-overexpressing Rat1a cells, we observed an increased rate of glycolytic extracellular medium acidification by cells overexpressing the MYC R323 mutant and, to a lesser extent, the R149 mutant (Fig. 6A; Supplemental Fig. S6A–C). For the R323 mutant cells, this coincided with a decreased basal and ATP-linked oxygen consumption rate (OCR), with no effect on maximal respiratory capacity (Fig. 6B; Supplemental Fig. S6D). Interestingly, MYC R323 transformed Rat1a cells had reduced expression of the Pdha1 gene encoding the E1α catalytic subunit of the pyruvate dehydrogenase complex (see below) and were resistant to cell death mediated by UK5099, an inhibitor of the mitochondrial pyruvate carrier (Fig. 6C). Notably, MYC R323-overexpressing cells had an increased glutamine oxidation dependency and capacity (Fig. 6D; data not shown). Accordingly, cells overexpressing the MYC R323 mutant had a reduced viability (as adherent cultures) in growth media containing low glutamine concentrations (Fig. 6E). The other R mutants had little (R149) or no (R158) effect on glycolysis, and neither R149 nor R158 affected OCR/respiration or glutamine oxidation. Thus, compared with MYC WT, the R323 mutant Rat1a cells had increased glycolysis and shifted their mitochondrial metabolic dependency from glucose/pyruvate to glutamine oxidation.
Figure 6.
Metabolic analyses of Rat1a and MCF10A cell lines overexpressing MYC WT or individual K-to-R mutants. (A,B) Rat1a cell line ECAR and OCR Seahorse analyses, respectively. (n.s.) Nonsignificant statistical difference (n = 5). (C) Cell viability measured by acid phosphatase assay after treatment with the mitochondrial pyruvate carrier inhibitor UK5099 for 72 h (n = 6). (D) Fraction of cellular OCR derived from glutamine oxidation (n = 8). (E) Viability of the cell lines (normalized to MYC WT cells) cultured for 72 h in medium with the indicated glutamine concentrations (n = 6). (F,G) MCF10A cell line ECAR and OCR Seahorse analyses, respectively, as above. The data (n = 5–8) were plotted as the means ± SD (A–E) or means ± SEM (F,G).
In MCF10A cells, glycolysis was significantly stimulated by the MYC R323 and R149 mutants (Fig. 6F; data not shown), similar to Rat1a cells (Fig. 6A). In stark contrast to Rat1a cells, however, all three MYC R mutant-overexpressing MCF10A cell lines had a dramatically reduced OCR/respiration (basal, maximal, and ATP-linked OCR) relative to MYC WT-overexpressing cells (Fig. 6G; Supplemental Fig. S6E). Altogether, these results indicate that the MYC AcK-to-R substitutions have different cell type-specific effects on MYC stability and impact MYC-dependent remodeling of cellular metabolism with similar effects on glycolysis but notable differences in OCR in Rat1a and MCF10A cells. These observations suggest that the similar transforming functions (anchorage-independent growth) of these MYC AcK residues in Rat1a and MCF10A cells could involve other MYC/AcK-dependent mechanisms conserved in these two cell types.
MYC-acetylated lysine residues have gene-selective functions and regulate conserved biological processes for cell adhesion-independent growth and survival
To identify the genetic programs and biological pathways that are dependent on the MYC AcK residues and potentially conserved in MYC-overexpressing Rat1a and MCF10A cells, we performed RNA sequencing (RNA-seq) analyses. Overexpression of MYC WT did not lead to a general amplification of total cellular RNA but altered expression of specific genes for known MYC-regulated biological processes, as described below.
In Rat1a cells, overexpression of MYC WT altered the expression of 2145 genes by at least 1.5-fold (800 genes up-regulated and 1345 genes down-regulated; FDR ≤ 0.05), while 904 genes were deregulated twofold or more (Supplemental Table S1), of which ∼42% are known direct targets of MYC (ENCODE; data not shown). Gene set enrichment analysis (GSEA) indicated that the top MYC-up-regulated pathways included MYC targets, E2F targets, G2/M checkpoint, spliceosome, and mRNA processing, while the top MYC-down-regulated pathways included epithelia–mesenchymal transition (EMT), myogenesis, TGFβ signaling, inflammatory response, p53 pathway, TNFα signaling via NFkB, extracellular matrix–receptor interaction, focal adhesion, and cell–cell communication (Supplemental Fig. S7). Gene ontology (GO) analyses of the most deregulated genes (twofold or greater) identified the biological processes “negative regulation of cell cycle arrest,” “lysine catabolic process” (MYC up-regulated), and “extracellular matrix organization” (MYC down-regulated), among others (Supplemental Fig. S8).
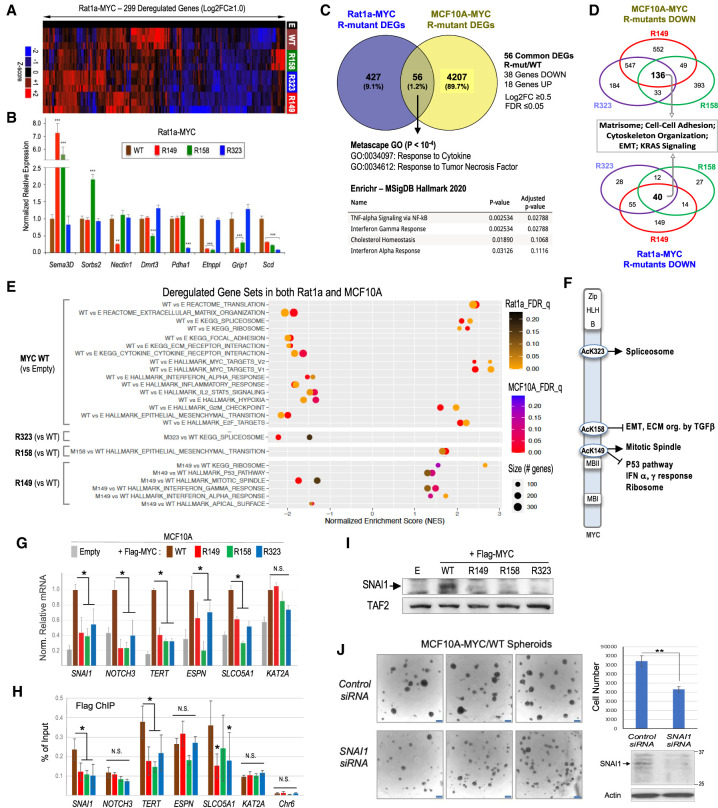
In Rat1a cells overexpressing the MYC R mutants, 299 genes were acutely deregulated (twofold or greater) relative to cells overexpressing MYC WT (Fig. 7A). Interestingly, the different R mutations affected distinct but also common sets of genes (Fig. 7A,B; Supplemental Fig. S9A). The R149 mutation deregulated the largest number of genes (200; 67%), and most genes altered by at least one R mutant were down-regulated (221 genes; 74%), suggesting a generally positive role of the AcK residues in MYC-dependent gene expression. Notably, the MYC R149 and R323 mutants shared the largest number of deregulated genes, and a significant number of genes (42 out of 299; 14%) were deregulated similarly by all three R mutants (Supplemental Fig. S9A).
