In this study, Wang et al. show that the inhibition of ATR kinase affords selective lethality to mismatch repair-deficient (MMR-d) cancer cells by promoting MUS81 nuclease- and replication-dependent DNA damage, increasing cytosolic DNA fragments, and activating cGAS signaling. The consequent interferon response potentially stimulates cytotoxic T-cell-dependent antitumor immunity and points to ATR inhibition as a notable strategy to augment PD-1 targeting immunotherapies against MMR-d cancers.
Keywords: ATR, immunotherapy, MMR, MSI, synthetic lethality
Abstract
The mismatch repair (MMR) deficiency of cancer cells drives mutagenesis and offers a useful biomarker for immunotherapy. However, many MMR-deficient (MMR-d) tumors do not respond to immunotherapy, highlighting the need for alternative approaches to target MMR-d cancer cells. Here, we show that inhibition of the ATR kinase preferentially kills MMR-d cancer cells. Mechanistically, ATR inhibitor (ATRi) imposes synthetic lethality on MMR-d cells by inducing DNA damage in a replication- and MUS81 nuclease-dependent manner. The DNA damage induced by ATRi is colocalized with both MSH2 and PCNA, suggesting that it arises from DNA structures recognized by MMR proteins during replication. In syngeneic mouse models, ATRi effectively reduces the growth of MMR-d tumors. Interestingly, the antitumor effects of ATRi are partially due to CD8+ T cells. In MMR-d cells, ATRi stimulates the accumulation of nascent DNA fragments in the cytoplasm, activating the cGAS-mediated interferon response. The combination of ATRi and anti-PD-1 antibody reduces the growth of MMR-d tumors more efficiently than ATRi or anti-PD-1 alone, showing the ability of ATRi to augment the immunotherapy of MMR-d tumors. Thus, ATRi selectively targets MMR-d tumor cells by inducing synthetic lethality and enhancing antitumor immunity, providing a promising strategy to complement and augment MMR deficiency-guided immunotherapy.
Mutations in DNA mismatch repair (MMR) genes are frequently found in patients of several cancer types and are particularly common in colorectal cancer (Cortes-Ciriano et al. 2017; Le et al. 2017). Loss of MMR function results in microsatellite instability (MSI), a type of genomic instability associated with insertions and deletions (indels) in DNA microsatellite repeats (Guan and Li 2023). Tumor cells with MMR deficiency or high MSI (MSI-H) tend to have an exceptionally high mutation rate, elevating the tumor mutation burden (TMB) (Cortes-Ciriano et al. 2017). Inactivation of MMR genes Mlh1 or Msh2 in mouse cancer cell lines increases mutational load in vitro and renders tumors responsive to immune checkpoint blockade (ICB) in syngeneic mouse tumor models (Germano et al. 2017; Mandal et al. 2019), leading to the model that MMR deficiency/MSI-H stimulate the antitumor immunity by increasing neoantigens. In cancer patients, MMR deficiency/MSI-H are associated with better responses to ICB (Le et al. 2017; André et al. 2020; Cercek et al. 2022). Pembrolizumab, an anti-PD-1 monoclonal antibody, has been approved by the FDA for treatment of MMR-deficient (MMR-d)/MSI-H tumors. However, while the use of MMR deficiency/MSI-H as biomarkers has improved the efficacy of immunotherapy, half of the patients with MMR-d tumors do not respond to pembrolizumab (André et al. 2020), suggesting that MMR deficiency is not sufficient to elicit robust antitumor immunity upon ICB. Indeed, a recent mouse study showed that MMR deficiency did not increase T-cell infiltration and ICB response due to substantial intratumor heterogeneity of mutations (Westcott et al. 2023). These findings raise an important question as to whether other therapeutic strategies can be developed to target MMR-d tumors, which may complement and augment immunotherapy. Novel neoadjuvant therapies of patients with MMR-d/MSI-H tumors may generate deeper clinical responses and allow organ preservation.
Recent studies have revealed that loss of the WRN helicase is synthetically lethal in MSI cancer cells, providing a promising strategy to target MSI tumors (Chan et al. 2019; Kategaya et al. 2019; Lieb et al. 2019; Picco et al. 2021; Mengoli et al. 2023). Mechanistically, WRN functions to unwind non-B-form DNA secondary structures in TA dinucleotide repeats [(TA)ns] (van Wietmarschen et al. 2020; Mengoli et al. 2023). In MSI cancer cells, (TA)ns undergo large-scale expansions, generating high levels of DNA secondary structures in (TA)ns. In the absence of WRN, the DNA secondary structures in (TA)ns become susceptible to the MUS81-EME1 nuclease, giving rise to DNA double-strand breaks (DSBs). This model explains how MSI-associated expansion of (TA)ns confers a dependency on WRN for cell survival. Notably, loss of MMR does not confer a WRN dependency immediately (Lieb et al. 2019; van Wietmarschen et al. 2020). MMR-d cells only become WRN-dependent after large-scale expansion of (TA)ns, a process that occurs over time. These findings suggest that inhibition of WRN may be an effective way to selectively kill MSI cancer cells harboring expanded (TA)ns. Indeed, WRN-specific inhibitors are being developed and their effects on MSI tumors are being assessed (Morales-Juarez and Jackson 2022).
The ATR checkpoint kinase is a master regulator of the DNA replication stress response and is critical for replication fork stability (Yazinski and Zou 2016; Saldivar et al. 2017). When replication forks are stalled by DNA damage or other impediments, ATR is recruited to stalled forks, where it is activated to orchestrate local and distal responses. A large group of DNA replication and repair proteins, including several MMR proteins and WRN, are substrates of ATR (Pichierri et al. 2003; Matsuoka et al. 2007). One of ATR's functions at stalled replication forks is to restrict the activities of SLX4-associated nucleases, such as MUS81, thereby preventing excessive cleavage of stalled forks and fork collapse (Forment et al. 2011; Couch et al. 2013; Ragland et al. 2013). Loss of ATR leads to increased replication fork collapse at poly(dA/dT) tracts and other repeats prone to non-B-form DNA secondary structures (Shastri et al. 2018; Tubbs et al. 2018), suggesting that ATR is important for preventing nucleolytic cleavage upon collisions between replication forks and DNA secondary structures. Of note, some of the most prominent ATR inhibitor (ATRi)-induced DSBs were mapped to TA-rich repeats (Shastri et al. 2018). These tantalizing links between ATR and the stability of replication forks at non-B-form DNA secondary structures raise the question as to whether ATR inhibition affects fork stability at microsatellites and the survival of MMR-d/MSI-H cells. In addition, ATRi enhances the DNA damage-induced expression of interferon-stimulated genes (ISGs) and immunotherapy responses (Harding et al. 2017; Vendetti et al. 2018, 2023; Chen et al. 2020), prompting us to investigate whether ATRi affects these responses in the context of MMR-d/MSI-H tumors.
In this study, we investigated how ATRi affects MMR-d cells in vitro and MMR-d tumors in vivo. We found that ATRi preferentially kills MMR-d cells as opposed to MMR-proficient cells. Notably, acute depletion of MLH1 or MSH2 is sufficient to confer ATRi sensitivity without a substantial increase in MSI, suggesting that ATR inhibition is synthetic lethal with MMR deficiency but not the consequent MSI. Using syngeneic mouse models, we found that ATRi effectively reduces the growth of MMR-d tumors in vivo. Interestingly, the antitumor effects of ATRi are partially dependent on CD8+ T cells. In vitro, ATRi preferentially promotes the accumulation of nascent DNA fragments in cytoplasm and activation of the cGAS–STING pathway in MMR-d cells. In syngeneic mouse models, the combination of ATRi and anti-PD-1 reduced the growth of MMR-d tumors more effectively than either ATRi or anti-PD-1 alone. These results suggest that ATRi selectively targets MMR-d tumors through both synthetic lethality and antitumor immunity, providing a promising strategy to complement and augment the immunotherapy of MMR-d tumors.
Results
MSI cancer cells are increasingly dependent on ATR for survival
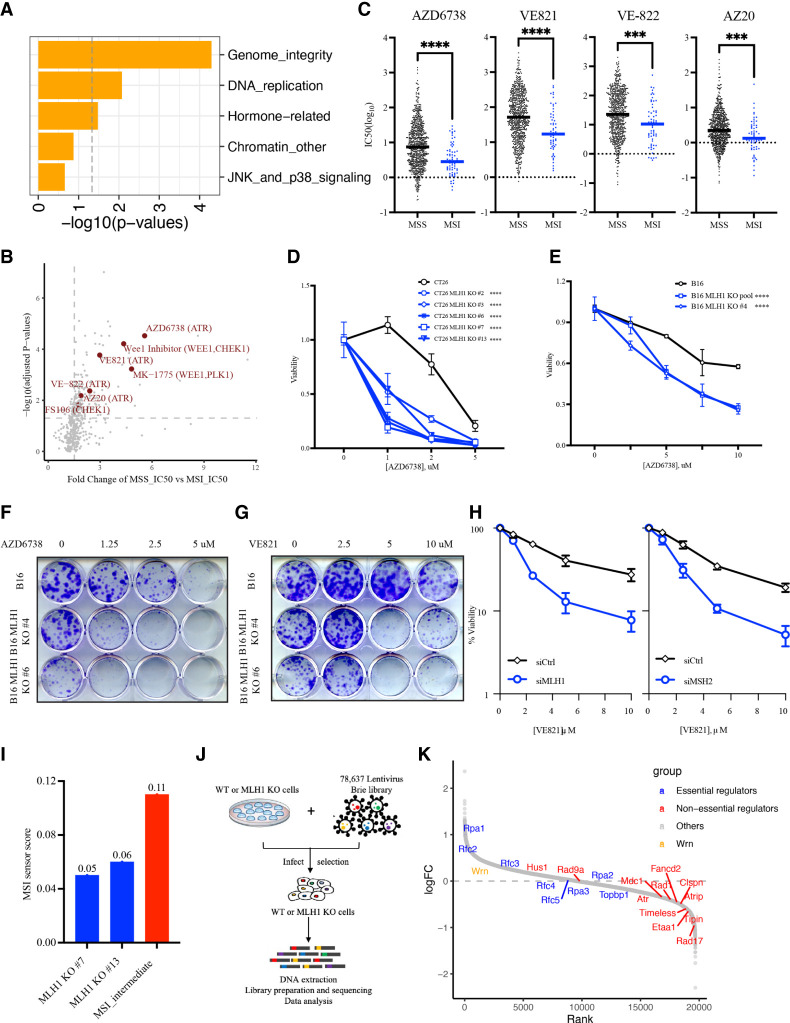
To investigate whether MSI cancer cells can be selectively killed by certain cancer drugs, we analyzed the drug sensitivity data of cancer cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) database. We separated the cell lines into microsatellite stability (MSS) and MSI groups according to the CCLE_MSI definition. Using information about the pathways targeted by various cancer drugs, we performed pathway enrichment analysis to understand which pathways are preferentially required for the survival of MSI cancer cells as opposed to MSS cancer cells. Only two pathways—genome integrity and DNA replication—were significantly associated with the preferential killing of MSI cancer cells by targeted drugs (Fig. 1A). This result is consistent with the idea that MSI cancer cells harbor high levels of genomic instability and replication stress and therefore are particularly susceptible to drugs exacerbating genomic instability and/or replication stress.