Figure 7.
MYC AcK-dependent genes and pathways in common in Rat1a and MCF10A cells and the role of AcK residues in MYC binding to select target genes. (A) Heat map with unsupervised hierarchical clustering of the top 299 genes deregulated twofold or more (FDR ≤ 0.07) in MYC R mutant Rat1a cells relative to cells overexpressing MYC WT. (B) RT-qPCR analysis of a select group of Rat1a genes deregulated by specific MYC R mutants. Gene mRNA expression was normalized to PPIA. (C) Venn diagram of deregulated genes (DEGs) in common in Rat1a and MCF10A cells (log2FC ≥ 0.5, FDR ≤ 0.05) affected by at least one MYC R mutant relative to MYC WT cells. Gene ontology (GO) terms (Metascape) and most enriched molecular signatures (Enrichr and MSigDB) for the 56 overlapping DEGs are shown. (D) Venn diagrams of genes down-regulated by 1.5-fold or more (FDR ≤ 0.05) in all three MYC R mutant cell lines (relative to MYC WT cells) and associated top GO pathways. (E) GSEA enrichment scores for gene sets deregulated similarly in Rat1a and MCF10A cell lines. (F) Summary of GSEA results shown in E, illustrating the gene sets/pathways enriched (arrows) or inhibited (blunt end arrows) in MYC WT relative to the AcK-to-R mutant cell lines. (G) RT-qPCR analysis of mRNAs of select MYC-activated genes in the indicated MCF10A cell lines. Expression values were normalized to TOP1 and UBC and are relative to expression in the FLAG-MYC WT cell line (set arbitrarily to 1.0). (H) ChIP analyses of FLAG-MYC WT and R mutants binding to the promoters of MYC-activated genes or to an unrelated locus in chromosome 6 (Chr6). (N.S.) No statistical significance (P > 0.05). N ≥ 3 biological replicates each in triplicates (error bars are SD). (I) Western blot analysis of SNAI1 expression in MCF10A-MYC WT and R mutant cell lines. (J) MCF10A-MYC/WT cells were transfected with control or SNAI1 siRNAs and, after 48 h, analyzed for SNAI1 protein expression by Western blot and suspension spheroid growth by microscopy (scale bar, 250 μm) and total spheroid cell counting (bar graph).
GSEA and GO analyses verified that the R substitutions altered only select biological processes and pathways that were either specific or shared by the R mutants (Supplemental Figs. S9B,C, S10, S11). Some of these altered pathways were consistent with the phenotypes of cells overexpressing specific MYC R mutants. For instance, the R158 mutant cells (which showed an increased apoptotic and cell adhesion phenotype) had increased expression of genes of the epithelial–mesenchymal transition (EMT), including cell adhesion and proapoptotic genes, while the R323 mutant cells (with an increased aerobic glycolysis and lower OCR) had reduced expression of Pdha1 (Fig. 7B) and genes involved in oxidative phosphorylation and respiration–electron transport–ATP synthesis (Supplemental Fig. S9B,C).
In MCF10A cells, overexpression of MYC WT altered the expression of 2726 genes by at least 1.4-fold (1129 genes up-regulated and 1597 genes down-regulated; FDR ≤ 0.05), of which 973 genes were deregulated twofold or more (Supplemental Table S2). MYC up-regulated only 82 genes (≥1.4-fold) in common between MCF10A and Rat1a cells; GO analyses identified ribosome biogenesis and MYC_Activ_Pathway (i.e., validated targets of MYC transcriptional activation) as the top two up-regulated pathways (P ≤ 10−7.5) (Supplemental Fig. S12A). In contrast, MYC down-regulated 225 genes in common between these two cell types, and the top down-regulated GO pathways included extracellular matrix (ECM) organization, regulation of cell adhesion, cell–substrate adhesion, and signaling by receptor tyrosine kinases (P ≤ 10−10) (Supplemental Fig. S12B).
Relative to MCF10A cells overexpressing MYC WT, cells overexpressing the R mutants had a total of 4263 genes deregulated by at least one MYC R mutant (≥1.4-fold; FDR ≤ 0.05), of which 56 genes (38 down-regulated and 18 up-regulated) were also deregulated by at least one R mutant in Rat1a cells (Fig. 7C). GO and pathway enrichment analyses for these common deregulated genes identified response to cytokine, response to tumor necrosis factor (TNF), TNFα signaling via NFkB, interferon α and γ response, and cholesterol homeostasis among the most significantly deregulated pathways (Fig. 7C; Supplemental Fig. S13). Among those 56 common deregulated genes, 34 genes were deregulated by the same R mutant(s) in both cell types, and ChIP-seq data from the ENCODE project indicated that most of them (31; 91%) had promoters bound by MYC in mammary MCF10A and/or MCF7 breast cancer cells (Supplemental Fig. S14A). Transcription factor enrichment analyses using Enrichr and the ChEA_2022 ChIP-seq database identified MYC and additional transcription factors as potential direct regulators of these 34 promoters (P ≤ 10−2), including SOX2, TET1, HAND2, NR3C2, UBTF (UBF1/2), SMAD3, SMAD4, CREB1, STAT4, ZFP281, SPI1, NFE2L2, FOSL1, CTCF, KDM2B, NFKB1, and TP63 (Supplemental Fig. S14B). These transcription factors form a significant MYC protein–protein interaction network (P = 1.56 × 10−11) and may contribute to coregulation of these core MYC/AcK site-dependent genes (Supplemental Fig. S14C), including their response to cytokines (e.g., SMAD3/SMAD4, STAT4, and NFKB1 transcription factors).