Figure 1.
Loss of MMR genes increases ATRi sensitivity. (A) Pathway enrichment analysis for drugs that preferentially kill MSI cancer cell lines in the Genomics of Drug Sensitivity in Cancer (GDSC) database. MSS and MSI cell lines were grouped according to the MSI definition in the CCLE database. (B) Volcano plot showing the distribution of drugs according to the ratio of IC50 in MSS lines and IC50 in MSI lines. The ATRi, Wee1i, and Chk1i that preferentially kill MSI cells are highlighted in dark red. (C) The IC50 of different ATR inhibitors in MSS and MSI cell lines from the GDSC database. Bars indicate the mean values of IC50 of ATR inhibitors in MSS or MSI cell line populations. (D) Relative viability of CT26 and CT26 Mhl1 KO cells (derived from single-cell clones) after 5 d of ATRi (AZD6738) treatment at the indicated concentrations. Cell viability was measured with CellTiter-Glo. (E) Relative viability of B16 and B16 Mhl1 KO cells (either cell pool or single-cell clones) after 5 d of ATRi (AZD6738) treatment at the indicated concentrations. (F,G) Clonogenic assay of B16 and B16 Mhl1 KO cells (derived from single-cell clones) after 9 d of ATRi (AZD6738 in F and VE-821 in G) treatment at the indicated concentrations. (H) Relative viability of U2OS cells treated with control, MLH1, or MSH2 siRNAs after ATRi (VE-821) treatment. (I) The MSI sensor scores of two CT26 Mhl1 KO clones (#7 and #13). The score of MSI intermediate CT26 Msh2 KO cells from a previous study (Mandal et al. 2019) is used as a reference (colored in red). (J) Scheme of the genome-wide CRISPR dropout screen to identify genes preferentially required for the survival of CT26 parental cells (MSS) or CT26 Mlh1 KO cells (clone #13; MSI). Cells were infected with lentivirus expressing the mouse genome-wide sgRNA library and analyzed by sgRNA sequencing 8 d after infection. (K) An S-curve plot showing the enrichment or depletion of sgRNAs targeting various genes in MSI or MSS cells. (Red) ATR, ATRIP, and nonessential regulators; (blue) essential replication genes involved in ATR regulation; (orange) WRN.
Next, we compared the IC50s of a large panel of cancer drugs between MSS and MSI lines and searched for those with lower IC50 in MSI lines than in MSS lines. The ATRi AZD6738 emerged as one of the top hits, suggesting that it preferentially kills MSI cancer cells (Fig. 1B). In addition, several other inhibitors of ATR (VE-821, VE-822, and AZ20), Wee1 (Wee1 inhibitor; MK-1775), and Chk1 (FS106) also displayed lower IC50s in MSI lines than in MSS lines (Fig. 1B). Thus, compared with MSS cancer cells, MSI cancer cells are generally more dependent on ATR, Chk1, and Wee1 for survival. These results suggest that MSI cancer cells are vulnerable to the increase of replication stress induced by ATRi, Chk1i, and Wee1i or to the defective replication stress response caused by these drugs (Hopkins et al. 2022). To extend our analysis from cancer cell lines to patient tumor samples, we analyzed the gene expression data of 91 colorectal and endometrial tumors from a previous study (GSE146889). In this cohort, the expression of ATR and its regulators is generally higher in MSI tumors than in MSS tumors (Supplemental Fig. S1A). This observation is consistent with the possibility that the ATR pathway is transcriptionally up-regulated in MSI tumors to cope with genomic instability and help tumor cell survival.
To confirm the ability of ATRi to preferentially kill MSI cancer cells, we specifically analyzed the ATRi sensitivity data from the GDSC database. Again, cell lines were separated into MSS and MSI groups according to the MSI definition of CCLE. All four ATR inhibitors tested in the cancer cell line panel (AZD6738, VE-821, VE-822, and AZ20) displayed lower IC50s in MSI lines than in MSS lines (Fig. 1C). These data lend further support to the notion that ATRi preferentially kills MSI cancer cells.
Loss of MMR genes increases ATRi sensitivity
To directly test whether MMR deficiency or MSI-H confers ATRi sensitivity, we used CRISPR/Cas9 to knock out the Mlh1 gene in mouse colorectal cancer cell lines CT26 and MC38 and in mouse melanoma cell line B16 (Supplemental Fig. S1B,C). Several independent KO clones were generated and cultured for <30 d before being tested for ATRi sensitivity. Multiple CT26 Mlh1 KO clones displayed increased sensitivity to two distinct ATR inhibitors (AZD6738 and VE-821) compared with parental cells (Fig. 1D; Supplemental Fig. S1D). Similarly, B16 Mlh1 KO clones and cell pool were also more sensitive to the two ATR inhibitors than parental cells (Fig. 1E–G; Supplemental Fig. S1E). Interestingly, MC38 cells, which carry mutations in the MMR gene Msh3 (Efremova et al. 2018), were more sensitive to ATRi than CT26 and B16 cells, but knockout of Mlh1 in MC38 did not increase ATRi sensitivity further (Supplemental Fig. S1F). These results suggest that loss of MMR genes is sufficient to confer ATRi sensitivity.
Because MSI increases over time after loss of MMR, we asked whether MMR deficiency alone is sufficient to confer ATRi sensitivity before MSI accumulates to a high level. We used multiple independent siRNAs to knock down MLH1 or MSH2 in the human osteosarcoma cell line U2OS (Supplemental Fig. S1G). Knockdown of MLH1 or MSH2 resulted in increased sensitivity to both AZD6738 and VE-821 (Fig. 1H; Supplemental Fig. S1H), showing that acute loss of MMR proteins can confer ATRi sensitivity. Furthermore, knockdown of MLH1 in three other MSS human cancer cell lines (LS513, SW620, and MKN45) also increased ATRi sensitivity (Supplemental Fig. S1I,J). To check whether CT26 Mlh1 KO cells are sensitive to ATRi because of high levels of MSI, we performed whole-genome sequencing on parental CT26 cells and two CT26 Mlh1 KO clones (#7 and #13) (Fig. 1I). The genomic MSI scores of the Mlh1 KO clones were 0.05 and 0.06, which were much lower than that of an MSI intermediate Msh2 KO clone (0.11) generated in a previous study (Mandal et al. 2019). This result suggests that loss of MLH1 confers ATRi sensitivity before MSI rises to a significant level. Thus, while cancer cells that have both MMR deficiency and MSI are sensitive to ATRi, MMR deficiency alone is sufficient to confer ATRi sensitivity even when MSI is not high.
A CRISPR screen confirms the dependency of Mlh1 KO cells on the ATR pathway
To unbiasedly assess whether MMR-d cells are more dependent on the ATR pathway for survival, we carried out a genome-wide CRISPR sgRNA dropout screen using parental CT26 cells and an Mlh1 KO clone with a low level of MSI (clone #13) (Fig. 1J). In addition to the sgRNAs targeting ATR, those targeting ATRIP, the functional partner of ATR, were also preferentially depleted in Mlh1 KO cells (Fig. 1K), suggesting that the ATR–ATRIP complex is required for the fitness of MMR-d cells. Furthermore, sgRNAs targeting nonessential ATR regulators, such as Rad17, Rad1, Rad9, Claspin, Timeless, Tipin, FANCD2, MDC1, and ETAA1, were all preferentially depleted in Mlh1 KO cells. In contrast, sgRNAs targeting the essential DNA replication factors involved in ATR regulation, such as TopBP1, RFC2-5, and RPA1-3, did not show preferential depletion in Mlh1 KO cells. Using siRNAs and U2OS cells, we confirmed that codepletion of MLH1 and Rad17 or MLH1 and Tipin led to a further reduction in cell survival compared with depletion of MLH1, Rad17, or Tipin alone (Supplemental Fig. S1K,L). Of note, the ATR regulators were not among the top hits of the CRISPR screen (Supplemental Table S1), possibly because their loss also reduces the fitness of MMR-proficient cells. Nonetheless, the preferential depletion of gRNAs targeting multiple nonessential ATR regulators in Mlh1 KO cells suggests that MMR-d cells are more dependent on the ATR pathway for survival than MMR-proficient cells. Notably, sgRNAs targeting WRN were not preferentially depleted in Mlh1 KO cells, which is consistent with the notion that MSI-associated (TA)n expansion, but not MMR deficiency alone, confers WRN dependency (van Wietmarschen et al. 2020).