Given that each AcK-to-R mutation impaired the ability of MYC to induce anchorage-independent cell growth, we analyzed the pathways affected by all three R mutants in both Rat1a and MCF10A cells and focused on the down-regulated genes since the R mutations predominantly inhibited gene expression in both cell types. Although the specific genes down-regulated by all three R mutants (relative to MYC WT) were different in the two cell types, GO and pathway enrichment analyses identified specific processes in common that were impacted by all three R mutants in both cell types, including cell–cell adhesion, matrisome (ECM core and associated proteins), cytoskeleton organization, EMT, and KRAS signaling (Fig. 7D; Supplemental Fig. S15).
Since each AcK residue also had gene-specific functions in both cell types, we further investigated whether MYC/AcK site-specific pathways could be deregulated in common in both cell types by performing GSEA. We identified deregulated gene sets significantly enriched (FDR q-value ≤ 0.25) in MYC WT versus control cells (empty vector) and for each R mutant versus MYC WT cells (Fig. 7E; Supplemental Fig. S16). The most significant gene sets deregulated in cells overexpressing the MYC R mutants, relative to cells overexpressing MYC WT, were spliceosome (negatively affected in R323 cells), EMT gene set including genes involved in ECM organization and TGFβ regulation of ECM (positively correlated in R158 cells), and mitotic spindle (negatively impacted in R149 cells), as well as p53 pathway, interferon α and γ response, and ribosome (all positively correlated in R149 cells) (Fig. 7E). Thus, in addition to pathways that are dependent on all three AcK sites (Fig. 7D), each MYC AcK residue also preferentially regulates specific cell biological processes in common in both cell types (Fig. 7F), consistent with the gene-selective activity of each AcK residue. Note, however, that in a given cell type some of these processes can also be dependent on several AcK residues. For instance, in MCF10A cells, the spliceosome gene set is dependent on both the AcK149 and AcK323 residues of MYC (data not shown), which is consistent with the significant number of genes dependent on both the AcK149 and AcK323 sites and the fact that MYC could be found coacetylated at more than one AcK site in these cells (Supplemental Fig. S17).
To investigate the mechanisms of MYC AcK site-dependent gene activation relevant to MCF10A cell transformation, we focused on a select group of cancer-associated MYC target genes (SNAI1, NOTCH3, TERT, ESPN, and SLCO5A1) that were dependent on all three AcK sites, since all three AcK sites were important for anchorage-independent growth. The KAT2A gene (coding for GCN5) served as a control MYC-activated gene that was unaffected by the R mutations (Fig. 7G). Chromatin immunoprecipitation (ChIP) experiments revealed a reduced binding of FLAG-MYC R149, R158, and R323 mutants to the promoters of SNAI1, TERT, and SLCO5A1, while no significant difference in binding of MYC WT versus R mutants was observed on the promoters of NOTCH3, ESPN, and KAT2A (Fig. 7H). ChIP analyses of total RNA polymerase II (RNAPII) and the elongation-competent Ser2 phosphorylated form of RNAPII (P-Ser2 RNAPII) indicated that MYC via its AcK sites stimulated RNAPII association with the promoters and 3′ ends (3′ UTR) of SNAI1, NOTCH3, and TERT (Supplemental Fig. S18). These data suggest that MYC AcK residues may activate transcription via facilitating MYC binding to specific gene promoters and via enhancing RNAPII recruitment.
We further investigated whether the MYC AcK-dependent gene SNAI1 is important for the transformed phenotype of MYC-overexpressing MCF10A cells. Indeed, this gene codes for a key transcription regulator of the EMT and stem-like phenotypes of breast cancer cells (Mani et al. 2008; Gras et al. 2014). We confirmed that MYC WT but not the R mutants induced expression of the SNAI1 protein in MCF10A cells (Fig. 7I). Furthermore, RNAi-mediated knockdown of SNAI1 in MYC-overexpressing MCF10A cells significantly inhibited their spheroid formation abilities (Fig. 7J). Thus, SNAI1 is one important downstream mediator of MYC AcK-dependent oncogenic signaling in this in vitro mammary cell transformation system.
Altogether, these results indicate that although MYC AcK sites are important for regulation of only a limited number of identical MYC target genes in Rat1a and MCF10A cells (as would be expected given the distinct cell types), each AcK site nevertheless regulates specific sets of genes and biological processes/pathways similarly in both cell types: All three AcK sites contribute to regulation of the EMT, cell adhesion, and organization of the ECM and cytoskeleton and contribute to anchorage-independent growth and spheroid formation. In addition, the AcK323 site is particularly important for MYC activation of spliceosome-associated genes; the AcK158 residue controls EMT-associated genes, including ECM organization and regulation by TGFβ, and is critical for the bypass of contact inhibition; and the AcK149 site is important for genes involved in the mitotic spindle and attenuates MYC activation of ribosome biogenesis, tumor-suppressive p53, and stress/interferon response pathways. Mechanistically, the gene-selective functions of MYC AcK residues involve stimulation of MYC binding to specific promoters and facilitation of RNAPII recruitment.
Discussion
In this study, we identified the clustered lysine residues K143(144), K148(149), and K157(158) of MYC in humans (and mice, shown in parentheses) as the major targets of p300 and identified the K323 residue as the preferred target site of GCN5. The K148(149), K157(158), and K323 residues were found acetylated on endogenous MYC in diverse cancer cell lines and in MYC transformed fibroblastic and epithelial cells. While endogenous MYC was found acetylated at these sites in nonmalignant cells, oncogenic MYC overexpression enhanced its acetylation and/or altered acetylation of specific AcK sites in response to proteasome and HDAC inhibitors. Additionally, different HDAC inhibitors had distinct K site-specific effects on MYC acetylation. These findings may have important implications for the use of proteasome and HDAC inhibitors in cancer therapy (Nebbioso et al. 2017; Bi et al. 2022).