ATRi induces DNA damage in MMR-d cells in a replication- and MUS81-dependent manner
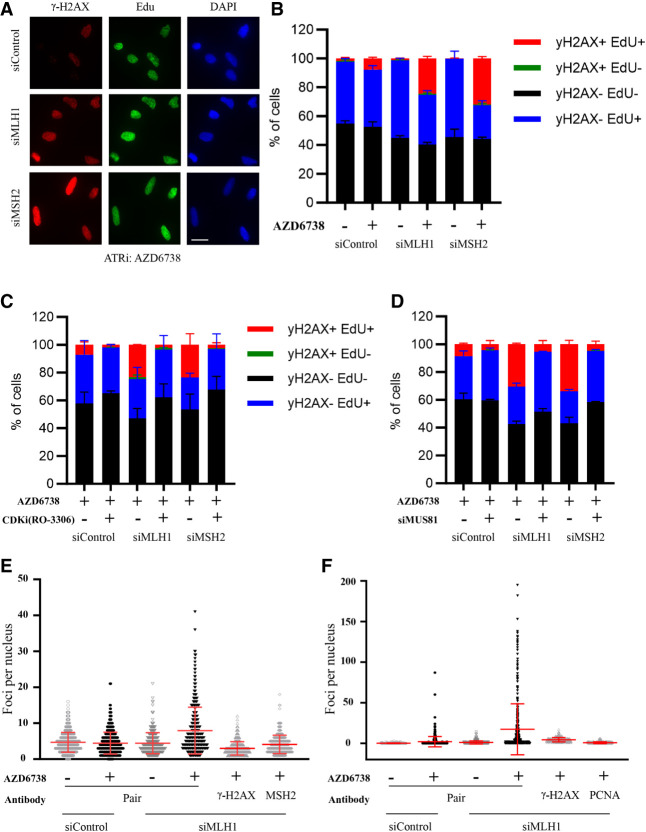
To understand how ATRi preferentially kills MMR-d cells, we treated CT26 parental and Mlh1 KO cells with ATRi (AZD6738) and followed the viability of cells over time (Supplemental Fig. S2A). ATRi progressively and preferentially reduced the viability of Mlh1 KO cells over time, leading to substantial killing of Mlh1 KO cells after 3–4 d. This result suggests that the effects of ATRi on MMR-d cells may be cell cycle-dependent. To test whether ATRi preferentially induces DNA damage in MLH1- and MSH2-depleted cells during DNA replication, we knocked down MLH1 or MSH2 with siRNAs in U2OS cells, pulse-labeled nascent DNA with EdU, and immunostained cells with anti-γH2AX antibody (Fig. 2A). ATRi induced more γH2AX in MLH1 and MSH2 knockdown cells than in cells treated with control siRNA (Fig. 2A,B). Importantly, this ATRi-induced increase of γH2AX specifically occurred in EdU+ cells (Fig. 2A,B), suggesting that ATRi induces DNA damage during replication. To test whether DNA replication is required for the induction of DNA damage by ATRi in MMR-d cells, we arrested control and MLH1 or MSH2 knockdown cells in G2 with the CDK1 inhibitor (CDK1i) RO-3360 and then treated them with ATRi (Fig. 2C). In control as well as MLH1 and MSH2 knockdown cell populations, CDK1i treatment reduced the fractions of EdU+ S-phase cells and the γH2AX-positive fractions of S-phase cells (Fig. 2C), suggesting that replication is needed for the induction of DNA damage by ATRi. These results are consistent with the idea that MMR-d cells are vulnerable during DNA replication if ATR function is compromised.
Figure 2.
ATRi preferentially induces DNA damage in MMR-d cells. (A,B) U2OS cells were transfected with control, MHL1, or MSH2 siRNA for 2 d; pulse-labeled with EdU for 20 min; and then treated with 2 µM ATRi (AZD6738) for 4 h. γ-H2AX and biotinylated EdU were analyzed by immunofluorescence. Representative images are shown in A. Fractions of cells positive for γ-H2AX and EdU were quantified as shown in B. Scale bar, 20 µm. (C) U2OS cells transfected with control, MLH1, or MSH2 siRNA were treated with 10 μM CDK1i (RO-3360) for 20 h followed by 2 μM ATRi (AZD6738) for 4 h. Fractions of cells positive for γ-H2AX and EdU were quantified. (D) U2OS cells were transfected with control, MLH1, MSH1, and MUS81 siRNAs as indicated and then treated with 2 μM ATRi (AZD6738) for 4 h. Fractions of cells positive for γ-H2AX and EdU were quantified. Scale bar, 20 µm. (E) U2OS cells were transfected with control or MLH1 siRNA and treated with 2 μM ATRi (AZD6738) for 4 h. Cells were analyzed by PLA using γ-H2AX antibody, MSH2 antibody, or both. The numbers of PLA foci in individual cells were quantified. (Red bars) Mean PLA foci per nucleus in cell populations. (F) U2OS cells were treated as in E and analyzed by PLA using γ-H2AX antibody, PCNA antibody, or both. The numbers of PLAs in individual cell foci were quantified as in E.
The instability of microsatellites in MMR-d cells is associated with the cleavage of non-B-form DNA secondary structures by the MUS81 nuclease (van Wietmarschen et al. 2021). Furthermore, ATR is known to restrict the activity of MUS81 at stalled replication forks (Forment et al. 2011; Couch et al. 2013; Ragland et al. 2013). Hence, we asked whether the ATRi-induced DNA damage in MMR-d cells is dependent on MUS81. We used siRNAs to knock down MUS81 alone or together with MLH1 or MSH2 in U2OS cells, pulse-labeled S-phase cells with EdU, and then treated them with ATRi (Fig. 2D; Supplemental Fig. S2B). Knockdown of MUS81 did not alter the fraction of EdU+ S-phase cells but significantly reduced the fraction of γH2AX-positive S-phase cells (Fig. 2D). These results suggest that ATRi preferentially induces DNA damage in MMR-d cells by allowing MUS81 to cleave DNA excessively.
ATRi induces DNA damage in MMR-d cells at sites of MMR and replication
The induction of excessive MUS81 cleavage of DNA by ATRi in MMR-d cells raises the possibility that DNA damage is generated in microsatellites in this context. To test this possibility, we performed proximity ligation assay (PLA) with anti-γH2AX and anti-MSH2 antibodies in ATRi-treated control U2OS and MLH1 knockdown cells (Fig. 2E). Because MLH1 functions downstream from MSH2 in the MMR pathway, MSH2 is expected to localize to sites where MMR initiates (e.g., microsatellites) in the absence of MLH1. Furthermore, if ATRi induces DNA damage at sites of MMR, then MSH2 and γH2AX should be in close proximity to each other, and PLA signals should be generated. Indeed, ATRi increased MSH2-γH2AX PLA foci in MLH1 knockdown cells but not in control U2OS cells (Fig. 2E; Supplemental Fig. S2C). Importantly, above-background levels of PLA foci in MLH1 knockdown cells were only detected when anti-MSH2 and anti-γH2AX antibodies were used together but not when either antibody was used alone (Fig. 2E), confirming the specificity of the PLA. Thus, in MMR-d cells, ATRi induces DNA damage at sites where MSH2 is located, likely including microsatellites.
Since the induction of DNA damage by ATRi in MMR-d cells is dependent on replication, we asked whether the ATRi-induced DNA damage occurs at replication forks. To test this possibility, we performed PLA with antibodies against γH2AX and PCNA, a component of replication forks (Fig. 2F; Supplemental Fig. S2D). When DNA mismatches are formed during replication, PCNA interacts with MHL1 and stimulates its endonuclease activity to promote MMR (Kadyrov et al. 2006). In MLH1 knockdown cells, PCNA is not engaged in MMR but serves as a marker for replication forks. ATRi specifically increased γH2AX-PCNA PLA foci in MLH1 knockdown cells but not in control U2OS cells. Again, above-background levels of PLA foci in MLH1 knockdown cells were only detected when both antibodies were used together but not when either antibody was used alone. These results suggest that the DNA damage induced by ATRi in MMR-d cells indeed occurs at replication forks.
Together, the results above suggest that in MMR-d cells, ATRi induces MUS81-generated DNA damage at sites where replication forks encounter non-B-form DNA structures that accumulate due to MMR deficiency. Since MMR proteins function at microsatellites to prevent MSI, MSH2 is likely present at microsatellites when downstream MMR events are defective. It is plausible that the ATRi-induced DNA damage is generated by MUS81 in DNA cruciforms at microsatellites upon collisions with replication forks. Alternatively, non-B-form DNA secondary structures can be formed at microsatellites during replication, providing substrates for MUS81. In addition to unwinding DNA cruciforms (Mengoli et al. 2023), MSH2 is also implicated in the suppression of G quadruplexes (Sakellariou et al. 2022), which can stall replication forks (Kumar et al. 2021). In all these scenarios, the ATRi-induced DNA damage at microsatellites or elsewhere is in close proximity to MSH2 and PCNA, just as observed in the γH2AX-MSH2 and γH2AX-PCNA PLA analyses.