Lysine acetylation is generally thought to increase MYC stability by antagonizing ubiquitination (or SUMOylation) and proteasomal degradation. However, previous studies did not directly analyze the stability of MYC acetylated at specific lysine residues. Here, with the use of site-specific MYC AcK antibodies, we found that MYC acetylated at K148(149) or K157(158) remained relatively unstable (half-life ∼30–37 min) in both cell types analyzed, while MYC acetylated at K323 was highly stable in MYC transformed Rat1a fibroblasts (half-life >2 h) but not in MCF10A cells (half-life ∼30 min). Hence, acetylation of MYC at different K residues by different HATs can have dramatically different effects on its turnover in a cell type-dependent manner, which may in part explain the discordant results in the literature (see above).
Despite differential effects on MYC stability, we found in both cell types tested that each AcK residue was required for MYC induction of anchorage-independent cell growth but not for the ability of overexpressed MYC to stimulate proliferation and colony formation of cells cultured under conventional adherent conditions. The latter is in agreement with previous observations that all or most K residues of MYC act redundantly in the stimulation of adherent cell proliferation via ubiquitin/proteasome-dependent degradation mechanisms (Jaenicke et al. 2016). Notably, MYC AcK148(149) and AcK157(158), but not AcK323, were also critical for the tumorigenic activity in vivo of MYC-overexpressing cells and for their increased proliferation as tumor spheroids in vitro in response to growth medium stimulation. Moreover, the AcK157(158) residue was uniquely important for MYC to (1) bypass contact inhibition in a p300 HAT activity-dependent manner and (2) attenuate apoptosis of MYC-overexpressing cells upon serum deprivation. Thus, the AcK148(149) and AcK157(158) residues, which are the main p300 target sites, appear to be most critical (compared with the AcK323 site) for the malignant transformation activity of MYC via, in part, regulating different cell biological pathways.
We found that the AcK residues of MYC regulate its metabolic rewiring functions albeit with notable differences in the two cell types tested. An inhibitory function of the AcK148(149) and AcK323 residues on glycolysis was identified in both Rat1a and MCF10A cells. In addition, the AcK323 site was required for basal and ATP-linked OCR/respiration (but not maximal OCR) in Rat1a cells, while all three AcK sites were required for mitochondrial respiration (basal, ATP-linked, and maximal OCR) in MCF10A cells. Thus, our results suggest that MYC acetylation by p300 and GCN5 may regulate the balance between glycolysis (inhibited by MYC AcK) and mitochondrial respiration (stimulated by MYC AcK). However, the observed cell type differences suggest that metabolic rewiring per se cannot fully explain the key roles of the AcK residues in the malignant transformation activity of MYC in both cell types.
Analyses of MYC AcK site-dependent genetic programs and biological pathways in MYC transformed Rat1a and MCF10A cells revealed gene-selective functions (predominantly positive but also negative) of each MYC AcK residue and identified a significant number of genes that are dependent on two or all three AcK sites. Although the MYC AcK-dependent genes identified were mostly distinct in each cell type (as would be expected form the distinct cell lineages), some of the pathways they regulate were conserved in both cell types, suggesting a functional/mechanistic conservation and potential roles of the identified genes and pathways in MYC AcK-driven malignant transformation.
Genetic programs involved in EMT, cell–cell adhesion, and organization of the ECM and cytoskeleton were dependent on all three MYC AcK residues in both Rat1a and MCF10A cells overexpressing MYC and thus are prime candidates for the observed requirement of each AcK site for cell anchorage-independent growth and suspension spheroid formation of MYC transformed cells. Indeed, all three MYC AcK sites were required in MCF10A cells for MYC activation of the EMT/stemness gene SNAI1 (SNAIL), and SNAI1 was required for MYC induction of spheroid formation in MCF10A cells. Furthermore, the EMT and regulation of the ECM were the only pathways significantly affected in both Rat1a and MCF10A cells by mutation of the AcK157(158) site, which regulated the smaller number of genes in both cell types; this may explain the observed key function of the AcK157(158) residue and p300 HAT activity in MYC-dependent bypass of cell–cell contact inhibition.
The MYC AcK148(149) residue was required for MYC-dependent regulation of the largest number of genes and biological processes in both cell types. The results suggest that MYC via its AcK148(149) site positively regulates the mitotic apparatus and negatively regulates tumor-suppressive (p53) and cytokine (e.g., IFNα and IFNγ) response pathways, as well as expression of many ribosomal protein genes. The latter may seem counterintuitive given the growth demands of cancer cells but could constitute an energy-saving or homeostatic survival mechanism that reduces the high energy cost of protein synthesis under conditions of limited nutrient availability (e.g., within tumors).
The genes dependent on the AcK323 residue were significantly enriched in components of the spliceosome in both cell types, suggesting that MYC acetylation at K323 by GCN5 may regulate cellular pre-mRNA splicing. This would be consistent with the known interactions of MYC and GCN5 (including human STAGA and yeast SAGA complexes) with splicing factors and their coregulation of splicing genes during somatic cell reprogramming (Martinez et al. 2001; Gunderson and Johnson 2009; Hirsch et al. 2015; Kalkat et al. 2018).
While the MYC AcK-dependent gene regulatory mechanisms remain to be fully delineated, our results indicate a function of the AcK residues in the stimulation of MYC binding to chromatin and recruitment of RNAPII to select target promoters (Fig. 7G,H; Supplemental Fig. S18) that does not involve regulation of MYC interaction with MAX, since mutation of all the AcK residues did not influence MYC:MAX complex formation (Supplemental Fig. S1B). Given the observed gene selectivity and lack of regulation of overall MYC turnover by the AcK-to-R mutants, a general transcription amplification mechanism involving a global ubiquitin-mediated proteasomal degradation of MYC is not involved in the cell systems studied here. It is, however, possible that acetylation of MYC at specific K residues may interfere with a gene-specific ubiquitination (or SUMOylation) mechanism and thereby may affect transcription of only select target genes. In support of such a possible PTM interplay, a recent study reported tumor-suppressive and transcription-inhibitory functions of TRAF6-mediated ubiquitination of MYC at K148(149), which inhibited acetylation at this site in acute myeloid leukemia cells (Muto et al. 2022). However, the fact that MYC K-to-Q substitutions at the K148(149) or K157(158) sites could not rescue growth of tumor spheroids suggests that acetylation may play a more active role and does not simply prevent other PTMs—or neutralize the charge—at these specific K sites.
The key roles of the p300-aceylated K148(149) and K157(158) residues in MYC transformation activity and their location immediately adjacent to the critically important MYC box II (MBII) are intriguing and might suggest a direct involvement in regulating MYC interaction with its transcription cofactors (Supplemental Fig. S19A). However, our results so far indicate that major MYC coactivators p300, GCN5, TRRAP, TIP49, and Mediator and the general basal transcription factor TBP can interact with MYC mutants with AcK-to-R substitutions (Supplemental Fig. S1B; data not shown), and we previously reported that p300-acetylated MYC can interact with MIZ1 (Zhang et al. 2005). It is thus likely that other still unknown cofactors mediate the AcK-dependent binding of MYC to chromatin and the stimulation of RNAPII recruitment observed here. Intriguingly, the differential growth stimulation and transformation activities of the distinct MYC family members (Barrett et al. 1992; Malynn et al. 2000) correlate with the conservation of the AcK residues in the order MYC > MYCN > MYCL (Supplemental Fig. S19B).
In conclusion, we have shown that oncogenic MYC overexpression deregulates its site-specific acetylation in transformed cells and identified key roles of individual MYC-acetylated lysine residues in the regulation of select MYC-dependent genetic programs and oncogenic pathways, in part via stimulation of MYC binding to chromatin and recruitment of RNAPII. We realize that the in vitro cell systems studied here do not fully or accurately model all of the processes that govern development of MYC-driven cancers in vivo. Hence, the identified MYC AcK site-dependent transformation and gene-selective regulatory activities warrant further mechanistic characterization and investigation of their possible roles in vivo and in human cancers (e.g., breast cancer). Indeed, uncovering the MYC AcK site-specific signaling pathways and mechanisms could offer new opportunities for selective therapeutic targeting of MYC oncogenic activities.
Materials and methods
Cell culture and generation of MYC-overexpressing cell lines
All cell lines were cultured in their recommended growth medium at 37°C with 5% CO2 in a humidified incubator (see the Supplemental Material). To generate stable Rat1a-MYC polyclonal cell lines, Rat1a cells (a gift from Dr. Michael Cole) were transfected with Lipofectamine 2000 (Invitrogen) and pMIG (empty) or pMIG-FLAG-mMYC (WT or R mutant) expression vectors together with pBABE-puro plasmid. After selection with 5 μg/mL puromycin, all resistant colonies were pooled and further selected for GFP expression by fluorescence-activated cell sorting (FACS) with a BD Biosciences FACSAria III. Two independent sets of stable polyclonal cell lines were generated similarly. Transient retroviral transduction of Rat1a cells with pMIG (empty) or pMIG-FLAG-mMYC (WT or mutant) is described in the Supplemental Material. The polyclonal MCF10A-MYC cell lines were obtained by transduction of MCF10A cells (ATCC) with retroviral particles encoding pMIG (empty) or pMIG-FLAG-mMYC (WT or R mutant) and pooling of GFP-positive clones as described in the Supplemental Material. Phase contrast and GFP/fluorescence microscopy images were obtained with a Keyence BZ-X710.
Antibodies, DNA plasmids, and RNA interference (RNAi)
The antibodies used were MYC Y69 from Abcam; MYC N-262, MYC C-33, MYC 9E10, MAX C-17, p300 N-15, GCN5 N-18, TRRAP T-17, and caspase-3 H-277 from Santa Cruz Biotechnology; cleaved caspase-3 and acetylated lysine from Cell Signaling Technology; vinculin, FLAG/M2, and FLAG M2 affinity resin from Sigma; TBP, which was a gift from Dr. Robert. G. Roeder; and TIP49, which was a gift from Dr. Bruno Amati. The affinity-purified site-specific MYC AcK antibodies were generated in rabbits by immunization with specific acetylated peptides (Supplemental Fig. S1E) in collaboration with EMD Millipore and are commercially available as acetyl-c-MYC (K148) ABE25, acetyl-c-MYC (K157) ABE27, and acetyl-c-MYC (K323) ABE26 (EMD Millipore). The plasmid DNAs are described in the Supplemental Material. For RNAi, HeLa cells were transfected in 10-cm plates with Lipofectamine 2000 and 7.5 μg of either pCbS-FLAG-mMYC WT or the indicated R mutants and with 400 pmol of either a specific p300 siRNA or a control siRNA (Dharmacon). MCF10A-MYC cells in six-well plates were transfected with Lipofectamine 3000 and either 60 pmol of SNAI1 siRNA (Santa Cruz Biotechnology sc-38398) or control siRNA-A (Santa Cruz Biotechnology sc-37007). For further details, see the Supplemental Material.
Immunofluorescence, immunoprecipitation, Western blot, and mass spectrometry
Immunofluorescence was performed with MYC N-262 and Alexa fluor 594 goat antirabbit IgG (Life Technologies) antibodies using an Eclipse TI inverted fluorescence microscope as described in the Supplemental Material. Preparation of whole-cell extracts (WCE), immunoprecipitation of endogenous MYC or ectopic FLAG-tagged MYC proteins, Western blot, and mass spectrometry analyses were performed essentially as previously described (Faiola et al. 2005). Chromatin immunoprecipitation (ChIP) is described in the Supplemental Material.
Cell proliferation, colony formation, and cell detachment assays
Logarithmic and postconfluency adherent cell proliferation and colony formation assays are described in the Supplemental Material. For detachment analysis, Rat1a cell lines were plated in six-well plates at low density (1.0 × 106 cells/well) in regular growth medium. At 24-h time intervals, plates with logarithmic growing cells for each time point were washed with 1× PBS on a rotary shaker at 60 rpm for 30 sec at room temperature. The detached cells were counted with a hemacytometer, the remaining cells attached to the plate were fixed with 0.5% glutaric dialdehyde in 1× PBS and stained with 0.1% crystal violet solution (Thermo Fisher Scientific), and the plates were scanned with a LaserJet Pro 300 color MFP M375nw (Hewlett-Packard). Alternatively, Rat1a cell lines were plated in six-well plates at high density. After 24 h, the medium was removed, and the cells were washed with 1× PBS and treated with 0.25% Trypsin and 2.21 mM EDTA (Corning). The plates were then gently rocked, and the times for the first visible sign of cell detachment and for complete detachment were recorded.