ATRi reduces growth of MMR-d tumors in vivo
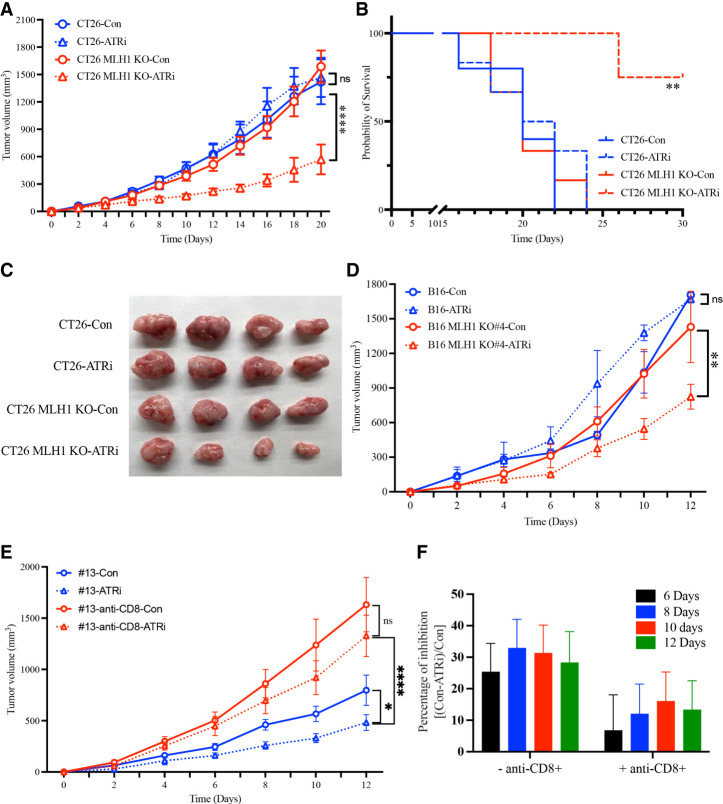
The preferential killing of MMR-d cells by ATRi in vitro prompted us to test whether ATRi can kill MMR-d cells in tumors and reduce the growth of MMR-d tumors in vivo. To test this possibility, we used CT26 parental cells and Mlh1 KO cells (clone #13) to generate xenograft tumors in immune-proficient mice and tested the effects of ATRi (AZD6738) on tumor growth (Fig. 3A). In the absence of ATRi, tumors generated by CT26 parental cells and Mlh1 KO cells grew at similar rates, the survival of mice was reduced by CT26 tumors and Mlh1 KO tumors similarly, and the sizes of CT26 tumors and Mlh1 KO tumors were similar (Fig. 3A–C). In the presence of ATRi, the growth of Mlh1 KO tumors was preferentially retarded, the survival of mice bearing Mlh1 KO tumors was preferentially prolonged, and the size of Mlh1 KO tumors was preferentially reduced (Fig. 3A–C). Similar results were obtained using syngeneic tumor models of B16 parental cells and multiple Mlh1 KO clones (Fig. 3D; Supplemental Fig. S3A–D). Together, these results demonstrate that ATRi can indeed reduce the growth of MMR-d tumors in vivo.
Figure 3.
ATRi reduces growth of MMR-d tumors in vivo. (A) CT26 and CT26 Mlh1 KO (clone #13) xenograft growth with or without treatment of 50 mg/kg ATRi (AZD6738) by oral gavage every 2 d after tumor size reached 70–100 mm3. (B) Curves for overall survival for mice bearing CT26 or CT26 Mhl1 KO tumors with or without treatment of 50 mg/kg ATRi (AZD6738) by oral gavage every 2 d. (C) Images for CT26 and CT26 Mlh1 KO tumors from the indicated mouse groups. (D) B16 and B16 Mlh1 KO (clone #4) xenograft growth with or without treatment of 50 mg/kg ATRi (AZD6738) by oral gavage every 2 d after tumor size reached 70–100 mm3. (E) CT26 Mlh1 KO (clone #13) xenograft growth after depletion of CD8+ T cells by injection of anti-CD8 antibody every 3–4 d for the duration of the experiment starting 1 d before tumor implantation. Mice were treated with or without 50 mg/kg ATRi (AZD6738) by oral gavage every 2 d after tumor size reached 70–100 mm3. (F) The percentage of inhibition of tumor growth by ATRi was quantified in the presence or absence of CD8+ T cells and after 6–12 d of ATRi treatment. Mice were treated with 50 mg/kg ATRi (AZD6738) as in A.
In cancer patients, MMR deficiency stimulates antitumor immunity, leading to the formation of hubs of interacting malignant cells and immune cells in tumors (Pelka et al. 2021). The use of syngeneic mouse models of MMR-d tumors gave us an opportunity to test whether immunity contributes to the effects of ATRi on tumor growth. To determine whether CD8+ T cells contribute to the inhibitory effects of ATRi on tumor growth, we first generated CT26 Mlh1 KO tumors (clone #13) and then treated mice with ATRi, anti-CD8, or ATRi and anti-CD8 (Fig. 3E). In the absence of ATRi, anti-CD8 significantly increased tumor growth (Fig. 3E; Supplemental Fig. S3E), suggesting that CD8+ T cells restrict the growth of MMR-d tumors. This result is consistent with the recent reports that MLH1 deficiency stimulates antitumor immunity (Germano et al. 2017; Guan et al. 2021; Lu et al. 2021). In the presence of ATRi alone, tumor growth was reduced as expected (Fig. 3E; Supplemental Fig. S3E). When ATRi and anti-CD8 were combined, tumor growth was increased to a level that was significantly higher than untreated tumors but still lower than tumors treated with anti-CD8 alone (Fig. 3E; Supplemental Fig. S3E). At the dose of ATRi used, the inhibition of tumor growth by ATRi was >30% in the presence of CD8+ T cells and <15% in the absence of CD8+ cells (Fig. 3E,F). Thus, ATRi becomes less effective at inhibiting MMR-d tumors when CD8+ T cells are depleted. However, ATRi still reduces tumor growth in the absence of CD8+ T cells, which is consistent with the synthetic lethal effect independent of immunity. Together, these results reveal that the inhibitory effects of ATRi on MMR-d tumors in vivo are partially due to the CD8+ T-cell-mediated antitumor immunity.
ATRi increases cytosolic DNA and interferon responses in MMR-d cells
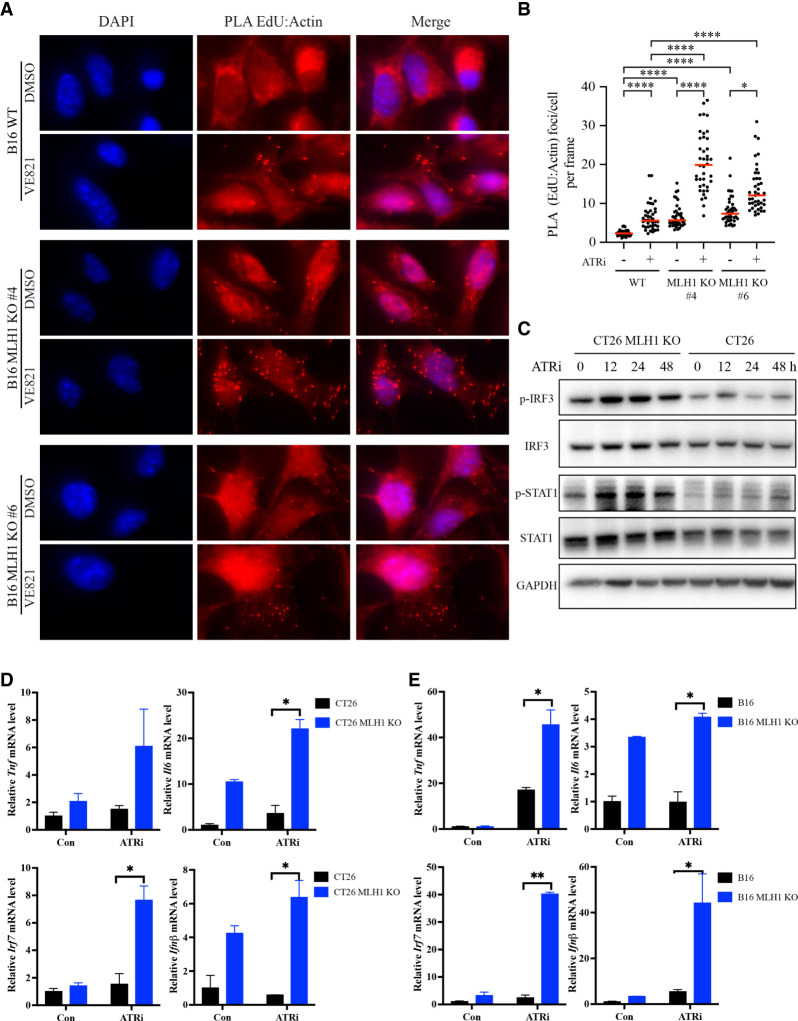
The contribution of CD8+ T cells to the antitumor effects of ATRi suggests that ATRi may stimulate the infiltration and/or function of T cells in tumor microenvironments. Recent studies have suggested that activation of the cGAS pathway by cytosolic DNA in tumor cells can stimulate the type I interferon response in dendritic cells, which in turn activates CD8+ T cells to eliminate tumor cells (Schadt et al. 2019). Furthermore, MLH1 deficiency stimulates cGAS in tumor cells and promotes antitumor immunity (Guan et al. 2021; Lu et al. 2021). The induction of DNA damage by ATRi in MMR-d cells raises the possibility that ATRi may preferentially increase cytosolic DNA in MMR-d cells to activate cGAS. Because ATRi likely induces cleavage of replication forks in microsatellites, we postulated that ATRi increases nascent DNA fragments and their accumulation in the cytoplasm. To test this possibility, we designed a PLA to monitor the accumulation of nascent DNA in the cytoplasm. In this assay, we pulse-labeled nascent DNA with EdU and then exposed cells to ATRi for 3 h. To detect the nascent DNA in the cytoplasm, we biotinylated EdU through a click reaction and then performed PLA with antibiotin and antiactin antibodies. As actin is mostly cytoplasmic, the accumulation of nascent DNA in the cytoplasm would bring it close to actin, generating PLA signals. Indeed, treatment of B16 cells with ATRi increased EdU-actin PLA foci (Fig. 4A,B), suggesting that ATRi increases the accumulation of nascent DNA fragments in the cytoplasm. Importantly, ATRi induced higher levels of EdU-actin PLA foci in two independent Mlh1 KO clones than in B16 parental cells (Fig. 4A,B), showing that ATRi preferentially induces cytosolic DNA in MMR-d cells. Similarly, ATRi preferentially increased the accumulation of nascent DNA in the cytoplasm of CT26 Mlh1 KO cells compared with parental cells (Supplemental Fig. S4A,B). The ATRi-induced PLA foci were detected only when both antibiotin and antiactin antibodies were used but not when one of these antibodies was used alone (Supplemental Fig. S4C). Furthermore, in MMR-d MC38 cells, ATRi induced EdU-actin PLA foci robustly (Supplemental Fig. S4D,E). These results suggest that ATRi indeed stimulates the accumulation of cytosolic DNA in MMR-d cells.