Apoptosis assays
Confluent Rat1a cell lines were incubated with medium containing reduced serum (0.1% FBS) for the indicated times (0–24 h). At each time point, both floating and attached cells were collected, stained with Trypan Blue solution (Corning), and counted with a Nexcelom Bioscience Cellometer Mini. To analyze apoptosis, the stable cell lines (or transiently transduced cells 48 h after viral infection) were serum-starved for ∼24 h as above, and cleaved caspase-3 was detected by Western blot. Alternatively, cells were serum-starved for 12 h as above and analyzed by flow cytometry with the annexin V-FITC apoptosis detection kit (Calbiochem/Millipore-Sigma), according to the manufacturer's instructions, with a FACSAria (BD Biosciences) (see also the Supplemental Material).
Focus formation assay
Confluent Rat1a cell lines were maintained in culture for 8 d with medium replacement every 2 d. The cells were then fixed with 0.5% glutaric dialdehyde in 1× PBS and stained with 0.1% crystal violet solution (Thermo Fisher Scientific). The cells were then partially destained with 1% acetic acid and air-dried before imaging.
Spheroid growth assays
For spontaneous spheroid formation, six-well plates were first coated with a thin layer of 1% agarose, and then 30,000 cells/well were plated in 2 mL of regular growth medium. The cells were left for 7 d at 37°C with 5% CO2 without further feeding or were fed on days 3 and 6 by addition of 500 μL of fresh medium where indicated. Spheroids were imaged with a Leica DMi8 microscope. After trypsinization, the cells were counted with a Cellometer Mini cell counter (Nexcelom Bioscience). Alternatively, spheroids were lysed with RIPA buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1.0% NP-40/Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA), and proteins were analyzed by Western blot.
Facilitated spheroid formation assays were performed in 96-well nonadherent U-bottom plates (Nunclon Sphera 96-well, Nunclon Sphera-treated, U-shaped-bottom microplate; Thermo Scientific) by aggregating 100 cells at the bottom of each well in regular growth medium. Plates were incubated for the indicated times at 37°C with 5% CO2. Images were taken every 24 h using a Leica DMi8 microscope and an Andor Technologies Zyla 4.2 sCMOS at 50× total magnification (see the Supplemental Material).
Soft agar colony formation assays
Soft agar assays were performed in triplicates in six-well plates. The bottom of each well contained a layer of 0.5% Nobel agar (Sigma-Aldrich A5431) in complete growth medium. The top layer contained the cell suspension in 0.35% Noble agar in complete growth medium (5 × 103 Rat1a cells/well or 6 × 104 MCF10A cells/well). Plates were incubated for ∼3 wk at 37°C in a humidified incubator with 5% CO2 with addition every other day of 100 μL of fresh complete growth medium. Colonies were stained with 0.005% crystal violet and imaged with a Bio-Rad ChemiDoc imager. Colonies ≥200 μm (Rat1a) or ≥100 μm (MCF10A) were counted (see also the Supplemental Material).
Mouse xenografts
Athymic nude male mice [Crl:NU(NCr)-Foxn1nu]; Charles River Laboratories] were injected at 7 wk old in the right rear flank with 5.0 × 106 cells in PBS containing 25% Matrigel (Corning). Tumor dimensions (length [L] and height [H]) were used to calculate tumor volumes using the modified ellipsoidal formula 1/2(L × H2). At the experimental end point, mice were euthanized with carbon dioxide. All procedures were approved by the University of California Riverside Institutional Animal Care and Use Committee (IACUC).
Metabolic analyses
The Seahorse Bioscience XF96 extracellular flux analyzer (Agilent) was used to measure glycolysis (glycolysis stress test assay), mitochondrial function (mito stress test assay), and mitochondrial fuel dependencies (mito fuel flex test assay) as recommended by the manufacturer; further details are in the Supplemental Material.
RNA-seq and RT-qPCR analyses
Stranded cDNA libraries (three biological replicates per sample) were constructed using the TruSeq stranded (dUTP) mRNA library preparation kit (Illumina) or the Kapa mRNA HyperPrep kit (Kapa Biosystems KR1352) and sequenced on an Illumina HiSeq 2000 (Rat1a) or NovaSeq 6000 (MCF10A) at the Core Genomic Facilities of University of California Riverside and City of Hope, respectively. Sequence reads were mapped to either the rat genome Rnor_6.0.82 (Rat1a cell lines) or the human reference genome hg38 (MCF10A cell lines). Differential gene expression was analyzed with Bioconductor edgeR. Additional details, reverse transcription quantitative PCR (RT-qPCR), and gene set enrichment analyses are provided in the Supplemental Material. RNA-seq data are available at GEO under accession number GSE220920.
Statistical analyses
All data were derived from at least three independent experiments (each in triplicates), and statistical analyses were performed using a t-test (two-tailed) with results shown as means ± standard deviation (SD) unless otherwise indicated. Statistically significant differences are indicated with asterisks as follows: P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Supplementary Material
Acknowledgments
We thank Dr. B. Amati, Dr. J.M. Bishop, Dr. M.D. Cole, Dr. D. Eick, Dr. R. Goodman, Dr. W.C. Greene, Dr. W. Hahn, Dr. S. Nasi, Dr. R.G. Roeder, Dr. F.M. Sladek, and Dr. I. Verma for generous gifts of materials. We also thank S. Sauer and the core Genomics and Bioinformatics Facilities at University of California Riverside and City of Hope for excellent technical assistance. M.H. was supported by a National Institutes of Health (NIH) predoctoral fellowship (1F31CA165776) from the National Cancer Institute. J.P. was supported by a predoctoral Graduate Assistance in Areas of National Need (GAANN) fellowship from the U.S Department of Education (P200A210136). R.G. and E.V. were supported by a NIH grant (T34GM062756) from the National Institute of General Medical Sciences. This study was supported by NIH grants from the National Cancer Institute (R01CA158540, R03CA230843, R03CA249489, and P20CA242620) and a University of California Riverside Academic Senate fellowship to E.M., and by an NIH grant from the National Cancer Institute (P20CA242619) to V.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
Author contributions: E.M. conceived and directed the project. M.H., J.P., K.J., M.M.A., M.V., W.I., Y.Z., R.G., E.V., M.J.H., F.F., S.P., Y.Q., and Y.-W.H. performed the experiments. D.A. and T.G. supervised the metabolic and systemPipeR analyses, respectively. M.H., J.P., K.J., and E.M. wrote the original draft of the manuscript. All authors reviewed and edited the manuscript. E.M. and V.S. acquired the resources and funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350736.123.