Figure 4.
ATRi increases cytosolic DNA and triggers interferon responses in MMR-d cells. (A,B) B16 or B16 Mlh1 KO cells were labeled with EdU for 2 h and then treated with 10 μM ATRi (VE-821) for 3 h. Cells were analyzed by PLA using antibodies to biotin (EdU) and actin. (A) Images of cells with or without EdU-actin PLA foci. (B) The numbers of PLA foci in multiple image frames were quantified, and the number of PLA foci per cell in each frame was determined (represented as individual dots). (Red bars) Mean number of PLA foci per cell in cell populations. (C) Western blots showing the levels of the indicated proteins and phosphorylated proteins in CT26 and CT26 Mlh1 KO cells after 2 μM ATRi (AZD6738) treatment at the indicated time points. (D,E) The mRNA levels of the indicated interferon-stimulated genes (ISGs) were analyzed by RT-qPCR in CT26 and CT26 Mlh1 KO cells (D) or in B16 and B16 Mlh1 KO cells (E) after mock or 2 μM ATRi (AZD6738) treatment for 2 d.
To test whether ATRi stimulates cGAS activation in MMR-d cells, we analyzed the levels of p-IRF3 and p-STAT1, two downstream phosphorylation events induced by cGAS activation. The levels of p-IRF3 and p-STAT1 were significantly higher in Mlh1 KO cells than in parental B16 cells and were further increased by ATRi (Fig. 4C), suggesting that cGAS is activated more robustly in MMR-d cells after ATRi treatment. In addition, we analyzed the mRNA levels of several ISGs known to be regulated by cGAS, including IL6, TNF, IRF7, and IFNβ. In both CT26 and B16 parental lines and their derived Mlh1 KO lines, ATRi preferentially stimulated the expression of ISGs in Mlh1 KO cells (Fig. 4D,E). These results lend further support to the notion that ATRi preferentially stimulates the interferon response in MMR-d cells by increasing the accumulation of nascent DNA fragments in the cytoplasm.
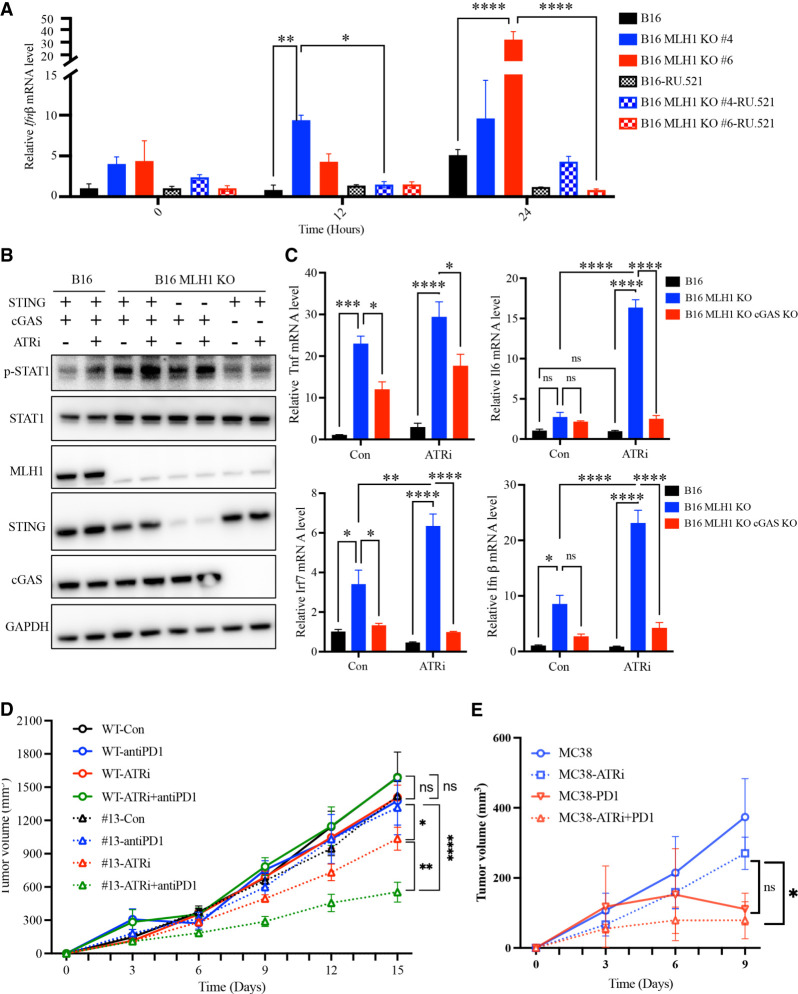
ATRi stimulates cGAS-mediated interferon responses in MMR-d cells
Next, we asked whether the ATRi-induced interferon response in MMR-d cells is dependent on the cGAS–STING pathway. First, we tested the effects of the cGAS inhibitor RU.251 on the ATRi-induced IFNβ expression in MMR-d cells. As expected, ATRi preferentially increased the mRNA of IFNβ in Mlh1 KO cells compared with parental B16 cells (Fig. 5A). The induction of IFNβ expression by ATRi was drastically reduced by RU.251 treatment, showing that this event is indeed cGAS-dependent. To test the roles of cGAS and STING in the ATRi-induced interferon response further, we knocked out cGAS and STING in B16 Mlh1 KO cells. Compared with parental B16 cells, Mlh1 KO cells displayed higher levels of p-STAT1 before and after ATRi treatment (Fig. 5B), supporting the idea that MMR deficiency stimulates the interferon response even when the response is enhanced by ATR inhibition. The levels of p-STAT in Mlh1 KO cells were significantly reduced by the loss of cGAS and modestly reduced by the loss of STING (Fig. 5B), showing that this is a largely cGAS-dependent and partially STING-dependent event. Of note, we cannot exclude the possibility that the partial STING dependency observed is due to the incomplete depletion of STING. Consistent with these findings, loss of cGAS in Mlh1 KO cells reduced the ATRi-induced expression of ISGs, including IL6, TNF, IRF7, and IFNβ (Fig. 5C). Together, these results suggest that ATRi preferentially stimulates the cGAS-mediated interferon response in MMR-d cells, providing a possible explanation for the T-cell-mediated antitumor effects induced by ATRi.
Figure 5.
ATRi stimulates cGAS-mediated interferon response and augments immunotherapy of MMR-d tumors. (A) B16 and B16 Mlh1 KO cells were treated with 2 μM ATRi (AZD6738) for the indicated lengths of time in the presence or absence of the cGAS inhibitor RU.521. The mRNA levels of INFβ were determined by RT-qPCR. (B) B16, B16 Mlh1 KO, B16 Mlh1;STING DKO (double KO), and B16 Mlh1;cGAS DKO cells were treated with ATRi for the indicated lengths of time. Levels of the indicated proteins and phosphorylated proteins were analyzed by Western blot. (C) B16, B16 Mlh1 KO, and B16 Mlh1;cGAS DKO cells were mock-treated or treated with 2 μM ATRi (AZD6738) for 2 d. The mRNA levels of TNF, IL6, IRF7, and INFβ were determined by RT-qPCR. (D) CT26 and CT26 Mlh1 KO (clone #13) xenograft growth with or without treatment with 35 mg/kg ATRi (AZD6738) by oral gavage every 3 d and with 3 mg/kg anti-PD-1 by intraparietal injection every 3–4 d. ATRi and anti-PD-1 were used either alone or in combination after tumor size reached 70–100 mm3. (E) MC38 xenograft growth with or without treatment with ATRi and anti-PD-1 as in D.
ATRi enhances the response of MMR-d tumors to immunotherapy
The ability of ATRi to stimulate CD8+ T-cell-mediated antitumor immunity against MMR-d tumors raises the possibility that ATRi can enhance the response of MMR-d tumors to immunotherapy. To test this possibility, we used parental CT26 cells and Mlh1 KO cells (clone #13) to generate xenograft tumors in immune-proficient mice and then treated the mice with ATRi, anti-PD-1, or a combination of both (Fig. 5D). The growth of CT26 tumors was not significantly affected by ATRi, anti-PD-1, or the combination (Fig. 5D; Supplemental Fig. S5A). Furthermore, the survival of the mice bearing CT26 tumors was not substantially extended by ATRi, anti-PD-1, or the combination (Supplemental Fig. S5B). In contrast, although Mlh1 KO tumors did not respond to anti-PD-1 alone, their growth was reduced by a low dose of ATRi and was further reduced by the combination of ATRi and anti-PD-1 (Fig. 5D; Supplemental Fig. S5A). Importantly, the survival of the mice bearing Mlh1 KO tumors was substantially extended by the combination of a low dose of ATRi and anti-PD-1 (Supplemental Fig. S5B). We also used MMR-d MC38 cells to generate tumors in a syngeneic model and tested the effects of ATRi and anti-PD-1. ATRi or anti-PD-1 alone reduced the growth of MC38 tumors, but the combination of ATRi and anti-PD-1 retarded tumor growth more efficiently than either drug alone (Fig. 5E), lending further support to the notion that the combination of ATRi and anti-PD-1 can target MMR-d tumors more effectively. These results show that ATRi, even when used at a low dose, can indeed complement and augment the immunotherapy of MMR-d tumors.
Discussion
MMR deficiency/MSI-H are clinically useful biomarkers for immunotherapy in multiple cancer types. Recent studies suggest that MSI-H may also be a good biomarker for WRN inhibitors (Chan et al. 2019; van Wietmarschen et al. 2020), which are still under development (Morales-Juarez and Jackson 2022). Our findings in this study suggest that MMR deficiency/MSI-H are also useful biomarkers to guide patient stratification for treatment with ATR inhibitors, which are already extensively tested in clinical trials (Sundar et al. 2017; Ngoi et al. 2022). Our findings are consistent with those from an independent study by the Nussenzweig laboratory (Zong et al. 2023), who also found that ATRi preferentially kills MMR-d/MSI-H tumor cells. Both studies suggest that the use of MMR deficiency/MSI-H as biomarkers to guide ATRi therapy may rapidly extend the use of ATRi in patients and improve the treatment of MMR-d/MSI-H tumors.
Although MMR deficiency/MSI-H are approved by the FDA as biomarkers to guide immunotherapy, half of the patients with MMR-d tumors do not respond to pembrolizumab (André et al. 2020). The lack of response of MMR-d tumors to immunotherapy could be a result of several mechanisms of immune evasion. For example, some MMR-d tumors may lack high levels of TMB or fail to generate neoantigens that are efficiently recognized by T cells (Westcott et al. 2021). Some MMR-d tumors may not have tumor microenvironments that favor the infiltration and function of immune cells (Guan et al. 2021; Lu et al. 2021). One way to complement the immunotherapy of MMR-d tumors is to induce synthetic lethality in MMR-d cells by inhibiting DNA repair or DNA damage signaling. Because this approach to selectively kill MMR-d tumor cells is independent of the immune system, it should work even in the MMR-d tumors that fail to respond to immunotherapy. It is conceivable that both ATR and WRN inhibitors can be useful for complementing the immunotherapy of MMR-d/MSI-H tumors, thus expanding the population of MMR-d patients who can benefit from biomarker-guided therapies.
It should be noted that ATR inhibition and WRN loss induce synthetic lethality in MMR-d/MSI-H cells in slightly different ways. While the synthetic lethality induced by WRN loss is dependent on MSI-H (Chan et al. 2019; Lieb et al. 2019; van Wietmarschen et al. 2020), MMR deficiency is sufficient to confer ATR dependency even when MSI is low. The mechanism underlying this difference between WRN loss and ATR inhibition is still unclear. Both WRN loss and ATR inhibition seem to induce MUS81-dependent DSBs in microsatellites during DNA replication, and their effects are likely exacerbated by the expansion of (TA)ns in MSI-H cells. It is possible that the substrates of MUS81 in (TA)ns are formed during DNA replication. Unlike WRN loss, ATR inhibition elevates origin firing and may increase the number of replication forks traversing microsatellites. The ability of ATRi, Chk1i, and Wee1i to increase origin firing may allow them to induce DSBs in (TA)ns efficiently even when the copy number of (TA)ns is relatively low. This property of ATRi, Chk1i, and Wee1i may be particularly useful for targeting the MMR-d tumors that lack high levels of MSI or TMB.
Another interesting finding of this study is that ATRi stimulates the CD8+ T-cell-mediated antitumor immunity against MMR-d tumors by activating the cGAS pathway. Because ATRi stimulates the cleavage of non-B-form DNA secondary structures by MUS81 in MMR-d cells during replication, fragments of nascent DNA are likely generated during this process. We found that ATRi increases the accumulation of nascent DNA fragments in the cytoplasm, explaining how cGAS is triggered by ATRi in MMR-d cells. It is plausible that the activation of cGAS by ATRi in MMR-d tumor cells induces a further type I interferon response through dendritic cells, which in turn stimulates CD8+ T cells to eliminate tumor cells (Schadt et al. 2019). It is particularly worth noting that ATRi selectively kills MMR-d tumor cells through two distinct mechanisms: It induces synthetic lethality by increasing DNA damage in microsatellites and stimulates the antitumor immunity by activating the cGAS pathway. These complementary effects of ATRi may render it particularly effective in the therapy of MMR-d/MSI-H tumors even when used at low doses, reducing its hematological toxicities (Martorana et al. 2022). It will be exciting to test the combination of ATRi and ICB in patients with MMR-d/MSI-H tumors, especially those that do not respond to ICB therapy alone. Our findings on the effects of ATRi in MMR-d tumors provide an example for targeting cancer cells through both synthetic lethality and antitumor immunity, suggesting a promising strategy that can be widely adapted to the development of future therapies.
Materials and methods
Cell lines
CT26 is a murine colorectal carcinoma cell line that was derived from BALB/c mice. MC38 is a murine colon adenocarcinoma cell line that was derived from C57BL/6J mice. B16F10 is a murine melanoma cell line that was derived from C57BL/6J mice. U2OS is an osteosarcoma cell line with epithelial morphology.
For cell lines with genes knocked out, guide RNA sequences targeting mouse MLH1 and cGAS were inserted into the CRISPR/Cas9 system plasmid PX458. PX458-MLH1 sg1 and PX458-MLH1 sg2 were transfected into the CT26, MC38, and B16F10 cell lines with Lipofectamine 3000. Forty-eight hours following transfection, GFP+ cells were seeded at one cell per well in 96-well plates by flow cytometry. Single-cell clones were expanded, and depletion of MLH1 was confirmed by immunoblotting (BD Pharmingen purified mouse antihuman MLH-1). The MLH1 KO cell lines were cultured and passaged for 4 wk before freezing. PX458-cGAS cells were transfected into the CT26 MLH1 KO or B16F10 MLH1 KO cell lines with Lipofectamine 3000. Forty-eight hours following transfection, GFP+ cells were seeded at one cell per 96-well by flow cytometry. Single-cell clones were expanded, and depletion of cGAS was confirmed by immunoblotting (cGAS [D3O8O] rabbit mAb 31659). B16 MLH1 KO or CT26 MLH1 KO single clones were infected with lentiCRISPR-V2 STING virus (a gift from Jun Tian). Forty-eight hours following infection, the cells were treated with puromycin for 5 d. STING KO cell lines were expanded, and depletion of cGAS was confirmed by immunoblotting (STING [D2P2F] rabbit mAb).
Cell culture and RNAi
U2OS cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). For RNAi experiments, U2OS cells were plated into six-well dishes, and small interfering RNA (siRNA) against MLH1, MSH2, and MUS81 was transfected using Lipofectamine RNAiMax (Thermo). The siRNA sequences used were MLH1-1 (GAAAUUGGAUGUGAGGAUA), MLH1-2 (CCACCAAGCUUAGUGAAGATT), MSH2-1 (GGAUAUUACUUUCGUGUAA), MSH2-2 (GCAAACAGAUUAAACUGGATT), RAD17 (CUUUAUGCCUCCAAACUCATT), TIPIN (GCAAAGCUGCUGAGUAAUATT) (Leung et al. 2023), and MUS81 (CAGCCCUGGUGGAUCGAUA) (Ouyang et al. 2015; Matos et al. 2020). Cells were harvested for Western blotting or stained for immunofluorescence 72 h after transfection.
Western blotting
For Western blotting, cells were washed twice with cold PBS, and whole-cell proteins were extracted in lysis buffer (40 mM Tris at pH 8.0, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM sodium orthovanadate) containing phosphatase inhibitor cocktail. The protein concentration was normalized using a protein assay dye reagent. Samples were boiled for 5 min at 95°C and loaded onto SDS-PAGE. After transfer, membranes were blocked with 5% milk or BSA in TBST buffer (0.1% Tween 20) for 1 h. Primary antibodies were diluted and incubated overnight at 4°C. After three washes with TBST buffer, antirabbit or antimouse secondary antibodies and HRP conjugates were incubated with membranes for 1 h. After three washes, membranes were covered with the ECL reagent and imaged.
Immunofluorescence
Cells transfected with the indicated siRNAs or treated for 20 h with 10 µM RO-3306 (Selleck Chemicals) were incubated for 20 min with 10 µM EdU (Thermo) followed by 2 µM ATRi AZD6738 (Selleck Chemicals) for 4 h. Cells were subsequently fixed, permeabilized, and stained for nascent DNA using click chemistry (Thermo) and γ-H2AX (Cell Signaling Technologies). Cells were imaged using an Axioimager wide-field microscope (Zeiss).
Cell viability assay
For the MLH1 KO cell viability assay, cells were cultured in ATRi AZD6738 or VE-821 (Selleck Chemicals) for 4–5 d before cell viability was examined using CellTiter-Glo (Promega) according to the manufacturer's instructions. For the siRNA transfected cell viability assay, cells transfected with the indicated siRNAs were cultured in ATRi AZD6738 or VE-821 (Selleck Chemicals) for 5 d before cell viability was examined using CellTiter-Glo (Promega) according to the manufacturer's instructions.
Clonogenic survival assay
Seven-hundred cells were seeded into 12-well plates and subsequently propagated for 9 d in the medium with or without AZD6738 or VE-821. For crystal violet staining, cells were fixed with methanol for 10 min at room temperature and subsequently stained with 250 μL of 0.1% crystal violet in 70% ethanol per well for 10 min at room temperature. To remove unbound crystal violet, cells were washed with deionized water three times for 5 min each.
Proximity ligation assay (PLA)
For the γ-H2AX-MSH2 PLA, U2OS cells transfected with control or MLH1 siRNA were incubated with 2 µM ATRi AZD6738 for 4 h. Subsequently, cells were fixed in 4% paraformaldehyde and permeabilized in 0.5% Triton X, and PLA was performed using the Duolink PLA kit (Sigma) according to the manufacturer's instructions using the γ-H2AX (Cell Signaling Technologies) and MSH2 (Abcam) antibodies. For the γ-H2AX-PCNA PLA, U2OS cells transfected with control or MLH1 siRNA were incubated with ATRi AZD6738 for 4 h. Subsequently, cells were fixed in ice-cold methanol and permeabilized in 0.5% Triton X, and PLA was performed using the Duolink PLA kit (Sigma) according to the manufacturer's instructions using the γ-H2AX (Cell Signaling Technologies) and PCNA (Santa Cruz Biotechnologies) antibodies. For the EdU-actin PLA, B16, MC38, and CT26 cells were labeled with 10 mM EdU for 2 h and treated with DMSO or 10 mM ATRi VE-821 for 3 h. Next, the cells were treated with permeabilization solution (0.1% Tween 20 in PBS) and primary antibodies to biotin (Jackson Laboratories) and actin (Cell Signaling Technologies).