Competing interest statement
The authors declare no competing interests.
References
- Allevato M, Bolotin E, Grossman M, Mane-Padros D, Sladek FM, Martinez E. 2017. Sequence-specific DNA binding by MYC/MAX to low-affinity non-E-box motifs. PLoS One 12: e0180147. 10.1371/journal.pone.0180147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. 1992. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359: 423–426. 10.1038/359423a0 [DOI] [PubMed] [Google Scholar]
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. 1993. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72: 233–245. 10.1016/0092-8674(93)90663-B [DOI] [PubMed] [Google Scholar]
- Barrett J, Birrer MJ, Kato GJ, Dosaka-Akita H, Dang CV. 1992. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol 12: 3130–3137. 10.1128/mcb.12.7.3130-3137.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Zhang Y, Malmrose PK, Losh HA, Newtson AM, Devor EJ, Thiel KW, Leslie KK. 2022. Blocking autophagy overcomes resistance to dual histone deacetylase and proteasome inhibition in gynecologic cancer. Cell Death Dis 13: 1–10. 10.1038/s41419-022-04508-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN. 1991. Max: a helix–loop–helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251: 1211–1217. 10.1126/science.2006410 [DOI] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. 2010. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol 17: 471–482. 10.1016/j.chembiol.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Dalton S. 2013. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb Perspect Med 3: a014381. 10.1101/cshperspect.a014381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T-Y, Hart GW, Dang CV. 1995. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. Journal of Biological Chemistry 270: 18961–18965. 10.1074/jbc.270.32.18961 [DOI] [PubMed] [Google Scholar]
- Dang CV. 2012. MYC on the path to cancer. Cell 149: 22–35. 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pretis S, Kress TR, Morelli MJ, Sabò A, Locarno C, Verrecchia A, Doni M, Campaner S, Amati B, Pelizzola M. 2017. Integrative analysis of RNA polymerase II and transcriptional dynamics upon MYC activation. Genome Res 27: 1658–1664. 10.1101/gr.226035.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini LM, Chen S, Marhin WW, Lear JN, Penn LZ. 1997. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-myc P2 minimal promoter. Mol Cell Biol 17: 100–114. 10.1128/MCB.17.1.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, Farina A, Martinez E. 2005. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol 25: 10220–10234. 10.1128/MCB.25.23.10220-10234.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiola F, Wu Y-T, Pan S, Zhang K, Farina A, Martinez E. 2007. Max is acetylated by p300 at several nuclear localization residues. Biochem J 403: 397–407. 10.1042/BJ20061593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell AS, Sears RC. 2014. MYC degradation. Cold Spring Harb Perspect Med 4: a014365. 10.1101/cshperspect.a014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay M, Li Y, Felsher DW. 2014. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 4: a014241. 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanz P, Quintanilla A, Lafita MC, Moreno-Bueno G, García-Gutierrez L, Tabor V, Varela I, Shiio Y, Larsson L-G, Portillo F, et al. 2014. Sin3b interacts with Myc and decreases Myc levels. J Biol Chem 289: 22221–22236. 10.1074/jbc.M113.538744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Prieto R, Cuijpers SA, Kumar R, Hendriks IA, Vertegaal AC. 2015. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 14: 1859–1872. 10.1080/15384101.2015.1040965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16: 653–699. 10.1146/annurev.cellbio.16.1.653 [DOI] [PubMed] [Google Scholar]
- Gras B, Jacqueroud L, Wierinckx A, Lamblot C, Fauvet F, Lachuer J, Puisieux A, Ansieau S. 2014. Snail family members unequally trigger EMT and thereby differ in their ability to promote the neoplastic transformation of mammary epithelial cells. PLoS One 9: e92254. 10.1371/journal.pone.0092254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson FQ, Johnson TL. 2009. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet 5: e1000682. 10.1371/journal.pgen.1000682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR. 2006. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol 16: 288–302. 10.1016/j.semcancer.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Hirsch CL, Coban Akdemir Z, Wang L, Jayakumaran G, Trcka D, Weiss A, Hernandez JJ, Pan Q, Han H, Xu X, et al. 2015. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev 29: 803–816. 10.1101/gad.255109.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro T, Ohata H, Sato A, Yamawaki K, Enomoto T, Okamoto K. 2017. Tumor-derived spheroids: relevance to cancer stem cells and clinical applications. Cancer Sci 108: 283–289. 10.1111/cas.13155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke LA, von Eyss B, Carstensen A, Wolf E, Xu W, Greifenberg AK, Geyer M, Eilers M, Popov N. 2016. Ubiquitin-dependent turnover of MYC antagonizes MYC/PAF1C complex accumulation to drive transcriptional elongation. Mol Cell 61: 54–67. 10.1016/j.molcel.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Kadota M, Yang HH, Gomez B, Sato M, Clifford RJ, Meerzaman D, Dunn BK, Wakefield LM, Lee MP. 2010. Delineating genetic alterations for tumor progression in the MCF10A series of breast cancer cell lines. PLoS One 5: e9201. 10.1371/journal.pone.0009201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkat M, Resetca D, Lourenco C, Chan P-K, Wei Y, Shiah Y-J, Vitkin N, Tong Y, Sunnerhagen M, Done SJ, et al. 2018. MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol Cell 72: 836–848.e7. 10.1016/j.molcel.2018.09.031 [DOI] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. 2003. Skp2 regulates Myc protein stability and activity. Mol Cell 11: 1177–1188. 10.1016/S1097-2765(03)00173-4 [DOI] [PubMed] [Google Scholar]
- Laurenti E, Wilson A, Trumpp A. 2009. Myc's other life: stem cells and beyond. Curr Opin Cell Biol 21: 844–854. 10.1016/j.ceb.2009.09.006 [DOI] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67. 10.1016/j.cell.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco C, Kalkat M, Houlahan KE, De Melo J, Longo J, Done SJ, Boutros PC, Penn LZ. 2019. Modelling the MYC-driven normal-to-tumour switch in breast cancer. Dis Model Mech 12: dmm038083. 10.1242/dmm.038083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco C, Resetca D, Redel C, Lin P, MacDonald AS, Ciaccio R, Kenney TMG, Wei Y, Andrews DW, Sunnerhagen M, et al. 2021. MYC protein interactors in gene transcription and cancer. Nat Rev Cancer 21: 579–591. 10.1038/s41568-021-00367-9 [DOI] [PubMed] [Google Scholar]