Real-time PCR
Cells were collected 24 h after treatment and lysed with Trizol (Invitrogen). RNA was isolated by phenol–chloroform extraction and converted to cDNA using a SuperScript III first-strand synthesis system (Invitrogen). qPCR was performed on a Roche LightCycler 480 Instrument II using SYBR Green master mix (Bio-Rad). To measure the transcription level, primers recognizing the transcripts of genes of interest and GAPDH (which served as the internal control) were designed, and the RNA level of each target gene was normalized to that of GAPDH.
GDSC database analysis
We retrieved drug sensitivity data from the Genomics of Drug Sensitivity in Cancer (GDSC) database and categorized the cell lines into MSS and MSI groups based on the CCLE_MSI definition. Subsequently, we calculated the IC50 variations between the MSS and MSI groups using the Wilcoxon test for the statistical test and the Benjamini–Hochberg (BH) procedure for adjusting P-values.
Pathway enrichment of drug sensitivity data sets
We performed drug sensitivity pathway enrichment using a methodology akin to gene set enrichment analysis. In brief, we compiled data sets comprising pathways related to drugs. Next, R package enrichR (https://github.com/wjawaid/enrichR) was used for enrichment analysis for drugs exhibiting significant differences in sensitivity between MSS and MSI cell lines.
RNA-seq analysis of MSS and MSI tumors
We obtained the gene expression matrix from GSE146889 for a total of 91 colorectal and endometrial tumors, comprising 19 MSS and 72 MSI tumors. Next, the average RPKM (reads per kilobase per million mapped reads) values of each gene in both MSS and MSI tumors were calculated. The R package ComplexHeatmap was used to construct a heat map illustrating the expression levels of ATR regulators in both MSS and MSI tumors.
Calculation of MSI score
MSIsensor2 (https://github.com/niu-lab/msisensor2) was used to access the genomic MSI status. The MSI sensor score for MLH1 KO mice was determined using tumor-paired modules, with CT26 wild-type mice serving as the control group. Additionally, we obtained the raw data from a previous study (Mandal et al. 2019), which provided the MSI intermediate score for comparative purposes.
Genome-wide CRISPR screening
Approximately 140 million CT26 or CT26 Mlh1 KO cells were dissociated into single cells and infected with the lentiviral Brie genome-wide library at an MOI of 0.3 in a reaction volume of 140 mL. Cells were subsequently seeded into seven 15-cm dishes and cultured in fresh medium overnight after the infection. On day 2, 10 µg/mL puromycin was used to select the infected cells. On day 8, ∼40 million (∼500× coverage) infected cells were collected for genomic DNA extraction and library construction, followed by sgRNA library sequencing. Differential sgRNA representation between the CT26 and CT26 Mlh1 KO cells was analyzed using MAGeCK 0.5.9.5 (Li et al. 2014). The MAGeCK count command was used to generate counts for individual small guide RNAs, while the MAGeCK test command was used for conducting sample comparisons. A single data set was analyzed.
Mice
C57BL/6J and BALB/c mice were purchased from the Jackson Laboratory. All mice were maintained in a specific-pathogen-free animal facility, and experiments were conducted in compliance with Institutional Animal Care and Use Committee-approved animal protocols according to Massachusetts General Hospital institutional guidelines.
CT26 cell line xenografts
To establish a murine colon adenocarcinoma cancer model, 10 × 105 to 15 × 105 CT26 or CT26 Mlh1 KO cells were subcutaneously implanted into the right flanks of female BALB/c mice. Tumor size and body weight were measured three times weekly. Tumor sizes were measured and calculated by the formula (length) × (width)2/2. For ATR inhibitor treatment, when the mean tumor volume reached ∼70–100 mm3, animals were randomized into two groups (group A [control] and group B [ATRi]) and dosed with 50 mg/kg ATRi treatment by oral gavage every 2 d. Mice were monitored for 15 min after administration to ensure no difficulty breathing. For ATR inhibitor and anti-PD-1 combination treatment, when the mean tumor volume reached ∼100–200 mm3, animals were randomized into four groups (group A [control], group B [ATRi], group C [anti-PD-1], and group D [ATRi+ anti-PD-1]) and dosed. Each group needed more than four mice. Antimouse PD-1 (3 mg/kg) was administered intraperitoneally every 3–4 d, and mice received 35 mg/kg ATRi treatment by oral gavage every 1–3 d. Mice were monitored for 15 min after administration to ensure no difficulty breathing. During the observation period, animals bearing oversized tumors >2000 mm3 were sacrificed by CO2 asphyxiation.
B16 cell line xenografts
To establish a murine cancer model, 3 × 105 to 10 × 105 B16 or B16 Mlh1 KO cells in 100 µL of PBS were subcutaneously implanted into the right flanks of female C57BL/6 mice. Tumor size and body weight were measured three times weekly. Tumor sizes were measured and calculated by the formula (length) × (width)2/2. When the mean tumor volume reached ∼70–100 mm3, animals were randomized into two groups (group A [control] and group B [ATRi]) and dosed with 50 mg/kg ATRi treatment by oral gavage every 2 d. Mice were monitored for 15 min after administration to ensure no difficulty breathing. During the observation period, animals bearing oversized tumors >2000 mm3 were sacrificed by CO2 asphyxiation.
MC38 cell line xenografts
To establish a murine cancer model, 3 × 105 to 10 × 105 MC38 cells in 100 µL of PBS were subcutaneously implanted into the right flanks of female C57BL/6 mice. Tumor size and body weight were measured three times weekly. Tumor sizes were measured and calculated by the formula (length) × (width)2/2. When the mean tumor volume reached ∼70–100 mm3, animals were randomized into four groups (group A [control], group B [ATRi], group C [anti-PD-1], and group D [ATRi+ anti-PD-1]) and dosed. Each group needed more than four mice. Antimouse PD-1 (3 mg/kg; clone 29F.1A12, Bio X Cell) was administered intraperitoneally every 3–4 d, and mice received 35 mg/kg ATRi treatment by oral gavage every 2 d. Mice were monitored for 15 min after administration to ensure no difficulty breathing. During the observation period, animals bearing oversized tumors >2000 mm3 were sacrificed by CO2 asphyxiation.
CD8+ T-cell depletion
To establish a murine cancer model, 3 × 105 to 10 × 105 CT26 MMR-d cells in 100 µL of PBS were subcutaneously implanted into the right flanks of 6- to 8-wk-old female BALB/c mice. Tumor size and body weight were measured three to four times weekly. Tumor sizes were measured and calculated by the formula (length) × (width)2/2. When the mean tumor volume reached ∼70–100 mm3, animals were randomized into three groups (group A [control], group B [ATRi], and group C [ATRi+ anti-CD8b]) for treatment. Each group included at least four mice. For depletion experiments, C57BL/6J mice were treated with 200 μg of anti-CD8b (clone 53-5.8, Bio X Cell) via intraperitoneal injection every 3–4 d for the duration of the experiment starting 1 d before tumor implantation. Mice were monitored for 15 min after administration to ensure no difficulty breathing. During the observation period, animals bearing oversized tumors >2000 mm3 were sacrificed by CO2 asphyxiation.
Supplementary Material
Acknowledgments
We thank members of the Zou, Dyson, Lan, Mostoslavsky, and Elia laboratories for helpful discussions, and Dr. Andre Nussenzweig and Dr. Dali Zong for communicating unpublished data. L.Z. was the James and Patricia Poitras Endowed Chair in Cancer Research at Massachusetts General Hospital (MGH). This work is supported by National Institutes of Health grants CA248526 and CA263934 to L.Z., CA249726 to X.-F.W., and CA274158 to J.O., and by a Rullo Family Innovation Award to M.S.L.
Author contributions: M.W. and L.Z. designed the study. M.W., X.R., W.L., A.K., S.S., J.O., P.S.P., Y.D., and T.Y. performed the experiments and data analysis. R.T.M. provided critical reagents and intellectual input. J.S., L.L., X.-F.W., M.S.L., and L.Z. supervised the study. M.W., X.R., and L.Z. wrote the manuscript with help from the other authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.351084.123.
Competing interest statement
The authors declare no competing interests.
References
- André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, et al. 2020. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med 383: 2207–2218. 10.1056/NEJMoa2017699 [DOI] [PubMed] [Google Scholar]
- Cercek A, Lumish M, Sinopoli J, Weiss J, Shia J, Lamendola-Essel M, El Dika IH, Segal N, Shcherba M, Sugarman R, et al. 2022. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N Engl J Med 386: 2363–2376. 10.1056/NEJMoa2201445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Shibue T, McFarland JM, Gaeta B, Ghandi M, Dumont N, Gonzalez A, McPartlan JS, Li T, Zhang Y, et al. 2019. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568: 551–556. 10.1038/s41586-019-1102-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Harding SM, Natesan R, Tian L, Benci JL, Li W, Minn AJ, Asangani IA, Greenberg RA. 2020. Cell cycle checkpoints cooperate to suppress DNA- and RNA-associated molecular pattern recognition and anti-tumor immune responses. Cell Rep 32: 108080. 10.1016/j.celrep.2020.108080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ciriano I, Lee S, Park WY, Kim TM, Park PJ. 2017. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun 8: 15180. 10.1038/ncomms15180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Bétous R, Carroll CM, Jung SY, Qin J, Cimprich KA, et al. 2013. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev 27: 1610–1623. 10.1101/gad.214080.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M, Rieder D, Klepsch V, Charoentong P, Finotello F, Hackl H, Hermann-Kleiter N, Löwer M, Baier G, Krogsdam A, et al. 2018. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat Commun 9: 32. 10.1038/s41467-017-02424-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment JV, Blasius M, Guerini I, Jackson SP. 2011. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS One 6: e23517. 10.1371/journal.pone.0023517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germano G, Lamba S, Rospo G, Barault L, Magri A, Maione F, Russo M, Crisafulli G, Bartolini A, Lerda G, et al. 2017. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552: 116–120. 10.1038/nature24673 [DOI] [PubMed] [Google Scholar]
- Guan J, Li GM. 2023. DNA mismatch repair in cancer immunotherapy. NAR Cancer 5: zcad031. 10.1093/narcan/zcad031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J, Lu C, Jin Q, Lu H, Chen X, Tian L, Zhang Y, Ortega J, Zhang J, Siteni S, et al. 2021. MLH1 deficiency-triggered DNA hyperexcision by exonuclease 1 activates the cGAS–STING pathway. Cancer Cell 39: 109–121.e5. 10.1016/j.ccell.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548: 466–470. 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins JL, Lan L, Zou L. 2022. DNA repair defects in cancer and therapeutic opportunities. Genes Dev 36: 278–293. 10.1101/gad.349431.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. 2006. Endonucleolytic function of MutLα in human mismatch repair. Cell 126: 297–308. 10.1016/j.cell.2006.05.039 [DOI] [PubMed] [Google Scholar]
- Kategaya L, Perumal SK, Hager JH, Belmont LD. 2019. Werner syndrome helicase is required for the survival of cancer cells with microsatellite instability. iScience 13: 488–497. 10.1016/j.isci.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C, Batra S, Griffith JD, Remus D. 2021. The interplay of RNA:DNA hybrid structure and G-quadruplexes determines the outcome of R-loop-replisome collisions. Elife 10: e72286. 10.7554/eLife.72286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. 2017. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357: 409–413. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W, Simoneau A, Saxena S, Jackson J, Patel PS, Limbu M, Vindigni A, Zou L. 2023. ATR protects ongoing and newly assembled DNA replication forks through distinct mechanisms. Cell Rep 42: 112792. 10.1016/j.celrep.2023.112792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15: 554. 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb S, Blaha-Ostermann S, Kamper E, Rippka J, Schwarz C, Ehrenhöfer-Wölfer K, Schlattl A, Wernitznig A, Lipp JJ, Nagasaka K, et al. 2019. Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells. Elife 8: e43333. 10.7554/eLife.43333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Guan J, Lu S, Jin Q, Rousseau B, Lu T, Stephens D, Zhang H, Zhu J, Yang M, et al. 2021. DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell 39: 96–108.e6. 10.1016/j.ccell.2020.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R, Samstein RM, Lee KW, Havel JJ, Wang H, Krishna C, Sabio EY, Makarov V, Kuo F, Blecua P, et al. 2019. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 364: 485–491. 10.1126/science.aau0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorana F, Da Silva LA, Sessa C, Colombo I. 2022. Everything comes with a price: the toxicity profile of DNA-damage response targeting agents. Cancers 14: 953. 10.3390/cancers14040953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos DA, Zhang JM, Ouyang J, Nguyen HD, Genois MM, Zou L. 2020. ATR protects the genome against R loops through a MUS81-triggered feedback loop. Mol Cell 77: 514–527.e4. 10.1016/j.molcel.2019.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166. 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- Mengoli V, Ceppi I, Sanchez A, Cannavo E, Halder S, Scaglione S, Gaillard PH, McHugh PJ, Riesen N, Pettazzoni P, et al. 2023. WRN helicase and mismatch repair complexes independently and synergistically disrupt cruciform DNA structures. EMBO J 42: e111998. 10.15252/embj.2022111998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Juarez DA, Jackson SP. 2022. Clinical prospects of WRN inhibition as a treatment for MSI tumours. NPJ Precis Oncol 6: 85. 10.1038/s41698-022-00319-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoi NYL, Westin SN, Yap TA. 2022. Targeting the DNA damage response beyond poly(ADP-ribose) polymerase inhibitors: novel agents and rational combinations. Curr Opin Oncol 34: 559–569. 10.1097/CCO.0000000000000867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Garner E, Hallet A, Nguyen HD, Rickman KA, Gill G, Smogorzewska A, Zou L. 2015. Noncovalent interactions with SUMO and ubiquitin orchestrate distinct functions of the SLX4 complex in genome maintenance. Mol Cell 57: 108–122. 10.1016/j.molcel.2014.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka K, Hofree M, Chen JH, Sarkizova S, Pirl JD, Jorgji V, Bejnood A, Dionne D, Ge WH, Xu KH, et al. 2021. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 184: 4734–4752.e20. 10.1016/j.cell.2021.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco G, Cattaneo CM, van Vliet EJ, Crisafulli G, Rospo G, Consonni S, Vieira SF, Rodriguez IS, Cancelliere C, Banerjee R, et al. 2021. Werner helicase is a synthetic-lethal vulnerability in mismatch repair-deficient colorectal cancer refractory to targeted therapies, chemotherapy, and immunotherapy. Cancer Discov 11: 1923–1937. 10.1158/2159-8290.CD-20-1508 [DOI] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F, Franchitto A. 2003. Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene 22: 1491–1500. 10.1038/sj.onc.1206169 [DOI] [PubMed] [Google Scholar]
- Ragland RL, Patel S, Rivard RS, Smith K, Peters AA, Bielinsky AK, Brown EJ. 2013. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev 27: 2259–2273. 10.1101/gad.223180.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakellariou D, Bak ST, Isik E, Barroso SI, Porro A, Aguilera A, Bartek J, Janscak P, Peña-Diaz J. 2022. Mutsβ regulates G4-associated telomeric R-loops to maintain telomere integrity in ALT cancer cells. Cell Rep 39: 110602. 10.1016/j.celrep.2022.110602 [DOI] [PubMed] [Google Scholar]
- Saldivar JC, Cortez D, Cimprich KA. 2017. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol 18: 622–636. 10.1038/nrm.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt L, Sparano C, Schweiger NA, Silina K, Cecconi V, Lucchiari G, Yagita H, Guggisberg E, Saba S, Nascakova Z, et al. 2019. Cancer-cell-intrinsic cGAS expression mediates tumor immunogenicity. Cell Rep 29: 1236–1248.e7. 10.1016/j.celrep.2019.09.065 [DOI] [PubMed] [Google Scholar]
- Shastri N, Tsai YC, Hile S, Jordan D, Powell B, Chen J, Maloney D, Dose M, Lo Y, Anastassiadis T, et al. 2018. Genome-wide identification of structure-forming repeats as principal sites of fork collapse upon ATR inhibition. Mol Cell 72: 222–238.e11. 10.1016/j.molcel.2018.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar R, Brown J, Ingles Russo A, Yap TA. 2017. Targeting ATR in cancer medicine. Curr Probl Cancer 41: 302–315. 10.1016/j.currproblcancer.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Tubbs A, Sridharan S, van Wietmarschen N, Maman Y, Callen E, Stanlie A, Wu W, Wu X, Day A, Wong N, et al. 2018. Dual roles of poly(dA:dT) tracts in replication initiation and fork collapse. Cell 174: 1127–1142.e19. 10.1016/j.cell.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wietmarschen N, Sridharan S, Nathan WJ, Tubbs A, Chan EM, Callen E, Wu W, Belinky F, Tripathi V, Wong N, et al. 2020. Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature 586: 292–298. 10.1038/s41586-020-2769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wietmarschen N, Nathan WJ, Nussenzweig A. 2021. The WRN helicase: resolving a new target in microsatellite unstable cancers. Curr Opin Genet Dev 71: 34–38. 10.1016/j.gde.2021.06.014 [DOI] [PubMed] [Google Scholar]
- Vendetti FP, Karukonda P, Clump DA, Teo T, Lalonde R, Nugent K, Ballew M, Kiesel BF, Beumer JH, Sarkar SN, et al. 2018. ATR kinase inhibitor AZD6738 potentiates CD8+ T cell-dependent antitumor activity following radiation. J Clin Invest 128: 3926–3940. 10.1172/JCI96519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendetti ML, Esther Moon SJ, Imes CC, Hergenroeder A, Sciurba F, Lendermon E, Pilewski J, Ren D, Parmanto B, Dewhirst B, et al. 2023. Design of lung transplant Go (LTGO): a randomized controlled trial evaluating the efficacy of a telerehabilitation behavioral exercise intervention to improve physical activity, physical function, and blood pressure control after lung transplantation. Contemp Clin Trials Commun 33: 101097. 10.1016/j.conctc.2023.101097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PMK, Sacks NJ, Schenkel JM, Ely ZA, Smith O, Hauck H, Jaeger AM, Zhang D, Backlund CM, Beytagh MC, et al. 2021. Low neoantigen expression and poor T-cell priming underlie early immune escape in colorectal cancer. Nat Cancer 2: 1071–1085. 10.1038/s43018-021-00247-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PMK, Muyas F, Hauck H, Smith OC, Sacks NJ, Ely ZA, Jaeger AM, Rideout WM, Zhang D, Bhutkar A, et al. 2023. Mismatch repair deficiency is not sufficient to elicit tumor immunogenicity. Nat Genet 55: 1686–1695. 10.1038/s41588-023-01499-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazinski SA, Zou L. 2016. Functions, regulation, and therapeutic implications of the ATR checkpoint pathway. Annu Rev Genet 50: 155–173. 10.1146/annurev-genet-121415-121658 [DOI] [PubMed] [Google Scholar]
- Zong D, Koussa NC, Cornwell JA, Pankajam AV, Kruhlak MJ, Wong N, Chari R, Cappell SD, Nussenzweig A. 2023. Comprehensive mapping of cell fates in microsatellite unstable cancer cells supports dual targeting of WRN and ATR. Genes Dev (this issue). 10.1101/gad.351085.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.