- Lynch JT, Somerville TDD, Spencer GJ, Huang X, Somervaille TCP. 2013. TTC5 is required to prevent apoptosis of acute myeloid leukemia stem cells. Cell Death Dis 4: e573. 10.1038/cddis.2013.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, de Alboran IM, O'Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW. 2000. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev 14: 1390–1399. 10.1101/gad.14.11.1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall GM, Liu PY, Gherardi S, Scarlett CJ, Bedalov A, Xu N, Iraci N, Valli E, Ling D, Thomas W, et al. 2011. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet 7: e1002135. 10.1371/journal.pgen.1002135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol 21: 6782–6795. 10.1128/MCB.21.20.6782-6795.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Lüscher B, Larsson L-G, Hermeking H. 2012. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci 109: E187–E196. 10.1073/pnas.1105304109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T, Guillamot M, Yeung J, Fang J, Bennett J, Nadorp B, Lasry A, Redondo LZ, Choi K, Gong Y, et al. 2022. TRAF6 functions as a tumor suppressor in myeloid malignancies by directly targeting MYC oncogenic activity. Cell Stem Cell 29: 298–314.e9. 10.1016/j.stem.2021.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento EM, Cox CL, MacArthur S, Hussain S, Trotter M, Blanco S, Suraj M, Nichols J, Kübler B, Benitah SA, et al. 2011. The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis. Nat Cell Biol 13: 1395–1405. 10.1038/ncb2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebbioso A, Carafa V, Conte M, Tambaro FP, Abbondanza C, Martens J, Nees M, Benedetti R, Pallavicini I, Minucci S, et al. 2017. c-Myc modulation and acetylation is a key HDAC inhibitor target in cancer. Clin Cancer Res 23: 2542–2555. 10.1158/1078-0432.CCR-15-2388 [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79. 10.1016/j.cell.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen C-J, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, et al. 2004. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol 24: 10826–10834. 10.1128/MCB.24.24.10826-10834.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellanda P, Dalsass M, Filipuzzi M, Loffreda A, Verrecchia A, Castillo Cano V, Thabussot H, Doni M, Morelli MJ, Soucek L, et al. 2021. Integrated requirement of non-specific and sequence-specific DNA binding in Myc-driven transcription. EMBO J 40: e105464. 10.15252/embj.2020105464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli V, Fagnocchi L, Fasciani A, Cherubini A, Mazzoleni S, Ferrillo S, Miluzio A, Gaudioso G, Vaira V, Turdo A, et al. 2018. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun 9: 1024. 10.1038/s41467-018-03264-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo MM, Ko B, Kim J, Brady R, Heatley HC, He J, Harrod CK, Barnett B, Ratner L, Lairmore MD, et al. 2015. Acetylation of the c-MYC oncoprotein is required for cooperation with the HTLV-1 p30II accessory protein and the induction of oncogenic cellular transformation by p30II/c-MYC. Virology 476: 271–288. 10.1016/j.virol.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabò A, Amati B. 2014. Genome recognition by MYC. Cold Spring Harb Perspect Med 4: a014191. 10.1101/cshperspect.a014191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabò A, Doni M, Amati B. 2014. SUMOylation of Myc-family proteins. PLoS One 9: e91072. 10.1371/journal.pone.0091072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, Zhao W, Zhang X, Ventura A, Liu Y, et al. 2018. Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst 6: 282–300.e2. 10.1016/j.cels.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SI, Freedman VH, Risser R, Pollack R. 1975. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc Natl Acad Sci 72: 4435–4439. 10.1073/pnas.72.11.4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, de Lange T, Ramsay G, Jakobovits E, Bishop JM, Varmus H, Lee W. 1987. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol 7: 1697–1709. 10.1128/mcb.7.5.1697-1709.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Pelz C, Huang T, Torkenczy K, Wang X, Cherry A, Daniel CJ, Liang J, Nan X, Dai M-S, et al. 2018. Post-translational modification localizes MYC to the nuclear pore basket to regulate a subset of target genes involved in cellular responses to environmental signals. Genes Dev 32: 1398–1419. 10.1101/gad.314377.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X-X, Chen Y, Su Y, Wang X, Chauhan KM, Liang J, Daniel CJ, Sears RC, Dai M-S. 2018. SUMO protease SENP1 deSUMOylates and stabilizes c-Myc. Proc Natl Acad Sci 115: 10983–10988. 10.1073/pnas.1802932115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tansey WP. 2014. Mammalian MYC proteins and cancer. New J Sci 2014: e757534. 10.1155/2014/757534 [DOI] [Google Scholar]
- Tworkowski KA, Salghetti SE, Tansey WP. 2002. Stable and unstable pools of Myc protein exist in human cells. Oncogene 21: 8515–8520. 10.1038/sj.onc.1205976 [DOI] [PubMed] [Google Scholar]
- Vervoorts J, Lüscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Lüscher B. 2003. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep 4: 484–490. 10.1038/sj.embor.embor821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoorts J, Lüscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Lüscher B. 2003. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep 4: 484–490. 10.1038/sj.embor.embor821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoorts J, Lüscher-Firzlaff J, Lüscher B. 2006. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem 281: 34725–34729. 10.1074/jbc.R600017200 [DOI] [PubMed] [Google Scholar]
- Vita M, Henriksson M. 2006. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol 16: 318–330. 10.1016/j.semcancer.2006.07.015 [DOI] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al. 2003. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 11: 1189–1200. 10.1016/S1097-2765(03)00193-X [DOI] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. 2014. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511: 483–487. 10.1038/nature13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Faiola F, Martinez E. 2005. Six lysine residues on c-Myc are direct substrates for acetylation by p300. Biochem Biophys Res Commun 336: 274–280. 10.1016/j.bbrc.2005.08.075 [DOI] [PubMed] [Google Scholar]
- Zhang N, Ichikawa W, Faiola F, Lo S-Y, Liu X, Martinez E. 2014. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim Biophys Acta 1839: 395–405. 10.1016/j.bbagrm.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.